Abstract
Background: We previously identified a panel of five miRNAs associated with prostate cancer recurrence and metastasis. Expression of one of the down-regulated miRNAs, miR-139-5p, was significantly associated with a lower incidence of biochemical recurrence and metastasis. Transcriptome profiling of miR-139-expressing prostate cancer cells revealed up-regulation of genes involved in interferon (IFN) stimulation. The association between miR-139 and IFN-β was further explored in this study. Materials and Methods: We examined miR-139 transfected PC3, Du145 and LNCaP cells and the associated IFN response by transcriptome sequencing, immunoblotting and pulldown assays. Results: Treatment of prostate cancer cells by miR-139 resulted in the up-regulation of IFN-related genes. Specifically, miR-139 induced expression of the IFN-β protein. The ability of miR-139 to induce IFN-β was due to its binding to RIG-1 and the induction of IFN-related genes was found to be dependent on RIG-1 expression. Conclusion: miR-139 acts as an immune agonist of RIG-1 to enhance IFN-β response in prostate cancer cells.
Keywords: microRNA, interferon, RIG-1, prostate cancer, NGS, transcriptome
Prostate cancer (PCa) is the second most commonly diagnosed cancer and the sixth leading cause of cancer death among men worldwide (1). PCa is a heterogenous and multifactorial disease ranging from asymptomatic to a rapidly fatal systemic malignancy. Although progress has been made in the early detection and treatment of prostate adenocarcinoma, the metastatic lesions from this tumor are incurable. Despite the fact that the 5-year survival rate for most men diagnosed with PCa is greater than 90%, patients with metastatic disease have a survival rate of just 29.0% (2). Thus, it is crucial to distinguish between clinically indolent PCas and aggressive tumors with potentially lethal outcomes. Identifying biomarkers that can differentiate between indolent or aggressive forms of PCa is key in preventing unnecessary treatment effects which may compromise patients’ quality of life. miRNAs have been analyzed as diagnostic and prognostic biomarkers of PCa (3). miRNAs are small noncoding RNAs that function to regulate gene expression through modulation of cellular differentiation, proliferation and apoptosis (4-6).
Through whole miRNome sequencing we identified a panel of five miRNAs from 33 miRNAs that were associated with PCa recurrence and metastasis following prostatectomy (7). Three of these miRNAs were up-regulated while two were down-regulated in metastatic PCa patient tumors compared to tumors from patients with no metastasis and no recurrence. We previously determined the mechanism of action of some of these miRNAs (8-10). Investigation of one of the down-regulated miRNAs, miR-139-5p, revealed that its tumor suppressive function in PCa was dependent on inhibition of two different targets, AXL and IGF1R, leading to growth inhibition and cell cycle arrest (10). Further investigation identified that miR-139 also induced incomplete autophagy leading to apoptosis in PCa cells, in part through inhibition of the mTOR signaling pathway (19).
Transcriptome sequencing of miR-139 treated PCa cells revealed that many genes involved in Interferon (IFN) type I and III signaling were up-regulated, compared to treatment with the negative control. The IFNs are a family of natural glycoproteins consisting of IFN-α, -β, -γ, and -λ. The antiviral activity of IFNs originally led to their discovery (11), but later studies revealed that they also control cell growth and differentiation (12), inhibit expression of oncogenes (13), and activate T lymphocytes, natural killer cells and macrophages (14,15). The growth inhibitory effect of IFNs on PCa has been previously observed. IFN-β treatment of PCa cells has been shown to inhibit their proliferation (16,17). Furthermore, the down-regulation of the expression of IFN-inducible proteins in prostate epithelial cells is associated with the development and progression of prostate cancer (18). In this study we examined the connection between miR-139 and IFN-β in PCa cells.
Materials and Methods
Cell lines. Prostate cancer cell-lines were purchased from American Type Culture Collection (ATCC). PC3 prostate cancer cells were maintained in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) (Wisent Bioproducts, St-Bruno, QC, Canada) while LNCaP prostate cancer cells were maintained in Roswell Park Memorial Institute (RPMI) media (Wisent), and DU145 prostate cancer cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent), all supplemented with 10% FBS at 37˚ C in 5% CO2. HEK-293T cells (ATCC) were maintained in DMEM media supplemented with 10% FBS.
Transient transfection. PC3, Du145 or LNCaP cells were seeded in six-well plates for 24 h. Immediately before transfection, regular media was replaced with media containing 5% FBS. Transfections were performed using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s recommendations. Transfections were performed with 25 pmol/well of either miR-139-5p mimic (MSY0000250; Qiagen, Hilden, Germany), miR-301a mimic (MSY0000688, Qiagen), miR-652 mimic (MSY0003322, Qiagen), or negative control mimic (AllStars Negative Control siRNA; Qiagen). For blocking experiments, 50 pmol/well of anti-hsa-miR-139-5p miScript miRNA inhibitor (Qiagen) were added per well. Recombinant human IFN-β (rhIFN) (LSBio) was added to cells at 200 IU/ml. Transfection of RIG-1 plasmid (DDX58, RC217615, Origene) was performed with 2 ug DNA/well, using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s recommendations.
