Abstract
Macrophages are desirable targets for gene therapy of cancer and other diseases. Cationic diblock copolymers of polyethylene glycol (PEG) and poly-L-lysine (PLL) or poly{N-[N-(2-aminoethyl)-2-aminoethyl]aspartamide} (pAsp(DET)) are synthesized and used to form polyplexes with a plasmid DNA (pDNA) that are decorated with mannose moieties, serving as the targeting ligands for the C type lectin receptors displayed at the surface of macrophages. The PEG-b-PLL copolymers are known for its cytotoxicity, so PEG-b-PLL-based polyplexes are cross-linked using reducible reagent dithiobis(succinimidyl propionate) (DSP). The cross-linked polyplexes display low toxicity to both mouse embryonic fibroblasts NIH/3T3 cell line and mouse bone marrow-derived macrophages (BMMΦ). In macrophages mannose-decorated polyplexes demonstrate an ≈8 times higher transfection efficiency. The cross-linking of the polyplexes decrease the toxicity, but the transfection enhancement is moderate. The PEG-b-pAsp(DET) copolymers display low toxicity with respect to the IC-21 murine macrophage cell line and are used for the production of non-cross-linked pDNA-contained polyplexes. The obtained mannose modified polyplexes exhibit ca. 500-times greater transfection activity in IC-21 macrophages compared to the mannose-free polyplexes. This result greatly exceeds the targeting gene transfer effects previously described using mannose receptor targeted non-viral gene delivery systems. These results suggest that Man-PEG-b-pAsp(DET)/pDNA polyplex is a potential vector for immune cells-based gene therapy.
Keywords: cationic block copolymer, macrophage transfection, mannose, targeted gene delivery
1. Introduction
Gene therapy holds promise to improve therapeutic outcomes by replacing a defective gene or introducing a new gene that encodes a specific therapeutic protein.[1,2] This approach has attracted much attention for its possible application in treating human disorders.[3–6] More recently, macrophages have emerged as a target for gene therapy, in particular, in cancer. Macrophages are recruited to both the primary tumors and the metastatic sites with the tumor-associated macrophages (TAMs) comprising up to 50% of the tumor cells.[7] A mannose receptor (CD206) can be exploited to target TAMs since this receptor is abundantly presented in the tissue macrophages and dendritic cells and is highly expressed in the tumor microenvironment.[8] The receptor belongs to the lectin group (C type)[9] and is well-characterized.[10] It is shown to bind various carbohydrates and oligosaccharides, in particular, mannose either in a free form or conjugated to polymer scaffolds.[11] As of today, there is an extensive body of work using mannose as a targeting moiety to mannose macrophage receptor (MMR), in which various nanocarriers were employed for the delivery of small molecules,[12–16] proteins,[17,18] nucleic acids,[19–23] and structures like nanoparticles of different types[24–26] or supramolecular aggregates.[27] As far as the delivery of the plasmid DNA (pDNA) is concerned the most suitable vehicles are either lipoplexes or polyplexes. For example, in an early publication Ferkol used mannose poly-L-lysine (PLL) conjugates to produce polyplexes with pDNA for macrophage transfection, albeit the transfection efficacy was relatively low.[28] More recently a grafted polyethyleneimine (PEI) decorated by mannose was employed with a greater success for in vitro transfection of macrophages.[29] The mannose-conjugated branched PEI is currently marketed as an in vitro macrophage transfection reagent (under the brand name “jetPEI®-Macrophage”). Hashida et al. applied mannosylated PEI-coated liposomes for gene delivery in macrophages.[30] Cho et al. used mannosylated chitosan-graft-PEI polymer for a gene transfer in the same type of blood cells.[31] Linear PEI conjugated to mannan[32] and branched PEI conjugated to polyaspartamide derivatives modified with mannose and folic acid[33] were also explored as gene vectors. However, PEI homopolymer is not a good choice for in vivo transfection due to its poor bioavailability and toxicity. The polyethylene glycol (PEG) conjugated polycations or cationic block copolymers are a better alternative since the pDNA polyplexes formed by these copolymers and more stable in biological milieu.[34] For example, David et al. reported on the delivery of polyion complexes formed by DNA and PEG-PEI block-copolymer in dendritic cells aiming via targeted mannose.[35] The transfection efficiency in macrophages has proven difficult, and the methods used are of low transfection efficiency or of high cellular toxicity.[36,37]
The essential issue in gene delivery is the rapid degradation of nucleic acid in the extracellular and intracellular spaces, which decreases the efficiency and duration for protein expression. The stabilization of the polyplexes prevents them from dissociation upon dilution or change of solution conditions. The disulfide cross-linking was successfully applied for improving the tolerability of the polyplexes against nucleases as well as stability against polyion exchange.[38] The introducing of the covalent cross-links decreases the charge density of the polycation and reduces its cytotoxicity. Previous studies demonstrated the reduced cytotoxicity of cross-linked branched PEI-based polyplexes compared to non-cross-linked particles.[39,40] The polyplexes formed from PEGylated poly(N-(2-hydroxypropyl)methacrylamide) (pHPMA) demonstrated reduced toxicity and excellent stability after introducing covalent cross-links.[41] The cellular toxicity of poly(2-hydroxypropilene imine) (PHPI)-based polyplexes was also reduced with the introduction of disulfide linkages.[42]
The DNA-contained polyplexes formed from cationic copolymers composed of PEG and PLL blocks were well-studied.[43] Previous works provided a deep insight into the DNA condensation behavior of PEG-b-PLL/pDNA polyplexes.[44,45] While the PEGylated PLL block copolymers were successfully applied for the transfection in vitro[46] and in vivo,[47] certain toxicity was demonstrated in each case. The employment of the reducible cross-linker could result in a decrease in toxicity of the PEG-b-PLL/pDNA polyplexes, making them a proper material for the designing of a platform for immune cells-based gene delivery.
Another way of reducing the cellular toxicity of the polyplexes is the use of highly biocompatible materials. A promising cationic block copolymer PEG-poly{N-[N-(2-aminoethyl)-2-aminoethyl]aspartamide} (PEG-b-pAsp(DET)) was developed in K. Kataoka’s lab.[48,49] This polymer has very low toxicity and is biodegradable. Because of its biocompatibility PEG-b-pAsp(DET)-based polyplexes can be applied at high ratios of the primary amino groups of the polymer to the phosphate groups of pDNA (N/P ratios), which usually leads to a more effective transfection.
In this study, we report on the synthesis and properties of macrophage-targeted transfection systems. Two mannosylated cationic block copolymers (PEG-b-PLL and PEG-b-pAsp(DET)), were synthetized by anionic ring opening polymerization using telechelic PEG amines as initiators. Mannose was further introduced as a targeting moiety at the free end of PEG. The resulting mannose-conjugated and non-conjugated polymers were characterized via 1H-NMR and gel permeation chromatography (GPC). These polycations were further used to condense pDNA to form polyplexes. The PEG-b-PLL-based polyplexes were cross-linked with dithiobis(succinimidyl propionate) (DSP). The PEG-b-pAsp(DET)-contained polyplexes were further used without any cross-links. The polyplexes were characterized and applied for targeted and non-targeted transfection.
2. Results
2.1. Synthesis of Man-PEG-b-PLL
A cationic block copolymer, PEG-b-PLL was synthetized by anionic ring opening polymerization of N-ε-benzyloxycarbonyl-L-lysine-N-carboxyanhydride 1 using telechelic precursor, propargyl-PEG-NH2 3 as a macroinitiator (Scheme 1). The synthetic procedure involves the formation of benzyloxycarbonyl-protected polymer followed by a deprotection step resulting in propargyl-PEG-b-PLL 4. A targeting ligand 2′-azidoethyl-O-α-D-mannopyranoside 6 was introduced via copper(I)-catalyzed alkyne-azide cycloaddition to obtain the final mannose modified product Man-PEG-b-PLL 7.
Scheme 1.
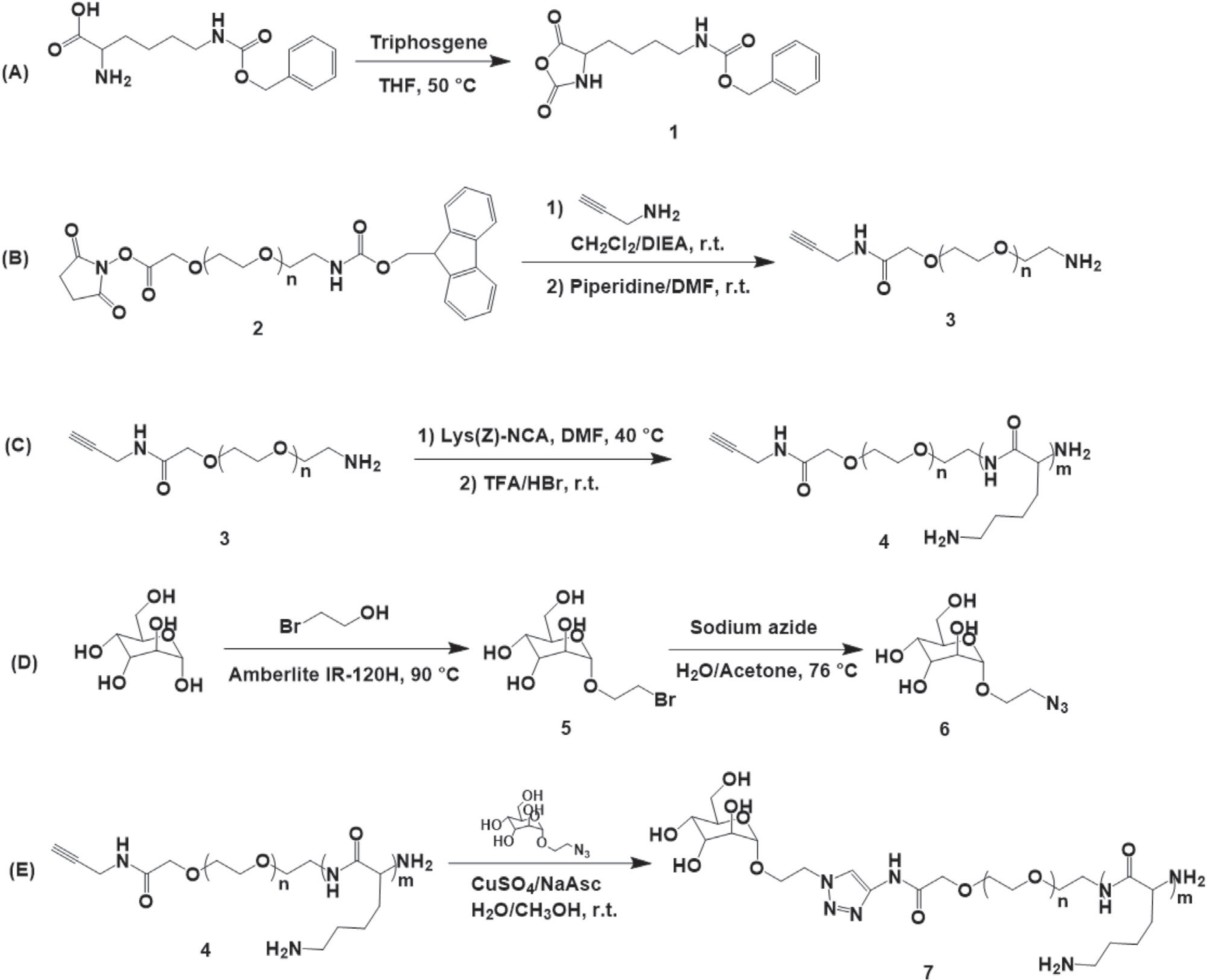
A) Synthesis of N-ε-benzyloxycarbonyl-L-lysine-N-carboxyanhydride. B) Synthesis of propargyl functionalized polyethylene glycol amine. C) Synthesis of propargyl-PEG-b-PLL block copolymer. D) Synthesis of 2′-azidoethyl-O-α-D-mannopyranoside. E) Conjugation of propargyl-PEG-b-PLL and 2′-azidoethyl-O-α-D-mannopyranoside via click reaction.
The DP of PLL block of polymer 4 was determined by 1H-NMR (Figure S1, Supporting Information) and calculated as the ratios of the integral intensity of methylene protons -CH2- δ = 3.6 of PEG to the intensity of benzyl protons -COOCH2C6H5 δ = 7.2 of the protective group before it was removed. The 4 was analyzed by GPC (Figure S2, Supporting Information) that determined the molecular weight and molecular distribution of this polymer. 1H-NMR analysis of 7 (Figure S3, Supporting Information) revealed the appearance of a characteristic signal of 1,2,3-triazole proton at δ = 7.8 ppm along with the disappearance of the peak corresponding to the propargyl proton of propargyl-PEG-b-PLL δ = 2.3. Based on these data we estimated the degree of conversion of 4 to 7 (Table 1). Combining the GPC and 1H-NMR analyses allowed us to assign the respective structures to 7 (Table 1).
Table 1.
Characterization of Man-PEG-b-PLL block copolymers.
| Composition | PDIa) | Mannose conjugation degreeb) [%] |
|---|---|---|
| Man-PEG114-b-PLL62 | 1.27 | 87 |
| Man-PEG114-b-PLL150 | 1.25 | 72 |
| Man-PEG114-b-PLL206 | 1.19 | 95 |
GPC PDI equals Mw/Mn
Conjugation degree is the fraction of block copolymers that contain mannose residue expressed on a percentage base of the total block copolymer amount.
