Abstract
Background:
While sentinel lymph node (SLN) biopsy is a standard procedure used to identify patients at risk for melanoma recurrence, it fails to accurately risk stratify certain patients. Since processes in SLNs regulate anti-tumor immune responses, we hypothesize that SLN gene expression may be used for risk stratification.
Methods:
The Nanostring nCounter® PanCancer Immune Profiling Panel was used to quantify expression of 730 immune-related genes in sixty SLN specimens (31 positive [pSLN], 29 negative [nSLN]) from a retrospective melanoma cohort. A multivariate prediction model for recurrence-free survival (RFS) was created by applying stepwise variable selection to Cox regression models; risk scores calculated using the model were used to stratify patients into low- and high-risk groups. Predictive power of the model was assessed using the Kaplan-Meier and log-rank tests.
Results:
At a median follow up of 6.3 years, 20 patients (33.3%) developed recurrence (14/31 [45.2%] pSLN and 6/29 [20.7%] nSLN, p=0.0445). A fitted Cox regression model incorporating twelve genes accurately predicted RFS (C-index 0.9919); Improved RFS was associated with increased expression of TIGIT (p = 0.0326), an immune checkpoint, and decreased expression of CXCL16 (p = 0.0273), a cytokine important in promoting dendritic and T cell interactions. Independent of SLN status, our model was able to stratify patients into cohorts at high- and low-risk for recurrence (log-rank p<0.001).
Conclusions:
SLN gene expression profiles are associated with melanoma recurrence, and may be able to identify patients as high or low risk regardless of SLN status, potentially enhancing patient selection for adjuvant therapy.
Introduction:
While surgical resection of the primary tumor and associated tumor draining lymph nodes via sentinel lymph node biopsy (SLNB) is potentially curative for localized melanoma, a significant proportion of patients go on to develop recurrence.1,2 Multiple effective adjuvant therapies are now available, which can significantly reduce recurrence risk in certain patients. Early adjuvant therapy may eradicate residual disease, prevent metastatic spread, and improve recurrence-free and overall survival, but these therapies come with potential for myriad side effects and significant financial cost to patients and the health system.3–8 Appropriate risk stratification is needed to identify patients at risk for recurrence, and therefore guide the use of adjuvant therapy to optimize benefit and minimize risks.
Current risk stratification and recommendations for consideration of adjuvant therapy are largely based on histopathologic assessment of the sentinel lymph nodes (SLN) for presence of tumor metastases.9 Patients with evidence of metastatic disease in the SLN are classified as having American Joint Committee on Cancer (AJCC) Stage III disease and considered eligible for adjuvant therapies. Conversely, patients without evidence of tumor spread are classified as AJCC Stage II and are currently ineligible for adjuvant therapies. Additional factors that mediate the risk of recurrence and are considered when deciding whether to pursue adjuvant therapy include extranodal extension and size of the metastatic tumor deposit, as well as primary tumor Breslow depth and presence or absence of ulceration.9,10 It is now widely accepted that patients with >1–2 mm tumor burden in the sentinel node (Stage III) should be considered for adjuvant therapy, as patients with smaller deposits and thin (<2 mm) non-ulcerated primary tumors are at lower risk for recurrence.9 However, this current strategy still fails to definitively risk stratify certain patients, particularly those with low-risk Stage III disease (Stage IIIA) and high-risk Stage II (Stage IIB/C) patients who actually have a worse prognosis than those with Stage IIIA disease.11
In order to better inform prognosis and maximize therapeutic benefits of additional therapy while minimizing toxicities, more precise risk stratification is needed for patients with locoregional melanoma. As an initial site of antigen presentation as well as early metastasis, SLNs serve as an interface between tumor cells and the immune system.12 Immunologic processes occurring at this interface can lead to tumor control and elimination, or allow disease progression and spread, and are critical to development of an effective anti-tumor immune response.13,14 Characterization of these processes may allow for more precise risk stratification of patients undergoing SLNB. In this study, we characterize immunologic gene profiles of SLN in melanoma patients and create a risk-stratification model to predict recurrence based on these immunologic gene profiles.
Methods:
Patient Selection
This study was approved by the Duke University Institutional Review Board. Patients who underwent SLNB, had their care managed at Duke University Hospital between 2001 and 2012, and had SLN formalin-fixed, paraffin-embedded (FFPE) blocks available in an institutional tissue bank were identified retrospectively. This time period pre-dates modern adjuvant therapy, and was chosen to avoid potential confounding from adjuvant therapy use in patients with positive SLN (pSLN). Patients were chosen from this cohort who had clinical data available, adequate follow up time, and a SLN specimen that was grossly at least 1 cm in size to allow for three 10 mm slides to be cut without using the entire block. Finally, the sample size was chosen to allow for relatively equal numbers of positive and negative SLNs (nSLN).
Sixty-seven SLN specimen candidates were identified that met the above criteria. Each SLN specimen was confirmed to have nodal tissue on the slide and confirmed as positive or negative for presence of tumor by a trained dermatopathologist using hematoxylin and eosin (H&E) staining of slides cut from the FFPE blocks (Figure 1). Deidentified FFPE samples were then delivered to the NanoString core facility and RNA was isolated from the FFPE blocks using the Maxwell® 16 MDx system. Sample quality control was performed using the Agilent TapeStation 2200 system. Sixty-two of the 67 RNA samples were considered of adequate quality and were hybridized for immune profiling using NanoString nCounter® technology.
