Abstract
A cascade formed by phosphorylation events of mitogen‐activated protein kinases (MAPKs) takes part in plant stress responses. However, the roles of these MAPKs in resistance of potato (Solanum tuberosum) against Phytophthora pathogens is not well studied. Our previous work showed that a Phytophthora infestans RXLR effector targets and stabilizes the negative regulator of MAPK kinase 1 of potato (StMKK1). Because in Arabidopsis thaliana the AtMPK4 is the downstream phosphorylation target of AtMKK1, we performed a phylogenetic analysis and found that potato StMPK4/6/7 are closely related and are orthologs of AtMPK4/5/11/12. Overexpression of StMPK4/7 enhances plant resistance to P. infestans and P. parasitica. Yeast two‐hybrid analysis revealed that StMPK7 interacts with StMKK1, and StMPK7 is phosphorylated on flg22 treatment and by expressing constitutively active StMKK1 (CA‐StMKK1), indicating that StMPK7 is a direct downstream signalling partner of StMKK1. Overexpression of StMPK7 in potato enhances potato resistance to P. infestans. Constitutively active StMPK7 (CA‐StMPK7; StMPK7D198G, E202A) was found to promote immunity to Phytophthora pathogens and to trigger host cell death when overexpressed in Nicotiana benthamiana leaves. Cell death triggered by CA‐StMPK7 is SGT1/RAR1‐dependent. Furthermore, cell death triggered by CA‐StMPK7 is suppressed on coexpression with the salicylate hydroxylase NahG, and StMPK7 activation promotes salicylic acid (SA)‐responsive gene expression. We conclude that potato StMPK7 is a downstream signalling component of the phosphorelay cascade involving StMKK1 and StMPK7 plays a role in immunity to Phytophthora pathogens via an SA‐dependent signalling pathway.
Keywords: MAPK, oomycete, Phytophthora infestans, plant resistance, potato
Potato StMPK7 is a downstream signalling component of StMKK1 and it enhances plant resistance to Phytophthora pathogens by activating the salicylic acid‐mediated signalling pathway.
1. INTRODUCTION
The oomycete plant pathogen Phytophthora infestans causes the most notorious disease on potato, known as potato late blight, which under humid and cool conditions can devastate a susceptible crop within 2 weeks (Fry, 2008). Although for many years potato breeders have brought resistance genes from wild potato varieties into cultivated potatoes, potato late blight disease is still the number one problem in potato production, and control of the disease is still largely dependent on fungicide sprays. Potato resistance genes against late blight are all from the nucleotide‐binding leucine‐rich repeat (NLR) gene family (Du et al., 2015). NLR genes confer race‐specific resistance against late blight, and each NLR is proposed to recognize a corresponding RXLR effector of P. infestans and triggers effector‐triggered immunity (ETI). Consequently, resistance provided by NLR genes is easily broken by P. infestans isolates that have lost the corresponding RXLR effector gene (Vleeshouwers et al., 2011). For example, the NLR gene R1 resistance is broken by P. infestans isolates that have deleted AVR1 in their genome (Du et al., 2015), whereas R3a resistance is broken by a point mutation in the gene encoding the AVR3a effector (Engelhardt et al., 2012). Currently, in potato production the loss of resistance of originally resistant cultivars is becoming a serious problem. Plant basal resistance is an important part of microbe‐associated molecular pattern (MAMP)‐triggered immunity (MTI), which is conferred by plasma membrane‐localized receptor‐like proteins (RLPs), lacking a cytoplasmic kinase domain, or receptor‐like kinases (RLKs) proteins that do have such a domain, together known as pattern recognition receptors (PRRs) (Jones & Dangl, 2006).
Mitogen‐activated protein kinase (MAPK) cascades exist widely in eukaryotes, and in plants they participate in responses to biotic and abiotic stresses, hormones, cell differentiation, and developmental processes. On immune activation, MAPK cascades transduce signals through phosphorylation by upstream MAPKKKs/MEKKs to their downstream targets, eventually activating the immune response in the nucleus as a result of transcription factor phosphorylation (Pitzschke, Schikora, et al., 2009). A MAPK cascade normally comprises a MAP kinase kinase kinase (MAPKKK, or MEKK), a downstream MAP kinase kinase (MAPKK), and a further downstream MAP kinase (MPK) (Kong et al., 2012), and the activation of upstream MAPKKKs results in the sequential phosphorylation of downstream MKKs and MPKs. On MTI activation, plants activate at least two different MAPK cascades, which are the MKKK‐MKK4/5‐MPK3/6 cascade and the MEKK1‐MKK1/2‐MPK4 cascade (Zhang et al., 2017). Activated MPK3/6 subsequently enhance the expression of defence‐related genes, such as the gene encoding the fructokinase FRK1 and the transcription factors WRKY22 and WRKY29, thereby mounting immunity to bacteria and fungi (Asai et al., 2002; Wang et al., 2018). MAPK cascades also participate in ETI. For instance, tomato LeMPK1, LeMPK2, and LeMPK3 all participate in the hypersensitive response triggered upon recognition of the Avr4 effector of the fungal pathogen Cladosporium fulvum by the Cf‐4 resistance protein of tomato (Stulemeijer et al., 2007), potato MAP3K proteins StMAP3Kε and StMAP3Kβ2 function in plant immunity to P. infestans (King et al., 2014; Ren et al., 2019), whereas rice OsMAPK6 participates in Pit‐mediated resistance to rice blast disease (Kawano et al., 2010). Furthermore, the MEKK1‐MKK1/2‐MPK4 cascade in the model plant Arabidopsis thaliana is monitored by the NLR protein SUMM2, and perturbation of this cascade activates SUMM2‐mediated immunity (Zhang et al., 2012).
The Arabidopsis genome encodes 20 MPK proteins and phylogenetic analysis of these MPKs shows that they can be divided into four clades, labelled from A to D (Ichimura et al., 2002). The conserved motif of MPKs, which is phosphorylated by upstream MPKKs, TxY, of which the tyrosine (Y) residue is phosphorylated, can be classified into two subtypes, the TEY and TDY subtypes. The TEY subtype is classified into clades A, B, and C, whereas the TDY subtype forms clade D. The clade B MPKs participate in plant growth and development, and responses to biotic and abiotic stress. For example, rice OsMPK6 negatively regulates resistance to Xanthomonas oryzae (Yuan et al., 2007), whereas tomato SlMPK4 positively regulates resistance to Botrytis cinerea (Virk et al., 2013) and Nicotiana attenuata MPK4 participates in the response to insects (Hettenhausen et al., 2013).
MAPK cascades were also found to participate in plant hormone signalling to modulate immunity to insects and microbial pathogens (Hou et al., 2013; Jagodzik et al., 2018). For example, tomato MPKs are known to regulate jasmonic acid (JA) biosynthesis and thereby JA‐related gene expression to modulate immunity against herbivorous insects (Kandoth et al., 2007). Arabidopsis MPK3 was found to suppress flg22‐induced salicylic acid (SA) accumulation (Frei dit Frey et al., 2014). There are also MPK(K)s that participate in both JA and SA signalling, that is, AtMPK4, AtMPK6, SlMKK4, and SlMKK9 (Chai et al., 2014; Li et al., 2014; Pitzschke, Djamei, et al., 2009).
MAPK cascade proteins have been extensively studied in model plants. However, there are only a few studies described that concern the role of MAPK proteins in immunity in crop plants, for instance potato. The StMEK1‐StMPK1/StWIPK (the orthologs of Arabidopsis AtMKK4/5‐AtMPK6/3) cascade is reported in potato to participate in immunity to P. infestans (Yamamizo et al., 2006). Our previous work showed that a P. infestans RXLR effector targets and stabilizes the potato negative immune regulator StMKK1 (Du et al., 2021). AtMPK4 is the downstream signalling target of AtMKK1/2 in immunity (Zhang et al., 2012), while AtMPK5 was known to be activated by AtMKK2 under stress conditions (Teige et al., 2004). To investigate the downstream immune signalling target of potato StMKK1, we BLAST‐searched the potato genome with the AtMPK4 amino acid sequence and performed a phylogenetic analysis of potato clade B MPKs. We identified three orthologous MPK proteins in potato, and named them StMPK4/6/7 according to their tomato orthologs (Virk et al., 2013), as orthologs of AtMPK4/5/11/12. Inoculation assays showed that StMPK4 and StMPK7 participate in immunity to Phytophthora pathogens, while StMPK6 does not show such a role. Silencing of the NbMPK4/6/7 genes in N. benthamiana also showed that NbMPK7 and NbMPK4 participate in resistance to Phytophthora pathogens. The potato stable overexpression (OE) transformants StMPK7‐OE lines show enhanced resistance to P. infestans. To determine whether StMPK7 functions as a downstream signalling target of StMKK1 in plant immunity, we confirmed that an interaction takes place between StMPK7 and StMKK1 and observed that StMPK7 is phosphorylated by a constitutively active form of StMKK1 (CA‐StMKK1). A constitutively active form of StMPK7 (CA‐StMPK7) was found to promote immunity to P. infestans and Phytophthora parasitica and to trigger cell death when overexpressed in N. benthamiana leaves. Taken together, we conclude that potato StMPK7 is a downstream signalling component of StMKK1 and plays a role in immunity to Phytophthora pathogens.
2. RESULTS
2.1. Identification and phylogenetic analysis of StMPK7
To identify the potato orthologs of MPK4/5, we BLAST‐searched the predicted proteomes of potato, tomato, and N. benthamiana (sol genomics network, https://solgenomics.net/tools/blast/) with the AtMPK4 protein sequence as a query and identified several homologous proteins. Because AtMPK4 and AtMPK5 are closely related proteins that belong to the clade B MPKs, we performed a phylogenetic analysis of clade B MPKs using the MPK protein sequences of various Solanaceae, Populus, Oryza, and Arabidopsis. Our phylogenetic tree showed that there are two proteins of potato (StMPK4, PGSC0003DMP400000144 and StMPK6, PGSC0003DMP400037535) that are orthologous proteins of Arabidopsis AtMPK4/11/12, whereas StMPK7 (PGSC0003DMP400021542) is the orthologous protein of AtMPK5 (Figure 1). Phylogenetic analysis showed that potato MPK4/6/7 are closely related homologous proteins belonging to subclade B1 of the clade B MPKs. These subclade B1 MPKs contain four MPKs MPK4/5/11/12 from Arabidopsis, four from Populus, two from Oryza, and three MPKs from tomato and potato. It seems that the duplication of solanaceous subclade B1 MPKs occurred twice independently, once in the common ancestor of Arabidopsis, Populus, and Solanaceae, and once in the common ancestor of solanaceous plants (Figure 1).