2.2. Synthesis of Man-PEG-b-pAsp(DET)
A cationic block copolymer, PEG-b-pAsp(DET) was synthetized by an anionic ring opening polymerization of BLA-NCA 8 using an amino-terminated mPEG-NH2 9a as a macroinitiator (Scheme 2) as previously described by Kataoka et al.[49,51,52] To allow terminal conjugation of a targeting moiety a telechelic polymer alkyne-PEG-NH2 9b was also used (Scheme 2). The synthetic procedure involves the formation of intermediate benzyl-protected polymers, mPEG-b-PBLA 10a and alkyne-PEG-b-PBLA 10b, that was further modified by reacting with DET to obtain final polymers mPEG-b-pAsp(DET) 11a or alkyne-PEG-b-pAsp(DET) 11b. In the case of alkyne derivative 11b a targeting ligand was introduced by coupling it with α-D-Man-TEG-N3 via copper(I)-catalyzed alkyne-azide cycloaddition to obtain the final mannose modified product ManPEG-b-pAsp(DET) 12.
Scheme 2.
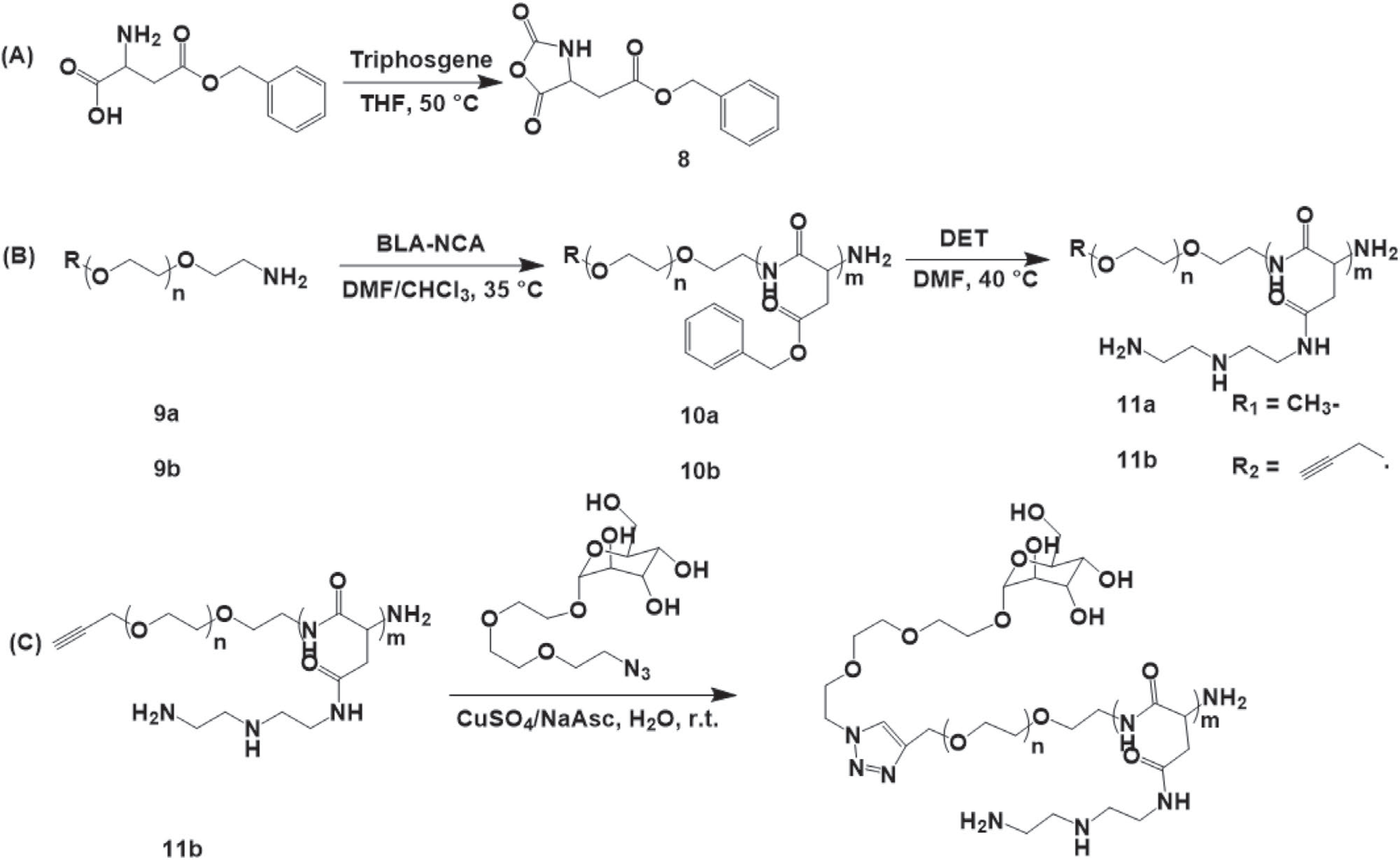
A) Synthesis of β-benzyl-L-aspartate-N-carboxyanhydride. B) Synthetic of mPEG-b-pAsp(DET) (R1) and alkyne-PEG-b-pAsp(DET) (R2). C) Conjugation of alkyne-PEG-b-pAsp(DET) and α-D-Man-TEG-N3 via click reaction.
The DP of the PBLA blocks of both synthesized polymers 10a and 10b was determined by 1H-NMR (Figure S4, Supporting Information) and calculated as the ratios of integral intensity of PBLA methylene protons -COOCH2C6H5 δ = 5.0 to the intensity of -CH3 protons of mPEG δ = 3.3 (10a) or alkyne proton of alkyne-PEG δ = 2.1 (10b). The effective 1H-NMR DP values of PBLA blocks were ≈56 and ≈40 for 10a and 10b, respectively. The results of GPC analysis (Figure S5, Supporting Information) of 10a and 10b showed that both polymers contain fractions of the main products with essentially the same Mn 15 360 Da and Mw/Mn 1.030 for 10a and 1.207 for 10b, and minor fractions of PBLA homopolymers (Mn of 2 480 and 3 290 Da, respectively).
The homopolymer admixture was completely removed after the conversion of the intermediates 10a and 10b to cationic block copolymers 11a and 11b by dialysis against a solution of 0.01 n HCl and distilled water using 10000 MWCO membrane. The 11a and 11b were analyzed again after the dialysis by GPC (Figure S7, Supporting Information) that: 1) confirmed the removal of the homopolymer admixtures; and 2) determined Mn 14 690 Da, Mw/Mn 1.018 for 11a and Mn 14 740 Da, Mw/Mn 1.036 for 11b. The full conversion of PBLA to pAsp(DET) in these copolymers was demonstrated by the complete disappearances of the peak “c” δ = 5.0 in the 1H-NMR spectra of 11a and 11b (Figure S6, Supporting Information). Based on these GPC and 1H-NMR analyses 11a and 11b were assigned the respective structures CH3-(CH2CH2O)114-p[Asp(DET)]50 and HC≡C-CH2O-(CH2CH2O)114-p[Asp(DET)]50.
Finally, 1H-NMR analysis of 12 (Figure S8, Supporting Information) revealed the appearance of a characteristic signal of 1,2,3-triazole proton at δ = 7.6 ppm along with the disappearance of a peak corresponding to the alkyne proton of alkyne-PEG δ = 2.3. Based on these data the degree of conversion of 11b to 12 was estimated as ≈90%.
2.3. Preparation and Characterization of PEG-b-PLL/pDNA Polyplexes
The length of cationic blocks of the synthetized PEG-b-PLL poly mers was 62, 150, and 206 repeating units. One could estimate that at the same polycation to pDNA ratios the polyplexes prepared using the block copolymer with PLL DP 62 would display 2.4 and 3.3 times more PEG chains per particle compared to those polyplexes with PLL DP 150 and 206, respectively. The stability of the PEGylated particles in aqueous dispersions increases as the molar fraction of the PEG block per particle increases.[53] The density of PEG corona influences the plasma protein adsorption of the particles.[54] A previous report suggested that the PEG chains crowding defines the shape of the pDNA-containing polyplex, leading to the formation of either the rod- or the globule-like structure. Rod-shaped polyplexes are formed when the tethered PEG chains covering pDNA in a pre-condensed state are dense enough to overlap one another.[55] The steric repulsion of PEG prevents the transition of a rod-shape to a globule with minimal surface area.[56] The parameter of a reduced tethering density (RTD) could be used for a depiction of PEG crowding.[57] This quantity illustrates the extent of PEG overlapping (RTD > 1; overlapping). Moreover, rod-shaped polyplexes exhibited significantly higher gene expression efficacies in a cell-free system compared to the globular polyplexes.[55] The RTD values for studied PEG-b-PLL copolymers are given in Table S1, Supporting Information. According to these values, only the polymer with PLL DP 62 tends to form a rod-shape polyplex with pDNA, while polyplexes produced using polymers with the length of cationic blocks 150 and 206 should have a globular structure. Particle shape influences the cell uptake and the rod-like nanoparticles are more favorable for drug and gene delivery applications.[58,59] The shape of the particle strictly impacts on its internalization by macrophage.[60,61] A recent study suggested that rods are taken up by the cells more rapidly than spherical particles.[62]
Guided by the above consideration the cationic copolymers PEG114-b-PLL62 4 and Man-PEG114-b-PLL62 7 (Scheme 1) were used for the production mannose-free and mannose-decorated polyplexes, respectively. The polyplexes were produced by a simple mixing the cationic copolymer with pDNA gWiz-GFP at various N/P ratios. Formation of the complexes pDNA with the cationic copolymers was demonstrated using an EtBr displacement assay. Intercalation of the EtBr into pDNA increases the fluorescence quantum yield of the dye. Upon pDNA compaction, EtBr is displaced from the complex, causing a decrease in the fluorescence signal. As previously documented the complete displacement of the dye in this assay is observed at the stoichiometric conditions at N/P equal 1 (Figure 1A). The leveling off of the signal at a stoichiometric mixing ratio occurs when all sterically accessible phosphate groups participated in an ion-pair formation with the PLL segment of the PEG-b-PLL.
Figure 1.
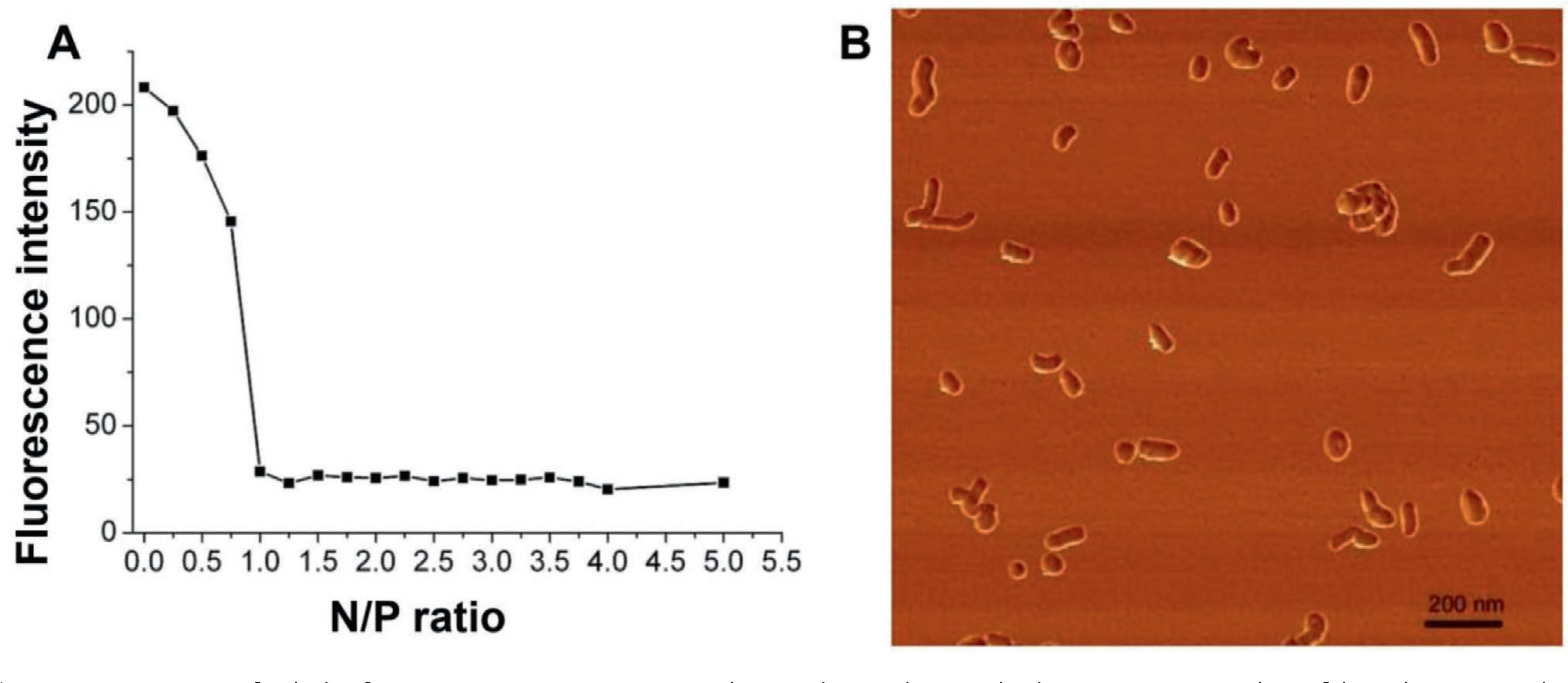
A) Detection of polyplex formation upon mixing pDNA and PEG114-b-PLL62 by EtBr displacement assay. Binding of the polycation results in decreases in the absolute fluorescence intensity of pDNA-EtBr complex depending on the polyplex N/P ratio. The polyplexes were formed in 10 mm HEPES buffer, pH 7.4, incubated for 30 min at 25 °C and the absolute fluorescence was determined (λex 520 nm; λem 590 nm). B) Atomic force microscopy images of cross-linked Man-PEG114-b-PLL62/pDNA polyplexes formed at N/P = 3 at 75% degree of cross-linking. Polyplexes were deposited on mica surface for 2 min, followed by surface drying using stream of argon. The scale bar stands for 200 nm.
