Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) recently emerged and is responsible for coronavirus disease 19 (COVID-19). Diagnostic tests have been developed, mainly based on reverse-transcriptase PCR (RT-PCR). Most RT-PCR assays target at least two SARS-CoV-2 genes. In some cases, only one target gene is detected; the interpretation of such cases remains unclear.
Objectives
Our objective was to analyse one target positive (OPT) RT-PCR results, using two RT-PCR assays: the Xpert® Xpress SARS-CoV-2 (Cepheid diagnosis, “Cepheid”) and the Cobas® 6800 SARS-CoV-2 Test (Roche Molecular Diagnostics, “Roche”).
Methods
All SARS-CoV-2 RT-PCR results performed on respiratory samples with the Roche or the Cepheid tests, from 23rd March to 6th August 2020 were collected. A patient with an OPT result was classified as “probable COVID-19” if they met at least one of the three following criteria: (i) history of a two gene-positive SARS-CoV-2 RT-PCR result, (ii) anti-SARS-CoV-2 antibody (IgG) detection or (iii) compatible chest computed tomography scan (CT-scan).
Results
A total of 18,630 and 1189 SARS-CoV-2 RT-PCR tests were performed with the Roche and Cepheid tests, respectively. Among the positive SARS-CoV-2 RT-PCR, 293 samples – corresponding to 264 patients – were OPT (11% of the positive samples). Of these patients, 180 (68%) had at least one of the three criteria listed above and were classified as probable COVID-19.
Conclusions
Sixty-eight percent of the patients with an OPT result were classified as probable COVID-19 and are probably at a late stage of infection. Serology and imaging can be helpful to confirm diagnosis.
Keywords: SARS-CoV-2, COVID-19, Molecular biology, One target positive RT-PCR result
1. Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread throughout the world [1,2], leading to the rapid development of commercial assays for diagnostic testing. During the outbreak, the diagnosis of SARS-CoV-2 is mainly based on reverse-transcriptase PCR (RT-PCR) on naso-pharyngeal swab. Correct interpretation and knowledge of the limitations of commercially available SARS-CoV-2 RT-PCR assays are necessary to identify infectious patients and take appropriate measures for infection control [3]. Most currently available RT-PCR assays target at least two SARS-CoV-2 genes. However, in some cases of positive RT-PCR results, only one target gene is detected; the interpretation of such cases remains unclear.
2. Objective
Our objective was to analyse all one target positive (OPT) RT-PCR results in our laboratory, using the two following commercial RT-PCR assays, the Xpert® Xpress SARS-CoV-2 (Cepheid diagnosis, “Cepheid”) and the Cobas® 6800 SARS-CoV-2 Test (Roche Molecular Diagnostics, “Roche”).
3. Study design
We retrospectively collected all SARS-CoV-2 RT-PCR results performed on respiratory samples from 23rd March to 6th August 2020. The Cepheid assay, targeting the viral envelope (E) and nucleocapsid (N) 2 genes, has a specified limit of detection of 250 copies per mL [4], measured at 100 copies/ml by some authors [5]. The Roche test, targeting the E and ORF1ab genes has a limit of detection of 100 copies per mL [4].
To classify patients with an OPT RT-PCR result as SARS-CoV-2 infected subjects, we collected results of (i) previous two gene positive SARS-CoV-2 RT-PCR. (ii) anti-SARS-CoV-2 antibody (IgG) detection (Abbott Architect®), (iii) chest computed tomography scan (CT-scan) report performed at the time of the OPT RT-PCR result. Thus, a patient with an OPT result and at least one of the three criteria above was classified as “probable COVID-19”.
The overall positivity rate (number of positive tests/number of tests) and the single target positivity rate (number of OPT/number of positive tests) were calculated on the whole period and per 15-day period (until 31st July) to evaluate their evolution with time.
The present study is a non-interventional retrospective study with no additional sampling for the patients beyond that needed for standard diagnostics and no impact on management.
4. Results
On the whole period, a total of 18,630 and 1189 SARS-CoV-2 RT-PCR tests were performed on Roche and Cepheid, respectively. Among the positive SARS-CoV-2 RT-PCR, a total of 293 samples were OPT, representing 11% (293/2584) of the positive samples and 1.5% (293/19819) of all RT-PCR results. Among them were 266 nasopharyngeal swabs, 12 broncho-alveolar lavage, 10 aspirates (2 nasopharyngeal, 7 tracheal and 1 bronchial) and 5 sputa. Using Roche assay, the OPT were the ORF1ab gene for 24 samples (median (IQR) cycle threshold (Ct) 34.4 [29.9–37.3]) and the E gene for 233 samples (37.5 [34.4–41.5]). Using the Cepheid assay, the OPT were the N2 gene for 35 samples (41.8 [38.3–44.8]) and the E gene for one sample (43.6).
