Abstract
Aim: To identify novel diagnostic markers for renal cell carcinoma (RCC), we analyzed miRNAs in serum extracellular vesicles (EVs). Materials and Methods: EVs were purified from serum of healthy controls and patients with localized and advanced RCC using T-cell immunoglobulin domain and mucin domain-containing protein 4 conjugated to magnetic beads. miRNA profiling of EVs was conducted by microarray analysis. miRNA expression was examined by quantitative reverse transcription-polymerase chain reaction. Lastly, proteomic analysis of RCC cells transfected with a miRNA inhibitor was performed to identify its potential targets. Results: Microarray analysis revealed that nine miRNAs were increased by more than 1.5-fold in EVs from patients with RCC. Among them, miRNA-4525 was significantly elevated; miRNA-4525 expression was higher in RCC tissue than in the adjacent normal tissue. Proteomic analysis identified alpha fetoprotein and albumin as its potential targets. Conclusion: These findings suggest the potential of miRNA-4525 in serum EVs as a novel biomarker for advanced RCC.
Keywords: Renal cell carcinoma, extracellular vesicle, microarray analysis, mirna-4525, proteomic analysis
Recent advances in cancer research have provided new options to treat advanced and metastatic renal cell carcinoma (RCC). Indeed, targeted therapies and immune check point inhibitors have improved survival outcomes in RCC patients, but the mortality still remains high (1-3). Currently, reliable biomarkers for diagnosis and prognosis of RCC are unavailable.
MicroRNA (miRNA) that post-transcriptionally regulates protein expression plays important roles in physiology and pathology, including cancer development and progression, and have potential as diagnostic markers and therapeutic targets (4-6). miRNAs released from cells are present in the blood bound to proteins such as high-density lipoprotein cholesterol and are also packaged in extracellular vesicles (EVs) (7-9). There have been many reports describing miRNAs in whole serum or plasma that are potentially useful for diagnosis and prognosis of cancer (5,8). EVs isolated from serum or plasma have also been subjected to global gene-expression profiling and candidate miRNA markers have been identified (7). However, EVs isolated from serum or plasma by conventional methods, such as serial centrifugation followed by ultracentrifugation, gel-filtration, immunoaffinity-based and polymer-based precipitation methods, are heavily contaminated with serum or plasma proteins, making it difficult to eliminate protein-bound miRNAs and investigate EVs-associated miRNAs in isolation. Nakai et al. reported a novel EV isolation method using T-cell immunoglobulin domain and mucin domain-containing protein 4/human Fc chimera recombinant protein (TIM4), by which highly-purified EVs are then amenable to several downstream analyses, including proteomic analysis (10). We recently reported that protein markers on urine EVs isolated by TIM4-conjugated magnetic beads are useful for differential diagnosis of urinary tract infection and asymptomatic bacteriuria (11). To date, as far as we are aware, no report has been published on miRNA profiling of EVs isolated using TIM4.
In the present study, we performed miRNA microarray analysis of EVs isolated from pooled serum of controls and patients with RCC using TIM4-conjugated beads in an attempt to profile EV-associated miRNAs with minimal contamination of serum protein-bound miRNAs. After confirmation of the miRNA expression levels in serum EV from each patient, as well as in RCC and its adjacent normal tissue, by quantitative reverse transcription-polymerase chain reaction (RT-PCR), we identified miRNA-4525 in serum EVs as a potential diagnostic marker for RCC progression. Lastly, we performed a cell-culture study to identify potential targets of miRNA-4525.
Materials and Methods
Cell lines. Human RCC cell line Caki-1 (JCRB0801) obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) was maintained in Dulbecco’s modified Eagle medium (Wako Pure Chemical Industries, Osaka Japan) containing penicillin, streptomycin and 10% fetal bovine serum (Equitech-Bio, Kerrville, TX, USA). Human OS-RC-2 cell line (RCB0735) was obtained from RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan, and maintained in RPMI 1640 containing the aforementioned antibiotics and serum. All experiments were carried out using cells with fewer than 10 passages after receiving them.
