Abstract
Phytophthora infestans is a destructive pathogen of potato and a model for investigations of oomycete biology. The successful application of a CRISPR gene editing system to P. infestans is so far unreported. We discovered that it is difficult to express CRISPR/Cas9 but not a catalytically inactive form in transformants, suggesting that the active nuclease is toxic. We were able to achieve editing with CRISPR/Cas12a using vectors in which the nuclease and its guide RNA were expressed from a single transcript. Using the elicitor gene Inf1 as a target, we observed editing of one or both alleles in up to 13% of transformants. Editing was more efficient when guide RNA processing relied on the Cas12a direct repeat instead of ribozyme sequences. INF1 protein was not made when both alleles were edited in the same transformant, but surprisingly also when only one allele was altered. We discovered that the isolate used for editing, 1306, exhibited monoallelic expression of Inf1 due to insertion of a copia‐like element in the promoter of one allele. The element exhibits features of active retrotransposons, including a target site duplication, long terminal repeats, and an intact polyprotein reading frame. Editing occurred more often on the transcribed allele, presumably due to differences in chromatin structure. The Cas12a system not only provides a tool for modifying genes in P. infestans, but also for other members of the genus by expanding the number of editable sites. Our work also highlights a natural mechanism that remodels oomycete genomes.
Keywords: CRISPR/Cas12a, functional genomics, genome editing, late blight disease, oomycete, transposable element
Successful editing of the INF1 gene by CRISPR/Cas12a in the late blight pathogen Phytophthora infestans expands the CRISPR toolkit for oomycetes.
1. INTRODUCTION
Tools for functional genomics are critical for understanding the biology of plant pathogens. This is especially true for the oomycete genus Phytophthora, which causes numerous devastating crop diseases. The infamous pathogen Phytophthora infestans, for example, is a limiting factor in potato production and can destroy fields in little more than a week (Leesutthiphonchai et al., 2018). Studies of P. infestans have shed light on oomycete metabolism, spore biology, and pathogenesis, thus helping to advance strategies for battling disease (Abrahamian et al., 2016; Blum et al., 2010; Jahan et al., 2015; Leesutthiphonchai & Judelson, 2018). Yet, the genome of Phytophthora spp. can evolve rapidly, losing sensitivity to chemical agents and overcoming resistance in its hosts. Plasticity of the genome is mediated by a high content of repetitive DNA, which includes long terminal repeat (LTR) retroelements and DNA transposons (Dong et al., 2015; Haas et al., 2009).
Historically, the diploidy of oomycetes has limited the utility of classic strategies for genetics such as mutagenesis using chemicals or radiation. Traditional methods for gene knockouts or replacements have failed due to a low rate of homologous recombination. Stable and transient gene silencing has proved useful for assessing gene function but such methods may have drawbacks (Leesutthiphonchai & Judelson, 2018; Vu et al., 2019; Whisson et al., 2005).
CRISPR‐based genome editing has revolutionized the functional genomics of many organisms. The most‐described system involves CRISPR‐associated endonuclease Cas9, which complexes with a single‐guide RNA (sgRNA) to recognize and cleave a DNA target (Anzalone et al., 2020). This causes indels through nonhomologous end joining or homology‐directed repair. However, applying CRISPR/Cas9 to some taxa has been challenging. For example, Cas9 cannot be expressed well in many organisms due to toxicity (Foster et al., 2018; Markus et al., 2019). In some species, the lack of Pol III promoters for expressing the sgRNA necessitated alternatives such as using ribozymes to cleave the sgRNA from a Pol II transcript of the CRISPR RNA (crRNA) precursor (Markus et al., 2019). Not long ago, CRISPR/Cas9 editing was adapted to Phytophthora sojae, but this required the identification of an oomycete nuclear localization signal (Fang & Tyler, 2016). This method has proved to be effective in several oomycetes (Li et al., 2020; Pettongkhao et al., 2020; Situ et al., 2020). However, many groups, including ours, have had a lack of success using that system in P. infestans (van den Hoogen & Govers, 2019).
In this paper we report an editing system for P. infestans. Our data suggested that Cas9 was toxic, causing us to concentrate efforts on CRISPR/Cas12a (Cpf1), which reportedly has fewer off‐targets (Zhang et al., 2019). Unlike Cas9, Cas12a has both DNase and RNase activity. The latter enables Cas12a to form its own sgRNA by cleaving at direct repeats (DR) in the crRNA (Fonfara et al., 2016). For a target in our experiments, we selected the gene encoding INF1, a sterol‐binding protein that induces defence responses in certain nonhost plants (Du et al., 2015; Kamoun, van West, et al., 1998). We succeeded in achieving editing, with the best success obtained using the intrinsic RNAse activity of Cas12a rather than ribozymes to process the crRNA. Although we could observe events in which both alleles of Inf1 were modified, one allele was more refractory to editing. This was attributed to the insertion of a copia‐like retroelement that blocked transcription and presumably altered local chromatin structure. In summary, we not only describe the development of CRISPR/Cas12a as an editing tool for oomycetes but also the occurrence of a natural process that shapes oomycete genomes.
2. RESULTS
2.1. Expression of Cas9 may be problematic in P. infestans
Following the development of a Cas9 editing system for P. sojae (Fang & Tyler, 2016), we and others (van den Hoogen & Govers, 2019) attempted to use that method to modify genes in P. infestans. However, no success was observed against several targets. To troubleshoot, we first investigated whether the P. sojae RPL41 promoter used to express the guide RNA functioned in P. infestans, as this had not been established previously. A fusion of this promoter to a hygromycin resistance gene yielded drug‐resistant transformants at a frequency similar to that obtained using promoters commonly used in P. infestans, such as one from the Ham34 gene of Bremia lactucae (Judelson et al., 1992). This suggested that the lack of editing in P. infestans by Cas9 was not attributable to a failure to transcribe the guide RNA precursor.
Next, we tested whether Cas9 protein was produced and delivered to nuclei. In the system developed by Fang and Tyler (2016), the Ham34 promoter is used to express a human codon‐optimized form of S. pyogenes Cas9 modified to contain a P. sojae nuclear localization signal (PsNLS) in a plasmid containing the nptII marker gene, which enables selection of transformants on G418. Following some failures to detect Cas9 in transformants by immunoblotting, we conducted more tests using a plasmid expressing a PsNLS‐Cas9‐green fluorescent protein (GFP) fusion. No expression was detected in more than 85 G418‐resistant transformants based on fluorescence microscopy and immunoblot analysis. In P. infestans transformants obtained using plasmids bearing multiple genes, it is not uncommon for only the selectable marker to be expressed, presumably due to some form of epigenetic silencing affecting the other transcription units (Ah‐Fong & Judelson, 2011). If Cas9 expression were deleterious, this would be expected to select for transformants in which the Cas9 transcription unit had been inactivated.
Further experimentation further suggested that Cas9 was toxic to P. infestans. Unlike catalytically active Cas9 for which expression was never observed, a catalytically inactive variant (PsNLS‐dCas9‐GFP) was expressed in the majority of P. infestans transformants. The protein was detected both by immunoblotting (Figure 1a) and fluorescence microscopy (Figure 1b). The latter also indicated that the PsNLS delivered the protein to P. infestans nuclei.
FIGURE 1.
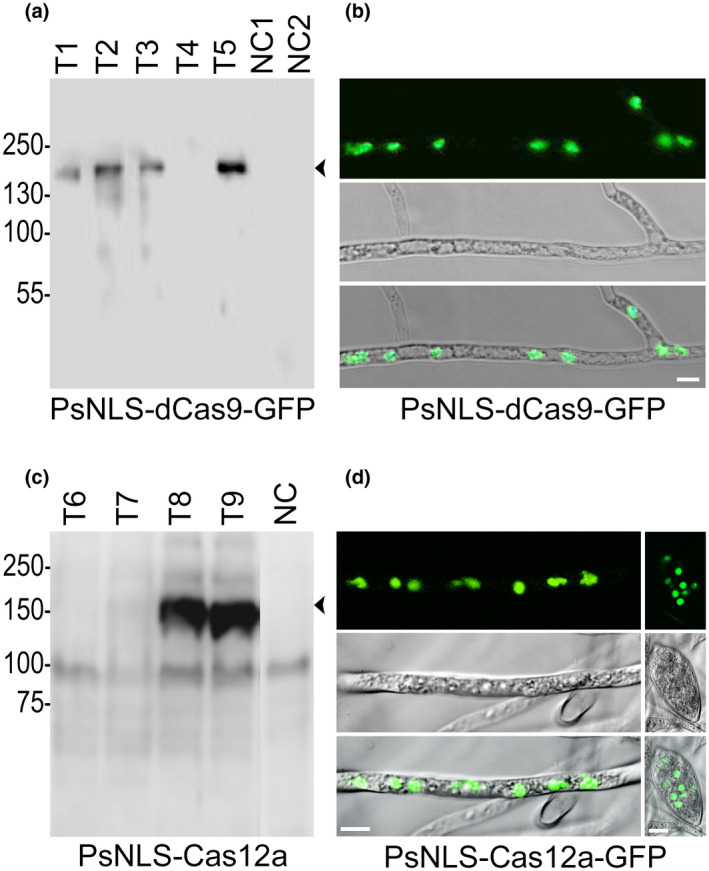
Expression of Cas endonucleases in Phytophthora infestans transformants. (a) Immunoblot of five representative strains transformed with the plasmid encoding the green fluorescent protein (GFP)‐tagged catalytically inactive form of the nuclease, PsNLS‐dCas9‐GFP, probed with anti‐GFP. NC1 and NC2 are negative controls, namely a strain expressing another protein and untransformed 1306, respectively. The expected size of the protein is 194 kDa. (b) Fluorescent micrograph of transformant expressing PsNLS‐dCas9‐GFP, showing localization of the protein to nuclei within a hypha. GFP, bright field, and merged channels are shown, top to bottom, with the scale bar equalling 10 µm. (c) Immunoblot of strains transformed with the plasmid encoding PsNLS‐Cas12a‐GFP, probed with anti‐Cas12a. NC is the untransformed progenitor strain. An empty lane between T9 and NC was deleted from the image. The expected size of the protein is 153 kDa. (d) Confocal image of transformant expressing PsNLS‐Cas12a‐GFP, showing a hypha on the left and a sporangium on the right. GFP, bright field, and merged channels are shown, top to bottom, with the scale bar equalling 10 µm
In contrast to Cas9, expression of catalytically active Lachnospiraceae bacterium Cas12a (formerly called LbCpf1) in P. infestans transformants did not appear to be problematic. This involved expressing a version modified to contain a consensus oomycete Kozak sequence and the ‐PsNLS. A protein of the expected size was detected by immunoblotting in many transformants (Figure 1c). In similar experiments but using a C‐terminal GFP tag, the protein was detected in nuclei (Figure 1d).
