Abstract
COVID-19 can result in severe disease characterized by significant immunopathology that is spurred by an exuberant, yet dysregulated, innate immune response with a poor adaptive response. A limited and delayed interferon I (IFN-I) and IFN-III response results in exacerbated proinflammatory cytokine production and in extensive cellular infiltrates in the respiratory tract, resulting in lung pathology. The development of effective therapeutics for patients with severe COVID-19 depends on our understanding of the pathological elements of this unbalanced innate immune response. Here, we review the mechanisms by which SARS-CoV-2 both activates and antagonizes the IFN and inflammatory response following infection, how a dysregulated cytokine and cellular response contributes to immune-mediated pathology in COVID-19, and therapeutic strategies that target elements of the innate response.
Keywords: SARS-CoV-2, innate immunity, interferon, inflammatory, cytokines, immune evasion, immune antagonism, coronavirus
Lowery et al. review the mechanisms by which SARS-CoV-2 activates and antagonizes the interferon and inflammatory response following infection, how a dysregulated cytokine and cellular response contributes to immune-mediated pathology in COVID-19, and therapeutic strategies that target elements of the innate response.
Introduction
In December 2019, a novel respiratory disease, later named coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome (SARS) coronavirus (CoV) 2 (SARS-CoV-2) was recognized in Wuhan, China (Zhou et al., 2020b). As of May 7, 2021, 156 million cases worldwide with a mortality global rate of 2.2% have been identified (WHO, 2021). The SARS-CoV-2 pandemic was preceded by two other CoV epidemics, SARS in 2002 and Middle East respiratory syndrome (MERS) in 2012, with the latter continuing to circulate in camels throughout Asia and Africa. COVID-19 is characterized by both upper and lower respiratory tract infections, leading to symptomatology that varies from asymptomatic to lethal infection (Huang et al., 2020). Patients with mild disease have non-specific symptoms, including fever and cough, while those with moderate or severe illness experience decreased oxygen saturation, requiring hospitalization or ICU care in the most affected individuals (NIH, 2021).
Activation of the inflammatory response, including of the interferon (IFN) response, contributes to SARS-CoV-2 symptomatology, including fever and dyspnea (Huang et al., 2020). Severe disease can entail inflammatory-driven multi-organ failure, acute respiratory distress syndrome (ARDS), sepsis, secondary infections, and severe pneumonia (NIH, 2021). Disease severity is linked to a highly dysregulated innate immune response, which is broadly characterized by a delayed interferon (IFN) response relative to symptom onset and possibly peak virus replication, and the production of an exuberant inflammatory response. These events together facilitate robust viral replication and inflammatory damage to tissues. Currently, many SARS-CoV-2 vaccines have been rolled out worldwide, but global morbidity and mortality due to COVID-19 remain substantial. Furthermore, the ongoing viral evolution of SARS-CoV-2 has caused concern over the efficacy of current vaccines. For these reasons, we need to better understand the innate immune response to SARS-CoV-2 in order to develop new and improved therapeutic options and enhance our understanding of pathogenesis. In this review, we discuss the current data on SARS-CoV-2 pathology, innate immune antagonism, and treatments. We show that both interferon expression and signaling, as well as pro-inflammatory cytokine production, if not properly temporally regulated, contribute to disease severity.
Innate immunity toward CoVs
Distinguishing an appropriate but robust innate immune response from one that is dysregulated has proven difficult. It is generally believed that the innate immune response was dysregulated in both SARS and severe MERS (Arabi et al., 2020; Cameron et al., 2007; Channappanavar et al., 2016, 2019). In SARS-CoV-infected mice, a delayed type I IFN (IFN-I) response resulted in rapid virus replication, the recruitment of inflammatory monocyte-macrophages (IMMs) to the lungs, and abnormally heightened cytokine and chemokine responses, resulting in high mortality (Channappanavar et al., 2016). These pathogenic responses were prevented by treating mice with IFN-I prior to the peak of virus replication or by depleting IMMs (Channappanavar et al., 2016). Similarly, when mice were treated with IFN-I prior to being infected with MERS-CoV, it protected them from infection, while delaying IFN-I treatment until 2 days post-infection (dpi) resulted in a heightened inflammatory state characterized by IMM and neutrophil recruitment to the lungs and an elevated inflammatory cytokine expression (Channappanavar et al., 2019). In SARS patients, a prolonged IFN response resulted in a delayed adaptive immune response, the continued upregulation of inflammatory chemokines, and in distinct IFN-stimulated gene (ISG) expression, indicating that IFN production needs to be resolved to initiate and generate a protective adaptive immune response (Cameron et al., 2007). Furthermore, treatment with IFN-I was effective only if administered early during MERS-CoV infection (Arabi et al., 2020). Current evidence indicates that some patients with severe COVID-19 present with delayed or no induction of IFN-I and -III (Galani et al., 2021). In the absence of effective IFN expression, SARS-CoV-2 is able to replicate to higher titers, resulting in an exaggerated inflammatory response. Given the importance of IFN expression and signaling in COVID-19 pathogenesis, we discuss IFN in more detail below.
Interferon response
Interferon production and signaling during viral infection
IFNs are a broad group of cytokines that are divided into three families—IFN-I, IFN-II, and IFN-III—and that are critical for immune defense against microbial pathogens. The IFN-I family is well studied in viral infections and include IFN-α, IFN-Β, IFN-ε, IFN-ω, and IFN-κ, while the IFN-II and IFN-III families include IFN-γ and IFN-l, respectively (Hertzog et al., 2016). Upon respiratory CoV infection, signaling cascades are induced that result in IFN production that acts in both an autocrine and paracrine manner to activate IFN signaling pathways. Epithelial cells, endothelial cells, alveolar macrophages (AMs), natural killer (NK) cells, dendritic cells (DCs), and IMMs are the primary IFN producers in respiratory virus infection. Plasmacytoid DCs (pDCs) in particular are thought to secrete most of the IFN-I during infection with respiratory CoVs, such as SARS-CoV (Channappanavar et al., 2016; Newton et al., 2016). Viral ssRNA and dsRNA intermediates are recognized by pattern recognition receptors (PRRs), with the cytosolic receptor MDA5 (melanoma differentiation-association protein 5) considered the most important for sensing CoV RNA (Cervantes-Barragan et al., 2007; Roth-Cross et al., 2008). Activated PRRs engage downstream adaptors, such as the mitochondrial antiviral-signaling (MAVS) protein or myeloid differentiation primary response 88 (MyD88) (Lazear et al., 2019; Lee and Ashkar, 2018). These adaptors activate interferon-regulatory factors (IRF3 and 7) and their nuclear translocation, where they bind IFN promoters. IFN-I and -III bind their respective receptors and activate the JAK/STAT signaling cascade to assemble the ISG factor 3 (ISGF3) complex, which ultimately binds to IFN-stimulated response element (ISRE) promoters, resulting in broad ISG production. IFN-II signaling results in STAT1 homodimer formation and in the production of a partially overlapping set of ISGs (Lee and Ashkar, 2018). These signaling cascades are essential for inducing an antiviral state on viral infection.