Patients. This study was approved by the Medical Review Board of Gifu University, Graduate School of Medicine (No. 29-6, 29-410) and Tokyo Metropolitan Institute of Gerontology (No. 6177). Written-informed consent was obtained from all participants. Eighteen patients with RCC that were diagnosed between July 2015 and May 2019 were enrolled in this study. Serum of seven healthy doners collected in February 2018 was used as control. Serum was sequentially centrifuged at 1,800 × g for 10 min to remove cells and at 16,500 × g for 20 min to remove debris and other contaminants. Samples were then stored at –80˚C until use. Tissue samples were obtained from surgical specimens and stored in RNAlater Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, USA) at –20˚C until use.
EV isolation. The EV isolation method using TIM4 was described elsewhere (11). Briefly, biotinylated mouse TIM4/human Fc region of human IgG1 chimera recombinant protein (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) was conjugated to streptavidin-magnetic beads (Thermo Fisher Scientific). Serum was thawed and centrifuged at 1,800 × g for 10 min before use. One milliliter of serum was incubated for 1 h at 4˚C with TIM4-conjugated beads in the presence of 2 mM CaCl2. The captured EVs were washed with phosphate-buffered saline (PBS) with 2 mM CaCl2 twice and released in 50 μl of PBS and tris-buffered saline containing 2 mM ethylenediaminetetra-acetic acid for proteomic analysis and RT-PCR, respectively.
Microarray analysis. RNA was extracted from EVs isolated from 1 ml of pooled serum from four individuals in each group (control, localized RCC and advanced RCC) using 3D-Gene RNA extraction reagent (Toray Industries, Tokyo, Japan) according to the manufacturer’s instructions. Extracted total RNA was checked by Bioanalyzer (Agilent, Santa Clara, CA, USA) and labeled with 3D-Gene miRNA labeling kit (Toray Industries). The half volume of labeled RNAs were hybridized onto 3D-Gene Human miRNA Oligo chip (Toray Industries). The annotation and oligonucleotide sequences of the probes were conformed to the miRBase miRNA data base (http://microrna.sanger.ac.uk/sequences/). After stringent washes, fluorescent signals were scanned with a 3D-Gene Scanner (Toray Industries) and analyzed using 3D-Gene Extraction software (Toray Industries). The raw data of each spot was normalized by subtraction of a background signal determined as the mean intensity of all blank spot signals. Spots with signal intensities greater than two standard deviations of the background signal intensity were considered to be valid. The relative expression level of a given miRNA was calculated by comparing the signal intensities of the valid spots throughout the microarray experiments. The normalized data were globally normalized per array, such that the median of the signal intensity was adjusted to 25.
Quantitative RT-PCR. Total RNA including miRNA was extracted from EVs using miRNeasy Serum/Plasma kit (Qiagen GmbH, Hilden, Germany). From RCC cells and tissues, miRNAs were isolated using miReasy Mini kit (Qiagen GmbH). Two microliters of total extracted RNA eluted in 14 μl of water were reverse transcribed using TaqMan Advanced miRNA cDNA Synthesis kit for miRNA-4516, 4525 and 4534. TaqMan MicroRNA Reverse Transcription kit was also used for miRNA-4525 and miRNA-4534 to achieve better sensitivity (Thermo Fisher Scientific). For this reaction, 5 μl of total RNA (28 μl) extracted from each sample were used. PCR was performed using TaqMan Advanced miRNA assay (Assay; 478303_mir for miRNA-4516, 480690_mir for miRNA-4525, 480832_mir for miRNA-4534 and 478293_mir for cel-miR-39-3p as a spike-in control, respectively) and TaqMan MicroRNA assay (Assay; 462485_mat for miRNA-4525, 462024_mat for miRNA-4534 and 000200 for cel-miR-39-3p as a spike-in control, respectively). Five out of 50 μl total cDNA and 0.67 out of 15 μl total cDNA were used in each quantitative PCR reaction with TaqMan Advanced miRNA assay and TaqMan MicroRNA assay, respectively. All reactions were performed in duplicate.
Western blot analysis. EVs were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by western blot analysis as described in previous reports (11,12). Rabbit anti-CD9 and mouse anti-CD63 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA) and Santa Cruz Biotechnology (Dallas, TX, USA), respectively. The immunoreactive proteins were detected using horseradish peroxidase-conjugated anti-rabbit or -mouse antibody (Cell Signaling Technology) and ImmunoStar (FUJIFILM Wako Pure Chemical).
