Abstract
The SARS-CoV-2 pandemic has caused unparalleled disruption of global behavior and significant loss of life. To minimize SARS-CoV-2 spread, understanding the mechanisms of infection from all possible routes of entry is essential. While aerosol transmission is thought to be the primary route of spread, viral particles have been detected in ocular fluid, suggesting that the eye may be a vulnerable point of viral entry. To this end, we confirmed SARS-CoV-2 entry factor and antigen expression in post-mortem COVID-19 patient ocular surface tissue and observed productive viral replication in cadaver samples and eye organoid cultures, most notably in limbal regions. Transcriptional analysis of ex vivo infected ocular surface cells and hESC-derived eye cultures revealed robust induction of NF-κB in infected cells as well as diminished type I/III interferon signaling. Together these data suggest that the eye can be directly infected by SARS-CoV-2 and implicate limbus as a portal for viral entry.
Keywords: COVID-19, SARS-CoV-2, cornea, limbus, stem cells, eye, ACE2, NF-κB, interferon, scRNA-seq
Graphical abstract
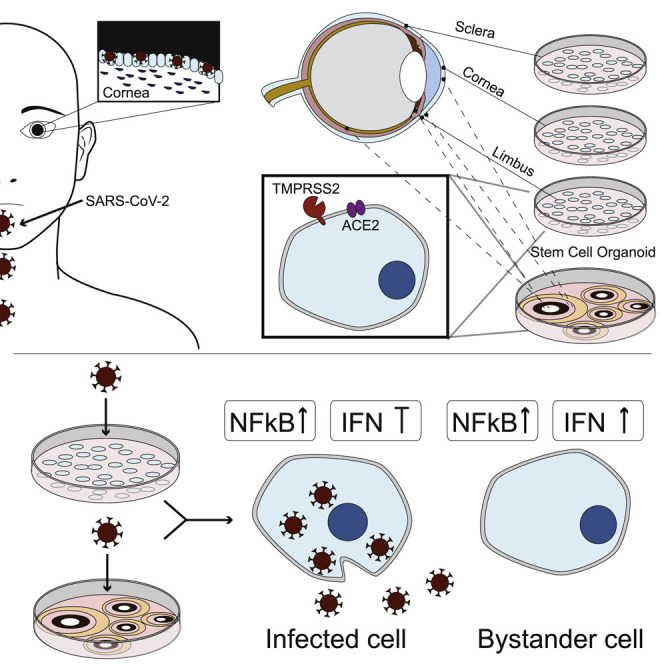
Eriksen et al. show that SARS-CoV-2 antigens are expressed in COVID-19 patient samples and find robust infection of cadaveric ocular surface cells. Limbus cells derived from human donors and a hPSC whole-eye differentiation model seem more prone to infection and exhibit robust chemokine production and an impaired IFN-I/-III response.
Introduction
One of the first widely reported deaths by coronavirus disease 2019 (COVID-19) in early January 2020 originated from an ophthalmologist in Wuhan, who reportedly contracted severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from an asymptomatic glaucoma patient (Lai et al., 2020). Coronaviruses (CoVs) are a large family of RNA viruses common among vertebrates (Masters and Perlman, 2013) that can be divided into four subtypes termed alpha, beta, gamma, and delta, of which NL63 and 229E (alpha CoVs) and OC43 and HKU1 (beta CoVs) are the most common (Bchetnia et al., 2020). CoVs were once thought to result only in mild upper respiratory infections, but the emergence of SARS-CoV-1 (a beta CoV) in 2002 was the first evidence that this family of viruses could elicit more severe disease, infecting >8,000 individuals worldwide, with a fatality rate of ~10%. Ten years later, another beta CoV, the Middle East respiratory syndrome-related CoV (MERS-CoV) emerged and infected ~2,500 people with a case-fatality rate of >35% since 2012 (de Wit et al., 2016). Although neither SARS-CoV-1 nor MERS-CoV resulted in a pandemic, the emergence of SARS-CoV-2 in 2019 has already infected more than 125 million individuals worldwide, killing more than 2.7 million people (Dong et al., 2020). In an effort to slow the spread of the virus, the use of face masks was recommended, but face masks fail to protect the eyes from contacting the virus. It is unclear whether the virus can enter from the ocular tissue or migrate to the eye from the respiratory tract, but RNA from SARS-CoV-1, MERS-CoV, and SARS-CoV-2 has been detected in the eye (Aiello et al., 2020).
Animal studies suggest that the eye can serve as a site of replication for numerous respiratory infections. Indeed, a study comparing various routes of entry in Rhesus monkeys confirmed the ability to detect virus in the nasolacrimal and pulmonary system upon SARS-CoV-2 infection via the conjunctiva (Deng et al., 2020), and infection via the ocular route has also been demonstrated to lead to infection of the respiratory system in golden hamsters (Imai et al., 2020; Hoagland et al., 2021). Detection of SARS-CoV-2 in conjunctival tear film from patients both with and without ocular secretions has been documented (Li et al., 2020; Xia et al., 2020), though the percentage of patients testing positive for SARS-CoV-2 in the ocular fluid is low (Wu et al., 2020; Zhang et al., 2020). In one study that analyzed ocular secretions during the progression of SARS-CoV-2 infection and during the peak of the infection, SARS-CoV-2 was detected in the ocular secretions and progressively declined to undetectable levels during recovery (Chen et al., 2020).
Despite accumulating evidence of SARS-CoV-2 in tears, it is unknown whether this is due to ocular surface cells’ becoming directly infected through respiratory droplets from an infected person. A potential source of viral material in ocular secretions could be systemic infection, but this is unlikely, as there have, been no transfusion-related transmission events, suggesting that the virus does not appear to enter the blood stream (Cappy et al., 2020), and in fact, evidence suggests that direct ocular infection is possible. Studies of normal human ocular surface tissue have found that ocular surface tissues including cornea, limbus, and conjunctival regions express both the entry receptor ACE2 and associated host protease TMPRSS2 (Hoffmann et al., 2020), shown by immunohistochemistry, RNA sequencing (RNA-seq), western blot, and assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Collin et al., 2021; Mencucci et al., 2021); hence, the necessary machinery for SARS-CoV-2 entry is present. Whether particular surface ocular tissues are more prone to infection than other ocular cells is also unknown. TMPRSS2 is lowly expressed in ocular tissues, while other serine protease family members are more highly expressed. Recently, a new list of genes associated with SARS-CoV-2 entry was published (Singh et al., 2020; Daniloski et al., 2021), and expression of proteases other than TMPRSS2 may be used by SARS-CoV-2 infectivity in the eye.