FIGURE 1.
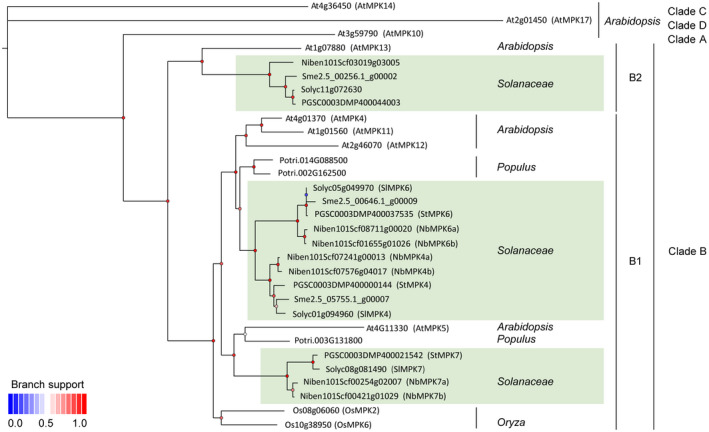
Phylogenetic analysis of clade B MPK proteins from Arabidopsis, Populus, various Solanaceae, and Oryza. The phylogenetic tree was inferred by the maximum‐likelihood method, using PhyML. The SH‐like support values of branches that represent the lower or higher values are indicated by dots from blueto red. The homologous proteins were BLAST‐searched against the genomes of Arabidopsis, Populus, Solanaceae, and Oryza
2.2. StMPK7 and StMPK4 promote plant resistance to Phytophthora pathogens
We cloned all three genes encoding the potato MPK4/6/7 homologs. To study the role of potato StMPK4/6/7 in plant defence to P. infestans, green fluorescent protein (GFP)‐β‐glucuronidase (GUS) and GFP‐StMPK7 were transiently expressed in the left and right panels of N. benthamiana leaves, respectively, and P. infestans was inoculated onto the leaves at 1 day after agroinfiltration. The results showed that, when compared to the GFP‐GUS‐expressing leaf halves, GFP‐StMPK7 expression resulted in significantly smaller lesions (Figure 2a–c), indicating that StMPK7 positively regulates resistance to P. infestans. To investigate whether StMPK7 has a similar role in resistance to other Phytophthora pathogens, P. parasitica was also inoculated onto the leaves. The results showed that StMPK7 also promotes resistance to P. parasitica (Figure 2d–f). The roles of StMPK4 and StMPK6 in plant immunity were also investigated in a similar way. It was observed that StMPK4 promotes plant resistance to both P. infestans and P. parasitica, while StMPK6 does not appear to play a role in immunity (Figure S1). The accumulation of the StMPK7/4/6 proteins and the controls GFP‐GUS and GUS‐myc was confirmed by western blotting, as shown in Figure S1j.
FIGURE 2.
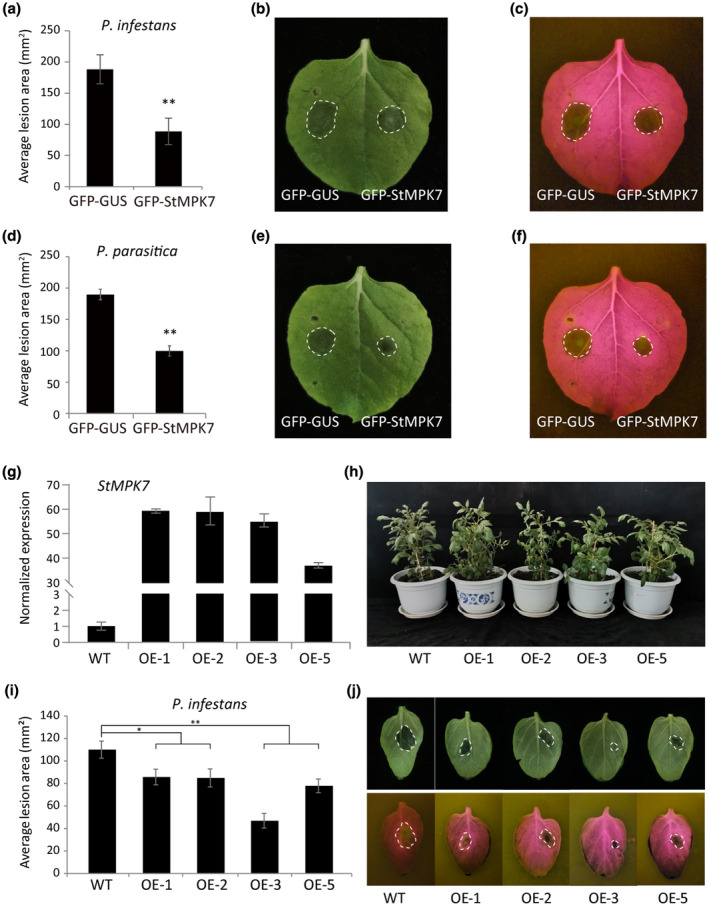
Overexpression of potato StMPK7 promotes resistance to Phytophthora pathogens. GFP‐StMPK7 (right) and control GFP‐GUS (left) were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day postinfiltration (dpi), the Phytopthora infestans isolate 14–3‐GFP (a, b, c) and Phytophthora parasitica isolate 1121 (d, e, f) were inoculated onto the leaves. The lesion diameters were measured at 5 days after inoculation (dai) for P. infestans and at 2 dai for P. parasitica, and average lesion areas (in mm2) were calculated and are shown in (a) and (d), respectively. Representative images, taken under normal light (b, e) or blue light (c, f), show lesion development on GFP‐GUS‐ and GFP‐StMPK7‐expressing leaves. (g) Quantitative reverse transcription PCR analysis shows StMPK7 expression in wild‐type (WT) and StMPK7‐overexpressing (OE) plants. Lines OE‐1, OE‐2, OE‐3, and OE‐5 are independent transgenic lines. The expression level of StActin was used as an internal control and the WT value was used as the reference. (h) Plant morphology of 2‐month‐old WT and StMPK7‐OE transgenic potato lines. (i) Graphs show the average lesion areas in mm2 in WT and StMPK7‐OE lines at 4 dai with P. infestans. (j) Representative images taken under normal (upper panel) or blue light (lower panel), showing lesion development on WT and StMPK7‐OE lines at 4 dai. Error bars indicate the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (one‐sided Student's t test, *p < .05, **p < .01). The experiments were repeated three times with similar results
To further investigate the role of MPK4/6/7 in N. benthamiana immunity, we silenced NbMPK7 (Figure 3), NbMPK4, and NbMPK6 (Figure S2) in N. benthamiana and inoculated the silenced plants with P. infestans or P. parasitica. Detached leaf assays revealed that, when compared to the control TRV‐GUS, the NbMPK7‐ and NbMPK4‐silenced plants developed larger lesions, while the NbMPK6‐silenced plants did not show altered immunity to the P. infestans (Figures S2 and S3), which confirms the positive roles that MPK7 and MPK4 play in plant resistance to Phytophthora pathogens. NbMPK7 and MPK6 silencing does not alter plant morphology, while NbMPK4 silencing results in larger plants when compared to the control treatment (Figure S3).
FIGURE 3.
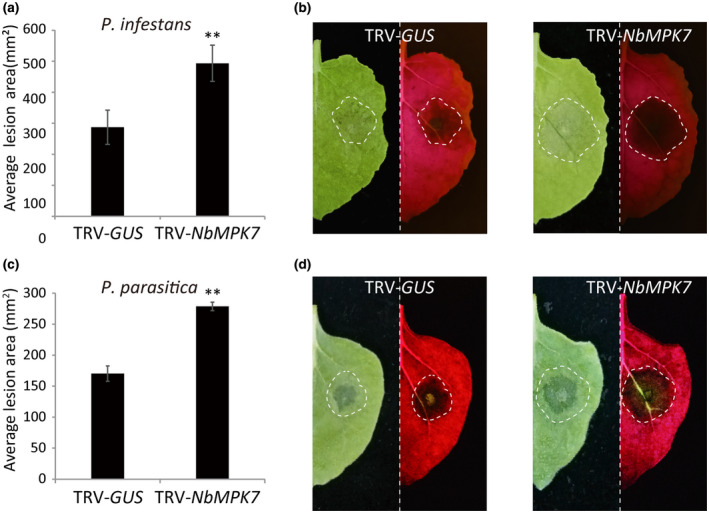
Silencing of NbMPK7 promotes colonization by Phytophthora pathogens. Average lesion areas at 5 days after inoculation (dai) for Phytophthora infestans (a) and at 2 dai for Phytophthora parasitica (c) on TRV‐GUS‐ and TRV‐NbMPK7‐inoculated plants are shown. The standard error from more than 10 technical replicates is indicated. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student's t test, p ≤ .01). (b, d) Representative images taken under normal (left panel) or blue light (right panel), showing the lesion development on TRV‐GUS‐ (left) and TRV‐NbMPK7‐ (right) inoculated leaves. The experiments were repeated three times with similar results
MPK4 has been extensively studied in Arabidopsis and is known as the downstream signalling target of StMKK1; however, the role of AtMPK5 and its potato ortholog StMPK7 is not well studied for its role in immunity. Thus, we chose StMPK7 for further investigation. To further confirm the role of StMPK7 in potato defence against P. infestans, GFP‐StMPK7 was transformed into the potato cultivar Desirée via Agrobacterium‐mediated transformation. Four independent transgenic lines were obtained and the enhanced expression of StMPK7 in transgenic lines was detected by quantitative reverse transcription PCR (RT‐qPCR) (Figure 2g). The transformants and control plants were grown over a period of 2 months and no developmental phenotypes were observed between the wild‐type (WT) and StMPK7‐overexpression (OE) lines (Figure 2h). The results of detached leaf assays showed that compared to the control plants, the StMPK7‐OE lines developed smaller lesions at 4 days after inoculation with P. infestans (Figure 2i,j). These results confirm that overexpression of StMPK7 in potato enhances plant immunity to the late blight pathogen P. infestans.