Transfection of miRNA inhibitor. An inhibitor for human miRNA-4525 and two negative controls were purchased from Bioneer Corporation (Daejeonm, Republic of Korea). Fifty picomoles of each inhibitor were transfected into Caki-1 and OS-RC-2 cells seeded in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Forty-eight hours after transfection, cells were collected after washing twice with ice-cold PBS and stored at –80˚C for further proteomic analysis.
Proteomic analysis. Digestion of proteins with trypsin was performed prior to proteomic analysis. Cells were dissolved in 0.1% Rapigest SF (Waters, Milford, MA, USA). Protein concentration was determined by Micro BCA protein assay kit (Thermo Fisher Scientific). Ten micrograms of proteins were reduced with 10 mM dithiothreitol in 50 mM triethylammonium bicarbonate (TEAB) at room temperature for 30 min, and alkylated with 55 mM iodoacetamide in 50 mM TEAB at room temperature for 30 min in the dark. The samples were then digested with trypsin (Promega, Madison, WI, USA) in 50 mM TEAB at 37˚C for 16 h. The cleaved peptides were desalted with a GL-Tip SDB column (GL Sciences, Tokyo, Japan), and evaporated in a SpeedVac vacuum concentrator (Thermo Fisher Scientific). The peptides were reconstituted in 0.1% formic acid (FA) and subjected to nano liquid chromatography-tandem mass spectrometry (LC-MS/MS). Protein identification was performed by nanoLC-MS-MS using UltiMate 3000 RSLCnano system (Thermo Fisher Scientific) coupled to a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer with nano electrospray ionization source (Thermo Fisher Scientific) as previously described (13). Peptides were separated using a 90-min gradient of water containing 0.1% FA (mobile phase A) and acetonitrile containing 0.1% FA (mobile phase B) at a flow rate of 300 nl/min. The elution gradient was set as follows: 0-3 min, 2-2% B; 3-93 min, 2-40% B; 93-95 min, 40-95% B; 95-105 min, 95-95% B; 105-107 min, 95-2% B; 107-120 min, 2-2% B. MS was operated in data-dependent acquisition mode. All data were analyzed for protein identification by Proteome Discoverer 2.4 software (Thermo Fisher Scientific).
Statistical analysis. Statistical analysis was performed using Graph Pad Prism 7 version 7.03 (Graph Pad Software, San Diego, CA, USA). Comparison among three groups was made using Kruskal–Wallis test with Dunn’s post hoc for multiple comparisons. Wilcoxon matched-pairs signed-rank test was used to compare paired tumor–normal samples. Differences were considered significant when p<0.05.
Results
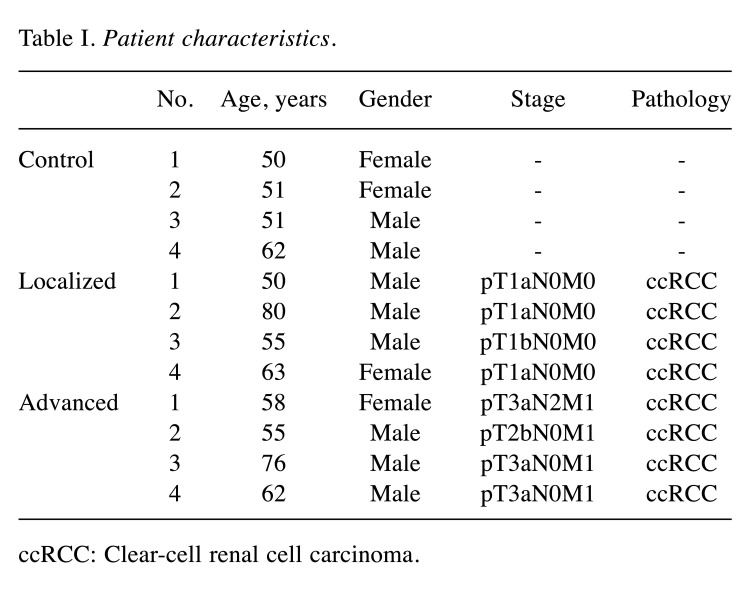
Identification of miRNAs elevated in serum EVs of patients with advanced RCC. In order to identify miRNAs increased in serum EVs of patients with advanced RCC, microarray analysis was performed. Patients’ characteristics are shown in Table I. Prior to microarray analysis, isolated EVs were subjected to western blot analysis to confirm successful EV isolation using antibodies against EV markers CD9 and CD63 (Figure 1).
