Abstract
STUDY QUESTION
Are genes known to be involved in somatic cell ageing, particularly related to longevity pathways, associated with the accelerated ageing process of the ovary?
SUMMARY ANSWER
Growth, metabolism, and cell-cycle progression-related pathways that are involved in somatic cell ageing are also associated with ovarian ageing.
WHAT IS KNOWN ALREADY
Ovarian ageing is characterized by a gradual decline in ovarian follicle quantity, a decline in oocyte quality, and lower chances of pregnancy. Genetic pathways modulating the rate of somatic cell ageing have been researched intensively. Ovarian ageing does not follow the same timeline as somatic cell ageing, as signs of ovarian ageing occur at a younger female age, while the somatic cells are still relatively young. It is not known whether the generally recognized somatic cell longevity genes also play a role during ovarian ageing. Looking at somatic cell longevity genes can lead to new hypotheses and possible treatment options for subfertility caused by ovarian ageing.
STUDY DESIGN, SIZE, DURATION
In this observational study, we analysed a dataset of individual gene expression profiles of 38 germinal vesicle (GV) oocytes from 38 women aged between 25 and 43 years. We correlated female age (calendar age in years) and biological age (factors known to be associated with ovarian ageing such as dosage of FSH needed for ovarian hyperstimulation, and antral follicle count (AFC)) with gene expression signatures of longevity pathways.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Transcripts of 38 GV oocytes were used for individual gene expression analysis. R version 3.5.1 was used to process and analyse data. The GeneAge database (build 19) was used to obtain mouse ageing-related genes. Human to mouse orthologues were obtained using the R package biomaRt. Correlations and significance between gene expression data and age were tested for using Pearson's product moment correlation coefficient using ranked expression data. Distributions were compared with an ANOVA, and the Tukey Honest Significant Difference method was used to control for the Type I error rate across multiple comparisons.
MAIN RESULTS AND THE ROLE OF CHANCE
Of the 136 genes in the GeneAge database, the expression of 15 anti-longevity genes identified in oocytes showed a positive correlation with female calendar age and FSH dosage administered during ICSI treatment, and a negative correlation with AFC. Expression of 32 pro-longevity genes was negatively correlated with calendar age and FSH dosage, and positively correlated with AFC. In general, anti- and pro-longevity genes changed in opposing directions with advancing maternal age in oocytes. Notably, the anti-longevity genes include many ‘growth’-related genes involved in the mechanistic target of rapamycin (mTOR) Complex 1 pathway, such as EIF5A2, EIF3H, EIF4E, and mTOR. The pro-longevity genes include many cell-cycle progression-related genes involved in DNA damage repair (e.g. XRCC6, ERCC2, and MSH2) or cell-cycle checkpoint regulation genes (e.g. ATM, BRCA1, TP53, TP63, TP73, and BUB1B).
LIMITATIONS, REASONS FOR CAUTION
Using mature oocytes instead of GV-stage oocytes discarded from ICSI treatments may provide different results. No correction for multiple testing was carried out on individual genes because a small set of longevity-related genes was selected a priori for the analysis. The global trend was corrected for multiple testing and remained significant. This work was an observational study and, as no additional experimental work was performed, the associations described do not directly demonstrate the involvement of such genes in oocyte ageing.
WIDER IMPLICATIONS OF THE FINDINGS
Growth, metabolism, and cell-cycle progression-related pathways that are known to be involved in somatic cell ageing were associated with ovarian ageing. If experimental data are obtained to support these associations, we suggest that interventions known to modulate these processes could benefit women suffering from ovarian ageing.
STUDY FUNDING/COMPETING INTEREST(S)
G.E.J. is supported by a VENI grant from ZonMw (https://www.zonmw.nl). Work in the Houtkooper group is financially supported by an ERC Starting grant (No. 638290), a VIDI grant from ZonMw (No. 91715305), and the Velux Stiftung (No. 1063). M.G. declares several research and educational grants from Guerbet, Merck and Ferring (all location VUmc), outside the scope of the submitted work. The other authors report no competing interest
TRIAL REGISTRATION NUMBER
N/A.
Keywords: ovarian ageing, oocytes, oocyte quality, transcriptome, longevity pathways, female fertility
WHAT DOES THIS MEAN FOR PATIENTS?