The behavior of pDNA containing polyplexes was also investigated by agarose gel electrophoresis. As demonstrated using an agarose gel electrophoresis assay addition of polycation to the pDNA results in the progressive incorporation of the pDNA into the polyplex that is immobile in the gel (Figure 2A). The intensity of the DNA-corresponding bands in the gel decreases as the N/P ratio increases. Such behavior indicates the presence of mixtures of free pDNA and almost fully neutralized DNA in the complex at various relative compositions of the mixture. As the N/P ratio increased, the fraction of the free pDNA decreased, which resulted in the reduction in the respective band intensity with no change in mobility.[63] At N/P ratio 1 there was no observable band of the free pDNA—, i.e., the pDNA binding with the polycation was complete. The DNA-corresponding bands could be observed after an incubation of the non-cross-linked polyplexes with heparin, indicating the effective substitution of pDNA by the polyanion (Figure 1B).
Figure 2.
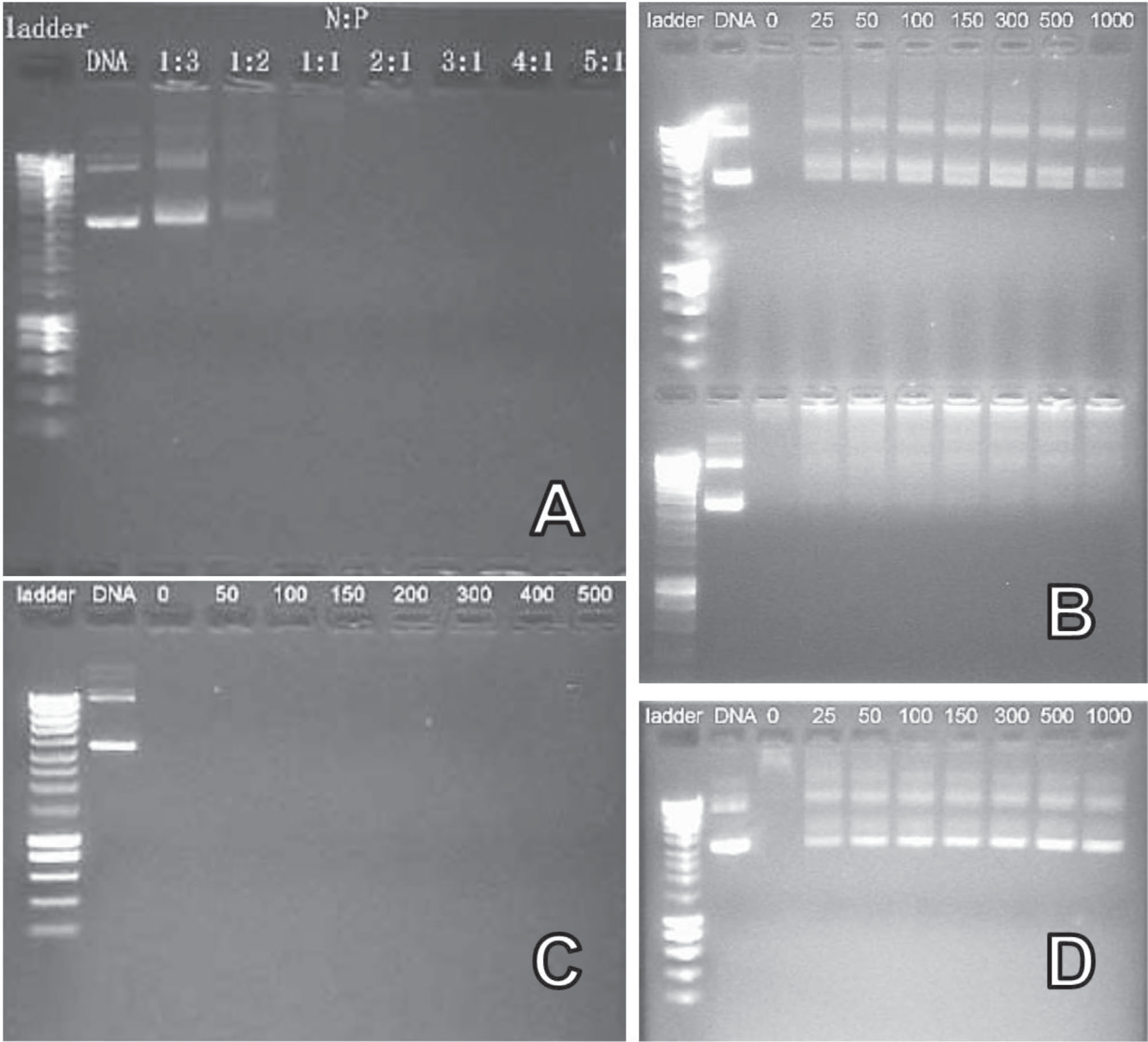
Agarose gel electrophoresis retardation analysis of non-cross-linked and cross-linked Man-PEG114-b-PLL62/pDNA polyplexes. A) Non-cross-linked polyplex, prepared at various N/P ratios. B) Non-cross-linked (upper gel) and cross-linked (bottom gel) polyplexes incubated with various concentrations of heparin (μg mL−1, as indicated above the lanes) for 75 min prior to gel electrophoresis; values represent the concentrations of heparin. C) Cross-linked polyplexes after 30 min incubation with sodium chloride solution (mm, as indicated above the lanes). D) Cross-linked polyplexes, incubated with DTT 10 mm for 1 h at 37 °C, and then incubated with varying concentrations of heparin (μg mL−1, as indicated above the lanes) for 75 min at room temperature. (B–D) Non-cross-linked and cross-linked polyplexes were prepared at N/P ratio 3. Cross-linking was carried out at targeted degree of cross-linking 75%. All experiments were conducted using HEPES buffer containing 10% (v/v) glycerol and were run for 60 min at 80 V and visualized under UV light.
For further experiments these polyplexes were produced at N/P equal to 3 and then cross-linked by the addition of various amounts of DSP stock solution calculated to target various cross-linking degrees. As the targeted cross-linking degree increased the particle size of the polyplexes decreased and polydispersity did not increase or slightly decreased (Table 2). There appeared to also be some changes in the particle charge with the non-cross-linked polyplexes being slightly positively charged and the cross-linked one essentially neutral with ζ-potentials close to zero. A likely reason for the surface charge decrease after cross-linking was the elimination of the excess of primary amino groups of PLL as a result of the reaction with DSP. The cross-linked polyplexes had a bean-like morphology and were relatively uniform as shown by AFM in tapping mode (Figure 1B).
Table 2.
Size and ζ-potential of Man-PEG114-b-PLL62/pDNA polyplexes (N/P = 3) at various targeted degrees of cross-linkinga).
| Targeted degree of cross-linking | Dz [nm] | PDIb) | ζ-potential [mV] |
|---|---|---|---|
| 0 | 158 ± 2 | 0.27 ± 0.04 | 4.5 ± 0.1 |
| 25% | 144 ± 4 | 0.21 ± 0.01 | 0.5 ± 0.7 |
| 50% | 114 ± 1 | 0.20 ± 0.01 | −0.3 ± 0.5 |
| 75% | 107 ± 1 | 0.19 ± 0.01 | −0.1 ± 0.1 |
Data are mean ± SD of n = 3 independent measurements;
DLS PDI equals (stddev/mean)2.
To examine whether the pDNA can be released from the polyplexes we carried out several experiments. First, we treated the cross-linked polyplexes with an increasing amount of sodium chloride and found that the polyplexes withstood at least 0.5 M NaCl without releasing the pDNA (Figure 2C). Second, we exposed the non-cross-linked and cross-linked polyplexes to heparin that serves as a competitor to pDNA and can displace the latter from a complex with the polycation. As seen in Figure 2B pDNA was released from the non-cross-linked polyplex but retained in the cross-linked polyplex. Therefore, cross-linking prevented the displacement of the pDNA by heparin. Finally, since the DSP cross-links contained the cleavable disulfide bond, we examined whether the reduction of this bond could facilitate the release of the pDNA by heparin. Indeed, after the treatment of the cross-linked polyplexes with 10 mM DTT for 1 h and then subjecting them to heparin the pDNA was released (Figure 2D), which demonstrated the critical role of the cross-links in the pDNA retention in the polyplex. Since the fluorescence of the reduced polyplexes was found to be similar to the fluorescence of free pDNA control, dissociation after removal of the stabilization seems to have occurred (Figure 2D). The increase in the cross-linking degree could be beneficial in the gene delivery due to the decrease of cytotoxicity.[64] Hence the polyplexes with the most cross-links were used for further investigation.
2.4. Cytotoxicity of the PEG-b-PLL/pDNA Polyplexes
The safety of the resulting polyplexes was examined by measuring their cytotoxicity in two different cellular models. First, the model was the NIH/3T3 cell line which is an immortalized mouse embryonic fibroblast cell line that does not express a mannose receptor. Second, the model was murine bone marrow derived monocytes cultured in the presence of M-CSF, a cytokine and growth factor to induce differentiation of these into macrophages (BMMΦ). All the tested polyplexes exhibited moderate toxicity to the NIH/3T3 cell line with the cell viability being in the range of ≈70% to 80% by MTT assay (Figure 3A). Interestingly, the polyplex based on Exgene 500 a linear PEI with an average molecular weight of 22 kDa, was the most toxic of the studied systems resulting in a nearly 50% decrease in the cell viability. A different result was obtained in the macrophage cell model (Figure 3A). In this case the cross-linked polyplexes, both targeted and non-targeted, were well-tolerated by BMMΦ. In contrast, the non-cross-linked mannose-conjugated polyplex was more cytotoxic to macrophages, resulting in a lower cell viability while the ExGen 500 based polyplex remained the most toxic with ≈56% cell viability.
Figure 3.
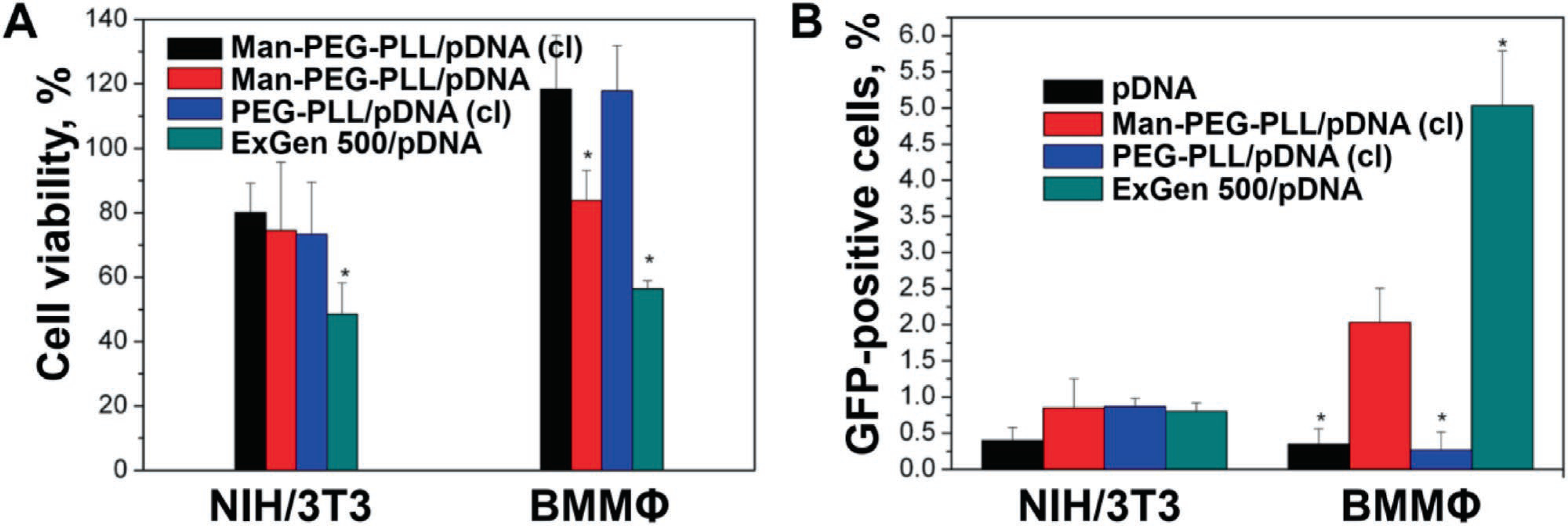
A) In vitro cytotoxicity of targeted and untargeted polyplexes to NIH/3T3 cells and mouse bone marrow macrophages (BMMΦ). Values are mean ± SEM (n = 8), cell viability was assessed by MTT assay. B) Transfection of NIH/3T3 cells and mouse bone marrow macrophages with targeted and non-targeted polyplexes carrying gWiz-GFP pDNA. The Man-PEG114-b-PLL62/pDNA and PEG114-b-PLL62/pDNA polyplexes were prepared at N/P = 3, and cross-linked (cl) at a targeted degree of cross linking of 75%. The polyplexes were diluted in 10 mm HEPES buffer (pH 7.4) and added to the cultured cells for 4 h followed by 48 h of incubation in fresh complete media containing 10% FBS and 1% P/S. The concentrations of pDNA and Man-PEG114-b-PLL62 or PEG114-b-PLL62 were 4.5 and 9.9 μg mL−1, respectively. After that, cells were harvested and bioluminescence was measured by flow cytometry. Values are mean ± SEM (n = 3).