2.3. StMPK7 interacts with potato StMKK1 and is phosphorylated by this MAPKK
Both StMPK7 and StMPK4 participate in plant immunity and their Arabidopsis orthologs AtMPK4 and AtMPK5 are reported to be activated by AtMKK1/2 (Teige et al., 2004). The interaction between StMPK7 and StMKK1 was confirmed by yeast two‐hybrid analysis. The results showed that StMPK7 strongly interacted with StMKK1 (Figure 4a). To further conform the interaction of StMKK1 with StMPK7, we performed coimmunoprecipitation assays (Co‐IP) and firefly luciferase complementation imaging (LCI) assays. Results showed that myc‐StMKK1 specifically coimmunoprecipitated with GFP‐StMPK7 but not the negative control GFP (Figure 4b). For LCI assays, the results showed that the combination of StMPK7‐nluc with cluc‐StMKK1 restored the luciferase catalytic activity (Figure 4c), similar to the positive control GmBZR2‐nluc and cluc‐PP2C (Lu et al., 2017). To investigate whether StMPK7 is phosphorylated by StMKK1, we cloned StMPK7 from the potato cultivar Tian11 into the pART‐27 vector and fused an N‐terminal GFP tag to the encoded protein. We transiently expressed the control GUS‐myc or constitutively active (CA) StMKK1‐myc (StMKK1T220D, T226E), together with GFP‐StMPK7, and checked the activation of the latter by phosphorylation, using anti‐pERK antibodies. CA‐StMKK1‐myc was indeed found to phosphorylate StMPK7 (Figure 4d), indicating that in potato StMPK7 is phosphorylated by StMKK1 on activation of the corresponding MAPK cascade. To investigate whether StMPK7 is activated on treatment with flg22, we transiently expressed StMPK7 in N. benthamiana leaves and treated the plants with flg22 and determined whether StMPK7 was activated as a result of phosphorylation. Results showed that StMPK7 was indeed phosphorylated upon treatment with flg22 (Figure 4e). Similarly, the interaction of StMKK1 with StMPK4 was proved by Co‐IP, and StMPK4 was found to be phosphorylated by the CA‐StMKK1‐myc (Figure S4a,b). The NbMPK4‐silenced plants significantly reduced SA‐related gene expression compared to control (Figure S4c).
FIGURE 4.
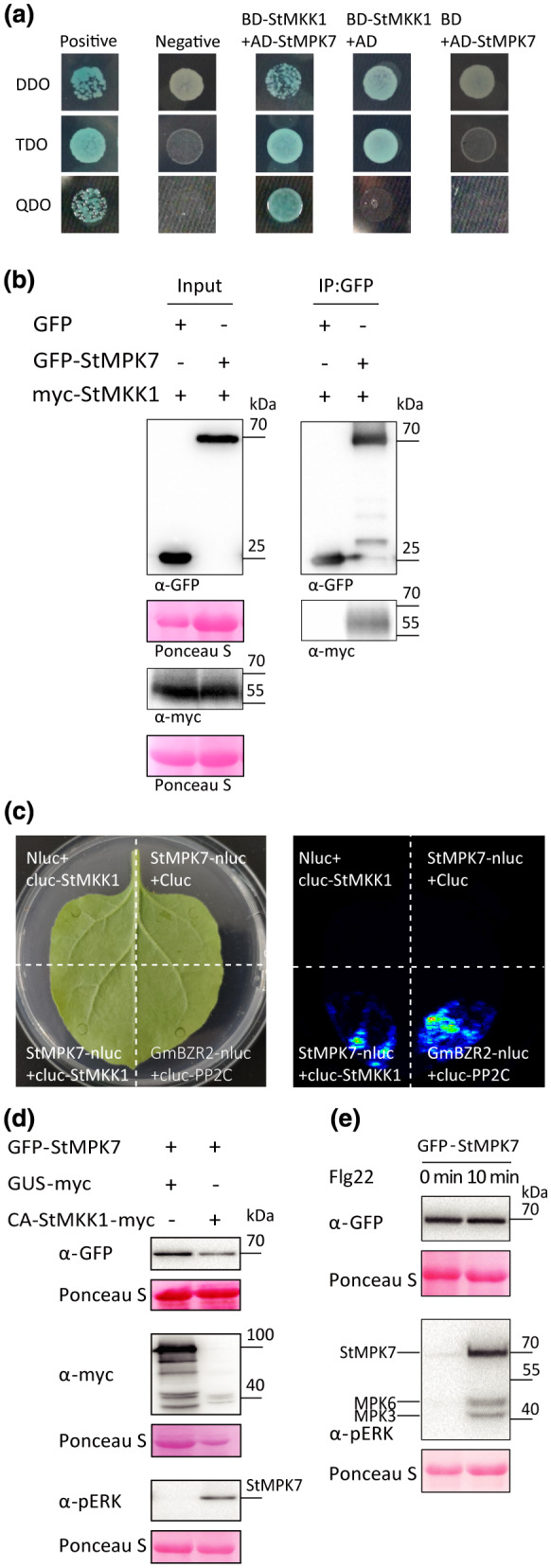
Potato StMPK7 interacts with StMKK1 and is phosphorylated by this MAPKK. (a) Yeast coexpressing BD‐StMKK1 and AD‐StMPK7, and the positive control, did grow on quadruple drop‐out (QDO) medium and yielded α‐galactosidase activity, while the negative control and yeast coexpressing the empty vector BD with AD‐StMPK7, or BD‐StMKK1 with the empty vector AD, did not. (b) Coimmunoprecipitation (Co‐IP) assays showed that StMKK1 associated with StMPK7. Green fluorescent protein (GFP) was used as negative control. Protein loading is indicated by Ponceau stain (Ponceau S). (c) Firefly luciferase complementation imaging (LCI) assays showed that StMPK7 interacts with StMKK1. GmBZR2‐nluc and cluc‐PP2C were used as positive control. Leaf samples were analysed and pictures were taken at 2−3 days postinfiltration using fluorescence imaging. (d) GFP‐StMPK7 was transiently coexpressed with either GUS‐myc or constitutively active CA‐StMKK1‐myc in Nicotiana benthamiana leaves. Total protein was extracted and the activation of GFP‐StMPK7 through phosphorylation was detected by western blot, using α‐pERK antibody. Ponceau S stain indicates the equal loading of the total protein extract. (c) Flg22‐induced MAPK activation in GFP‐StMPK7‐overexpressing plants was detected by western blot, using α‐pERK antibody. Leaves were treated with 10 µM flg22 and total proteins were extracted at 0 and 10 min after treatment
To investigate whether StMPK7 plays a role in the plant MTI response, we treated N. benthamiana plants, transiently overexpressing StMPK7, with flg22 and determined the expression levels of the MTI marker genes FRK1 and WRKY33. Results showed that, when compared to the GUS control, StMPK7 overexpression significantly enhanced FRK1 and WRKY33 gene expression (Figure S5). Furthermore, to investigate the subcellular localization of StMPK7, we transiently expressed GFP‐StMPK7 or GFP only in leaves of N. benthamiana, and at 2 days postinfiltration confocal microscopy was employed to observe the subcellular localization of StMPK7. The results showed that, similar to free GFP, GFP‐StMPK7 showed both a nuclear and a cytoplasmic localization (Figure S6a). The integrity of the GFP‐StMPK7 protein is shown in Figure S6b.
2.4. Kinase activity of StMPK7 is required for promoting resistance to P. infestans
The self‐activating mutation in the AtMPK4 protein (D198G, E202A), by which the protein becomes constitutively kinase‐active, has been reported before (Berriri et al., 2012), and we performed a sequence alignment of AtMPK4 with potato, tomato, and N. benthamiana MPK7, of which the results showed that the D198 and E202 sites are conserved among these homologous proteins (Figure S7). We then generated a constitutively active mutant of StMPK7, referred to as CA‐StMPK7‐myc (StMPK7D198G,E202A), to test whether the kinase activity of StMPK7 is required for defence against Phytophthora pathogens. The control GUS‐myc and CA‐StMPK7‐myc were agroinfiltrated into the left and right panels of N. benthamiana leaves, respectively, and at 1 dpi P. infestans or P. parasitica was inoculated onto these panels. CA‐StMPK7‐myc overexpression significantly promoted resistance to both P. infestans and P. parasitica (Figure S8). The resistance provided by the constitutively active mutant CA‐StMPK7‐myc was much stronger than that triggered by wild‐type StMPK7, and lesion formation by P. infestans was restricted to only the inoculation sites (Figure 5a,b). The accumulation of the myc‐ and GFP‐tagged proteins was confirmed by western blotting (Figure 5c).
FIGURE 5.
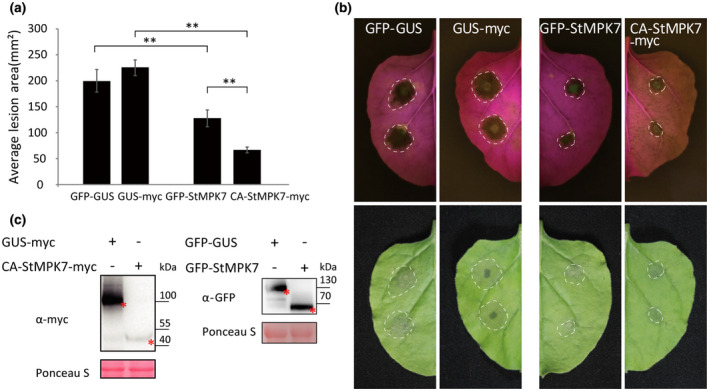
Overexpression of constitutively active (CA)StMPK7 confers stronger resistance to Phytophthora infestans than overexpression of wild‐type (WT) StMPK7. (a) Green fluorescent protein (GFP)‐β‐glucuronidase (GUS), GUS‐myc, GFP‐StMPK7, and CA‐StMPK7‐myc were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day postinfiltration (dpi), P. infestans isolate 14‐3‐GFP was inoculated at the agroinfiltrated sites. The lesion diameter was measured at 5 days after inoculation (dai) and average lesion areas were calculated and are shown in the graph. (b) Representative images taken under normal light (lower panel) or blue light (upper panel) show lesion development as a result of infection by P. infestans on N. benthamiana leaves. Error bars show the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student's t test, p ≤ .01). The experiment was repeated three times, with similar results. (c) The accumulation of CA‐StMPK7‐myc, GUS‐myc, GFP‐GUS, and GFP‐StMPK7 proteins is shown by western blot and the expected protein bands are marked with red asterisks
2.5. Constitutively active (CA) StMPK7 induces cell death in N. benthamiana leaves
We observed that transient expression of CA‐StMPK7‐myc in leaves of N. benthamiana triggered a mild cell death starting from 3 dpi, after which the infiltrated region had completely dried out at 5 dpi. At an OD600 of 0.7, CA‐StMPK7‐myc triggered 100% cell death at 5 dpi, while for an OD600 of 0.1, there was hardly any cell death observed (Figure 6a,b). To test whether transient expression of CA‐StMPK7‐myc induces reactive oxygen species (ROS) production, we performed 3,3′‐diaminobenzidine (DAB) staining to visualize H2O2 accumulation. Results showed that in the CA‐StMPK7‐myc‐expressing leaf half, significant amounts of H2O2 were generated when compared to the GUS‐myc control (Figure 6c). To check whether the different severities of the cell death triggered by CA‐StMPK7 were due to different levels of protein accumulation, we performed a western blot analysis with different OD600 of the infiltrated bacteria. Interestingly, wild‐type StMPK7 accumulated at much higher levels than CA‐StMPK7 at an OD600 of 0.1, and increasing the OD600 did not increase StMPK7 protein accumulation (Figure S9a). For CA‐StMPK7, at an OD600 of 0.1 a weak band was detected and on an increase in OD600, an enhanced accumulation of the CA‐StMPK7 protein was observed (Figure S9b). However, with an OD600 above 0.7, protein accumulation was less, probably due to an enhanced and faster cell death taking place in the leaves.
