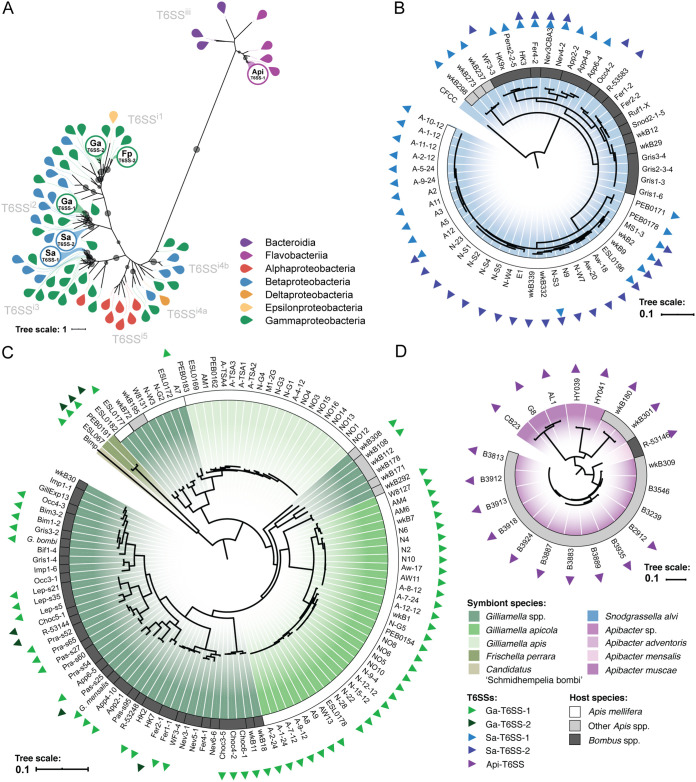
FIG 2.
The genomes of bee gut symbionts encode multiple conserved T6SSs. (A) A maximum-likelihood phylogeny of the TssB protein, a conserved structural component of T6SSs, shows evolutionary relationships between the T6SSs of bee gut symbionts and previously described T6SS subfamilies. Leaf colors indicate the bacterial class from which each TssB sequence was extracted (further described in Table S2 in the supplemental material). Labeled circles represent TssB proteins associated with T6SS loci found in bee symbionts, as follows: Sa-T6SS-1, Sa-T6SS-2, Ga-T6SS-1, Ga-T6SS-2, Fp-T6SS-2, and Api-T6SS-1. Gray points on branches indicate >70% support with 1,000 bootstraps. (B) S. alvi genome phylogeny constructed from 719 single-copy gene trees. Each leaf represents a single sequenced isolate genome. The color of the ring around the phylogeny indicates the host species from which each strain was isolated: White, Apis mellifera (Western honey bees); light gray, Apis spp. (other honey bees); dark gray, Bombus spp. (bumble bees). Colored triangles at the outer edge of each strain name indicate that the strain genome encodes one or more T6SS loci. (C) Gilliamella spp. genome phylogeny, constructed from 927 single-copy gene trees, with Frischella perrara and “Candidatus Schmidhempelia bombi” as outgroups. (D) Apibacter spp. genome phylogeny constructed from 1,279 single-copy gene trees. Shading of phylogenies highlights different species within each genus.