2.5. Transfection Efficiency of the PEG-b-PLL/pDNA Polyplexes
For the cell transfection experiments NIH/3T3 cells and BMMΦ were treated with the mannose-free and mannose-decorated cross-linked polyplexes for 4 h. The cells transfected with the naked pDNA or the ExGen 500 based polyplex were used as control groups. The results are presented in Figure 3B. The naked plasmid exhibited little if any ability for transfecting the cells. In this case the percent of GFP positive cells after transfection was nearly at the baseline ≈0.35% to 0.4%. Moreover in NIH/3T3 cells transfection was marginal with any polyplex in this study (≈0.80% to 0.85% GFP positive cells). In contrast in BMMΦ that expresses the mannose receptor[65,66] a clear differentiation in the transfection was observed for polyplexes. The best transfection result was achieved with an ExGen 500 based polyplex (≈5% of GFP positive cells) followed by the mannose decorated polyplex (≈2% GFP positive cells). The non-targeted polyplex was at the baseline level observed for the naked pDNA group.
2.6. Cell Toxicity of the PEG-b-pAsp(DET) Copolymers
The safety of the resulting cationic copolymers was examined by measuring their cytotoxicity in a mouse IC-21 macrophage cell line. As presented in Figure 4 no cell toxicity was observed up to ≈13 μm of the polymers with at least 80% of cells remaining viable. The IC50 values for mPEG-b-pAsp(DET) and Man-PEG-b-pAsp(DET) were estimated as 17.7 and 16.7 μm respectively. Therefore, in subsequent cell experiments the polymer concentrations did not exceed 13 μm.
Figure 4.
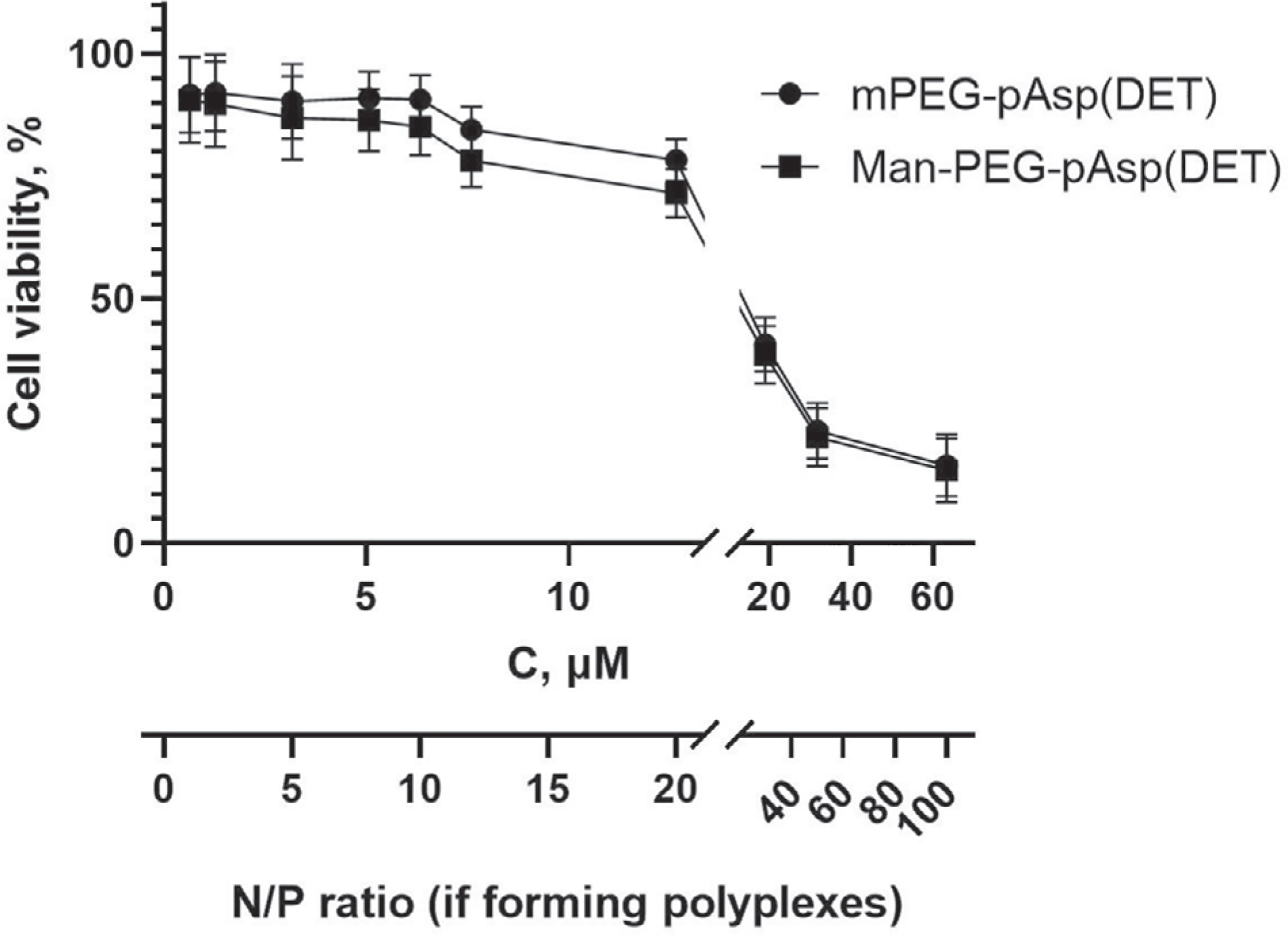
In vitro cytotoxicity of block copolymers to IC-21 macrophages. Cells were exposed to mPEG-b-pAsp(DET) and Man-PEG-b-pAsp(DET) at various concentrations for 24 h. Twenty-four hours later, cell viability was assessed by CCK-8 assay. Values shown are mean ± SEM. The additional X-axis relates the copolymer concentrations to the N/P ratios during the transfection of these cells with the polyplexes.
2.7. Preparation and Characterization of the PEG-b-pAsp(DET)/pDNA Polyplexes
The polyplexes were produced by simply mixing the methoxy- or mannose-terminated cationic copolymers 11a or 12 with pDNA gWiz-Luc at various N/P ratios. The polyplexes were small and relatively uniform and depending on the N/P ratio were either negatively or positively charged (Table 3). Incorporation of mannose in the polyplex resulted in the increase of the particle size which may be due to secondary interactions (hydrogen bonds, van der Waals interactions) of the mannose residues displayed at the ends of Man-PEG-b-pAsp(DET). Notably, at stoichiometric N/P ratio (=1) the complexes had a net negative charge suggesting that the polycation was only partially incorporated into the complex. At the two higher N/P ratios used the complexes had a net positive charge due to the incorporation of the excess polycation in the complex.
Table 3.
Size and zeta-potential of mPEG-b-pAsp(DET)/pDNA and Man-PEG-b-pAsp(DET)/pDNA polyplexes at various N/P ratiosa).
| N/P | Dz [nm] | PDIb) | ζ-potential [mV] |
|---|---|---|---|
| mPEG-b-pAsp(DET)/pDNA | |||
| 1 | 121 ± 3 | 0.16 ± 0.01 | −31 ± 1 |
| 8 | 161 ± 6 | 0.16 ± 0.02 | 29 ± 1 |
| 20 | 112 ± 2 | 0.11 ± 0.02 | 32 ± 1 |
| Man-PEG-b-pAsp(DET)/pDNA | |||
| 1 | 137 ± 8 | 0.27 ± 0.02 | −30 ± 1 |
| 8 | 190 ± 10 | 0.27 ± 0.03 | 28 ± 1 |
| 20 | 208 ± 9 | 0.16 ± 0.03 | 27 ± 1 |
Data are mean ± SD of n = 3 independent measurements;
DLS PDI equals (stddev/mean)2.
2.8. Transfection Efficiency of the PEG-b-pAsp(DET)/pDNA Polyplexes
For the cell transfection experiments the IC-21 macrophage cells were treated with the mannose-free and mannose-decorated non-cross-linked PEG-b-pAsp(DET)-based polyplexes at various N/P ratios for 4, 8, and 24 h and luciferase gene expression was determined by bioluminescence 24 h after the addition of the polyplexes. The results are presented in Figure 5 and Table S2, Supporting Information, as the luciferase expression normalized per the cell protein. The cell protein counts shown in the Table S4, Supporting Information, suggest that no cell toxicity was evident as the treatments did not result in a considerable cell protein loss.
Figure 5.
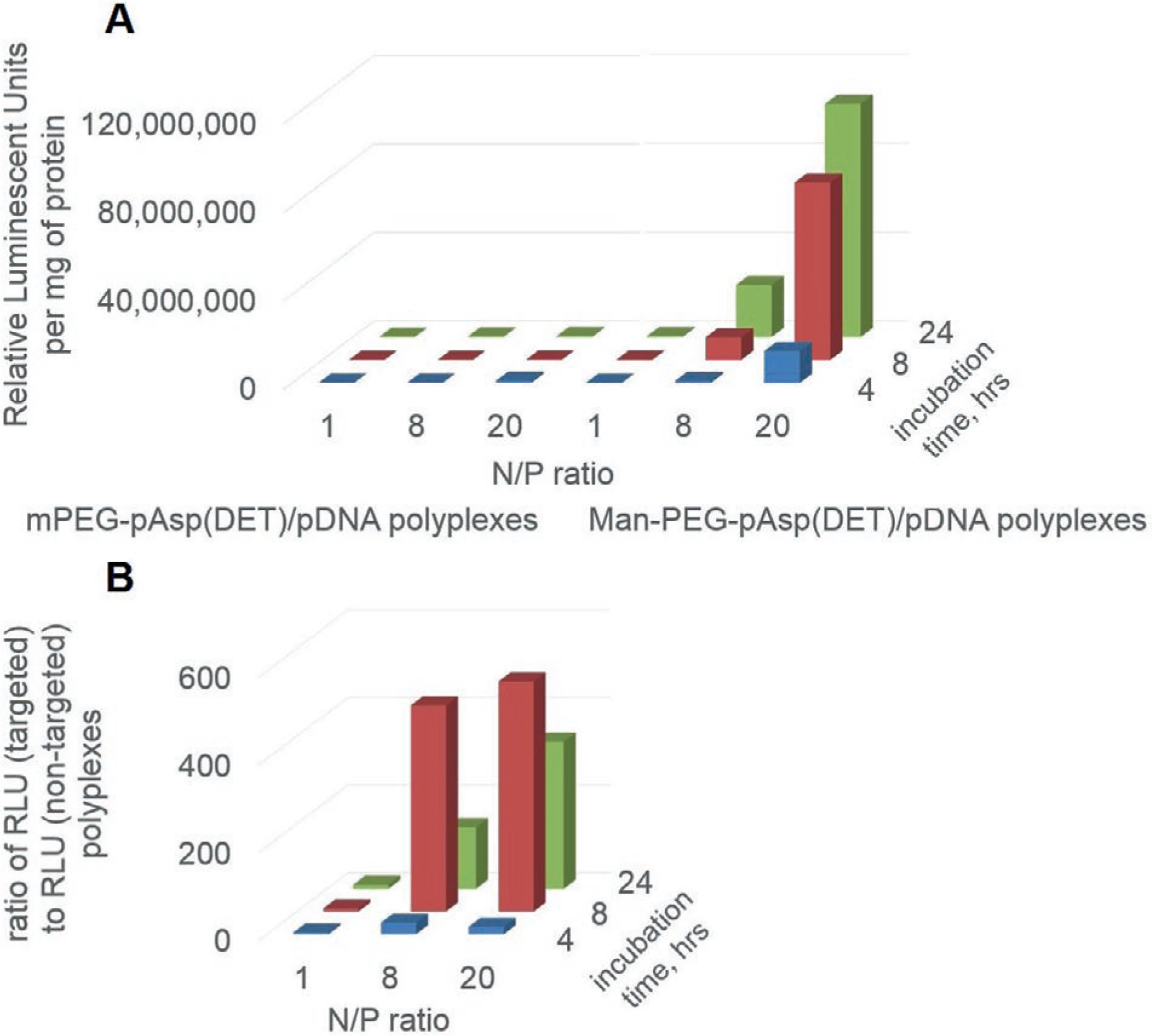
A) Transfection of IC-21 macrophages with targeted and non-targeted PEG-b-pAsp(DET)-based polyplexes carrying gWiz-Luc pDNA. The concentrations of pDNA were 4.5 μg mL−1. The cells were incubated for 4, 8, and 24 h with polyplexes in a serum-free media and then for 24 h in the complete media containing 10% FBS. After that the cells were lysed and the luciferase activity was measured and normalized per milligram of cell protein in the lysate. Polyplexes were prepared at various N/P ratios (1, 8, 20). Relative Luminescent Units (RLU) were normalized for protein content. B) Extent of enhancement of transfection as a result of attachment of the mannose targeting moieties to the polyplexes. For every N/P ratio and time point data in panel (A) are presented as the ratio of luciferase expression levels (RLU/mg protein) for the targeted versus non-targeted polyplexes.