Western blotting. Cell extracts were prepared by lysis in RIPA buffer in the presence of proteinase and phosphatase inhibitors. The cell lysates were collected by centrifugation at 20,000 g for 15 min at 4˚C and protein content in the supernatant was measured using the Bradford protein assay (BioRad, USA). Approximately 5-10 μg of protein were separated on either 8 or 15% SDS-polyacrylamide gels and electroblotted on to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk in TBS-T [20 mmol/l Tris-HCl (pH 8.0), 137 mmol/l NaCl and 0.1% Tween 20] or 5% BSA for one hour at RT, and incubated with the primary antibodies overnight at 4˚C. After washing with TBS-T, blots were probed with HRP-conjugated secondary antibodies for 1 hour at RT. The blots were then washed and treated with ECL reagent (1M Tris pH 8.5, 198 μM para-coumaric acid, 1.25 mM luminol, 0.009% H2O2) and the protein bands were visualized using Kodak XOMAT- AR film for autoradiography. The following monoclonal and polyclonal antibodies were used: anti-LC3B, anti-IFN-β, anti-IL28/29 (Cell Signaling Technology, Danvers, MA, USA); anti-RIG-1, anti-AXL, anti-p-STAT1, anti-STAT1, anti-DNMT1 (Santa Cruz Biotechnology, Dallas, TX, USA); anti-β-tubulin (Sigma-Aldrich), and HRP conjugated goat anti-mouse and goat anti-rabbit (Promega, Madison, WI, USA). Image J version 1.51 was used for densitometry of immunoblots.
Pulldown experiment. HEK-293T cells were transfected with RIG-1 (DDX58) plasmid or empty vector for 3 days, followed by lysis with RIPA buffer. 35 μg of lysate was incubated with 200 pmol of biotin-labeled mimic [miR-139-bio or neg-mimic-bio, Applied Biological Materials (ABM)] for 1 hour at 4˚C. Streptavidin-agarose beads (5 ul) were added for an additional 1 hour. Beads were then washed 4x with wash buffer containing 0.5% NP40, followed by removal of immunoprecipitated proteins by boiling in 2x sample buffer for 5 min prior to loading of 8% SDS-PAGE gels. Blots were probed with anti-DDK to detect DDK-tagged RIG-1, and β tubulin for loading controls. WB were blocked, probed and washed as described in the WB protocol.
Ion AmpliSeq transcriptome sequencing. The transcriptome sequencing of miR-139 over-expressing and empty vector transfected PC3, Du145 and LNCaP cell lines was performed on the Ion S5XL Next Generation Sequencing system with the Ion AmpliSeq Transcriptome Human Gene Expression Kit (Thermo Fisher Scientific). The cDNA was synthesized from 10 ng of total RNA by SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). The cDNA libraries were constructed by Ion AmpliSeq Library Kit Plus and the sequencing template preparation was done by using Ion Chef with Ion 540 Chef Kits. Sequencing was performed for 500 flows on an Ion S5XL Sequencer with the Ion 540 chip. The differential gene expression analysis between samples and pathway enrichment analysis of differentially expressed genes was performed by Transcriptome Analysis Console (TAC) Software 4.0.2 (Thermo Fisher Scientific).
Statistical analysis. All statistical analysis was conducted using Excel (version 2007). Student’s t-test was used to obtain p-values for comparison of two groups. All graphs show the mean±SD of triplicate experiments.
Results
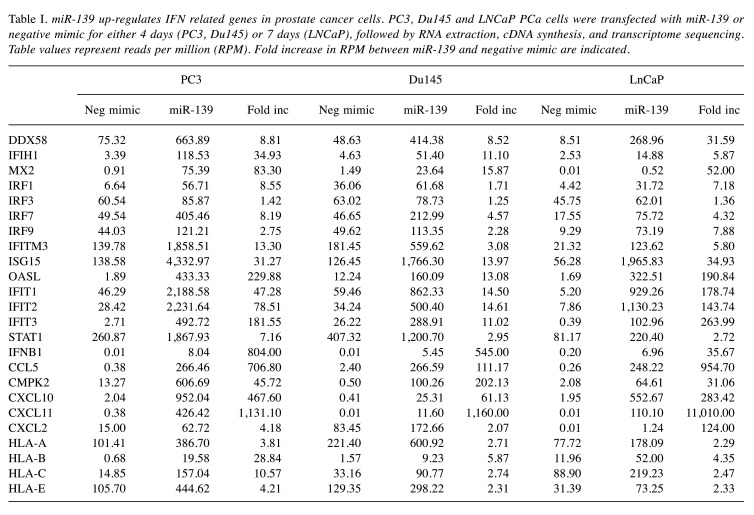
miR-139 up-regulates IFN-related genes in PCa cells. We had previously performed whole transcriptome analysis of miR-139 treated PC3, Du145 and LNCaP PCa cells, which showed that expression of certain genes, such as mTOR, were down-regulated, and likely contributed to growth inhibition and autophagy (19). Upon further assessment, we noticed a number of IFN-related genes up-regulated by miR-139 in all 3 cell lines (Table I). We focused our investigation on IFN-β rather than IFN-α or γ, since only IFN-β was significantly increased upon miR-139 treatment (Figure 1). miR-139 treated PC3 cells had the highest up-regulation of IFN-β expression (>800 fold over control) followed by Du145 (>500), and LNCaP (>35) (Table I). In order to confirm the RNA expression data, PCa cells were treated for various time points with miR-139, followed by Western blotting on both conditioned media and cell lysates. Results revealed that both PC3 and Du145 cell lines produce intracellular IFN-β as early as 24 h post miR-139 treatment, after which intracellular IFN-β expression declined around day 3 (Figure 1). Secreted IFN-β in conditioned medium accumulated from day 1 to day 3. In LNCaP cells IFN-β production took longer, starting at 3 day and continued to increase from days 6-12.
Table I. miR-139 up-regulates IFN related genes in prostate cancer cells. PC3, Du145 and LNCaP PCa cells were transfected with miR-139 or negative mimic for either 4 days (PC3, Du145) or 7 days (LNCaP), followed by RNA extraction, cDNA synthesis, and transcriptome sequencing. Table values represent reads per million (RPM). Fold increase in RPM between miR-139 and negative mimic are indicated.
