Florfenicol is widely used for the treatment of respiratory infections and as a feed additive in food animal production. As a foodborne pathogen, Campylobacter is constantly exposed to florfenicol, and resistance to this antimicrobial agent has increased in recent years.
KEYWORDS: Campylobacter, fexA, optrA, multidrug resistance
ABSTRACT
Previous studies indicated that Campylobacter has developed several mechanisms that confer resistance to florfenicol, which is used in food animal production. This study describes the coexistence of optrA and fexA in Campylobacter jejuni and Campylobacter coli isolates from pigs and poultry. Moreover, whole-genome sequencing data showed that the two genes are located in various multidrug resistance genomic islands within different regions of the Campylobacter genomes. The emergence of optrA and fexA may support the spread of florfenicol-resistant Campylobacter strains of animal origin.
IMPORTANCE Florfenicol is widely used for the treatment of respiratory infections and as a feed additive in food animal production. As a foodborne pathogen, Campylobacter is constantly exposed to florfenicol, and resistance to this antimicrobial agent has increased in recent years. Previous studies indicated that Campylobacter has developed several mechanisms that confer resistance to florfenicol. This study describes for the first time the coexistence of the florfenicol exporter FexA and the ribosomal protective protein OptrA in Campylobacter jejuni isolated from pigs. The two genes were located in various multidrug resistance genomic islands within different regions of the Campylobacter genomes. Although phenicols are not commonly used for the treatment of Campylobacter infections, the extensive use of florfenicol in food animals may play a role in the coselection of multidrug resistance genomic island (MDRGI)-carrying Campylobacter isolates which also exhibited resistance to critically important antimicrobial agents (macrolides, aminoglycosides, and tetracyclines) commonly used for the treatment of human campylobacteriosis.
OBSERVATION
Campylobacter is the leading bacterial pathogen that causes diarrheal illness worldwide, with most cases of campylobacteriosis being triggered by Campylobacter jejuni. As a foodborne pathogen, Campylobacter is constantly exposed to multiple antimicrobial agents used during food animal production. Thus, Campylobacter has developed various resistance mechanisms, including the formation of multidrug resistance genomic islands (MDRGIs), for fitness advantage upon exposure to multiple antimicrobial agents (1–4). Florfenicol is a fluorinated thiamphenicol derivative that was exclusively approved as a broad-spectrum antimicrobial agent for the treatment of animals raised for food (5). To date, several mechanisms of antibiotic resistance to florfenicol have been characterized, including the multidrug resistance protein Cfr(C), the multidrug efflux pump RE-CmeABC, and the recently described florfenicol exporter FexA and the ribosomal protective OptrA (4, 6–10). cfr(C), RE-cmeABC, and fexA were characterized in both C. jejuni and Campylobacter coli, whereas optrA was identified only in C. coli (4, 6–10).
The phenicol exporter gene fexA is responsible for florfenicol resistance. optrA not only confers resistance to phenicols but also results in elevated MICs of the oxazolidinone linezolid. Although these drugs are not commonly used for the treatment of Campylobacter infections, the extensive use of florfenicol in food animals may play a role in the coselection of MDRGI-carrying Campylobacter isolates, which also exhibit resistance to macrolides, aminoglycosides, and tetracyclines, commonly used for treating human campylobacteriosis (7). Linezolid represents one of the last-resort antimicrobial agents for the treatment of severe infections caused by methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus spp. Thus, the coexistence of these two drug-resistant genes in Campylobacter aggravates the spread of antimicrobial resistance and poses a threat to human health. In this study, the coexistence of optrA and fexA was identified in C. jejuni isolates from pig and C. coli isolates from chicken and duck, and whole-genome sequencing was used to characterize their genetic environment.
To determine the presence of optrA, the primers A–F (5′-AGGTGGTCAGCGAACTAA-3′) and A–R (5′-ATCAACTGTTCCCATTCA-3′) (11) were used for PCR analysis of 146 C. coli and 54 C. jejuni strains isolated from poultry and swine farms in Zhejiang and Hunan provinces, China. The optrA sequence was identified in two C. jejuni and five C. coli isolates. Of note, all the strains also contained fexA.