Figure 1:
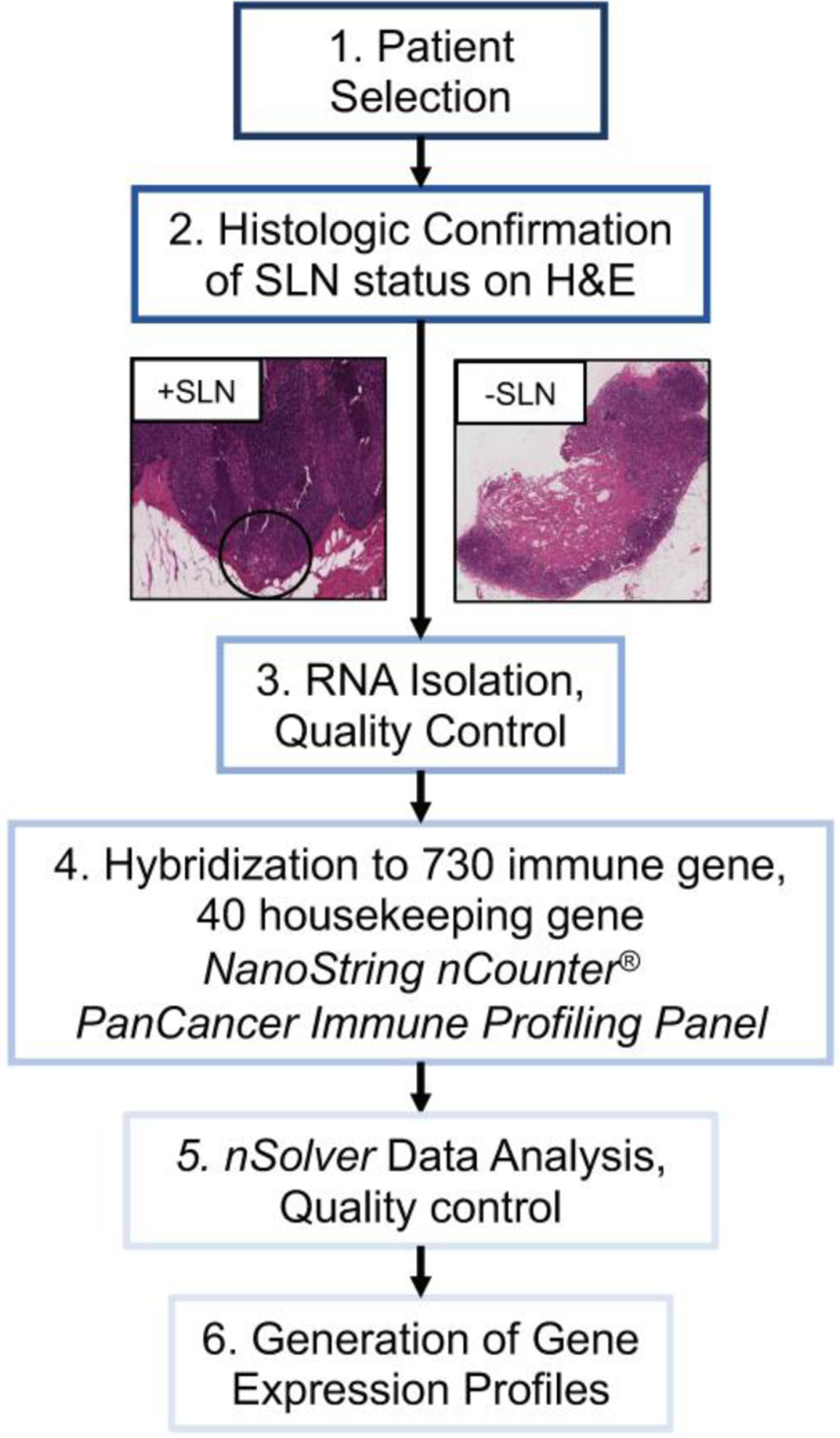
Trial schema depicting overall flow of study from patient selection through generation of individualized gene expression profiles. Patients selected had invasive melanoma diagnosed between 2001–2012 and underwent sentinel lymph node (SLN) biopsy. All patients had formalin-fixed paraffin-embedded SLN specimens available, which were used to confirm SLN status on hematoxylin and eosin (H&E) prior to RNA extraction and hybridization to the NanoString nCounter PanCancer Immune Profiling Panel. nSolver data analysis software was used to evaluate quality of hybridization and generate gene expression profiles for each patient.
Immune Profiling
NanoString nCounter® is an RNA-based technology that allows for digital quantification of multiplexed target molecules through the use of molecular barcodes without the need for amplification, allowing for quantification of target molecules on small amounts of tissue including FFPE tissue samples.15,16 The NanoString nCounter PanCancer Immune Profiling Panel (NanoString Technologies Inc, Seattle, Washington, USA; Full gene list available in Supplementary Table 1) is a highly multiplexed gene expression panel designed to quantify 730 genes related to immune cell profiling and function across the innate and adaptive immune systems and tumor specific antigens, in addition to 40 housekeeping genes that facilitate sample-to-sample normalization for quality assurance. Nanostring nCounter® technology and nSolver Analysis Software were used to quantify the gene expression of these 770 genes in the 62 isolated RNA samples. Two of the 62 samples had inadequate hybridization and were excluded, leading to the creation of 60 gene expression profiles: 31 from patients with a pSLN, and 29 with a nSLN. One of these 60 patients, with a pSLN, had borderline RNA quality as shown by assessment of the 40 housekeeping genes, however it was considered sufficiently adequate and it was decided to include this sample in the analysis.
Clinical Characteristics
Retrospective chart review was performed to collect demographic and clinical information for all sixty patients including age, gender, tumor thickness (Breslow depth), presence or absence of ulceration, SLN status (positive or negative), size of SLN metastasis (if applicable), presence or absence of extranodal extension, receipt of adjuvant therapy, type of adjuvant therapy, receipt of any additional treatment (yes or no) including adjuvant therapy or treatment for recurrence, type of additional treatment, development of recurrence (yes or no), interval of recurrence from SLNB, recurrence site (nodal or distant) and vital status. Chart review was blinded to genomic expression profile to limit bias.
Univariate Analysis
As an initial exploratory analysis, we aimed to use the gene expression data from the NanoString nCounter® PanCancer Immune Profiling Panel expression data to identify genes associated with recurrence-free survival (RFS). A univariate Cox proportional hazards regression model was used to identify any individual genes from the 730-gene NanoString nCounter® PanCancer Immune Profiling Panel were associated with disease recurrence. All gene expression data were log-transformed before this analysis. Genes with a two-sided p-value <0.2 are reported (Table 2).
