Abstract
An effective response to the ongoing coronavirus disease (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will involve a range of complementary preventive modalities. The current studies were conducted to evaluate the in vitro SARS-CoV-2 antiviral and virucidal (irreversible) activity of astodrimer sodium, a dendrimer with broad spectrum antimicrobial activity, including against enveloped viruses in in vitro and in vivo models, that is marketed for antiviral and antibacterial applications. We report that astodrimer sodium inhibits replication of SARS-CoV-2 in Vero E6 and Calu-3 cells, with 50% effective concentrations (EC50) for i) reducing virus-induced cytopathic effect of 0.002–0.012 mg/mL in Vero E6 cells, and ii) infectious virus release by plaque assay of 0.019–0.032 mg/mL in Vero E6 cells and 0.030–0.037 mg/mL in Calu-3 cells. The selectivity index (SI) in these assays was as high as 2197. Astodrimer sodium was also virucidal, irreversibly reducing SARS-CoV-2 infectivity by >99.9% (>3 log10) within 1 min of exposure, and up to >99.999% (>5 log10) shown at astodrimer sodium concentrations of 10–30 mg/mL in Vero E6 and Calu-3 cell lines. Astodrimer sodium also inhibited infection in a primary human airway epithelial cell line. The data were similar for all investigations and were consistent with the potent antiviral and virucidal activity of astodrimer sodium being due to irreversible inhibition of virus-host cell interactions, as previously demonstrated for other viruses. Further studies will confirm if astodrimer sodium binds to SARS-CoV-2 spike protein and physically blocks initial attachment of the virus to the host cell. Given the in vitro effectiveness and significantly high SI, astodrimer sodium warrants further investigation for potential as a topically administered agent for SARS-CoV-2 therapeutic applications.
Keywords: Astodrimer, COVID-19, Dendrimer, Antiviral, SARS-CoV-2, SPL7013
1. Introduction
The ongoing pandemic coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection, has resulted in unprecedented efforts to rapidly develop strategies to contain infection rates for the protection of vulnerable populations. An effective public health response to the current pandemic will involve currently available vaccines being complemented by supplementary preventive modalities.
SARS-CoV-2 receptors and coreceptors have been shown to be highly expressed in nasal epithelial cells (Sungnak et al., 2020). This finding is consistent with the virus infectivity or replication pattern along the respiratory tract, which peaks proximally (nasal cavity) and is relatively minimal in the distal alveolar regions (Hou et al., 2020). These findings suggest that nasal carriage of the virus is a key feature of transmission, and that nasally administered therapeutic modalities could be potentially effective in helping to prevent spread of infection.
Astodrimer sodium (SPL7013) is a generation-four lysine dendrimer with a polyanionic surface charge (McCarthy et al., 2005) that is active against several enveloped and non-enveloped viruses including human immunodeficiency virus-1 (HIV-1) (Lackman-Smith et al., 2008; Tyssen et al., 2010), herpes simplex virus (HSV)-1 and −2 (Gong et al., 2005), H1N1 and H3N2 influenza virus, human respiratory syncytial virus (HRSV), human papillomavirus (HPV), adenovirus and Zika virus (unpublished data). Astodrimer sodium also has antibacterial properties. Both size and surface charge contribute to the function of the compound (Tyssen et al., 2010), and when administered topically, astodrimer sodium is not absorbed systemically (Chen et al., 2009; O'Loughlin et al., 2010; McGowan et al., 2011).
Vaginally administered astodrimer sodium protected macaques from infection with chimeric simian-HIV-1 (SHIV)89.6P (Jiang et al., 2005), and mice and guinea pigs from HSV-2 infection (Bernstein et al., 2003) in vaginal infection challenge models. Astodrimer 1% Gel (10 mg/mL astodrimer sodium) administered vaginally has been shown to be safe and effective in phase 2 and large phase 3 trials for treatment and prevention of bacterial vaginosis (BV) (Chavoustie et al., 2020; Waldbaum et al., 2020; Schwebke et al., 2021) and is marketed in Europe, Australia, New Zealand and several countries in Asia.
The current studies were conducted to assess the antiviral and virucidal (irreversible) activity of astodrimer sodium against SARS-CoV-2 in vitro, to determine its potential for investigation as a reformulated agent against SARS-CoV-2 infection.
2. Materials and methods
2.1. Virus, cell culture, astodrimer sodium and controls
SARS-CoV-2 hCoV-19/Australia/VIC01/2020 was a gift from Melbourne's Peter Doherty Institute for Infection and Immunity (Melbourne, Australia). Virus stock was generated at 360Biolabs (Melbourne, Australia) by two passages in Vero cells in virus growth media, which comprised Minimal Essential Medium (MEM) without L-glutamine supplemented with 1% (w/v) L-glutamine, 1.0 μg/mL of L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington Biochemical, NJ, USA), 0.2% bovine serum albumin (BSA) and 1% insulin-transferrin-selenium (ITS).
SARS-CoV-2 2019-nCoV/USA-WA1/2020 strain was isolated from an oropharyngeal swab from a patient with a respiratory illness who developed clinical disease (COVID-19) in January 2020 in Washington, US, and sourced from BEI Resources (NR-52281). Virus was derived from African green monkey kidney Vero E6 cells or lung homogenates from human angiotensin converting enzyme 2 (hACE2) transgenic mice.
Vero E6 and human Calu-3 cell lines were cultured in MEM without L-glutamine supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% (w/v) L-glutamine. Vero E6 and Calu-3 cells were passaged for a maximum of 10 passages for antiviral and virucidal studies. Hank's balanced salt solution (HBSS) with 2% FBS was used for infection. The 2019-nCoV/USA-WA1/2020 strain antiviral assays were performed with a multiplicity of infection (MOI) of 0.1.
The virus inoculums for virucidal assays were 104, 105, and 106 pfu/mL. After defined incubation periods, the solution was pelleted through a 20% sucrose cushion (Beckman SW40 Ti rotor) and resuspended in 1.5 mL MEM, which was then added to 2.5 × 104 cells/well.
Primary human bronchial epithelial cells (HBEpC) (Sigma-Aldrich, MO, USA) were grown and maintained in HBEpC/HTEpC growth medium (Cell Applications, CA, USA). These primary cells express the ACE2 receptor and are permissive to SARS-CoV-2 infection. These cells were used to determine the antiviral effect of astodrimer sodium against SARS-CoV-2 in a primary human airway epithelial cell line. Cells were infected with SARS-CoV-2 2019-nCoV/USA-WA1/2020 at 103 pfu/mL with 1 mL added to 2.5 × 104 cells/well. The positive control was addition of 10 μg/mL of SARS-CoV-2 spike protein antibody (pAb, T01KHuRb) (ThermoFisher, MA, USA) at the time of infection.