FIGURE 6.

Overexpression of constitutively active (CA) StMPK7 triggers an SGT1/RAR1‐dependent cell death in Nicotiana benthamiana leaves. (a) Cell death triggered by CA‐StMPK7‐myc in leaves of N. benthamiana. The number next to the infiltrated zone indicates the OD600 of the agrobacterial suspension used for agroinfiltration. (b) Cell death triggered by CA‐StMPK7‐myc is visible as dark spots in the leaves from which the chlorophyll was removed by treatment with ethanol. (c) 3,3′‐diaminobenzidine (DAB) staining revealing reactive oxygen species accumulation at the CA‐StMPK7‐myc‐infiltrated sites. (d) SGT1 and RAR1 are required for CA‐StMPK7‐myc‐triggered cell death. Three weeks after inoculation to initiate virus‐induced silencing of SGT1 or RAR1, the negative control GUS‐myc, the cell death inducer INF1, and CA‐StMPK7‐myc were agroinfiltrated at an OD600 of 1.0 into the middle leaves of silenced plants. Pictures were taken at 5 days postinfiltration (dpi). The ratios next to the infiltrated zones show the number of infiltration sites showing cell death versus the total amount of infiltrated sites from two independent experiments at 5 dpi
To investigate on which type of signalling components the cell death triggered by CA‐StMPK7‐myc depends, we silenced the Suppressor of the G2 allele of Skp1 (SGT1, a chaperone‐like protein generally required for R gene‐mediated immunity), Required for Mla12 resistance (RAR1, encoding a co‐chaperone of HSP90 required for ETI), the gene encoding the F‐box protein Col1 (involved in JA activation), and the gene encoding the MPK4 downstream signalling component WRKY33, and subsequently infiltrated CA‐StMPK7‐myc. The cell death‐inducer P. infestans elicitin INF1 and GUS‐myc were included as controls. The results showed that CA‐StMPK7‐myc‐triggered cell death was abolished in SGT1‐ and RAR1‐silenced plants, and was partially suppressed in WRKY33‐silenced plants, whereas Col1 appeared to play a less important role in the activation of cell death by CA‐StMPK7 (Figure 6d).
2.6. StMPK7 promotes potato resistance to Phytophthora pathogens via SA‐related immune signalling
Although SGT1 and RAR1 participate in multiple cellular processes, they are essential for R gene‐mediated immunity that largely depends on the hormone salicylic acid (SA) (Zhang & Li, 2019). Indeed, silencing of SGT1 has been reported earlier to reduce SA‐responsive gene expression (Rong et al., 2010). We thus investigated whether CA‐StMPK7‐myc‐triggered cell death depends on SA. SA is a key plant hormone regulating defence to Phytophthora pathogens (Zhou et al., 2018). We tested SA‐related marker gene expression in N. benthamiana leaves transiently expressing StMPK7. For this, GFP‐StMPK7 and the control GFP‐GUS were transiently expressed in N. benthamiana leaves, and at 2 days after agroinfiltration the leaves were harvested for RNA extraction. Expression of the SA‐related genes Pathogenesis‐related (PR) protein‐1 and PR‐2 was detected by RT‐qPCR, and we observed that overexpression of StMPK7 promoted NbPR‐1 and NbPR‐2 gene expression in N. benthamiana (Figure 7a). The SA‐related marker gene expression in potato StMPK7‐OE lines was also tested by RT‐qPCR, and results showed that in potato StMPK7‐OE transgenic lines the StPR1 and StPR2 gene expression was significantly enhanced compared to the control plants (Figure 7a). To test whether the cell death triggered by CA‐StMPK7 also requires SA‐related signalling, we coexpressed the salicylate hydroxylase NahG gene with CA‐StMPK7 and checked whether the cell death response was altered. Again, the P. infestans elicitin INF1 was used as a cell death control. Results showed that coexpression of NahG significantly suppressed the CA‐StMPK7‐triggered cell death, whereas the INF1‐triggered cell death was not altered by NahG coexpression (Figure 7b). The H2O2 accumulation at the sites agroinfiltrated with CA‐StMPK7, either in combination with GUS or with NahG, was measured by DAB staining. We observed that a significant amount of H2O2 was generated at the sites where CA‐StMPK7 and GUS were coexpressed and at the sites were INF1 and GUS/NahG were coexpressed, whereas the expression of NahG significantly repressed the CA‐StMPK7‐triggered H2O2 accumulation (Figure 7b). The repression of CA‐StMPK7‐triggered cell death by NahG was quantified by ion leakage assays. The reduced ion leakage that we observed confirmed that NahG significantly repressed CA‐StMPK7‐triggered cell death (Figure 7c).
FIGURE 7.
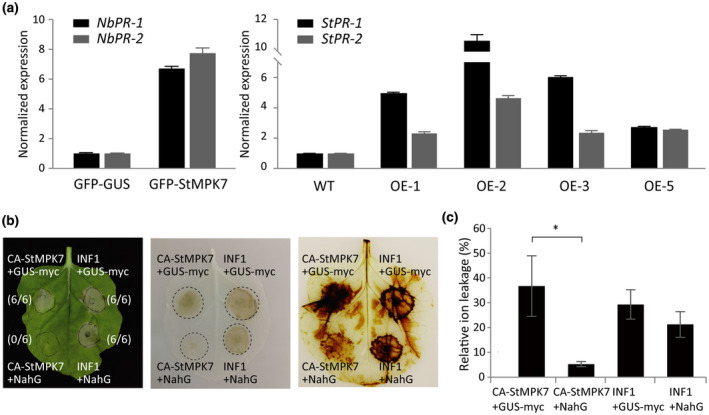
StMPK7 promotes plant resistance via salicylic acid (SA)‐related immune signalling. (a) StMPK7 promotes plant SA‐related gene expression. Graphs show the relative expression of NbPR‐1, NbPR‐2 in green fluorescent protein (GFP)‐β‐glucuronidase (GUS)‐, and GFP‐StMPK7‐overexpressing (OE) Nicotiana benthamiana leaves and StPR‐1, StPR‐2 in wild‐type (WT) and StMPK7‐OE stable transgenic potato lines. The expression of the NbActin gene and the StActin gene was used for normalization. Gene expression levels in GFP‐StMPK7‐expressing plants and stable StMPK7‐OE transgenic potato plants were significantly higher than those in the control plants. The experiments were repeated twice, with similar results. (b) CA‐StMPK7‐myc‐triggered cell death is compromised by coexpression with the salicylate hydroxylase NahG. The left picture shows cell death triggered by CA‐StMPK7‐myc and INF1 when coexpressed with GUS‐myc or NahG. The ratios next to the infiltrated zones show the amount of infiltrated sites showing cell death versus the total amount of infiltrated sites from two independent experiments at 5 days postinoculation (dpi). Pictures were taken at 5 dpi. The middle picture shows cell death as dark spots in the leaves from which chlorophyll was removed by treatment with ethanol. The right picture shows the H2O2 accumulation at the infiltrated sites with 3,3′‐diaminobenzidine (DAB) staining. (c) Quantification of cell death triggered by CA‐StMPK7 and INF1 by ion leakage assays. CA‐StMPK7 and INF1 were coexpressed with either GUS‐myc or NahG, and the amount of cell death was quantified by ion leakage assays. The relative ion leakage values were measured at 5 dpi. Error bars indicate the SD
3. DISCUSSION
In this study, we describe the identification and characterization of potato StMPK7 as a downstream signalling target of StMKK1 (Figure 1). We show that the StMKK1 interacted with and phosphorylated StMPK7 (Figure 4). StMPK7 and StMPK4 both positively regulated immunity against Phytophthora pathogens (Figures 2, 3, S1, and S2) and the constitutively active mutant CA‐StMPK7 enhanced resistance to Phytophthora pathogens (Figures 5 and S8) and triggered an SGT1/RAR1‐dependent cell death (Figure 6). Further investigations revealed that overexpression of StMPK7 enhanced pathogenesis‐related gene expression (Figure 7), whereas CA‐StMPK7‐induced cell death was strongly repressed by the SA hydroxylase NahG (Figure 7). Thus, we conclude that potato StMPK7 and StMPK4 function as downstream signalling components of the phosphorelay cascade initiated by StMKK1, which enhances resistance to Phytophthora pathogens by triggering SA‐dependent immunity. Phylogenetic analysis of StMPK7 showed that Arabidopsis AtMPK4/5/11/12 group together with three MPKs of Populus, two of Oryza, and three of tomato and potato. These subclade B MPKs have undergone complicated gene duplication and gene loss processes. StMPK7 and StMPK4 are functional orthologs of AtMPK4/5/11/12 (Figure 1). Because StMKK1 interacted with and phosphorylated StMPK7 (Figure 4), just like Arabidopsis AtMKK1 interacts with and phosphorylates AtMPK4 (Bi et al., 2018), we conclude that StMPK7 is a downstream signalling component of StMKK1. Thus, potato probably has a StMKK1‐StMPK7 phosphorelay cascade, similar to the Arabidopsis MKK1‐MPK4 cascade. StMPK7 was phosphorylated on MTI activation, and overexpression of StMPK7 promoted MTI‐related gene expression (Figure S5), indicating that the StMKK1‐StMPK7 cascade positively regulates MTI.