When women become older, their chances of becoming pregnant and having a healthy live-birth decrease. However, when older women receive young donor oocytes, their pregnancy chances are the same as the donor’s age. We, therefore, investigated the differences between old and young oocytes. We compared a gene expression database of genes that are known to be involved in general ageing with the gene expression profiles of young and old oocytes. It appeared that processes that are involved in general ageing of somatic cells and tissues, like growth, metabolism and cell-cycle progression, are also associated with the lowered chance of pregnancy in women above 35 years of age. This knowledge can potentially be used, following further confirmatory research, to develop treatments to increase the chances of women of older age to have a healthy live-birth.
Introduction
Ageing affects both physiological and pathophysiological changes in the human body (Niccoli and Partridge, 2012). The mechanisms involved in somatic cell ageing are well studied and include nine known hallmarks: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, cellular senescence, stem cell exhaustion, altered intercellular communication, and mitochondrial dysfunction (Lopez-Otin et al., 2013). Several genetic pathways involved in the hallmarks of ageing are known to influence age, in that they can accelerate ageing (anti-longevity) or decelerate ageing (pro-longevity) and, therefore, decrease or increase lifespan. Studying these anti- and pro-longevity pathways has led to the development of well-studied interventions in somatic cell ageing. These interventions include, for example, a caloric restriction diet and inhibition of the mechanistic target of rapamycin (mTOR) pathway, which are commonly believed to optimize longevity (Fontana et al., 2010; Johnson et al., 2013). However, the same interventions are considered to be suboptimal for fecundity (Fontana et al., 2010; Bouwland-Both et al., 2013; Johnson et al., 2013). This is in line with the disposable soma theory of ageing, which suggests that an evolutionary trade-off between somatic maintenance and reproduction exists (Kirkwood, 1977, 2002). This apparent contradiction could be caused by a different ageing mechanism in the germline compared to somatic cells.
In humans, the decline in quantity and quality of the oocytes, often referred to as ovarian ageing, generally becomes apparent as early as the female age of approximately 35 years, which is much sooner than pathologies generally manifest themselves in somatic cell ageing (Navot et al., 1991; Li et al., 2012). This suggests a different ageing mechanism, but it could also be that oocytes undergo a kind of accelerated ageing or that the effects of ageing assert themselves more strongly in these cells. The effect of ovarian ageing is a decrease in pregnancy chances, while the chances of pregnancy loss and chromosomal aberrations in pregnancy increase (Schieve et al., 2003; Hassold and Hunt, 2009; McLernon et al., 2016). Mechanisms involved in ovarian ageing have been less thoroughly investigated, and are, therefore, less well understood, compared to somatic cell ageing. For these reasons, we wondered what the ovarian ageing field could learn from the well-studied field of somatic cell ageing.
The present observational study investigates the possible commonalities between ovarian ageing and somatic cell ageing by searching for anti- and pro-longevity pathways known for their roles in somatic cell ageing, by comparing a database containing individual transcriptome datasets of single human germinal vesicle (GV) oocytes from women aged 25–43 years (Smits et al., 2018) with a database of genes known to be involved in somatic cell ageing (Tacutu et al., 2013).
Materials and methods
Sample collection and processing
In 2018, our research group collected and analysed the transcriptional profiles in immature GV oocytes (Smits et al., 2018). Briefly, RNA was isolated and converted to cDNA. The cDNA was labelled and hybridized to microarrays (Agilent Technologies, Santa Clara, CA, USA, https://www.agilent.com/, design ID 028004, 4 × 180 K format containing ∼42 000 unique probes) and these were scanned and processed. Microarray data were then analysed with the Limma package (Bioconductor Limma package, R Foundation for Statistical Computing, Vienna, Austria, https://bioconductor.org/packages/limma/). Probes were annotated with the annotation file of the array designed by the MicroArray Department (MAD) of the Microarray facility of the University of Amsterdam, The Netherlands, and EntrezIDs and Ensemble GeneIDs were obtained with BioMart using the Gene Symbols as input. This provided final oocyte transcriptome data comprising the relative abundance values of transcripts annotated at the gene level, which were further analysed in the present study.
The final oocyte transcriptome dataset included 38 different GV oocytes belonging to women with an age range of 25–43 years. These were donated after ICSI treatment according to local protocol in the Amsterdam UMC in the Netherlands. Data were available on oocyte donors’ calendar age (in years), and factors that are associated with biological age. These factors included the dosage of FSH administered during ovarian hyperstimulation as part of the ICSI treatment and the antral follicle count (AFC) measured by ultrasonography before ovarian hyperstimulation.