Based on these results, we shifted our efforts to developing a Cas12a‐based editing system. We concentrated on LbCas12a instead of its Acidaminococcus ortholog because LbCas12a reportedly has a broader temperature range (Moreno‐Mateos et al., 2017). The rationale was that LbCas12a might be useful for the many Phytophthora species such as P. infestans that grow optimally at relatively cool temperatures.
2.2. Design of single transcription unit Cas12a vectors
To test whether Cas12a was adaptable to P. infestans, we chose an appropriate recipient strain for experimentation, selected a target gene for our proof‐of‐concept studies, and designed vectors. Isolate 1306 was chosen for analysis because it is pathogenic on tomato and potato, sporulates well, and is diploid. The latter was established through genome‐wide single nucleotide polymorphism (SNP) analysis (Figure 2a). The average frequency of alternate alleles was 50%, which signals diploidy.
FIGURE 2.
Targets and plasmids used for editing. (a) Genome‐wide allele ratio analysis of isolate 1306. (b) Copy number of Inf1 relative to single‐copy control genes (= 1.0), determined based on read depth in DNA library. (c) Backbones of Cas12a editing plasmids pSTU‐1 and pSTU‐2. All use the promoter and transcriptional terminator from the constitutive Ham34 gene (Ham) to express a single transcript encoding LbCas12a, fused to a Phytophthora sojae nuclear localization signal (NLS), a 73‐nucleotide adenine tract, and a BsaI cloning site for guide RNAs. Also present is the nptII gene for G418 selection. Guide RNA processing is enabled by the native Cas12a direct repeat (DR), hammerhead (HH) ribozyme, and/or hepatitis delta virus (HDV) ribozyme sequences. (d) Binding sites of sg33 and sg156 guide RNAs in the Inf1 open reading frame. (e) Plasmids used in transformation and frequency of editing
As a target, we selected the gene encoding the INF1 elicitor protein (PITG_12551). Previous studies showed that Inf1 is not required for growth and thus editing would not be lethal (Ah‐Fong et al., 2008). We confirmed that PITG_12551 was a single‐copy gene in isolate 1306 based on read depth analysis (Figure 2b).
In the editing system developed for P. sojae (Fang & Tyler, 2016), Cas9 and sgRNA were expressed from two separate transcriptional units. We and others have observed that some Phytophthora transformants containing plasmids with two or three separate transcription units fail to express all genes (Gamboa‐Melendez & Judelson, 2015; Judelson & Whittaker, 1995; van West et al., 1999). To raise the likelihood of editing we therefore designed vectors in which Cas12a and sgRNA were driven by a single promoter, mimicking a strategy used in plants (Tang et al., 2019). Our constructs utilized the constitutive Ham34 promoter to transcribe an RNA encoding LbCas12a, an array of 73 adenines following the TAA stop codon to promote translation, a cassette for forming sgRNA, and finally the Ham34 transcription terminator (Figure 2c). The LbCas12a variant was the same expressed in the experiment shown in Figure 1d. While the Ham34 promoter is recognized by Pol II, such promoters do allow the crRNA to be processed into sgRNA (Zhong et al., 2017).
Vectors were designed to form the sgRNA through either of two mechanisms. pSTU‐1 exploits the ribonuclease activity of Cas12a by flanking the sgRNA sequence with the 21‐bp short direct repeats (DRs) of the native Cas12a scaffold (Figure 2c). pSTU‐2 processes the sgRNA with ribozymes using the hammerhead (HH) and hepatitis delta virus (HDV) sequences employed by Fang and Tyler (2016).
2.3. Configuration of guide RNA cassettes
Two sgRNAs matching the 357 nucleotide (nt) Inf1 coding sequence were designed as detailed in Experimental Procedures (Figure 2d and Table S2). Using a TTTV protospacer adjacent motif (PAM), targets were identified having their 3′ ends at nt 33 and 156 of the open reading frame (ORF) (Figure 2d). The corresponding guide RNAs were named sg33 and sg156. The efficiency of sg156 was predicted to be slightly higher by the Deep‐Cpf1 program (Kim et al., 2018). We designed 23 nt targets because this was optimal for editing in prior studies (Gao et al., 2018; Zetsche et al., 2015).
Five vectors containing these sgRNAs were constructed (Figure 2e). In pSTU‐1A and pSTU‐2A, sgRNA156 was cloned between either DR or ribozyme sequences, respectively. In pSTU‐1B, a variant of sgRNA156 was tested in which the crRNA region included 20 nt of the target plus four 3′ thymidines. A uridine‐rich‐tail has been thought to facilitate the maturation and folding of crRNA by some Cas12a proteins (Moon et al., 2018). We also tested arrays of sg156 and sg33 in pSTU‐1C and pSTU‐2B, using either DR or ribozyme plus DR sequences for processing. A DR was included at the 3′ terminus of the expression cassette in most of our vectors as this may facilitate sgRNA maturation (Zhong et al., 2017).
2.4. Cas12a enables editing in P. infestans
Editing was observed in transformants using four of the five vectors, based on sizing and sequencing a PCR fragment spanning the target sites. Most events generated heterozygotes containing wild‐type and edited alleles, as illustrated in Figure 3a, where the indicated transformant contains both the 522‐nt wild‐type allele and a smaller edited band. Such events resulted in double peaks in Sanger sequencing chromatograms, as illustrated in Figure 3c for transformant T90 (obtained using sg33) and Figure 3d for T74 and T251 (obtained using sg156). We observed 18 cases of heterozygous events (i.e., one wild‐type and one mutated allele) out of 292 independent transformants.
FIGURE 3.
Detection of editing. (a) Representative PCR assay that identified a heterozygous event. The arrow points out transformant T180, which has a deletion in one allele of Inf1 (WT/Δ). (b) Example of biallelic editing. Marked by the arrow is T171, which contains deletions in both alleles (Δ/Δ). (c) Sequencing chromatograms of Inf1 in strains edited using sg33. Shown is wild‐type isolate 1306 (unedited), transformant T90, with double peaks indicating a mixture of wild‐type (WT) and edited alleles, and two edited alleles from T171 (Δ1, Δ2). The data from 1306 and T90 comes from uncloned PCR products, while T171 data comes from cloned alleles. The wild‐type and sgRNA sequence are shown at the top of the panel; arrows indicate the 5′ border of the deletion. (d) Same as panel c but using sg156, showing transformants T251 and T74
Only in about 6% of the edited transformants were both alleles altered. An example is shown in Figure 3b, where transformant T171 lacks the wild‐type 522‐nt PCR band. Shown in Figure 3c is sequence analysis of its two alleles, which contain deletions at different sites. This is unlike the situation observed frequently in P. sojae where both alleles contained the same mutation, probably due to gene conversion occurring after a single editing event (Fang & Tyler, 2016). As will be discussed later, the Inf1 locus in isolate 1306 has unusual features that may suppress the frequency of editing and/or gene conversion.
In about one‐third of cases, the peaks in the sequencing chromatograms signalling editing represented less than 50% of the total signal, which suggested heterokaryosis. Such heterokaryons presumably occur when editing takes place after the first nuclear division after transformation. Based on comparing areas under the peaks in the chromatograms, the ratio of edited to unedited nuclei was calculated to range from 1:1 to 6:1, with a median of 2.6:1. In several cases heterokaryosis was confirmed by single‐zoospore (i.e., single‐nuclear) derivatives of the primary transformant.
The rates at which editing occurred with each vector are recorded in Figure 2e. These results pool data obtained from transformations performed on two separate days. Most events were generated by vectors that used the self‐processing ability of Cas12a with the DR sequences, not the ribozyme system. For example, three out of 62 transformants obtained with DR‐based sg156‐containing pSTU‐1A (5%) were edited compared to zero of 67 with ribozyme‐based pSTU‐2A. Also, while six edited events were obtained with pSTU‐2B, five involved sg156 which was flanked by DRs, and only one involved sg33 which relied on the HDV sequence for its maturation. The difference in the frequency of editing between DR and ribozyme‐based constructs was significant (p = .02). We cannot exclude the possibility that sequences within the sg33 or sg156 regions impaired ribozyme activity, as flanking sequences are known to influence their function (Wang, Wang, et al., 2018).
The highest frequency of editing (13%) was obtained using pSTU‐1C, which expressed both guide RNAs and relied on the DR for crRNA processing. A lower rate (5%) was observed using pSTU‐1A, which only contained sg156. Whether the rate of success using two versus one sgRNAs was significantly different was borderline based on statistical tests, however (p = .12).
The presence of a four‐base uridine tail of the sgRNA did not cause a large increase in editing. More mutated transformants were generated by pSTU‐1B compared to pSTU‐1A (7% versus 5%). However, the distinction was not statistically significant (p = .46).