Interferon antagonism by SARS-CoV-2 genes
CoV genomes encode 4 major structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), and 16 non-structural proteins (Nsps) that are critical for replication (Figure 1 ). CoV replication and transcription takes place in double-membrane vesicles (Figure 2), which may function to shield viral RNA from PRR recognition (Versteeg et al., 2007; Zhou and Perlman, 2007). In addition, a varying number of accessory proteins are interspersed between the structural genes. These proteins are dispensable for virus replication but contribute to immune antagonism and pathogenesis (Figure 2 ). Many of these genes are known to antagonize or evade the interferon response, which may contribute to the delayed IFN expression seen in COVID-19 patients. These proteins act both upstream of IFN production, through evasion and antagonism of PRR recognition and signaling, and downstream by directly antagonizing the IFN signaling pathway, facilitating early rapid virus replication (Figure 2; Table 1 ). Thus, CoVs have developed many ways to subvert the IFN response.
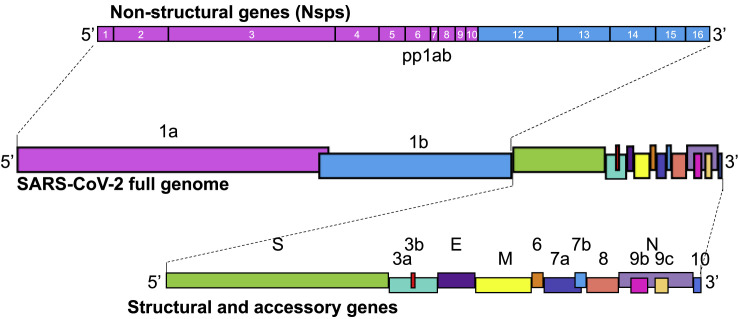
Figure 1.
Schematic of the SARS-CoV-2 genome
SARS-CoV-2 is a 30 kb, positive-strand RNA virus with a genome divided into non-structural genes (Nsps) on the 5′ end (pink and blue boxes representing contributions from ORF1a and ORF1b [together, polyprotein 1ab, pp1ab] with numbered Nsps) and structural and accessory genes interspersed on the 3′ end. Nsps 1–16 are critical for viral replication and have some immune evasion functions, while the major structural genes spike (S), membrane (M), envelope (E), and nucleocapsid (N) make up the virion. Accessory genes 3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10 are not necessary for viral replication, but may play a structural or immune evasion role, as seen in other CoV accessory proteins. Dotted lines represent magnified genome portions.
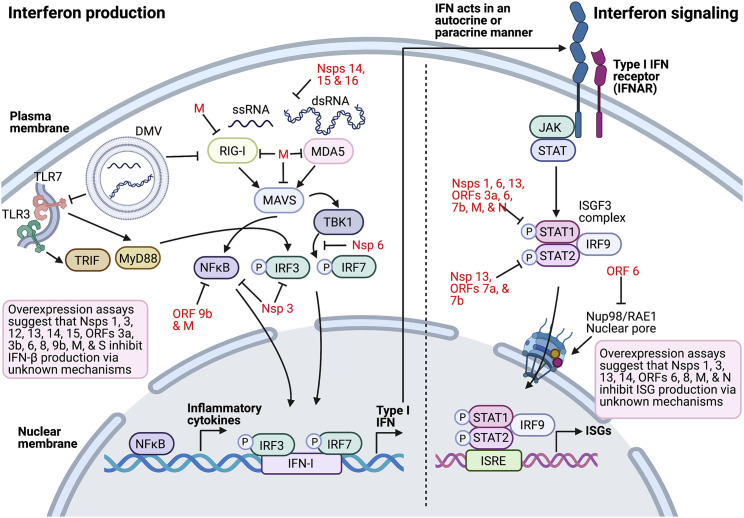
Figure 2.
Innate immune antagonism by SARS-CoV-2 proteins
Coronavirus replication occurs in double-membraned vesicles (DMVs), which can shield viral RNA (ssRNA and dsRNA) from recognition by PRRs such as TLR3, TLR7, RIG-I, and MDA5. These PRRs activate adaptors TRIF and MyD88 downstream of TLRs, and MAVS and TBK1 downstream of RIG-I and MDA5 beginning at the interferon production pathway. Activation of IRF3, IRF7, or NFκB results in their nuclear translocation and in the transcriptional activation of immune genes including inflammatory cytokines and IFN. The dotted line represents the transition to IFN signaling beginning with IFN-I binding IFNAR to initiate JAK/STAT signaling and formation of the ISGF3 complex STAT1/STAT2/IRF9, which translocates into the nucleus to activate ISRE transcription. SARS-CoV-2-encoded proteins (red) inhibit multiple aspects of these pathways, resulting in decreased IFN and altered proinflammatory cytokine expression. Many of the SARS-CoV-2 IFN antagonists have been identified by overexpression assays in vitro, and thus await in vivo confirmation of their role in the virus’s replication and pathogenesis (black).
Table 1.