Transient expression of StMPK7 and CA‐StMPK7 in N. benthamiana enhanced resistance to Phytophthora pathogens (Figures 2, 5, and S8). Furthermore, resistance conferred by CA‐StMPK7 was much stronger than that provided by wild type StMPK7 (Figure 5). These results indicate that the kinase activity of StMPK7 is important for plant immunity. In Arabidopsis, an mpk4 knock‐out mutant shows an autoimmune phenotype and, therefore, AtMPK4 was suggested to be a negative regulator of immunity (Kong et al., 2012). However, later it was reported to be a positive regulator of immunity through participating in the MTI response (Bi et al., 2018; Zhang et al., 2017). The autoimmune response of the mpk4 knock‐out mutant was in fact caused by the activation of the NLR protein SUMM2, which guards the MEKK1‐MKK1/2‐MPK4 proteins taking part in this phosphorelay cascade and confers R gene‐mediated immunity when this cascade is being manipulated, either by microbial pathogens or by mutations (Zhang et al., 2012, 2017). This finding is supported by the observation that the Pseudomonas syringae effector HopAI1 triggers plant immunity by inactivating MPK4 (Zhang et al., 2012). However, in our study on silencing of NbMPK4/7 the plants become more susceptible to Phytophthora pathogens, instead of more resistant (Figure 3). This observation indicates that in solanaceous plants there is probably no SUMM2‐like NLR protein guarding the StMKK1‐StMPK7 cascade.
Both StMPK7 and StMPK4 promote resistance, which was reflected by the reduced lesion areas in leaf tissue overexpressing these MAPKs when compared to overexpression of the GFP‐GUS control (Figures 2 and S1). StMKK1 interacted with and phosphorylated both StMPK7 and StMPK4 (Figures 4 and S4). StMPK7 was activated by flg22 treatments (Figure 4) just as reported for MPK4 (Gao et al., 2008), and both StMPK7 and MPK4 participated in SA‐related signalling pathway (Figures 7 and S4c). Taken together with the fact that silencing either NbMPK4 or NbMPK7 hampered plant immunity to Phytophthora pathogens (Figures 3 and S2), this indicates that they are not simply functional redundant genes. Instead, probably StMPK4 and StMPK7 function collaboratively downstream of StMKK1. Potato has six MKKs and 21 MPK proteins (Iftikhar et al., 2017), so probably several MPKs work downstream of one MKK. In our research we identified that besides StMPK4, StMPK7 also works downstream of StMKK1, and StMPK4 and StMPK7 show overlapping but nonredundant functions in plant immunity. Probably plants employ multiple MPK proteins to amplify the immune signalling. Our results showed that resistance conferred by CA‐StMPK7 was stronger than that provided by StMPK7, as is apparent from the severely restricted lesions that developed at the inoculation sites (Figure 5). Moreover, CA‐StMPK7 triggered a mild cell death at 3 dpi when agroinfiltrated at an OD600 of 0.3 or higher (Figure 6). Thus, the cell death triggered by CA‐StMPK7 is probably due to the overactivation of immune signalling downstream of StMPK7.
AtMPK4 has been reported to sequester the transcription factor WRKY33 to the cytoplasm in the resting state, whereas on activation of MTI, WRKY33 is released from AtMPK4, allowing it to move to the nucleus and to activate a number of genes that is required for camalexin biosynthesis (Zhou et al., 1999). Our results showed that WRKY33 played a minor role in CA‐StMPK7‐triggered cell death (Figure 6d), which may be due to the fact that this transcription factor is probably not the key signalling component downstream of StMPK7 in regulating host immunity. WRKY33 was not accountable for the MPK4‐regulated hormone‐responsive gene expression (Qiu et al., 2008), and AtMPK4 has multiple putative substrates besides WRKY33 (Zhang, Chhajed, et al., 2019; Zhang, Schneider, et al., 2019). Furthermore, there is a plethora of WRKY genes in plants, and the minor involvement of WRKY33 in CA‐StMPK7‐triggered cell death may be due to redundancy of the various WRKYs.
SGT1 and RAR1 are involved in multiple cellular processes. However, most importantly, they are involved in R gene‐mediated immunity, which largely depends on SA‐signalling (Zhang & Li, 2019). We observed that overexpression of StMPK7 significantly enhanced PR1 and PR2 gene expression (Figure 7a), typically reflecting SA‐related gene expression, and the presence of the salicylate hydroxylase NahG significantly repressed CA‐StMPK7‐triggered cell death (Figure 7b,c), supporting the notion that StMPK7‐mediated immune signalling requires the accumulation of SA. MAPK cascades are generally known to participate in plant MTI and ETI responses and trigger the alteration of plant hormone levels, such as that of SA (Kawano et al., 2010; Zhang et al., 2012, 2018). This also supports our findings that the StMKK1‐StMPK7 cascade enhances plant immunity via SA‐dependent signalling.
In summary, our research identified the potato MPK4/5‐orthologous genes StMPK4/6 and StMPK7, and we have provided evidence that a functional StMKK1‐StMPK7 phosphorelay cascade exists in potato and functions in MTI. StMPK7 was shown to activate SA‐related immunity, and cell death triggered by constitutively active StMPK7 depends on SA signalling. Our work adds to the studies on MAPK cascades in crop plants and provides evidence for the functioning of MAPK cascades in SA‐related immunity.
4. EXPERIMENTAL PROCEDURES
4.1. Plasmid construction
The open reading frames (ORFs) of StMPK7 (PGSC0003DMC400021542), StMPK6 (PGSC0003DMP400037535), and StMPK4 (PGSC0003DMP400000144) were cloned by performing a PCR employing the primer pairs shown in Table S1, using a cDNA library from potato cultivar Tian11 as a template. The amplified fragments were inserted into the pART27‐NGFP, pART27‐Nmyc, pART27‐CGFP, and pART27‐Cmyc vectors using EcoRI and XbaI sites present in the primers to generate the GFP‐StMPK7, GFP‐StMPK6, myc‐StMKK1, StMPK4‐GFP, and StMPK4‐myc plasmids. The constitutively active (CA) mutants CA‐StMPK7‐myc (StMPK7D198G,E202A) and CA‐StMKK1‐myc (StMKK1T220D,T226E) were generated by overlap extension PCR using GFP‐StMPK7 and GFP‐StMKK1 as templates, respectively, with the primers shown in Table S1, and cloned into the pART27‐cmyc vector using XhoI and EcoRI sites (Fan et al., 2018). For yeast two‐hybrid assays, StMPK7 was cloned into the pGADT7 vector using the EcoRI and BamHI sites to generate the pGADT7:StMPK7 plasmid. For firefly luciferase complementation imaging (LCI) assays, StMPK7 and StMKK1 were cloned into pCAMBIA‐nLuc and pCAMBIA‐cLuc vectors using KpnI and SalI sites to form the StMPK7‐nluc and cluc‐StMKK1 plasmids, respectively.
4.2. Plant growth conditions and detached leaf assays
N. benthamiana and potato plants were grown in a climate chamber with a 16 hr light and 8 hr dark cycle at 25 °C, and with a relative humidity (RH) of 45%. The middle two or three fully expanded leaves from 4‐ to 5‐week‐old N. benthamiana plants were detached and used for infection assays. For this, P. infestans 14‐3‐GFP and P. parasitica isolate 1121 were grown as described by Li et al. (2019) and zoospores were harvested and used for inoculation. Ten microlitres of a 1 × 105/ml zoospore suspension of P. infestans and 10 µl of a 2 × 105/ml zoospore suspension of P. parasitica were used for inoculation. P. infestans‐ and P. parasitica‐inoculated leaves were incubated at 100% RH at 18 °C for 5 or 6 days, and at 23 °C for 2 or 3 days, respectively, before the diameters of the lesions were measured.
4.3. Agroinfiltration
GFP‐StMPK7 and CA‐StMPK7‐myc were transformed into the A grobacterium strain C58C1, and the bacteria were gown at 28 °C in liquid Luria‐Bertani medium with the antibiotics tetracycline and spectinomycin for 2 days. Then the bacterial suspension was centrifuged at 1,000 × g, the cells were resuspended in infiltration medium (Champouret et al., 2009), and the density was adjusted to an OD600 of 0.1 for assays involving confocal microscopy and infection assays, OD600 of 0.3 for coimmunoprecipitations (Co‐IPs), and OD600 of 0.7 for cell death assays.
4.4. Virus‐induced gene silencing
To select the target sequence of NbMPK7 for virus‐induced gene silencing (VIGS) assays, we BLAST searched the PGSC0003DMC400021542 gene sequence in the Sol genomics network database (https://solgenomics.net/) and identified two genes, Niben101Scf00254g02007.1 (NbMPK7a) and Niben101Scf00421g01029.1 (NbMPK7b), both encoding a NbMPK7 ortholog in N. benthamiana. We selected one cDNA fragment targeting both genes for generating the tobacco rattle virus (TRV)‐NbMPK7 vector. The TRV‐NbMPK4/6 constructs were generated in a similar way using the primer pairs shown in Table S1. TRV‐GUS was used as a negative control in our VIGS assays. Agrobacterium strain C58C1, containing either TRV1 (TRV‐RNA1) or TRV2 (TRV‐RNA2) (Ryu et al., 2004), was cultured, centrifuged, and resuspended in infiltration medium before being mixed in a 1:1 ratio to a final OD600 of 1. The first two leaves of 2‐week‐old N. benthamiana plants were used for agroinfiltration of the TRV‐NbMPK7/4/6 constructs. Three weeks after infiltration, the fifth and sixth leaves counted from the bottom were selected for infection assays.
4.5. Potato transformation
Agrobacterium strain GV3101, harbouring the GFP‐StMPK7 plasmid, was transformed into potato cultivar Desirée as described by Sun et al. (2016). Rooted transformants were transferred to Murashige & Skoog plates without antibiotics and were cultured for 3 weeks in a climate chamber with a 16 hr light and 8 hr dark cycle at 23 °C before being planted in potting soil.