Besides editing events suggestive of repair by nonhomologous end‐joining, one transformant included a 121‐nt insertion within the site targeted by the sgRNA. The insert matched part of the plasmid used for transformation. This suggested that the repair event initiated by Cas12a had incorporated a fragment of plasmid DNA generated by P. infestans nucleases after the transformation procedure. Such events have been described in other systems, being particularly common in Chlamydomonas (Jiang et al. 2014).
To assist users of the system, the sequences of pSTU‐1 and pSTU‐2 are presented in Appendix S1, and the DNA fragments used for cloning the crRNA regions are listed in Table S1. In addition, a more detailed description of crRNA design in our optimal constructs is given in Figure S1. We also made counterparts using the hygromycin phosphotransferase (hpt) gene for hygromycin selection, which is popular for some members of the genus. Editing of Inf1 was observed in an experiment using the hpt‐encoding counterpart of pSTU‐1A (pSTUH‐1), confirming the function of that vector.
2.5. Variation in size of editing events
Studies in other systems indicated that mutations caused by Cas12a tend to be larger than those resulting from Cas9 (Kim et al., 2016). The same seems to hold true for P. infestans. Based on the detailed analysis of 23 mutations generated with the nptII and hpt vectors, insertions and deletions represented 10% and 90% of the events, respectively. These ranged in size from 1 to 140 nt with a median of 13 nt (Figure 4a). An alignment of these events is shown in Figure S1.
FIGURE 4.
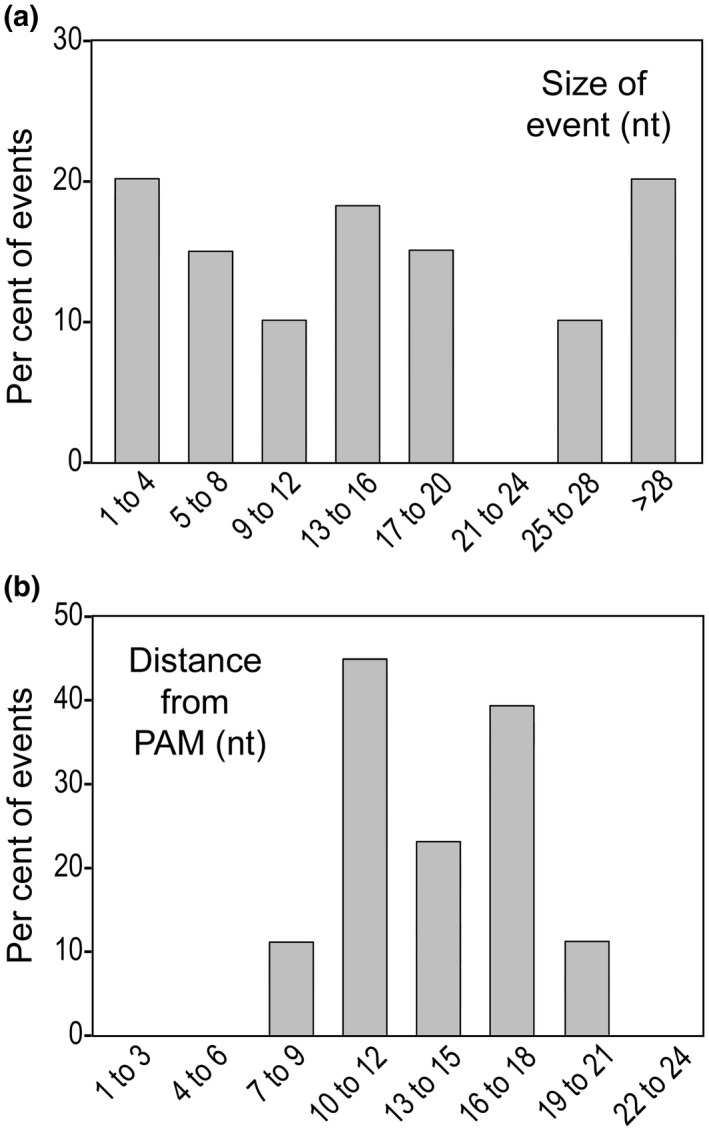
Summary of editing events. (a) Size of indels in nucleotides. (b) Distance of the 5′ boundary of the mutation from the protospacer adjacent motif (PAM) sequence, in nucleotides. Represented in each panel are 23 events that include those listed in Figure 2, and data from transformants obtained using a hpt‐containing plasmid
One difference in the effects of Cas12a in P. infestans compared to other organisms involved the site of target cleavage. This was typically 18–26 bp downstream of the PAM in previously studied species using sgRNAs ≥ 20 nt (Zhang et al., 2019). In contrast, in P. infestans we witnessed editing 7 to 21 nt from the PAM, with a median of 14 nt (Figure 4b).
No difference was obvious in the size or location of editing events involving sg156 and sg33. However, editing was more common with sg156 compared to sg33, which was consistent with the relative efficiencies of the two sgRNAs predicted by the Deep‐Cpf1 algorithm. This may also explain why transformants obtained with pSTU‐1C and pSTU‐2B usually showed evidence of editing by sg156 but not sg33.
2.6. Confirmation of editing at the protein level
We verified editing by examining silver‐stained gels of extracellular proteins. This was possible because INF1 is the major protein secreted by P. infestans, being translated as a 118 amino acid preprotein that is processed to 98 amino acids after removal of the signal peptide. As expected, the wild‐type 10. 2 kDa INF1 band was not detected in transformant T171 in which editing mutated both alleles (Figure 5a). These two mutations should have resulted in the production of proteins of 3.2 and 5.5 kDa, which would have run off the bottom of the gel. Also consistent with expectations, small in‐frame mutations resulted in slightly smaller INF1 variants. This is illustrated in Figure 5b, where T140 and T203 contain mutations that removed six and two amino acids from the middle of the wild‐type protein, respectively.
FIGURE 5.
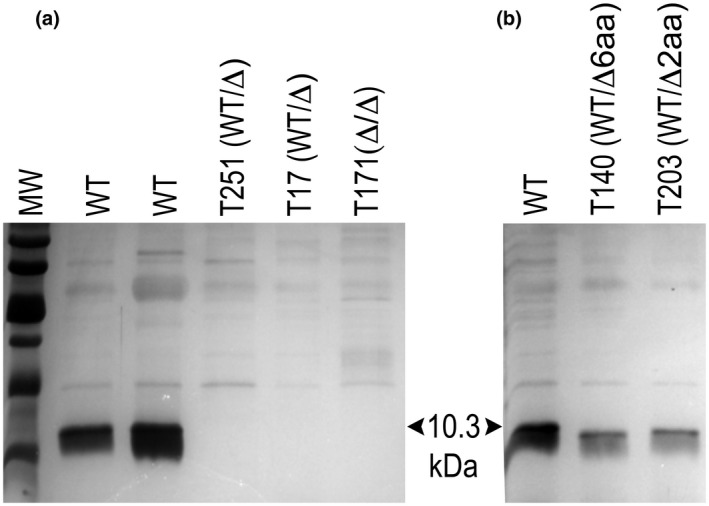
Detection of INF1 protein. (a) Sodium dodecyl sulphate (SDS)‐polyacrylamide gel electrophoresis of secreted proteins from two unedited Phytophthora infestans strains (WT), transformants T251 and T17 that contain one wild‐type allele and frameshift mutations in the other allele (WT/Δ), and transformant T171 that has deletions in both alleles (Δ/Δ). (b) Secreted proteins from wild‐type P. infestans and two transformants heterozygous for editing. T140 and T203 contain in‐frame deletions in one Inf1 allele that remove six and two amino acids, respectively
An unexpected finding was that transformants bearing one wild‐type and one edited allele also failed to make INF1. This is illustrated by the two transformants labelled WT/Δ in Figure 5a. We also saw that transformants such as T140 or T203 did not express a broader INF1 band (or doublet), which was anticipated if both normal and slightly truncated versions were being produced. This led us to hypothesize that expression of Inf1 in isolate 1306 is monoallelic, and that editing had occurred on the expressed allele. As described below, this hypothesis proved to be correct.
2.7. SNP analysis indicates that one Inf1 allele is not transcribed in 1306
To investigate why transformants bearing only one edited copy of Inf1 lacked detectable levels of the protein, we used SNPs to test if only one allele was transcribed in isolate 1306. As stated previously, copy number analysis based on read depth was consistent with Inf1 having two alleles in 1306 (Figure 2b). To identify SNPs, Illumina DNA reads were aligned to the Inf1 region of our 1306 consensus genome assembly (Pan et al., 2018). An A/C SNP in the 3′ UTR was discovered 482 nt from the start codon. This served to distinguish alleles that we name A and B (Figure 6b). As expected for a diploid, the alternate bases occurred at similar frequencies in the library (24 A, 28 C).
FIGURE 6.
Only one Inf1 allele is expressed. (a) Inf1 alleles in isolates 1306 and 618 as defined using single nucleotide polymorphisms (SNPs) in the promoter (nucleotide 306), 5′ untranslated region (UTR) (−47), and 3′ UTR (+482; this is 121 nucleotides from the stop codon). Allele B is inferred to be silent based on the absence of transcripts with SNP 482C. (b) Frequency of the alleles in DNA and RNA libraries of 1306 and 618. (c) Segregation of Inf1 genotypes and RNA level in hyphae of 1306, 618, and their progeny. Bars are coded to represent the 1306 A (white) and B alleles (black)
Reads containing cytidine at nt 482 were never detected in Illumina RNA libraries from isolate 1306. For example, the ratio of the A:C SNPs in hyphal libraries was counted at 148:0. This suggested that allele B was not transcribed. We considered whether mRNA from allele B was produced but degraded, possibly through a small RNA pathway. However, no small RNAs containing the C SNP were detected in a library of 33 million reads.