Data analysis
R version 3.5.1 was used to process and analyse data (R Core Team, 2018) (R Foundation for Statistical Computing, Vienna, Austria; https://www.r-project.org/). The GeneAge database build 19 (release date 24 June 2017) was used to obtain mouse ageing-related genes and ‘pro’ and ‘anti’ longevity genes were selected from the ‘Longevity Influence’ data column (Tacutu et al., 2013). Human to mouse orthologues were obtained using the R package biomaRt (Durinck et al., 2009). Correlations and significance between gene expression data and female calendar age, FSH dosage administered during treatment and AFC were tested for using Pearson's product moment correlation coefficient using ranked expression data. Distributions were compared with an ANOVA, and the Tukey Honest Significant Difference method was used to control for the Type I error rate across multiple comparisons. Results were deemed significant with a P value less than 0.05.
Ethical approval
All experiments were performed according to rules and regulations of the local ethical committee.
Results
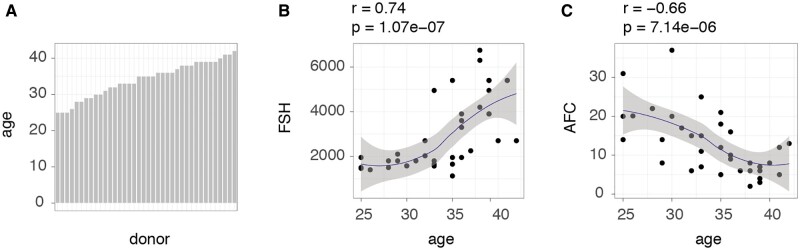
For this study, we used an oocyte gene expression dataset obtained from a population of women aged 25–43 years (Fig. 1A) (Smits et al., 2018). In this study population, we found advanced female age to be associated with an increase in dosage of FSH (Pearson’s correlation r = 0.74, P-value 1.07e−07) administered during ovarian hyperstimulation (Fig. 1B), and with a decline in AFC (Pearson’s correlation r −0.66 and P-value 7.14e−06) (Fig. 1C), two parameters associated with ovarian ageing (Podfigurna et al., 2018).
Figure 1.
Demographics and age-related changes of study participants. (A) Stratification of ages of participants, and (B, C) how donor age is interrelated with FSH (IU) and antral follicle count (AFC). FSH indicates the dosage needed for ovarian hyperstimulation. Age designates the donor’s age in years. Correlations (r) and P values (P) between individuals’ ages and either FSH or AFC levels were assessed using Pearson's product moment correlation coefficient.
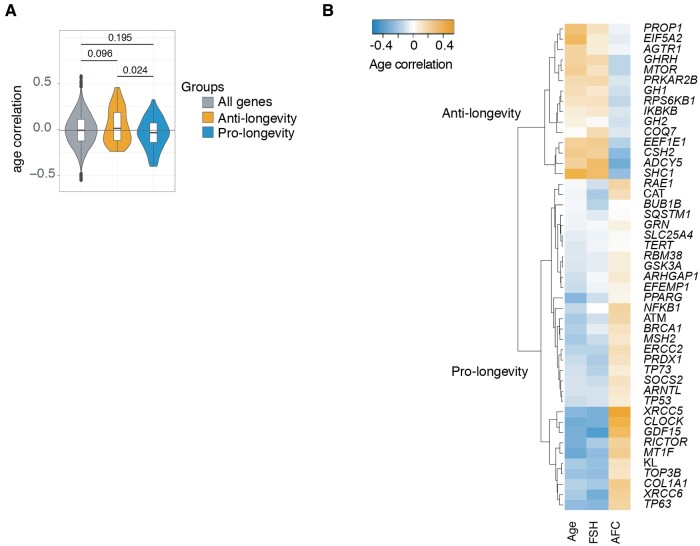
To assess how known longevity regulators are differentially expressed in relation to female age, FSH dosage administered, and AFC, we accessed a repository of age-related genes using the GeneAge database (Tacutu et al., 2013). We focused on mammalian genes, specifically using observations made in mice, since they had clear annotations and references to the literature for genes that were either pro-longevity (e.g. extending mouse lifespan when overexpressed) or anti-longevity (e.g. extending mouse lifespan when knocked out). Mouse genes present in GeneAge were converted to their human orthologues and compared with our human transcriptome datasets. We found that 116 of these were present in our transcriptome data, of which 72 were associated with a pro-longevity phenotype, while 39 were associated with an anti-longevity phenotype (Supplementary Table SI). The remaining five genes had no longevity phenotype, although are known to play a role during somatic ageing in mammals.
