Abstract
Background/Aim: c-Met (mesenchymal-epithelial transition factor) facilitates cancer progression and is recognized as a promising drug target. The molecular target of gigantol from Dendrobium draconis in suppressing cancer metastasis is largely unknown. Materials and Methods: Proteins affected by gigantol treatment were subjected to proteomic and bioinformatic analysis. Protein–Protein interaction (PPI) networks were constructed by the Search Tool for the Retrieval of Interacting Genes (STRING). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database and hub gene were used to enrich the dominant pathways. Western blot analysis and immunofluorescence were used to validate the effect of gigantol on the target protein and signaling. Results: Gigantol down-regulates 41 adhesion proteins and 39-migratory proteins, while it up-regulates 30 adhesion-related proteins and 22 proteins controlling cell migration. The key components of our constructed PPI network comprised 41 proteins of cell adhesion enriched in 40 nodes with 25 edges, 39 proteins of cell migration enriched in 39 nodes with 76 edges in down-regulated proteins, 30 proteins of cell adhesion enriched in 30 nodes with 21 edges, and 22 proteins of cell migration enriched in 22 nodes with 22 edges in up-regulated protein. c-Met was identified as a central protein of the PPI network in the largest degree. KEGG mapper further suggested that c-Met, PI3K, and AKT were the regulatory proteins affected by gigantol. To confirm, the effects of gigantol on c-Met, the p-PI3K, PI3K, p-AKT, and AKT proteins were investigated by western blotting and the results showed a consistent effect of gigantol in the suppression of the c-Met/PI3K/AKT signal. Next, immunofluorescence showed a dramatic decrease in c-Met, PI3K and AKT activation in response to gigantol. Conclusion: c-Met is an important target of gigantol treatment in lung cancer cells. Gigantol suppresses metastasis-related cell motility through decreasing c-Met resulting in PI3K/AKT signaling disruption.
Keywords: Gigantol, c-Met, metastasis, lung cancer, PI3K, proteomics
Lung cancer is one of the leading causes of worldwide mortality and accounts for approximately 27% of all cancer-related deaths (1). Metastasis is responsible for the majority of lung cancer deaths and most lung cancer patients are found with metastasis at the time of first diagnosis. Cancer metastasis involves a multi-step process, including cell detachment, cell migration, cell invasion, survival in the circulation, and establishment of new tumors (2). Cell adhesion plays a crucial role in cell to cell and cell to extracellular matrix (ECM) interactions through a group of cell adhesion molecules (CAMs), such as cadherins, integrins or selectins and lymphocyte homing receptors (CD44). Therefore, reduction of these cell-cell adhesions and cell-ECM adhesions is an early step in cancer cell dissemination (3). In addition, cell migration plays an important prerequisite process in a loose ECM attachment and loss of polarity, which facilitates cancer cell invasion and metastasis (4). Regarding the upstream signaling of migratory control, c-Met, also known as MET (mesenchymal-epithelial transition factor), a receptor tyrosine kinase encoded by the MET gene, has been shown to facilitate several metastatic behaviors. The binding of hepatocyte growth factor (HGF) to c-Met triggers the receptor activation and transmits the HGF/c-Met axis through the interaction and activation of other tyrosine kinases, resulting in the induction of PI3K/AKT, Ras/MAPK, JAK/STAT, SRC, and Wnt/β-catenin (5-8). The activation of signals downstream of c-Met drive several cellular processes that stimulate cell proliferation, migration, invasion, and angiogenesis. In lung cancer, overexpression of the c-Met protein has been reported and found to be linked with a high metastasis potential, increased tumor growth, and drug resistance (9,10). Recently, c-Met has been considered an important target for therapeutic approaches (11).
Gigantol, a bibenzyl compound from Dendrobium draconis, has attracted increasing attention in recent years as it exhibits several potential effects on cancer cells. One study reported that gigantol inhibits lung cancer proliferation through induction of GSK3β-mediated MYC ubiquitin-proteasome degradation (12). In addition, gigantol was shown to inhibit the expression of Slug, the regulator of epithelial–to–mesenchymal transition (EMT), and diminished cancer stem cell properties through ATP-dependent tyrosine kinase (AKT) inhibition (13,14). However, the complete profile of how gigantol can control cell migration and adhesion is currently unknown. Consequently, the present study aimed to unravel the cellular pathways affected by gigantol treatment in lung cancer cells, focusing on cell migration and cell adhesion, as these behaviors are related to the metastasis potential of cancer. We performed proteomic analysis on gigantol-treated lung cancer cells and used bioinformatics to examine the affected pathways. These findings might be useful for the development of this compound for lung cancer treatment as well as might benefit the further prediction of its potential pharmacological activities based on its cellular mechanism of action.
Materials and Methods
Cell cultures. Human non-small cell lung cancer cells (H460) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium at 37˚C in a 5% CO2 humidified incubator. The medium was supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 units/ml penicillin, and streptomycin.
Chemicals and reagents. Gigantol was purchased from Sigma chemical, Inc. (Chemical Express, Bangkok, Thailand). RPMI-1640 medium, phosphate buffer saline, glutamine, penicillin, and streptomycin were purchased from Gibco Company (Grand Island, NY, USA). Fetal bovine serum (FBS), and agarose were obtained from Bio-Rad Laboratories (Hercules, CA, USA). Primary antibodies against c-Met, p-AKT, AKT, p-PI3K, PI3K, and β-actin and specific horseradish peroxidase (HRP)-link secondary antibody were obtained from Cell Signaling Technology, Inc. (Danver, MA, USA).