To assess if strains exhibiting monoallelic expression were common in P. infestans, we examined Illumina data generated by our laboratory for strains representing the US‐1, US‐8, US‐11, US‐22, US‐23, and US‐24 clonal lineages as well as isolates 511, 550, and 618 from Mexico, the presumed centre of origin of the species. We also analysed strains for which both DNA and RNA reads were available in the NCBI Short Read Archive, and the T30‐4 reference assembly. This analysis was usually uninformative due to a lack of SNPs in the transcribed region. In nearly all strains the ORF, 564 nt of sequences 5′ of the ORF, and 482 nt of sequences 3′ of the ORF were identical to allele A of isolate 1306. Regions farther up‐ or downstream were often polymorphic but these were composed of repetitive DNA.
Only for one isolate was it possible to score both alleles for expression. Mexican isolate 618, a diploid (Matson, 2018), contained an A/G SNP in its 5′ untranslated region (UTR) (alleles C and D, Figure 6a). Both occurred at similar frequencies in DNA and RNA data sets, which indicated that isolate 618 bears two transcribed Inf1 alleles.
In other organisms, monoallelic transcription does not always affect mRNA level due to dosage compensation (Eckersiey‐Maslin & Spector, 2014). To test whether this was the case for Inf1, its transcript abundance in hyphae of isolates 1306 and 618 was scored by RNA‐Seq. The level of Inf1 RNA in 1306 was about half that of 618, indicating a lack of dosage compensation (Figure 6c). A similar conclusion came from studying 30 F1 hybrids of 1306 and 618, which we genotyped using the SNPs in Figure 6a. Although variation was observed within each genotypic class, the average mRNA level of progeny with two functional alleles, that is, genotypes AC or AD, was about twice that of the progeny that had inherited the nontranscribed B allele. An examination of RNA‐Seq reads from the progeny indicated that allele B (SNP 482C) was never expressed, identical to the situation in the 1306 parent.
We also exploited the A482C SNP to assess which allele had been altered in transformants having one edited and one normal Inf1 allele. Based on Sanger sequencing, only the expressed allele (allele A) had been mutagenized. This presumably reflects the fact that transcribed loci are more amenable to editing in P. infestans, as has been reported in plants and animals (Kim et al., 2017).
2.8. Monoallelic expression can be explained by a copia‐like element
The possibility that the transcriptional dormancy of allele B resulted from a SNP in a transcription factor binding site was considered. We did identify SNPs between the A and B alleles at nt 376 and 418. However, based on past studies of P. infestans promoters we suspected that these were too far upstream to affect transcription (Ah‐Fong et al., 2007; Tani & Judelson, 2006). As an alternative, we speculated that B might contain a large indel not found in our earlier analysis that relied on mapping DNA reads to the 1306 consensus assembly. Indeed, a search of PacBio reads revealed major heteromorphism in the Inf1 promoter: a 6,310‐nt insert resided 224 bases 5′ of the transcription start site in the B allele (Figure 7a). The existence of the insert was confirmed by identifying Inf1‐copia hybrid reads in an Illumina DNA library. It is reasonable to assume that this large insertion would interfere with the transcription of Inf1.
FIGURE 7.
Copia‐like element in Inf1 promoter. (a) Location of the element in allele B. Indicated are the 5′ untranslated region (UTR), open reading frame (ORF), and 3′ UTR of Inf1; 5‐nucleotide (nt) target site duplication (TSD) flanking the copia‐like insertion; features of the element including the 216‐nt long terminal repeat (LTR), polypurine tract (PPT), and translated region (grey) that includes GAG (pfam14223), GAG‐pre‐integrase (GP; pfam13976), integrase (INT, pfam00665), reverse transcriptase (RT; pfam07727), and RNAse H (RH; pfam00075) domains. (b) Mapping of RNA‐Seq reads across the element, placed in 75‐nt bins. RNA for this analysis was taken from mycelia of isolate 1306 grown in rye‐sucrose broth. (c) Expression level determined by RNA‐Seq in nonsporulating hyphae from rye medium, preinfection stages (sporangia, chilled sporangia, zoospores, germinating cysts), and infected potato tubers and tomato leaves. The plant samples are from presporulation (1.5 day postinoculation [dpi] tubers, 2 dpi leaves) and postsporulation stages (4 dpi tubers, 5 dpi leaves). (d) Mapping of small RNA reads to the element. (e) Size and orientation of small RNAs matching the element
Sequence analysis indicated that the 6.3‐kb insert was a Ty1/copia‐like retrotransposon. It contained 216‐nt long terminal repeats (LTR) and an intact ORF encoding a 1,527 amino acid polyprotein characteristic of functional copia‐like elements including all expected pfam domains (Figure 7a). Also present was polypurine tract adjacent to the right LTR, which is used to prime reverse transcription. The retrotransposon was bounded by a target site duplication of 5‐nt (TGCAG), which is the same size reported for Ty1 in yeast (Curcio et al., 2015).
A search of the genome indicated that the element belonged to a small family. Besides the element upstream of Inf1, the family included one 6.3‐kb sequence that had >99% identity to the Inf1‐linked element and an intact ORF. Four other 6.3‐kb sequences were detected but these appeared crippled due to multiple internal stop codons. Also detected were 24 loci containing only the 216‐nt LTR. Such sites are notable because LTRs can drive transcription of flanking sequences (Curcio et al., 2015), and because solo LTRs are hallmarks of retroelement excision through intrachromosomal recombination. When compared to 14 copia‐like families described previously for P. infestans isolate T30‐4 (Haas et al., 2009), the insertion in Inf1 most closely resembled copia_LTR_12, with 74% nucleotide identity. We consequently name the element copia_LTR_12B.
2.9. Copia_LTR_12B is ancient but the insertion is recent
Not counting the Inf1‐linked copy, other isolates of P. infestans contain a similar complement of copia_LTR_12B sequences based on investigating 46 strains represented in the NCBI Short Read Archive. However, the disruption of Inf1 is unique to isolate 1306 based on searching for chimeric copia‐Inf1 reads. One strain lacking the insert was DDR7602, which does not produce INF1 despite containing Inf1 coding sequences (Kamoun, van der Lee, et al., 1998); its failure to make the protein thus seems due to a phenomenon other than disruption by this transposable element. Because only 1306 contained the copia_LTR_12B insert and few SNPs had accumulated in the promoter or ORF of the nonfunctional allele, the apparent retrotransposition event is likely to have occurred relatively recently in the history of the species.
Several relatives of P. infestans bear sequences resembling copia_LTR_12B, suggesting that it predates the expansion of the genus. In Phytophthora Clade 1, which includes P. infestans (Yang et al., 2017), we detected the element in Phytophthora mirabilis and Phytophthora ipomoea. Their LTRs have 99% and 98% identity, respectively, with those of P. infestans; LTRs are commonly used in phylogenetic studies of retroelements due to their relatively fast evolution (SanMiguel et al., 1998). The element was also found in another Clade 1 species, Phytophthora cactorum, but similarity to the P. infestans LTR was limited to its right‐most 132‐nt. Another Clade 1 species, P hytophthora parasitica, contained copia‐like sequences but their LTRs did not resemble copia_LTR_12B. In other clades, the element had a limited distribution. Nevertheless, Phytophthora cinnamomi, a member of the fairly distant Clade 7, a relative was observed with an LTR 68% identical to that of P. infestans.
In P. infestans, transcripts matching the copia‐like element were detected. A 10‐fold increase in mRNA was observed during early stages of tomato leaf and potato tuber infection compared to late infection, mycelia from artificial media, sporangia, zoospores, and germinating zoospore cysts (Figure 7c). However, nearly all reads from mRNA‐Seq libraries matched the 3′ half of the LTR, with few mapping internal to the element (Figure 7b). Reads from a small RNA library aligned across the retrotransposon albeit with a majority also corresponding to the LTR (Figure 7d). Most small RNAs ranged from 25 to 27 nt, similar to those matching transposons in prior genome‐wide studies (Fahlgren et al., 2013). Whether the reads in Figure 7b–d are from the Inf1 insertion or other loci cannot be determined due to sequence identity among members of the copia_LTR_12B family.
Because copia_LTR_12B may be transcribed, we considered whether additional retrotransposition events had occurred in the 1306 × 618 progeny. In other organisms, sexual hybridization can derepress transposable elements (Henault et al., 2020). However, no new insertion sites were detected in the progeny. This conclusion was drawn by mapping Illumina reads split between the LTR and other genomic DNA to P. infestans chromosomes; all locations in progeny were also in the parents.
3. DISCUSSION
To our knowledge, this is the first report of gene editing in P. infestans and the first using Cas12a in an oomycete. Central to our success was the use of Cas12a instead of Cas9, as the latter seemed to be toxic to P. infestans. Unlike the Cas9 system developed for P. sojae, which expresses the nuclease and sgRNA from separate promoters, our vectors express them in a single transcript to increase the likelihood that both would be produced in transformants. Another difference is that we also exploited the innate RNase activity of Cas12a to mature the sgRNA, instead of ribozyme sequences. Our vectors also allow a single plasmid to produce multiple sgRNAs. While not essential for our experiments with Inf1, the ability to multiplex sgRNAs can be useful because often several need to be tested in order to achieve editing (Wang, Mao, et al., 2018).
The apparent toxicity of Cas9 to P. infestans is not entirely surprising because this problem has also been reported in many other eukaryotes and prokaryotes (Foster et al., 2018; Jiang et al., 2014; Markus et al., 2019; Wang et al., 2019). The most common explanation given for toxicity is inability of the recipient cell to cope with DNA damage caused by off‐target cleavage. While high Cas9 expression is often correlated with more editing, an excess of the protein can cause cleavage at PAM sites in the absence of sgRNAs (Markus et al., 2019). Other factors may also cause toxicity based on problems reported with dCas9 in Chlamydomonas and bacteria (Jiang et al., 2014; Zhang & Voigt, 2018). Myriad strategies for reducing the deleterious effects of Cas9 have been developed. These include expressing the nuclease from an inducible promoter, using a transient expression system, regulating the level of active nuclease using a photoactivatable split protein system, and controlling the translation of Cas9 mRNA using a ligand‐binding riboswitch (Jiang et al., 2014; Nihongaki et al., 2015; de Solis et al., 2016; Wang et al., 2019). Another approach used to attenuate toxicity has been to boost ATP levels, ostensibly to support DNA repair (Wang et al., 2019). Toxicity has also been avoided by delivering Cas9 in a ribonucleoprotein complex or using base editor versions of the protein (Anzalone et al., 2020). Our strategy of expressing the sgRNA and nuclease in a single transcription unit may also reduce toxicity by balancing their expression (Markus et al., 2019).