Astodrimer sodium was prepared as 100 mg/mL in water and stored at 4 °C. Astodrimer sodium has a molecular weight of 16581.57 g/mol and the structure is described and illustrated in Tyssen et al. (2010). The purity of the compound used in these studies was assessed by ultra-high-performance liquid chromatography (UPLC) to be 98.79%.
Remdesivir (MedChemExpress, NJ, USA) was used as a positive control in the virus-induced cytopathic effect (CPE) inhibition and plaque assays.
Iota-carrageenan (Sigma-Aldrich, MO, USA) was used in the primary epithelial cell nucleocapsid and plaque assays to compare the antiviral activity of this substance with astodrimer sodium. Concentrations used are those reported to show activity against SARS-CoV-2 (Bansal et al., 2020).
2.2. Virus-induced cytopathic effect inhibition assay
Vero E6 (ATCC-CRL1586) cell stocks were generated in cell growth medium, which comprised MEM without L-glutamine supplemented with 10% (v/v) heat-inactivated FBS and 1% (w/v) L-glutamine. Vero E6 cell monolayers were seeded in 96-well plates at 2 × 104 cells/well in 100 μL growth medium (MEM supplemented with 1% (w/v) L-glutamine, 2% FBS) and incubated overnight at 37 °C in 5% CO2. SARS-CoV-2 infection was established by using an MOI of 0.05 to infect cell monolayers.
Astodrimer sodium or remdesivir were serially diluted 1:3, 9 times and each compound concentration was assessed for both antiviral efficacy and cytotoxicity in triplicate. Astodrimer sodium was added to Vero E6 cells 1 h prior to infection or 1 h post-infection with SARS-CoV-2. Cell cultures were incubated at 37 °C in 5% CO2 for 4 days prior to assessment of CPE. The virus growth media was MEM supplemented with 1% (w/v) L-glutamine, 2% FBS, and 4 μg/mL TPCK-treated trypsin. On Day 4, viral-induced CPE and cytotoxicity of the compound were determined by measuring the viable cells using the methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay (MP Biomedicals, NSW, Australia). Absorbance was measured at 540–650 nm on a plate reader.
2.3. Antiviral plaque assay evaluation and nucleocapsid ELISA
For the antiviral evaluation, astodrimer sodium was added to cells 1 h prior to, at the time of, and 1 h after exposing the cells to virus. For both the antiviral and virucidal assays, at 6 h after infection, cells were washed to remove astodrimer sodium and/or any virus remaining in the supernatant.
Following initial infection, cell cultures were incubated and supernatants recovered after 16 h or 4 days. The amount of virus in the supernatants was determined by plaque assay (plaque forming unit [pfu]) and by nucleocapsid enzyme-linked immunosorbent assay (ELISA). The plaque assay used was as described in van den Worm et al. (2012), utilizing 2% sodium carboxymethyl cellulose overlay, fixation of cells by 4% paraformaldehyde and staining with 0.1% crystal violet. The nucleocapsid ELISA assay was as described by Bioss Antibodies, USA (BSKV0001).
The assessment of astodrimer sodium cytotoxicity occurred on Day 4 by measuring lactate dehydrogenase (LDH) activity in the cytoplasm using an LDH detection kit (Cayman Chemical), with 0.5% saponin used as the positive cytotoxic control.
2.4. Virucidal assay
For the virucidal evaluation, concentrations of astodrimer sodium (0.0046–30 mg/mL) were incubated with SARS-CoV-2 2019-nCoV/USA-WA1/2020 for times ranging from 5 s to 2 h. To neutralize the effect of astodrimer sodium, unbound compound was separated from the astodrimer:virus mixture by pelleting the preincubated mixture through a 20% sucrose cushion (Beckman SW40 Ti rotor). The astodrimer sodium-containing supernatant was removed (i.e., neutralising the effect of SPL7013) and then the pelleted virus was gently resuspended and added to Vero E6 or Calu-3 cell cultures. Virus infection, cell culture and cytotoxicity assessment was as described for the plaque assay described in Section 2.3.
All antiviral and virucidal assays were performed in triplicate, except where indicated in the results.
2.5. Determination of 50% effective concentration (EC50) and cytotoxicity (CC50)
The concentration of compound that gives a 50% reduction in viral-induced CPE, infectious virus (pfu/mL), or secreted viral nucleocapsid (EC50) was calculated using the formula of Pauwels et al. (1998).
The concentration of compound that resulted in a 50% reduction in cell viability (CC50) after 4 days of culture was also calculated by the formula of Pauwels et al. (1998).
2.6. Primary epithelial cell assay
Astodrimer sodium (0, 1.1, 3.3 and 10 mg/mL) or iota-carrageenan (0, 6, 60 and 600 μg/mL) were added to HBEpC cells 1 h prior to infection with SARS-CoV-2. Cells were cultured for 4 days post-infection and the cell supernatant was analysed for the amount of secreted SARS-CoV-2 nucleocapsid by ELISA, and infectious virus was quantitated by plaque assay, as described in Section 2.3.
3. Results
3.1. Virus-induced cytopathic effect inhibition
In two independent virus-induced CPE inhibition assays, astodrimer sodium inhibited SARS-CoV-2 (hCoV-19/Australia/VIC01/2020) replication in Vero E6 cells in a dose dependent manner (Table 1 ). Astodrimer sodium inhibited viral replication when added either 1 h prior to infection, or 1 h post-infection with SARS-CoV-2.
Table 1.
Antiviral efficacy, measured by a reduction in CPE in virus-infected cells at Day 4 post-infection, and selectivity of astodrimer sodium against SARS-CoV-2 (hCoV-19/Australia/VIC01/2020) infection of Vero E6 cells.
| Compound/Assay Type | Repeat | EC50 (mg/mL) | CC50 (mg/mL) | SI |
|---|---|---|---|---|
| Astodrimer sodium added 1-h pre-infection | Assay 1 | 0.004 | 2.93 | 793 |
| Assay 2 | 0.012 | >0.863 | >70 | |
| Mean ± SD | 0.008 ± 0.006 | NC | >431a | |
| Astodrimer sodium added 1-h post-infection | Assay 1 | 0.002 | 3.29 | 2197 |
| Assay 2 | 0.011 | >0.863 | >80 | |
| Mean ± SD | 0.006 ± 0.007 | NC | >1138a | |
| Remdesivir added 1-h post-infection | Assay 1 | 0.59 μM | >20 μM | >33 |
| Assay 2 | 0.61 μM | >20 μM | >33 |
EC50 = 50% effective concentration; CC50 = 50% cytotoxic concentration; SI = selectivity index (CC50/EC50).