Proteomics sample preparation. The H460 cells treated with 20 μM of gigantol for 24 h were dissolved with 0.5% SDS and protein concentration was measured by the Lowry’s method. An equal amount of pooled protein from control or gigantol-treated cells (50 μg each) were subjected to in-solution digestion. Samples were completely dissolved in 10 mM ammonium bicarbonate (NH4HCO3) containing 10 mM dithiothreitol (DTT) for 1 hour at 60˚C and alkylated with 15 mM iodoacetamide (IAA) in 10 mM NH4HCO3 for 45 min in the dark, and then and digested with 1 ng protein per 20 ng sequencing grade trypsin (Promega, Walldorf, Germany) at 37˚C overnight. The trypsiniszed peptides were protonated with 0.1% formic acid before injection into LC-MS/MS.
LC/MS-MS analysis. LC/MS-MS for protein quantitation and identification from the digested samples was conducted using the Ultimate 3000 Nano/Capillary LC System (Thermo Scientific, Gloucester, UK) coupled to a Hybrid quadrupole Q-Tof impact II™ (Bruker Daltonics, Coventry, UK) equipped with a Nano-captive spray ion source with a nanocolumn (PepMap100, C18, 300 μm i.d.×5 mm). Solvent A contained 0.1% formic acid and solvent B contained 80% ACN in water containing 0.1% formic acid were used to elute peptides. Peptide separation was achieved with a linear gradient from 5% to 55% of solvent B at a constant flow rate of 0.30 μL/min for 30 minutes including a regeneration and an equilibration step at 90% and 10% eluent B, respectively. Next, the LC-MS/MS raw data were converted into a software file format mzXML (Compass 1.9 software, Bruker Daltonics). The MS and MS/MS spectra which over the range 150-2200 m/z were generated for further protein identification against database search.
Proteomics data and bioinformatics analysis. The MS/MS spectra were searched using MaxQuant 1.6.6.0 to identify proteins and peptides. MS/MS spectra were searched by the Andromeda search engine against the Uniprot Homo sapiens database as following parameters: maximum of two miss cleavages, mass tolerance of 20 ppm for main search, enzyme (trypsin), variable modifications (carbamidomethylation of cysteine as fixed modification, oxidation of methionine and acetylation of the protein N-terminus). For protein identification, only proteins with at least two peptides and at least one unique peptide corresponding to a p-value of less than 0.05 was considered and used for data analysis. In order to determine an appropriate statistical method to assess the differential abundance of proteins, protein fold-changes between untreated control cells and gigantol-treated cells were computed using a comparison of the averaged log2 fold change. The list comparison of identified proteins between the control and treated group were displayed in Venn diagrams (http://jvenn.toulouse.inra.fr/app/index.html). By comparing the presence of detected proteins between gigantol-treated and control non-treated cells, the proteins that were only detected in gigantol-treated cells were assigned as up-regulated proteins. The proteins that were detectable only in the non-treated cells were assigned as down-regulated proteins. Protein Analysis through Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org/) were used to for GO term underlying ‘cell adhesion (GO: 0007155)’ and ‘cell migration (GO: 0016477)’. The Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/cgi/input.pl; version: 11.0) was used to construct a protein–protein interaction (PPI)) network of the involved mechanism in process underlying to GO term ‘cell adhesion (GO: 0007155)’ and ‘cell migration (GO: 0016477)’. The hub genes were constructed by Cytoscape software 3.7.2 (https://cytoscape.org/).agre KEGG mapper analysis was utilized the important signaling pathways in down-regulated protein related to cell migration (https://www.genome.jp/kegg/mapper.html).
Western blot analysis. Cells were treated with gigantol at non-toxic concentrations (0-20 μM) for 24 h. After treatment, the cells were washed with cold PBS twice cells were incubated on ice for 40 min with RIPA buffer, 1% Triton X-100, 100 mM PMSF, and a protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN, USA). Cell lysates were analysed for protein content using the BCA protein assay kit from Pierce Biotechnology (Rockford, IL, USA). Equal amounts of protein samples (60 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) (Bio-Rad Laboratories Inc., CA, USA). The transferred membranes were blocked for 2 h with 5% (w/v) non-fat dry milk (Merck, DA, Germany) in TBS-T (Tris-buffer saline with 0.1% Tween containing 25 mM Tris-HCl (pH 7.5), 125 mM NaCl, and 0.1% Tween 20) and were incubated with specific primary antibodies against c-Met, p-AKT, AKT, p-PI3K, PI3K, and β-actin (Cell Signaling, Danvers, MA, USA) at 4˚C overnight. Then, membranes were washed three times with TBS-T and incubated with appropriate horseradish peroxidase (HRP)-labeled secondary antibodies for 2 h at room temperature. The immune complexes were detected by enhanced chemiluminescence (Supersignal West Pico; Pierce, Rockford, IL, USA) and quantified using ImageJ software (NIH, Bethesda, MD, USA).
Immunofluorescence assay. Cells were seeded onto 96-well plates at a density of 1×104 cells/well and treated with non-toxic concentrations (0-20 μM) of gigantol for 24 h. The cells were washed twice with 1x PBS and fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton-x in PBS for 20 min and blocked with 4% BSA in 1xPBS for 30 min at room temperature. The cells were incubated with primary antibodies against c-Met, p-AKT, p-PI3K overnight at 4˚C. Cells were washed twice with 1x PBS and subsequently incubated with Alexa Fluor 488 or 594 conjugated secondary antibodies for 1 h in the dark at room temperature. The cells were washed with 1x PBS and incubated with Hoechst 33342 (Sigma, St. Louis, MO, USA) for 20 min in the dark, rinsed with 1x PBS and mounted by 50% glycerol (Merck, DA, Germany). Images were assessed under fluorescence microscope with a 40x objective lens (Nikon ECLIPSE Ts2, Tokyo, Japan) and the analysis was assessed by ImageJ software.