To further characterize these seven optrA+ fexA+ isolates and the genetic environment of the genes, a hybrid sequencing strategy using Illumina short-read and MinION long-read technology was used to generate the complete genomes, as previously described (12). Five complete and two draft genome sequences were obtained for further mining (Table 1). In silico multilocus sequence typing of the whole-genome sequencing data showed that these seven isolates belonged to three sequence types (ST), including ST825, ST828, and a new ST (aspA_8, glnA_620, gltA_292, glyA_28, pgm_1072, tkt_668, and uncA_23). Acquired antimicrobial resistance genes can explain the resistance phenotype, including florfenicol resistance, determined by the broth microdilution method (Table 2). Only three and one single nucleotide polymorphisms were detected in the optrA and fexA sequences, respectively, in these seven strains (see Fig. S1 in the supplemental material).
TABLE 1.
Isolation and genomic information of optrA+ fexA+ strains
| Isolate | Host | Species | Genome length (bp) | GC content (%) | MLSTa | Quality | MDRGI length (bp) | MDRGI GC content (%) | Accession no. |
|---|---|---|---|---|---|---|---|---|---|
| CC19DZ036 | Duck | C. coli | Chromosome: 1,761,335 | 31.37 | ST828 | Completed | 11,195 | 36.22 | CP068565 |
| CC19DZ037 | Duck | C. coli | Chromosome: 1,761,334 | 31.37 | ST828 | Completed | 11,195 | 36.22 | CP068566 |
| CC19PF050 | Pig | C. jejuni | 1,798,069 | 30.21 | Unknown | Draft | JAESVI000000000 | ||
| CC19PF065 | Pig | C. jejuni | Chromosome: 1,681,082 | 30.57 | Unknown | Completed | 18,223 | 34.79 | CP068567 |
| pPF065-186: 186,647 | 26.41 | CP068568 | |||||||
| pPF065-3: 3,395 | 31.37 | CP068569 | |||||||
| CC19CH074 | Chicken | C. coli | 1,831,137 | 31.16 | ST825 | Draft | JAESVJ000000000 | ||
| CC19CH075 | Chicken | C. coli | Chromosome: 1,781,472 | 31.47 | ST825 | Completed | 18,553 | 36.36 | CP068581 |
| pCH075-80: 80,135 | 26.07 | CP068582 | |||||||
| pCH075-4: 4,944 | 29.57 | CP068583 | |||||||
| pCH075-3: 3,405 | 31.01 | CP068584 | |||||||
| pCH075-2: 2,426 | 25.89 | CP068585 | |||||||
| CC19CH076 | Chicken | C. coli | Chromosome: 1,781,471 | 31.47 | ST825 | Completed | 18,553 | 36.36 | CP068586 |
| pCH076-80: 80,131 | 26.09 | CP068587 | |||||||
| pCH076-4: 4,944 | 29.57 | CP068588 | |||||||
| pCH076-3: 3,405 | 31.01 | CP068589 | |||||||
| pCH076-2: 2,426 | 25.89 | CP068590 | |||||||
| ZS007 | Duck meat | C. jejuni | Chromosome: 1,658,567 | 30.55 | ST10317 | Completed | 22,697 | 38.01 | CP048771 |
| 1712SZ1KX20C | Chicken | C. coli | 1,713,884 | 31.36 | ST825 | Draft | 9,611 | 36.79 | JAATKE000000000 |
MLST, multilocus sequence type.
TABLE 2.