In an effort to determine the susceptibility and potential virus-host interactions between SARS-CoV-2 and the human eye, we used a whole-eye organoid model from human embryonic stem cells (hESCs) that included the differentiation of retina, retinal pigment epithelium (RPE), ciliary margin, iris, lens, and cornea (Hayashi et al., 2016). We also examined adult human ocular cells isolated from non-infected, as well as COVID-19-positive cadaver donors for comparison. We found cells that express known corneal and corneal endothelium markers also express both ACE2 and TMPRSS2 and, upon exposure to SARS-CoV-2, become infected. Moreover, transcriptional profiling of these cultures identified an antiviral response not unlike what is found in SARS-CoV-2-infected lung or airway epithelia (Blanco-Melo et al., 2020). These results suggest that the eye can be infected directly by SARS-CoV-2 and supports virus replication, as the observed host response is triggered by pathogen-associated molecular patterns (PAMPs), which are the products of productive replication. Moreover, our studies suggest that the limbal region is particularly prone to infection and may serve as a potential entry route for the virus.
Results
Assessment of surface ocular tissue from SARS-CoV-2-positive post-mortem donors
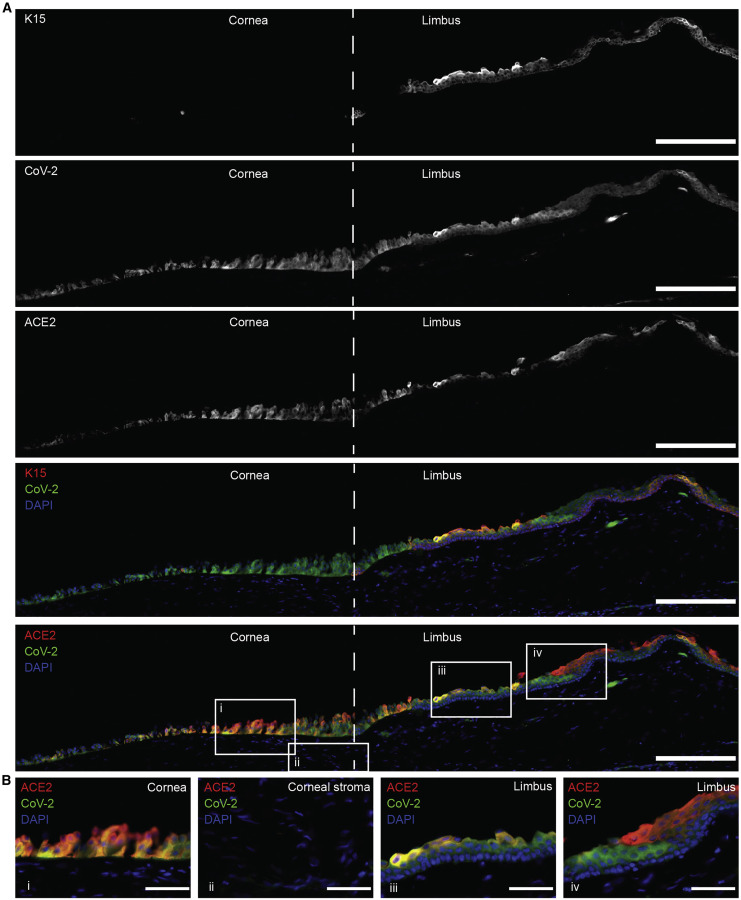
We obtained three donations from the Eye Bank for Sight Restoration from patients who passed and were SARS-CoV-2 positive on the basis of nasal swab qPCR. The anterior segments were obtained, which included part of the sclera, limbus, and cornea. We tested these tissues for the presence of SARS-CoV-2 antigen by immunofluorescence and co-stained with markers of cornea- and limbus-associated keratins (KRTs) (Figures 1 and S1–S4). Ocular surface tissues from all three donors were positive for SARS-CoV-2 spike protein (S). KRT 15 is preferentially expressed in the limbal region, and SARS-CoV-2 S could be detected in cells both positive and negative for KRT15. Moreover, S-protein detection was predominantly limited to the epithelium, and S protein was not detected in the corneal stroma. Co-staining of SARS-CoV-2 S and ACE2 showed strong co-localization, but we also detected S protein in some cells negative for ACE2, likely illustrating the internalization of the receptor following SARS-CoV-2 entry (Figure 1, crop iv) (Monteil et al., 2020). SARS-CoV-2 protein could also be detected together with KRT12, suggesting that infection was not limited to the limbus (Figure S2). SARS-CoV-2 nucleocapsid (N) protein was also present in ocular surface cells positive for tumor protein 63 (p63), specifically at the limbus (Figures S3A and S3B). Last, we also observed co-staining of SARS-CoV-2 (N) with ACE2 and TMPRSS2 (Figures S3C and S3D). Data from these samples could not show whether ocular surface tissue was originally infected or if the infection began in another organ.
Figure 1.
Limbal region of post-mortem human SARS-CoV-2-positive cadaver donor tissue
Post-mortem human ocular surface tissue stained for limbal epithelial marker cytokeratin 15 (K15) and co-staining with antibodies to SARS-CoV-2 (S) and ACE2. Scale bar, 200 μm in tile-scan images. Scale bar, 50 μm in zoomed in images; iso-type controls can be seen in Figure S1. Images are representative from one donor (donor A); images of tissue from two additional donors can be seen in Figure S2 (iso-type controls for Figure S2 can be found in Figures S4A and S4B).