Table I. Patient characteristics.
ccRCC: Clear-cell renal cell carcinoma.
Figure 1. Confirmation of isolation of extracellular vesicles (EVs) from serum. EVs were isolated by using T-cell immunoglobulin domain and mucin domain-containing protein 4/human Fc chimera recombinant protein-conjugated magnetic beads from pooled serum of healthy controls, and patients with localized or advanced renal cell carcinoma (n=4 in each group).
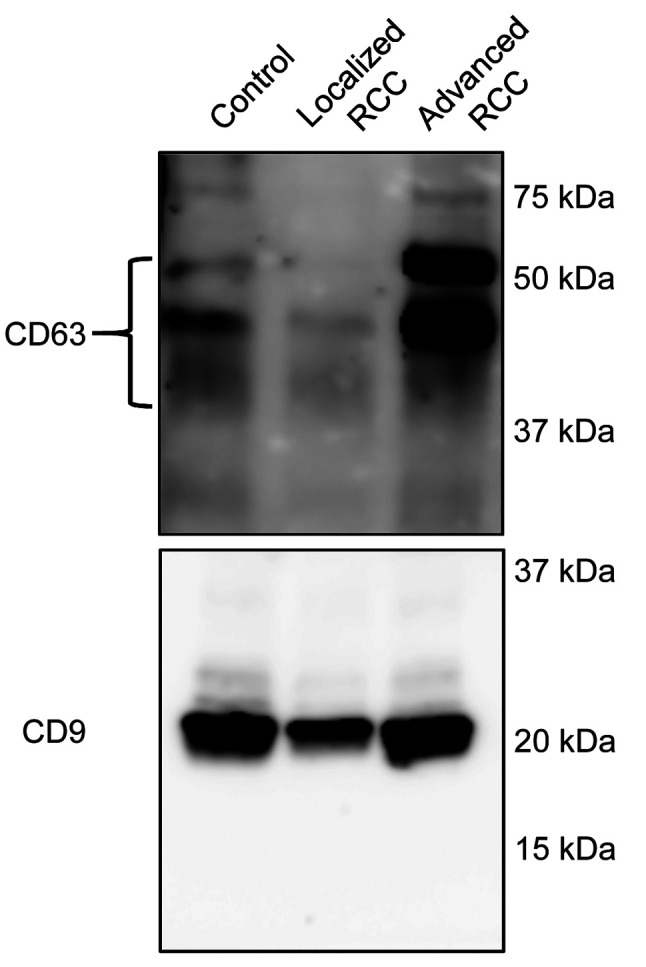
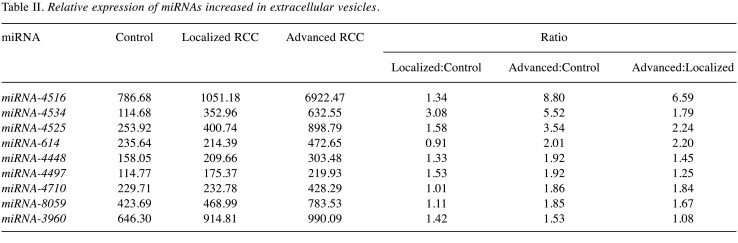
A total of 1,797 miRNAs were detected (data not shown) and 254 miRNAs had a signal intensity of more than 100. Among them, nine were found to be up-regulated by more than 1.5-fold in EVs isolated from patients with either localized or advanced RCC compared with controls (Table II). Three out of nine miRNAs, namely miRNA-4516, 4534 and 4525, with higher expression change and basal level were selected for further analysis by quantitative RT-PCR.
Table II. Relative expression of miRNAs increased in extracellular vesicles.