Of the 264 patients corresponding to the 293 samples, 124 (47%) had a previous positive RT-PCR assay for two genes – among which were all patients who had 2 or more OPT results. In most cases, the OPT sample and the two-gene positive samples were the same type; when the sample type was different, it was either lower or upper in the respiratory tract. Among the 140 other patients, 36 (26%) had a known positive SARS-CoV-2 serology – 19 had a negative serology. Among the 104 remaining patients, 20 (19%) had a chest CT-scan in favour of COVID-19 – 18 had a negative CT-scan. Globally, among the 264 patients with an OPT result, 180 were classified as probable COVID-19 (68%).
The Ct values were not statistically different between patients classified as "probable COVID-19" and the other OPT patients.
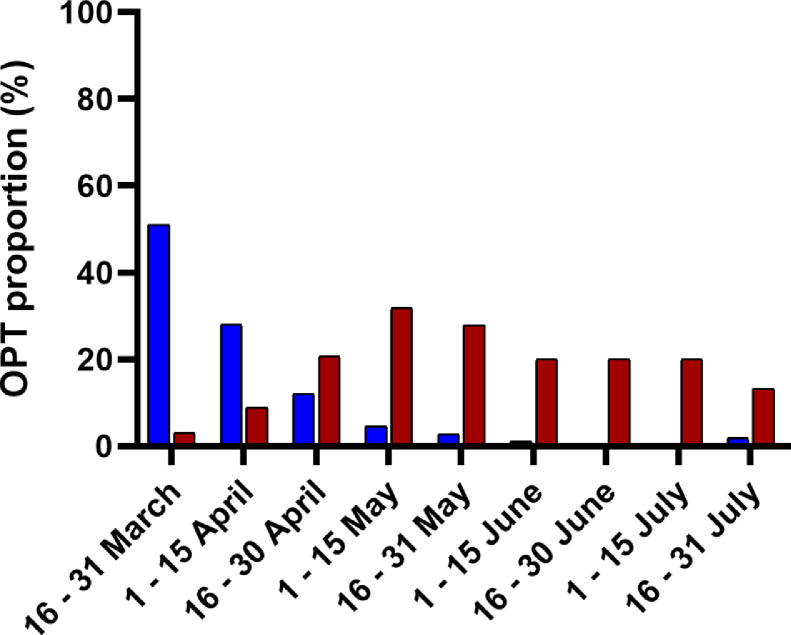
The overall positivity and single gene positivity rates varied according to the different stages of the epidemic ( Fig. 1). The overall positivity rate was of 51% in the second half of March and progressively dropped to 1.9% in the second half of July. The single gene positivity rate was of 3.1% in the second half of March, increased up to 31.8% in the first half of May and remained between 13.3 and 27.9% until July.
Fig. 1.
Overall positivity and single gene positivity rates by 15-day period. Blue vertical bars: overall positivity rate, red vertical bars: single target positivity rate.
5. Discussion
The main RT-PCR assays used for SARS-CoV-2 diagnostic amplify at least two viral target genes, but in some cases only one gene is amplified. The frequency of OPT results was 11% of positive RT-PCR samples and 1.5% of all RT-PCR results done between March and August 2020.
Sixty-eight percent of the patients with an OPT result were classified as probable COVID-19, found mainly by a previously double target positive RT-PCR and were probably at a late stage of infection with a low viral excretion. SARS-CoV-2 RNA shedding can be evidenced until day 83 in some patients [6]. Indeed, our results show that the single target positivity rate increases during the study period with 3.1% at the beginning of the epidemic and 31.8% at the end of the first wave. Tests were performed mainly on symptomatic patients at the beginning of the epidemic. Later on, the number of tests performed on asymptomatic patients increased in a context of large testing. OPT results on Cepheid were recently studied [7]: 34.1% of studied patients had a previous diagnosis of COVID-19, 18% had COVID-19 symptoms and some had abnormalities at chest CT-scan. The authors mention the fact that OPT results are very often and probably associated with prolonged infection.