Comparison of ocular cell types to SARS-CoV-2 infection
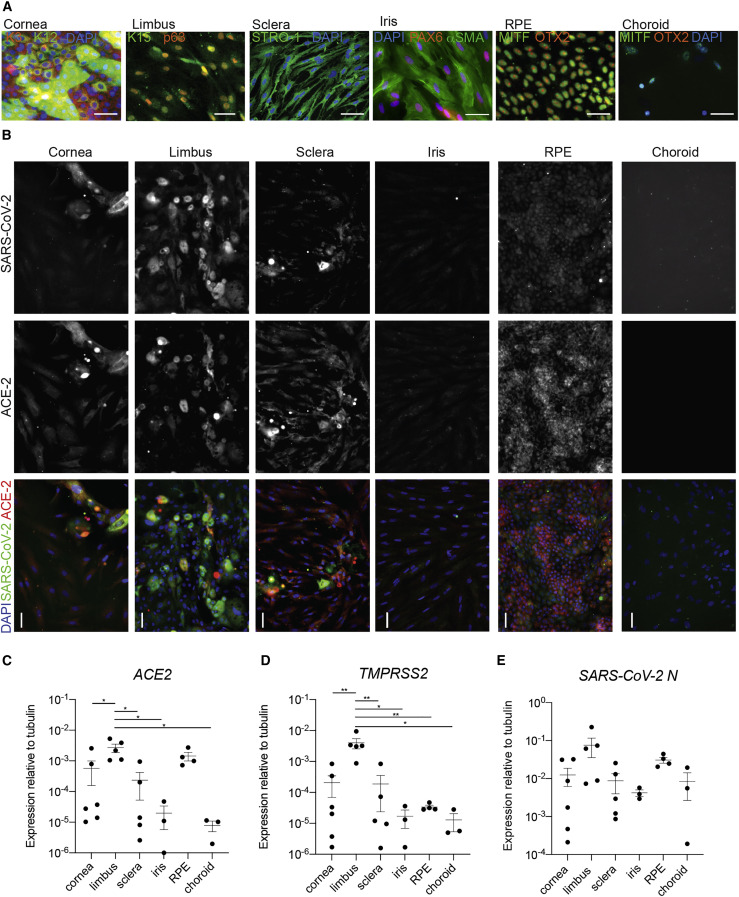
Susceptibility to SARS-CoV-2 infection requires the expression of receptors and proteases that enable viral entry and cleavage, most notably ACE2 and TMPRSS2. In order to evaluate which ocular cells express ACE2 and whether expression of this entry factor is consistent with susceptibility to SARS-CoV-2 infection, we cultured cell types isolated from six eye tissues isolated from healthy adult human cadaver donor globes, including cornea, limbus, sclera, iris, RPE, and choroid. Cells dissected from these eye regions were first stained with markers specific to their respective cell types to verify cellular identity (Figure 2 A). Cells were then infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 1.0 and incubated for 24 h. We found SARS-CoV-2 S protein expression in cornea, limbus, sclera, and RPE, with no detectable expression in iris or choroid (Figure 2B). This cell type-specific infection was consistent with the expression of ACE2 and TMPRSS2 mRNA in the respective tissues, which on average was lower in choroid and iris compared with the other tissues, albeit not statistically significant (Figures 2C and 2D). To further evaluate viral gene expression, qRT-PCR was conducted on mRNA from infected cells, and we observed SARS-CoV-2 subgenomic N expression in cornea, sclera, limbus, iris, RPE, and choroid (Figure 2E). Expression of ACE2 and TMPRSS2 mRNA was significantly higher in the limbus compared with all other tissues, with the exemption of ACE2 expression between limbus and RPE (Figures 2C and 2D). As we were unable to obtain all tissues from genetically matched donors, this genetic background/lifestyle heterogeneity may mask genuine cellular infectivity differences.
Figure 2.
Expression of ACE2, TMPRSS2, and SARS-CoV-2 in primary ocular tissues isolated from adult human eye
Post-mortem globes were dissected and cells cultured.
(A) Confirmation of cell identity was conducted using antibodies to cell-specific markers keratin 3 (K3) and 12 (K12) for cornea, keratin 15 (K15) and tumor protein p63 (p63) for limbus, STRO-1 for sclera, co-expression of PAX6 and αSMA for iris muscle, co-expression of MITF and OTX2 for RPE, and expression of MITF and negative expression of OTX2 for choroid melanocytes. See also Figure S5A for ISO-type controls.
(B) Cells were exposed to SARS-CoV-2 for 24 h, then fixed and stained for antibodies to SARS-CoV-2 (S) and ACE2. See also Figure S5B for ISO-type controls.
(C) qRT-PCR for ACE2 on total RNA extracted from healthy ocular tissues; p = 0.0062.
(D) Same as (C) for TMPRSS2; p = 0.0018 (Vidal et al., 2015).
(E) qRT-PCR for SARS-CoV-2 subgenomic N transcript on total RNA extracted from ocular tissues infected with SARS-CoV-2 as in (B).
For (C)–(E), n = 6 for cornea, n = 5 for limbus and sclera, n = 4 for RPE, and n = 3 for iris and choroid. Adjusted p < 0.05 and ∗∗adjusted p < 0.01. Horizontal bars denote mean ± standard error of the mean.
SARS-CoV-2 infection elicited an NF-κB-mediated chemokine response in human ocular cells
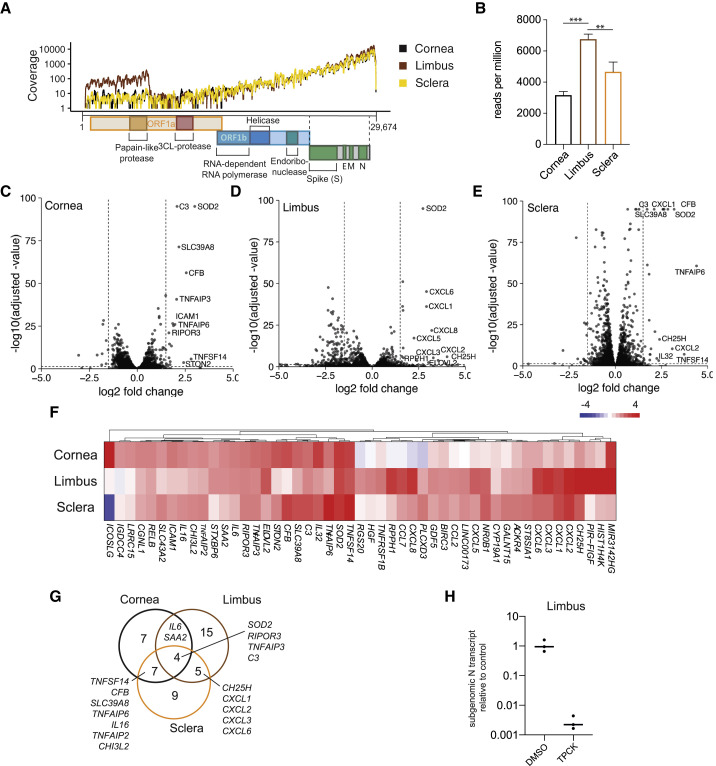
Given the observed infectivity of several ocular tissues, we sought to conduct a more careful analysis between ocular surface cells from genetically matched individuals and examine the host response to infection. To this end, we infected cells from the cornea, limbus, and scleral regions from uninfected post-mortem donor tissue with SARS-CoV-2 for 24 h and evaluated responses by whole-transcriptome RNA-seq. Reads were first mapped to the viral genome and were found in all three tissues, with the limbus exhibiting greater coverage (Figure 3 A). Quantification of SARS-CoV-2 reads relative to the read depth confirmed that the limbal cells were significantly more infected than corneal or scleral cells (Figure 3B), in line with the higher expression of TMPRSS2 and ACE2 observed above. Next, we investigated the host response to viral infection. Volcano plots of the host response in the three ocular tissues revealed a response similar in magnitude, and the limbus cells were characterized by being the only cell type with a relatively large number of genes that showed decreased expression upon infection (Figures 3C–3E; Table S1). Several chemokines, including CXCL1-3 and -6 as well as TNF-α-induced genes such as TNFAIP2, -3, and -6 and signaling components IL6 and RELB, were among the genes with the highest increase in expression, indicative of a response mediated by nuclear factor κB (NF-κB) (Figure 3F) (Mitchell et al., 2016). When comparing the genes with significant increases in gene expression (log2 fold change [L2FC] > 1.5, adjusted p < 0.05), chemokines seemed to be more induced in limbus and sclera compared with cornea. The induction of IL6 and SAA2 was attenuated in sclera, and SOD2, RIPOR3, TNFAIP3, and C3 were strongly induced in all three cell types, revealing unique and shared aspects of the transcriptional response to infection in cornea, limbus, and sclera (Figures 3F and 3G). In order to test whether the infection was in fact dependent on serine proteases, as it has been suggested, we exposed limbal cells to SARS-CoV-2 in the presence of N-p-Tosyl-L-phenylalanine chloromethyl ketone (TPCK). TPCK is an inhibitor of serine proteases that include the TMPRSS family. Pre-treatment with TPCK almost completely abrogated SARS-CoV-2 infection, confirming both viral entry and dependency on serine proteases for infection (Figure 3H). Interestingly, given that the limbus is a site of corneal and conjunctival stem cells, it is possible that these cells serve as a site of productive virus replication.