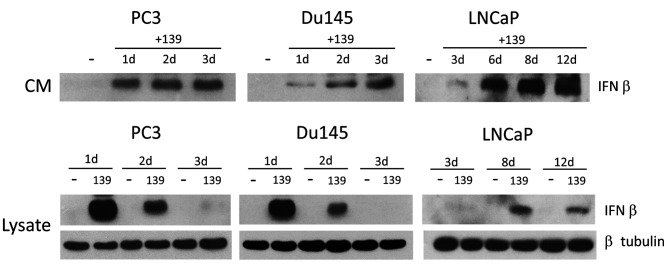
Figure 1. miR-139 induces IFN-β expression in prostate cancer cells. PC3, Du145 and LNCaP cells were transfected with either miR-139 or negative mimic for days 1, 2 and 3 (PC3, Du145) or days 3, 6, 8 and 12 (LNCaP). Conditioned media (CM) was collected and analyzed by Western blot probed with anti-IFN-β. Lysates were also blotted for IFN-β, and β tubulin for loading controls.
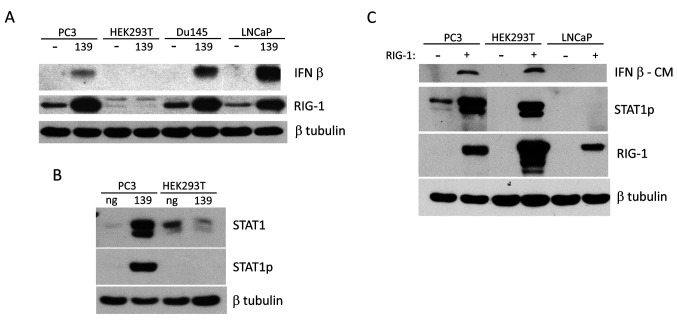
miR-139-mediated IFN-β response depends on RIG-1 expression. IFN-β up-regulation by miR-139 occurred in all 3 PCa cell lines, PC3, Du145 and LNCaP (Figure 2A). However, miR-139 was unable to induce IFN-β in HEK293T cells. HEK293T cells do not express RIG-1 (Figure 2A). RIG-1 triggers the downstream transcription of IFN-β and its associated genes (20). Thus, it is no surprise that HEK293T cells are not capable of up-regulating STAT1p/STAT1 (Figure 2B). Transfection of RIG-1 into HEK293T cells rescued their ability to produce IFN-β and the related downstream STAT1p expression (Figure 2C). Transfection of RIG-1 alone in PC3 cell line is also sufficient to up-regulate IFN-β and associated downstream signaling proteins. These results reveal that miR-139 treatment of PCa cells is very similar to RIG-1 transfection, suggesting that miR-139 may be upstream of RIG-1 in the up-regulation of IFN-β and associated pathways.
Figure 2. The miR-139-mediated IFN-β response is dependent on RIG-1 expression. (A) PC3, HEK293T, Du145 and LNCaP cells were treated for 3 days (PC3, HEK293T, Du145) or 7 days (LNCaP) with either miR-139 or negative mimic, followed by WB with antibodies against IFN-β or RIG-1, with β tubulin as loading control. (B) PC3 or HEK293T cells were treated with either miR-139 or negative mimic for 3 days, followed by WB with anti-STAT1p, stripped and reprobed with anti-STAT1 total and β tubulin for loading control. (C) PC3, Du145, HEK293T and LNCaP cells were transfected with RIG-1 plasmid for 3 days. WB of conditioned medium (CM) for IFN-β is shown. WB of lysates with the indicated antibodies are also shown. WB were stripped and reprobed for loading controls with anti-β tubulin. Two alternatively spliced isoforms of STAT1 exist, the antibody used detects both of them.
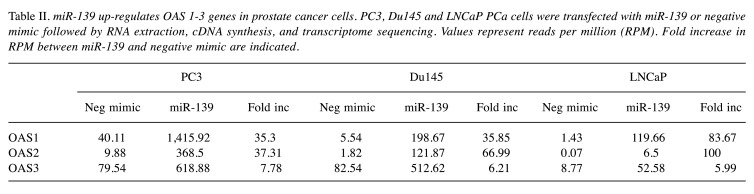
miR-139 mimic is a RIG-1 agonist. Several miRNAs have been recognized as ligands of RIG-1 or TLR3, thereby contributing to immune enhancement. miR-136 was determined to be an immune agonist of RIG-1, causing IL-6 and IFN-β accumulation (21). The observation that transfection of either miR-139, IFN-β or RIG-1 into PCa cells induces similar downstream pathways suggests a close association between them. Based on these results, we speculated that RIG-1 may be a potential receptor for miR-139. We performed pulldown experiments to test this hypothesis. HEK293T cells were transfected with RIG-1 expression vectors for 3 days, followed by incubation of lysates with biotin-labeled-miR-139 or biotin-negative-mimic control, followed by streptavidin-coated Sepharose beads. Immunoprecipitated proteins were separated on SDS-PAGE followed by blotting with anti-DDK, corresponding to the RIG-1 associated tag. Results revealed that biotin-labeled miR-139 pulled down a strong band corresponding to the RIG-1 protein (Figure 3). In contrast, the biotin-negative-mimic was associated with a very faint band, suggesting possible background RIG-1 binding to Sepharose beads. Addition of unlabeled miR-139 mimic together with biotin-labeled-miR-139 mimic, resulted in significant inhibition of RIG-1 pulldown compared to biotin-miR-139 alone, which was likely due to specific removal of RIG-1 by the unlabeled miR-139, demonstrating the specificity of miR-139 for RIG-1. These results strongly suggest that RIG-1 acts as a receptor for miR-139 recognition.
Figure 3. miR-139 binds to RIG-1. HEK293T cells were treated with RIG-1 plasmid for 3 days. 35 μg of lysate were incubated with either biotin-labeled miR-139 mimic, biotin-labeled negative mimic, or biotinlabeled miR-139 mimic together with unlabeled miR-139 mimic followed by streptavidin Sepharose beads. Lysates were also incubated with streptavidin-sepharose beads alone. After washing, protein bound to beads were removed by boiling, then analyzed by WB for RIG-1 expression using an anti-DDK antibody. For input, 0.5 μg of lysate is also shown on WB, with β tubulin as loading control.