As expected, the transfection results for both, targeted and non-targeted polyplexes strongly depended on the N/P ratios. There was also a dependence of the gene expression on the duration of the cells exposure to the polyplexes. At N/P = 1 for non-targeted polyplexes the luciferase expression was ≈104–105 RLU/mg for all exposure times, suggesting that these negatively charged polyplexes were practically inactive (Figure 5A and Table S2, Supporting Information). As the N/P ratio increased the gene expression also increased. However, for the non-targeted polyplexes the maximal luciferase count observed at N/P = 20 was less than 106 RLU/mg. In contrast, for the targeted polyplexes at each N/P ratio and exposure time there was a major increase in the transfection efficiency versus non-targeted treatment groups, resulting in the luciferase counts as high as ≈107–108 RLU/mg (Figure 5B and Table S3, Supporting Information). Although the total yields of the luciferase count were higher at N/P = 20, the maximal transfection efficiencies were observed at N/P = 8. The best effect of a targeted delivery was observed at 8 h of the incubation, transfection was ≈400 (N/P = 8) and ≈500 (N/P = 20) times more successful.
3. Discussion
The goal of this study was to develop polyion complex constructs for the delivery of pDNA to macrophages that carry the mannose receptor. We used a well-known cationic block copolymer principle[67–69] to produce core—shell polyplexes with the polycation—pDNA condensed in the core, and PEG shell surrounded the core. To target polyplexes to the cell receptors we attached mannose residue to the polyplexes at the free ends of the PEG chains. Our synthetic method for mannose attachment was streamlined by using a quantitative one-step click reaction, compared to some previous mannose conjugation procedures that required multiple steps and had a lower yield.[70–74] For this purpose, we evaluated two different cationic block copolymer constructs.
Initially we used PEG-b-PLL to produce polyplexes. We synthesized propargyl- and mannose-terminated PEG-b-PLL, and used these polymers to prepare pDNA polyplexes. To attach targeting moiety to the polyplexes we synthesized 2′-azidoethyl-O-α-D-mannopyranoside and attached it to the copolymer via a high-yield click reaction. We then formed the polyplexes by a standard self-assembly of the cationic copolymers with the pDNA. The pDNA molecule undergoes a coil-globule transition in the process of complexation to take a compact conformation.[63–66] The process of the pDNA condensation can be assessed from a change in the fluorescence intensity of the intercalating dye EtBr, because EtBr is excluded from the double-helical strand of pDNA with the progress of the DNA condensation, resulting in a decreased fluorescent intensity. A previous report demonstrated that the decrease in the fluorescent signal is in line with a trend of the diameter change due to pDNA compaction.[45]
Since PEG-b-PLL copolymers display relatively high cell toxicity, and the toxicity can be decreased by cross-linking the polyplexes to eliminate free polycation[75] we used the excess PLL amino groups present in the polyplex to introduce cross-links. The cross-linking led to the particle size reduction, indicating that no significant amount of covalent bonds between different complex particles were formed in the process. This was confirmed by AFM observation. The particle charge also decreased as a result of the cross-linking probably due to the reaction of the cross-linker with the PLL amino groups, which resulted in electroneutral amide bonds. However, the cross-linked particles did not coagulate even in the absence of surface charge, presumably due to steric shielding of the polyplexes by the PEG corona. The cross-links contained bio-reducible disulfide bonds that are known to be cleaved inside the cells,[76] which was part of our design to enable a pDNA release from the polyplexes. We have shown that the cross-links protected the polyplexes in the presence of 0.5 m NaCl, which normally disintegrates the non-cross-linked polyplexes, and even more importantly prevents the release of pDNA by the exchange with heparin. The latter, however, was reversed when the cross-links were cleaved. After removal of the cross-links by the reduction of disulfide bonds with DTT the pDNA could be released from the polyplex by displacement with heparin.
The cross-linking decreased the toxicity of these polyplexes to primary mouse macrophages and allowed us to proceed with the transfection experiments. In these experiments we obtained evidence that mannose receptor targeted polyplexes formed by the PEG-b-PLL copolymer were substantially more active in transfecting macrophages than the non-targeted polyplex, although their activity was less than that of the Exgene 500 based polyplex. Interestingly all polyplexes, targeted, non-targeted, or even Exgene 500 based were not active in the fibroblast cells, and Exgene 500 was also quite toxic to the fibroblast cell line. Both mannose-free and mannose-decorated cross-linked polyplexes produced a similar transfection of the cultured NIH/3T3 cells. A previous study demonstrated, that the uptake of mannosylated BSA was not different from that of galactosylated BSA in NIH/3T3 cells as opposed to MMR-expressing macrophages.[77] In BMMΦ, mannosylated cross-linked PEG-b-PLL/pDNA polyplexes demonstrated an 8.2 times higher transfection efficiency as opposed to those without the targeting moiety. The difference is explained by the expression of the mannose receptor on monocytes. Therefore, mannosylated cross-linked PEG-b-PLL/pDNA polyplexes could represent a reasonable strategy and construct for the targeted delivery of pDNA to macrophages.
The second type of cores-shell constructs we evaluated for further development of the platform for gene delivery to macrophages did not require cross-linking. Here, to produce polyplexes we have selected mPEG-b-pAsp(DET) as a building block previously described by Kataoka’s group.[48,49,51,52] The choice of this cationic block copolymer was dictated by its safety to the cells.[78] We synthesized methoxy- and mannose-terminated PEG-b-pAsp(DET) polymers, used these polymers to prepare pDNA polyplexes and evaluated the ability of these polyplexes to transfect a macrophage cell line IC-21. The polyplexes were prepared by the simple mixing of the pDNA and the cationic block copolymer and were electrostatically bound without any cross-linking of the copolymer. The cell viability was assessed using a WTS-8 reagent, which is transformed into the water-soluble product of cellular reduction.[79] This allowed us to evaluate the cytotoxicity of the polymers to the cells after long-term incubation.
The cytotoxicity of both targeted and untargeted block copolymers in the IC-21 macrophages was relatively low. The IC50 values for mPEG-b-pAsp(DET) and Man-PEG-b-pAsp(DET) were very close to each other (≈17.7 and ≈16.7 μm, respectively) suggesting that attachment of the mannose residue to the PEG terminus of the block copolymer does not alter the copolymer cytotoxicity. Also, these IC50 values were in reasonably good agreement with the IC50 values previously reported for mPEG-pAsp(DET) in different cell lines (≈14.7 μm for immortalized mouse motor neurons NSC-34 and ≈104.9 μm, for brain microvascular endothelial cells hCMEC/D3).[80]
Complexes formed at the stoichiometric N/P ratio were negatively charged, while those formed at higher N/P ratios (8 and 20) displayed a high positive charge. A previous report suggested that the charge of the polyplexes formed by pDNA and mPEG-b-pAsp(DET) increases as the N/P ratio increases and eventually reaches some close-to-constant value at high N/P ratios.[49] The leveling off of the polyplex charge may be due to the saturation binding of the polycation to the DNA as well as some shielding effect of the PEG chains surrounding the poly ion complex core.
The Man-PEG-b-pAsp(DET)/DNA polyplexes produced an efficient transfection of the cultured IC-21 macrophages. Interestingly, after a 4 h exposure to the cells neither untargeted nor targeted polyplexes produced considerable levels of gene expression. Increasing the incubation time from 4 to 8 h resulted in dramatic increases in the gene expression for the targeted polyplexes comparted to the untargeted ones: ≈400 and ≈500 for N/P = 8 and N/P = 20, respectively. At a longer incubation there was a decline in the luciferase levels, along with an obvious cytotoxic effect to the cells as evidenced by the over twofold decrease in the protein count. The observed transfection enhancements as a result of mannose decoration of our polyplexes are superior compared to those reported in several other studies. For example, mannose decorated pAsp(DET)/DNA polyplex at N/P = 5 exhibited a ≈2.5-fold increase in transfection in the Raw 264.7 cells compared to untargeted polyplex.[33] A mannose-targeted chitosan-g-PEI-based polyplex formed at N/P = 14 demonstrated a 9.4 times higher transfection efficiency in the same cell line as opposed to those without the targeting moiety.[31] Mannosylated PEG-b-PEI particles reveal a 15-fold increase in transfection efficacy in dendritic cells at N/P = 8.[35] pDNA in mannosylated PEI-containing cholesteryl-based liposomes displayed as much as a ≈40-fold increase in the transfection in mouse macrophages compared to the pDNA formulated with mannose-free liposomes of the same composition.[30] The polyplexes formed by pDNA and spermin-mannan conjugates, at N/P = 5 exhibited a 28.5-fold increase in the transfection of Raw 264.7 macrophages compared to the similar polyplexes that were produced using spermin-pullulan conjugates.[32]
The exposure time is also an important factor in the mannose-targeted transfection of the cells. The observed time dependence, in particular an increase in the transfection of the cells with the mannose decorated polyplexes observed after 8 h, could be due to the turnover time of the mannose receptor in the cells. The turnover time was previously estimated to be between 30 and 60 min, and was shown to increase with the increase in the size and multivalence of the ligand.[81,82] At each given time point less than 30% of the mannose receptor is displayed at the cell surface while the rest is intracellular,[82,83] as it was shown for other receptors of the same family.[84,85] The receptor half-life was estimated as ≈6 h.[86] Therefore, the turnover time should affect the amount of the polyplexes that are trafficked from the external solution to the cells. Since, there was practically no difference in transfection for polyplexes prepared at N/P = 8 and N/P = 20, we propose to further use the lower N/P ratio to minimize the excess of the mannose-terminated cationic polymer. We estimate that our polyplex particles contain at least 240 mannose residues per one pDNA molecule. That may be an important parameter to consider in future work because the more mannose residues are located nearby, the better is their binding to the receptor (due to the cooperative effect as it was shown for lectins[87,88] and for MMR itself[35]).
A number of studies reported on the high cellular toxicity of the PEG-b-PLL copolymers. The PEG-b-PLL copolymer exerted an apparent toxicity in the NIH/3T3 cells at 10 μg mL−1.[89] The IC50 values for PEG-b-PLL copolymer in hCMEC/D3 and NSC-34 cell lines were 3.3 and 2.0 μm, respectively. The PEG-b-pAsp(DET) was reported to be well-tolerated by both cell lines. In the hCMEC/D3 cell line it was about 3227% less toxic than PEG-b-PLL.[90]
Previous studies reported on a similarity between the primary bone marrow macrophages and IC-21 macrophage cell line in the cell size, morphology, adhesion, and spreading behaviors. Among all secondary-derived macrophage cell lines (IC-21, J774A.1, RAW 264.7), IC-21 represents the most physiologically relevant cell line to bone marrow macrophages.[91] Both IC-21 and BMMΦ shift towards an M2 phenotype in absence of any extern.al signaling when cultured extendedly, as was assessed by the decrease of a toll-like receptor 4 (TLR-4) presentation and a simultaneous increase of the expression of MMR and a cluster of differentiation 14 (CD14).[65] Even at standard culturing both cell lines are shown to be producing MMR at a significantly higher level than monocytic phenotype cells (J774A.1, RAW 264.7).[66]
Two different plasmids (gWiz-GFP and gWiz-Luc) were used in this study. The genes of GFP and luciferase were expressed under the control of an identical cytomegalovirus (CMV) promoter. Both plasmids were successfully applied for in vitro and in vivo transfection.[92] The efficacy of gWiz-GFP transfection was assessed by the flow cytometry, counting the percentage of the GFP-expressing cells. The gWiz-Luc gene expression was determined by the bioluminescence in the cell lysate. The former method qualitatively determines the number of the GFP-positive cells and cannot be used for the evaluation of the gene expression level. The latter method evaluates the total amount of the protein in lysate, which is a qualitative measure of the gene expression. The luciferase bioluminescence assay requires an additional reagent (D-luciferin) and has a lower detection limit in comparison to the GFP fluorescence assay.[93]
In both experiments was evaluated the transfection enhancement as a result of the mannose decoration of the polyplexes. The PEG-b-pAsp(DET)-based polyplexes exhibited a higher gain of gene transmission as a result of mannosylation, compared to the PEG-b-PLL-based particles. The delivered gene expression indicates the uptake of the polyplex. The uptake of mannose-decorated particles was observed much more in MMR-positive bone marrow macrophages and IC-21 cells but was negligible in the MMR-negative NIH/3T3 cell line. The enhancement of the transfection of the cells with the mannosylated polyplexes went up with the increase of the exposure time. The observed time dependence provided an indirect evidence of the involvement of receptor-mediated endocytosis.