Figure 3.
Whole-transcriptome analysis of human ocular surface cells infected by SARS-CoV-2
(A) Read coverage of the SARS-CoV-2 genome in human donor cornea, limbus, and sclera tissues infected with SARS-CoV-2 (MOI = 1.0, 24 h) and subjected to mRNA sequencing (mRNA-seq).
(B) Quantification of reads mapped in (A), indicated as mean reads per million. Error bars indicate SD (n = 3/condition); ∗∗adjusted p = 0.0024 and ∗∗∗adjusted p = 0.0001 (one-way ANOVA).
(C–E) Differential gene expression of all genes plotted by the log2 fold change and the statistical significance as −log10 of the adjusted p value the experiment in (A) and (B) in (C) cornea, (D) limbus, and (E) sclera. The ten genes with highest increases in gene expression that also reached statistical significance (adjusted p < 0.05) were named. Horizontal line indicates adjusted p = 0.05, and vertical lines indicate log2 fold change = ±1.5.
(F) Heatmap comparing the differential gene expression of genes that showed increased gene expression upon infection (log2 fold change > 1.5, adjusted p < 0.05) in at least one of the tissues as in (C)–(E).
(G) Venn diagram showing the overlap of genes with increased expression (log2 fold change > 1.5, adjusted p < 0.05) in response to SARS-CoV-2 infection as in (C)–(F).
(H) Relative expression of subgenomic SARS-CoV-2 N gene RNA in infected human donor limbus tissue (MOI = 1.0, 24 h) pre-treated with TPCK or vehicle control (DMSO) quantified by qRT-PCR. Relative expression = 1 is the average expression in the vehicle control (n = 3/condition).
See also Table S1.
To better understand why limbal cells may be more susceptible to infection than corneal cells, we examined the genes with higher than 2-fold expression in limbal cells compared with corneal cells and evaluated their known function. In order to conduct a system-wide profiling of genes, we used the integrative web-based application Enrichr (Chen et al., 2013). We searched for genes associated with SARS-CoV-2 infection among those higher expressed in limbus and identified ACE2, TMPRSS2, TMPRSS4, TMPRSS11E, SPINT1, and SPINT2, which are genes previously associated with SARS-CoV-2 infection and also significantly more expressed in limbal cells than corneal cells (Table S2Ai) (Singh et al., 2020). We also compared the list of genes enriched in limbus with Gene Ontology datasets and found cornification morphogenesis of epithelium and regulation of water loss to be represented, which is consistent with the role of limbus as a stem cell niche contributing cells to the conjunctiva and cornea (Tables S2Aii and S2Aiii). MGI Mammalian Phenotype datasets that correlated significantly with the list of limbal cell genes included abnormal hair, small intestine, stomach mucosa, esophageal epithelium, and absent epidermis stratum corneum, all of which express similar genes and perform similar functions in their respective organs as the secretory mucosa in the corneal limbal region (Table S2Aiv). Interestingly, cornification morphogenesis signatures have also been reported in the context of SARS-CoV-2 infection of primary lung epithelial cultures (Blanco-Melo et al., 2020). We next compared Gene Ontology datasets with the genes exhibiting increased expression in response to infection in limbal cells. This gene signature was significantly correlated to TNF-α via NF-κB, inflammatory response, IL-6, and interferon (IFN) gamma, in that order (Table S2Bi). We also observed stress pathways, including epithelial-to-mesenchymal transition (EMT), complement pathway, KRAS, and UV responses. COVID-19-related gene datasets significantly correlated with infected limbus genes, including upregulated genes in SARS-CoV-2-infected Vero E6, Calu-3, bronchial epithelial, and cardiomyocytes (Table S2Bii). Jensen COMPARTMENTS datasets (Binder et al., 2014) that significantly correlated with infected limbal cell expression included complement pathway, Rad51, telomere, inflammasome, and TNF superfamily complexes (Table S2Biii). As mentioned previously, decreased expression of a subset of genes was observed in limbal cells in response to infection. We compared these genes with the BioPlanet dataset (Huang et al., 2019) and found that protein-modifying processes significantly correlated, including O-glycan biosynthesis, neuroregulin receptor degradation, glycosylation of mucins, tight junction interactions, glycosphingolipid biosynthesis, and more, which are normal limbal functions (Table S2Ci). When comparing these genes with COVID-19-related gene sets, we observed significant correlations with genes downregulated by SARS-CoV-2 in ferret nasal wash, intestinal organoids, Calu-3 cells, and infected lung (Table S2Cii) (Blanco-Melo et al., 2020; Han et al., 2021). Finally, Jensen COMPARTMENT datasets that correlated with downregulated infected limbal cell genes included sweat gland, stratified epithelium, keratinocyte, tear, and conjunctiva (Table S2Ciii). The genes responsible for normal limbal function were suppressed.