SD = standard deviation; NC = not calculated.
Average of Assay 1 and Assay 2 SI.
Astodrimer sodium was initially tested in the range of 0.0013–8.63 mg/mL. In the repeat set of assays, astodrimer sodium was tested in the range of 0.0001–0.86 mg/mL to help further characterize the lower end of the dose response curve. The effective and cytotoxic concentrations, and selectivity indices from the assays are shown individually and as means in Table 1 for CPE determined readouts.
The selectivity index (SI) for astodrimer sodium against SARS-CoV-2 in the CPE studies ranged from 793 to 2197 for the initial assays where compound was added 1 h prior to infection and 1 h after infection, respectively, and was >70 to >80 in the repeat assays, in which cytotoxicity was not observed up to the highest concentration tested (0.86 mg/mL).
The positive control, remdesivir, was also active in the CPE inhibition assay, with a SI of >33.
3.2. Antiviral efficacy
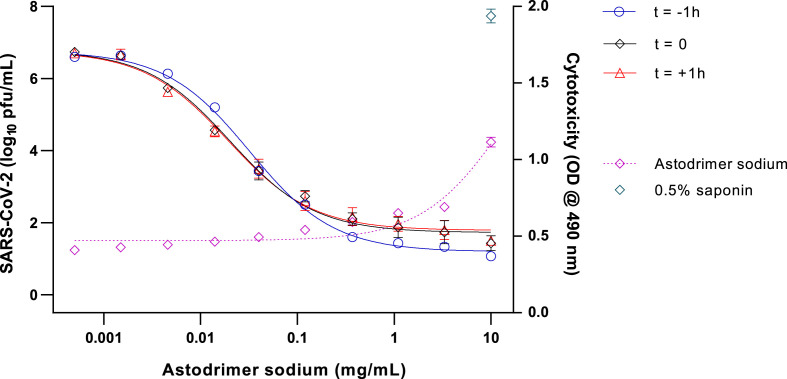
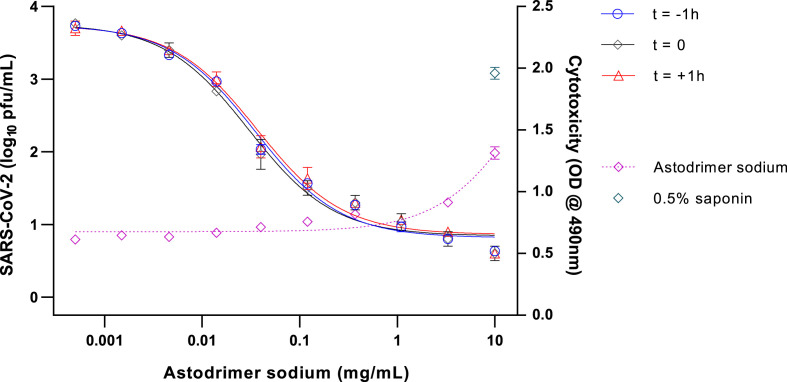
To determine the ability of astodrimer sodium to inhibit globally diverse SARS-CoV-2 strains, the compound was evaluated against the 2019-nCoV/USA-WA1/2020 virus in Vero E6 cells and human Calu-3 cells. Antiviral readouts were based on virological endpoints of infectious virus or viral nucleocapsid released into the supernatant post-infection. As shown in Table 2 and Fig. 1, Fig. 2 , astodrimer inhibited the 2019-nCoV/USA-WA1/2020 strain with an EC50 0.019–0.032 mg/mL and 0.030–0.037 mg/mL for infectious virus release as determined by plaque assay in Vero E6 cell for Calu-3 cells, respectively. These data are consistent with the inhibition by astodrimer of the replication of the Australian SARS-CoV-2 isolate in vitro. The dose response data for the nucleocapsid released into the supernatant by ELISA were similar to the infectious virus release data in each cell line (data not shown). The positive control, remdesivir, was also active in the plaque assay.
Table 2.
Antiviral efficacy, measured by a reduction in mean infectious virus (Log10 pfu/mL), and selectivity of astodrimer sodium against SARS-CoV-2 (2019-nCoV/USA-WA1/2020) on Day 4 post-infection.
| Compound/Assay Type | Cell Line | EC50 (mg/mL) | CC50 (mg/mL) | SI |
|---|---|---|---|---|
| Astodrimer sodium added 1-h pre-infection | Vero E6 | 0.032 | 15.09 | 472 |
| Calu-3 | 0.037 | 21.76 | 588 | |
| Astodrimer sodium added at time of infection | Vero E6 | 0.020 | 15.09 | 755 |
| Calu-3 | 0.035 | 21.76 | 622 | |
| Astodrimer sodium added 1-h post-infection | Vero E6 | 0.019 | 15.09 | 794 |
| Calu-3 | 0.030 | 21.76 | 725 | |
| Remdesivir added 1-h post-infection | Vero E6 | 0.791 μM | N/A | N/A |
| Calu-3 | 0.589 μM | N/A | N/A |
EC50 = 50% effective concentration; CC50 = 50% cytotoxic concentration; SI = selectivity index (CC50/EC50); N/A = not applicable.
Fig. 1.
Dose-response analysis of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) antiviral activity of astodrimer sodium in Vero E6 cells, as measured by infectious virus release (Log10 pfu/mL), and cytotoxicity on Day 4 post-infection.
Astodrimer sodium (0.0005–10 mg/mL) was added to cell cultures 1 h prior to (t = -1 h), at the time of (t = 0), and 1 h post-infection (t = +1 h). Cytotoxicity was assessed by LDH detection (OD @ 490 nm), with 0.5% saponin used as the positive cytotoxic control. Points and error bars represent mean ± SD of triplicate readings.
Fig. 2.
Dose-response analysis of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) antiviral activity of astodrimer sodium in Calu-3 cells, as measured by infectious virus release (Log10 pfu/mL), and cytotoxicity on Day 4 post-infection.