Despite precedents of toxicity in other taxonomic groups, our apparent difficulty with Cas9 in P. infestans was unanticipated considering its successful use in some other members of the genus. This might reflect biological differences between the species. For example, Cas9 could be more toxic to P. infestans if its DNA repair system was less efficient, or if its genome contained more off‐target cleavage sites. Variation in the nature of DNA‐mediated transformation may also be to blame. Cas9 protein might be more abundant and thus more toxic in P. infestans if the Ham34 promoter was more active, if transgene copy numbers were higher, or if transgene expression was more durable. If true, then modifications to the gene transfer procedure or vector might make Cas9 more serviceable in P. infestans. We do not claim that using Cas9 in P. infestans is impossible; we only state that we and others (van den Hoogen & Govers, 2019) failed to achieve editing after screening several hundred transformants using multiple sgRNAs, and Cas9 toxicity is a plausible explanation.
Even for Phytophthora spp. for which success with Cas9 is reported, our Cas12a system provides a useful alternative. Cas12a recognizes a different PAM motif, thus providing additional sites for editing (Zetsche et al., 2015). Cas12a is more sensitive to mismatches in the guide RNA than Cas9 and thus off‐target cleavage is lower (Kim et al., 2016). Compared to Cas9, the Cas12a crRNA is shorter (c.44 nt) and does not require a trans‐activating crRNA (tracrRNA); this makes constructing crRNAs less expensive and facilitates the assembly of multiplex editing arrays. The larger deletions caused by Cas12a may also augment the frequency of loss‐of‐function events (Kim et al., 2016). Moreover, LbCas12a is known to function at a broader temperature range, which may make it more useful for some species (Fernandez et al., 2018; Moreno‐Mateos et al., 2017).
Meaningful comparisons of the frequency of editing that we observed with Cas12a to those witnessed in other species with Cas9 are challenging because different genes were targeted and due to the unusual nature of Inf1 in isolate 1306. Still, the 13% rate obtained with our optimal vector was within ranges reported for LbCas12a in other organisms (Tang et al., 2019; Wolter & Puchta, 2019). One notable difference between our results and those described for P. sojae is that the latter often resulted in homozygous mutants (Fang & Tyler, 2016), while here only Inf1 allele A was usually altered. We propose that the infrequent modification of allele B is attributable to inaccessible chromatin resulting from its lack of transcription or adjacency to the retroelement. Nucleosomes are known to impair digestion by editing nucleases (Isaac et al., 2016). Moreover, a lack of transcription is understood to suppress gene conversion (Kim & Jinks‐Robertson, 2012), which appears to contribute to the homozygous edits in P. sojae (Fang & Tyler, 2016). Although conversion events have been described for P. infestans (Matson et al., 2015), how the overall rate of gene conversion in P. infestans and P. sojae compares is unknown. It is nevertheless interesting to speculate that methods known to stimulate targeted gene conversion in other species (Liu et al., 2009) might prove to be a useful addition to editing studies in Phytophthora. This could be tested in future experiments along with variables associated with editing such as incubation temperature, and whether Cas12a will enable homology‐directed repair in P. infestans. Apparently due to its ability to produce staggered breaks in DNA, Cas12a was shown to promote homology‐directed repair more than nonhomologous end‐joining in plants and animal cells (Alok et al., 2020; Moreno‐Mateos et al., 2017).
While the original goal of this study was to develop a tool for editing genes in P. infestans, we also encountered a natural phenomenon that alters genomes: insertional mutagenesis by mobile DNA. In other organisms, transposons affect genes by disrupting coding or promoter sequences and less directly via small RNAs that target elements adjacent to genes (Hollister et al., 2011). Transposon‐like sequences represent about 74% of the P. infestans genome (Haas et al., 2009). Most are Gypsy retroelements (about 29% of the genome), DNA transposons (17%), and copia‐like sequences (3.5%). Repeated DNA is thought to have enabled Phytophthora genomes to expand, and transposon‐like fragments have been found adjacent to or within many genic sequences (Dong et al., 2015; Jiang et al., 2005; Qutob et al., 2009). Most mutagenic events caused by repeats in Phytophthora are believed to have resulted from illegitimate recombination and not transposition, as evidence for recent transpositions is scant. However, a copia‐like element flanked by target site duplications, PSCR, was discovered near Avr4/6 of P. sojae; while the polyprotein gene contained several frameshift mutations, these could have arisen after transposition (Basnayake et al., 2009). Transposon‐derived transcripts were observed to increase during the growth of Phytophthora ramorum on certain hosts, and was correlated with chromosome instability (Kasuga et al., 2016). Whether transposition was occurring was unknown, however.
The copia‐like element identified here (which lacks significant nucleotide identity with PSCR of P. sojae) has maintained all of the structural features of an active element. Nevertheless, we detected little evidence of ongoing retrotransposition by comparing the genomes of 1306 × 618 progeny with the parents. While copia_LTR_12B appears to be capable of retrotransposition, the abundance of small RNAs that map to the element signals that its activity may be suppressed by the RNA interference system of P. infestans. It was interesting, nevertheless, to note that mRNAs matching the element rose 10‐fold during the biotrophic stages of plant infection. The derepression of transposable elements during plant infection has also been reported in fungi (Fouche et al., 2020).
A final technical point from this study is the importance of having comprehensive genomic resources for the organism of interest. When we chose Inf1 and isolate 1306 for our studies, we were careful to examine the copy number of the gene and ploidy of the isolate. Such checks are important because in Phytophthora many genes have near‐identical paralogs, and polyploidy and trisomy are common (Aguayo et al., 2016; Ah‐Fong et al., 2017; Yoshida et al., 2013); both would magnify the number of targets to edit. We also confirmed that alternate alleles of the Inf1 coding sequences lacked polymorphisms that would prevent sgRNA binding. That one allele had been rendered inactive by the copia‐like element was unforeseen, however. Such events would typically not be evident in a consensus genome assembly. While the frequencies of interallelic structural and transcriptional polymorphisms in Phytophthora are not well‐characterized, there have been reports of hemizygous regions in several species (Dobrowolski et al., 2002; Jiang et al., 2006; Lamour et al., 2012; Martin et al., 2013; Randall et al., 2003). Studying these on a genome‐wide basis could yield more insight into the origins of variation.
4. EXPERIMENTAL PROCEDURES
4.1. Manipulations of P. infestans
P. infestans strains were cultured at 18 °C on rye‐sucrose medium. Isolate 1306, an A1 mating type strain, originated from infected tomato in San Diego County, California in 1982. Isolate 618 was of the A2 mating type and had been isolated from potato in Mexico in 1987 (Goodwin et al., 1994). Progeny from a 1306 × 618 cross were obtained and their hybrid nature confirmed by scoring SNPs as described (Matson et al., 2015). Single‐zoospore purifications of transformants (to reduce heterokaryons to homokaryons) involved stimulating sporangia to produce zoospores by chilling, removal of sporangia by passage through 15 µm nylon mesh, and plating the purified zoospores at low density on rye‐sucrose medium.
Attempts to detect expression of Cas9 and Cas12a used both the protoplast and electroporation transformation methods (Ah‐Fong & Judelson, 2011; Ah‐Fong et al., 2018). For the data shown in Figure 2e using Cas12a in a G418 resistance backbone, we pooled transformants from two independent experiments using the protoplast method. The electroporation approach was used to test Cas12a in a hygromycin resistance backbone. While mutations in Inf1 were observed, the volume of data is insufficient to indicate if the protoplast or electroporation procedures is superior for editing.
4.2. Vectors for transformation
Vectors were constructed using the plasmids, oligonucleotides, dsDNAs, and PCR primers listed in Table S1. pYF2‐PsNLS‐Cas9‐GFP was altered to express a catalytically inactive Cas9‐GFP fusion by replacing the SpeI and KflI fragment encoding Cas9 with a PCR‐amplified fragment from pAC154‐dual‐dCas9VP160‐sgExpression. The latter contains mutations that block activity, and was obtained using primers dCASF and R. Our Cas12a‐GFP expression plasmid, pYF2‐Cas12a‐GFP, was made by amplifying the PsNLS from pYF515 using primers PsNLSF and R followed by cloning into the SacII/SpeI sites of pYF2. These primers also add a Kozak sequence. We then used primers Cas12F and R to amplify human codon‐optimized LbCas12a from pSQT1665, and cloned that fragment into AflII/SpeI sites downstream of PsNLS.
To construct vectors with Cas12a and the crRNA system in a single transcript, PsNLS‐Cas12a sequences were excised from pYF2‐Cas12a‐GFP using XmaI and AflII and inserted in the StuI/AflII sites of pYF515 to create pY515‐1. This was modified to remove the Ham34 promoter, neomycin phosphotransferase gene (nptII), and Ham34 terminator by digestion with BsrG1 and BamHI and religating the ends. The nptII gene was then amplified from pTOR using NPTF and R and cloned into the NheI/AgeI sites of pY515‐1, downstream of the Rpl41 promoter. Subsequent modifications generated vectors where the target sequences are flanked by direct repeats (pSTU‐1) or ribozymes (pSTU‐2). This entailed inserting synthetic dsDNA fragments PolyA1 and PolyA2 into the AflII/ApaI sites of pY515‐1. These contain polyA‐BsaI‐BsaI‐DR and polyA‐HH‐BsaI‐BsaI‐HDV sequences, respectively. The final vector sequences are provided in Appendix S1. Similar vectors containing the hygromycin resistance marker were constructed by amplifying hpt from pGFPH (Ah‐Fong & Judelson, 2011) with HPTF and R, which was cloned into the NheI/AgeI sites of STU‐1 and STU‐2 to replace nptII.