The particular mechanism of mannose-mediated pathways of the polyplexes is the subject of the further exploration. However, a previous study reported on a significant decrease of the intracellular uptake of mannosylated particles in macrophages in the presence of free α-D-mannose and anti-MMR antibody.[20,22,27,35] The mannose pretreated TAM cells did not show a significant uptake as well due to saturating of the receptors.[12] By combining the results of all assays, it can be concluded that mannosylation drastically enhances the ability of the polyplexes to transfect selectively the macrophages. The PEG-b-pAsp(DET) block copolymer shown to be a better candidate for in vivo gene delivery applications is because of low toxicity.
4. Conclusions
Two mannose-conjugated cationic block copolymers (PEG-PLL and PEG-pAsp(DET)) were synthetized and characterized via 1H-NMR and GPC and were used to produce pDNA-containing polyplexes. PEG-b-PLL-based polyplexes were cross-linked to reduce its toxicity in NIH/3T3 and BMMΦ. While in macrophages mannose-decorated polyplexes demonstrated a higher transfection efficiency than in the NIH/3T3, the overall transfection enhancement produced by cross-linked PEG-b-PLL polyplexes was moderate. The PEG-b-pAsp(DET) copolymers displayed a low toxicity with respect to an IC-21 macrophage cell line and were used for the production of non-cross-linked pDNA-contained polyplexes. The obtained polyplexes exhibited an efficient transfection in the cultured IC-21 macrophages. The obtained mannose modified polyplexes exhibited a ca. 500-times greater transfection activity compared to the mannose-free polyplexes. The results suggest that ManPEG-b-pAsp(DET)/DNA polyplexes is a potential vector for the targeted delivery of the pDNA to macrophages and can be in future used in designing the in vivo gene delivery platform.
5. Experimental Section
Materials and Equipment: Reagents:
Methoxy(polyethylene glycol) amine (mPEG-NH2) (Mw 5 kDa), alkyne functionalized poly(ethylene glycol) amine (alkyne-PEG-NH2) (Mw 5 kDa), and Fmoc-NH-PEG-NHS (Mw 5 kDa) were purchased from Creative PEGWorks (Durham, NC, USA). Dithiobis(succinimidyl propionate) (DSP) and DL-Dithiothreitol (DTT) were purchased from Pierce Inc. Triphosgene. N-ε-benzyloxycarbonyl-L-lysine (Lys(Z)), β-benzyl-L-aspartic acid (BLA), N,Ndimethylformamide (DMF), dimethyl sulfoxide (DMSO), tetrahydrofuran (THF), n-hexane, diethyl ether, ethyl acetate, diethylenetriamine (DET), mannose azide, copper(II) sulfate pentahydrate (CuSO4*5H2O), sodium ascorbate, hydrochloric acid (HCl), acetic acid, lithium chloride (LiCl), and acetone were products of Sigma-Aldrich Chemical Company (St. Louis, MO, USA). 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), bovine serum albumin (BSA), heparin, and Exgene 500 were purchased from Fermentas (Burlington, ON, Canada). 1-O-(2-(2-(2-Azidoethoxy)ethoxy)ethoxy)-α-D-mannopyranoside (α-D-Man-TEG-N3) was purchased from Iris Biotech GmbH (Marktredwitz, Germany). A luminescent substrate, D-luciferin, was purchased in Caliper Life Sciences (Hopkinton, MA, USA). Hank’s Balanced Salt Solution (HBBS), Roswell Park Memorial Institute medium (RPMI), trypsin and ethylenediaminetetraacetic acid solution (trypsin-EDTA), and phosphate buffered saline (PBS) were products of Gibco Life Technologies, Inc. (Grand Island, NY, USA). Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), and penicillin/streptomycin (P/S) were purchased in Invitrogen (Carlsbad, CA, USA). Macrophage colony-stimulating factor (M-CSF) was a gift from Dr. Howard Gendelman.
Plasmids:
The gWiz high expression vectors encoding the reporter genes luciferase (gWiz-Luc) and green fluorescent protein (GFP) (gWiz-GFP) both under control of an optimized human cytomegalovirus (CMV) promoter followed by intron A from the CMV immediate-early (IE) gene were purchased from Genlantis (San Diego, CA, USA).
Cells:
NIH/3T3 mouse embryonic fibroblasts were purchased from ATCC (Manassas, VA, USA) and were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin at 37 °C and 5% CO2. Adherent murine IC-21 macrophages were purchased from ATCC (Manassas, VA, USA) and were maintained in an RPMI medium supplemented with 10% FBS at 37 °C and 5% CO2. Bone marrow cells were extracted from femurs of C67Bl/6 male mice 6–7 weeks of age (Charles River Laboratories, Wilmington, MA, USA) according to previously published protocols[50] and cultured for 12 days in a DMEM medium supplemented with a 1000 U mL−1 macrophage colony-stimulating factor. All animal experiments were carried out with the approval of the University of Nebraska Medical Center (UNMC) and University of North Carolina at Chapel Hill (UNC-CH) Institutional Animal Care and Use Committee and in accordance with NIH Guide for Laboratory Animal Use. The animals were kept five per cage with an air filter cover under a light- (12-h light/dark cycle) and temperature-controlled (22 ± 1 °C) environment. Food and water were given ad libitum. Differentiated bone-marrow derived macrophages (BMMΦ) were cultured in DMEM + 10% FBS + 0.5% P/S + 0.1% M-CSF at 37 °C and 5% CO2.
Synthesis of Mannosylated PEG-b-PLL (Man-PEG-b-PLL): Synthesis of N-ε-benzyloxycarbonyl-L-lysine-N-carboxyanhydride (Lys(Z)-NCA):
N-ε-benzyloxycarbonyl-L-lysine (Lys(Z)) (5.0 g, 17.8 mmol) was dispersed in anhydrous THF (60 mL) and was heated to 50 °C. Triphosgene (2.0 g, 6.7 mmol) was dissolved in anhydrous THF (15 mL) and was slowly added to the above suspension till a clear solution was formed. After 3 h, the reaction mixture was poured into hexane (225 mL) and the resulting suspension was stored at −20 °C overnight to assure complete precipitation. The product was recrystallized from a THF/hexane mixture (1:3 v/v). The yield was 4.87 g (89%).
Synthesis of Propargyl Functionalized PEG Amine (propargyl-PEG-NH2):
Fmoc-NH-PEG-NHS (0.4 g, 75 μmol) was dissolved in dichloromethane (5 mL). Propargylamine (32 μL, 78 mmol) and N,N′-diisopropylethylamine (DIEA) (87 μL, 0.5 mmol) were added to the solution. The reaction mixture was stirred at room temperature, and after 24 h was poured into diethyl ether and the precipitate was collected by filtration. The Fmoc group was removed according a common procedure using 20% piperidine in the DMF solution. The yield was 0.36 g (95%).
Synthesis of Propargyl Functionalized PEG-b-PLL (propargyl-PEG-b-PLL):
Propargyl-PEG-NH2 (1.0 g, 0.19 mmol) was dissolved in anhydrous DMF (10 mL) in a water-free and oxygen-free Schlenk flask. The precalculated amount of Lys(z)-NCA (based on desired DP) was dissolved in anhydrous DMF and added to the mixture. After three freeze-pump-thaw cycles, the reaction mixture was stirred for 72 h at 40 °C. DMF was removed from the reaction mixture by a rotary evaporator, and the residue was dissolved in dichloromethane. The solution was poured in a tenfold volume of diethyl ether at 0 °C. The precipitate was collected by filtration, washed with diethyl ether, dried in vacuo, and lyophilized.
The benzyloxycarbonyl protecting group (Z) was removed according the general procedure. Propargyl-PEG-b-PLL(Z) (0.8 g) was dissolved in TFA (8 mL), and a solution of 33% hydrobromic acid in acetic acid (4 times excess to the Z groups) was added to the polymer solution. The mixture was stirred at room temperature for 1 h, and then poured into an excess of cold diethyl ether. The precipitate was collected by filtration, dissolved in methanol, and precipitated using diethyl ether twice. For different chain lengths and scales, yields ranged from 81% to 85%.
Synthesis of 2′-bromoethyl-α-D-mannopyranoside:
Amberlite IR-120H (1.5 g) was dispersed in 2-bromoethanol (11.5 mL, 0.16 mol). The mixture was heated up to 90 °C for 30 min, and then D-mannose (1.5 g, 8.3 mmol) was added to the mixture. The reaction was allowed to proceed for 3 h at 90 °C, then the reaction mixture was filtered to remove Amberlite IR-120H. The precipitate was washed with 2-bromoethanol (2 mL). The excess of 2-bromoethanol was distilled off under a reduced pressure, the resulting sticky residue was dissolved in methanol followed with an addition of SiO2 (5.0 g). The reaction mixture was loaded onto the column previously filled with SiO2 and pre-eluted with ethyl acetate/methanol (19:1 v/v). Mobile phase of the same composition was used for elution. The solvent from the collected fractions was removed under reduced pressure to give a product in the form of a light-yellow oil. The yield was 2.17 g (91%).
Synthesis of 2′-Azidoethyl-O-α-D-mannopyranoside:
Sodium azide (0.91 g, 14 mmol) and 2′-bromoethyl-α-D-mannopyranoside (2.0 g, 7 mmol) were dissolved in water (5 mL), followed by the addition of acetone (30 mL). The reaction mixture was stirred at 76 °C for 20 h. After the removal of acetone the resulting sticky residue was dissolved in methanol followed with the addition of SiO2 (6.0 g). The reaction mixture was loaded onto the column previously filled with SiO2 and pre-eluted with ethyl acetate/methanol (19:1 v/v). The mobile phase of the same composition was used for elution. The solvent from the collected fractions was removed under a reduced pressure to give a product in the form of a light-yellow oil. The yield was 1.55 g (89%).
Synthesis of Man-PEG-b-PLL by Click Chemistry:
Propargyl-PEG-b-PLL was introduced into copper(I)-catalyzed azide alkyne cycloaddition (CuAAC) reaction with 2′-azidoethyl-O-α-D-mannopyranoside in the presence of Cu+ ions generated via reduction of Cu2+ by ascorbate in situ. To the water–methanol mixture (1:1 v/v) contained 2′-azidoethyl-O-α-D-mannopyranoside and propargyl-PEG-PLL (molar ratio 1:10) was added 150 μL of mixture containing 10 mm CuSO4*5H2O, 10 mm pentamethyldiethylenetriamine (PMDETA), and 50 mm ascorbic acid dissolved in water. The reactions were allowed to proceed for 24 h at room temperature, then the reaction mixtures were dialyzed against deionized water for 48 h and freeze-dried. The yields ranged from 91% to 95%.
Synthesis of Mannosylated PEG-b-pAsp(DET) (Man-PEG-b-pAsp(DET)): Synthesis of β-Benzyl-L-aspartate-N-carboxyanhydride (BLA-NCA):
β-benzyl-L-aspartic acid (10 g, 45 mmol) was dispersed in anhydrous THF (100 mL), heated to 50 °C, and then triphosgene (4.45 g, 15 mmol) in anhydrous THF (40 mL) was slowly added till a clear solution was formed. After three hours, the reaction mixture was poured into hexane (300 mL), and the resulting suspension was stored at −20 °C overnight to assure complete precipitation. The product was recrystallized from ethyl acetate/hexane mixture (1:2 v/v). The yield was 8.07 g (72%).
Synthesis of Alkyne Functionalized PEG-b-poly(benzyl-L-aspartate) (alkyne-PEG-b-pBLA):
BLA-NCA (0.9 g, 3.3 mmol) was dissolved in DMF (1.36 mL), followed by an addition of distilled chloroform (6.8 mL). mPEG-NH2 (Mw 5kDa) or alkyne-PEG-NH2 (Mw 5kDa) (each 0.3 g, 0.06 mmol) were dissolved in distilled chloroform (3 mL) and added to the solution of BLA-NCA. The reaction mixture was stirred at 35 °C under a dry argon atmosphere for 24 h until BLA-NCA was completely dissolved. Then, the reaction mixture was poured into a tenfold volume of diethyl ether at 0 °C, and a precipitate was collected by filtration and washed with diethyl ether, dried in vacuo, and lyophilized. The yields were 0.82 (89%) and 0.79 g (86%) for mPEG-b-pBLA and alkyne-PEG-b-pBLA, respectively.
Synthesis of Alkyne-Functionalized PEG-b-pAsp(DET) (alkyne-PEG-b-pAsp(DET)):
Lyophilized mPEG-b-pBLA or alkyne-PEG-b-pBLAn(each 0.3 g, 19.6 μmol) were dissolved in DMF (10 mL), of DET (5.04 g, 49 mmol, 50 equivalent of benzyl group of PBLA segment) were added under anhydrous conditions at 40 °C. After 24 h, the reaction mixture was added dropwise into a solution of acetic acid (10% v/v, 40 mL) and dialyzed against 0.01 n HCl using a membrane with molecular weight cut-off (MWCO) 3 500 Da. The final product was lyophilized to obtain the polymer as the chloride salt form with a yield of ≈90%. Polymers were stored under dry nitrogen. The yields were 0.24 (84%) and 0.25 g (88%) for mPEG-b-pAsp(DET) and alkyne-PEG-b-pAsp(DET), respectively.