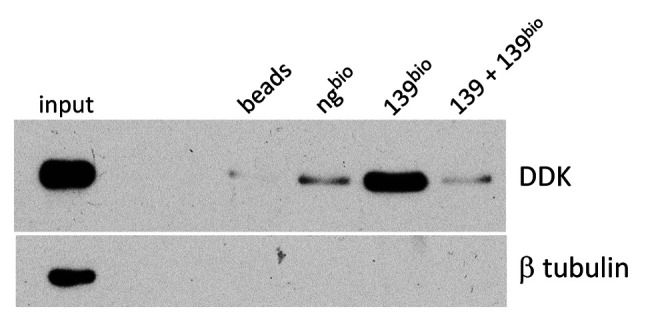
miR-139 treatment activates RIG-1 with IRF-3 phosphorylation. Our data suggested that miR-139 is upstream of RIG-1 signaling. It also suggests that miR-139 activates RIG-1 to induce downstream signaling leading to IFN-β production. To verify this, the activation of RIG-1 was examined indirectly through examination of phospho-IRF-3 levels in miR-139-treated PC3 cells. After RIG-1 activation by recognition of dsRNA, downstream signaling converge at the recruitment of IRF-3, which when phosphorylated, becomes activated, translocates to the nucleus, and initiates transcription of IFN-1 genes (22-27). PC3 cells treated with miR-139 for 1 to 2 days followed by western blotting revealed that IRF-3 is phosphorylated at day 1 of treatment (Figure 4). This data indicates that miR-139 treatment results in activation of IRF-3, which is likely responsible for the observed IFN-β production. Both the phosphorylation of IRF-3 and IFN-β expression peak at day 1 of miR-139 treatment in PC3 cells.
Figure 4. miR-139 activates RIG-1 through IRF3-phosphorylation. PC3 was transfected with either miR-139 or negative mimic. WB using antibodies for IFN-β, RIG-1 or phospho-IRF3 expression are shown. β tubulin expression is shown for loading controls.
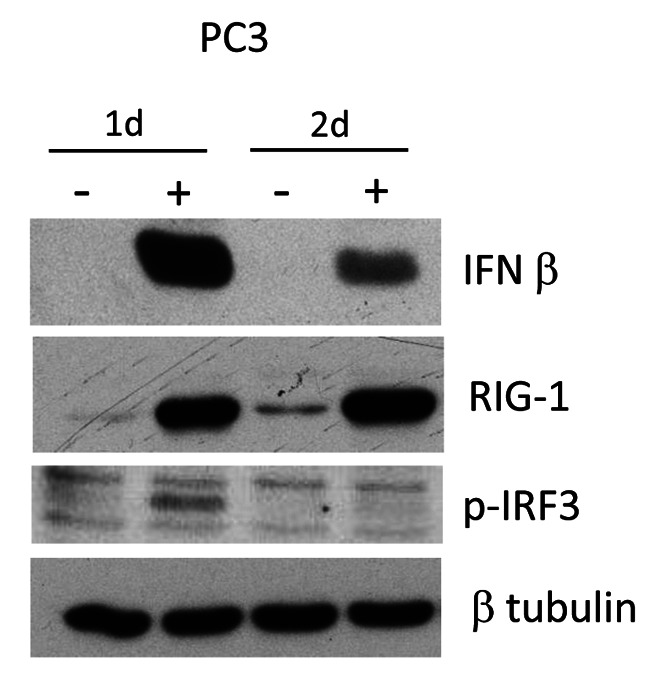
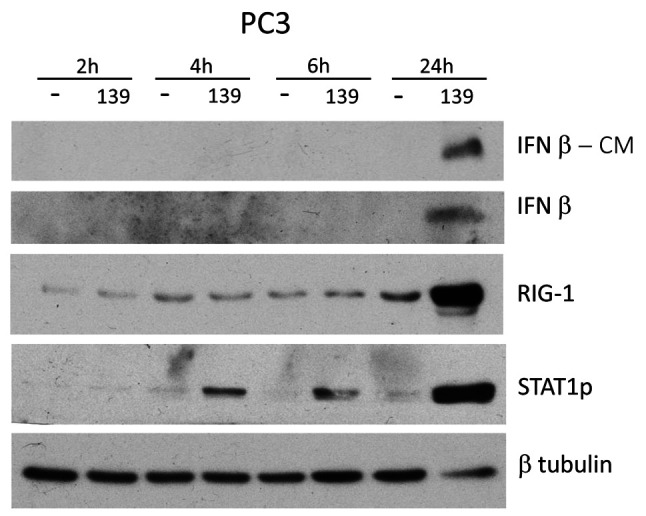
miR-139 initially activates STAT1p, followed by RIG-1 and IFN-β. In order to identify the initial signaling pathways that are activated by miR-139, a time course of miR-139 treatment of PC3 cells was evaluated by western blotting. Results revealed that STAT1p is the earliest signaling protein expressed after miR-139 mimic exposure at 4 h post treatment (Figure 5). The rest of the proteins, RIG-1 and IFN-β are induced at 24 h post treatment.