Astodrimer sodium (0.0005–10 mg/mL) was added to cell cultures 1 h prior to (t = -1 h), at the time of (t = 0), and 1 h post-infection (t = +1 h). Cytotoxicity was assessed by LDH detection (OD @ 490 nm), with 0.5% saponin used as the positive cytotoxic control. Points and error bars represent mean ± SD of triplicate readings.
3.3. Virucidal efficacy
Virucidal assays investigated if astodrimer sodium could reduce viral infectivity by irreversibly inactivating SARS-CoV-2 prior to infection of Vero E6 cells and human airway Calu-3 cells. Following incubation of virus with astodrimer for up to 2 h and neutralization of astodrimer, the astodrimer-exposed virus was added to cell cultures. After either 16 h or 96 h (Day 4), the cell culture supernatant was collected for assessment of progeny viral infectivity as determined by the amount of secreted infectious virus and nucleocapsid. The SARS-CoV-2 replication lifecycle is completed in approximately 8 h (Ogando et al., 2020) and in these studies, we sampled at 16 h (2 lifecycles) or Day 4 (12 lifecycles) post-infection.
Enabling a possible 12 rounds of infection, the Day 4 (96 h) sampling time point identified that exposure of 106 pfu/mL SARS-CoV-2 to astodrimer sodium for 1–2 h resulted in a dose-dependent reduction in viral infectivity, with 10–30 mg/mL astodrimer sodium achieving up to >99.999% (>5 log10) reduced infectivity in Vero E6 cells and >99.9% (>3 log10) reduced infectivity in Calu-3 cells (data not shown) compared to untreated virus. SARS-CoV-2 infectivity was also reduced by up to >99.999% in Vero E6 cells when the incubation time of astodrimer (10–30 mg/mL) with 106 pfu/mL virus was reduced to 15–30 min (data not shown).
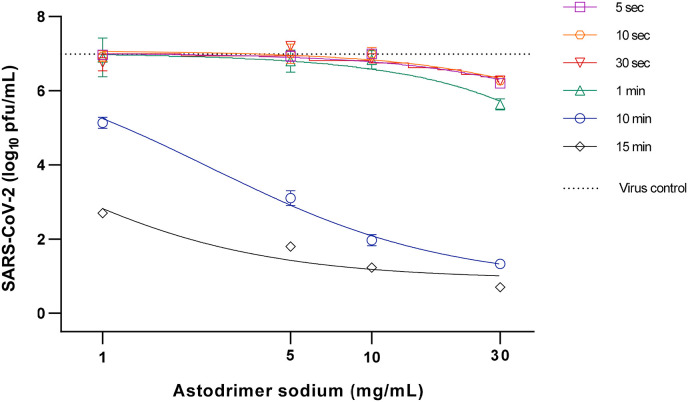
Incubation of astodrimer sodium with viral inoculums of 104, 105 and 106 pfu/mL for as little as 5 s resulted in evidence of reduced infectivity, with 10–15 min exposure being sufficient to achieve >99.9% reduction in virus infectivity, and greater reduction achieved with lower viral inoculum (>99.999%, 104 pfu/mL viral inoculum, 10–30 mg/mL astodrimer sodium, and 10–15 min incubation time) (Table 3 , Fig. 3 ).
Table 3.
Virucidal efficacy of 10 mg/mL astodrimer sodium against SARS-CoV-2 (2019-nCoV/USA-WA1/2020), measured by a reduction in mean infectious virus (Log10 pfu/mL), at 96 h post-infection.
| Viral Load (pfu/mL) | Virus:Astodrimer Incubation Time |
Reduction vs. Virus Control (Log10 ± SD) | Reduction vs. Virus Control (%) |
|---|---|---|---|
| 106 | 5 s | 0.10 ± 0.20 | 20.567 |
| 10 s | 0.03 ± 0.06 | 7.388 | |
| 30 s | 0.10 ± 0.10 | 20.567 | |
| 1 min | 0.33 ± 0.12 | 53.584 | |
| 10 min | 2.20 ± 0.10 | 99.369 | |
| 15 min | 3.67 ± 0.23 | 99.979 | |
| 105 | 5 s | 0.33 ± 0.21 | 53.584 |
| 10 s | 0.23 ± 0.06 | 41.566 | |
| 30 s | 0.30 ± 0.17 | 49.881 | |
| 1 min | 0.47 ± 0.21 | 65.855 | |
| 10 min | 3.70 ± 0.26 | 99.980 | |
| 15 min | 4.60 ± 0.10 | 99.998 | |
| 104 | 5 s | −0.13 ± 0.21 | −35.936 |
| 10 s | 0.07 ± 0.29 | 14.230 | |
| 30 s | 0.10 ± 0.10 | 20.567 | |
| 1 min | 0.10 ± 0.00 | 20.567 | |
| 10 min | 5.07 ± 0.25 | >99.999 | |
| 15 min | 5.83 ± 0.12 | >99.999 |
Virus control = untreated virus, 0 mg/mL astodrimer sodium; SD = standard deviation.
Fig. 3.
Virucidal efficacy of astodrimer sodium against SARS-CoV-2 (2019-nCoV/USA-WA1/2020) measured by a reduction in mean infectious virus (Log10 pfu/mL), at 96 h post-infection in Vero E6 cells.
Astodrimer sodium (1–30 mg/mL) was incubated with SARS-CoV-2 (2019-nCoV/USA-WA1/2020) for 5 s up to 15 min. Treated virus was added to Vero E6 cells and the amount of infectious virus in the supernatant was determined by plaque assay 96 h post-infection. Graph shows dose-response of astodrimer sodium virucidal activity using 104 pfu/mL virus inoculum. Points and error bars represent mean ± SD of triplicate readings. Dotted line indicates level of mean infectious virus when untreated virus was added to Vero E6 cells (virus control).
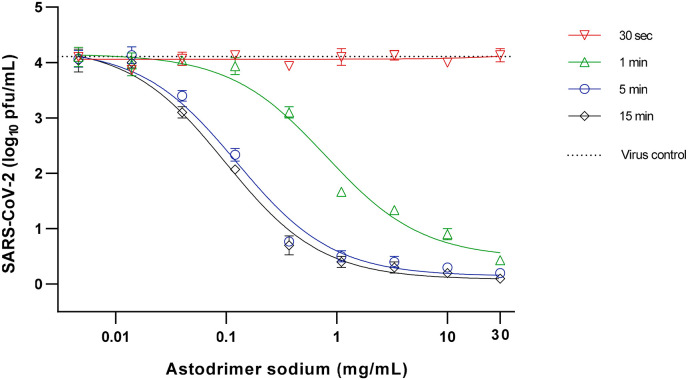
When assessed 16 h post-infection of cells with astodrimer-exposed virus, it was found that ≥10 mg/mL astodrimer sodium inactivated >99.9% SARS-CoV-2 (104 pfu/mL) within as little as 1 min of exposure (Table 4 , Fig. 4 ).