SARS-CoV-2 antagonistic and evasion proteins
| Protein | Purported antagonism/evasion function | Demonstrated in SARS-CoV-2? | Common mechanism known in other CoVs? | References |
|---|---|---|---|---|
| Nsp1 | suppresses host protein expression inhibits IFNβ production | yes (live virus)a | similar function across CoVs, though mechanism varies | Lei et al., 2020; Lin et al., 2021; Xia et al., 2020 |
| Nsp3 | inhibits IRF3, NFκB signaling via papain-like protease activity | no | broadly conserved across CoVs | Lei et al., 2020 |
| Nsp6 | antagonizes TBK1 phosphorylation of IRF3inhibits STAT1 phosphorylation | yes (overexpression)b | likely | Xia et al., 2020 |
| Nsp12 | inhibits IFNβ production | yes (overexpression)b | likely | Lei et al., 2020 |
| Nsp13 | inhibits IFNβ productioninhibits STAT1/STAT2 phosphorylation | yes (overexpression)b | likely | Lei et al., 2020; Xia et al., 2020; Yuen et al., 2020 |
| Nsp14 | N7-methyltransferase activity involved in RNA capping to evade PRR detection | no | broadly conserved across CoVs | Lei et al., 2020; Yuen et al., 2020 |
| Nsp15 | endonuclease activity degrades viral RNA to evade PRR detection | no | broadly conserved across CoVs | Yuen et al., 2020 |
| Nsp16 | 2’-O-methyltransferase activity involved in RNA capping to evade PRR detection | no | broadly conserved across CoVs | DeDiego et al., 2014 |
| S | inhibits IFNβ production | yes (overexpression)b | no | Yuen et al., 2020 |
| M | inhibits PRR signalinginhibits activation of IFNβ and NFκB promotersinhibits STAT1 phosphorylation | yes (overexpression)b | SARS- and MERS-CoV | Fu et al., 2021; Xia et al., 2020; Zheng et al., 2020 |
| N | inhibits ISG production | yes (overexpression)b | SARS- and MERS-CoV | Mu et al., 2020 |
| ORF3a | inhibits IFNβ productioninhibits STAT1 phosphorylation | yes (overexpression)b | SARS-CoV | Lei et al., 2020; Xia et al., 2020 |
| ORF3b | inhibits IFNβ production | yes (overexpression)b | SARS-CoV | Konno et al., 2020 |
| ORF6 | inhibits nuclear translocation of STAT1 via NUP98/RAE1 inhibition | yes (overexpression)b | SARS-CoV | Lei et al., 2020; Miorin et al., 2020; Xia et al., 2020; Yuen et al., 2020 |
| ORF7a | inhibits STAT2 phosphorylation | yes (overexpression)b | no | Xia et al., 2020 |
| ORF7b | inhibits STAT1/STAT2 phosphorylation | yes (overexpression)b | no | Xia et al., 2020 |
| ORF8 | inhibits ISG productioninhibits IRF3 nuclear translocation | yes (overexpression)b | no | Lei et al., 2020; Rashid et al., 2021 |
| ORF9b | disrupts MAVS/TRAF3/TRAF6 signalosome | no | SARS- and MERS-CoV | Wu et al., 2021 |
Experimentally validated using clinical isolates and genetically modified live SARS-CoV-2 containing deletion mutations in Nsp1.
Demonstrated via overexpression of the protein in vitro in the absence of SARS-CoV-2 infection.
SARS-CoV-2 induction of IFN pathways in human patients
The IFN response during COVID-19 varies dramatically by patient and by disease severity, but it is clear that an early IFN response can be protective during acute infection. Individuals with genetically disrupted IFN production or signaling genes are at greater risk of developing severe COVID-19 (Bastard et al., 2020; Pairo-Castineira et al., 2021; Zhang et al., 2020). Additionally, anti-IFN-I autoantibodies were found in 10% of patients with severe COVID-19, whereas none of these antibodies were found in patients with mild/asymptomatic disease (Bastard et al., 2020). These data suggest that understanding the dynamics of the IFN response in mild and severe COVID-19 is critical. In order to address this, a longitudinal study of the innate immune response of 113 COVID-19 patients has been performed. All COVID-19 patients were found to express elevated IFN-α, interleukin-1α (IL-1α), IL-1β, IL-17A, and IL-12 p70 in blood plasma relative to healthy controls, comprising a core COVID-19 inflammatory signature, while IFN-III was elevated only in severe patients (Lucas et al., 2020). Elevation of IFN-⍺ and/or IFN-III correlated with duration of hospitalization and mortality. In contrast, others observed decreased IFN-I and -III in COVID-19 patients relative to healthy controls, and an increase in IFN-II correlated with disease severity (Galani et al., 2021; Hadjadj et al., 2020). Several studies also reported a similar upregulation of IFN-II in critically ill patients, with a concomitant elevation of pro-inflammatory cytokines, such as IL-6 and TNF (Huang et al., 2020; Yao et al., 2021; Zhou et al., 2020a). This poor IFN-I and -III response did not, however, result in similarly poor ISG expression, as the ISGs OAS1 and MX1 (2’-5′-oligoadenylate synthetase 1 and MX dynamin like GTPase 1) were upregulated in critically ill patients, suggesting either that a minimal IFN response was sufficient to establish ISG expression or that this expression occurred in an IFN-I/III-independent manner (Galani et al., 2021). Overall, these data suggest that there is interpatient variability in the IFN response in COVID-19, with severe disease characterized by prolonged IFN-I production in some instances and by a paucity of IFN-I expression in others. Further work will be needed to reconcile these results, which may reflect differences in patient populations under study. Patients with mild or moderate disease develop IFN-I and IFN-III responses, which decline as virus is cleared, coinciding with recovery (Figure 3 A).
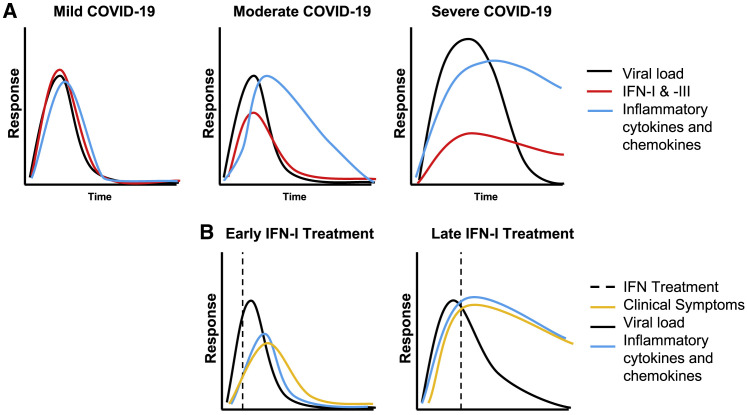
Figure 3.
Kinetics of the innate immune response to COVID-19
(A) A mild disease course is generally characterized by an IFN response that is robust, coincides with the peak of viral replication, results in rapid viral clearance, and is the resolution of IFN and of other inflammatory responses. As disease severity increases, the IFN-I and -III response is milder and delayed relative to viral replication. This allows for prolonged viral replication and for the prolonged expression of IFN and inflammatory cytokines that contribute to immune-mediated pathogenesis.
(B) Based on animal models and available clinical data, IFN-I treatment at early time points (relative to viral replication and symptom onset) is expected to enhance virus clearance and the rapid resolution of the inflammatory response and clinical symptoms. Late IFN-I treatment is expected not to enhance viral clearance, nor reduce inflammatory cytokine and chemokine responses, producing no change or possibly exacerbated clinical symptoms.