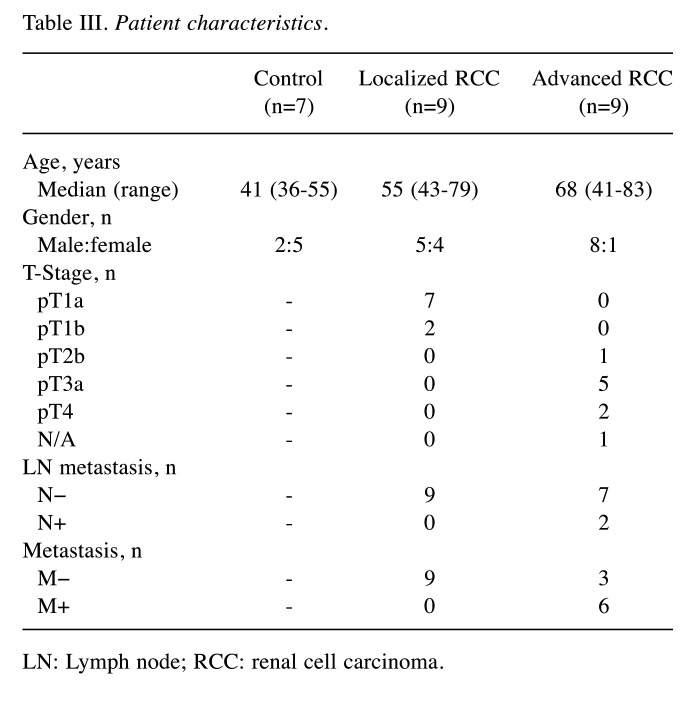
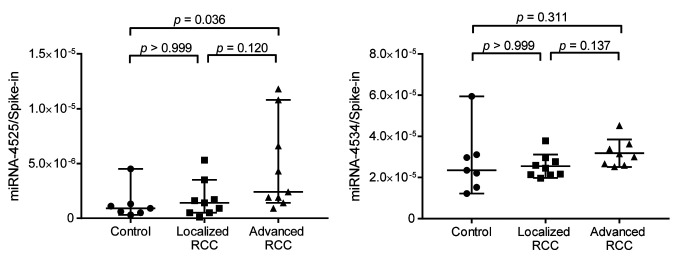
Confirmation of increased miRNA-4525 in serum EVs of patients with advanced RCC by quantitative RT-PCR. Among the three candidate miRNAs, miRNA-4534 and miRNA-4525 were barely detectable by quantitative RT-PCR using TaqMan Advanced miRNA cDNA Synthesis kit and TaqMan Advanced miRNA assay but were successfully detected using TaqMan MicroRNA Reverse Transcription kit and TaqMan MicroRNA Assay. We therefore analyzed the ratio of these two miRNAs to that of cel-miR-39-3p spike-in control for another set of patients including controls, and patients with localized and advanced RCC. Patient characteristics are listed in Table III. All serum samples were taken before surgery or biopsy prior to initial treatment. All patients underwent partial nephrectomy or radical nephrectomy, except one who underwent only biopsy, and all tumors were pathologically diagnosed as clear-cell RCC according to the 2004 World Health Organization renal tumor classification (14). Localized RCC was defined as pathologically proven RCC of T-stage 1a-2b without any metastasis, whereas advanced RCC was defined as pathologically proven RCC of T-stage T3a-4 with/without metastasis. As shown in Figure 2, there was a significant difference in the level of miRNA-4525 in serum EVs among the three groups (p=0.0259). The miRNA-4525 level in serum EVs was significantly increased in the group with advanced RCC compared with the control group (p=0.0357) but not when compared with those with localized RCC. For miRNA-4534, however, no statistical difference was found among the three groups. These results suggest that miRNA-4525 in serum EVs might be a marker for advanced RCC.
Table III. Patient characteristics.
LN: Lymph node; RCC: renal cell carcinoma
Figure 2. Expression levels of miRNA-4525 and miRNA-4534 in extracellular vesicles (EVs) isolated from patients with renal cell carcinoma (RCC). miRNA-4525 expression in serum EVs significantly was increased in patients with advanced RCC compared with healthy controls, whereas miRNA-4534 expression was not. Cel-miR-39-3p was used as spike-in control for standardization. Bars and lines represent the median and 95% confidence interval, respectively.