Table 4.
Virucidal efficacy of 10 mg/mL astodrimer sodium against SARS-CoV-2 (2019-nCoV/USA-WA1/2020), measured by a reduction in mean infectious virus (Log10 pfu/mL), at 16 h post-infection.
| Viral Load (pfu/mL) | Virus:Astodrimer Incubation Time | Reduction vs. Virus Control (Log10 ± SD) | Reduction vs. Virus Control (%) |
|---|---|---|---|
| 105 | 30 s | 0.00 ± 0.36 | 0.000 |
| 1 min | 2.63 ± 0.15 | 99.767 | |
| 5 min | 4.63 ± 0.31 | 99.998 | |
| 15 min | 4.60 ± 0.10 | 99.998 | |
| 104 | 30 s | 0.20 ± 0.20 | 36.904 |
| 1 min | 3.17 ± 0.12 | 99.932 | |
| 5 min | 3.67 ± 0.21 | 99.979 | |
| 15 min | 4.00 ± 0.10 | 99.990 |
Virus control = untreated virus, 0 mg/mL astodrimer sodium; SD = standard deviation.
Fig. 4.
Virucidal efficacy of astodrimer sodium against SARS-CoV-2 (2019-nCoV/USA-WA1/2020) measured by a reduction in mean infectious virus (Log10 pfu/mL), at 16 h post-infection in Vero E6 cells.
Astodrimer sodium (0.0046–30 mg/mL) was incubated with SARS-CoV-2 (2019-nCoV/USA-WA1/2020) for 30 s, 1 min, 5 min and 15 min. Treated virus was added to Vero E6 cells and the amount of infectious virus in the supernatant was determined by plaque assay 16 h post-infection. Graph shows dose-response of astodrimer sodium virucidal activity using 104 pfu/mL virus inoculum. Points and error bars represent mean ± SD of triplicate readings. Dotted line indicates level of mean infectious virus when untreated virus was added to Vero E6 cells (virus control).
3.4. Antiviral efficacy in primary human airway epithelial cells
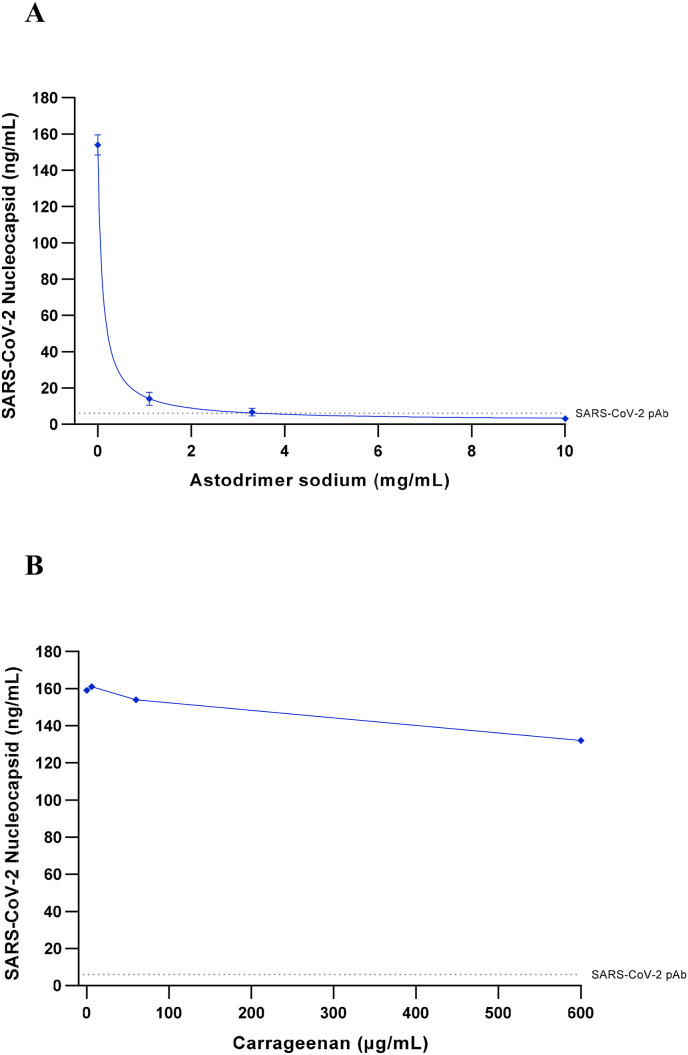
To determine the ability of astodrimer sodium to prevent SARS-CoV-2 infection of primary human epithelial cells, the compound was evaluated against the 2019-nCoV/USA-WA1/2020 strain in HBEpC cell culture. Astodrimer sodium was found to reduce infection of HBEpC primary cells by SARS-CoV-2 by up to 98% vs virus control by nucleocapsid ELISA (Fig. 5 A), and by up to 95% in the plaque assay (data not shown). In contrast, treatment with iota-carrageenan had minimal antiviral effect against SARS-CoV-2 in this cell line, with the highest concentration tested reducing infection by just 17% by nucleocapsid ELISA (Fig. 5B), and just 21% in the plaque assay (data not shown). The maximum level of inhibition with astodrimer sodium was comparable to inhibition achieved with the SARS-CoV-2 spike protein antibody (pAb, T01KHuRb) positive control (see Fig. 5A and B).
Fig. 5.
Antiviral efficacy of astodrimer sodium and iota-carrageenan against SARS-CoV-2 (2019-nCoV/USA-WA1/2020) measured by a reduction in nucleocapsid (ng/mL), at Day 4 post-infection in human bronchial epithelial primary cells (HBEpC).
Astodrimer sodium (0, 1.1, 3.3 and 10 mg/mL) or iota-carrageenan (0, 6, 60 and 600 μg/mL) were added to cell cultures 1 h prior to infection.
A. Dose-response of astodrimer sodium antiviral activity. Points and error bars represent mean ± SD of triplicate readings. B. Dose-response of carrageenan antiviral activity. Points represent one replicate.