For all these 116 genes, we checked the correlation of their gene expression levels from the Smits et al. (2018) microarray data with the age of the women, by assessing ranked expression data using Pearson's product moment correlation coefficient r, which is the measure of linear correlation between the expression data and individuals’ ages (Supplementary Table SI). We furthermore tested if anti- and pro-longevity genes, annotated for their influence in general somatic ageing, also correlated with ovarian ageing. This was achieved by comparing the correlations with age of all 39 anti- and 72 pro-longevity genes (Fig. 2A, orange and blue). We found that when we compare the gene expression trend of anti- and pro-longevity genes to the general gene expression trend of all 15 357 genes of the dataset (Smits et al., 2018), anti-longevity genes tended to increase in abundance with ovarian ageing (P = 0.096) while pro-longevity genes appeared to decrease with age (P = 0.195), and it was clear that, in relation to age, anti- and pro-longevity genes changed significantly in opposing directions (P = 0.024) (Fig. 2A).
Figure 2.
Ageing regulation of anti- and pro-longevity genes in human oocyte transcriptomes. (A) Distributions of correlations of all genes (grey), GeneAge anti-longevity genes (orange) and GeneAge pro-longevity genes (blue) and their significant differences. Distributions were compared with an ANOVA, and the Tukey Honest Significant Difference method was used to control for the Type I error rate across multiple comparisons. The 0.024 P value denotes a significant difference between how anti-longevity genes and pro-longevity genes associate with age, which is in opposing directions. The changes in direction are opposite to what would be required for a longevity benefit. (B) Selection of GeneAge anti- and pro-longevity genes that correlated with age in a manner opposite to what would be required for a longevity benefit. Correlations between individuals’ ages and ranked expression data were assessed using Pearson's product moment correlation coefficient.
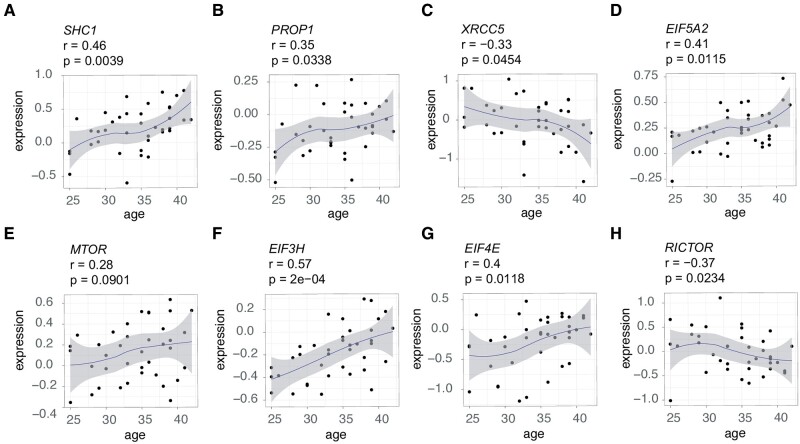
Strikingly, not only did we observe a correlation with increasing calendar age but also we observed a positive correlation with the dosage of FSH administered during treatment, and a negative correlation with AFC in 15 anti-longevity genes, and we observed the opposite trend in 32 pro-longevity genes, which were negatively correlated with calendar age and FSH dosage, and positively correlated with AFC (Fig. 2B). Below, we highlight some of the most interesting genes identified in this analysis. For example, the anti-longevity gene SHC-transforming protein 1 (SHC1) increased in abundance with increasing female age (r = 0.46, P = 0.0039; Fig. 3A). Likewise, the anti-longevity gene PROP paired-like homeobox 1 (PROP1) was also increasing with age (r = 0.35, P = 0.0338; Fig. 3B). The reverse was also true, as we found that the expression of pro-longevity genes positively correlated with female calendar age and FSH dosage administration, and negatively with AFC (Fig. 2B). For example, we found that the pro-longevity gene X-ray repair cross complementing 5 (XRCC5) decreased with advancing female age (r = −0.33, P = 0.0454; Fig. 3C).
Figure 3.