Acquired drug resistance genes and MIC of optrA+ fexA+ isolates
| Isolate | Acquired drug resistance genes | MIC (μg/ml) ofa: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | NAL | GEN | TET | CLI | ERY | AZM | TEL | FFC | ||
| CC19DZ036 | tet(L), tet(O), optrA, fexA, catA9, blaOXA-61 | 16 | 64 | 0.5 | 32 | 4 | >64 | >32 | 32 | >32 |
| CC19DZ037 | tet(L), tet(O), optrA, fexA, catA9, blaOXA-61 | 16 | 128 | 0.5 | 32 | 4 | >64 | >32 | 32 | >32 |
| CC19PF050 | aac(6′)-aph(2′′), aph(3′)-III, aph(2′′)-If, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-465, erm(B) | 16 | >128 | >64 | >64 | >32 | >64 | >32 | 32 | >32 |
| CC19PF065 | aac(6′)-aph(2′′), aph(3′)-III, aph(2′′)-If, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-465, erm(B) | 16 | 128 | >64 | >64 | 16 | >64 | >32 | 32 | >32 |
| CC19CH074 | aac(6′)-aph(2′′), aph(3′)-III, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-61, erm(B) | 32 | 64 | >64 | 64 | >32 | 64 | 32 | 32 | >32 |
| CC19CH075 | aac(6′)-aph(2′′), aph(3′)-III, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-61, erm(B) | 32 | 64 | >64 | 64 | >32 | 64 | 16 | 16 | >32 |
| CC19CH076 | aac(6′)-aph(2′′), aph(3′)-III, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-61, erm(B) | 32 | 64 | 64 | 64 | >32 | >64 | 32 | 16 | >32 |
Abbreviations: CIP, ciprofloxacin; NAL, nalidixic acid; GEN, gentamicin; TET, tetracycline; CLI, clindamycin; ERY, erythromycin; AZM, azithromycin; TEL, telithromycin; FFC, florfenicol. CLSI- or NARMS-approved breakpoint concentrations for resistance, in micrograms per milliliter, are as follows: CIP, 4; NAL, 32; GEN, 4; TET, 16; CLI, 8; ERY, 32; AZM, 1; TEL, 8; FFC, 8.
Comparison of nucleic acid homology of fexA and optrA genes in different strains. Download FIG S1, DOCX file, 0.3 MB (339.5KB, docx) .
Copyright © 2021 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
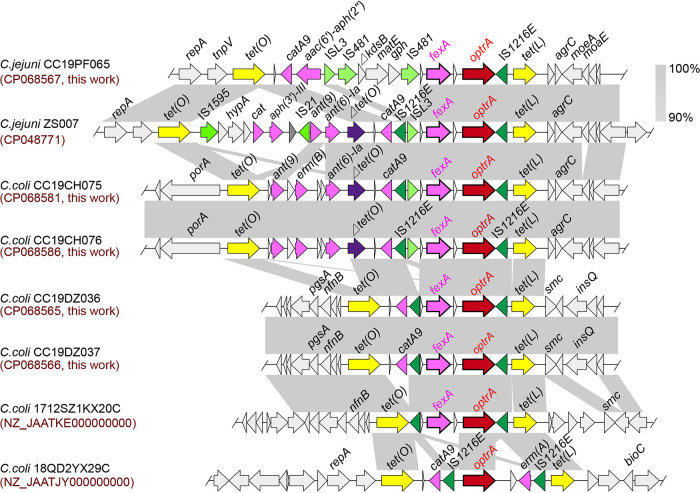
C. jejuni CC19PF065 belongs to a new ST (the nearest STs are 8670, 8672, and 10317), and the complete genome was 1,681,082 bp in length, with a GC content of 30.57% (accession no. CP068567). optrA along with its upstream phenicol exporter fexA and the gene hp, which encodes a hypothetical protein, were located within a 18,223-bp MDRGI with 34.79% GC content. The formed the MDRGI tet(O)-hp-catA9-aac(6′)-aph(2′)-ISL3-IS481-ΔkdsB-matE-gph-IS481-hp-fexA-hp-optrA-IS1216E-tet(L) (Fig. 1) was inserted into the C. jejuni housekeeping genes, between repA and agrC. In addition to fexA and optrA, this MDRGI also contained other genes conferring resistance to tetracycline and aminoglycosides. Moreover, ΔkdsB (3-deoxy-manno-octulosonate cytidylyltransferase), matE (multiantimicrobial extrusion protein), and gph (phosphoglycolate phosphatase) were flanked by the insertion sequence IS481 in the same orientation. The segment IS481-ΔkdsB-matE-gph-IS481 (3,709 bp; 36.88% GC content) exhibited 78.3% nucleotide sequence identity to the corresponding region of Helicobacter cholecystus strain NCTC 13205 (accession no. LR134518), with matE encoding a novel MATE family efflux transporter, most likely from Helicobacter (Fig. S2).