SARS-CoV-2 induction of IFN pathways in vitro
Several studies have revealed that SARS-CoV-2 is highly sensitive to pretreatment with IFN-I and -III, more so than SARS-CoV (Katsura et al., 2020; Lokugamage et al., 2020; Miorin et al., 2020). As expected, based on the several IFN-antagonistic proteins it encodes, SARS-CoV-2 disrupts the IFN response by reducing or delaying IFN expression, particularly when compared with viruses such as influenza A virus (IAV) and Sendai virus (Blanco-Melo et al., 2020; Lei et al., 2020). Treatment with recombinant IFN following infection did not reverse this reduction in IFN and ISG expression, further demonstrating that IFN signaling, not just IFN production, is antagonized (Miorin et al., 2020). Similarly, infection of ex vivo human lung cultures did not result in IFN-I, -II, or -III expression in type 1 and 2 pneumocytes and AMs (Chu et al., 2020). Thus, while AMs produce large amounts of IFN upon IAV infection, SARS-CoV-2, like SARS-CoV, inhibited IFN expression in these cells, disrupting a key antiviral defense (Cheung et al., 2005; Chu et al., 2020). Consistent with these data, in infected patients, IFN-I expression by AMs was prolonged (and possibly delayed), contributing to a prolonged inflammatory response (Grant et al., 2021).
In contrast to these results demonstrating defects in IFN signaling, others have found that IFNs and ISGs are upregulated following the infection of primary organoid cultures with SARS-CoV-2. In human lung stem-cell-based organoids (alveolospheres), transcriptome profiling at 48 h post-infection (hpi) revealed increased expression of IFNs, STATs, ISGs, and chemokines relative to mock infection (Katsura et al., 2020). A similar model system showed robust ISG expression despite minimally elevated IFN-I at 72 hpi, indicating that ISG expression occurred in an IFN-I-independent manner (Youk et al., 2020). These apparently contradictory results indicate that a number of factors, including differences in cell types, in size of virus inoculum, and in the timing of the analyses need to be considered. In addition, further studies will be necessary to understand how ISGs are produced outside of traditional IFN signaling and will be key to understanding the antiviral state against SARS-CoV-2 infection.
SARS-CoV-2 response to IFN in animal models
Animal models that recapitulate features of COVID-19 and the human immune responses to SARS-CoV-2 are needed to study COVID-19 pathogenesis and to develop therapeutics to treat it. Immunocompetent wild-type (WT) mice are resistant to infection with original strains of SARS-CoV-2, such the Wuhan-Hu-1 strain, because the virus cannot use mouse ACE2 to enter cells. Multiple models were developed to overcome this, including transducing mice with adenovirus vectors that express human ACE2, the generation of human ACE2 transgenic mice, and infecting mice with mouse-adapted (MA) virus (Muñoz-Fontela et al., 2020). Each of these mouse models varies in pathogenesis and in their development of an antiviral immune response (Figure 4 ). The infection of mice lacking IFN-I and -II receptors with MA SARS-CoV-2 resulted in increased viral replication, loss of lung function, and exacerbated lung congestion, compared with WT mice (Leist et al., 2020). In contrast to the minimal IFN upregulation in mice that developed mild disease, infection of K18-hACE2 transgenic mice, in which human ACE2 expression is driven by the cytokeratin 18 promoter, is robust and results in increased IFN-I, -II, and -III that persists beyond the peak of virus replication, recapitulating the correlation of IFN expression with disease severity in COVID-19 patients (Oladunni et al., 2020; Zheng et al., 2021a). Prolonged IFN expression was detrimental in WT mice infected with MA SARS-CoV, as it contributed to an exacerbated and pathogenic inflammatory response (Channappanavar et al., 2016). Similarly, SARS-CoV-2-infected K18-hACE2 transgenic mice exhibited prominent immune cell recruitment to the lungs, including of DCs, IMMs, and CD4 and CD8 T cells, and showed substantial weight loss, morbidity, and mortality (Zheng et al., 2021a). As seen with SARS- and MERS-CoV infection, prophylactic and early therapeutic IFN treatment was protective against MA SARS-CoV-2 infection and disease in BALB/c mice (Dinnon et al., 2020). IFN-III-treated animals showed significantly reduced weight loss and reduced virus replication in their lungs relative to untreated mice (Dinnon et al., 2020). Overall, these data suggest that early IFN signaling is protective against SARS-CoV-2, while poor IFN induction due to immune evasion or antagonism results in minimal protection from viral infection.
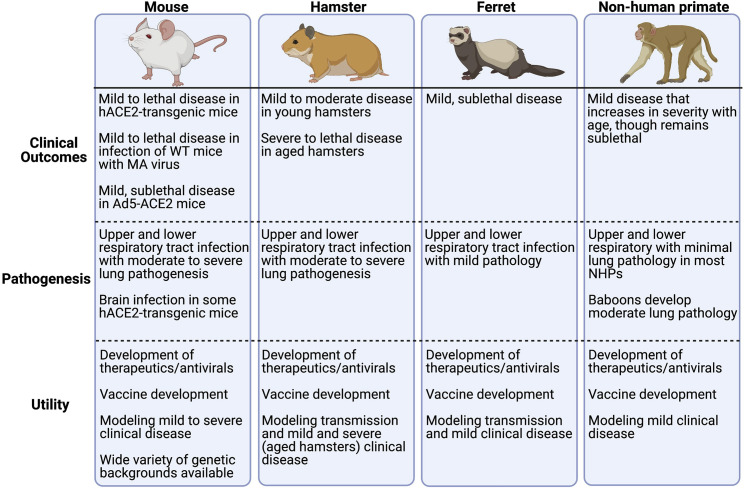
Figure 4.
Animal models of COVID-19
Various laboratory animals, particularly mice, hamsters, ferrets, and non-human primates, have been used to model different aspects of COVID-19 to develop antiviral drugs and to improve our understanding of COVID-19 disease in humans. Because mice are not naturally susceptible to initial isolates of SARS-CoV-2, due to the incompatibility of the virus with mouse ACE2, adenovirus-5-transduced human ACE2-transgenic mice (Ad5-hACE2) and hACE2-transgenic mice have been used to confer susceptibility. Wild-type (WT) mice can also be infected with mouse-adapted (MA) SARS-CoV-2. Hamsters and ferrets are naturally susceptible to SARS-CoV-2 infection and can transmit virus to uninfected animals, enabling studies of viral transmission. Non-human primates are also naturally susceptible to SARS-CoV-2 and, as the laboratory animal most closely related to humans, represent an important model for preclinical trials of therapeutics and vaccines (Muñoz-Fontela et al., 2020).