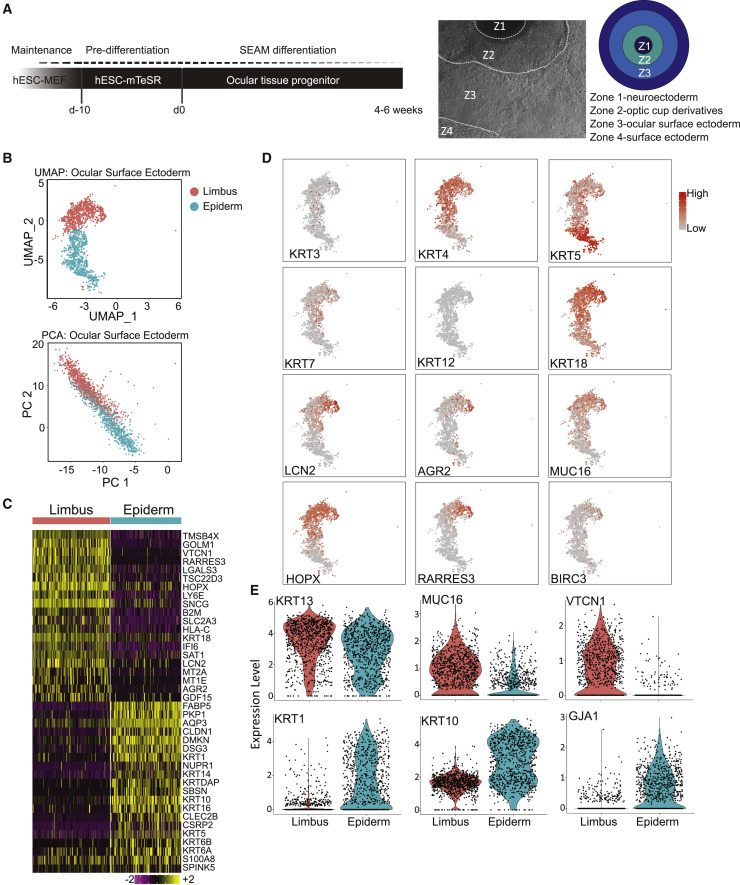
Coordinated generation of multiple ocular-like cell lineages
Human eye tissue is accessible only following organ donation from deceased patients, and this limited availability complicates the study of SARS-CoV-2 in the eye. To address this limitation, we adopted a pluripotent stem cell differentiation protocol that has shown cell fate specification resembling neuroectoderm, neural crest, and ocular surface ectoderm, termed SEAM, for self-formed ectodermal autonomous multizone (Hayashi et al., 2016) (Figure 4 A) as a model for SARS-CoV-2 infection in the eye. Original SEAM publications have shown that concentric rings of cells expressed markers with distinct morphological characteristics of cell types of the eye. These rings partially resembled the coordinating rings of cell types of the developing eye field and ocular surface ectoderm (Figure S6A). To validate our SEAM cultures, we confirmed distinct expression patterns of Pax6 as a marker of neuroectoderm and surface ectoderm in zones 2 and 3, respectively, 7 weeks into differentiation. These PAX6 expression patterns segregated with markers tubulin beta 3, CHX10, and OTX2 for the optic cup derivatives, and E-cadherin co-expressed with lower expression of PAX6 for the surface ectoderm (Figure S6B).
Figure 4.
Zone 3 in the SEAM whole-eye colonies is composed of cells expressing ocular surface ectoderm gene profile
(A) Schematic of SEAM culture differentiation and lineages of each SEAM zone. hESC-derived SEAM organoids were differentiated for 55 days, then processed for immunohistochemical analysis single-cell RNA-seq (n = 1 biological replicate). Unbiased clustering of SEAM culture was conducted using the Seurat package (Satija et al., 2015) in R.
(B) Cells possessing ocular surface ectoderm annotation were further clustered, presented as UMAP and principal-component analysis (PCA).
(C) Heatmap of genes distinguishing each cluster by relative expression.
(D) Expression pattern of genes associated with ocular surface ectoderm clusters.
(E) Violin plots of known markers of corneal, limbal, conjunctival, and epidermal lineages.
See also Figures S6 and S7.
Single-cell RNA-seq of SEAM colonies confirmed ocular cell fate expression
To further examine the cellular composition of SEAM colonies, we performed single-cell RNA-seq (scRNA-seq). In total, we identified 11 clusters. Using the Jensen TISSUES dataset in addition to a gene expression-based cell type clustergram, we were able to annotate all clusters with correlation significance greater than a p value of 0.001 (Figure S7). The SEAM culture was composed of surface ectoderm, ocular surface ectoderm, RPE, neural lineages, dorsal optic cup, and periocular mesenchyme. We also found non-ocular populations identified most closely with lymphatic cells, intestinal cells, bone marrow, and macrophages. Two distinct clusters were given ocular surface ectoderm annotation on the basis of differential gene expression analysis, as visualized by uniform manifold approximation and projection (UMAP) and principal-component analysis (Figure 4B). We generated lists of genes in which expression differed between these clusters. The top 20 genes conferring elevated expression for each cluster were displayed in a heatmap (Figure 4C). We compared these gene lists with the Human Cell Atlas (Regev et al., 2017), as well as our own bulk RNA-seq data from adult human ocular tissues, and identified one cluster that expressed genes of the limbus, based both on bulk RNA-seq data and on datasets from conjunctiva. KRT3 was expressed only in a subset of these cells, suggesting that these populations were immature (Figure 4D). KRT4 is specifically found in mucosal cells, which suggests that the top cluster may have been differentiating toward limbal cell fates. We detected KRT12 only sporadically, suggesting that these populations did not represent mature cornea, but we did find a number of genes uniquely found in limbus gene expression profile, including KRT7, 18, LCN2, AGR2, MUC16, HOPX, RARRES3, and BIRC3 (Figure 4D). The bottom ocular surface ectoderm sub-cluster expressed genes more associated with epidermis, which included KRT clusters.