Table 2.
Genes identified on univariate Cox regression whose expression was associated with the development of melanoma recurrence (p<0.2). Listed are the gene probe names from the NanoString nCounter® PanCancer Immune Profiling Panel, coefficient estimates, standard errors, chi-square statistics, p-values and their estimated hazard ratios. Hazard ratios of less than one indicate having high expression of the gene is associated with reduced risk of disease recurrence and vice versa.
| Gene Probe Name | Gene Name | Coefficient Estimate | Standard Error | Wald Chi-square | P-Value | Hazard Ratio |
|---|---|---|---|---|---|---|
| Gene Probes Positively Associated with Recurrence | ||||||
| MBL2 | Mannose-binding lectin (protein C) 2 | 2.64277 | 1.38206 | 3.6565 | 0.0559 | 14.052 |
| AICDA | Activation-induced cytidine deaminase | 0.93905 | 0.69521 | 1.8245 | 0.1768 | 2.558 |
| IL8 | C-X-C motif chemokine ligand 8 | 0.54307 | 0.41403 | 1.7205 | 0.1896 | 1.721 |
| Gene Probes Negatively Associated with Recurrence | ||||||
| NEFL | Neurofilament, light polypeptide | −1.6538 | 0.799 | 4.284 | 0.0385 | 0.191 |
| SEMG1 | Semenogelin I | −2.1099 | 1.19426 | 3.1211 | 0.0773 | 0.121 |
| CCR3 | Chemokine (C-C motif) receptor 3 | −1.4524 | 0.82534 | 3.0966 | 0.0785 | 0.234 |
| TPTE | Transmembrane phosphatase with tensin homology | −2.514 | 1.49594 | 2.8243 | 0.0928 | 0.081 |
| IL19 | Interleukin 19 | −1.658 | 1.0058 | 2.7172 | 0.0993 | 0.191 |
| KIR3DL3 | Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 | −2.1613 | 1.31425 | 2.7044 | 0.1001 | 0.115 |
| CD1E | CD1e molecule | −0.8081 | 0.4945 | 2.6708 | 0.1022 | 0.446 |
| MAGEC2 | Melanoma antigen family C, 2 | −2.0605 | 1.26815 | 2.6401 | 0.1042 | 0.127 |
| CD3G | CD3g molecule, gamma (CD3-TCR complex) | −0.5477 | 0.3379 | 2.6271 | 0.1051 | 0.578 |
| CCL24 | Chemokine (C-C motif) ligand 24 | −0.655 | 0.43404 | 2.2773 | 0.1313 | 0.519 |
| IL13 | Interleukin 13 | −2.0541 | 1.37108 | 2.2445 | 0.1341 | 0.128 |
| ZAP70 | Zeta-chain (TCR) associated protein kinase 70kDa | −0.608 | 0.40806 | 2.2197 | 0.1363 | 0.544 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | −0.6698 | 0.46725 | 2.0547 | 0.1517 | 0.512 |
| CD7 | CD7 molecule | −0.5888 | 0.4215 | 1.9514 | 0.1624 | 0.555 |
| LAIR2 | Leukocyte-associated immunoglobulin-like receptor 2 | −0.5642 | 0.41283 | 1.8679 | 0.1717 | 0.569 |
| SPN | Sialophorin | −0.4945 | 0.36416 | 1.8441 | 0.1745 | 0.61 |
| ELANE | Elastase, neutrophil expressed | −1.7578 | 1.29881 | 1.8316 | 0.1759 | 0.172 |
| ARG1 | Arginase, liver | −1.1456 | 0.85266 | 1.8051 | 0.1791 | 0.318 |
| CD3D | CD3d molecule, delta (CD3-TCR complex) | −0.4163 | 0.31366 | 1.7614 | 0.1845 | 0.659 |
| CRP | C-reactive protein, pentraxin-related | −1.5624 | 1.1773 | 1.7611 | 0.1845 | 0.21 |
| FLT3LG | Fms-related tyrosine kinase 3 ligand | −0.5333 | 0.40423 | 1.7405 | 0.1871 | 0.587 |
| CTAGE1 | Cutaneous T-cell lymphoma-associated antigen 1 | −1.8823 | 1.42907 | 1.7348 | 0.1878 | 0.152 |
| RORC | RAR-related orphan receptor C | −0.7594 | 0.57951 | 1.7172 | 0.1901 | 0.468 |
| MASP2 | Mannan-binding lectin serine peptidase 2 | −2.4156 | 1.86224 | 1.6826 | 0.1946 | 0.089 |
| HLA_DRB4 | Major histocompatibility complex, class II, DR beta 4 | −0.2954 | 0.22937 | 1.6582 | 0.1979 | 0.744 |
Multivariate analysis
We then developed multivariate models for RFS. Using the gene expression data of the 730 genes quantified by the NanoString nCounter PanCancer Immune Profiling Panel, a model was created by selecting genes whose expression was associated with recurrence; this was done by applying stepwise variable selection to Cox regression models for RFS which allowed selection of relevant genes (variables) from the 730 genes quantified by the panel, using alpha levels of 0.05 for insertion and 0.1 for deletion. The resulting model incorporated twelve of the 730 genes and allowed for the calculation of risk scores for each patient based on the linear combination of regression estimates and their quantified expression of the 12 selected genes. These risk scores were used to stratify patients as high risk (above the median) or low risk (below the median) for recurrence. Kaplan-Meier analysis, log rank test and pairwise comparison were used to evaluate performance of the model and compare RFS for patients with high versus low risk scores. Finally, an additional multivariate Cox regression model was created in the same way, but selecting variables from the 730 genes as well as five relevant clinical characteristics, specifically Breslow depth, presence of ulceration, gender, age at time of surgery, and receipt of additional therapy. Harrell’s c-statistic was used to evaluate the performance of the fitted regression models for RFS. Patients who did not develop recurrence or were lost to follow-up were censored at time of their last known survival status.