4.3. sgRNA design and cloning
Cas12a targets within Inf1 were identified using the EuPaGDT, CRISPOR, and Deep‐Cpf1 programs (Concordet & Haeussler, 2018; Kim et al., 2018; Peng & Tarleton, 2015). Criteria used to select sgRNAs included using the more restrictive PAM TTTV instead of TTTN, a GC content of 30%–70%, and high on‐target efficiency. Candidates were checked against the assembly of isolate 1306 to avoid off‐targets. We also tried to avoid sgRNA candidates in which more than three consecutive bases were paired as assessed using RNAStructure (Bellaousov et al., 2013).
DNA oligonucleotides encoding the crRNAs were synthesized, annealed, and cloned into the BsaI sites of STU‐1 and STU‐2. These fragments consisted of the Cas12a scaffold followed by the target sequence (Table S1). A more detailed guide for assembling the sgRNA regions is in Appendix S2.
4.4. Detection of editing
Genomic DNA was extracted from transformants by resuspending 1 cm2 of tissue scraped from a 9‐day culture in 300 μl of 0.2 M Tris pH 8.5, 0.25 M NaCl, 25 mM EDTA, 2% sodium dodecyl sulphate (SDS). After boiling for 5 min, 300 μl of 1:1 phenol:chloroform was added. The mixture was vortexed for 5 min, spun at 18,000 × g for 5 min, and 0.75 vol of isopropanol was added to the supernatant. The tube was spun for 10 min, the supernatant discarded, and then pellet washed with 70% alcohol and air‐dried for 15 min. The pellet was resuspended in 30 μl of 10 mM Tris pH 7.5 containing 0.1 mM EDTA, heated at 65 °C for 5 min, and used for PCR using primers INFR2 and F1. Editing was identified by electrophoresis on 1.5% agarose or Sanger sequencing of the amplicons. To confirm editing, some PCR products were cloned and sequenced.
4.5. Protein analysis
INF1 protein was detected as described (Ah‐Fong et al., 2008). In brief, hyphae were grown for 14 days in modified Plich medium at 18 °C. Culture supernatants were then resolved by electrophoresis on an SDS‐15% polyacrylamide gel and visualized by silver staining.
Immunoblots were performed essentially as described (Abrahamian et al., 2016). In brief, protein was extracted by grinding mycelia from rye‐sucrose medium in extraction buffer (40 mM phosphate pH 7.0, 5 mM EDTA, 20% glycerol, 0.1% Triton X‐100, 1 mM dithiothreitol [DTT]) containing protease inhibitors (Sigma P9599), spin‐clarified, resolved on a SDS‐8% polyacrylamide gel, and electroblotted to nitrocellulose. The membranes were then incubated with primary antibody followed by horseradish peroxidase‐conjugated secondary antibody, developed using chemiluminescence, and digitally imaged. Primary antibodies were against Cas9 (Novus Biologicals NBP2‐36440), GFP (Novus Biologicals NB100‐56401), or LbCas12a (Millipore Sigma MABE1824).
Microscopy was performed using hyphae from young rye‐sucrose broth cultures. Assays used Leica SP5 confocal or Evos fluorescence microscopes (Thermo Fisher). For the former, we used the fluorescein isothiocyanate (488 nm) channel for excitation and an emission window of 510–535 nm. For the latter, we used the manufacturer's GFP light cube package.
4.6. RNA‐Seq, DNA‐Seq, and SNP detection
RNA was extracted from cultures grown in rye broth by grinding tissue to a powder under liquid nitrogen, followed by extraction using the Spectrum Total Plant RNA kit (Sigma). Four biological replicates were used for 1306 and 618 and two for progeny analysis. Indexed libraries were prepared using the Illumina Truseq kit v. 2. Paired‐end libraries were multiplexed and sequenced on an Illumina NextSeq550 to generate 75‐nt single‐end reads with 25 million per sample. Data were analysed using systemPipeR (Backman & Girke, 2016). This filtered and trimmed reads using ShortRead, aligned reads to the P. infestans genome allowing for one mismatch using HiSat2, calculated the reads mapped to the P. infestans genome, and made expression calls with edgeR. Transcription levels of the copia‐like family were determined by counting hits using BlastN, followed by normalization to library size taking into account the fraction of reads mapped to the genome.
For small RNA analysis, RNA was extracted using phenol/chloroform, size‐selected by polyacrylamide‐urea gel electrophoresis, and used to generate a library using the Small RNA Library Prep Kit (New England Biolabs). Using a HiSeq2500, 31.6 million 50‐bp single‐end reads were obtained and submitted to Trim Galore to remove adaptors and low‐quality reads. The resulting reads were mapped to the copia‐like element using BlastN.
DNA was extracted from rye broth cultures using phenol/chloroform or the GeneJET Plant Genomic DNA Purification Kit (Thermo Fisher). Libraries were prepared using the Illumina DNA PCR‐free kit. For 1306 and 618, sequencing was performed to >50‐fold coverage using 50‐ and 250‐nt paired‐end reads from the Illumina HiSeq2500 and MiSeq, respectively. Progeny were sequenced to 15‐ to 30‐fold coverage using 100‐ or 150‐nt paired‐end reads on a HiSeq4000 or NextSeq500, respectively.
Genome‐wide SNPs were identified as follows. Illumina DNA‐Seq files were trimmed using Sickle (github.com/najoshi/sickle) and aligned to the 1306 assembly using bwa‐mem (Li & Durbin, 2009) with default settings. Variants were called using GATK UnifiedGenotyper, selecting loci if they had a total read depth >8, and an indel size of fewer than 6 nt. This identified 275,041 heterozygous loci. Allele read ratios were generated by dividing the alternate read depth into the total read depth per site. The distributions were modelled using fitdistrplus and mixtools (Delignette‐Muller & Dutang, 2015), and plotted using ggplot2 (Benaglia et al., 2009). SNPs in Inf1 were identified by matching its sequence against reads in RNA‐Seq and DNA‐Seq libraries.
Other genomic resources included Phytophthora sequences deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). Searches for copia‐like sequences were executed using BLAST utilities within Fungidb (https://fungidb.org) and Ensemble Protists (https://www.ensemble.org).
Supporting information
FIGURE S1 Sequence alignments showing editing events obtained using sg33 and sg156
TABLE S1 Plasmids and other DNAs used for cloning
APPENDIX S1 Sequence of pSTU‐1 and pSTU‐2
APPENDIX S2 Guidelines for vector construction
ACKNOWLEDGEMENTS
This work was supported by grants to H.S.J. from the National Science Foundation of the United States and the National Institute of Food and Agriculture of the United States Department of Agriculture. The authors thank Meenakshi Kagda, Laetitia Poidevin, and Heber Gamboa‐Melendez for assistance in generating libraries for RNA and DNA sequencing, and Andrea Vu for sharing preliminary results. The authors also thank Ian Wheeldon and Suomeng Dong for helpful discussions.
Ah‐Fong AMV, Boyd A, Matson MEH, Judelson HS. A Cas12a‐based gene editing system for Phytophthora infestans reveals monoallelic expression of an elicitor. Mol Plant Pathol. 2021;22:737–752. 10.1111/mpp.13051
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the NCBI Short Read Archive at https://www.ncbi.nlm.nih.gov/sra under Bioproject PRJNA407960, and in Genbank at https://www.ncbi.nlm.nih.gov/genbank/ as accession MW535258. Plasmids are available upon request.