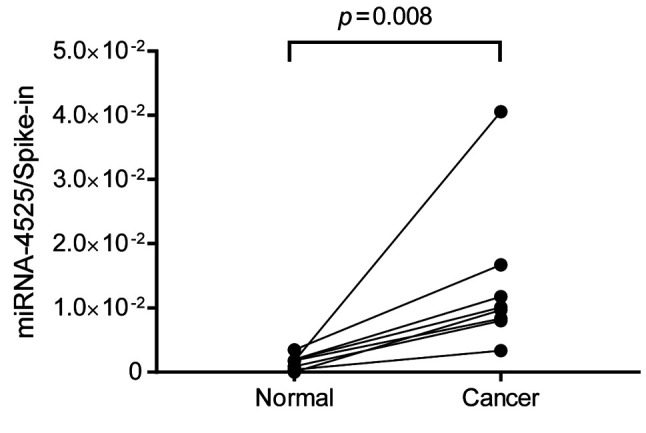
Increased miRNA-4525 in RCC tissue. Since EVs are secreted from various cells, an increased level of miRNA-4525 in serum EVs may not be a consequence of its increased expression in RCC but may arise from other causes, such as tumor-associated inflammation. In order to define the mechanism involved, RCC tissue and its adjacent normal tissue in surgically resected specimen were subjected to quantitative RT-PCR for miRNA-4525. The results showed that miRNA-4525 expression was significantly increased in RCC tissue compared to adjacent normal tissue (Figure 3).
Figure 3. miRNA-4525 expression in renal cell carcinoma (RCC) tissue. miRNA-4525 expression was significantly increased in RCC tissue compared to that in adjacent normal tissue. Cel-miR-39-3p was used as spike-in control for standardization.
Proteomic analysis of RCC cells transfected with miRNA-4525 inhibitor. In an attempt to define the role of miRNA-4525 in RCC, proteomic analysis of two RCC cell lines transfected with miRNA-4525 inhibitor were performed. Prior to the analysis, miRNA-4525 expression in Caki-1 and OS-RC-2 RCC cell lines was evaluated by quantitative RT-PCR (1.77×109 and 1.64×109 copies in 0.054 μg of total RNA, respectively). Successful transfection of miRNA inhibitor was confirmed by the reduced expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in Caki-1 and OS-RC-2 cells by parallel transfection with a positive control miRNA inhibitor known to inhibit GAPDH (data not shown). Proteomic analysis showed that 32 and 45 proteins were up-regulated by more than 1.5-fold in Caki-1 and OS-RC-2 cells, respectively, transfected with miRNA-4525 inhibitor compared with those transfected with two negative control miRNA inhibitors (data not shown). Among proteins increased by transfection of miRNA-4525 inhibitor in RCC cell lines, albumin and alpha fetoprotein (AFP) were found to be common to both.
Discussion
Although metastatic and advanced RCC remains largely incurable, no reliable diagnostic or prognostic markers have been reported. EVs have enormous potential as biomarkers for diseases, including cancer, an effective EV isolation method suitable for comprehensive analysis, such as microarray and proteomic analyses, has not been established. In our previous report, we isolated EVs from urine by TIM4-conjugated beads for differential diagnosis of urinary tract infection and asymptomatic bacteriuria (11). In the present study, in order to obtain miRNA profiling data of serum EVs with minimal contamination of serum protein-bound miRNAs, we performed microarray analysis of EVs isolated by TIM4-conjugated beads, resulting in detection of more than 1,700 miRNAs.
Among the top three miRNAs found to be increased in patients with advanced RCC by microarray analysis, miRNA-4534 and miRNA-4525 were only successfully detectable by quantitative RT-PCR. This discrepancy may, in part, be due to specificity and sensitivity of the primers used for RT-PCR. We also showed that miRNA-4525 expression was increased in RCC tissue compared to normal tissue in surgically resected specimen, which suggests that increased miRNA-4525 in serum EVs in patients with advanced RCC may be caused by its increased expression in tumor. In any case, our pilot study demonstrated the potential of miRNA-4525 in serum EVs as a biomarker for advanced RCC.