The infection of other animal models with SARS-CoV-2, including of hamsters, ferrets, and non-human primates, results in mild clinical disease, with a lethal infection only observed in aged hamsters (Selvaraj et al., 2021). The infection of hamsters and ferrets with SARS-CoV-2 does not result in an IFN-β response (Bessière et al., 2021; Blanco-Melo et al., 2020; Hoagland et al., 2021), although ISG15 expression was observed after infection and remained elevated to 8 dpi, by which time the virus had already fully cleared (Hoagland et al., 2021). These data indicate that non-classical, IFN-independent induction of ISGs occurred in these animals after SARS-CoV-2 infection. When hamsters were treated prophylactically with IFN-⍺, it reduced inflammation and viral titers in their lungs. Moreover, when uninfected hamsters that were cohoused with infected animals were treated with IFN-⍺ prophylactically, it resulted in fewer infections among the uninfected, pretreated group. The IFN-pretreated animals that did become infected had only limited virus titers, indicating that IFN pre-treatment reduced viral transmission and the infectivity of treated animals (Hoagland et al., 2021). Meanwhile, treating hamsters with IFN-⍺ at 3 dpi, coincident with symptom onset, did not change clinical outcomes (Bessière et al., 2021). By contrast, when non-human primates were infected with SARS-CoV-2, it resulted in significant upregulation of IFN-I and -II expression in bronchoalveolar lavage (BAL) fluid and induced robust ISG expression in the lungs, indicating that non-human primates mount a stronger IFN response to SARS-CoV-2 infection than that observed in other animal models (Chandrashekar et al., 2020; Singh et al., 2021). More studies are needed to understand why non-human primates develop a more robust IFN response relative to other infected animals. Of note, prophylactic IFN-⍺ treatment of non-human primates followed by SARS-CoV infection resulted in reduced viral load and lung damage compared to untreated animals, again indicating that the IFN-I response is protective (Haagmans et al., 2004). Overall, these data indicate that hamster and ferret models recapitulate the minimal IFN-I expression with induction of an ISG response phenotype that is observed in some COVID-19 patients. They also demonstrate that the prophylactic treatment of hamsters with IFN-I reduced transmission and viral load, warranting further study of this treatment for use as an intervention in high-risk scenarios.
Therapeutic administration of IFN
As a result of the significant benefit observed in animals experimentally infected with SARS-CoV, MERS-CoV, and SARS-CoV-2, in response to being treated with IFN early in the course of disease, multiple clinical trials and retrospective studies are now being conducted to evaluate IFN treatment in human COVID-19 disease. A retrospective study of IFN-a2b treatment of COVID-19 patients in China revealed that treatment via nebulizer decreased the duration of hospitalization in critical patients in comparison to patients treated with antiviral protease inhibitors (Wang et al., 2020). Interestingly, this study found that IFN-a2b treatment did not benefit moderately ill patients; however, whether this was because the IFN response in these patients was already sufficiently robust is unknown. IFN-a2b treatment was also only beneficial when provided early, within 5 days of post-hospital admission, while those that received IFN treatment at later time points had extended hospital stays and slower recovery compared to patients treated early (Wang et al., 2020). A separate study using a similar treatment regimen found that IFN-a2b treatment reduced virus titers and viral RNA in the airways relative to untreated patients (Zhou et al., 2020c). Similarly, early combination treatment of MERS-CoV-infected patients with IFN-β1 and antiviral protease inhibitors within 7 days of symptoms resulted in decreased mortality, compared to placebo and those treated 7 days post-symptom onset (Arabi et al., 2020). IFN-β treatment was also found to reduce symptoms, as well as incidence of severe disease and mortality in patients with COVID-19 (Hung et al., 2020; Monk et al., 2021). Of note, the WHO Solidarity Trial Consortium clinical trial found that IFN-I treatment was not beneficial in treating SARS-CoV-2 infection (WHO, 2021), which might reflect the timing of treatment. The therapeutic use of IFN-III in patients with mild-moderate COVID-19 resulted in modest to no benefits (Feld et al., 2021; Jagannathan et al., 2021). Together, these results indicate that IFN-I will not be beneficial for all patients but may be useful for subsets of patients, such as those that are treated early during infection and in those with a minimal IFN response (Figure 3B). However, this treatment is unlikely to be useful in patients with pre-existing anti-IFN antibodies.
Cytokine and chemokine response to SARS-CoV-2
In addition to IFNs, many other cytokines and chemokines are produced once PRRs sense viral RNA, leading to the activation of signaling cascades, such as the NFkB pathway, which ultimately results in the expression of a wide variety of cytokines and chemokines. Activation of the NOD-like receptor pyrin domain containing 3 (NLRP3) inflammasome, which enables the cleavage and activation of cytokines such as IL-1β and IL-18 (Rodrigues et al., 2021). As with the IFN response, many of these responses are dysregulated in patients with severe COVID-19, with critically ill patients exhibiting a more substantially dysregulated response than patients with mild or moderate disease. Many studies have sought to identify the inflammatory mediators most linked to severe outcomes, and while an exact set of inflammatory markers cannot be established due to patient-to-patient variability, a general phenotype of elevated IL-6, IL-8, IL-10, and TNF, along with a mixture of elevated chemokines, including CCL2, CCL3, and CXCL8, has been observed (Blanco-Melo et al., 2020; Huang et al., 2020; Mulchandani et al., 2021; Zhou et al., 2020a).
In vitro, a similar pro-inflammatory cytokine response to SARS-CoV-2 has been observed in a wide variety of susceptible cell types and organoids, resulting in the upregulation of inflammatory cytokines such as IL-6 and TNF, as well as a robust chemotactic response that includes elevated levels of various chemokines, including CCL20, CXCL1, CXCL3, CXCL5, CXCL6, CXCL2, CXCL10, and CXCL16 (Katsura et al., 2020; Yang et al., 2020). Similarly, the infection of monocyte-derived macrophages with SARS-CoV-2, though not resulting in the production of infectious virus, resulted in the upregulation of TNF, IL-8, IP-10 (CXCL10), and IL-1β, indicating that while IFN expression was impaired in these cells, inflammatory cytokine and chemokine production was not (Hui et al., 2020; Zheng et al., 2021b). As with the IFN signaling pathways, SARS-CoV-2 proteins can antagonize other cytokine responses, such as those mediated by NFκB. An accessory protein of SARS-CoV-2, ORF9b, has been demonstrated to interact with NFκB essential modulator (NEMO) and to disrupt its K63-linked polyubiquitination, disrupting NFκB signaling and inhibiting the production of inflammatory cytokines such as IL-6 and TNF (Wu et al., 2021).