REFERENCES
- Abrahamian, M. , Ah‐Fong, A.M. , Davis, C. , Andreeva, K. & Judelson, H.S. (2016) Gene expression and silencing studies in Phytophthora infestans reveal infection‐specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Pathogens, 12, e1006097. 10.1371/journal.ppat.1006097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguayo, J. , Halkett, F. , Husson, C. , Nagy, Z.A. , Szigethy, A. , Bakonyi, J. et al. (2016) Genetic diversity and origins of the homoploid‐type hybrid Phytophthora × alni . Applied and Environmental Microbiology, 82, 7142–7153. 10.1128/AEM.02221-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah‐Fong, A.M. , Bormann‐Chung, C.A. & Judelson, H.S. (2008) Optimization of transgene‐mediated silencing in Phytophthora infestans and its association with small‐interfering RNAs. Fungal Genetics and Biology, 45, 1197–1205. 10.1016/j.fgb.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Ah‐Fong, A.M. & Judelson, H.S. (2011) Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biology, 115, 882–890. 10.1016/j.funbio.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Ah‐Fong, A.M.V. , Kagda, M. & Judelson, H.S. (2018) Illuminating Phytophthora biology with fluorescent protein tags. In: Ma, W. & Wolpert, T. (Eds.) Plant Pathogenic Fungi and Oomycetes: Methods and Protocols. Humana Press Inc., pp. 119–129. [DOI] [PubMed] [Google Scholar]
- Ah‐Fong, A.M. , Shrivastava, J. & Judelson, H.S. (2017) Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization. BMC Genomics, 18, 764. 10.1186/s12864-017-4151-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah‐Fong, A. , Xiang, Q. & Judelson, H.S. (2007) Architecture of the sporulation‐specific Cdc14 promoter from the oomycete Phytophthora infestans . Eukaryotic Cell, 6, 2222–2230. 10.1128/EC.00328-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alok, A. , Sandhya, D. , Jogam, P. , Rodrigues, V. , Bhati, K.K. , Sharma, H. et al. (2020) The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Frontiers in Plant Science, 11, 264. 10.3389/fpls.2020.00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone, A.V. , Koblan, L.W. & Liu, D.R. (2020) Genome editing with CRISPR‐Cas nucleases, base editors, transposases and prime editors. Nature Biotechnology, 38, 824–844. 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- Backman, T.W.H. & Girke, T. (2016) systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics, 17, 388. 10.1186/s12859-016-1241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnayake, S. , Maclean, D.J. , Whisson, S.C. & Drenth, A. (2009) Identification and occurrence of the LTR‐Copia‐like retrotransposon, PSCR and other Copia‐like elements in the genome of Phytophthora sojae . Current Genetics, 55, 521–536. 10.1007/s00294-009-0263-9 [DOI] [PubMed] [Google Scholar]
- Bellaousov, S. , Reuter, J.S. , Seetin, M.G. & Mathews, D.H. (2013) RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Research, 41, W471–W474. 10.1093/nar/gkt290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglia, T. , Chauveau, D. , Hunter, D.R. & Young, D.S. (2009) mixtools: An R package for analyzing finite mixture models. Journal of Statistical Software, 32, 1–29. 10.18637/jss.v032.i06 [DOI] [Google Scholar]
- Bin Moon, S.U. , Lee, J.M. , Kang, J.G. , Lee, N.‐E. , Ha, D.‐I. , Kim, D.Y. et al. (2018) Highly efficient genome editing by CRISPR‐Cpf1 using CRISPR RNA with a uridinylate‐rich 3′‐overhang. Nature Communications, 9, 3651. 10.1038/s41467-018-06129-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M. , Boehler, M. , Randall, E. , Young, V. , Csukai, M. , Kraus, S. et al. (2010) Mandipropamid targets the cellulose synthase‐like PiCesA3 to inhibit cell wall biosynthesis in the oomycete plant pathogen, Phytophthora infestans . Molecular Plant Pathology, 11, 227–243. 10.1111/j.1364-3703.2009.00604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet, J.P. & Haeussler, M. (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research, 46, W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M.J. , Lutz, S. & Lesage, P. (2015) The Ty1 LTR‐Retrotransposon of budding yeast, Saccharomyces cerevisiae . Microbiology Spectrum, 3, 1–35. 10.1128/microbiolspec.MDNA3-0053-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delignette‐Muller, M.L. & Dutang, C. (2015) fitdistrplus: an R package for fitting distributions. Journal of Statistical Software, 64, 1–34. 10.18637/jss.v064.i04 [DOI] [Google Scholar]
- Dobrowolski, M.P. , Tommerup, I.C. , Blakeman, H.D. & O’Brien, P.A. (2002) Non‐Mendelian inheritance revealed in a genetic analysis of sexual progeny of Phytophthora cinnamomi with microsatellite markers. Fungal Genetics and Biology, 35, 197–212. 10.1006/fgbi.2001.1319 [DOI] [PubMed] [Google Scholar]
- Dong, S.M. , Raffaele, S. & Kamoun, S. (2015) The two‐speed genomes of filamentous pathogens: Waltz with plants. Current Opinion in Genetics & Development, 35, 57–65. 10.1016/j.gde.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Du, J. , Verzaux, E. , Chaparro‐Garcia, A. , Bijsterbosch, G. , Keizer, L.C.P. , Zhou, J.I. et al. (2015) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nature Plants, 1, 15034. 10.1038/nplants.2015.34 [DOI] [PubMed] [Google Scholar]
- Eckersiey‐Maslin, M.A. & Spector, D.L. (2014) Random monoallelic expression: regulating gene expression one allele at a time. Trends in Genetics, 30, 237–244. 10.1016/j.tig.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Bollmann, S.R. , Kasschau, K.D. , Cuperus, J.T. , Press, C.M. , Sullivan, C.M. et al. (2013) Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS One, 8, e77181. 10.1371/journal.pone.0077181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y. & Tyler, B.M. (2016) Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Molecular Plant Pathology, 17, 127–139. 10.1111/mpp.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J.P. , Vejnar, C.E. , Giraldez, A.J. , Rouet, R. & Moreno‐Mateos, M.A. (2018) Optimized CRISPR‐Cpf1 system for genome editing in zebrafish. Methods, 150, 11–18. 10.1016/j.ymeth.2018.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara, I. , Richter, H. , Bratovic, M. , Le Rhun, A. & Charpentier, E. (2016) The CRISPR‐associated DNA‐cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature, 532, 517–521. 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
- Foster, A.J. , Martin‐Urdiroz, M. , Yan, X. , Wright, H.S. , Soanes, D.M. & Talbot, N.J. (2018) CRISPR‐Cas9 ribonucleoprotein‐mediated co‐editing and counterselection in the rice blast fungus. Scientific Reports, 8, 14355. 10.1038/s41598-018-32702-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouche, S. , Badet, T. , Oggenfuss, U. , Plissonneau, C. , Francisco, C.S. & Croll, D. (2020) Stress‐driven transposable element de‐repression dynamics and virulence evolution in a fungal pathogen. Molecular Biology and Evolution, 37, 221–239. 10.1093/molbev/msz216 [DOI] [PubMed] [Google Scholar]
- Gamboa‐Melendez, H. & Judelson, H.S. (2015) Development of a bipartite ecdysone‐responsive gene switch for the oomycete Phytophthora infestans and its use to manipulate transcription during axenic culture and plant infection. Molecular Plant Pathology, 16, 83–91. 10.1111/mpp.12161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z.L. , Herrera‐Carrillo, E. & Berkhout, B. (2018) Improvement of the CRISPR‐Cpf1 system with ribozyme‐processed crRNA. RNA Biology, 15, 1458–1467. 10.1080/15476286.2018.1551703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. , Cohen, B.A. & Fry, W.E. (1994) Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proceedings of the National Academy of Sciences of the United States of America, 91, 11591–11595. 10.1073/pnas.91.24.11591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- Henault, M. , Marsit, S. , Charron, G. & Landry, C.R. (2020) The effect of hybridization on transposable element accumulation in an undomesticated fungal species. eLife, 9, e60474. 10.7554/eLife.60474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister, J.D. , Smith, L.M. , Guo, Y.L. , Ott, F. , Weigel, D. & Gaut, B.S. (2011) Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata . Proceedings of the National Academy of Sciences of the United States of America, 108, 2322–2327. 10.1073/pnas.1018222108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac, R.S. , Jiang, F. , Doudna, J.A. , Lim, W.A. , Narlikar, G.J. & Almeida, R. (2016) Nucleosome breathing and remodeling constrain CRISPR‐Cas9 function. eLife, 5, 13450. 10.7554/eLife.13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan, S.N. , Asman, A.K. , Corcoran, P. , Fogelqvist, J. , Vetukuri, R.R. & Dixelius, C. (2015) Plant‐mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans . Journal of Experimental Botany, 66, 2785–2794. 10.1093/jxb/erv094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Dawe, A.L. , Weide, R. , Staveren, M.V. , Peters, S. , Nuss, D.L. et al. (2005) Elicitin genes in Phytophthora infestans are clustered and interspersed with various transposon‐like elements. Molecular Genetics and Genomics, 273, 20–32. 10.1007/s00438-005-1114-0 [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Weide, R. , de Vondervoort, P.J.I.V. & Govers, F. (2006) Amplification generates modular diversity at an avirulence locus in the pathogen Phytophthora . Genome Research, 16, 827–840. 10.1101/gr.5193806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Brueggeman, A.J. , Horken, K.M. , Plucinak, T.M. & Weeks, D.P. (2014) Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii . Eukaryotic Cell, 13, 1465–1469. 10.1128/EC.00213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. , Tyler, B.M. & Michelmore, R.W. (1992) Regulatory sequences for expressing genes in oomycete fungi. Molecular Genetics and Genomics, 234, 138–146. 10.1007/BF00272355 [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. & Whittaker, S.L. (1995) Inactivation of transgenes in Phytophthora infestans is not associated with their deletion, methylation, or mutation. Current Genetics, 28, 571–579. 10.1007/BF00518171 [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van der Lee, T. , van der Berg‐Velthuis, G. , De Groot, K.E. & Govers, F. (1998) Loss of production of the elicitor protein INF1 in the clonal lineage US‐1 of Phytophthora infestans . Phytopathology, 88, 1315–1323. 10.1094/PHYTO.1998.88.12.1315 [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van West, P. , Vleshouwers, V.G.A.A. , De Groot, K.E. & Govers, F. (1998) Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. The Plant Cell, 10, 1413–1425. 10.1105/tpc.10.9.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T. , Bui, M. , Bernhardt, E. , Swiecki, T. , Aram, K. , Cano, L.M. et al. (2016) Host‐induced aneuploidy and phenotypic diversification in the sudden oak death pathogen Phytophthora ramorum . BMC Genomics, 17, 385. 10.1186/s12864-016-2717-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Kim, J. , Hur, J.K. , Been, K.W. , Yoon, S.H. & Kim, J.S. (2016) Genome‐wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nature Biotechnology, 34, 863–868. 10.1038/nbt.3609 [DOI] [PubMed] [Google Scholar]