4.6. Firefly LCI assay
The firefly LCI assays were performed according to the protocol described by Chen et al. (2008).
4.7. Flg22 treatments and ROS production assays
A 10 µM flg22 solution in water was infiltrated into the leaves of N. benthamiana. After 3 hrs of flg22 treatment, leaves were harvested for checking of MTI marker gene expression. ROS production assays were performed according to the method described by Li et al. (2016).
4.8. Kinase activation assay
GFP‐StMPK7 was coexpressed with CA‐StMKK1‐myc or the control GUS‐myc by transient transformation of N. benthamiana leaves. Proteins were extracted using an extraction buffer with phosphatase inhibitor cocktails 2 and 3 (Sigma). The activation of StMPK7 through phosphorylation was detected using anti‐pERK antibody (Phospho‐p44/42 MAPK [Erk1/2] [Thr202/Tyr204]; Cell Signaling Technology).
4.9. Gene expression assays
TRIzol reagent (Invitrogen) was used for total RNA extraction, and first‐strand cDNA was synthesized from 1 µg of total RNA according to the manufacturer's instructions (PrimeScript RT reagent Kit; TaKaRa). For each PCR, 5 µl of the 20 times diluted cDNA was used as a template and SYBR Green master mix (Roche) was used. An iQ7 Real‐Time Cycler (Life Technologies) was used to run RT‐qPCRs. Gene expression was quantified and normalized to the N. benthamiana housekeeping gene actin using the ΔΔC t method. Primer pairs for testing gene silencing efficiency were designed beyond the VIGS‐targeting sequence (Table S1).
4.10. Coimmunoprecipitation and western blot assays
Total proteins were extracted using GTEN buffer as described by Du et al. (2021). For Co‐IP assays, for each 1.6 ml of total protein extraction a 12 µl of GFP‐trap_A beads (Chromotek) were added, and the mixtures were incubated for 2 hr with gentle shaking at 4 °C before proteins were spun down. The beads were washed with protein extraction buffer five times before being boiled in protein loading buffer. For western blot assays, proteins were separated on 10% sodium dodecyl sulphate (SDS)‐polyacrylamide gels and subsequently transferred to Immune‐Blot PVDF membranes (Roche). The membranes were blocked in 10 ml of blocking buffer (TBST; Tris‐buffered saline pH 7.2, 0.05% Tween 20 [Sigma]) containing 5% of nonfat milk, with gentle shaking for 2 hr at room temperature. Subsequently, membranes were incubated with anti‐GFP (#AE012, ABclonal), anti‐myc (#AE010, ABclonal), or anti‐pERK antibody at the appropriate dilution for 1.5–2 hr with gentle shaking at room temperature. Then the membranes were washed five times and incubated with the secondary antibodies horseradish peroxidase‐conjugated goat anti‐rabbit IgG (H + L) antibody (#AS014, ABclonal) for another 1.5–2 hr. After this incubation, membranes were washed with TBST five times before proteins were detected (eECL western blot kit; CWBio).
4.11. Yeast two‐hybrid assays
The binding domain (BD) plasmid pGBKT7:StMKK1, or the empty vector pGBKT7, and the activation domain (AD) plasmid pGADT7:StMPK7, or empty vector pGADT7, were cotransformed into Saccharomyces cerevisiae AH109 (Li et al., 2019). Transformations were checked on selective drop‐out (SD)/−Trp−Leu medium and interactions were confirmed on QDO (SD/−Trp−Leu−His−Ade) medium, together with a gain of α‐galactosidase activity (α‐gal), by adding X‐α‐Gal in QDO medium. Yeast controls showing either positive or negative interaction were provided by the Matchmaker GAL4 Two‐Hybrid System 3 (Clontech).
4.12. Confocal microscopy
An IX83 confocal microscope (Olympus Life Science) was used to determine the subcellular localization of GFP‐StMPK7. GFP was excited at a wavelength of 488 nm and the emission was detected between 500 and 540 nm. Images were processed with Olympus Fluoview and figures were generated using Adobe Illustrator.
4.13. DAB staining and ion leakage assays
Leaves were incubated with a DAB staining solution (1 mg/ml, DAB dissolved in Milli‐Q water with addition of HCl to adjust the pH to 3.7) overnight, and then washed with water before pictures were taken. Ion leakage assays were performed as described by Bouwmeester et al. (2014).
AUTHOR CONTRIBUTIONS
Y.D. and W.S. designed the research. H.Z., F.L., Z.L., J.C., and X.C. performed the experiments. Y.D. and H.Z. analysed the data. Q.W. built the phylogenetic tree. Y.D., M.H.A.J.J., and W.S. wrote the manuscript. All authors reviewed the manuscript.
Supporting information
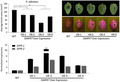
FIGURE S1 The role of StMPK4 and StMPK6 in immunity to Phytophthora pathogens. StMPK4‐myc (right) and GUS‐myc (left) or GFP‐StMPK6 (right) and GFP‐GUS (left) were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day postinfiltration (dpi), the Phytophthora infestans isolate 14‐3‐GFP (upper and lower three panels) and Phytophthora parasitica isolate 1121 (middle three panels) were inoculated onto the leaves. The lesion diameters were measured at 5 days after inoculation (dai) for P. infestans and at 2 dai for P. parasitica, and average lesion areas in mm2 were calculated and are shown in graphs (a), (d), and (g). Representative images taken under normal light (b, e, h) or blue light (c, f, i) show lesion development. Error bars show the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). The experiments were repeated three times with similar results. (j) The accumulation of StMPK7/4/6 and control proteins on agroinfiltration is shown by western blot. The expected protein bands are marked with red asterisks. (k) The accumulation of GFP‐StMPK7 proteins in wild‐type (WT) and StMPK7‐overexpression (OE) transgenic lines are shown by western blot, and the expected protein bands are marked with red asterisks
FIGURE S2 Silencing of NbMPK4 promotes Phytophthora infestans colonization. P. infestans isolate 14‐3‐GFP (upper and lower three panels) and Phytophthora parasitica isolate 1121 (middle three panels) were inoculated onto the TRV‐GUS‐ and TRV‐NbMPK4/6‐inoculated plants. The lesion diameters were measured at 5 days after inoculation (dai) for P. infestans and at 2 dai for P. parasitica, and average lesion areas in mm2 were calculated and are shown in graphs (a), (c), and (e). The standard error from more than 10 technical replicates is indicated. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). (b), (d), (f) Representative images taken under normal (left panel) or blue light or with trypan blue staining (right panel), showing the lesion development on TRV‐GUS‐ and TRV‐NbMPK4/6‐inoculated leaves. The experiments were repeated three times with similar results
FIGURE S3 Plant morphology and silencing efficiency of NbMPK4/6/7 in Nicotiana benthamiana plants inoculated with TRV‐GUS and TRV‐NbMPK4/6/7. (a), (c), (e) Morphology of TRV‐GUS‐, TRV‐NbMPK4/6/7‐, and TRV‐SGT1‐inoculated plants 3 weeks after inoculation. (b), (d), (f) Relative quantification (RQ) of NbMPK7, ‐4 and ‐6 expression in TRV‐GUS‐ and TRV‐NbMPK4/6/7‐inoculated plants. Error bars indicate the standard error from three technical replicates. The experiments were repeated at least five times, with eight replicates per construct, and similar patterns were consistently observed
FIGURE S4 Potato StMKK1 interacts with and phosphorylates StMPK4, which participates in salicylic acid (SA)‐related immunity. (a) Coimmunoprecipitation (Co‐IP) assays confirmed that myc‐StMKK1 associated with StMPK4‐GFP. Green fluorescent protein (GFP) was used as negative control. Protein loading is indicated by Ponceau stain (Ponceau S). (b) StMPK4‐GFP was transiently coexpressed with either GUS‐myc or constitutively activated CA‐StMKK1‐myc in Nicotiana benthamiana leaves. Total proteins were extracted and the phosphorylated StMPK4‐GFP was detected by western blot using α‐pERK antibody. Ponceau stain (Ponceau S) indicates the protein loading. (c) Silencing of NbMPK4 repressed plant SA‐related gene expression. Graphs show the relative expressions of NbPR‐1, NbPR‐2, NbPR‐5, NbNPR1, NbEDS1, and NbPAD4 in TRV‐GUS and TRV‐NbMPK4 plants. The expression of the NbActin gene was used for normalization. The experiments were repeated twice, with similar results
FIGURE S5 StMPK7 overexpression promotes MAMP‐triggered immunity (MTI)‐related gene expression. Graphs show the relative expression levels of NbFRK1 and NbWRKY33 in GFP‐GUS‐ and GFP‐StMPK7‐overexpressing leaves. Expression of the NbActin gene was used for normalization. NbFRK1 and NbWRKY33 expression levels in GFP‐StMPK7‐expressing leaves were significantly higher when compared to the control GFP‐GUS‐expressing plants. The experiments were repeated twice with similar results
FIGURE S6 GFP‐StMPK7 localizes to the cytoplasm and the nucleus. (a) GFP‐StMPK7 and GFP were agroinfiltrated into Nicotiana benthamiana leaves and pictures were taken using a confocal microscope at 2 days postinfiltration with a 63× magnification. The bars indicate 20 µm. (b) The western blot shows that GFP‐StMPK7 is not cleaved on its expression in planta
FIGURE S7 Sequence alignment of StMPK7 orthologous proteins from Arabidopsis and several solanaceous plants. Protein sequences of selected accessions were aligned using Vector NTI. The accession numbers of the proteins used for sequence alignment are as follows: StMPK7, PGSC0003DMP400021542; AtMPK4, AT4g01370; NbMPK7a, Niben101Scf00254g02007; NbMPK7b, Niben101Scf00421g01029; SlMAPK7, Solyc08g081490. The stars indicate the amino acids D and E that are mutated for generating a constitutively active variant of the MPK protein
FIGURE S8 Overexpression of potato CA‐StMPK7 promotes resistance to Phytophthora pathogens. CA‐StMPK7‐myc (right) and the control GUS‐myc (left) were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day after agroinfiltration (dai), the Phytophthora infestans isolate 14‐3‐GFP (upper three panels) or Phytophthora parasitica isolate 1121 (lower three panels) were inoculated onto the leaves. The lesion diameters were measured at 5 dai for P. infestans and at 2 dai for P. parasitica, and average lesion areas (in mm2) were calculated and are shown in graphs (a) and (d), respectively. Images taken under normal light (b, e) or blue light (c, f) show lesion development on GUS‐myc and CA‐StMPK7‐myc‐expressing leaves. Error bars show the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). The experiments were repeated three times with similar results
FIGURE S9 Western blots showing the different levels of protein accumulation for StMPK7 and CA‐StMPK7. Different densities of agrobacterium cultures driving expression of myc‐StMPK7 (a) or CA‐StMPK7‐myc (b) were agroinfiltrated into leaves of Nicotiana benthamiana. Proteins were isolated at 2 days after infiltration and western blotting was performed to reveal protein accumulation. The OD600 of the agrobacterial cultures that were infiltrated are indicated and the relative intensities of the protein bands, normalized to RuBisCO, are indicated by numbers
TABLE S1 Sequence of the primers used in this study
ACKNOWLEDGEMENTS
The authors thank the Life Science Research Core Services, the State Key Laboratory of Crop Stress Biology for Arid Areas, and the Horticulture Science Research Center (Northwest A&F University, Yangling, China) for experimental assistance. This work was supported by the National Natural Science Foundation of China (31701770, 32072401), the China Postdoctoral Science Foundation (2019T120956, 2016M600818), the Northwest A&F University Scientific Research Fund for Advanced Talents and the Young Talent Training Program (2452018028, 2452017069) from Northwest A&F University, and the Programme of Introducing Talents of Innovative Discipline to Universities (Project 111) from the State Administration of Foreign Experts Affairs (no. B18042). The authors declare no conflict of interest.