Results:
Patient Population
Of 67 patients initially identified who underwent SLNB, had their care managed at Duke University Hospital between 2001 and 2012 and had FFPE blocks available, adequate RNA specimens were isolated from 62 patients. Of these 62 RNA specimens, adequate hybridization and immune gene expression profiles were generated for 60 patients.
Among the 60 patients for whom adequate gene expression profiles were generated, 31 were confirmed on H&E to have a pSLN and 29 were confirmed to have a nSLN. Patients with positive and negative SLN were similar in age, gender, and presence of ulceration (Table 1). Patients with pSLN had thicker primary tumor Breslow depths (median 2.5 mm vs 1.4 mm, p=0.0019). Among the 31 patients with pSLN, 2 had isolated cells on pathologic analysis, 12 had nodal metastases ≤1 mm, 14 had nodal metastases >1 mm and 3 did not have available data. The median size of the nodal metastasis was 1.9 mm (Interquartile Range [IQR] 0.3–5.0 mm) and the minority had extranodal extension (24.1%). The majority of pSLN patients had a single pSLN (67.7%), while 8 (25.8%) had two pSLNs and 2 (6.5%) had three pSLNs. The majority of patients with pSLN (90.3%) underwent immediate completion lymph node dissection (CLND). Eleven patients (35.5%) with a pSLN received adjuvant therapy: nine with high-dose interferon, one with ipilimumab on a clinical trial and one with a vaccine trial. The remainder of the pSLN patients and all of the nSLN patients did not receive adjuvant therapy. Patients with pSLN more often received any type of additional therapy (including adjuvant therapy as noted above, chemotherapy, radiation therapy, targeted therapy or immune therapy for recurrence, 51.6% vs 6.9%, p=0.0002).
Table 1.
Background characteristics of study patients stratified by positive or negative sentinel lymph node (SLN) status
| Positive SLN (n=31) | Negative SLN (n=29) | |
|---|---|---|
| Age, years, median (IQR) | 56.6 (47.5–64.3) | 54.2 (47.5–60.2) |
| Sex, female | 14 (45.2%) | 18 (62.1%) |
| Breslow depth, mm (median, IQR) | 2.5 (2.0–4.1) | 1.4 (0.9–2.5) |
| Ulceration, yes | 12 (42.9%) | 11 (39.3%) |
| Size SLN metastasis, mm, median (IQR) | 1.9 (0.3–5.0) | n/a |
| Isolated cells | 2 (7.1%) | n/a |
| <=1mm | 12 (42.9%) | n/a |
| >1mm | 14 (50.0%) | n/a |
| # Positive SLN | ||
| 1 | 21 (67.7%) | n/a |
| 2 | 8 (25.8%) | n/a |
| 3 | 2 (6.5%) | n/a |
| Extranodal extension, yes | 7 (24.1%) | n/a |
| Immediate CLND, yes | 28 (90.3%) | 0 (0.0%) |
| None | 20 (64.5%) | 29 (100.0%) |
| Any additional treatment*, yes | 16 (51.6%) | 2 (6.9%) |
| Recurrence, yes | 14 (45.2%) | 6 (20.7%) |
| Death, yes | 11 (35.5%) | 3 (10.3%) |
IQR: Interquartile range, SLN: sentinel lymph node, CLND: Completion lymph node dissection.
Any additional treatment includes receipt of any additional treatment following sentinel lymph node biopsy other than surgery, including adjuvant therapy or treatment with immune therapy, targeted therapy, chemotherapy or radiation.
At a median follow up of 6.3 years, 20 patients (33.3%) developed melanoma recurrence (pSLN vs nSLN: 45.2% vs 20.7%, p=0.0445). Compared to patients who did not develop recurrence, patients who developed recurrence had thicker median tumor Breslow depth (2.5 mm vs 1.8 mm) and more frequently had ulceration present (45.0% vs 35.0%). Among those with a pSLN and recurrence, initial recurrence was in transit alone (3 of 14), nodal alone (2 of 14), pulmonary (2 of 14), subcutaneous (2 of 14) or at multiple simultaneous sites (5 of 14; pulmonary and cranial metastases [n=2], in transit with pelvic metastases [n=1]; nodal and hepatic metastases [n=1], subcutaneous and cranial metastases [n=1]). Among those with a nSLN and recurrence, initial recurrence was nodal (1 of 6), pulmonary (1 of 6), local (1 of 6), breast (1 of 6) or at multiple simultaneous sites (2 of 6: local and nodal, local and in transit). At a median follow up of 6.3 years, significantly more patients with pSLN had died (35.5% vs 10.3%, p=0.0214).
Univariate Analysis
The initial exploratory gene finding analysis identified twenty-eight genes whose expression was associated with the development of recurrence (p<0.2). Table 2 shows the gene names and associated test statistics for identified gene candidates associated with RFS. Positive coefficient estimates and hazard ratios (HR) >1 indicate that higher expression of that gene is associated with increased risk of recurrence, and vice versa.
Multivariate Analysis
When considering only the 730 immune-related genes from the gene expression profiles of all 60 patients, and no clinical characteristics, a model incorporating the expression of twelve immune genes accurately predicted risk of recurrence in this cohort regardless of SLN status (C-index = 0.9919, Figure 2A). The 12 genes and direction of expression included increased expression of TIGIT (T cell immunoreceptor with Ig and ITIM domains), a known inhibitory immune receptor, and decreased expression of CXCL16 (chemokine [C-X-C motif] ligand 16), a cytokine important in promoting interaction between dendritic and T cells. The individualized risk score equation produced by the model is represented here: risk score = 119.16549*CSF3 + 151.34767*CXCL1 − 54.14052*CXCL16 − 131.92101*FCGR3A − 144.39280*FOXJ1 − 141.61948*IL22 − 401.58362*LTA + 249.59812*MBL2 − 269.29974*NEFL + 114.25611*PLA2G6 + 116.55562*TIGIT + 260.53276*TLR6.