Dotted lines indicates level of inhibition achieved with positive control, SARS-CoV-2 pAb.
4. Discussion
Astodrimer sodium demonstrated potent antiviral activity against globally diverse SARS-CoV-2 strains in vitro. Antiviral activity was demonstrated by reduction in CPE, release of infectious virus and release of viral nucleocapsid protein. Antiviral activity was demonstrated when astodrimer sodium was added to cells prior to infection of cells and when the compound was added to cells already exposed to SARS-CoV-2. Irreversible antiviral activity (virucidal effect) was demonstrated when astodrimer sodium was mixed with virus for as little as 1 min.
Of note is a significantly high SI for astodrimer sodium in the antiviral assays relative to other antiviral compounds under investigation for SARS-CoV-2 activity (Pizzorno et al., 2020). While some cytotoxicity was observed in CPE and plaque assays, astodrimer sodium at a 10 mg/mL concentration has been shown to be non-cytotoxic, non-sensitizing and non-irritant in animal and human (Chavoustie et al., 2020; Waldbaum et al., 2020; Schwebke et al., 2021) studies of vaginal application, where an intact epithelium is more resilient than monolayer cells in culture.
Remdesivir was used as the antiviral positive control for the CPE inhibition and antiviral assays and the experimental EC50 was consistent with published data generated with a different clinical isolate of SARS-CoV-2 (Wang et al., 2020).
There was some variability noted between two CPE assays, where the EC50s were 3–5 times higher in the repeat assay compared with the initial assay. The difference could be due to the fact that these assays were conducted early in the course of the pandemic when virology laboratories were learning how to handle the new SARS-CoV-2 virus and optimize cell conditions. The EC50 of the repeat assay was comparable to the EC50s obtained in plaque assays at a second laboratory.
Astodrimer sodium inhibited infection of a human airway primary epithelial cell by SARS-CoV-2, whereas iota-carrageenan failed to provide significant inhibition at concentrations that have previously been shown to reduce SARS-CoV-2 infection in Vero E6 cells (Bansal et al., 2020). The unique structure of astodrimer sodium, a sulphonated, roughly spherical molecule with a core and densely packed branches radiating out from the core, appears to provide potential benefits over other polyanionic compounds such as iota-carrageenan and heparin, which are linear sulphated molecules with a distribution of molecular weight. The authors are not aware of data showing that iota-carrageenan irreversibly inactivates virus, while heparin has demonstrated a lack of irreversible, virucidal interaction with HSV virion components (Ghosh et al., 2009).
The antiviral data from addition of astodrimer sodium 1 h prior to, at the time of, or 1 h post-infection are consistent with astodrimer sodium being a potent inhibitor of early events in the virus lifecycle, including virus attachment and fusion that are ongoing in these timeframes. The virucidal assay data suggest that astodrimer sodium antiviral activity was consistent with the proposed mechanism of action of tight binding to virus envelope proteins, including the viral spike protein that is essential for attachment to the host cell receptor, thereby irreversibly inactivating virus and blocking infection.
The virucidal assay for astodrimer sodium demonstrated that it irreversibly inhibits the early phase of virus infection and replication. These findings suggest potent inhibition of viral attachment, fusion and entry of the virus, which prevents virus replication and release of infectious virus progeny.
Astodrimer sodium has been previously found to be an effective antiviral that exerts its inhibition in the early virus-host receptor recognition interactions (Tyssen et al., 2010; Telwatte et al., 2011), and its potential mechanism of action against SARS-CoV-2 is likely similar to that identified for other pathogens. Astodrimer sodium was found to bind to HIV-1 by strong electrostatic forces to positively charged clusters of highly conserved amino acids on HIV-1 gp120 protein and/or positively charged amino acid regions located between the stems of V1/V2 and V3 loops, which are exposed by conformational changes to gp120 after viral binding to the receptor/co-receptor complex (Tyssen et al., 2010; Connell and Lortat-Jacob, 2013).
Many viruses utilize negatively charged heparan sulfate proteoglycans (HS) on the cell plasma membrane as an initial means to scan the surface of the cell, and to attach in order to chaperone the virus onto the receptor complex prior to viral entry (Sarrazin et al., 2011; Connell and Lortat-Jacob, 2013). The receptor interactions occur in a sequential manner with virus-HS interactions preceding receptor/co-receptor binding, which combined leads to fusion of the viral envelope and the cell membrane.
Data indicate that astodrimer sodium-gp120 interaction may physically block initial HIV-1 association with HS and thereby block the subsequent virus-receptor complex functions. Virucidal assays of astodrimer sodium determined that it did not disrupt the HIV-1 particle or cause the loss of gp120 spike protein from the viral surface, suggesting that astodrimer sodium achieves irreversible inactivation of virus by tight binding to viral envelope proteins (Telwatte et al., 2011).
A report by Liu et al. (2020), described that densely glycosylated trimeric SARS-CoV-2 spike (S) protein subunit S1, which is important for receptor binding, binds to HS. To engage with the ACE2 receptor, the S protein undergoes a hinge-like conformational change that transiently hides or exposes the determinants of receptor binding (Wrapp et al., 2020). Recent studies have identified the binding of heparin to the receptor binding domain (RBD) of S1 resulting in a conformational change to the S protein (Mycroft-West et al., 2020a, b and c). Mutations in the S protein that are distal from the RBD also impact on viral transmission (Walls et al., 2020; Korber et al., 2020; Yuan et al., 2020). Non-RBD polybasic cleavage sites, including S1/S2 loop (Hoffmann et al., 2020a), have been described on SARS-CoV-2 S protein (Qiao and Olvera de la Cruz, 2020) and may also be a site of potential interaction with astodrimer sodium.
SARS-CoV-2 utilizes the ACE2 receptor for viral infection of host cells (Hoffmann et al., 2020b). Human CoV-NL63 also utilizes HS and ACE2 as its cellular receptor complex (Milewska et al., 2014). The importance of HS for viral infectivity was also demonstrated for close genetically related pseudo-typed SARS-CoV (Lang et al., 2011). The potential dependence of SARS-CoV-2 on HS for attachment and entry combined with antiviral data from other viruses suggest that negatively charged astodrimer sodium may have antiviral activity against SARS-CoV-2 in vitro by blocking the early virus-receptor recognition events.