Synthesis of Man-PEG-b-pAsp(DET) by Click Chemistry:
Alkyne-PEG-b-pAsp(DET) was introduced into copper(I)-catalyzed azide alkyne cycloaddition (CuAAC) reaction with α-D-Man-TEG-N3 in the presence of Cu+ ions generated via reduction of Cu2+ by ascorbate in situ. To the PBS (5 mL, pH 7.2) contained alkyne-PEG-pAsp(DET) (100 mg, 6.8 μmol) and α-D-Man-TEG-N3 (22.9 mg, 68 μmol, molar ratio 1:10) was added 150 μL of mixture containing 10 mm CuSO4*5H2O and 50 mm ascorbic acid dissolved in water. The reaction was conducted 4 h at room temperature, then the reaction mixture was dialyzed against 0.01 n HCl for 24 h and freeze-dried. The yield was 99.4 mg (97%).
Polymers Characterizations:
1H-NMR spectra were acquired using a Bruker 400 spectrometer at 25 °C. Dry polymer (≈30 mg) was dissolved in CDCl3, DMSO-d6, or D2O (≈500 μL). The spectra were calibrated using the solvent signals (CHCl3: 7.24 ppm, DMSO: 2.50 ppm, H2O: 4.80 ppm). The molecular weight and molecular weight distribution (Mw/Mn) were measured by GPC using Viscotek GPC system (Viscotek, Houston, TX, USA) against a narrow molecular weight distribution of polymethylmethacrylate in DMF at a flow rate of 1.0 mL min−1 at 45 °C with refraction index, laser light, and viscosity detectors.
Polyplex Formation: PEG-b-PLL/pDNA Cross-Linked Polyplexes:
The N/P ratio was calculated as a molar ratio of the primary amino groups of PEG-b-PLL (pKa = 10.5) to the phosphate groups of pDNA (gWiz-GFP, 5757 bp, 1.85*1015 P ug−1). To form the polyplexes, solution of pDNA and polymer in 10 mm HEPES buffer (pH 7.4) were mixed with various precalculated N/P ratios, immediately vortexed for 20 s, and then kept still for 30 min for further experimenting. The final concentration of pDNA was 15 μg mL−1. The precalculated amounts of DSP solution in DMSO were added to the polyplexes and were gently mixed. After 1 h at room temperature the solutions were dialyzed against 10 mm HEPES buffer (pH 7.4) to remove excess amount of cationic polymers.
PEG-b-pAsp(DET)/pDNA Polyplexes:
N/P ratio was calculated as a molar ratio of the primary amino groups of PEG-b-pAsp(DET) (Degree of polymerization/DP = 50, pKa1(DET) = 8.9) to the phosphate groups of pDNA (gWiz-Luc, 6732 bp, 1.86*1015 P ug−1). To form the polyplexes, solution of 1 μg pDNA in 150 μL serum-free media and different amounts of polymer in SFM were mixed with various precalculated N/P ratios up to 221 μL of total volume, immediately vortexed for 20 s, and then kept still for 30 min for further experiment.
Exclusion Assay of Ethidium Bromide:
PEG-b-PLL was added to the mixture of pDNA and ethidium bromide (EtBr) at pDNA:EtBr = 5:1 (w/w) at the precalculated N/P ratios and immediately vortexed for 20 s. The final volumes were adjusted with 10 mm HEPES buffer (pH 7.4). The mixtures were stored for 30 min, followed by the fluorescence intensity measurement at 25 °C using Shimadzu P5000 spectrofluorimeter (λex 520 nm, λem 590 nm).
Agarose Gel Electrophoresis:
PEG-b-PLL/pDNA polyplexes were mixed with glycerol (9:1 v/v, the concentrations of pDNA were 13.5 μg mL−1) and further analyzed by electrophoresis in a 1% agarose gel at 80 V for 60 min. DNA was visualized by UV illumination following staining of gels with ethidium bromide (0.5 μg mL−1) for 20 min at room temperature
Dynamic Light Scattering (DLS):
Hydrodynamic diameter (the intensity-mean Z-average diameter) and ζ-potential of the polyplex particles were determined using a Malvern Zetasizer instrument (Malvern Instruments Ltd., Malvern, UK) with a 4 mW He-Ne laser operating at 633 nm with a 173° scattering angle. All measurements were performed in automatic mode, at 25 °C. Software provided by the manufacturer was used to calculate the size, polydispersity indices (PDI), and ζ-potential. The values were calculated from the measurements performed at least in triplicate. For particle size measurement the PEG-b-PLL-based and the PEG-b-pAsp(DET)-based polyplexes were prepared in 10 mm HEPES buffer (pH 7.4) (at 15 μg mL−1 pDNA) and in SFM (at 4.53 μg mL−1 pDNA), respectively, and diluted to obtain count rates of ≈100–400 kcps for each sample; for ζ-potential measurements polyplexes were prepared in 10 mm HEPES buffer (pH 7.4) and were diluted with dH2O.
Atomic Force Microscopy (AFM):
The AFM imaging was performed for pDNA containing polyplexes using a Multimode NanoScope IV system (Veeco, Santa Barbara, CA, USA) operated in tapping mode. Polyplexes were deposited on mica surface for 2 min, followed by surface drying using the stream of argon. The images were processed and the widths and heights of the particles were determined using Femtoscan software (Advanced Technologies Center, Moscow, Russia).
In vitro Cytotoxicity Assay: The Cytotoxicity of PEG-b-PLL/pDNA Polyplexes:
The cytotoxicity of PEG-b-PLL/pDNA polyplexes was measured using MTT assay according to the protocol provided by the manufacturer. Briefly, NIH/3T3 and BMMΦ were seeded in 24-well plates (5000 cells/well and 100000 cells/well, respectively) and cultured in DMEM containing 10% FBS for 24 h and then the media was replaced for serum-free media. The polyplexes containing 1 μg of pDNA were added for 4 h. The media was then again replaced for the fresh complete media. After 48 h, complete media was removed and 120 μL of mixture (5:1 v/v) of fresh DMEM and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg mL−1) was added to the cells. Cells were incubated for a further 4 h, then DMSO (150 μL) was added and cells were shaken for 15 min at room temperature The absorbance at 562 nm was recorded in a plate reader (Molecular Devices, Sunnyvale, CA). ExGen 500 was used as a positive control. Each treatment was performed 8 times.
The Cytotoxicity of PEG-b-pAsp(DET)-Based Cationic Copolymers:
The cytotoxicity of mPEG-b-pAsp(DET) and Man-PEG-b-pAsp(DET) cationic copolymers was measured using CCK-8 assay according to the protocol provided by the manufacturer. Briefly, IC-21 macrophages were seeded in 96-well plates at 5000 cells/well and cultured in DMEM containing 10% FBS for 24 h and then the media was replaced for serum-free media containing cationic copolymers at different concentrations. After that the cells were incubated with the polymer for additional 24 h. The media was then again replaced for the fresh complete media and 10 μl of CCK-8 solution (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) was added to the cells. The absorbance at 450 nm was recorded in a plate reader after a 1 h incubation. Each treatment was performed in triplicate.
In vitro Gene Delivery: Transfection with PEG-b-PLL-Based Polyplexes:
For the in vitro transfection study, NIH/3T3 and BMMΦ were seeded into 24-well plates (5000 cells/well and 100000 cells/well, respectively) and then incubated in antibiotic-free DMEM containing 10% FBS for 24 h. Then the media was replaced for the serum-free media and the polyplexes containing 1 μg of pDNA (gWiz-GFP) were added for 4 h at 37 °C. The pDNA and ExGen 500 were used as negative and positive control, respectively. Cells were rinsed with serum-free media after and incubated in complete media containing 10% FBS and 1% P/S for 48 h. After washing with HBSS the cells were harvested using 0.05% Trypsin-EDTA, followed by addition of PBS with 10% FBS (1 mL). The cells were pelleted by centrifugation and resuspended in PBS + 1% BSA (500 μL). The percentage of cells expressing GFP was assessed by flow cytometry in triplicates using a Beckton Dickinson FACStarPlus device operating under Lysis II (San Jose, CA, USA), equipped with an argon ion laser (488 nm emission wavelength). The filter used for emission was a 530/30 nm band-pass. Results are expressed as % GFP transfected cells.
Transfection with PEG-b-pAsp(DET)-Based Polyplexes:
For the in vitro transfection study, IC-21 macrophages were seeded into 24-well plates at 100000 cells/well and then incubated in antibiotic-free DMEM containing 10% FBS for 24 h. Then the media was replaced for the serum-free media and the polyplexes containing 1 μg of pDNA (gWiz-Luc) were added for 4, 8, or 24 h at 37 °C. Cells were rinsed with serum-free media after and incubated in complete media containing 10% FBS for 24 h. The cells were treated with the lysis buffer for 2 h, and the bioluminescence was determined in the lysate in triplicates using a TD20/20 luminometer (Promega, Fitchburg, WI, USA) according to a standard procedure. The protein concentration was determined using BCA assay and the data were normalized for the amount of the protein.
Supplementary Material
Acknowledgements
This study was supported in part by the Eshelman Institute for Innovation grant “Systemic Targeting of Mononuclear Phagocytes for Parkinson’s Disease Gene Therapy” (AVK), National Cancer Institute grant 1R21CA220148 (AVK), Russian Foundation for Basic Research grants 17–54-33027 (NLK), and the Russian Science Foundation grant 20–63-46029 (AVK, NLK, AVL).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Data Availability Statement
Data available on request from the authors.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Anton V. Lopukhov, Laboratory for Chemical Design of Bionanomaterials Faculty of Chemistry M. V. Lomonosov Moscow State University, 1 Leninskie Gory, Moscow 117234, Russia.
Zigang Yang, Department of Pharmaceutical Sciences and Center for Drug Delivery and Nanomedicine, College of Pharmacy, University of Nebraska Medical Center, 985830 Nebraska Medical Center, Omaha, NE 68198, USA.
Matthew J. Haney, Division of Pharmacoengineering and Molecular Pharmaceutics, Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina, 125 Mason Farm Road, Chapel Hill, NC 27599, USA
Tatiana K. Bronich, Department of Pharmaceutical Sciences and Center for Drug Delivery and Nanomedicine, College of Pharmacy, University of Nebraska Medical Center, 985830 Nebraska Medical Center, Omaha, NE 68198, USA
Marina Sokolsky-Papkov, Division of Pharmacoengineering and Molecular Pharmaceutics, Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina, 125 Mason Farm Road, Chapel Hill, NC 27599, USA.
Elena V. Batrakova, Division of Pharmacoengineering and Molecular Pharmaceutics, Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina, 125 Mason Farm Road, Chapel Hill, NC 27599, USA
Natalia L. Klyachko, Laboratory for Chemical Design of Bionanomaterials Faculty of Chemistry M. V. Lomonosov Moscow State University, 1 Leninskie Gory, Moscow 117234, Russia; Division of Pharmacoengineering and Molecular Pharmaceutics, Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina, 125 Mason Farm Road, Chapel Hill, NC 27599, USA.
Alexander V. Kabanov, Laboratory for Chemical Design of Bionanomaterials Faculty of Chemistry M. V. Lomonosov Moscow State University, 1 Leninskie Gory, Moscow 117234, Russia; Division of Pharmacoengineering and Molecular Pharmaceutics, Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina, 125 Mason Farm Road, Chapel Hill, NC 27599, USA.