Gene expression levels in germinal vesicle oocytes from women of different ages. (A–D) Correlation of mRNA expression levels of selected genes with age. Genes were selected based on the observation that they changed in an opposite direction to what would be required for a longevity benefit. This includes (A) an increase in SHC adaptor protein 1 (SHC1), (B) an increase in PROP paired-like homeobox 1 (PROP1), (C) a decrease in the DNA repair related gene X-ray repair cross complementing 5 (XRCC5), and (D) an increase in the eukaryotic translation initiation factor 5A2 (EIF5A2). (E–H) Expression levels in oocytes with age of a selection of ‘growth’-related genes involved in ageing and longevity. Genes were selected to highlight that multiple other genes in the mechanistic target of rapamycin (mTOR) pathway are upregulated in ovarian ageing. This includes (E) MTOR itself, (F) EIF3H, (G) EIF4E, and (H) EIF5A2. Gene expression levels are relative expression from microarray data of oocytes, from Smits et al. (2018). Age designates the donor’s age in years. Correlations (r) and P values (P) between individuals’ ages and ranked expression data were assessed using Pearson's product moment correlation coefficient.
Furthermore, several anti-longevity genes of the mTOR pathway, involved in the regulation of growth, were increasing in abundance with female age (Fig. 2B). Among the genes was the eukaryotic translation initiation factor 5A2 (EIF5A2) (r = 0.41, P = 0.0115; Fig. 3D). MTOR itself showed a trend of increasing expression with female age, although this was not significant (r = 0.28, P = 0.0901; Fig. 3E). Furthermore, we found other elongation initiation factors in the transcriptome, e.g. EIF3H and EIF4E (Fig. 3F and G), to also increase with increasing female age (r = 0.57, P = 2e−04; r = 0.4, P = 0.0118; respectively). On the other hand, pro-longevity genes were decreasing with advancing female age. For example, RPTOR Independent Companion Of mTOR Complex 2 (RICTOR), which is part of the mTOR Complex 2 (mTORC2), decreased with ovarian ageing (r = −0.37, P = 0.0234; Fig. 3H).
We also noted that genes involved in DNA damage repair and cell cycle checkpoint regulation were decreasing in expression with increasing female age (Fig. 2B). These cell cycle progression genes are all pro-longevity genes (Fig. 2B). While the individual correlations of many of these genes were not statistically significant, we noted the presence of many cell cycle progression related genes here. These include, for instance, X-ray repair cross complementing 6 (XRCC6), excision repair gene 2 (ERCC2), and MutS homolog 2 (MSH2), as well as many cell-cycle checkpoint regulation genes (e.g. ATM serine/threonine kinase (ATM), BRCA1 DNA repair associated (BRCA1), tumour proteins P53, P63 and P73 (TP53, TP63 and TP73) and BUB1 mitotic checkpoint serine/threonine kinase (BUB1B)). The fact that so many genes involved in cell-cycle progression and checkpoint regulation changed into the opposite direction of longevity, suggests that these mechanisms play a role in ovarian ageing, alongside the significant changes observed in the mTOR pathway-related genes.
Discussion
This research focused on identifying longevity pathways known for their role in somatic cell ageing in the oocytes of women of different ages and therefore finding communalities between somatic cell and ovarian ageing. Several anti- and pro-longevity pathways known for their roles in somatic cell ageing also seem to be involved in ovarian ageing. Relative to all genes in the oocyte ageing transcriptome dataset (Smits et al., 2018), gene expression profiles of genes involved in pro-longevity pathways decreased with age, while genes involved in anti-longevity pathways increased with age. Here, we found a variety of changes in genes belonging to pathways such as growth, cell-cycle progression, and stress resistance to associate with ovarian ageing. Although more empirical research is required, these pathways together may contribute to the decrease in oocyte quality that occurs with increasing female age.
We observed an increase in the expression of several anti-longevity genes in relation to increasing female age. For example, the genes SHC1 and PROP1 whose knockout increased life-span in mice (Brown-Borg et al., 1996). Interestingly, reduced SHC1 is believed to increase oxidative stress resistance, therefore, the increase occurring in ovarian ageing may suggest a decreased ability to protect against oxidative stress in the ageing oocytes. This is in line with a recent study in ovarian ageing describing an inactivation of antioxidant genes in aged monkey and human ovaries (Wang et al., 2020). Also EIF5A2, a protein and ‘growth’-related gene, which in somatic cells leads to chromosomal instability, increased with female age (Chen et al., 2011; Janssens et al., 2015). Interestingly, PROP1 mutations inhibit the release of growth hormone and LH and FSH, which together are responsible for ovulation (Brown-Borg et al., 1996; Stelzer et al., 2016). On the other hand, pro-longevity genes were decreasing with increasing female age, for example XRCC5, a gene whose knockout in mice results in signs of an early onset of senescence (Vogel et al., 1999).