Figure 2:
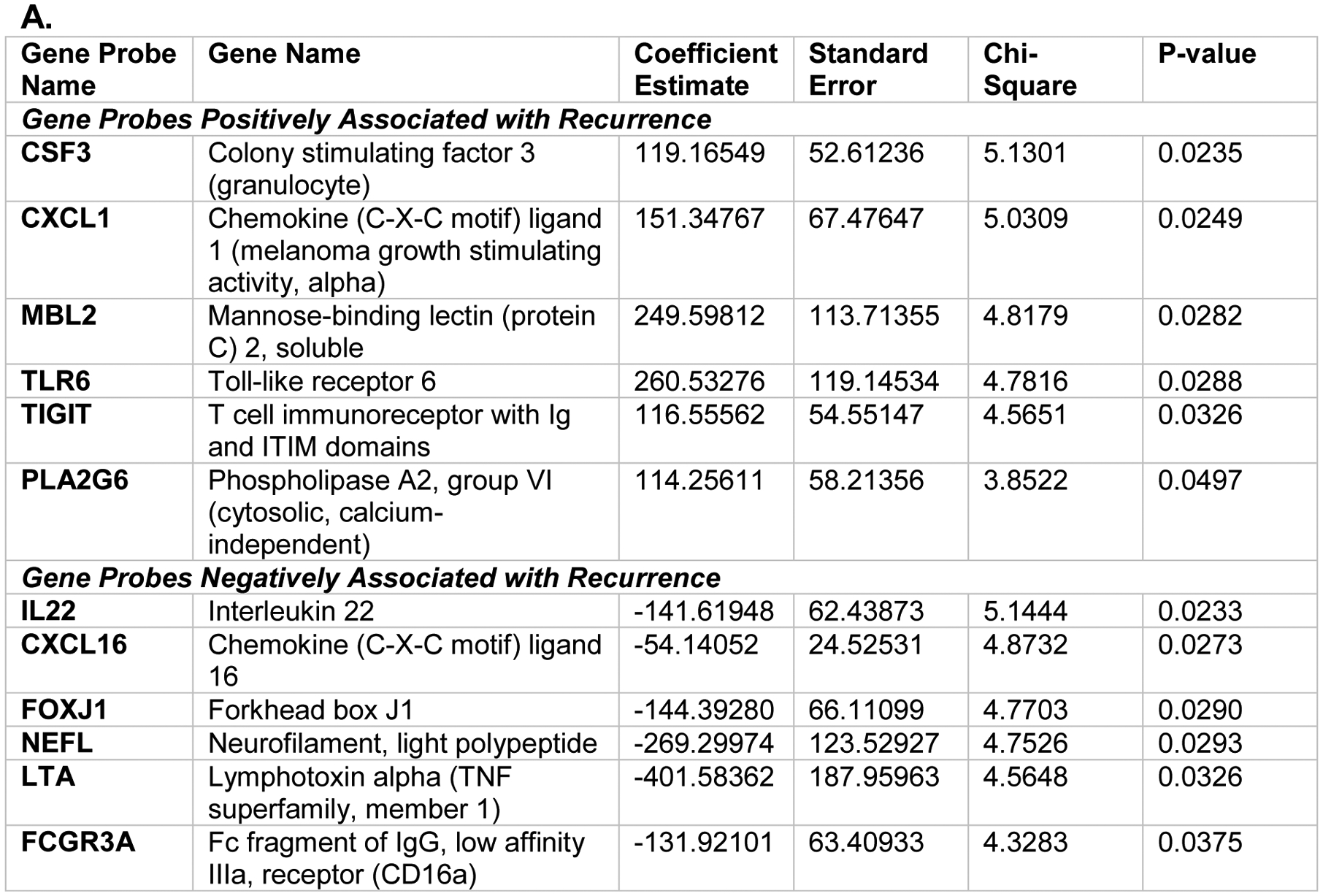
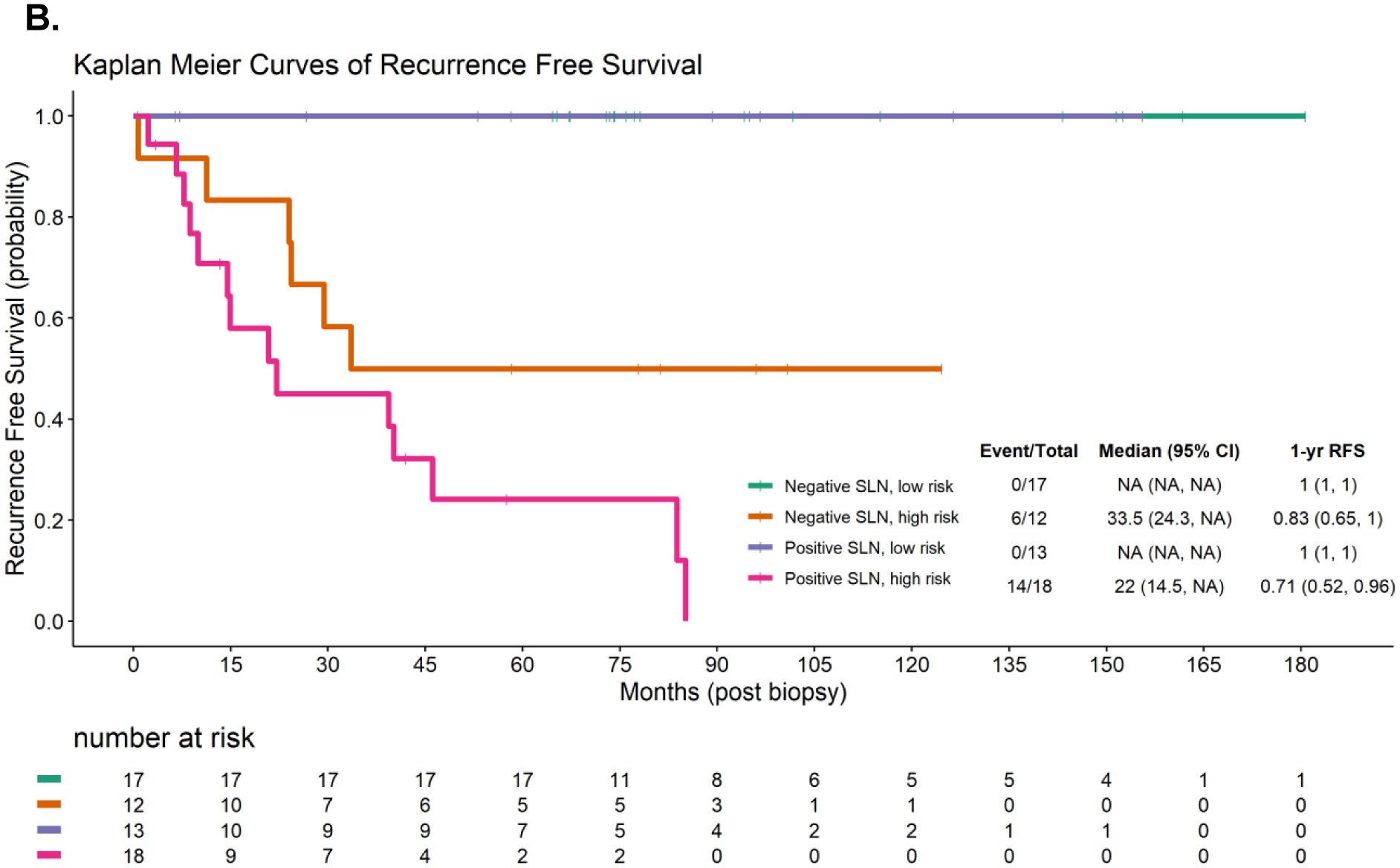
A: Multivariate Cox proportional hazards model incorporating the expression of 12 genes in the sentinel lymph node from the NanoString nCounter® PanCancer Immune Profiling Panel to predict melanoma recurrence free survival (RFS), C-index = 0.9919. Positive coefficient estimates indicate having high expression of the gene is associated with increased risk of disease recurrence and vice versa. B. Kaplan-Meier curves showing RFS in patients with positive or negative sentinel lymph node (SLN) status stratified by calculated recurrence risk score into high- (above median risk score) or low- (below median risk score) risk cohorts.
Independent of SLN status, the individual recurrence risk score calculated using these 12 genes was able to stratify patients into cohorts at high (above median) and low (below median) risk for recurrence with high concordance (Figure 2B, log-rank p<0.001). None of the patients characterized as having a low-risk score developed recurrence during the study period (median follow up 6.3 years); this included the nSLN, low-risk score cohort (n=17), and the pSLN, low-risk score cohort (n=13, 1-year RFS: 1.0, IQR: 1.0–1.0). Six of twelve patients with a nSLN and high-risk score developed recurrence during follow up (1-year RFS 0.83, IQR: 0.65–1.0), while 14 of 18 patients with a pSLN and high-risk score developed recurrence (1-year RFS: 0.71, IQR: 0.52–0.96). Pairwise comparisons of RFS between high- and low-risk score groups were all statistically significant with p-value <0.05 (Table 3: pSLN, low-risk vs nSLN, high-risk: p=0.0009; nSLN, low-risk vs pSLN, high-risk: p <0.0001; pSLN, low-risk vs pSLN, high-risk: p<0.0001; pSLN, low-risk vs nSLN, high-risk: p=0.0114). Pairwise comparisons between pSLN and nSLN groups controlling for same risk scores were not statistically significant (nSLN, low-risk vs pSLN, low-risk: p =1.0; nSLN, high-risk vs pSLN, high-risk: p = 0.0602).
Table 3.
Pairwise comparison of Kaplan Meier curves comparing melanoma recurrence free survival of cohorts stratified by sentinel lymph node (SLN) status (positive vs negative), as well as calculated recurrence risk score (high [above median] vs low [below median]) based on a Multivariate Cox proportional hazards model incorporating the expression of 12 genes in the SLN from the NanoString nCounter® PanCancer Immune Profiling Panel.
| Negative SLN, low risk | Negative SLN, high risk | Positive SLN, low risk | |
|---|---|---|---|
| Negative SLN, high risk | 0.0009 | - | - |
| Positive SLN, low risk | 1 | 0.0114 | - |