The SARS-CoV-2 serology contributed to classify the COVID-19 infection status in only 26% of patients that had not been detected by a positive double target RT-PCR but most of the patients had no available serology. The Abbott Architect sensitivity was reported at 82.4% 10 days after symptom onset and at 100% 17 days after symptom onset [8] and there is a percentage of false negative of about 20% in asymptomatic subjects [9].
Another issue is the contagiousness of the patient. A RT-PCR test only detects viral RNA, not correlated with infectivity. Many infected patients keep having positive RT-PCR tests during weeks [6,10], but contagiousness is doubtful at this point [6,[11], [12], [13]]. In contrast, an OPT result could also reflect a beginning viral replication before onset of symptoms, in which case the contagiousness will be high few days later. French guidelines about correlation between Ct value and probability of excreting infectious SARS-CoV-2 recommend considering results with a Ct value earlier than 33 (32 for Roche) as “strong excretion”, between 33 and 37 (32 and 35 for Roche) as “moderate excretion” and later than 37 (35 for Roche) as “weak excretion” [14]. Only one sample matched the “strong excretion” criteria, 7.5% the “moderate excretion” criteria and 92.1% “weak excretion”. A majority of OPT patients thus has a low probability of being contagious.
We have no argument enabling us to classify 84 patients as COVID-19 positive. The OPT result could be a false positive. In PCR methods, false positive results are known to occur and such results in SARS-CoV-2 RT-PCR are described: this can be due to the test conception (unoptimized primer sets [15]) – especially in a context of rapid development, a contamination of reagents or an improper sample handling [10]. The Cepheid and Roche methods have been shown to be highly sensitive and specific [5,16] and to have a high inter method agreement [17]. Another possibility is that these samples are true low positives, which might not be confirmed when another RT-PCR is performed.
To conclude, most patients with OPT result were probable COVID-19 cases. Clinical data were, in our study, uninformative. Some experts recommend that tests with a positive result on a single target be made negative, confirming that investigations should be continued.
Declaration of Competing Interest
None.
Acknowledgments
No funding.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldenberger D., Leuzinger K., Sogaard K.K., Gosert R., Roloff T., Naegele K., Cuénod A., Mari A., Seth-Smith H., Rentsch K., Hinić V., Hirsch H.H., Egli A. Brief validation of the novel GeneXpert Xpress SARS-CoV-2 PCR assay. J. Virol. Methods. 2020;284 doi: 10.1016/j.jviromet.2020.113925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broder K., Babiker A., Myers C., White T., Jones H., Cardella J., Burd E.M., Hill C.E., Kraft C.S. Test Agreement between Roche cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 assays at high cycle threshold ranges. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.01187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. The Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoshchehreh M., Wald-Dickler N., Holtom P., Butler-Wu S.M. A needle in the haystack? Assessing the significance of envelope (E) gene-negative, nucleocapsid (N2) gene-positive SARS-CoV-2 detection by the Cepheid Xpert Xpress SARS-COV-2 assay. J. Clin. Virol. 2020;133 doi: 10.1016/j.jcv.2020.104683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A., Jerome K.R., Mathias P.C., Greninger A.L. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koopmans M., Haagmans B. Assessing the extent of SARS-CoV-2 circulation through serological studies. Nat. Med. 2020;26:1171–1172. doi: 10.1038/s41591-020-1018-x. [DOI] [PubMed] [Google Scholar]
- 10.Motley M.P., Bennett-Guerrero E., Fries B.C., Spitzer E.D. Review of viral testing (polymerase chain reaction) and antibody/serology testing for severe acute respiratory syndrome-coronavirus-2 for the intensivist. Crit. Care Explor. 2020;2:e0154. doi: 10.1097/CCE.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scola B.La, Bideau M.Le, Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 14.Avis-SFM-valeur-Ct-excrétion-virale-_-Version-Finale-07102020-V3.pdf, (n.d.). https://www.sfm-microbiologie.org/wp-content/uploads/2020/10/Avis-SFM-valeur-Ct-excre%CC%81tion-virale-_-Version-Finale-07102020-V3.pdf (accessed December 18, 2020).
- 15.Park M., Won J., Choi B.Y., Lee C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020;52:963–977. doi: 10.1038/s12276-020-0452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cradic K., Lockhart M., Ozbolt P., Fatica L., Landon L., Lieber M., Yang D., Swickard J., Wongchaowart N., Fuhrman S., Antonara S. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am. J. Clin. Pathol. 2020;154:201–207. doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V., Love N. Detection of SARS-CoV-2 by Use Of The Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]