Statistical analysis. All of the experiments were repeated at least three times. Statistical differences between multiple groups were analyzed using an analysis of variance (ANOVA). The data was presented as the mean±standard error of the mean (SEM) of three independent experiments with three replicates per experiment. All p-values of less than 0.05 were considered as statistically significant.
Results
Proteomic analysis of the down- and up-regulated proteins in gigantol-treated cells. Lung cancer H460 cells were treated with 20 μM gigantol for 24 h. Proteins were collected and subjected to LC-MS/MS analysis. From the proteomic analysis, the differentially expressed proteins were compared between groups, and 4,140 differentially expressed proteins, including 2,373 down-regulated proteins and 1,767 up-regulated proteins in the gigantol-treated cells were identified. Then, the down-regulated and up-regulated proteins were classified according to the GO term ‘cell adhesion (GO: 0007155)’ and ‘cell migration (GO: 0016477)’, which are related to cancer metastasis ability. The results showed that gigantol caused the down-regulation of 80 proteins including 41 proteins related to cell adhesion and 39 proteins related to cell migration. In addition, 52 up-regulated proteins including 30 proteins related to cell adhesion and 22 proteins related to cell migration, were detected.
Subsequently, the protein–protein interaction (PPI) networks and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were constructed. Down-regulated and up-regulated proteins were enrolled in the Search Tool for the Retrieval of Interacting Genes (STRING) to construct a PPI network associated with the down-regulated and up-regulated proteins in the gigantol-treated cells. The PPI network illustrates the nodes and the edges represent the proteins and the interactions/connections between them, respectively. Further, from KEGG pathway map analysis, a protein interaction diagram was generated with genomic information and this was used to illustrate pathways of the proteins affected by gigantol treatment.
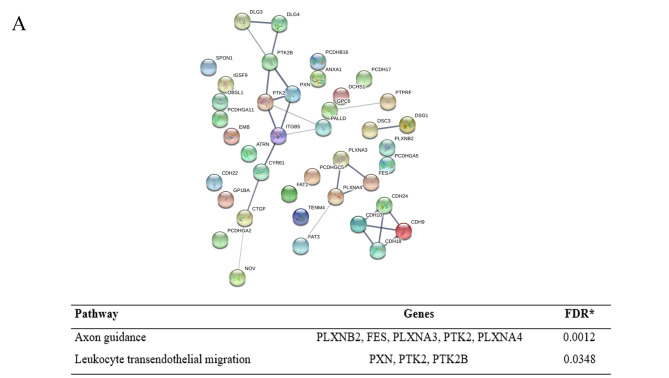
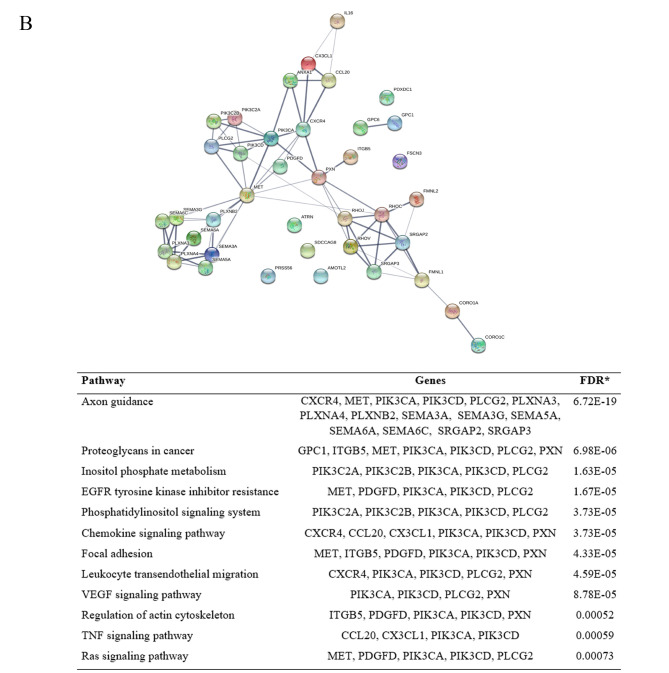
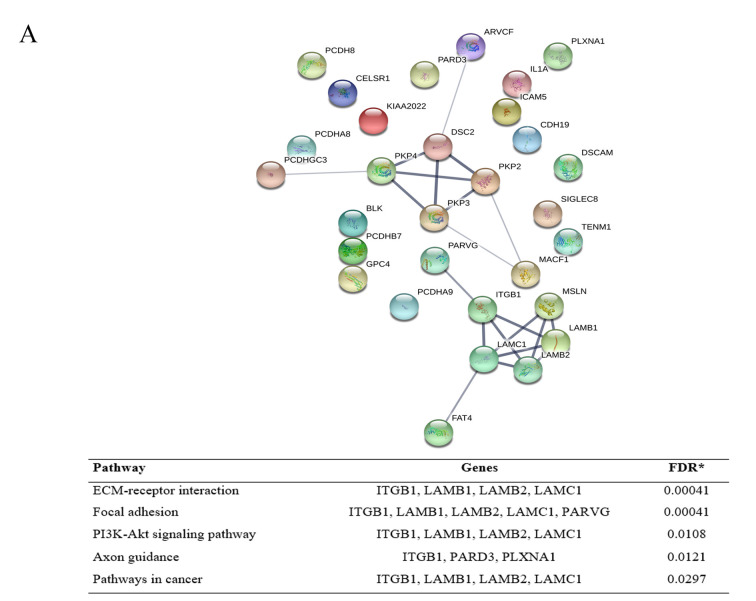
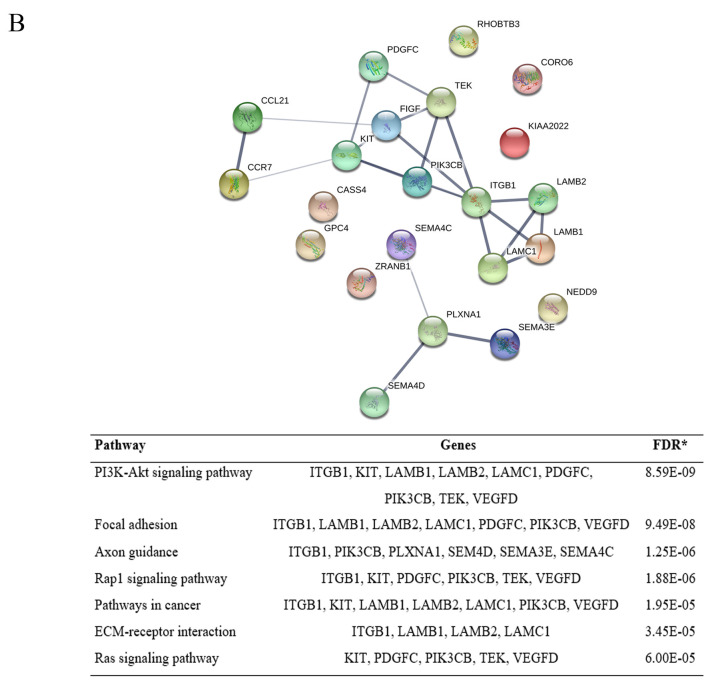