Intrigued by seeing that EIF5A2 was upregulated in ovarian ageing, we looked further to see if we could identify other protein translation-related genes changing with ovarian ageing. We found MTOR, the canonical regulator of growth and ageing, increasing with female age, although not significant. Furthermore, we found other EIFs (EIF3H and EIF4E) to significantly increase with female age and noted that the opposite happened to the pro-longevity gene RICTOR, part of the mTORC2 complex and whose deletion shortens lifespan in male mice (Lamming et al., 2014). These observations hint at a possibility of ‘growth’ pathway activation during ovarian ageing. In mice, mTOR activates the primordial follicles and, in this way, depletes the primordial follicle pool via the Akt pathway (Shah et al., 2018). In human, it is known that there is a relation between the depleted follicle pool and misbalance in sex hormones in women of older age (O'Connor et al., 2001). Together, these findings warrant further investigation into how the mTOR pathway could be involved in human ovarian ageing.
Several pro-longevity genes were cell-cycle progression related genes involved in DNA damage repair (e.g. XRCC6, ERCC2, and MSH2) or cell-cycle checkpoint regulation (e.g. ATM, BRCA1, TP53, TP63, TP73, and BUB1B). XRCC6, encoding the Ku70 protein, is a single-stranded DNA- and ATP-dependent helicase involved in repairing DNA double-strand breaks (Rathmell and Chu, 1994). The product of the ERCC2 gene is also an ATP-dependent DNA helicase. Polymorphisms of both the XRCC6 and ERCC2 genes give rise to several cancer types (Sturgis et al., 2002; Kiyohara and Yoshimasu, 2007). A study in oocytes of female rats showed a significant age-dependent decline in mRNA expression levels of several DNA repair genes, including ERCC2 (Govindaraj et al., 2015). Lastly, MSH2, which decreased with increasing female age, is responsible for nuclear mismatch repair (Wang et al., 1999). MutL homologs, for example, Mlh3, bind to MutS homologs like MSH2. One study in mice showed that Mlh3−/− mice are sterile, unable to complete Meiosis 1 in oocytes, suggesting that this pathway is essential in meiosis (Lipkin et al., 2002). While the individual correlations of these genes to age were not significant, together they may represent a decreased ability to achieve genomic stability.
Cell-cycle checkpoint regulation genes that were decreasing with increasing female age were involved in the ATM-dependent double strand breaks repair pathway. Recently a review was published on this pathway as an important driver for ovarian ageing (Turan and Oktay, 2020). ATM is a kinase that, when phosphorylated, activates many downstream targets, such as BRCA1 and TP53, TP63, and CHK2, all of which were found to be decreasing in GV oocytes with increasing female age (Wells et al., 2005). BRCA1 mutations are common in women suffering from ovarian and breast cancer, and these women also show an accelerated ovarian ageing phenotype (Turan and Oktay, 2020). TP53, TP63, and TP73 are homologs of tumour suppressor genes and all play a role in meiotic checkpoint regulation. Both TP53 and TP63 are activated by CHK2 as a result of meiotically induced double-stranded breaks, which is shown to result in oocyte apoptosis in mice (Bolcun-Filas et al., 2014). Mice with a knock down of the P53 and P73 genes show oocyte maturation problems, an increase of chromosomal abnormalities in offspring and decrease in pregnancy rates and live births, or they are sterile (Hu et al., 2007; Tomasini et al., 2009). TP73 seems to also regulate BUB1 and other genes in the spindle assembly checkpoint complex (Tomasini et al., 2009). BUB1, which also decreased, checks if the chromatids are correctly attached to the spindle microtubules (Marston and Wassmann, 2017). If this is not done correctly, this can give rise to chromosome aggregation errors, for example, non-disjunction related aneuploidies (Cimadomo et al., 2018). Together, the trends observed in the cell-cycle progression genes changing with ovarian ageing could account for the increase in female age-related aneuploidies observed in clinically recognized pregnancies, pregnancy loss tissues, and pre-implantation embryos (Regan and Rai, 2000; Hassold and Hunt, 2009; Nagaoka et al., 2012).