FIG 1.
Genetic environment of optrA in the genomes of C. jejuni isolates and comparison of the optrA-carrying regions. Arrows indicate the transcription direction. Regions of >90% homology are marked with gray shading. Genes are differentiated by different colors.
(A) Comparative analysis of ΔkdsB-matE-gph regions between strain Z65 CC19PF065 and Helicobacter cholecystus NCTC 13205. (B) Phylogenetic analysis of MatE protein of C. jejuni Z65 CC19PF065. Download FIG S2, DOCX file, 0.3 MB (328.7KB, docx) .
Copyright © 2021 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. jejuni ZS007 belonged to ST10317, with a complete genome 1,658,567 bp in length and a GC content of 30.55% (accession no. CP048771). This strain was previously reported to contain fexA. Here, the optrA containing MDRGI was found to be 22,697 bp in length (38.01% GC content) (Table 1). The order of gene content was tet(O)-IS1595-hp-hypA-hp-cat-aph(3′)-III-hp-IS21-ant(9)-ant(6)-Ia-Δtet(O)-hp-catA9-IS1216E-ISL3-hp-fexA-hp-optrA-IS1216E-tet(L), which was inserted between repA and agrC. The fragment hp-fexA-hp-optrA-IS1216E-tet(L) was the same as that from C. jejuni CC19PF065. The segment IS1595-hp-hypA-hp-cat-aph(3′)-III-hp-IS21 showed 98.92% nucleotide sequence identity to the corresponding region of the C. jejuni strain BC chromosome (accession no. CP032522).
In addition, a novel optrA-containing MDRGI was also characterized in C. coli by comparing the results from published studies (7, 9). According to the sequencing results, C. coli CC19CH075 and CC19CH076 belonged to ST825, and the complete genome sequences were 1,781,472 bp (accession no. CP068581) and 1,781,471 bp (accession no. CP068586) in length, with a GC content of 31.47% (Table 1). In total, 60 single nucleotide polymorphisms and seven gaps were found between the two strains. The order of gene content was tet(O)-hp-ant(9)-hp-erm(B)-hp-hp-ant(6)-Ia-Δtet(O)-hp-catA9-IS1216E-ISL3-hp-fexA-hp-optrA-IS1216E-tet(L), which was inserted between porA and agrC. By comparing with the segment from C. jejuni ZS007, hp-ant(9)-hp-erm(B)-hp-hp was found to be replaced by IS1595-hp-hypA-hp-cat-aph(3′)-III-hp-IS21-ant(9) in C. coli strains CC19CH075 and CC19CH076.
C. coli CC19DZ036 and CC19DZ037 belong to ST828, and the complete genomes were 1,761,335 and 1,761,334 bp in length, with a GC content of 31.37% (Table 1) (accession no. CP068565 and CP068566, respectively). In total, only three gaps difference were found between the two strains. The order of gene content was tet(O)-hp-catA9-IS1216E-hp-fexA-hp-optrA-IS1216E-tet(L), which was inserted between nfnB and smc.