For the down-regulated proteins, the network revealed 41 proteins of cell adhesion were enriched in 40 nodes with 25 edges, while 39 proteins of cell migration were enriched in 39 nodes with 76 edges. KEGG pathway enrichment analysis of the down-regulated proteins affected by gigantol treatment involving cell adhesion and cell migration are shown in Figure 1. For the up-regulated proteins, 30 proteins of cell adhesion were enriched in 30 nodes with 21 edges and 22 proteins of cell migration were enriched in 22 nodes with 22 edges. KEGG pathway enrichment analysis of the down-regulated proteins affected by gigantol treatment involving cell adhesion and cell migration is shown in Figure 2.
Figure 1. Protein–Protein interaction network and KEGG pathway enrichment analysis of the down-regulated proteins affected by gigantol treatment. (A) The down-regulated proteins affected by gigantol treatment involving cell adhesion. (B) The down-regulated proteins affected by gigantol treatment involving cell migration.
Figure 2. Protein–Protein interaction network and KEGG pathway enrichment analysis of the up-regulated proteins affected by gigantol treatment. (A) The up-regulated proteins affected by gigantol treatment involving cell adhesion. (B) The up-regulated proteins affected by gigantol treatment involving cell migration.
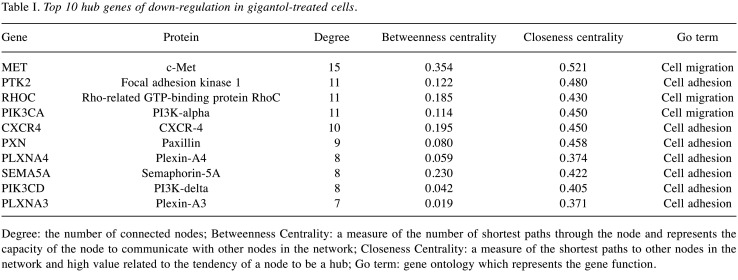
The potential hub genes affected by gigantol. The Cytoscape software 3.7.2. (https://cytoscape.org/) was used to visualize the protein interaction relationship network and provide the hub proteins, which are important nodes with many interaction partners. The protein nodes from the PPI networks were analyzed by Cytoscape software 3.7.2. The results showed that the down-regulated hub genes of cell adhesion were PTK2B, PXN, PTK2, ITGB5, CYR61, DLG4, DLG3, CDH10, CDH9, CDH18, DSC3, DSG1, CDH24, PLXNA4, FES, PLXNA3, CTGF, PALLD, GPC6, PTPRF, FAT3 and NOV. The down-regulated hub genes of cell migration were PLXNA4, SEMA6A, SEMA3A, PLXNA3, FMNL2, RHOC, SRGAP2, FMNL1, SEMA3G, MET, PIK3CA, CXCR4, CX3CL1, CCL20, SRGAP3, SEMA6C, SEMA5A, ITGB5, PXN, PLCG2, PIK3CD, RHOV, RHOJ, ANXA1, PIK3C2B, GPC6, GPC1, PLXNB2, CORO1C, CORO1A, PIK3C2A, PDGFD and IL16. To identify the mechanism of gigantol in the suppression of cancer cell migration, the top 10 hub genes were selected and further analyzed by degree, betweenness centrality and closeness centrality (Table I).
Table I. Top 10 hub genes of down-regulation in gigantol-treated cells.
Degree: the number of connected nodes; Betweenness Centrality: a measure of the number of shortest paths through the node and represents the capacity of the node to communicate with other nodes in the network; Closeness Centrality: a measure of the shortest paths to other nodes in the network and high value related to the tendency of a node to be a hub; Go term: gene ontology which represents the gene function.