| Positive SLN, high risk | <0.0001 | 0.0602 | <0.0001 |
When a separate multivariate model was created by repeating variable selection from the 730 genes as well as relevant clinical characteristics (gender, age at time of surgery, Breslow depth, ulceration and receipt of additional therapy), the only clinical factor that was selected to be included in the model was receipt of additional therapy. Compared to the multivariate model incorporating the 12 genes alone without clinical characteristics (Figure 2A and 2B), the resulting model similarly predicted RFS, but did not improve the prediction accuracy of the model (C-index = 0.9839).
Discussion:
Melanoma recurrence after surgical resection is associated with significant morbidity and mortality, though this risk can be decreased with appropriate use of effective modern adjuvant therapies. In this study evaluating the immunologic gene profiles of SLN in 60 patients with melanoma, we showed that these SLN profiles differ between those at high- versus low-risk for recurrence. Furthermore, we found that characterization of a 12-gene expression profile can successfully stratify patients into cohorts at high- and low-risk for recurrence with high concordance (log-rank p<0.001).
Prior to 2015, the only adjuvant therapy approved for melanoma was high-dose interferon-a 2b (IFN), which was associated with a modest benefit in disease-free survival but came at the cost of frequent and significant adverse events.17,18,19 In 2011, a new era of systemic therapies began with the approval of the checkpoint inhibitor ipilimumab for advanced melanoma, followed shortly by the approval of nivolumab and pembrolizumab, as well as a variety of BRAF/MEK inhibitors. Randomized controlled trials soon showed that these modern therapies provided improvements in recurrence-free and overall survival in patients with resected melanoma and have been approved for the adjuvant setting for resected stage III and IV melanoma starting in 2015.4,5,7,8 The patients in this study underwent SLNB and were treated prior to the advent of modern, effective adjuvant therapy: only 11 pSLN patients in our study received adjuvant therapy (9 with IFN, 1 with ipilimumab, 1 with a vaccine). Now that adjuvant therapies are more effective, the importance of identifying patients at high risk is more pronounced, as the potential benefit is higher. Similarly, though the modern adjuvant therapies have relatively better side effect profiles compared to IFN, they have a broad range of toxicities, with grade 3 or 4 events occurring in approximately 15% of patients.4,5 Thus, identifying patients who can safely forego adjuvant therapy is equally important.