Figure 5. miR-139 initially activates STAT1p followed by RIG-1 and IFN-β. PC3 cells were transfected with either miR-139 or negative mimic for 2, 4, 6, and 24 h, followed by WB of conditioned medium (CM) and lysates for the indicated proteins. β-tubulin is represented as loading control for the various antibodies.
miR-139 and IFN-β induce identical downstream signaling responses. The production of IFN-β protein by miR-139 together with up-regulation of IFN-responsive genes, led to examination of IFN-β mediated downstream signaling protein production by miR-139 treatment. PCa cells were stimulated for 3 days with miR-139 mimic or negative mimic, or IFN-β, followed by Western blotting of downstream signaling pathways. The RNA helicase, RIG-1, is activated by dsRNA molecules, triggering the downstream transcription of IFN-β, which positively feeds back to up-regulate RIG-1 itself (20). Binding of IFN-β to its receptor leads to the phosphorylation of STAT1 and STAT2, which subsequently result in the transcription of IFN-stimulated genes (28,29). miR-139 treatment results in significant up-regulation of RIG-1, STAT1p and STAT1, similar to IFN-β stimulation (Figure 6A). Interestingly, IFN-β treatment of both PC3 and Du145 cells results in down-regulation of AXL, which we previously reported to be a down-regulated target of miR-139 (10). This suggests that down-regulation of AXL may be a consequence of stimulation of IFN-β related proteins. We previously reported that another consequence of miR-139 treatment was induction of autophagy, with up-regulation of LC3B-II (19). IFN-β treatment of PC3 cells for 3 days resulted in up-regulation of LC3B-II, although with a slightly weaker response than miR-139 treatment (Figure 6B). Interestingly, autophagy has been reported to be an antiviral mechanism utilized by IFN-β (30,31). Treatment of PC3 cells with anti-miR-139 antagomir and miR-139 mimic abrogated IFN-β protein expression observed upon miR-139 mimic alone (Figure 6C). However, RIG-1 expression was not affected by anti-miR-139 antagomir treatment, suggesting that miR-139 may be acting after IFN-β-mediated feedback up-regulation of RIG-1 expression.
Figure 6. Both miR-139 and IFN-β induce identical downstream signaling responses. (A) PC3 or Du145 cells were transfected for 3 days with either miR-139 mimic, negative mimic, IFN-β or no stimulation (Ø). WBs were performed using the specified antibodies, followed by stripping and reprobing with β-tubulin for loading control. Two alternatively spliced isoforms of STAT1 exist, the antibody used detects both of them. (B) PC3 cells were transfected for 3 days with either miR-139 mimic, negative mimic or IFN-β followed by WB with anti-LC3B or β-tubulin. (C) PC3 cells were transfected for 3 days with either miR-139 mimic, negative mimic, or miR-139 mimic together with anti-miR-139 antagomir. WB for IFN-β and RIG-1 are shown, with β-tubulin as loading control.
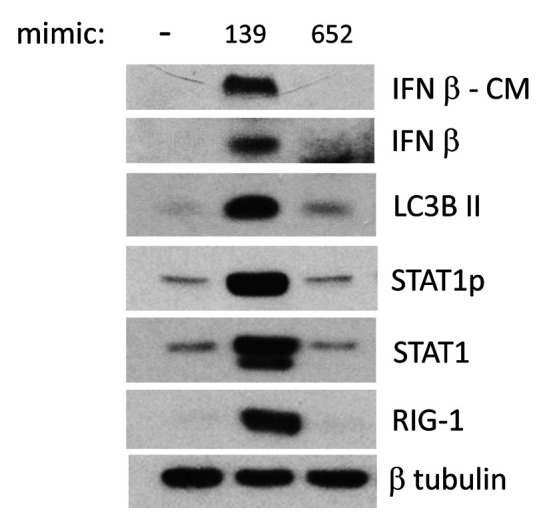
IFN-β response is specifically associated with miR-139 treatment. The induction of IFN-β and associated signaling pathways was compared upon PC3 treatment with miR-139, negative mimic or the nonspecific miR-652 mimic by western blotting. Only miR-139 was able to induce IFN-β production and secretion, as well as downstream up-regulation of IFN-β pathway components, i.e. RIG-1, STAT1p, and STAT1 (Figure 7). These results confirm the specificity of miR-139 to up-regulate IFN-β and downstream pathways.
Figure 7. IFN-β response is specifically associated with miR-139 treatment. PC3 cells were treated for 3 days with either miR139, miR-652 or negative mimic. Conditioned medium (CM) was analyzed by WB for IFN-β, and lysates were examined by WB using antibodies for the specified proteins, followed by β-tubulin for loading control. Two alternatively spliced isoforms of STAT1 exist, the antibody used detects both of them.
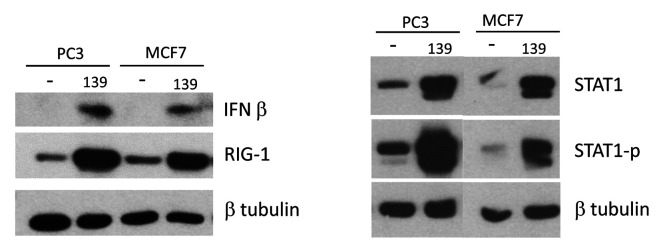
Induction of IFN-β by miR-139 is not limited to PCa cells. The ability of miR-139 to up-regulate the IFN-β pathway in PCa cells prompted us to evaluate whether this phenomenon is exclusive to PCa cells or whether it is a common pathway in other cancer cells. A breast cancer cell line was treated with miR-139, followed by examination of the IFN-β signaling pathway by western blotting. Results revealed that miR-139 was indeed able to up-regulate RIG-1, STAT1p, STAT1 and IFN-β in MCF-7 (Figure 8). Thus, induction of IFN-β pathways by miR-139 is not limited to PCa cells, but also occurs in breast cancer cells.
Figure 8. miR-139-mediated IFN-β response is not limited to PCa cells. PC3 or MCF-7 cells were treated for the indicated time points with either miR-139 or negative mimic, followed by WB for the indicated proteins. β tubulin shows equivalent protein loading. Two alternatively spliced isoforms of STAT1 exist, the antibody used detects both of them.