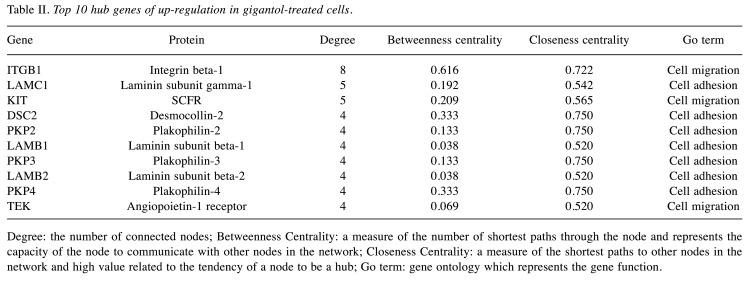
On the other hand, the up-regulated hub genes of cell adhesion were DSC2, PKP2, LAMC1, LAMB1, PKP3, LAMB2, PKP4, ITGB1, MSLN, PARVG, FAT4, MACF1, PCDHGC3, and ARVCF. The up-regulated hub genes of cell migration were CCL21, CCR7, LAMC1, LAMB1, LAMB2, PLXNA1, SEMA3E, ITGB1, TEK, PIK3CB, SEMA4D, FIGF, KIT, PDGFC, and SEMA4C. The top 10 up-regulated hub genes are also summarized in Table II.
Table II. Top 10 hub genes of up-regulation in gigantol-treated cells.
Degree: the number of connected nodes; Betweenness Centrality: a measure of the number of shortest paths through the node and represents the capacity of the node to communicate with other nodes in the network; Closeness Centrality: a measure of the shortest paths to other nodes in the network and high value related to the tendency of a node to be a hub; Go term: gene ontology which represents the gene function.
c-Met was identified as a key protein target of gigantol. We retrieved the top 10 hub genes in gigantol-treated cells and computed the properties of the network, such as its degree, betweenness centrality, and closeness centrality to select the dominant protein. Proteins with a large degree were identified as hub proteins (15). It was suggested that hub proteins tend to be the central or dominant protein of the network (16). The betweenness centrality is a measure of the number of shortest paths through the node and represents the capacity of the node to communicate with other nodes in the network (15). A node with higher closeness centrality is recognized as a center node of a network and is related to the tendency of a node to be a hub. According to its large degree as well as closeness centrality value, we identified c-Met as an important protein hub in response to gigantol treatment.
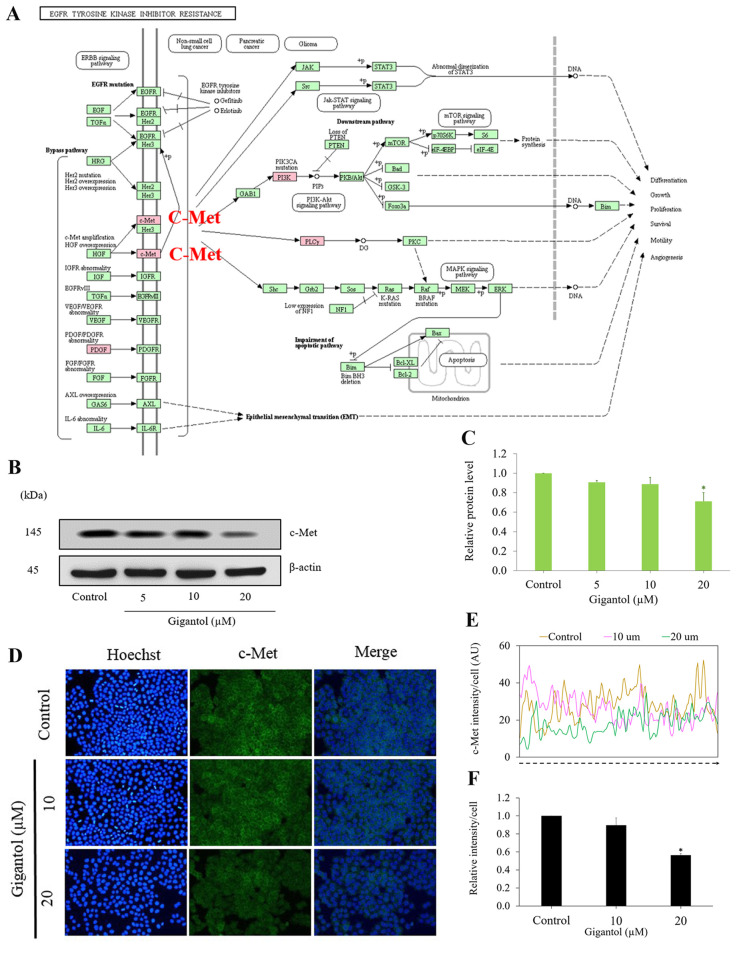
To further reveal the affected pathway of gigantol treatment, we utilized the KEGG mapper (https://www.genome.jp/kegg/mapper.html) to construct the signaling pathway affected by gigantol. The top 20 proteins of down-regulated and up-regulated proteins, after gigantol treatment, involved with cell adhesion and the cell migration were mapped in the KEGG mapper to construct the 2nd pathways. Consistent with the above analysis, the KEGG mapper also suggested that c-Met was a key player in the mechanism of action of gigantol in the suppression of cell migration through EGFR tyrosine kinase inhibitor resistance (Figure 3A).
Figure 3. The c-Met-dependent pathway is a target of gigantol. (A) Effect of gigantol on the down-regulated proteins related to cell migration through EGFR tyrosine kinase inhibitor resistance obtained from the KEGG Pathway Database (https://www.genome.jp/kegg/mapper.html). The red box represents the down-regulated proteins, including c-Met: Met; PI3K: PIK3CA, PIK3CD; PLC gamma: PLCG1, and PLCG2. (B) The c-Met protein expression was determined by western blotting. H460 cells were treated with 0-20 μM gigantol for 24 h. β-actin was used to confirm the equal loading of each protein sample. (C) The band intensities of the treatment groups were quantified by densitometry, and the results are presented as a relative protein level. Data represent the mean±SD (n=3; *p<0.05 compared with the untreated control). (D-F) The c-Met protein signal was determined by immunofluorescence. The fluorescence intensity was analyzed by ImageJ software. Values are the mean±S.E.M. (n=3). *p<0.05 compared to untreated cells.