The current patient selection strategy for adjuvant therapy includes histopathologic analysis of SLN via SLNB, and it is currently accepted that patients with nodal metastases greater than 1 mm in size should be considered for adjuvant therapy.9 However, this strategy fails to identify stage II patients at high-risk for recurrence who may benefit from adjuvant therapy, as well as stage III patients who are at low-risk and may safely forego additional treatment, avoiding potentially serious toxicities and significant cost.1,11 Numerous strategies have been suggested to improve risk-stratification and prognosis for this patient population including histopathologic details of nodal burden such as nodal metastasis size and number of metastatic foci, positive-to-total SLN burden, location of tumor deposits and presence of extranodal extension.20 Additionally, ongoing trials are evaluating the role of adjuvant therapy in patients with high-risk stage II disease.21 In the current study, we showed the proof of principle that gene expression profiles of the SLN, and particularly the immune microenvironment of the SLN, correlate with risk for recurrence in patients with melanoma. In the future, gene expression profiles may play a role in identifying which patients should be considered for adjuvant therapies to reduce recurrence risk.
Gene expression profiles evaluating the primary tumor using reverse transcription polymerase-chain reaction (RT-PCR) have been used with some success to predict risk of SLN metastases and relapse, though unlike in breast cancer, this approach has not yet been applied in widespread clinical practice and is not currently recommended as part of national guidelines.22–26 One potential benefit of using gene expression profiles from primary tumors could be that it would provide prognostic information on patients who do not undergo SLNB, as well as the theoretical benefit of eventually eliminating the need for SLNB for prognostic purposes. However, primary tumor tissue is often limited in quantity, particularly after a biopsy has been done prior to surgical excision; SLN tissue may be more readily available, making SLN an attractive practical target for additional analyses. Furthermore, compared to RT-PCR techniques, NanoString has the advantage of quantifying gene expression without the need for amplification, which limits the need for large amounts of tissue available for processing.27 Characterization of SLN gene expression profiles may provide insights into the mechanisms of metastatic spread, allow for more precise risk stratification, and potentially identify therapeutic targets in molecular pathways.12
Tarhini et al. recently evaluated gene expression in the SLN associated with SLN positivity using RT-PCR and microarray analysis, and identified a set of 25 genes that were differentially expressed in pSLNs.28 They were able show that the SLN gene expression differs based on the presence of absence of SLN metastases, however, their planned secondary analyses of correlating gene signatures with clinical outcomes of survival and recurrence were limited by a relatively short follow-up period and are awaited. Similar to our study, many genes identified in their analysis as being associated with SLN positivity appear to be involved in immunosuppressive pathways. Both studies support the theory that local immunologic changes, and not just tumor burden and biology, are permissive of or contribute to the development of metastases and may lead to inferior outcomes.28 Related work by our group has shown that SLN dendritic cell expression of indoleamine 2,3-dioxygenase (IDO) and beta-catenin genes was associated with metastatic potential.13 These studies suggest the SLN microenvironment may play a role in disease outcomes. Future studies should continue to investigate and delineate these mechanisms to in order to provide an opportunity to better understand mechanisms of metastasis and recurrence, as well the potential for use of SLN gene expression for prognostic purposes.
Limitations:
Our study has a number of limitations. Due to its single-center, retrospective nature, we are limited by selection of patients treated at our tertiary-care, academic institution; this small cohort may not be representative of the overall population of patients undergoing SLNB for melanoma in the United States. Furthermore, we are limited by availablity and quality of specimens and clinical data; We attempted to control for specimen quality by normalizing samples using 40 housekeeping genes in the Nanostring gene panel, however there was one patient who had borderline specimen quality included in the analysis. Our sample size is relatively small, and we evaluated a relatively large number of genes, however we have accounted for this by controlling for the false discovery rate (FDR) in the univariable analysis. We also attempted to control for potential confounders by creating an additional multivariate model that incorporated genes as well as clinical factors, however the addition of the clinical factors did not improve the success of our prediction models for RFS. This may be due to the small number of patients in the study, and should not imply that gene expression profiles will replace the role of clinical characteristics known to be important in prognosis for patients with melanoma. Rather, we envision these profiles may in the future serve as an adjunct that could be used together with clinical characteristics to guide treatment decisions. Finally, and importantly, our findings have not yet been validated outside of this patient cohort, and may not be applicable to a broader population. As this is an exploratory study, it shows proof of principle and can be used to identify potential gene targets which will need to be validated with larger retrospective or prospective studies.
Conclusion:
In this retrospective, single-institution study characterizing immunologic gene expression profiles of SLN in patients with melanoma, we show that patients who develop recurrence, regardless of SLN status, have unique gene expression profiles that can be used to stratify patients at high and low risk for recurrence. Gene expression profiles of the SLN may be used to better inform prognosis for melanoma patients undergoing SLNB, and may enhance selection for adjuvant therapy.
Supplementary Material
Synopsis:
Current risk stratification for adjuvant therapy in melanoma fails to identify certain patients at high or low risk. In this study, gene expression profiles of sentinel lymph nodes were associated with recurrence and may enhance patient selection for adjuvant therapy.