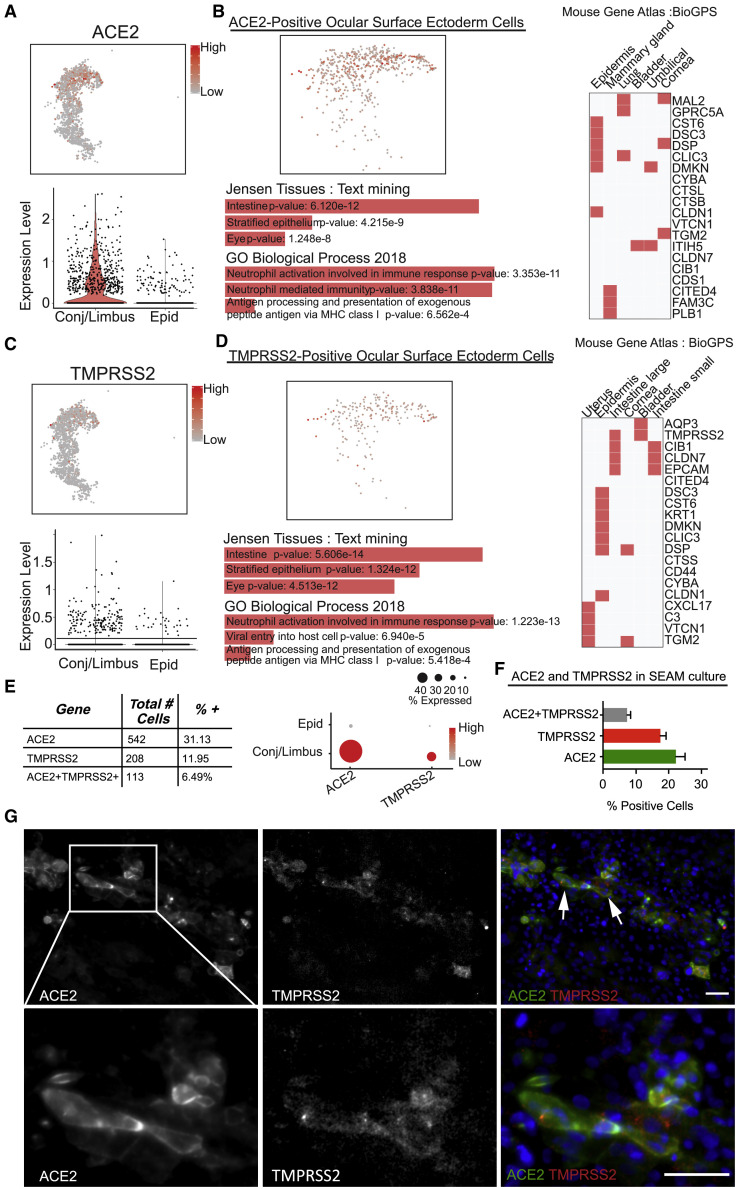
We next asked if SARS-CoV-2 entry factors were present in the SEAM and more specifically in the ocular surface ectoderm cell populations. We found that 11.7% of cells (6,268 cells in total) expressed ACE2 above baseline, and UMAP expression analysis of ACE2 expression in the whole SEAM culture illustrated that greater than 73.7% (542) of ACE2-positive cells are within in the ocular surface ectoderm annotated clusters, predominantly in the conjunctiva/limbus population (Figure 5 A). Jensen TISSUES text mining of gene expression data solely from ACE2-positive cells identified functional categories, including intestine, stratified epithelium, and eye (Figure 5B). These categories connect ACE2-positive cells of eye origin to expression of cells of intestine and other stratified epithelia. Gene Ontology analysis identified genes involved in immune responses, which were shared with ACE2-expressing cells (Figure 5B). Mouse Gene Atlas results implicated additional epidermal cell types, including the cornea (Figure 5B). Therefore, consistent with adult human donor data and our previous study (Hamashima et al., 2020), a subset of ocular surface ectoderm-like cells expressed ACE2.
Figure 5.
Presumptive corneal cell clusters from SEAM whole-eye colonies express ACE2 and TMPRSS2
(A) Relative expression of ACE2 in corneal clusters from SEAM colonies presented as UMAP and violin plot.
(B) ACE2-positive cells evaluated by Jensen TISSUES, Mouse Gene Atlas, and Gene Ontology analyses.
(C) Relative expression of TMPRSS2 in corneal clusters from SEAM colonies presented as UMAP and violin plot.
(D) TMPRSS2-positive cells evaluated by Jensen TISSUES, Mouse Gene Atlas, and Gene Ontology analyses.
(E) Table showing total cell number and percentage of the corneal cells from SEAM colonies expressing potential SARS-CoV-2 targets.
(F) Bar graph illustrating the percentage of ACE2, TMPRSS2, and ACE2+TMPRSS2-positive cells quantified by immunofluorescence (IF) staining of zone 3 in SEAM colonies (SEAM differentiated for 31 days). Error bars indicate standard error of mean (SEM) from six images counted, with an average total number of cells of per image of 916.67 ± 91.68 (mean ± SEM).
(G) Immunohistochemistry of antibodies targeting ACE2 and TMPRSS2 in SEAM colonies. White arrows indicate double-positive cells. Blue is DAPI staining illustrating nuclei in merged image, and ACE2 and TMPRSS2 are shown in green and red, respectively. Scale bars, 50 μm.
We next examined the expression of TMPRSS2 (Matsuyama et al., 2010). Similar to ACE2, we found 208 of 266 of TMPRSS2-expressing cells (78.2%) within ocular surface ectoderm annotated clusters (Figure 5C). Again, the conjunctiva/limbus cluster contained the highest numbers of cells expressing TMPRSS2. Jensen TISSUES text-mining analysis of TMPRSS2-positive cornea cells identified terms associated with nerve, eye, and stratified epithelium (Figure 5D). Mouse Gene Atlas similarly identified epithelial tissue types, including the cornea (Figure 5D). Gene Ontology analysis of TMPRSS2-positive cells identified terms including epidermis development, positive regulation of viral entry into host cell, and negative regulation of epithelial proliferation (Figure 5D). Thirty-one percent of the total ocular surface ectoderm population expressed ACE2. TMPRSS2 was expressed in 208 cells or 11.95% of ocular surface ectoderm cells. Co-expression of ACE2 and TMPRSS2 was found in 113 cells (6.49%) (Figure 5B). Furthermore, we were able to confirm co-expression using immunohistochemistry on SEAM colonies and found a subpopulation of cells expressing both ACE2 and TMPRSS2 organized somewhat linearly in the colony in zone 3, and quantification of ACE2- and TMPRSS2-positive cells resulted in similar numbers as found using scRNA-seq (Figures 5F and 5G).