Having shown that c-Met was suggested to be a key player in the gigantol effect, and that the signaling of PI3K and AKT was suggested to be the mechanism of the compound action, we then sought to confirm the effect of gigantol on the protein and activation of such signals by western blot analysis. Cells were treated with non-toxic concentrations of gigantol (0, 5, 10, and 20 μM) for 24 h and the protein and activated proteins related to c-Met/PI3K/AKT were determined. Figure 3B and C shows that c-Met expression was dramatically reduced in response to gigantol treatment. The cellular expression of c-Met was further detected by immunofluorescence. The results indicated that in the untreated control, the cells exerted a strong signal from c-Met. Following treatment with gigantol at 10 and 20 μM, the signals of c-Met staining were found to be depleted (Figure 3D-F).
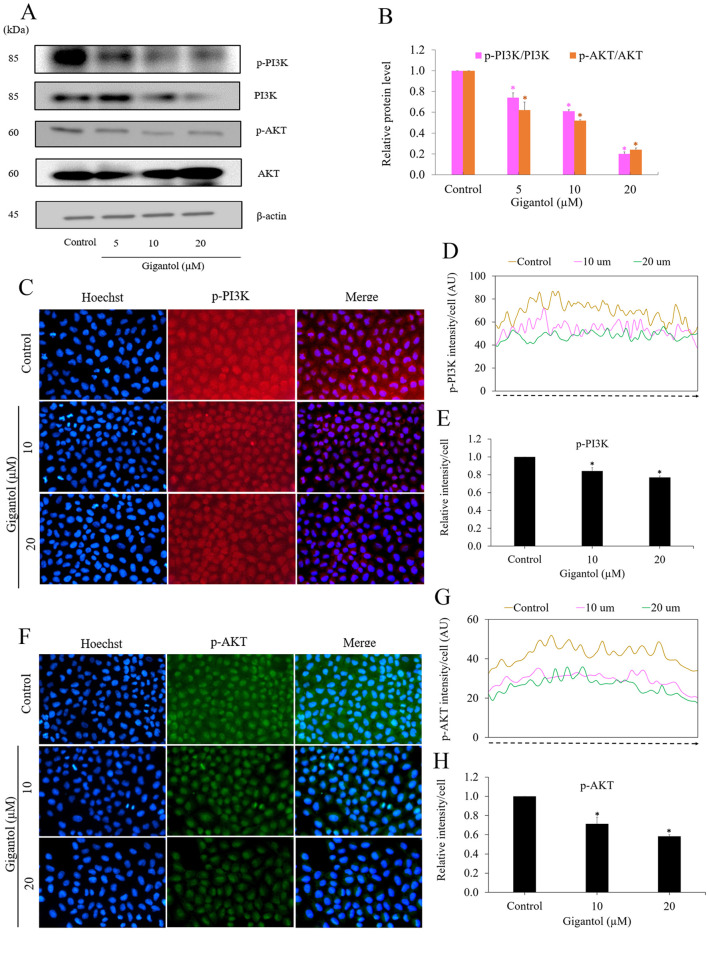
Gigantol suppresses cell metastasis-related behaviors through c-Met-dependent PI3K/AKT suppression. c-Met is known to exert its function via the PI3K/AKT pathway and our analysis of KEGG mapping indicated that c-Met/PI3K/AKT was the mechanism of gigantol action. To further determine the c-Met downstream signaling, we performed western blot protein analysis for the protein and phosphorylated protein determination. The levels of AKT, phosphorylated AKT (p-AKT), PI3K, and phosphorylated PI3K (p-PI3K) were determined after gigantol treatment. The results revealed that the treatment of the cells with gigantol caused a significant decrease in p-PI3K and total PI3K in a dose-dependent manner. Also, gigantol significantly decreased the levels of p-AKT, while it had only a minimal effect on AKT in comparison to those of the untreated control (Figure 4A and B). Immunofluorescence staining with specific antibodies against p-AKT and p-PI3K further confirmed that treatment of the lung cancer cells with gigantol resulted in a suppression of the PI3K/AKT signals (Figure 4C-H).
Figure 4. Gigantol suppresses c-Met-dependent PI3K/AKT signaling. (A) Representative blots of PI3K, p-PI3K, AKT, and p-AKT proteins detected by western blot analysis. H460 cells were treated with 0-20 μM gigantol for 24 h. β-actin antibody was used to confirm the equal loading of each protein sample. (B) The band intensities of the treatment groups were quantified by densitometry, and the results are presented as a relative protein level. Data represent the mean±SD (n=3; *p<0.05 compared with the untreated control). (C-H) The cells were stained with p-PI3K (red fluorescence), p-AKT (green fluorescence) and Hoechst 33342 (blue fluorescence). The expression of p-PI3K and p-AKT were examined using immunofluorescence. The fluorescence intensity was analyzed by ImageJ software. Values are the mean±S.E.M. (n=3). *p<0.05 compared with untreated cells.
Discussion
Gigantol a natural product derived compound from Dendrobium draconis exerts several anti-cancer activities, such as anti-proliferation, apoptosis induction, and anti-migration (17-19). As the key molecular target of gigantol in the suppression of cancer cell movement is largely unknown, the present study utilized proteomics and bioinformatics for identifying the major protein target and its downstream regulatory mechanism.