As in COVID-19 patients and in vitro models, animal models of SARS-CoV-2 infection present with a similar pro-inflammatory cytokine and chemokine profile with genes encoding TNF, IL-6, CXCL10, CCL2, CCL5, and IFN-II being the most commonly upregulated across multiple studies and linked to severe outcomes (Blanco-Melo et al., 2020; Boudewijns et al., 2020; Dinnon et al., 2020; Leist et al., 2020; Oladunni et al., 2020; Singh et al., 2021; Sun et al., 2020; Zheng et al., 2021a). TNF and IFN-II have been shown to synergistically promote PANoptosis in SARS-CoV-2-infected mice (PANoptosis is a form of inflammatory-mediated cell death characterized by the collective activation of the pyroptosis, apoptosis, and necroptosis pathways). When these mice were treated with neutralizing antibodies against these cytokines, disease was ameliorated and mortality reduced (Karki et al., 2021). Supporting the notion that immune-mediated pathology is a significant driver of disease severity, infection of baboons with SARS-CoV-2 resulted in a more robust inflammatory cytokine and chemokine profile than that seen in SARS-CoV-2-infected rhesus macaques and was reflected in a more severe lung pathology in the baboons. Interestingly, the inflammatory and IFN responses in rhesus macaques peaked and resolved with kinetics similar to those of the virus. Resolution was coupled with a robust T cell response, suggesting that a coordinated innate immune and T cell response is critical for preventing immunopathological changes (Singh et al., 2021).
Since a dysregulated inflammatory response appears to occur during COVID-19 disease progression, many have suggested that SARS-CoV-2 causes a prolonged cytokine storm that leads to morbidity and mortality in patients. While “cytokine storm” does not have a precise definition, it is broadly characterized as being an excessive inflammatory response with substantial elevation of many pro-inflammatory cytokines and chemokines. Classic examples of cytokine storms are found in sepsis, ARDS, and sometimes as a side effect of chimeric antigen receptor (CAR) T cell therapy; viral diseases caused by influenza, SARS, and MERS have also been similarly labeled, although not with unanimous agreement. Data suggest that while inflammatory mediators are significantly upregulated in COVID-19 patients relative to healthy patients, they are not substantially differentially regulated relative to influenza patients (Galani et al., 2021; Mudd et al., 2020). For example, IL-6 and IL-8 are similarly elevated in COVID-19 and influenza patients, while many other inflammatory genes were actually decreased in expression in COVID-19 compared to influenza patients (Mudd et al., 2020). Interestingly, when comparing these responses to classical cytokine storms, such as those seen in ARDS and sepsis, IL-6 concentrations are 5 to 100 times higher in these conditions than in patients with COVID-19 (Leisman et al., 2020). Similar results are seen for IL-8, TNF, and IFN-II, all of which are known significant drivers of inflammation (Leisman et al., 2020). It may therefore be incorrect to characterize COVID-19 disease as a cytokine storm, and significant inflammatory dysregulation may be a more accurate characterization of the immune response in these patients.
Cellular innate immune response to SARS-CoV-2
In addition to the innate cytokine response, SARS-CoV-2 infection results in the extensive recruitment of macrophages, monocytes, DCs, and neutrophils to the lungs; pathological findings indicate that neutrophil chemoattractants such as CXCL8, CXCL1, and CXCL2, and the monocyte chemoattractants CCL2 and CCL7, are also induced and upregulated (Boudewijns et al., 2020; Zheng et al., 2021a). The infiltration of these cells into the lungs has been demonstrated to contribute to the pathology observed in SARS-CoV-infected mice; when IMMs were depleted in these mice with antibodies, they were protected from lethal infection and showed improved virus-specific T cell responses (Channappanavar et al., 2016). Phenotypic studies of infiltrating monocytes in the BAL of COVID-19 patients have revealed extensive differentiation of monocytes with an inflammatory phenotype, potentially driven by granulocyte-macrophage colony-stimulating factor (GM-CSF)-expressing T cells (Chevrier et al., 2020; Zhou et al., 2020d). These inflammatory monocytes are a significant source of IL-6 and other inflammatory cytokines, contributing to pathogenesis (Chevrier et al., 2020; Zhou et al., 2020d). Neutrophils are known to produce reactive oxygen species (ROS) and neutrophil extracellular traps (NETs) that promote cell death, and these NETs have been characterized in the lungs and tracheal aspirates of COVID-19 patients (Veras et al., 2020). ROS and NET production by neutrophils may exacerbate the deleterious response to infection; in vitro data suggest that NETs produced by neutrophils following SARS-CoV-2 infection promote lung epithelial cell death (Veras et al., 2020). NK cells participate in the innate immune response by producing cytokines, such as IFN-II and TNF, and by killing virus-infected cells. However, it is widely reported that overall NK cell numbers are substantially reduced in the blood of COVID-19 patients relative to healthy controls (Maucourant et al., 2020). Those NK cells that are present show robust activation with high levels of granzyme B and perforin, proteins involved in the cell lysis activity of T and NK cells. The expression levels of these proteins correlated with increased IL-6 expression and with markers of organ failure, suggesting that cytokine-producing NK cells may play a role in exacerbating immunopathology in COVID-19 patients.
Pathology of COVID-19
SARS-CoV-2, similar to SARS- and MERS-CoV, causes severe tissue damage in the lungs and other organs, leading to vascular damage and progression to ARDS, multi-organ failure, and other clinical manifestations classified as critical in patients. Post-mortem lung histology of SARS patients has revealed the presence of alveolar damage, the loss of bronchial epithelial cell layers and cilia, abnormal squamous cells, hyaline membrane formation, and edema. Extensive monocyte/macrophage accumulation in the lungs was also found, which likely drove a robust inflammatory cytokine response, contributing to the observed pathology (Nicholls et al., 2003). Unsurprisingly, SARS-CoV-2 infection results in similar pathological manifestations. An early autopsy report of a COVID-19 patient with early-phase ARDS found pneumocyte desquamation and hyaline membrane formation, as well as significant inflammatory mononuclear cell infiltration, including a large number of lymphocytes (Barton et al., 2020; Liu et al., 2020; Tian et al., 2020; Xu et al., 2020). An autopsy of a COVID-19 patient who had recovered from mild respiratory disease but succumbed to a sudden cardiac event identified SARS-CoV-2 particles remaining in the lungs of the patient, despite negative PCR testing of nasopharyngeal swabs (Yao et al., 2020). Alveolar damage, desquamation of pneumocytes, hyaline membrane formation, and inflammatory mononuclear infiltrates were also observed in the lungs; however, pulmonary edema was not seen (Yao et al., 2020). This suggests that infection of lung tissue and immunopathology may remain extensive in recovered patients, potentially contributing to unknown long-term effects (Yao et al., 2020). Consistent with this, 22%–56% of COVID-19 patients had long-term pulmonary sequelae when assessed 6 months after resolution of acute COVID-19 (Huang et al., 2021).