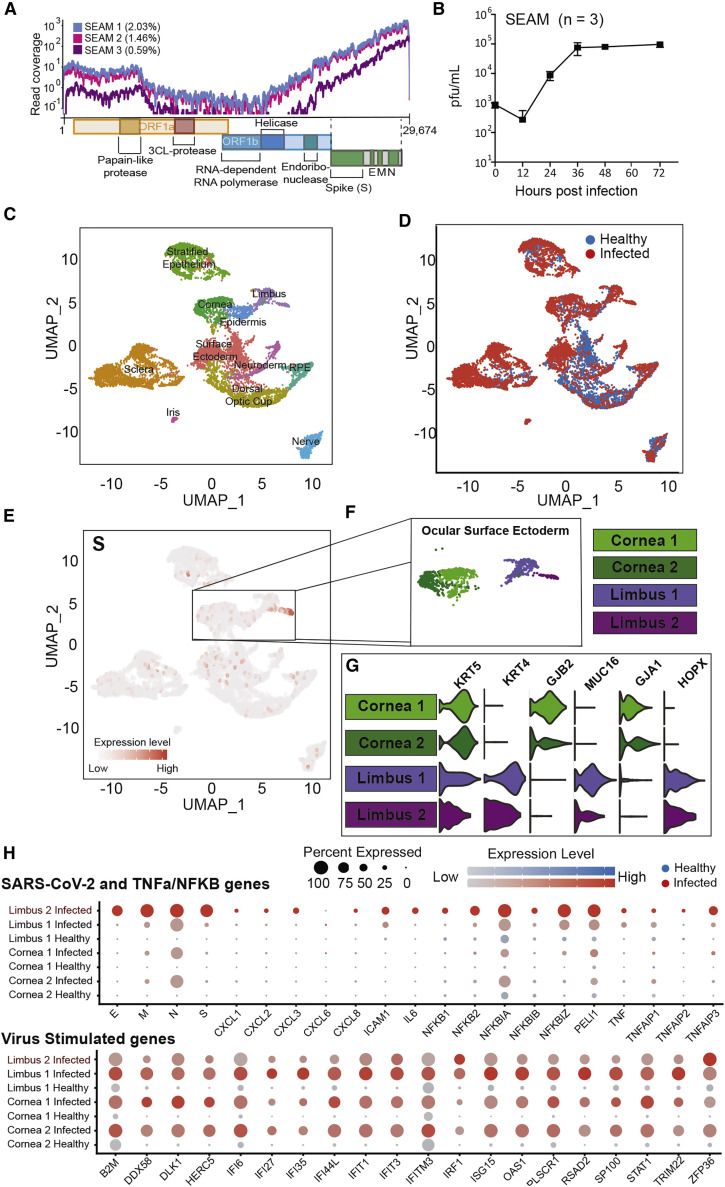
SARS-CoV-2-infected conjunctival/limbal-like cells of SEAM colonies
On the basis of the presence of viral entry factors, we hypothesized that SEAM cultures may be susceptible to SARS-CoV-2 infection and could serve as a suitable model to study infection of the eye. To this end, we infected SEAM cultures with SARS-CoV-2 at an MOI of 1.0 for 24 h and were able to detect viral reads using RNA-seq, demonstrating viral entry and transcription (Figure 6 A). We next quantified virus in the supernatant of infected SEAM cultures by plaque assay to assess whether the infection was productive. SARS-CoV-2 infectious virions increased by 100-fold compared with input virus as a result of productive infection when quantified using plaque assay. Viral load in the supernatant reached a plateau after 24 h that was maintained for at least 3 days (Figure 6B). Considering the fact that ACE2 and TMPRSS2 expression in SEAM cultures was found predominantly in cells with conjunctival/limbal annotations, we hypothesized that conjunctival/limbal cells would be preferentially infected. To this end, we performed scRNA-seq to compare uninfected (healthy) with SARS-CoV-2-infected SEAM cultures through integration of the two datasets. Cells from both the infected and healthy SEAM were represented in every major cluster, with the surface ectoderm partly shifted in the UMAP, verifying that both SEAM cultures were similar in complexity and cellular composition (Figures 6C and 6D), with the exemption of a second cluster also annotated as limbus (limbus 2) present only in the infected SEAM. Next, we examined the expression of SARS-CoV-2 genes. UMAP representation of the viral S-transcript was preferentially expressed in the cornea and limbal cluster of the infected SEAM (Figure 6E). The cluster annotated as limbus (limbus 2) seemed to be most infected, corroborating our finding that the limbus cells from post-mortem donors exhibit elevated infection rates than other ocular cells (Figures 2C and 3A). Interestingly, the host response to infection as well as the abundant viral transcripts were among the defining genes of the limbus 2 cluster because of their high abundance, which may have been sufficient to migrate infected cells away from the other limbus cluster (limbus 1) (Figure 6F). To validate the identity of the corneal/limbal clusters, we assessed the distinct cell-specific marker expression patterns, which were consistent with previous data (Figure 6G). The viral genes E, M, N, and S were all expressed to high levels in nearly all cells in limbus 2, indicating that the cells in limbus 2 were infected. The viral gene expression was in stark contrast to low background expression levels in the other clusters, with the exemption of the N transcript (Figure 6H). This low expression could originate from abundant N mRNA released into the media by cells dying from infection, consistent with the findings of others (Hoagland et al., 2021). Next, we sought to characterize the antiviral response of the infected cells through a comparison of the NF-κB and type I IFN (IFN-I) signaling pathways in the corneal and limbal clusters. We observed an increased expression of NF-κB- and TNF-α-mediated genes in the infected limbus 2 cluster compared with the uninfected clusters, similar to what was observed for the infected post-mortem donor cells. It is noteworthy that expression of these genes was most elevated in cells that also had high viral reads and not in the bystander cells, indicating that this gene signature was mediated by the virus itself and not by cytokines released into the media by infected cells (Figure 6H).
Figure 6.
SARS-CoV-2 infection of SEAM whole-eye colonies
(A) Read coverage of the SARS-CoV-2 genome from next-generation sequencing of RNA from three different infected SEAM cultures (MOI = 1.0, 24 h).
(B) Virus growth in SEAM cultures shown as plaque-forming units per milliliter of supernatant at the time points indicated. Error bars show SD (n = 3).
(C) UMAP based on unbiased clustering of integrated scRNA-seq data from a non-infected SEAM culture (healthy [SEAM differentiated for 31 days]) and a SEAM infected with SARS-CoV-2 (MOI = 1.0, 24 h) (infected [SEAM differentiated for 54 days]), resolution 0.75.
(D) UMAP of integrated scRNA-seq data colored by origin of data (healthy [blue] versus infected [red]).
(E) Feature plot showing the location of cells expressing the SARS-CoV-2 spike gene (S) in red.
(F) Zoom in of the cornea and limbal clusters made up of four sub-clusters.
(G) Violin plots showing some differences in genes expression between the corneal and limbal sub-clusters used for sub-annotation of cell-type identity.
(H) Dot plots showing differential expression of inflammation related genes between the corneal and limbal sub-clusters split by data origin, healthy (blue) versus infected (red). Top: SARS-CoV-2 genes and TNF-α/NF-κB pathway-related gens. Bottom: SARS-CoV-2-stimulated genes.
Next, we investigated the relative expression of IFN-stimulated genes (ISGs) induced by either IFN-I or IFN-III (Figure 6H, bottom panel). The cornea clusters in the infected SEAM exhibited a robust ISG response compared with cornea clusters in the healthy SEAM, and a similar response was observed in the limbus cluster (limbus 1 infected) from which the virus was absent. The ISG signature was most likely a result of paracrine IFN-I/-III signaling induced by the production and/or spill-over of PAMPs generated during infection (Figure 6H). When comparing the limbus cluster with high levels of virus reads in the infected SEAM to the limbus cluster from the same SEAM with low reads, the expression of ISGs was diminished compared with all other clusters from the infected SEAM, albeit still higher than the background expression in the healthy SEAM. This provides support for the hypothesis that SARS-CoV-2 antagonizes the induction of ISGs in infected cells. Moreover, aside from IRF1 and ZFP36, the pathway as a whole was less induced compared with the response in bystander cells, suggesting that SARS-CoV-2 antagonism may occur at a major node of the IFN-I/-III response (Figure 6H), which has also been described in the literature (Konno et al., 2020; Miorin et al., 2020; Thoms et al., 2020).