Several considerations are warranted with our study. First, the database used in this analysis contained transcripts of single GV oocytes (Smits et al., 2018). The use of mature Metaphase II oocytes (MII), rather than GV oocytes, might have produced different results. MII oocytes are used for treatment, and, therefore, we were not able to use them for research purposes. Furthermore, when we previously compared transcripts of single GV oocytes in relation to age and corrected for multiple-hypothesis testing across the entire genome, we could not identify significantly differentially expressed genes (Smits et al., 2018). In this article, we selected with a priori knowledge only anti- and pro-longevity genes known to play a role in somatic cell ageing and, therefore, did not correct for multiple testing. The hypotheses we generate in this article therefore serve to point towards possible links between general somatic and ovarian ageing, and require further investigation. Notably, our work demonstrates associations with, rather than causal involvement of the genes assessed. Nonetheless, our results have provided novel insights that may generate new hypotheses that can be tested in future research.
Our study shows that longevity pathways that have been extensively researched in somatic cell ageing are also associated with ovarian ageing. We suggest that the totality of small changes in anti- and pro-longevity genes may aggregate into an acceleration of the ovarian ageing phenotype. Besides that, GV oocytes are in dictate arrest, which could make them extra vulnerable to age-related changes. Therefore, following additional research to confirm our findings, interventions that are known to modulate somatic cell ageing may, thus, be made of use to benefit ovarian ageing. Different longevity pathways were up- or downregulated in a way that was detrimental for longevity, including the regulation of growth: the MTOR pathway and cell-cycle regulation pathways involved in preventing chromosomal errors. These pathways could provide an interesting avenue for future research, with experiments that can directly confirm our observational findings. It could also be interesting for future research in ovarian ageing to look more into the recent developments in the somatic cell ageing field, since the mechanisms behind both might not be so different.
Supplementary data
Supplementary data are available at Human Reproduction Open online.
Data availability
The transcriptomics data underlying this article are available through the Smits et al. (2018) publication in Mol Hum Reprod, 24(10):469–477. https://doi.org/10.1093/molehr/gay036.
Supplementary Material
Acknowledgements
The authors acknowledge Kai Mee Wong and Cindy M. Korver for all effort put into preparing the oocytes and would specifically like to thank Kai Mee Wong for building the GV gene expression database.
Authors’ roles
M.A.J.S. and G.E.J. wrote the manuscript, and G.E.J. analysed the data. All authors were involved in the design of the study and reviewing of the manuscript.
Funding
G.E.J. is supported by a VENI grant from ZonMw (No. 09150161810014, https://www.zonmw.nl). Work in the Houtkooper group is financially supported by an ERC Starting grant (No. 638290), a VIDI grant from ZonMw (No. 91715305), and the Velux Stiftung (No. 1063).
Conflict of interest
M.G. declares several research and educational grants from Guerbet, Merck and Ferring (all location VUmc), outside the scope of the submitted work.
References
- Bolcun-Filas E, Rinaldi VD, White ME, Schimenti JC.. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 2014;343:533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwland-Both MI, Steegers-Theunissen RP, Vujkovic M, Lesaffre EM, Mook-Kanamori DO, Hofman A, Lindemans J, Russcher H, Jaddoe VW, Steegers EA.. A periconceptional energy-rich dietary pattern is associated with early fetal growth: the Generation R study. BJOG 2013;120:435–445. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A.. Dwarf mice and the ageing process. Nature 1996;384:33. [DOI] [PubMed] [Google Scholar]
- Chen M, Huang JD, Deng HK, Dong S, Deng W, Tsang SL, Huen MS, Chen L, Zan T, Zhu GX. et al. Overexpression of eIF-5A2 in mice causes accelerated organismal aging by increasing chromosome instability. BMC Cancer 2011;11:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L.. Impact of maternal age on oocyte and embryo competence. Front Endocrinol 2018;9:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD.. Extending healthy life span—from yeast to humans. Science 2010;328:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraj V, Keralapura Basavaraju R, Rao AJ.. Changes in the expression of DNA double strand break repair genes in primordial follicles from immature and aged rats. Reprod Biomed Online 2015;30:303–310. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P.. Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr Opin Pediatr 2009;21:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ.. p53 regulates maternal reproduction through LIF. Nature 2007;450:721–724. [DOI] [PubMed] [Google Scholar]
- Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V, Bischoff R, Wit EC, Veenhoff LM, Heinemann M.. Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 2015;4:e08527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M.. mTOR is a key modulator of ageing and age-related disease. Nature 2013;493:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB. Evolution of ageing. Mech Age Dev 2002;123:737–745. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. Evolution of ageing. Nature 1977;270:301–304. [DOI] [PubMed] [Google Scholar]
- Kiyohara C, Yoshimasu K.. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci 2007;4:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Mihaylova MM, Katajisto P, Baar EL, Yilmaz OH, Hutchins A, Gultekin Y, Gaither R, Sabatini DM.. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell 2014;13:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leridon H. Can assisted reproduction technology compensate for the natural decline in fertility with age? A model assessment. Hum Reprod 2004;19:1548–1553. [DOI] [PubMed] [Google Scholar]
- Li Q, Geng X, Zheng W, Tang J, Xu B, Shi Q.. Current understanding of ovarian aging. Sci China Life Sci 2012;55:659–669. [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED. et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 2002;31:385–390. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G.. The hallmarks of aging. Cell 2013;153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Wassmann K.. Multiple duties for spindle assembly checkpoint kinases in meiosis. Front Cell Dev Biol 2017;5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLernon DJ, Steyerberg EW, Te VE, Lee AJ, Bhattacharya S.. Predicting the chances of a live birth after one or more complete cycles of in vitro fertilisation: population based study of linked cycle data from 113 873 women. BMJ 2016;355:i5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA.. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 2012;13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navot D, Bergh PA, Williams MA, Garrisi GJ, Guzman I, Sandler B, Grunfeld L.. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet 1991;337:1375–1377. [DOI] [PubMed] [Google Scholar]
- Niccoli T, Partridge L.. Ageing as a risk factor for disease. Curr Biol 2012;22:R741–752. [DOI] [PubMed] [Google Scholar]
- O'Connor KA, Holman DJ, Wood JW.. Menstrual cycle variability and the perimenopause. Am J Hum Biol 2001;13:465–478. [DOI] [PubMed] [Google Scholar]
- Podfigurna A, Lukaszuk K, Czyzyk A, Kunicki M, Maciejewska-Jeske M, Jakiel G, Meczekalski B.. Testing ovarian reserve in pre-menopausal women: why, whom and how? Maturitas 2018;109:112–117. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing.Vienna, Austria: Foundation for Statistical Computing, 2018. [Google Scholar]
- Rathmell WK, Chu G.. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci USA 1994;91:7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan L, Rai R.. Epidemiology and the medical causes of miscarriage. Baill Best Pract Res Clin Obst Gynaecol 2000;14:839–854. [DOI] [PubMed] [Google Scholar]
- Schieve LA, Tatham L, Peterson HB, Toner J, Jeng G.. Spontaneous abortion among pregnancies conceived using assisted reproductive technology in the United States. Obst Gynecol 2003;101:959–967. [DOI] [PubMed] [Google Scholar]
- Shah JS, Sabouni R, Cayton Vaught KC, Owen CM, Albertini DF, Segars JH.. Biomechanics and mechanical signaling in the ovary: a systematic review. J Assist Reprod Genet 2018;35:1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits MAJ, Wong KM, Mantikou E, Korver CM, Jongejan A, Breit TM, Goddijn M, Mastenbroek S, Repping S.. Age-related gene expression profiles of immature human oocytes. Mol Hum Reprod 2018;24:469–477. [DOI] [PubMed] [Google Scholar]
- Stelzer GRR, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Iny ST, Nudel R, Lieder I, Mazor Y, Kaplan S. et al. The GeneCards suite: from gene data mining to disease genome sequence analysis. Curr Protoc Bioinformatics2016;54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- Sturgis EM, Dahlstrom KR, Spitz MR, Wei Q.. DNA repair gene ERCC1 and ERCC2/XPD polymorphisms and risk of squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg 2002;128:1084–1088. [DOI] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhaes JP.. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res 2013;41:D1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Rufini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G. et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA 2009;106:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan V, Oktay K.. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum Reprod Update 2020;26:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P.. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA 1999;96:10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N.. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA 1999;96:13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, Liu Z, Min Z, Hu H, Jing Y. et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell 2020;180:585–600.e519. [DOI] [PubMed] [Google Scholar]
- Wells D, Bermudez MG, Steuerwald N, Thornhill AR, Walker DL, Malter H, Delhanty JD, Cohen J.. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum Reprod 2005;20:1339–1348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomics data underlying this article are available through the Smits et al. (2018) publication in Mol Hum Reprod, 24(10):469–477. https://doi.org/10.1093/molehr/gay036.