This study revealed that the emerging gene optrA is associated with various MDRGIs in C. jejuni. Moreover, the core segment fexA-hp-optrA-IS1216E was identified in both C. jejuni and C. coli, which agrees with previous reports (7). Of note, the optrA-containing MDRGIs varied from 9,611 to 22,697 bp and were inserted into different regions over the genomes of Campylobacter, all of which contained tet(O) and tet(L) at the two ends. The GC content of these MDRGIs ranged from 34.79% to 38.01%, which is different from the GC content of the Campylobacter genome (∼31.0%), suggesting that Campylobacter might have obtained these MDRGIs from other species. There were 12 antimicrobial resistance genes in MDRGI, including aac(6′)-aph(2′′), aph(3′)-III, aph(2′′)-If, ant(6)-Ia, tet(L), tet(O), optrA, fexA, cat, catA9, blaOXA-465, and erm(B), which were resistant to aminoglycosides, tetracyclines, phenicol, and macrolides. All of these antibiotics were used for the prevention and treatment of infections in farm animals in China. In addition, the GC content within several MDRGIs was not evenly distributed, and the presence of multiple insertion sequences suggested that their integration may have occurred through a multistep process. Therefore, MDRGI in Campylobacter was likely to be the product of multiple-antibiotic coselection. Due to the use of florfenicol in livestock and poultry production, the emergence of fexA and optrA could confer a fitness advantage under selection pressure, which will support the spread of fexA and optrA and their associated MDRGIs through Campylobacter natural transformation.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2017YFC1601501), the Key Research and Development Program of Zhejiang Province (2020C02031), State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-ZZ2008), and National Natural Science Foundation of China (31761133004, 31722057, and 31700007).
REFERENCES
- 1.Liu D, Deng F, Gao Y, Yao H, Shen Z, Wu C, Wang Y, Shen J. 2017. Dissemination of erm(B) and its associated multidrug-resistance genomic islands in Campylobacter from 2013 to 2015. Vet Microbiol 204:20–24. doi: 10.1016/j.vetmic.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Liu D, Liu W, Lv Z, Xia J, Li X, Hao Y, Zhou Y, Yao H, Liu Z, Wang Y, Shen J, Ke Y, Shen Z. 2019. Emerging erm(B)-mediated macrolide resistance associated with novel multidrug resistance genomic islands in Campylobacter. Antimicrob Agents Chemother 63:e00153-19. doi: 10.1128/AAC.00153-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, Wu C, Wang S, Zhang J, Shen J. 2014. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother 69:964–968. doi: 10.1093/jac/dkt492. [DOI] [PubMed] [Google Scholar]
- 4.Tang Y, Dai L, Sahin O, Wu Z, Liu M, Zhang Q. 2017. Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J Antimicrob Chemother 72:1581–1588. doi: 10.1093/jac/dkx023. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Liu D, Li X, Wang Y, Schwarz S, Shen J. 2020. Emergence of the phenicol exporter gene fexA in Campylobacter coli and Campylobacter jejuni of animal origin. Antimicrob Agents Chemother 64:e00240-20. doi: 10.1128/AAC.00240-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Yang D, Liu X, Li X, Feßler AT, Shen Z, Shen J, Schwarz S, Wang Y, 2020. Detection of the enterococcal oxazolidinone/phenicol resistance gene optrA in Campylobacter coli. Vet Microbiol 246:108731. doi: 10.1016/j.vetmic.2020.108731. [DOI] [PubMed] [Google Scholar]
- 8.Tang B, Tang Y, Zhang L, Liu X, Chang J, Xia X, Yang H, Shen Z. 2020. Emergence of fexA in mediating resistance to florfenicols in Campylobacter. Antimicrob Agents Chemother 64:e00260-20. doi: 10.1128/AAC.00260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Lai Y, Wang X, Lei C, Li C, Kong L, Wang Y, Wang H. 2020. Novel insertion sequence ISChh1-like mediating acquisition of optrA gene in foodborne pathogen Campylobacter coli of swine origin. Vet Microbiol 252:108934. doi: 10.1016/j.vetmic.2020.108934. [DOI] [PubMed] [Google Scholar]
- 10.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, Wu C, Yu EW, Zhang Q, Shen J. 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 12.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of nucleic acid homology of fexA and optrA genes in different strains. Download FIG S1, DOCX file, 0.3 MB (339.5KB, docx) .
Copyright © 2021 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Comparative analysis of ΔkdsB-matE-gph regions between strain Z65 CC19PF065 and Helicobacter cholecystus NCTC 13205. (B) Phylogenetic analysis of MatE protein of C. jejuni Z65 CC19PF065. Download FIG S2, DOCX file, 0.3 MB (328.7KB, docx) .
Copyright © 2021 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.