Discussion
In this study, we demonstrate that miR-139 treatment is associated with the production of IFN-β and associated downstream signaling pathways and genes. Transcriptome sequencing from PCa cells treated with miR-139 revealed the up-regulation of a significant number of IFN-stimulated genes. The anti-tumor effects of type I IFNs are well documented. IFN-β inhibits cell growth and down-regulates oncogene expression in breast cancer cells, while it enhances the antiproliferative activity of the antiestrogen tamoxifen and of the progestin medroxyprogesterone acetate (32-34). Down-regulated expression of proteins induced by IFNs in human prostate epithelial cells has been reported to be associated with development and progression of PCa (35,36). IFN-β1a showed significant anti-proliferative activity in androgen-resistant cell lines (37). Recombinant human IFN-β (rHuIFN-β) reduced the motility and invasiveness of poorly differentiated PCa cells and prevented the acquisition of a neuroendocrine phenotype (38). Highly metastatic human PCa cells engineered to constitutively produce IFN-β, upon injection in nude mice were unable to develop tumors and metastases, or inhibited angiogenesis and progression of orthotopic tumors (39,40).
Previously, we had shown that miR-139 inhibited PCa cell growth through down-regulation of targets, IGF-1 and AXL (10). Subsequently, we identified that PCa cells are forced to undergo incomplete autophagy followed by apoptosis in response to miR-139 treatment (19). The discoveries identified in this study, consolidate our previous results into a mechanism initiated by miR-139-mediated up-regulation of the innate immune response through production of IFN-β. Previous studies have shown that the presence of AXL was associated with prevention of viral-induced activation of type I interferon signaling-genes (41). The inhibition of AXL through IFN-β treatment of PCa cells in our study further confirmed that down-regulation of AXL is an IFN-β-mediated response. miR-139 treatment may be inhibiting AXL production both directly through targeting the 3’UTR of AXL, and indirectly by inducing IFN-β.
We show that miR-139 treatment of PCa cells induced IFN-β secretion. Type I IFN has also been shown to induce autophagy in different cancer cells (31,42-44). In agreement with this, we showed that IFN-β treatment also induces autophagy, with up-regulation of LC3B-II by western blot (Figure 2B). Oligoadenylate synthase (OAS) genes act as pathogen recognition receptors that sense dsRNAs and activate the synthesis of 5’-phosphorylated 2’-5’ linked oligoadenylates from ATP (54). The 2’-5’ linked oligoadenylates act as second messengers which bind monomeric RNase L, activating its dimer formation (45). Active RNase L cleaves cellular and viral RNAs, and the RNA degradation directly and indirectly activates several cellular signaling pathways including autophagy (46-48). Supporting this mechanism, our transcriptome sequencing data revealed up-regulation by miR-139 of OASs1-3 in PCa cells (Table II). Mechanistically, autophagosomes have been shown to deliver virus-derived pathogen-associated molecular patterns (PAMPs) to pattern recognition receptors (PRRs) such as RIG-1, which initiates the IFN responses of antiviral immunity (49,50). In our study, the autophagosomes may be delivering miR-139 to RIG-1 to initiate IFN-β responses.
Table II. miR-139 up-regulates OAS 1-3 genes in prostate cancer cells. PC3, Du145 and LNCaP PCa cells were transfected with miR-139 or negative mimic followed by RNA extraction, cDNA synthesis, and transcriptome sequencing. Values represent reads per million (RPM). Fold increase in RPM between miR-139 and negative mimic are indicated.
Our data show that IFN-β treatment modified the same downstream proteins as miR-139 treatment in PCa cells. Both miR-139 and IFN-β up-regulated RIG-1, STAT1p/STAT1, LC3B-II, and down-regulated AXL protein expression. Transfection of RIG-1 plasmid alone in PCa cells also induced IFN-β and downstream signaling. miR-139 treatment was unable to induce IFN-β expression or its downstream effector, STAT1, in HEK293T cells since they lack RIG-1. RIG-1 transfection in HEK293T cells rescued the IFN-β signaling pathway, suggesting that miR-139 requires RIG-1 expression to induce IFN-β production. We showed that RIG-1 can be pulled down with miR-139 (Figure 3). Other studies have shown that endogenous miRNAs can function as ligands of PPRs such as RIG-1, leading to serial signaling activation. miR-145 was shown to induce immune responses through RIG-1 recognition (51). Similarly, miR-136 was also identified as an immune agonist of RIG-1 leading to IFN-β accumulation (21). Based on our data, miR-139 appears to be an agonist of RIG-1 as well. The effect observed with miR-139 is unlikely to be nonspecific or a consequence of the introduction of dsRNA in the cell, since transfection of other mimics, such as miR-652 (Figure 7), in PCa cells did not up-regulate the IFN-β pathway. In addition, the up-regulation of the IFN-β pathway by miR-139 treatment was not confined to PCa cells, since a similar response was also induced by miR-139 in breast cancer cells (Figure 8).
Conclusion
We identified a novel mechanism of action by miR-139 in arresting the growth of PCa cells. miR-139 treatment up-regulated IFN-β, and its downstream signaling pathways, evidenced at both the protein and RNA level. The down-regulation of AXL and up-regulation of autophagy-associated LC3B-II by IFN-β itself suggested that the modulation of both these proteins by miR-139, which we previously published, is a downstream consequence of the miR-139-mediated IFN-β pathway. Furthermore, our results suggest that miR-139 may be functioning as an agonist of RIG-1, by directly binding RIG-1 to initiate an IFN-β response. This response was specific to miR-139, but not limited to PCa cells. Taken together, these results suggest that by binding and activating RIG-1, miR-139 up-regulated IFN-β signaling pathways, resulting in antitumor, growth regulatory pathways, which may ultimately recruit the innate and adaptive immune system in a clinical setting.
Conflicts of Interest
All Authors have no conflicts of interest to declare.