Loss of cell-cell and cell-ECM adhesion and increasing cell migration play key roles in facilitating cancer cell metastasis. Also, differentially expressed proteins were identified in the gigantol-treated cells compared to control cells. We demonstrated that gigantol treatment affected many proteins related to cell adhesion (GO: 0007155) as well as cell migration (GO: 0016477).
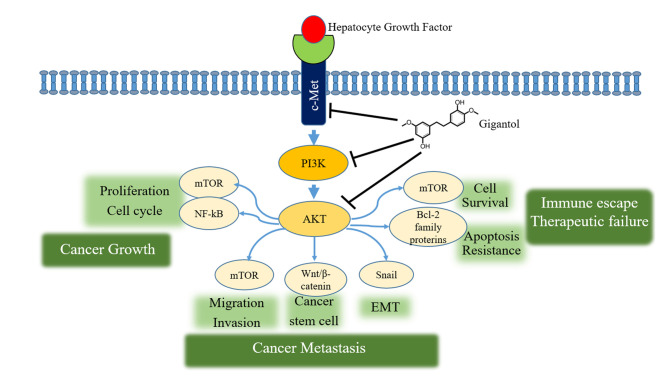
By analyzing protein–protein interactions and identifying the key protein target of the treatment, we could identify the top proteins with the highest degree of protein interaction. MET, RHOC, and PIK3CA were the genes with the highest degree of hub gene in response to gigantol, implying that gigantol may exert its anti-migratory action through a c-Met-dependent pathway. Mesenchymal-epithelial transition factor (c-Met) is a tyrosine kinase receptor at the cell membrane that can be activated by the hepatocyte growth factor (HGF), resulting in initiation of cellular signals controlling survival, and is associated with progression and metastasis of many cancers (20). The activation of c-Met has been demonstrated to promote tumor generation, growth, and metastasis (21). In a lung cancer study, the stimulation of c-Met resulted in an increase in cancer cell movement (22). The inhibition of c-Met has been proposed as a potential therapy for lung cancer (23,24). The stimulation of c-Met causes activation of several cell signals, including the phosphatidylinositol-3-kinase (PI3K)/AKT. For mechanism, HGF interacts with c-Met and induces autophosphorylation of the receptor, allowing an open docking site for PI3K. PI3K then binds to the adaptor protein and initiates the activation of the phosphatidylinositol intermediates. PI3K facilitates the conversion of PIP2 to PIP3. PIP3 then form a complex with PDK1 and AKT and mediates the phosphorylation at Thr-308 and at Ser-473 on the AKT molecule by the function of PDK1 (25). AKT can activate downstream survival protein targets, such as, Bcl-2 and NF-kB (26) and the phosphorylates mTOR (27), resulting in the proliferation, migration, and survival of cancer cells.
In the module analysis, MET was found to be enriched in several pathways, including the ‘regulation of chemotaxis’, ‘focal adhesion’, ‘leukocyte transendothelial migration’, and ‘EGFR tyrosine kinase inhibitor resistance’. The ‘regulation of chemotaxis’ plays a role in regulating cell migration, which contributes to cancer metastasis (28). The pathways associated with RHOC and PIK3CA were found to be ‘inositol phosphate metabolism’, ‘phosphatidylinositol signaling system’ and ‘VEGF signaling pathway’. ‘Inositol phosphate metabolism’ plays a role in the regulation of cell proliferation, migration, and phosphatidylinositol-3-kinase (PI3K)/AKT signaling. A previous study reported that ‘inositol phosphate metabolism’ was closely related with lung cancer (29). ‘Phosphoinositide signaling’ is closely related to PI3K and phosphatase and tensin homolog deleted on chromosome 10 (PTEN) through regulation of cell survival, proliferation, cell growth and cancer progression (30).
Conclusion
Herein, we utilized proteomic and bioinformatic tools to identify the up-stream molecular mechanism by which gigantol suppresses c-Met/PI3K/AKT in lung cancer cells (Figure 5). The down-regulation of c-Met and suppression of its down-stream signaling were confirmed by protein level determination and immunofluorescence. The results from this study may be beneficial for the further development of gigantol for anti-cancer approaches as well as provide detailed information regarding the proteins affected by the compounds.
Figure 5. Schematic diagram summarizing the underlying mechanism of gigantol inhibiting lung cancer metastatic potential through c-Met suppression and inhibition of the PI3K/AKT signaling pathway.
Conflicts of Interest
The Authors declare that they have no competing interests.
Authors’ Contributions
Conceptualization: P.C.; Research design: N.A. and P.C.; Conducted experiments: N.A., N.L., S.R and S.T.; Performed data analysis: P.C., S.R. and N.A.; Contributed to the writing, review, and/or revision of the manuscript of the manuscript: P.C. and N.A.
Acknowledgements
The Authors would like to thank the Office of the Ministry of Higher Education, Science, Research and Innovation and the National Research Council of Thailand (NRCT) for the support grant.