Financial Disclosures:
Dr. Beasley was a one-time consultant for Regeneron (2019). Dr. Beasley is supported by the Society of Surgical Oncology Young Investigator award (2019) and is supported by NIH K08 CA237726-01A1. Dr. Farrow is supported by a National Institutes of Health T-32 grant (T32-CA093245) for translational research in surgical oncology.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Thomas DC, Han G, Leong SP, et al. Recurrence of Melanoma After a Negative Sentinel Node Biopsy: Predictors and Impact of Recurrence Site on Survival. Annals of surgical oncology. 2019;26(7):2254–2262. [DOI] [PubMed] [Google Scholar]
- 2.Landow SM, Gjelsvik A, Weinstock MA. Mortality burden and prognosis of thin melanomas overall and by subcategory of thickness, SEER registry data, 1992–2013. Journal of the American Academy of Dermatology. 2017;76(2):258–263. [DOI] [PubMed] [Google Scholar]
- 3.Eggermont AMM, Dummer R. The 2017 complete overhaul of adjuvant therapies for high-risk melanoma and its consequences for staging and management of melanoma patients. European journal of cancer (Oxford, England : 1990). 2017;86:101–105. [DOI] [PubMed] [Google Scholar]
- 4.Eggermont AMM, Blank CU, Mandala M, et al. Adjuvant Pembrolizumab versus Placebo in Resected Stage III Melanoma. N Engl J Med. 2018;378(19):1789–1801. [DOI] [PubMed] [Google Scholar]
- 5.Weber J, Mandala M, Del Vecchio M, et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N Engl J Med. 2017;377(19):1824–1835. [DOI] [PubMed] [Google Scholar]
- 6.Oh A, Tran DM, McDowell LC, et al. Cost-Effectiveness of Nivolumab-Ipilimumab Combination Therapy Compared with Monotherapy for First-Line Treatment of Metastatic Melanoma in the United States. Journal of managed care & specialty pharmacy. 2017;23(6):653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggermont AM, Chiarion-Sileni V, Grob JJ, et al. Prolonged Survival in Stage III Melanoma with Ipilimumab Adjuvant Therapy. N Engl J Med. 2016;375(19):1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long GV, Hauschild A, Santinami M, et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med. 2017;377(19):1813–1823. [DOI] [PubMed] [Google Scholar]
- 9.Coit DG, Thompson JA, Albertini MR, et al. Cutaneous Melanoma, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(4):367–402. [DOI] [PubMed] [Google Scholar]
- 10.Balch CM, Gershenwald JE, Soong SJ, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J Clin Oncol. 2010;28(14):2452–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gershenwald JE, Scolyer RA, Hess KR, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(6):472–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochran AJ, Huang RR, Lee J, Itakura E, Leong SP, Essner R. Tumour-induced immune modulation of sentinel lymph nodes. Nat Rev Immunol. 2006;6(9):659–670. [DOI] [PubMed] [Google Scholar]
- 13.Holtzhausen A, Zhao F, Evans KS, et al. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotolerance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer immunology research. 2015;3(9):1082–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Hout M, Koster BD, Sluijter BJR, et al. Melanoma Sequentially Suppresses Different DC Subsets in the Sentinel Lymph Node, Affecting Disease Spread and Recurrence. Cancer immunology research. 2017;5(11):969–977. [DOI] [PubMed] [Google Scholar]
- 15.Tsang HF, Xue VW, Koh SP, Chiu YM, Ng LP, Wong SC. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert review of molecular diagnostics. 2017;17(1):95–103. [DOI] [PubMed] [Google Scholar]
- 16.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. [DOI] [PubMed] [Google Scholar]
- 17.Eggermont AM, Suciu S, MacKie R, et al. Post-surgery adjuvant therapy with intermediate doses of interferon alfa 2b versus observation in patients with stage IIb/III melanoma (EORTC 18952): randomised controlled trial. Lancet. 2005;366(9492):1189–1196. [DOI] [PubMed] [Google Scholar]
- 18.Mocellin S, Lens MB, Pasquali S, Pilati P, Chiarion Sileni V. Interferon alpha for the adjuvant treatment of cutaneous melanoma. Cochrane Database Syst Rev. 2013(6):CD008955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauschild A, Gogas H, Tarhini A, et al. Practical guidelines for the management of interferon-alpha-2b side effects in patients receiving adjuvant treatment for melanoma: expert opinion. Cancer. 2008;112(5):982–994. [DOI] [PubMed] [Google Scholar]
- 20.Sinnamon AJ, Song Y, Sharon CE, et al. Prediction of Residual Nodal Disease at Completion Dissection Following Positive Sentinel Lymph Node Biopsy for Melanoma. Annals of surgical oncology. 2018;25(12):3469–3475. [DOI] [PubMed] [Google Scholar]
- 21.Luke JJ, Ascierto PA, Carlino MS, et al. KEYNOTE-716: Phase III study of adjuvant pembrolizumab versus placebo in resected high-risk stage II melanoma. Future Oncol. 2020;16(3):4429–4438. [DOI] [PubMed] [Google Scholar]
- 22.Zager JS, Gastman BR, Leachman S, et al. Performance of a prognostic 31-gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer. 2018;18(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller J, Schwartz TL, Lizalek JM, et al. Prospective validation of the prognostic 31-gene expression profiling test in primary cutaneous melanoma. Cancer Med. 2019;8(5):2205–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaral TMS, Hoffmann MC, Sinnberg T, et al. Clinical validation of a prognostic 11-gene expression profiling score in prospectively collected FFPE tissue of patients with AJCC v8 stage II cutaneous melanoma. European journal of cancer (Oxford, England : 1990). 2020;125:38–45. [DOI] [PubMed] [Google Scholar]
- 25.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. [DOI] [PubMed] [Google Scholar]
- 26.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. [DOI] [PubMed] [Google Scholar]
- 27.Veldman-Jones MH, Brant R, Rooney C, et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res. 2015;75(13):2587–2593. [DOI] [PubMed] [Google Scholar]
- 28.Tarhini AA, Floros T, Lin HM, et al. A unique gene expression signature is significantly differentially expressed in tumor-positive or tumor-negative sentinel lymph nodes in patients with melanoma. Melanoma Res. 2017;27(5):429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