References
- [1].Chew SA, Hacker MC, Saraf A, Raphael RM, Kasper FK, Mikos AG, Biomacromolecules. 2009, 10, 2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schaffert D, Wagner E, Gene Ther. 2008, 15, 1131. [DOI] [PubMed] [Google Scholar]
- [3].Hao J, Li SK, Kao WWY, Liu CY, Brain Res. Bull. 2010, 81, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Long HA, McMillan JR, Qiao HJ, Akiyama M, Shimizu H, Curr. Gene Ther. 2009, 9, 487. [DOI] [PubMed] [Google Scholar]
- [5].Koeberl DD, Kishnani PS, Curr. Gene Ther. 2009, 9, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Choi D, Biotechnol. Bioprocess Eng. 2007, 12, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wayne EC, Long C, Haney MJ, Batrakova EV, Leisner TM, Parise LV, Kabanov AV, Adv. Sci. 2019, 6, 1900582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lahmar Q, Keirsse J, Laoui D, Movahedi K, Van Overmeire E, Van Ginderachter JA, Biochim. Biophys. Acta, Rev. Cancer 2016, 1865, 23. [DOI] [PubMed] [Google Scholar]
- [9].Isacke C, East L, Bioch. Biophys. Acta 2002, 1572, 364. [DOI] [PubMed] [Google Scholar]
- [10].Heise CT, Taylor ME, Feinberg H, Park-Snyder S, Kolatkar AR, Weis WI, J. Bio. Chem. 2000, 275, 21539. [DOI] [PubMed] [Google Scholar]
- [11].Taylor ME, Bezouska K, Drickamer K, J. Bio. Chem. 1992, 267, 1719. [PubMed] [Google Scholar]
- [12].Gao X, Mao D, Zuo X, Hu F, Cao J, Zhang P, Sun JZ, Liu J, Liu B, Tang BZ, Anal. Chem. 2019, 91, 6836. [DOI] [PubMed] [Google Scholar]
- [13].Yin L, Chen Y, Zhang Z, Yin Q, Zheng N, Cheng J, Macromol. Rapid Commun. 2015, 36, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Su FY, Srinivasan S, Lee B, Chen J, Convertine AJ, West TE, Ratnera DM, Skerrett SJ, Stayton PS, J. Controlled Release 2018, 287, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Filatova LY, Klyachko NL, Kudryashova EV, Russ. Chem. Rev. 2018, 87, 374. [Google Scholar]
- [16].Francis AP, Jayakrishnan A, J. Biomater. Sci., Polym. Ed. 2019, 30, 1471. [DOI] [PubMed] [Google Scholar]
- [17].Smith J, Lowsberg D, bioRxiv 2019, 620906. [Google Scholar]
- [18].De Vlaeminck Y, Lecocq Q, Giron P, Heirman C, Geeraerts X, Bolli E, Goyvaerts C, J. Controlled Release 2019, 299, 107. [DOI] [PubMed] [Google Scholar]
- [19].Hattori Y, Kawakami S, Nakamura K, Yamashita F, Hashida M, J. Pharmacol. Exp. Ther. 2006, 318, 828. [DOI] [PubMed] [Google Scholar]
- [20].Yu SS, Lau CM, Barham WJ, Onishko HM, Nelson CE, Li H, Giorgio TD, Mol. Pharmaceutics 2013, 10, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fukuda I, Mochizuki S, Sakurai K, Biomed Res. Int. 2015, 2105, 350580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Glass EB, Masjedi S, Dudzinski SO, Wilson AJ, Duvall CL, Yull FE, Giorgio TD, ACS Omega 2019, 4, 16756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zang X, Zhang X, Zhao X, Hu H, Qiao M, Deng Y, Chen D, Mol. Pharmaceutics 2019, 16, 1714. [DOI] [PubMed] [Google Scholar]
- [24].He H, Yuan Q, Bie J, Wallace RL, Yannie PJ, Wang J, LancinaIII MG, Zolotarskaya OY, Korzun W, Yang H, Ghosh S, Transl. Res. 2018, 193, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gan J, Dou Y, Li Y, Wang Z, Wang L, Liu S, Li Q, Yu H, Liu C, Han C, Huang Z, Zhang J, Wang C, Dong L, Biomaterials 2018, 178, 95. [DOI] [PubMed] [Google Scholar]
- [26].Costa A, Sarmento B, Seabra V, Eur. J. Pharm. Sci 2018, 114, 103. [DOI] [PubMed] [Google Scholar]
- [27].Shibaguchi K, Tamura A, Terauchi M, Matsumura M, Miura H, Yui N, Molecules 2019, 24, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ferkol T, Perales JC, Mularo F, Hanson RW, Proc. Natl. Acad. Sci. USA 1996, 93, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nguyen HK, Lemieux P, Vinogradov SV, Gebhart CL, Guerin N, Paradis G, Kabanov AV, Gene Ther. 2000, 7, 126. [DOI] [PubMed] [Google Scholar]
- [30].Sato A, Kawakami S, Yamada M, Yamashita F, Hashida M, J. Drug Targeting 2001, 9, 201. [DOI] [PubMed] [Google Scholar]
- [31].Jiang HL, Kim YK, Arote R, Jere D, Quan JS, Yu JH, Cho CS, Int. J. Pharm. 2009, 375, 133. [DOI] [PubMed] [Google Scholar]
- [32].Ruan GX, Chen YZ, Yao XL, Du A, Tang GP, Shen YQ, Gao JQ, Acta Biomater. 2014, 10, 1847. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Y, Wang Y, Zhang C, Wang J, Pan D, Liu J, Feng F, ACS Appl. Mater. Interfaces 2016, 8, 3719. [DOI] [PubMed] [Google Scholar]
- [34].Mishra S, Webster P, Davis ME, Eur. J. Cell Biol. 2004, 83, 97. [DOI] [PubMed] [Google Scholar]
- [35].Raviv L, Jaron-Mendelson M, David A, Mol. Pharmaceutics 2015, 12, 453. [DOI] [PubMed] [Google Scholar]
- [36].Romoren K, Thu BJ, Bols NC, Evensen O, Biochim. Biophys. Acta, Biomembr. 2004, 1663, 127. [DOI] [PubMed] [Google Scholar]
- [37].Dokka S, Toledo D, Shi XL, Ye JP, Int. J. Pharm 2000, 206, 97. [DOI] [PubMed] [Google Scholar]
- [38].Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K, J. Am. Chem. Soc 2004, 126, 2355. [DOI] [PubMed] [Google Scholar]
- [39].Wang Y, Chen P, Shen J, Biomaterials 2006, 27, 5292. [DOI] [PubMed] [Google Scholar]
- [40].Aravindan L, Bicknell KA, Brooks G, Khutoryanskiy VV, Williams AC, Macromol. Biosci 2013, 13, 1163. [DOI] [PubMed] [Google Scholar]
- [41].Novo L, van Gaal EV, Mastrobattista E, van Nostrum CF, Hennink WE, J. Controlled Release 2013, 169, 246. [DOI] [PubMed] [Google Scholar]
- [42].Bockuviene A, Slavuckyte K, Vareikis A, Zigmantas S, Zaliauskiene L, Makuska R, Macromol. Biosci 2016, 16, 1497. [DOI] [PubMed] [Google Scholar]
- [43].Harada A, Kataoka K, Macromolecules 1995, 28, 5294. [Google Scholar]
- [44].Yamasaki Y, Katayose S, Kataoka K, Yoshikawa K, Macromolecules 2003, 36, 6276. [Google Scholar]
- [45].Itaka K, Yamauchi K, Harada A, Nakamura K, Kawaguchi H, Kataoka K, Biomaterials 2003, 24, 4495. [DOI] [PubMed] [Google Scholar]
- [46].Vanderkerken S, Vanheede T, Toncheva V, Schacht E, Wolfert MA, Seymour L, Urtti A, J. Bioact. Compat. Polym 2000, 15, 115. [Google Scholar]
- [47].Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, Pasumarthy MK, Mol. Ther 2003, 8, 936. [DOI] [PubMed] [Google Scholar]
- [48].Kanayama N, Fukushima S, Nishiyama N, Itaka K, Jang WD, Miyata K, Yamasaki Y, Chung UI, Kataoka K, ChemMedChem 2006, 1, 439. [DOI] [PubMed] [Google Scholar]
- [49].Miyata K, Fukushima S, Nishiyama N, Yamasaki Y, Kataoka K, J. Controlled Release 2007, 122, 252. [DOI] [PubMed] [Google Scholar]
- [50].Dou H, Destache CJ, Morehead JR, Mosley RL, Boska MD, Kingsley J, Werling J, Blood 2006, 108, 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Itaka K, Ishii T, Hasegawa Y, Kataoka K, Biomaterials 2010, 31, 3707. [DOI] [PubMed] [Google Scholar]
- [52].Yokoyama M, Kwon GS, Okano T, Sakurai Y, Seto T, Kataoka K, Bioconjugate Chem. 1992, 3, 295. [DOI] [PubMed] [Google Scholar]
- [53].Bronich TK, Popov AM, Eisenberg A, Kabanov VA, Kabanov AV, Langmuir 2000, 16, 481. [Google Scholar]
- [54].Gref R, Lück M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Müller RH, Colloids Surf., B 2000, 18, 301. [DOI] [PubMed] [Google Scholar]
- [55].Takeda KM, Osada K, Tockary TA, Dirisala A, Chen Q, Kataoka K, Biomacromolecules 2017, 18, 36. [DOI] [PubMed] [Google Scholar]
- [56].Osada K, Polym. J. 2019, 51, 381. [Google Scholar]
- [57].Chen WY, Zheng JX, Cheng SZ, Li CY, Huang P, Zhu L, Lotz B, Phys. Rev. Lett. 2004, 93, 028301. [DOI] [PubMed] [Google Scholar]
- [58].Fan MM, Zhang WZ, Cheng C, Liu Y, Li BJ, Sun X, Zhang S, Part. Part. Syst. Charact. 2014, 31, 994. [Google Scholar]
- [59].Dirisala A, Osada K, Chen Q, Tockary TA, Machitani K, Osawa S, Uchida S, Biomaterials 2014, 35, 5359. [DOI] [PubMed] [Google Scholar]
- [60].Garapaty A, Champion JA, Bioeng. Transl. Med. 2017, 2, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sharma G, Valenta DT, Altman Y, Harvey S, Xie H, Mitragotri S, Smith JW, J. Controlled Release 2010, 147, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nowacek AS, Balkundi S, McMillan J, Roy U, Martinez-Skinner A, Mosley RL, Gendelman HE, J. Controlled Release 2011, 150, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dey D, Kumar S, Banerjee R, Maiti S, Dhara D, J. Phys. Chem. B 2014, 118, 7012. [DOI] [PubMed] [Google Scholar]
- [64].Oupický D, Carlisle RC, Seymour LW, Gene Ther. 2001, 8, 713. [DOI] [PubMed] [Google Scholar]
- [65].Chamberlain LM, Holt-Casper D, Gonzalez-Juarrero M, Grainger DW, J. Biomed. Mater. Res., Part A 2015, 103, 2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chamberlain LM, Godek ML, Gonzalez-Juarrero M, Grainger DW, J. Biomed. Mater. Res., Part A 2009, 88, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kabanov AV, Vinogradov SV, Suzdaltseva YG, Alakhov VY, Bioconjugate Chem. 1995, 6, 639. [DOI] [PubMed] [Google Scholar]
- [68].Kataoka K, Togawa H, Harada A, Yasugi K, Matsumoto T, Katayose S, Macromolecules 1996, 29, 8556. [Google Scholar]
- [69].Vinogradov SV, Bronich TK, Kabanov AV, Bioconjugate Chem. 1998, 9, 805. [DOI] [PubMed] [Google Scholar]
- [70].Erbacher P, Roche A, Hum. Gene Ther. 1996, 7, 721. [DOI] [PubMed] [Google Scholar]
- [71].Diebold S, Zenke M, J. Bio. Chem. 1999, 274, 19087. [DOI] [PubMed] [Google Scholar]
- [72].Hashimoto M, Sato T, Biotechnol. Lett. 2006, 28, 815. [DOI] [PubMed] [Google Scholar]
- [73].Cho C, Jiang H, Biomaterials 2008, 29, 884.18031806 [Google Scholar]
- [74].Yu W, Xu W, Pharm. Res. 2010, 27, 1584. [DOI] [PubMed] [Google Scholar]
- [75].Manickam DS, Brynskikh AM, Kopanic JL, Sorgen PL, Klyachko NL, Batrakova EV, Kabanov AV, J. Controlled Release 2012, 162, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klein PM, Wagner E, Antioxid. Redox Signaling 2014, 21, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Martinez-Pomares L, Reid DM, Brown GD, Taylor PR, Stillion RJ, Linehan SA, Wong SY, J. Leukocyte Biol. 2003, 73, 604. [DOI] [PubMed] [Google Scholar]
- [78].Miyata K, Nishiyama N, Kataoka K, Chem. Soc. Rev 2012, 41–7, 2562. [DOI] [PubMed] [Google Scholar]
- [79].Tominaga H, Ishiyama M, Ohseto F, Sasamoto K, Hamamoto T, Suzuki K, Watanabe M, Anal. Commun 1999, 36, 47. [Google Scholar]
- [80].Jiang Y, Arounleut P, Rheiner S, Bae Y, Kabanov AV, Milligan C, Manickam DS, J. Controlled Release 2016, 231, 38. [DOI] [PubMed] [Google Scholar]
- [81].Stahl P, Schlesinger PH, Sigardson E, Rodman JS, Lee YC, Cell 1980, 19, 207. [DOI] [PubMed] [Google Scholar]
- [82].Steinman RM, Mellman IS, Muller WA, Cohn ZA, Cell Biol J 1983, 96, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Stahl PD, Am. J. Respir. Cell Mol. Biol. 1990, 2, 317. [DOI] [PubMed] [Google Scholar]
- [84].Sheikh H, Yarwood H, Ashworth A, Isacke CM, J. Cell Sci. 2000, 113, 1021. [DOI] [PubMed] [Google Scholar]
- [85].Storrie B, Exp. Cell Res. 1979, 118, 135. [DOI] [PubMed] [Google Scholar]
- [86].Fiani ML, Beitz J, Turvy D, Blum JS, Stahl PD, J. Leukocyte Biol. 1998, 64, 85. [DOI] [PubMed] [Google Scholar]
- [87].Lee YC, Lonn H, J. Bio. Chem 1983, 258, 199. [PubMed] [Google Scholar]
- [88].Dam T, Brewer C, J. Bio. Chem 2000, 275, 14223. [DOI] [PubMed] [Google Scholar]
- [89].Katakura H, Harada A, Kataoka K, Furusho M, Tanaka F, Wada H, Ikenaka K, J. Gene Med. 2004, 6, 471. [DOI] [PubMed] [Google Scholar]
- [90].Jiang Y, Arounleut P, Rheiner S, Bae Y, Kabanov AV, Milligan C, Manickam DS, J. Controlled Release 2016, 231, 38. [DOI] [PubMed] [Google Scholar]
- [91].Godek ML, Michel R, Chamberlain LM, Castner DG, Grainger DW, J. Biomed. Mater. Res., Part A 2009, 88A, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gaymalov ZZ, Yang Z, Pisarev VM, Alakhov VY, Kabanov AV, Biomaterials 2009, 30, 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Close DM, Hahn RE, Patterson SS, Ripp SA, Sayler GS, Baek SJ, J. Biomed. Opt 2011, 16, 047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