Zhang H, Li F, Li Z, et al. Potato StMPK7 is a downstream component of StMKK1 and promotes resistance to the oomycete pathogen Phytophthora infestans . Mol Plant Pathol. 2021;22:644–657. 10.1111/mpp.13050
Houxiao Zhang and Fangfang Li contributed equally to this work.
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415 977–983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Berriri, S. , Garcia, A.V. , Frei dit Frey, N. , Rozhon, W. , Pateyron, S. , Leonhardt, N. et al. (2012) Constitutively active mitogen‐activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. The Plant Cell, 24, 4281–4293. 10.1105/tpc.112.101253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, G. , Zhou, Z. , Wang, W. , Li, L. , Rao, S. , Wu, Y. et al. (2018) Receptor‐like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen‐activated protein kinase cascades in Arabidopsis. The Plant Cell, 30, 1543–1561. 10.1105/tpc.17.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester, K. , Han, M. , Blanco‐Portales, R. , Song, W. , Weide, R. , Guo, L. et al. (2014) The Arabidopsis lectin receptor kinase LecRK‐I.9 enhances resistance to Phytophthora infestans in solanaceous plants. Plant Biotechnology Journal, 12, 10–16. 10.1111/pbi.12111 [DOI] [PubMed] [Google Scholar]
- Chai, J. , Liu, J. , Zhou, J. & Xing, D. (2014) Mitogen‐activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. Journal of Experimental Botany, 65, 6513–6528. 10.1093/jxb/eru369 [DOI] [PubMed] [Google Scholar]
- Champouret, N. , Bouwmeester, K. , Rietman, H. , van der Lee, T. , Maliepaard, C. , Heupink, A. et al. (2009) Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi‐blb1 potato. Molecular Plant‐Microbe Interactions, 22, 1535–1545. 10.1094/MPMI-22-12-1535 [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. et al. (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology, 146, 368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , Berg, J. , Govers, F. & Bouwmeester, K. (2015) Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytologist, 207, 735–747. 10.1111/nph.13355 [DOI] [PubMed] [Google Scholar]
- Du, Y. , Chen, X. , Guo, Y. , Zhang, X. , Zhang, H. , Li, F. et al. (2021) Phytophthora infestans RXLR effector PITG20303 targets a potato MKK1 protein to suppress plant immunity. New Phytologist, 229, 501–515. 10.1111/nph.16861 [DOI] [PubMed] [Google Scholar]
- Engelhardt, S. , Boevink, P.C. , Armstrong, M.R. , Ramos, M.B. , Hein, I. & Birch, P.R.J. (2012) Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. The Plant Cell, 24, 5142–5158. 10.1105/tpc.112.104992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, G. , Yang, Y. , Li, T. , Lu, W. , Du, Y.U. , Qiang, X. et al. (2018) A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum‐mediated immunity. Molecular Plant, 11, 1067–1083. 10.1016/j.molp.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Frei dit Frey, N. , Garcia, A. , Bigeard, J. , Zaag, R. , Bueso, E. , Garmier, M. et al. (2014) Functional analysis of Arabidopsis immune‐related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biology, 15, R87. 10.1186/gb-2014-15-6-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, W. (2008) Phytophthora infestans: The plant (and R gene) destroyer. Molecular Plant Pathology, 9, 385–402. 10.1111/j.1364-3703.2007.00465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , Liu, J. , Bi,, D. , Zhang, Z. , Cheng, F. & Chen, S. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen‐activated protein kinase cascade to regulate innate immunity in plants. Cell Research, 18, 1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Hettenhausen, C. , Baldwin, I.T. & Wu, J. (2013) Nicotiana attenuata MPK4 suppresses a novel jasmonic acid (JA) signaling‐independent defense pathway against the specialist insect Manduca sexta, but is not required for the resistance to the generalist Spodoptera littoralis . New Phytologist, 199, 787–799. 10.1111/nph.12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Ding, L. & Yu, H. (2013) Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Reports, 32, 1067–1074. 10.1007/s00299-013-1423-4 [DOI] [PubMed] [Google Scholar]
- Ichimura, K. , Shinozaki, K. , Tena, G. , Sheen, J. , Henry, Y. , Champion, A. et al. (2002) Mitogen‐activated protein kinase cascades in plants: A new nomenclature. Trends in Plant Science, 7, 301–308. 10.1016/S1360-1385(02)02302-6 [DOI] [PubMed] [Google Scholar]
- Iftikhar, H. , Naveed, N. , Virk, N. , Bhatti, M.F. & Song, F. (2017) In silico analysis reveals widespread presence of three gene families, MAPK, MAPKK and MAPKKK, of the MAPK cascade from crop plants of Solanaceae in comparison to the distantly‐related syntenic species from Rubiaceae, coffee. PeerJ, 5, e3255. 10.7717/peerj.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagodzik, P. , Tajdel‐Zielinska, M. , Ciesla, A. , Marczak, M. & Ludwikow, A. (2018) Mitogen‐activated protein kinase cascades in plant hormone signaling. Frontiers in Plant Science, 9, 1387. 10.3389/fpls.2018.01387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kandoth, P.K. , Ranf, S. , Pancholi, S.S. , Jayanty, S. , Walla, M.D. , Miller, W. et al. (2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin‐mediated defense response against herbivorous insects. Proceedings of the National Academy of Sciences of the United States of America, 104, 12205–12210. 10.1073/pnas.0700344104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, Y. , Akamatsu, A. , Hayashi, K. , Housen, Y. , Okuda, J. , Yao, A.I. et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host & Microbe, 7, 362–375. 10.1016/j.chom.2010.04.010 [DOI] [PubMed] [Google Scholar]
- King, S.R. , McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Bukharova, T. , Sukarta, O. et al. (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. The Plant Cell, 26, 1345–1359. 10.1105/tpc.113.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q. , Qu, N. , Gao, M. , Zhang, Z. , Ding, X. , Yang, F. et al. (2012) The MEKK1‐MKK1/MKK2‐MPK4 kinase cascade negatively regulates immunity mediated by a mitogen‐activated protein kinase kinase kinase in Arabidopsis . The Plant Cell, 24, 2225–2236. 10.1105/tpc.112.097253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Wang, Q. , Feng, R. , Li, L. , Ding, L. , Fan, G. et al. (2019) Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a‐like effectors. New Phytologist, 10.1111/nph.16139 [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Huang, L. , Ouyang, Z. , Hong, Y. , Zhang, H. et al. (2014) Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea . BMC Plant Biology, 14, 166. 10.1186/1471-2229-14-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Chang, Y. , Zhao, C. , Yang, H. & Ren, D. (2016) Expression of the inactive ZmMEK1 induces salicylic acid accumulation and salicylic acid‐dependent leaf senescence. Journal of Integrative Plant Biology, 58, 724–736. 10.1111/jipb.12465 [DOI] [PubMed] [Google Scholar]
- Lu, X. , Xiong, Q. , Cheng, T. , Li, Q.‐T. , Liu, X.‐L. , Bi, Y.‐D. et al. (2017) A PP2C‐1 allele underlying a quantitative trait locus enhances soybean 100‐seed weight. Molecular Plant, 10, 670–684. 10.1016/j.molp.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Pitzschke, A. , Djamei, A. , Bitton, F. & Hirt, H. (2009) A major role of the MEKK1‐MKK1/2‐MPK4 pathway in ROS signalling. Molecular Plant, 2, 120–137. 10.1093/mp/ssn079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Schikora, A. & Hirt, H. (2009) MAPK cascade signalling networks in plant defence. Current Opinion in Plant Biology, 12, 421–426. 10.1016/j.pbi.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Qiu, J.‐L. , Zhou, L.U. , Yun, B.‐W. , Nielsen, H.B. , Fiil, B.K. , Petersen, K. et al. (2008) Arabidopsis mitogen‐activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiology, 148, 212–222. 10.1104/pp.108.120006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Armstrong, M. , Qi, Y. , McLellan, H. , Zhong, C. , Du, B. et al. (2019) Phytophthora infestans RXLR effectors target parallel steps in an immune signal transduction pathway. Plant Physiology, 180, 2227–2239. 10.1104/pp.18.00625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, W. , Feng, F. , Zhou, J. & He, C. (2010) Effector‐triggered innate immunity contributes Arabidopsis resistance to Xanthomonas campestris . Molecular Plant Pathology, 11, 783–793. 10.1111/j.1364-3703.2010.00642.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.M. , Anand, A. , Kang, L. & Mysore, K.S. (2004) Agrodrench: A novel and effective agroinoculation method for virus‐induced gene silencing in roots and diverse solanaceous species. The Plant Journal, 40, 322–331. 10.1111/j.1365-313X.2004.02211.x [DOI] [PubMed] [Google Scholar]