Authors’ Contributions
T.B. performed molecular experiments, analyzed the data and wrote the paper. R.N. designed the study, data analysis and acquisition of funding. Y.A. did the next-generation RNA-seq experiments and analysis. A.S. provided the conceptual and technical guidance, designed the study, analyzed the data and revised it critically for important intellectual content.
Acknowledgements
This study was funded, in part, by the Edmond Odette Foundation, the Ajmera Family Chair in Urologic Oncology, and a generous philanthropic contribution from Archie and Betty McCallum.
References
- 1.Culp MB, Soerjomataram I, Efstathiou JA, Bray F, Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol. 2020;77(1):38–52. doi: 10.1016/j.eururo.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Steele CB, Li J, Huang B, Weir HK. Prostate cancer survival in the United States by race and stage (2001-2009): Findings from the CONCORD-2 study. Cancer. 2017;24 (123 Suppl):5160–5177. doi: 10.1002/cncr.31026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeb S, Bjurlin MA, Nicholson J, Tammela TL, Penson DF, Carter HB, Carroll P, Etzioni R. Overdiagnosis and overtreatment of prostate cancer. Eur Urol. 2014;65(6):1046–1055. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.DeVere White RW, Vinall RL, Tepper CG, Shi XB. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27(3):307–311. doi: 10.1016/j.urolonc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witte JS. Prostate cancer genomics: Towards a new understanding. Nat Rev Genet. 2009;10(2):77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam RK, Amemiya Y, Benatar T, Wallis CJ, Stojcic-Bendavid J, Bacopulos S, Sherman C, Sugar L, Naeim M, Yang W, Zhang A, Klotz LH, Narod SA, Seth A. Identification and validation of a five MicroRNA signature predictive of prostate cancer recurrence and metastasis: A cohort study. J Cancer. 2015;6(11):1160–1171. doi: 10.7150/jca.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam RK, Benatar T, Wallis CJ, Amemiya Y, Yang W, Garbens A, Naeim M, Sherman C, Sugar L, Seth A. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate. 2016;76(10):869–884. doi: 10.1002/pros.23177. [DOI] [PubMed] [Google Scholar]
- 9.Nam RK, Benatar T, Amemiya Y, Wallis CJD, Romero JM, Tsagaris M, Sherman C, Sugar L, Seth A. MicroRNA-652 induces NED in LNCaP and EMT in PC3 prostate cancer cells. Oncotarget. 2018;9(27):19159–19176. doi: 10.18632/oncotarget.24937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam RK, Benatar T, Wallis CJD, Kobylecky E, Amemiya Y, Sherman C, Seth A. MicroRNA-139 is a predictor of prostate cancer recurrence and inhibits growth and migration of prostate cancer cells through cell cycle arrest and targeting IGF1R and AXL. Prostate. 2019;79(12):1422–1438. doi: 10.1002/pros.23871. [DOI] [PubMed] [Google Scholar]
- 11.Baron S, Dianzani F. The interferons: A biological system with therapeutic potential in viral infections. Antiviral Res. 1994;24(2-3):97–110. doi: 10.1016/0166-3542(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 12.Hertzog PJ, Hwang SY, Kola I. Role of interferons in the regulation of cell proliferation, differentiation, and development. Mol Reprod Dev. 1994;39(2):226–232. doi: 10.1002/mrd.1080390216. [DOI] [PubMed] [Google Scholar]
- 13.Barrasso R, Gross G. Male HPV-associated lesions: Epidemiology and diagnostic criteria. Genital Papillomavirus. 2020;Infections:23–33. [Google Scholar]
- 14.Krown SE. Interferons in malignancy: Biological products or biological response modifiers. J Natl Cancer Inst. 1988;80(5):306–309. doi: 10.1093/jnci/80.5.306. [DOI] [PubMed] [Google Scholar]
- 15.Thomas H, Balkwill FR. Effects of interferons and other cytokines on tumors in animals: A review. Pharmacol Ther. 1991;52(3):307–330. doi: 10.1016/0163-7258(91)90030-p. [DOI] [PubMed] [Google Scholar]
- 16.Sica G, Dell’Acqua G, Iacopino F, Fattorossi A, Marchetti P, van der Kwast TH, Pavone-Macaluso M. Androgen receptors and hormone sensitivity of a human prostatic cancer cell line (PC-3) are modulated by natural beta-interferon. Urol Res. 1994;22(1):33–38. doi: 10.1007/BF00431546. [DOI] [PubMed] [Google Scholar]
- 17.Dicitore A, Grassi ES, Borghi MO, Gelmini G, Cantone MC, Gaudenzi G, Persani L, Caraglia M, Vitale G. Antitumor activity of interferon-β1a in hormone refractory prostate cancer with neuroendocrine differentiation. J Endocrinol Invest. 2017;40(7):761–770. doi: 10.1007/s40618-017-0631-0. [DOI] [PubMed] [Google Scholar]
- 18.Shou J, Soriano R, Hayward SW, Cunha GR, Williams PM, Gao WQ. Expression profiling of a human cell line model of prostatic cancer reveals a direct involvement of interferon signaling in prostate tumor progression. Proc Natl Acad Sci USA. 2002;99(5):2830–2835. doi: 10.1073/pnas.052705299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam RK, Benatar T, Amemiya Y, Sherman C, Seth A. Mir-139 regulates autophagy in prostate cancer cells through Beclin-1 and mTOR signaling proteins. Anticancer Res. 2020;40(12):6649–6663. doi: 10.21873/anticanres.14689. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Zhu J, Zhou H, Zhao Z, Zou Z, Liu X, Lin X, Zhang X, Deng X, Wang R, Chen H, Jin M. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci Rep. 2015;5:14991. doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiscott J, Lin R. IRF-3 releases its inhibitions. Structure. 2005;13(9):1235–1236. doi: 10.1016/j.str.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Zhao B, Shu C, Gao X, Sankaran B, Du F, Shelton CL, Herr AB, Ji JY, Li P. Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc Natl Acad Sci USA. 2016;113(24):E3403–E3412. doi: 10.1073/pnas.1603269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39(6):1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanai H, Negishi H, Taniguchi T. The IRF family of transcription factors: Inception, impact and implications in oncogenesis. Oncoimmunology. 2012;1(8):1376–1386. doi: 10.4161/onci.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13(9):1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10(11):913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- 28.de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: Biochemistry and biological functions. J Biol Chem. 2007;282(28):20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 29.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 30.Tian Y, Wang ML, Zhao J. Crosstalk between autophagy and type I interferon responses in innate antiviral immunity. Viruses. 2019;11(2):132. doi: 10.3390/v11020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmeisser H, Bekisz J, Zoon KC. New function of type I IFN: Induction of autophagy. J Interferon Cytokine Res. 2014;34(2):71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sica G, Angelucci C, Marini L, Milazzo F, Donini S. Oncogene expression is modulated by recombinant human interferon-beta in human breast-cancer cells. Int J Cancer. 1996;67(3):441–446. doi: 10.1002/(SICI)1097-0215(19960729)67:3<441::AID-IJC21>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Sica G, Natoli V, Stella C, Del Bianco S. Effect of natural beta-interferon on cell proliferation and steroid receptor level in human breast cancer cells. Cancer. 1987;60(10):2419–2423. doi: 10.1002/1097-0142(19871115)60:10<2419::aid-cncr2820601011>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 34.Sica G, Marini L. Potentiation of the antitumor activity of tamoxifen by interferons: A mini-review. Molecular Oncology and Clinical. 2019;Applications:391–394. doi: 10.1007/978-3-0348-5663-8_46. [DOI] [Google Scholar]
- 35.Hobeika AC, Subramaniam PS, Johnson HM. IFN alpha induces the expression of the cyclin-dependent kinase inhibitor p21 in human prostate cancer cells. Oncogene. 1997;14(10):1165–1170. doi: 10.1038/sj.onc.1200939. [DOI] [PubMed] [Google Scholar]
- 36.Cheon H, Borden EC, Stark GR. Interferons and their stimulated genes in the tumor microenvironment. Semin Oncol. 2014;41(2):156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dicitore A, Grassi ES, Borghi MO, Gelmini G, Cantone MC, Gaudenzi G, Persani L, Caraglia M, Vitale G. Antitumor activity of interferon-β1a in hormone refractory prostate cancer with neuroendocrine differentiation. J Endocrinol Invest. 2017;40(7):761–770. doi: 10.1007/s40618-017-0631-0. [DOI] [PubMed] [Google Scholar]
- 38.Angelucci C, Iacopino F, Ferracuti S, Urbano R, Sica G. Recombinant human IFN-beta affects androgen receptor level, neuroendocrine differentiation, cell adhesion, and motility in prostate cancer cells. J Interferon Cytokine Res. 2007;27(8):643–652. doi: 10.1089/jir.2006.0120. [DOI] [PubMed] [Google Scholar]
- 39.Dong Z, Greene G, Pettaway C, Dinney CP, Eue I, Lu W, Bucana CD, Balbay MD, Bielenberg D, Fidler IJ. Suppression of angiogenesis, tumorigenicity, and metastasis by human prostate cancer cells engineered to produce interferon-beta. Cancer Res. 1999;59(4):872–879. [PubMed] [Google Scholar]
- 40.Lee J, Wang A, Hu Q, Lu S, Dong Z. Adenovirus-mediated interferon-beta gene transfer inhibits angiogenesis in and progression of orthotopic tumors of human prostate cancer cells in nude mice. Int J Oncol. 2006;29(6):1405–1412. [PubMed] [Google Scholar]
- 41.Chen J, Yang YF, Yang Y, Zou P, Chen J, He Y, Shui SL, Cui YR, Bai R, Liang YJ, Hu Y, Jiang B, Lu L, Zhang X, Liu J, Xu J. AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat Microbiol. 2018;3(3):302–309. doi: 10.1038/s41564-017-0092-4. [DOI] [PubMed] [Google Scholar]
- 42.Schmeisser H, Fey SB, Horowitz J, Fischer ER, Balinsky CA, Miyake K, Bekisz J, Snow AL, Zoon KC. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy. 2013;9(5):683–696. doi: 10.4161/auto.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambjørn M, Ejlerskov P, Liu Y, Lees M, Jäättelä M, Issazadeh-Navikas S. IFNB1/interferon-β-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013;9(3):287–302. doi: 10.4161/auto.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Zhu H, Zeng X, Fan J, Qian X, Wang S, Wang Z, Sun Y, Wang X, Wang W, Ju D. Suppression of autophagy enhanced growth inhibition and apoptosis of interferon-β in human glioma cells. Mol Neurobiol. 2013;47(3):1000–1010. doi: 10.1007/s12035-013-8403-0. [DOI] [PubMed] [Google Scholar]
- 45.Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF, Silverman RH. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem. 1994;269(19):14153–14158. [PubMed] [Google Scholar]
- 46.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31(1):49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. RNase L triggers autophagy in response to viral infections. J Virol. 2012;86(20):11311–11321. doi: 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiqui MA, Malathi K. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J Biol Chem. 2012;287(52):43651–43664. doi: 10.1074/jbc.M112.399964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richetta C, Faure M. Autophagy in antiviral innate immunity. Cell Microbiol. 2013;15(3):368–376. doi: 10.1111/cmi.12043. [DOI] [PubMed] [Google Scholar]
- 50.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 51.Karlsen TA, Brinchmann JE. Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I. Mol Ther. 2013;21(6):1169–1181. doi: 10.1038/mt.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]