- Kim, H.K. , Min, S. , Song, M. , Jung, S. , Choi, J.W. , Kim, Y. et al. (2018) Deep learning improves prediction of CRISPR‐Cpf1 guide RNA activity. Nature Biotechnology, 36, 239–241. 10.1038/nbt.4061 [DOI] [PubMed] [Google Scholar]
- Kim, H.K. , Song, M. , Lee, J. , Menon, A.V. , Jung, S. , Kang, Y.‐M. et al. (2017) In vivo high‐throughput profiling of CRISPR‐Cpf1 activity. Nature Methods, 14, 153–159. 10.1038/nmeth.4104 [DOI] [PubMed] [Google Scholar]
- Kim, N. & Jinks‐Robertson, S. (2012) Transcription as a source of genome instability. Nature Reviews Genetics, 13, 204–214. 10.1038/nrg3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour, K. , Mudge, J. , Gobena, D. , Hurtado‐Gonzales, O.P. , Schmutz, J. , Kuo, A. et al. (2012) Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici . Molecular Plant‐Microbe Interactions, 25, 1350–1360. 10.1094/MPMI-02-12-0028-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leesutthiphonchai, W. & Judelson, H.S. (2018) A MADS‐box transcription factor regulates a central step in sporulation of the oomycete Phytophthora infestans . Molecular Microbiology, 110, 562–575. 10.1111/mmi.14114 [DOI] [PubMed] [Google Scholar]
- Leesutthiphonchai, W. , Vu, A.L. , Ah‐Fong, A.M.V. & Judelson, H.S. (2018) How does Phytophthora infestans evade control efforts? modern insight into the late blight disease. Phytopathology, 108, 916–924. 10.1094/PHYTO-04-18-0130-IA [DOI] [PubMed] [Google Scholar]
- Li, H. & Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Liu, Y. , Tan, X. , Li, D. , Yang, X. , Zhang, X. et al. (2020) The high‐affinity phosphodiesterase PcPdeH is involved in the polarized growth and pathogenicity of Phytophthora capsici . Fungal Biology, 124, 164–173. 10.1016/j.funbio.2020.01.006 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Nairn, R.S. & Vasquez, K.M. (2009) Targeted gene conversion induced by triplex‐directed psoralen interstrand crosslinks in mammalian cells. Nucleic Acids Research, 37, 6378–6388. 10.1093/nar/gkp678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus, B.M. , Bell, G.W. , Lorenzi, H.A. & Lourido, S. (2019) Optimizing systems for Cas9 expression in Toxoplasma gondii . mSphere, 4, e00386‐19. 10.1128/mSphere.00386-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.D. , Cappellini, E. , Samaniego, J.A. , Zepeda, M.L. , Campos, P.F. , Seguin‐Orlando, A. et al. (2013) Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nature Communications, 4, 2172. 10.1038/ncomms3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, M.E.H. (2018) A chromosome‐scale reference assembly facilitates new insights into fungicide sensitivity and genome plasticity in Phytophthora infestans. Ph.D. thesis, University of Calfiornia. [Google Scholar]
- Matson, M.E.H. , Small, I.M. , Fry, W.E. & Judelson, H.S. (2015) Metalaxyl resistance in Phytophthora infestans : assessing role of RPA190 gene and diversity within clonal lineages. Phytopathology, 105, 1594–1600. 10.1094/PHYTO-05-15-0129-R [DOI] [PubMed] [Google Scholar]
- Moreno‐Mateos, M.A. , Fernandez, J.P. , Rouet, R. , Vejnar, C.E. , Lane, M.A. , Mis, E. et al. (2017) CRISPR‐Cpf1 mediates efficient homology‐directed repair and temperature‐controlled genome editing. Nature Communications, 8, 2024. 10.1038/s41467-017-01836-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki, Y. , Kawano, F. , Nakajima, T. & Sato, M. (2015) Photoactivatable CRISPR‐Cas9 for optogenetic genome editing. Nature Biotechnology, 33, 755–760. 10.1038/nbt.3245 [DOI] [PubMed] [Google Scholar]
- Pan, W. , Wanamaker, S.I. , Ah‐Fong, A.M.V. , Judelson, H.S. & Lonardi, S. (2018) Novo&Stitch: accurate reconciliation of genome assemblies via optical maps. Bioinformatics, 34, i43–i51. 10.1093/bioinformatics/bty255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, D. & Tarleton, R. (2015) EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microbial Genomics, 1, e000033. 10.1099/mgen.0.000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettongkhao, S. , Navet, N. , Schornack, S. , Tian, M. & Churngchow, N. (2020) A secreted protein of 15 kDa plays an important role in Phytophthora palmivora development and pathogenicity. Scientific Reports, 10, 2319. 10.1038/s41598-020-59007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Tedman‐Jones, J. , Dong, S. , Kuflu, K. , Pham, H. , Wang, Y. et al. (2009) Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a . PLoS One, 4, e5066. 10.1371/journal.pone.0005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, T.A. , Ah‐Fong, A. & Judelson, H. (2003) Chromosomal heteromorphism and an apparent translocation detected using a BAC contig spanning the mating type locus of Phytophthora infestans . Fungal Genetics and Biology, 38, 75–84. 10.1016/S1087-1845(02)00512-1 [DOI] [PubMed] [Google Scholar]
- SanMiguel, P. , Gaut, B.S. , Tikhonov, A. , Nakajima, Y. & Bennetzen, J.L. (1998) The paleontology of intergene retrotransposons of maize. Nature Genetics, 20, 43–45. 10.1038/1695 [DOI] [PubMed] [Google Scholar]
- Situ, J. , Jiang, L. , Fan, X. , Yang, W. , Li, W. , Xi, P. et al. (2020) An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Molecular Plant Pathology, 21, 415–428. 10.1111/mpp.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Solis, C.A. , Ho, A. , Holehonnur, R. & Ploski, J.E. (2016) The development of a viral mediated CRISPR/Cas9 system with doxycycline‐dependent gRNA expression for inducible in vitro and in vivo genome editing. Frontiers in Molecular Neuroscience, 9, 70. 10.3389/fnmol.2016.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Ren, Q. , Yang, L. , Bao, Y. , Zhong, Z. , He, Y. et al. (2019) Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnology Journal, 17, 1431–1445. 10.1111/pbi.13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, S. & Judelson, H.S. (2006) Activation of zoosporogenesis‐specific genes in Phytophthora infestans involves a 7‐nucleotide promoter motif and cold‐induced membrane rigidity. Eukaryotic Cell, 5, 745–752. 10.1128/EC.5.4.745-752.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen, J. & Govers, F. (2019) Attempts to implement CRISPR/Cas9 for genome editing in the oomycete Phytophthora infestans . bioRxiv, 274829 [preprint]. 10.1101/274829 [DOI] [Google Scholar]
- van West, P. , Reid, B. , Campbell, T.A. , Sandrock, R.W. , Fry, W.E. , Kamoun, S. et al. (1999) Green fluorescent protein (GFP) as a reporter gene for the plant pathogenic oomycete Phytophthora palmivora . FEMS Microbiology Letters, 178, 71–80. 10.1111/j.1574-6968.1999.tb13761.x [DOI] [PubMed] [Google Scholar]
- Vu, A.L. , Leesutthiphonchai, W. , Ah‐Fong, A.M.V. & Judelson, H.S. (2019) Defining transgene insertion sites and off‐target effects of homology‐based gene silencing informs the application of functional genomics tools in Phytophthora infestans . Molecular Plant‐Microbe Interactions, 32, 915–927. 10.1094/MPMI-09-18-0265-TA [DOI] [PubMed] [Google Scholar]
- Wang, K. , Zhao, Q.‐W. , Liu, Y.‐F. , Sun, C.‐F. , Chen, X.‐A. , Burchmore, R. et al. (2019) Multi‐layer controls of Cas9 activity coupled with ATP synthase over‐expression for efficient genome editing in Streptomyces . Frontiers in Bioengineering and Biotechnology, 7, 304. 10.3389/fbioe.2019.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Mao, Y. , Lu, Y. , Wang, Z. , Tao, X. & Zhu, J.K. (2018) Multiplex gene editing in rice with simplified CRISPR‐Cpf1 and CRISPR‐Cas9 systems. Journal of Integrative Plant Biology, 60, 626–631. 10.1111/jipb.12667 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, Z. , Liu, T. , Gong, S. & Zhang, W. (2018) Effects of flanking regions on HDV cotranscriptional folding kinetics. RNA, 24, 1229–1240. 10.1261/rna.065961.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Avrova, A.O. , van West, P. & Jones, J.T. (2005) A method for double‐stranded RNA‐mediated transient gene silencing in Phytophthora infestans . Molecular Plant Pathology, 6, 153–163. 10.1111/j.1364-3703.2005.00272.x [DOI] [PubMed] [Google Scholar]
- Wolter, F. & Puchta, H. (2019) In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant Journal, 100, 1083–1094. 10.1111/tpj.14488 [DOI] [PubMed] [Google Scholar]
- Yang, X. , Tyler, B.M. & Hong, C. (2017) An expanded phylogeny for the genus Phytophthora . IMA Fungus, 8, 355–384. 10.5598/imafungus.2017.08.02.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Schuenemann, V.J. , Cano, L.M. , Pais, M. , Mishra, B. , Sharma, R. et al. (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife, 2, e00731. 10.7554/eLife.00731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B. , Gootenberg, J. , Abudayyeh, O. , Slaymaker, I. , Makarova, K. , Essletzbichler, P. et al. (2015) Cpf1 is a single‐RNA‐guided endonuclease of a Class 2 CRISPR‐Cas system. Cell, 163, 759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. & Voigt, C.A. (2018) Engineered dCas9 with reduced toxicity in bacteria: implications for genetic circuit design. Nucleic Acids Research, 46, 11115–11125. 10.1093/nar/gky884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, Y. & Qi, Y. (2019) Plant gene knockout and knockdown by CRISPR‐Cpf1 (Cas12a) systems. Methods in Molecular Biology, 1917, 245–256. 10.1007/978-1-4939-8991-1_18 [DOI] [PubMed] [Google Scholar]
- Zhong, G. , Wang, H. , Li, Y. , Tran, M.H. & Farzan, M. (2017) Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nature Chemical Biology, 13, 839–841. 10.1038/nchembio.2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Sequence alignments showing editing events obtained using sg33 and sg156
TABLE S1 Plasmids and other DNAs used for cloning
APPENDIX S1 Sequence of pSTU‐1 and pSTU‐2
APPENDIX S2 Guidelines for vector construction
Data Availability Statement
The data that support the findings of this study are openly available in the NCBI Short Read Archive at https://www.ncbi.nlm.nih.gov/sra under Bioproject PRJNA407960, and in Genbank at https://www.ncbi.nlm.nih.gov/genbank/ as accession MW535258. Plasmids are available upon request.