There have been two reports describing the potential of miRNA-4525 as a biomarker one of increased miRNA-4525 in plasma EVs for high-risk pancreatic ductal adenocarcinoma (15), and the other showing reduced circulating miRNA-4525 for acute myocardial infarction (16). However, the functional role of miRNA-4525 remains unknown. In order to understand the role of miRNA-4525 in RCC, we performed a cell-culture study. We transfected miRNA-4525 inhibitor rather than its mimic because it was inherently highly expressed in RCC cells. Proteomic analysis identified AFP and albumin as proteins up-regulated in both Caki-1 and OS-RC-2 cells by inhibition of miRNA-4525. AFP is produced by the embryonic yolk sac and the fetal liver, and its level in blood has been widely used as a tumor marker to diagnose and monitor hepatocellular carcinoma, germ-cell tumors and yolk-sac tumor, however, its increase in patients with RCC has only been shown in case reports (17,18). The roles of AFP have been well documented, mainly as a tumor promoter mediating inhibition of apoptosis and promoting cell proliferation and metastasis (19). Albumin is synthesized in liver and its serum level is commonly used for evaluating nutritional status. Hypoalbuminemia, an abnormally low level of albumin in blood, reflects malnutrition, hypercatabolism and systemic inflammation in patients with cancer, therefore many studies have demonstrated the relationship of reduced serum albumin with shorter overall survival or cancer-specific survival (20). In patients with RCC, Chen et al. reported the prognostic role of serum albumin level before treatment by meta-analysis (21). On the other hand, it has been well described that miRNAs in EVs may promote disease pathogenesis via intercellular crosstalk [reviewed in (7)]. Thus, one possible explanation for the role of miRNA-4525 in EVs would be as follows. EVs with increased miRNA-4525 content are secreted from RCC tumor itself into blood and delivered to hepatic cells, where miRNA-4525 may function as an inhibitor of ALB synthesis, resulting in hypoalbuminemia, which is consistent with a previous report showing that hypoalbuminemia correlates with poor prognosis in RCC (21). However, further research is needed to corroborate this hypothesis.
One limitation of this study is the relatively small number of patients with RCC for miRNA-4525 analysis as a diagnostic marker and the lack of prognostic data and of direct evidence for the role of miRNA-4525.
In conclusion, we demonstrated that a TIM4-based EV isolation method is suitable for microarray analysis of miRNAs contained in EVs and that increased miRNA-4525 in EVs has potential as a diagnostic marker for advanced RCC.
Conflicts of Interest
The Authors declare that they have no conflicts of interest in regard to this study.
Authors’ Contributions
Conception and design: KM, and MI. Performed the experiments: KM, KK, and YMM. Data analysis and interpretation: KM, KK, YF, MT, DK, KN, TKa, TT, TKo, YMi, and MI. Manuscript writing: YMM, KM, KK, and MI. Approval of manuscript: All Authors.
Acknowledgements
The Authors are grateful to Ayako Nagasawa for her invaluable contributions to data acquisition. This work was partially supported by JSPS KAKENHI Grant Number 16K20130 (KM).
References
- 1.Rini BI, Escudier B, Tomczak P, Kaprin A, Szczylik C, Hutson TE, Michaelson MD, Gorbunova VA, Gore ME, Rusakov IG, Negrier S, Ou YC, Castellano D, Lim HY, Uemura H, Tarazi J, Cella D, Chen C, Rosbrook B, Kim S, Motzer RJ. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): A randomised phase 3 trial. Lancet. 2011;378(9807):1931–1939. doi: 10.1016/S0140-6736(11)61613-9. [DOI] [PubMed] [Google Scholar]