Astodrimer sodium is a polyanionic dendrimer that has potential advantages over other technologies, including its lack of systemic absorption following topical application (Chen et al., 2009; O'Loughlin et al., 2010; McGowan et al., 2011). In addition, the SI of astodrimer sodium for SARS-CoV-2 is high and in a vaginal gel formulation (10 mg/mL), the compound has been shown to be safe and effective in phase 2 and large phase 3 trials for treatment and prevention of BV (Chavoustie et al., 2020; Waldbaum et al., 2020; Schwebke et al., 2021) and is now marketed in Europe, Australia, New Zealand and several countries in Asia. However, the gel formulation of astodrimer sodium is not appropriate for use to protect against SARS-CoV-2 infection.
5. Conclusions
Data from the current studies, taken together with studies of astodrimer sodium antiviral activity against HIV-1, and HSV-1 and -2, indicate that the compound exerts its antiviral activity against geographically diverse SARS-CoV-2 isolates by interfering with the early virus-cell recognition events. Astodrimer sodium is a potent virucidal agent that irreversibly reduces the infectivity of SARS-CoV-2 by >99.9% after 1 min of exposure to the virus, likely via tight binding to attachment proteins. These studies support astodrimer sodium being able to prevent early virus entry steps such as attachment, thereby reducing or preventing viral infection or cell-cell spread.
A topical antiviral agent such as astodrimer sodium that blocks binding of the virus to target cells could potentially be used against SARS-CoV-2 to augment other protective and therapeutic strategies.
The potent antiviral and irreversible virucidal activity of astodrimer sodium against SARS-CoV-2 warrants further investigation.
Funding
The research was funded by Starpharma Pty Ltd, which was responsible for study design, interpretation of data, writing the manuscript and decision to submit the article for publication. Matched funding for part of the research was also provided to Starpharma Pty Ltd by the Australian Medical Research Future Fund (MRFF) Biomedical Translation Bridge (BTB) Program, Award No. BTBR300093, delivered by MTPConnect in partnership with UniQuest. These groups had no role in the design, conduct, interpretation, write-up or publication of the research.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:J.R.A.P., J.K.F. and G.P.H. are paid employees of Starpharma Pty Ltd. A.C. and C.A.L. are paid consultants to Starpharma Pty Ltd.
Acknowledgments
The authors were fully responsible for the content, editorial decisions, and opinions expressed in the current article. The authors would like to acknowledge 360Biolabs Pty Ltd (Melbourne, Australia) for the conduct of the CPE assays and Scripps Research Institute for the conduct of the antiviral and virucidal assays. The authors would also like to thank the Peter Doherty Institute for Infection and Immunity (Melbourne, Australia) for the gift to 360Biolabs of SARS-CoV-2 hCoV-19/Australia/VIC01/2020 used in these studies.
Glossary
- ACE2
angiotensin converting enzyme 2
- BSA
bovine serum albumin
- BV
bacterial vaginosis
- CC50
50% cytotoxic concentration
- COVID-19
coronavirus disease 2019
- CPE
cytopathic effect
- EC50
50% effective concentration
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- HBEpC
human bronchial epithelial primary cells
- HIV-1
human immunodeficiency virus type 1
- HPV
human papillomavirus
- HRSV
human respiratory syncytial virus
- HS
heparan sulfate proteoglycan
- HSV
herpes simplex virus
- HTEpC
human tracheal epithelial primary cells
- ITS
insulin-transferrin-selenium
- LDH
lactate dehydrogenase
- MEM
minimal essential medium
- MOI
multiplicity of infection
- MTT
methylthiazolyldiphenyl-tetrazolium bromide
- NC
not calculated
- OD
optical density
- pfu
plaque forming unit
- RBD
receptor binding domain
- S protein
spike protein
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SD
standard deviation
- SHIV
simian-human immunodeficiency virus
- SI
selectivity index
- TPCK
L- (tosylamido-2-phenyl) ethyl chloromethyl ketone
- UPLC
ultra-high-performance liquid chromatography
References
- Bernstein D.I., Stanberry L.R., Sacks S., Ayisi N.K., Gong Y.H., Ireland J., Mumper R.J., Holan G., Matthews B., McCarthy T., Bourne N. Evaluations of unformulated and formulated dendrimer-based microbicide candidates in mouse and Guinea pig models of genital herpes. Antimicrob. Agents Chemother. 2003;47(12):3784–3788. doi: 10.1128/aac.47.12.3784-3788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S., Jonsson C.B., Taylor S.L., Figueroa J.M., Vanesa A.D., Palacios C., Vega J.C. Iota-carrageenan and xylitol inhibit SARS-CoV-2 in cell culture. 2020. bioRxiv 2020.08.19.225854. [DOI] [PMC free article] [PubMed]
- Chavoustie S.E., Carter B.A., Waldbaum A.S., Donders G.G.G., Peters K.H., Schwebke J.R., Paull J.R.A., Price C.F., Castellarnau A., McCloud P., Kinghorn G.R. Two phase 3, double-blinded, placebo-controlled studies of the efficacy and safety of Astodrimer 1% Gel for the treatment of bacterial vaginosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020;245:13–18. doi: 10.1016/j.ejogrb.2019.11.032. [DOI] [PubMed] [Google Scholar]
- Chen M.Y., Millwood I.Y., Wand H., Poynten M., Law M., Kaldor J.M., Wesselingh S., Price C.F., Clark L.J., Paull J.R., Fairley C.K. A randomized controlled trial of the safety of candidate microbicide SPL7013 gel when applied to the penis. J. Acquir. Immune Defic. Syndr. 2009;50(4):375–380. doi: 10.1097/QAI.0b013e318198a7e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T., Chattopadhyay K., Marschall M., Karmakar P., Mandal P., Ray B. Focus on antivirally active sulfated polysaccharides: from structure-activity analysis to clinical evaluation. Glycobiology. 2009;19(1):2–15. doi: 10.1093/glycob/cwn092. [DOI] [PubMed] [Google Scholar]
- Gong E., Matthews B., McCarthy T., Chu J., Holan G., Raff J., Sacks S. Evaluation of dendrimer SPL7013, a lead microbicide candidate against herpes simplex virus. Antivir. Res. 2005;68(3):139–146. doi: 10.1016/j.antiviral.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446. doi: 10.1016/j.cell.2020.05.042. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.H., Emau P., Cairns S., Flanary L., Morton W.R., McCarthy T., Tsai C.C. SPL7013 gel as a topical microbicide for prevention of vaginal transmission of SHIV89.6P in macaques. AIDS Res. Hum. Retrovir. 2005;21(3):207–213. doi: 10.1089/aid.2005.21.207. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D., Partridge D.G., Evans C.M., de Silva T.I., LaBranche C.C., Montefiori D.C. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020. [DOI]