- Stulemeijer, I.J. , Stratmann, J.W. & Joosten, M.H.A.J. (2007) Tomato mitogen‐activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf‐4/Avr4‐induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiology, 144, 1481–1494. 10.1104/pp.107.101063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, K. , Wolters, A.‐M. , Vossen, J.H. , Rouwet, M.E. , Loonen, A.E.H.M. , Jacobsen, E. et al. (2016) Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Research, 25, 731–742. 10.1007/s11248-016-9964-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige, M. , Scheikl, E. , Eulgem, T. , Dóczi, R. , Ichimura, K. , Shinozaki, K. et al. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis . Molecular Cell, 15, 141–152. 10.1016/j.molcel.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Virk, N. , Liu, B.O. , Zhang, H. , Li, X. , Zhang, Y. , Li, D. et al. (2013) Tomato SlMPK4 is required for resistance against Botrytis cinerea and tolerance to drought stress. Acta Physiologiae Plantarum, 35, 1211–1221. 10.1007/s11738-012-1160-2 [DOI] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Raffaele, S. , Vossen, J.H. , Champouret, N. , Oliva, R. , Segretin, M.E. et al. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology, 49, 507–531. 10.1146/annurev-phyto-072910-095326 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Schuck, S. , Wu, J. , Yang, P. , Döring, A.C. , Zeier, J. et al. (2018) A MPK3/6‐WRKY33‐ALD1‐pipecolic acid regulatory loop contributes to systemic acquired resistance. The Plant Cell, 30, 2480–2494. 10.1105/tpc.18.00547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizo, C. , Kuchimura, K. , Kobayashi, A. , Katou, S. , Kawakita, K. , Jones, J.D.G. et al. (2006) Rewiring mitogen‐activated protein kinase cascade by positive feedback confers potato blight resistance. Plant Physiology, 140, 681–692. 10.1104/pp.105.074906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, B. , Shen, X. , Li, X. , Xu, C. & Wang, S. (2007) Mitogen‐activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta, 226, 953–960. 10.1007/s00425-007-0541-z [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Chhajed, S. , Schneider, J.D. , Feng, G. , Song, W.Y. & Chen, S. (2019) Proteomic characterization of MPK4 signaling network and putative substrates. Plant Molecular Biology, 101, 325–339. 10.1007/s11103-019-00908-9 [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Chiang, Y.H. , Toruño, T.Y. , Lee, D. , Ma, M. , Liang, X. et al. (2018) The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host & Microbe, 24,379–391.e5. 10.1016/j.chom.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Schneider, J.D. , Lin, C. , Geng, S. , Ma, T. , Lawrence, S.R. et al. (2019) MPK4 phosphorylation dynamics and interacting proteins in plant immunity. Journal of Proteome Research, 18, 826–840. 10.1021/acs.jproteome.8b00345 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. & Li, X. (2019) Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Current Opinion in Plant Biology, 50, 29–36. 10.1016/j.pbi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Liu, Y. , Huang, H. , Gao, M. , Wu, D. , Kong, Q. et al. (2017) The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Reports, 18, 292–302. 10.15252/embr.201642704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Wu, Y. , Gao, M. , Zhang, J. , Kong, Q. , Liu, Y. et al. (2012) Disruption of PAMP‐induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB‐LRR protein SUMM2. Cell Host & Microbe, 11, 253–263. 10.1016/j.chom.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Zhou, N. , Tootle, T.L. & Glazebrook, J. (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. The Plant Cell, 11, 2419–2428. 10.1105/tpc.11.12.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.T. , Jia, L.J. , Wang, H.Y. , Zhao, P. , Wang, W.Y. , Liu, N. et al. (2018) The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. The Plant Journal, 95, 1055–1068. 10.1111/tpj.14010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 The role of StMPK4 and StMPK6 in immunity to Phytophthora pathogens. StMPK4‐myc (right) and GUS‐myc (left) or GFP‐StMPK6 (right) and GFP‐GUS (left) were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day postinfiltration (dpi), the Phytophthora infestans isolate 14‐3‐GFP (upper and lower three panels) and Phytophthora parasitica isolate 1121 (middle three panels) were inoculated onto the leaves. The lesion diameters were measured at 5 days after inoculation (dai) for P. infestans and at 2 dai for P. parasitica, and average lesion areas in mm2 were calculated and are shown in graphs (a), (d), and (g). Representative images taken under normal light (b, e, h) or blue light (c, f, i) show lesion development. Error bars show the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). The experiments were repeated three times with similar results. (j) The accumulation of StMPK7/4/6 and control proteins on agroinfiltration is shown by western blot. The expected protein bands are marked with red asterisks. (k) The accumulation of GFP‐StMPK7 proteins in wild‐type (WT) and StMPK7‐overexpression (OE) transgenic lines are shown by western blot, and the expected protein bands are marked with red asterisks
FIGURE S2 Silencing of NbMPK4 promotes Phytophthora infestans colonization. P. infestans isolate 14‐3‐GFP (upper and lower three panels) and Phytophthora parasitica isolate 1121 (middle three panels) were inoculated onto the TRV‐GUS‐ and TRV‐NbMPK4/6‐inoculated plants. The lesion diameters were measured at 5 days after inoculation (dai) for P. infestans and at 2 dai for P. parasitica, and average lesion areas in mm2 were calculated and are shown in graphs (a), (c), and (e). The standard error from more than 10 technical replicates is indicated. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). (b), (d), (f) Representative images taken under normal (left panel) or blue light or with trypan blue staining (right panel), showing the lesion development on TRV‐GUS‐ and TRV‐NbMPK4/6‐inoculated leaves. The experiments were repeated three times with similar results
FIGURE S3 Plant morphology and silencing efficiency of NbMPK4/6/7 in Nicotiana benthamiana plants inoculated with TRV‐GUS and TRV‐NbMPK4/6/7. (a), (c), (e) Morphology of TRV‐GUS‐, TRV‐NbMPK4/6/7‐, and TRV‐SGT1‐inoculated plants 3 weeks after inoculation. (b), (d), (f) Relative quantification (RQ) of NbMPK7, ‐4 and ‐6 expression in TRV‐GUS‐ and TRV‐NbMPK4/6/7‐inoculated plants. Error bars indicate the standard error from three technical replicates. The experiments were repeated at least five times, with eight replicates per construct, and similar patterns were consistently observed
FIGURE S4 Potato StMKK1 interacts with and phosphorylates StMPK4, which participates in salicylic acid (SA)‐related immunity. (a) Coimmunoprecipitation (Co‐IP) assays confirmed that myc‐StMKK1 associated with StMPK4‐GFP. Green fluorescent protein (GFP) was used as negative control. Protein loading is indicated by Ponceau stain (Ponceau S). (b) StMPK4‐GFP was transiently coexpressed with either GUS‐myc or constitutively activated CA‐StMKK1‐myc in Nicotiana benthamiana leaves. Total proteins were extracted and the phosphorylated StMPK4‐GFP was detected by western blot using α‐pERK antibody. Ponceau stain (Ponceau S) indicates the protein loading. (c) Silencing of NbMPK4 repressed plant SA‐related gene expression. Graphs show the relative expressions of NbPR‐1, NbPR‐2, NbPR‐5, NbNPR1, NbEDS1, and NbPAD4 in TRV‐GUS and TRV‐NbMPK4 plants. The expression of the NbActin gene was used for normalization. The experiments were repeated twice, with similar results
FIGURE S5 StMPK7 overexpression promotes MAMP‐triggered immunity (MTI)‐related gene expression. Graphs show the relative expression levels of NbFRK1 and NbWRKY33 in GFP‐GUS‐ and GFP‐StMPK7‐overexpressing leaves. Expression of the NbActin gene was used for normalization. NbFRK1 and NbWRKY33 expression levels in GFP‐StMPK7‐expressing leaves were significantly higher when compared to the control GFP‐GUS‐expressing plants. The experiments were repeated twice with similar results
FIGURE S6 GFP‐StMPK7 localizes to the cytoplasm and the nucleus. (a) GFP‐StMPK7 and GFP were agroinfiltrated into Nicotiana benthamiana leaves and pictures were taken using a confocal microscope at 2 days postinfiltration with a 63× magnification. The bars indicate 20 µm. (b) The western blot shows that GFP‐StMPK7 is not cleaved on its expression in planta
FIGURE S7 Sequence alignment of StMPK7 orthologous proteins from Arabidopsis and several solanaceous plants. Protein sequences of selected accessions were aligned using Vector NTI. The accession numbers of the proteins used for sequence alignment are as follows: StMPK7, PGSC0003DMP400021542; AtMPK4, AT4g01370; NbMPK7a, Niben101Scf00254g02007; NbMPK7b, Niben101Scf00421g01029; SlMAPK7, Solyc08g081490. The stars indicate the amino acids D and E that are mutated for generating a constitutively active variant of the MPK protein
FIGURE S8 Overexpression of potato CA‐StMPK7 promotes resistance to Phytophthora pathogens. CA‐StMPK7‐myc (right) and the control GUS‐myc (left) were agroinfiltrated into Nicotiana benthamiana leaves. At 1 day after agroinfiltration (dai), the Phytophthora infestans isolate 14‐3‐GFP (upper three panels) or Phytophthora parasitica isolate 1121 (lower three panels) were inoculated onto the leaves. The lesion diameters were measured at 5 dai for P. infestans and at 2 dai for P. parasitica, and average lesion areas (in mm2) were calculated and are shown in graphs (a) and (d), respectively. Images taken under normal light (b, e) or blue light (c, f) show lesion development on GUS‐myc and CA‐StMPK7‐myc‐expressing leaves. Error bars show the standard error from more than 10 technical replicates. Significant differences are indicated with asterisks (n ≥ 10; one‐sided Student’s t test, p ≤ .01). The experiments were repeated three times with similar results
FIGURE S9 Western blots showing the different levels of protein accumulation for StMPK7 and CA‐StMPK7. Different densities of agrobacterium cultures driving expression of myc‐StMPK7 (a) or CA‐StMPK7‐myc (b) were agroinfiltrated into leaves of Nicotiana benthamiana. Proteins were isolated at 2 days after infiltration and western blotting was performed to reveal protein accumulation. The OD600 of the agrobacterial cultures that were infiltrated are indicated and the relative intensities of the protein bands, normalized to RuBisCO, are indicated by numbers
TABLE S1 Sequence of the primers used in this study
Data Availability Statement
Research data are not shared.