- 2.Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P, Porta C, George S, Powles T, Donskov F, Neiman V, Kollmannsberger CK, Salman P, Gurney H, Hawkins R, Ravaud A, Grimm MO, Bracarda S, Barrios CH, Tomita Y, Castellano D, Rini BI, Chen AC, Mekan S, McHenry MB, Wind-Rotolo M, Doan J, Sharma P, Hammers HJ, Escudier B, CheckMate 214 Investigators. Nivolumab plus Ipilimumab versus Sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, Pouliot F, Alekseev B, Soulières D, Melichar B, Vynnychenko I, Kryzhanivska A, Bondarenko I, Azevedo SJ, Borchiellini D, Szczylik C, Markus M, McDermott RS, Bedke J, Tartas S, Chang YH, Tamada S, Shou Q, Perini RF, Chen M, Atkins MB, Powles T, KEYNOTE-426 Investigators Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019;380(12):1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 4.Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7(10):e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong S, Golpon H, Zardo P, Borlak J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. 2020;Res:S1931. doi: 10.1016/j.trsl.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Adil MS, Khulood D, Somanath PR. Targeting Akt-associated microRNAs for cancer therapeutics. Biochem. 2020;Pharmacol:114384. doi: 10.1016/j.bcp.2020.114384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Liu J. Potential of cancer cell-derived exosomes in clinical application: a review of recent research advances. Clin Ther. 2014;36(6):863–872. doi: 10.1016/j.clinthera.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez-Avalos JA, Fernández-Macías JC, González-Palomo AK. Circulating exosomal MicroRNAs: New non-invasive biomarkers of non-communicable disease. Mol Biol Rep. 2021;48(1):961–967. doi: 10.1007/s11033-020-06050-w. [DOI] [PubMed] [Google Scholar]
- 9.Lakshmi S, Hughes TA, Priya S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur J Cancer. 2021;144:252–268. doi: 10.1016/j.ejca.2020.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, Naruse K, Sadamura Y, Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizutani K, Kawakami K, Horie K, Fujita Y, Kameyama K, Kato T, Nakane K, Tsuchiya T, Yasuda M, Masunaga K, Kasuya Y, Masuda Y, Deguchi T, Koie T, Ito M. Urinary exosome as a potential biomarker for urinary tract infection. Cell Microbiol. 2019;21(7):e13020. doi: 10.1111/cmi.13020. [DOI] [PubMed] [Google Scholar]
- 12.Mizutani K, Terazawa R, Kameyama K, Kato T, Horie K, Tsuchiya T, Seike K, Ehara H, Fujita Y, Kawakami K, Ito M, Deguchi T. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34(7):3419–3423. [PubMed] [Google Scholar]
- 13.Enomoto A, Fukasawa T, Tsumoto H, Karube M, Nakagawa K, Yoshizaki A, Sato S, Miura Y, Miyagawa K. Prevention of calpain-dependent degradation of STK38 by MEKK2-mediated phosphorylation. Sci Rep. 2019;9(1):16010. doi: 10.1038/s41598-019-52435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eble JN, Sauter G, Epstein JI, Sesterhenn IA. IARC Press. Lyon. 2004. Pathology and Genetics of Tumors of the Urinary System and Male Genital Organs. [Google Scholar]
- 15.Kawamura S, Iinuma H, Wada K, Takahashi K, Minezaki S, Kainuma M, Shibuya M, Miura F, Sano K. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J Hepatobiliary Pancreat Sci. 2019;26(2):63–72. doi: 10.1002/jhbp.601. [DOI] [PubMed] [Google Scholar]
- 16.Zhong Z, Wu H, Zhong W, Zhang Q, Yu Z. Expression profiling and bioinformatics analysis of circulating microRNAs in patients with acute myocardial infarction. J Clin Lab Anal. 2020;34(3):e23099. doi: 10.1002/jcla.23099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall C, Enzerra M, Rahnemai-Azar AA, Ramaiya NH. Serum tumor markers and testicular germ cell tumors: A primer for radiologists. Abdom Radiol (NY) 2019;44(3):1083–1090. doi: 10.1007/s00261-018-1846-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HZ, Wang SY. AFP-producing Xp11 translocation renal cell carcinoma: Case report and review of the literature. Curr Probl Cancer. 2020;45(3):100689. doi: 10.1016/j.currproblcancer.2020.100689. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y, Zhu M, Li M. Effects of alpha-fetoprotein on the occurrence and progression of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2020;146(10):2439–2446. doi: 10.1007/s00432-020-03331-6. [DOI] [PubMed] [Google Scholar]
- 20.Gupta D, Lis CG. Pretreatment serum albumin as a predictor of cancer survival: A systematic review of the epidemiological literature. Nutr J. 2010;9:69. doi: 10.1186/1475-2891-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Shao Y, Wang K, Cao W, Xiong Y, Wu R, Luo S, Xu X, He X. Prognostic role of pretreatment serum albumin in renal cell carcinoma: A systematic review and meta-analysis. Onco Targets Ther. 2016;9:6701–6710. doi: 10.2147/OTT.S108469. [DOI] [PMC free article] [PubMed] [Google Scholar]