- Lackman-Smith C., Osterling C., Luckenbaugh K., Mankowski M., Snyder B., Lewis G., Paull J., Profy A., Ptak R.G., Buckheit R.W., Jr., Watson K.M., Cummins J.E., Jr., Sanders-Beer B.E. Development of a comprehensive human immunodeficiency virus type-1 screening algorithm for discovery and preclinical testing of topical microbicides. Antimicrob. Agents Chemother. 2008;52(5):1768–1781. doi: 10.1128/AAC.01328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PloS One. 2011;6(8) doi: 10.1371/journal.pone.0023710. e23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Chopra P., Li X., Wolfert M.A., Tompkins S.M., Boons G.J. SARS-CoV-2 spike protein binds heparan sulfate in a length- and sequence-dependent manner. 2020. bioRxiv. [DOI]
- McCarthy T.D., Karellas P., Henderson S.A., Giannis M., O'Keefe D.F., Heery G., Paull J.R.A., Matthews B.R., Holan G. Dendrimers as drugs: discovery and preclinical and clinical development of dendrimer microbicides for HIV and STI prevention. Mol. Pharm. 2005;2(4):312–318. doi: 10.1021/mp050023q. [DOI] [PubMed] [Google Scholar]
- McGowan I., Gomez K., Bruder K., Febo I., Chen B.A., Richardson B.A., Husnik M., Livant E., Price C., Jacobson C. Phase 1 randomized trial of the vaginal safety and acceptability of SPL7013 gel (VivaGel®) in sexually active young women (MTN-004) AIDS. 2011;25(8):1057–1064. doi: 10.1097/QAD.0b013e328346bd3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88(22):13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C., Su D., Elli S., Li Y., Guimond S., Miller G., Turnbull J., Yates E., Guerrini M., Fernig D., Lima M., Skidmore M. The 2019 coronavirus (SARS-CoV-2) surface protein (spike) S1 receptor binding domain undergoes conformational change upon heparin binding. 2020. bioRxiv. [DOI]
- Mycroft-West C.J., Su D., Li Y., Guimond S., Rudd T.R., Elli S., Miller G., Nunes Q.M., Procter P., Bisio A., Forsyth N.R., Turnbull J.E., Guerrini M., Fernig D., Yates E.A., Lima M.A., Skidmore M.A. SARS CoV-2 spike S1 receptor binding domain undergoes conformational change upon interaction with low molecular weight heparins. 2020. bioRxiv. [DOI]
- Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Guimond S., Miller G., Meneghetti M.C.Z., Nader H.B., Li Y., Nunes Q.M., Procter P., Mancini N., Clementi M., Forsyth N.R., Turnbull J.E., Guerrini M., Fernig D., Vicenzi E., Yates E.A., Lima M.A., Skidmore M.A. Heparin inhibits cellular invasion of SARS-CoV-2: structural dependence on the interaction of the surface protein (spike) S1 receptor binding domain with heparin. 2020. bioRxiv 2020.2004.2028.066761. [DOI] [PMC free article] [PubMed]
- O'Loughlin J., Millwood I.Y., McDonald H.M., Price C.F., Kaldor J.M., Paull J.R.A. Safety, tolerability, and pharmacokinetics of SPL7013 gel (VivaGel®): a dose ranging phase 1 study. Sex. Transm. Dis. 2010;37(2):100–104. doi: 10.1097/OLQ.0b013e3181bc0aac. [DOI] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., Sidorov I., Snijder J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaption and cytopathology. J. General Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels R., Balzarini J., Baba M., Snoeck R., Schols D., Herdewijn P., Desmyter J., De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods. 1988;20(4):309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Dubois J., Julien T., Traversier A., Dulière V., Brun P., Lina B., Rosa-Calatrava M., Terrier O. In Vitro Evaluation of Antiviral Activity of Single and Combined Repurposable Drugs against SARS-CoV-2. Advance online publication; 2020. Antiviral Res. 104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao B., Olvera de la Cruz M. The distal polybasic cleavage sites of SARS-CoV-2 spike protein enhance spike protein-ACE2 binding. bioRxiv June. 2020;10 doi: 10.1101/2020.06.09.142877. [DOI] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W.C., Esko J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011;3(7) doi: 10.1101/cshperspect.a004952. a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwebke J.R., Carter B.A., Waldbaum A.S., Price C.F., Castellarnau A., Paull J.R.A., McCloud P., Kinghorn G.R. A phase 3, randomized, controlled trial of Astodrimer 1% Gel for preventing recurrent bacterial vaginosis. Eur. J. Obstet. Gynecol. Reprod. Biol. X. 2021;10 doi: 10.1016/j.eurox.2021.100121. 100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telwatte S., Moore K., Johnson A., Tyssen D., Sterjovski J., Aldunate M., Gorry P.R., Ramsland P.A., Lewis G.R., Paull J.R.A., Sonza S., Tachedjian G. Virucidal activity of the dendrimer microbicide SPL7013 against HIV-1. Antivir. Res. 2011;90(3):195–199. doi: 10.1016/j.antiviral.2011.03.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssen D., Henderson S.A., Johnson A., Sterjovski J., Moore K., La J., Zanin M., Sonza S., Karellas P., Giannis M.P., Krippner G., Wesselingh S., McCarthy T., Gorry P.R., Ramsland P.A., Cone R., Paull J.R.A., Lewis G.R., Tachedjian G. Structure activity relationship of dendrimer microbicides with dual action antiviral activity. PloS One. 2010;5(8) doi: 10.1371/journal.pone.0012309. e12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Worm S.H., Eriksson K.K., Zevenhoven J.C., Weber F., Zust R., Kuri T., Dijkman R., Chang G., Siddell S.G., Snijder E., Thiel V., Davidson A.D. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PloS One. 2012;7(3) doi: 10.1371/journal.pone.0032857. e32857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbaum A.S., Schwebke J.R., Paull J.R.A., Price C.F., Edmondson S.R., Castellarnau A., McCloud P., Kinghorn G.R. A phase 2, double-blind, multicenter, randomized, placebo-controlled, dose-ranging study of the efficacy and safety of Astodrimer Gel for the treatment of bacterial vaginosis. PloS One. 2020;15(5) doi: 10.1371/journal.pone.0232394. e0232394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M./., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]