Availability of Data and Material
The datasets used during the present study are available from the corresponding author upon reasonable request.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat Res. 2011;728(1-2):23–34. doi: 10.1016/j.mrrev.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li DM, Feng YM. Signaling mechanism of cell adhesion molecules in breast cancer metastasis: Potential therapeutic targets. Breast Cancer Res Treat. 2011;128(1):7–21. doi: 10.1007/s10549-011-1499-x. [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh AM, Shiri S, Farsinejad S. Metastasis review: From bench to bedside. Tumour Biol. 2014;35(9):8483–8523. doi: 10.1007/s13277-014-2421-z. [DOI] [PubMed] [Google Scholar]
- 5.Imura Y, Nakai T, Yamada S, Outani H, Takenaka S, Hamada K, Araki N, Itoh K, Yoshikawa H, Naka N. Functional and therapeutic relevance of hepatocyte growth factor/c-MET signaling in synovial sarcoma. Cancer Sci. 2016;107(12):1867–1876. doi: 10.1111/cas.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Y, Liu J, Liu T, Yang X. Anti-c-Met antibody bioconjugated with hollow gold nanospheres as a novel nanomaterial for targeted radiation ablation of human cervical cancer cell. Oncol Lett. 2017;14(2):2254–2260. doi: 10.3892/ol.2017.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilotto S, Carbognin L, Karachaliou N, Ma PC, Rosell R, Tortora G, Bria E. Tracking MET de-addiction in lung cancer: A road towards the oncogenic target. Cancer Treat Rev. 2017;60:1–11. doi: 10.1016/j.ctrv.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Wu JC, Wang CT, Hung HC, Wu WJ, Wu DC, Chang MC, Sung PJ, Chou YW, Wen ZH, Tai MH. Heteronemin is a novel c-Met/STAT3 inhibitor against advanced prostate cancer cells. Prostate. 2016;76(16):1469–1483. doi: 10.1002/pros.23230. [DOI] [PubMed] [Google Scholar]
- 9.Go H, Jeon YK, Park HJ, Sung SW, Seo JW, Chung DH. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol. 2010;5(3):305–313. doi: 10.1097/JTO.0b013e3181ce3d1d. [DOI] [PubMed] [Google Scholar]
- 10.De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, Gabriele P, Comoglio PM, Boccaccio C. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103(8):645–661. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 11.Trusolino L, Bertotti A, Comoglio PM. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11(12):834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 12.Losuwannarak N, Roytrakul S, Chanvorachote P. Gigantol targets MYC for ubiquitin-proteasomal degradation and suppresses lung cancer cell growth. Cancer Genomics Proteomics. 2020;17(6):781–793. doi: 10.21873/cgp.20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unahabhokha T, Chanvorachote P, Sritularak B, Kitsongsermthon J, Pongrakhananon V. Gigantol inhibits epithelial to mesenchymal process in human lung cancer cells. Evid Based Complement Alternat Med. 2016;2016:4561674. doi: 10.1155/2016/4561674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhummaphan N, Chanvorachote P. Gigantol suppresses cancer stem cell-like phenotypes in lung cancer cells. Evid Based Complement Alternat Med. 2015;2015:836564. doi: 10.1155/2015/836564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azuaje F, Devaux Y, Wagner DR. Coordinated modular functionality and prognostic potential of a heart failure biomarker-driven interaction network. BMC Syst Biol. 2010;4:60. doi: 10.1186/1752-0509-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charoenrungruang S, Chanvorachote P, Sritularak B, Pongrakhananon V. Gigantol, a bibenzyl from dendrobium draconis, inhibits the migratory behavior of non-small cell lung cancer cells. J Nat Prod. 2014;77(6):1359–1366. doi: 10.1021/np500015v. [DOI] [PubMed] [Google Scholar]
- 18.Klongkumnuankarn P, Busaranon K, Chanvorachote P, Sritularak B, Jongbunprasert V, Likhitwitayawuid K. Cytotoxic and antimigratory activities of phenolic compounds from dendrobium brymerianum. Evid Based Complement Alternat Med. 2015;2015:350410. doi: 10.1155/2015/350410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Huang Y, Huang J, Lin L, Wei G. Gigantol attenuates the proliferation of human liver cancer HepG2 cells through the PI3K/Akt/NF-ĸB signaling pathway. Oncol Rep. 2017;37(2):865–870. doi: 10.3892/or.2016.5299. [DOI] [PubMed] [Google Scholar]
- 20.Boccaccio C, Comoglio PM. Invasive growth: A MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6(8):637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 21.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4(12):915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 22.Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R. Role of the hepatocyte growth factor receptor, c-Met, in oncogenesis and potential for therapeutic inhibition. Cytokine Growth Factor Rev. 2002;13(1):41–59. doi: 10.1016/s1359-6101(01)00029-6. [DOI] [PubMed] [Google Scholar]
- 23.Goździk-Spychalska J, Szyszka-Barth K, Spychalski L, Ramlau K, Wójtowicz J, Batura-Gabryel H, Ramlau R. C-MET inhibitors in the treatment of lung cancer. Curr Treat Options Oncol. 2014;15(4):670–682. doi: 10.1007/s11864-014-0313-5. [DOI] [PubMed] [Google Scholar]
- 24.Sattler M, Reddy MM, Hasina R, Gangadhar T, Salgia R. The role of the c-Met pathway in lung cancer and the potential for targeted therapy. Ther Adv Med Oncol. 2011;3(4):171–184. doi: 10.1177/1758834011408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7(4):261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 26.Mortenson M, Schlieman M, Virudalchalam S, Bold R. Overexpression of BCL-2 results in activation of the AKT/NF-kB Cell survival pathway. Journal of Surgical Research. 2019;114(2):302. doi: 10.1016/j.jss.2003.08.103. [DOI] [Google Scholar]
- 27.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 28.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan J, Yu CY, Wang ZH, Chen HY, Guan J, Chen YX, Fang JY. Genetic variants in the inositol phosphate metabolism pathway and risk of different types of cancer. Sci Rep. 2015;5:8473. doi: 10.1038/srep08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunney TD, Katan M. Phosphoinositide signalling in cancer: Beyond PI3K and PTEN. Nat Rev Cancer. 2010;10(5):342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.