Anti-inflammatory therapeutic strategies
The robust inflammatory response to SARS-CoV-2 has led to many clinical trials that are testing the efficacy of anti-inflammatory drugs on COVID-19. Most prominently, the large RECOVERY trial in the UK found that treatment with dexamethasone reduced mortality if given to patients receiving mechanical ventilation or oxygen support, though it had no effect or even negative effects when administered to patients with mild disease (Horby et al., 2021). Several clinical trials of glucocorticoids have further corroborated their efficacy in reducing the duration of mechanical ventilation and mortality and are now the standard treatment for critically ill COVID-19 patients (Cavalli et al., 2021; Angus et al., 2020; Tomazini et al., 2020). In addition to the broadly acting glucocorticoids, inhibitors of specific cytokines known to be upregulated in patients with moderate and critical COVID-19 have been employed as well, such as the anti-IL-6 receptor monoclonal antibody, tocilizumab. While some trials have reported a lack of improvement in COVID-19 patient mortality upon IL-6 inhibitor treatment, several others have demonstrated an amelioration of lung pathology and lymphopenia, and a reduction in the time spent in the ICU and in overall mortality, especially if administered with corticosteroids (Cavalli et al., 2021; The REMAP-CAP Investigators, 2021). Similarly, IL-1β signaling antagonists, such as the anti-IL-1β monoclonal antibody canakinumab or the IL-1 receptor antagonist anakinra have been shown to reduce COVID-19 patients’ need for supplemental oxygen and to reduce fever, time in ICU, and mortality as well (Cavalli et al., 2021; Della-Torre et al., 2020). Other therapeutic strategies have also targeted signaling molecules downstream of IFN, including JAKs and STATs, in order to inhibit pathogenesis related to a dysregulated IFN response. COVID-19 patients treated with the JAK/STAT inhibitor ruxoilitinib show similar improvements to those reported for the aforementioned immunomodulatory therapies, indicating that a reduction in IFN signaling reduces overall inflammation in these patients (D’Alessio et al., 2021). Other potential inflammatory targets for therapeutics include cytokines such as TNF, neutrophil NET production, and components of the complement cascade, as excessive complement activation may contribute to the thrombosis and endothelialitis that is a hallmark of severe COVID-19 (Ackermann et al., 2020).
Conclusions
In conclusion, these studies show that COVID-19 pathogenesis is a manifestation both of direct virus replication and the resultant host response. Many different pro-inflammatory mediators appear to have a role in poor outcomes, although thus far, no single cytokine or chemokine has been shown to be the most important. Most likely, the actions of a group of cytokines, lack of an early IFN-I and -III response, and innate inflammatory cells contribute to severe disease (Figure 3). The challenge going forward will be to determine which of these inflammatory mediators are the most important in individual COVID-19 patients with severe disease and, consequently, which patients would benefit from the therapies described above. As highlighted in this review, the pathogenic mechanisms of this viral disease are similar but not the same in all patients.
Acknowledgments
We thank Lok-Yin Roy Wong for critical review of this manuscript. Figures 2 and 4 were created using BioRender. This work was supported in part by grants from the NIH (PO1:AI060699 and RO1 AI129269) to S.P. and (T32 AI007533) to S.A.L.
Declaration of interests
The authors declare no competing interests.
References
- Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus D.C., Derde L., Al-Beidh F., Annane D., Arabi Y., Beane A., van Bentum-Puijk W., Berry L., Bhimani Z., Bonten M., et al. Writing Committee for the REMAP-CAP Investigators Effect of Hydrocortisone on Mortality and Organ Support in Patients With Severe COVID-19: The REMAP-CAP COVID-19 Corticosteroid Domain Randomized Clinical Trial. JAMA. 2020;324:1317–1329. doi: 10.1001/jama.2020.17022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Asiri A.Y., Assiri A.M., Balkhy H.H., Al Bshabshe A., Al Jeraisy M., Mandourah Y., Azzam M.H.A., Bin Eshaq A.M., Al Johani S., et al. Saudi Critical Care Trials Group Interferon Beta-1b and Lopinavir-Ritonavir for Middle East Respiratory Syndrome. N. Engl. J. Med. 2020;383:1645–1656. doi: 10.1056/NEJMoa2015294. [DOI] [PubMed] [Google Scholar]
- Barton L.M., Duval E.J., Stroberg E., Ghosh S., Mukhopadhyay S. COVID-19 Autopsies, Oklahoma, USA. Am. J. Clin. Pathol. 2020;153:725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessière P., Wasniewski M., Picard-Meyer E., Servat A., Figueroa T., Foret-Lucas C., Coggon A., Lesellier S., Boué F., Cebron N., et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. Biorxiv. 2021 doi: 10.1101/2021.02.09.430458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., et al. Canadian SARS Research Network Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Larcher A., Tomelleri A., Campochiaro C., Della-Torre E., De Luca G., Farina N., Boffini N., Ruggeri A., Poli A., et al. Interleukin-1 and interleukin-6 inhibition compared with standard management in patients with COVID-19 and hyperinflammation: a cohort study. Lancet Rheumatol. 2021;3:E253–E261. doi: 10.1016/S2665-9913(21)00012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L.M., Ng I.H.Y., Luk W., Sia S.-F., Wu M.H.S., Chan K.-H., Yuen K.-Y., Gordon S., Guan Y., Peiris J.S. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S., Zurbuchen Y., Cervia C., Adamo S., Raeber M.E., de Souza N., Sivapatham S., Jacobs A., Bachli E., Rudiger A., et al. A distinct innate immune signature marks progression from mild to severe COVID-19. Cell Rep Med. 2020;2:100166. doi: 10.1016/j.xcrm.2020.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F.-W., Wang Y., Yuen T.T.-T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., et al. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin. Infect. Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio A., Del Poggio P., Bracchi F., Cesana G., Sertori N., Di Mauro D., Fargnoli A., Motta M., Giussani C., Moro P., et al. Low-dose ruxolitinib plus steroid in severe SARS-CoV-2 pneumonia. Leukemia. 2021;35:635–638. doi: 10.1038/s41375-020-01087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Torre E., Della-Torre F., Kusanovic M., Scotti R., Ramirez G.A., Dagna L., Tresoldi M. Treating COVID-19 with colchicine in community healthcare setting. Clin. Immunol. 2020;217:108490. doi: 10.1016/j.clim.2020.108490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld J.J., Kandel C., Biondi M.J., Kozak R.A., Zahoor M.A., Lemieux C., Borgia S.M., Boggild A.K., Powis J., McCready J., et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir. Med. 2021;9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-Z., Wang S.-Y., Zheng Z.-Q., Yi Huang, Li W.-W., Xu Z.-S., Wang Y.-Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 2021;18:613–620. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.-E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., Abbott D.A., Donnelly H.K., Donayre A., Goldberg I.A., et al. NU SCRIPT Study Investigators Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A.M., Rimmelzwaan G.F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.-H., et al. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog P.J., Bourke N.M., de Weerd N.A., Mangan N.E. In: Encyclopedia of Immunobiology. Ratcliffe M.J.H., editor. Academic Press; 2016. New Interferons; pp. 501–508. [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54:557–570.e5. doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Huang L., Wang Y., Li X., Ren L., Gu X., Kang L., Guo L., Liu M., Zhou X., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Cheung M.-C., Perera R.A.P.M., Ng K.-C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.-W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan P., Andrews J.R., Bonilla H., Hedlin H., Jacobson K.B., Balasubramanian V., Purington N., Kamble S., de Vries C.R., Quintero O., et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J., et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K., USFQ-COVID19 Consortium SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J., Ashkar A.A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V., Mastroiani F., Turtle C.J., Harhay M.O., et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.W., Tang C., Wei H.C., Du B., Chen C., Wang M., Zhou Y., Yu M.X., Cheng L., Kuivanen S., et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021;29:489–502.e8. doi: 10.1016/j.chom.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020 doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., et al. Karolinska COVID-19 Study Group Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5:eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.-P., et al. Inhaled Interferon Beta COVID-19 Study Group Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd P.A., Crawford J.C., Turner J.S., Souquette A., Reynolds D., Bender D., Bosanquet J.P., Anand N.J., Striker D.A., Martin R.S., et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci. Adv. 2020;6:eabe3024. doi: 10.1126/sciadv.abe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulchandani R., Lyngdoh T., Kakkar A.K. Deciphering the COVID-19 cytokine storm: Systematic review and meta-analysis. Eur. J. Clin. Invest. 2021;51:e13429. doi: 10.1111/eci.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.-S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A.H., Cardani A., Braciale T.J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin. Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH Clinical Spectrum. 2021. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/
- Oladunni F.S., Park J.-G., Pino P.A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. GenOMICC Investigators. ISARIC4C Investigators. COVID-19 Human Genetics Initiative. 23andMe Investigators. BRACOVID Investigators. Gen-COVID Investigators Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Rashid F., Dzakah E.E., Wang H., Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj P., Lien C.Z., Liu S., Stauft C.B., Nunez I.A., Hernandez M., Nimako E., Ortega M.A., Starost M.F., Dennis J.U., et al. SARS-CoV-2 infection induces protective immunity and limits transmission in Syrian hamsters. Life Sci. Alliance. 2021;4:e202000886. doi: 10.26508/lsa.202000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Singh B., Ganatra S.R., Gazi M., Cole J., Thippeshappa R., Alfson K.J., Clemmons E., Gonzalez O., Escobedo R., et al. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat. Microbiol. 2021;6:73–86. doi: 10.1038/s41564-020-00841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The REMAP-CAP Investigators Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Xiong Y., Liu H., Niu L., Guo J., Liao M., Xiao S.-Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod. Pathol. 2020;33:1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazini B.M., Maia I.S., Cavalcanti A.B., Berwanger O., Rosa R.G., Veiga V.C., Avezum A., Lopes R.D., Bueno F.R., Silva M.V.A.O., et al. COALITION COVID-19 Brazil III Investigators Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients With Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., Schneider A.H., Caetité D., Tavares L.A., Paiva I.M., et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217:e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Bredenbeek P.J., van den Worm S.H.E., Spaan W.J.M. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO Coronavirus (COVID-19) Dashboard. 2021. https://covid19.who.int/
- Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., Zhong T., Tang H., Du W., Wang L., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Chu H., Hou Y., Chai Y., Shuai H., Lee A.C.-Y., Zhang X., Wang Y., Hu B., Huang X., et al. Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated With Viral Antagonism of STAT1 Phosphorylation. J. Infect. Dis. 2020;222:734–745. doi: 10.1093/infdis/jiaa356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Bora S.A., Parimon T., Zaman T., Friedman O.A., Palatinus J.A., Surapaneni N.S., Matusov Y.P., Cerro Chiang G., Kassar A.G., et al. Cell-Type-Specific Immune Dysregulation in Severely Ill COVID-19 Patients. Cell Rep. 2021;34:108590. doi: 10.1016/j.celrep.2020.108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.-H., He Z.-C., Li T.-Y., Zhang H.-R., Wang Y., Mou H., Guo Q., Yu S.-C., Ding Y., Liu X., et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;30:541–543. doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J., Kim T., Evans K.V., Jeong Y.-I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J., et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., Cai J.-P., Jin D.-Y., To K.K.-W., Chan J.F.-W., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhuang M.-W., Han L., Zhang J., Nan M.-L., Zhan P., Kang D., Liu X., Gao C., Wang P.-H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020;5:299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., Leidinger M.R., Knudson C.M., Meyerholz D.K., McCray P.B., Jr., Perlman S. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wang Y., Li K., Meyerholz D.K., Allamargot C., Perlman S. Severe Acute Respiratory Syndrome Coronavirus 2-Induced Immune Activation and Death of Monocyte-Derived Human Macrophages and Dendritic Cells. J. Infect. Dis. 2021;223:785–795. doi: 10.1093/infdis/jiaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Perlman S. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 2007;81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., MacArthur M.R., He X., Wei X., Zarin P., Hanna B.S., Wang Z.-H., Xiang X., Fish E.N. Interferon-α2b Treatment for COVID-19 Is Associated with Improvements in Lung Abnormalities. Viruses. 2020;13:44. doi: 10.3390/v13010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]