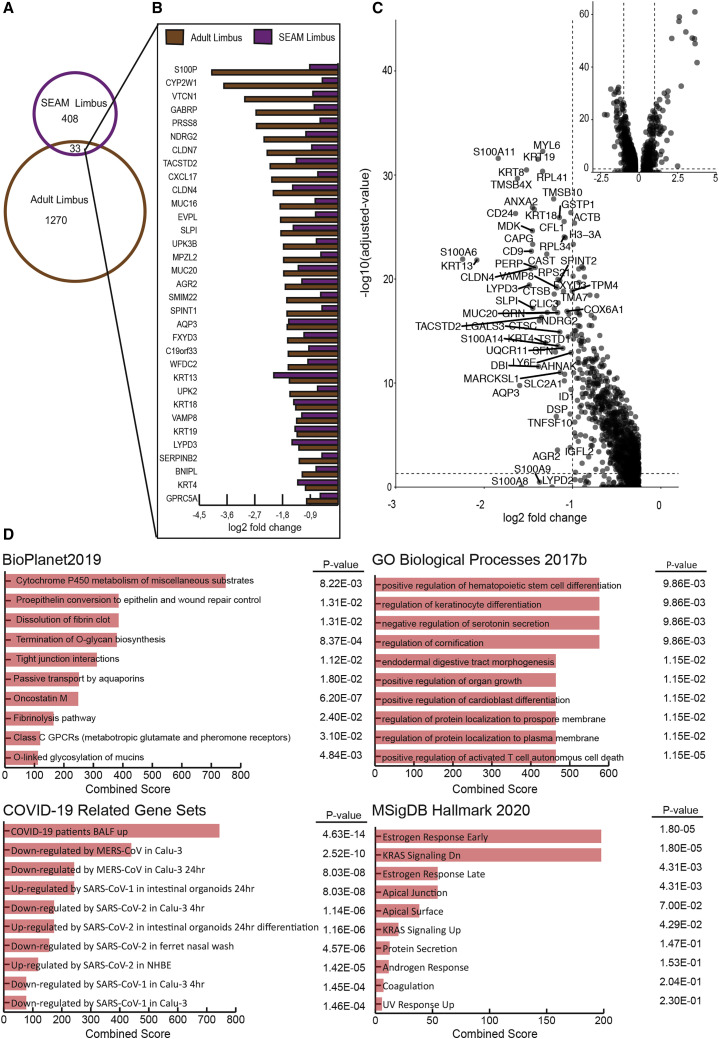
When considering SARS-CoV-2 infection in the eye, the notion of blocking normal cellular functions is alarming. We therefore further examined which affected genes were shared in the SEAM limbus and adult limbus. Of the total genes found in the SEAM limbus, 87% of genes were also expressed in adult limbus, while 80% of genes in the corneal clusters were expressed in adult cornea. In comparing common transcriptional signatures downregulated in both donor-derived and SEAM limbus, SARS-CoV-2 induces genes represented pathways involved in response to injury, immune cell recruitment, cell-cell communication, limbal/conjunctival function, and barrier function (Figures 7 B–7D).
Figure 7.
Genes downregulated in limbus cells after infection with Sars-CoV-2
(A) Venn diagram of genes downregulated after infection with SARS-CoV-2 in primary adult limbus cells (brown) and limbus cell cluster from scRNA-seq of infected SEAM (purple).
(B) Log2 fold change of overlapping downregulated genes after infection with SARS-CoV-2 at 1 MOI for 24 h in limbus cells isolated from human cadaver donors (bulk sequencing, DEseq2) and limbus clusters from SEAM organoid (scRNA-seq, Wilcoxon p < 0.05).
(C) Volcano plot showing log2 fold change of gene expression after infection on the basis of Wilcoxon test between limbus clusters (infected cluster and nearest neighbor cluster). The inset shows the full volcano plot. Genes with log2 fold change < −1 were labeled.
(D) Gene expression analysis on overlapping downregulated genes from BioPlanet2019, GO Biological Processes 2017b, COVID-19 Regulated Gene Sets and MSigDB Hallmark 2020. All databases accessed through Enrichr.
Discussion
Unprecedented research efforts have been undertaken to characterize SARS-CoV-2 in response to the COVID-19 pandemic. It has become clear that the symptoms of COVID-19 extend beyond the respiratory tract, which is the primary site of replication, with an expanding list of reports of post-acute sequalae of SARS-CoV-2 infection (PASC) that prompt an investigation of each affected body organ (Davis et al., 2020). The exposed position of the eye to aerosols and droplets as well as reports of ocular COVID-19 complications led us to characterize the susceptibility and host response of the eye to SARS-CoV-2 infection. In the present study, we provide evidence of viral antigen in the ocular surface ectoderm of patients who tested positive for SARS-CoV-2. In addition, we show that donor-derived ocular tissues become infected and respond to the virus when challenged experimentally and that SEAM cultures recapitulate both the composition of the eye and its response to virus, putting it forward as a useful model to further study these dynamics.
The current evidence of SARS-CoV-2 infection in the eyes of COVID-19 patients is mixed. Although virus can be detected in the conjunctiva of animal models for SARS-CoV-2 (Deng et al., 2020), human studies suggest that roughly 1%–5% of COVID-19 patients present with SARS-CoV-2 in tears (Guan et al., 2020; Mungmungpuntipantip and Wiwanitkit, 2020; Hu et al., 2020). A study of more than 25,000 patients found a significant correlation between the use of eye protection and reduced rates of infection (Chu et al., 2020), while others suggest that the risk for becoming infected through the eye is exaggerated (Zhou et al., 2020). The symptoms of COVID-19 are highly heterogeneous and may in part explain the sporadic detection of virus and complications in the eyes of patients (Williamson et al., 2020). Nevertheless, even a 1% incidence rate of ocular complications is more than 1 million people worldwide and should not be neglected. We show that the ocular surface cells, and to some extent posterior parts of the eye, possess the necessary machinery to support infection and become readily infected in an experimental setting. These findings may also be relevant as a site of viral reservoirs for the virus.
Limbal cells infected by SARS-CoV-2 seem to undergo an EMT alongside a decrease in cell junctions and an increase in injury response genes, which may in part be an attempt of the infected cell to recover. Many of the genes that contribute to normal limbus functions, including KRTs, mucins, aquaporins, and tight junctions, are also suppressed. Evidence suggests that SARS-CoV-2 hijacks the host transcriptional machinery to replicate its genome at the expense of normal cell functions (Miorin et al., 2020; Roy et al., 2020), which may be the reason for the decrease in genes characteristic for the limbus.
Our data suggest that ocular infection seems to preferentially occur in the limbus. In search of a mechanism for the increased infectivity, we evaluated the expression of alternative entry factors (Singh et al., 2020). Donor-derived limbus cells had an 11-fold higher expression of TMPRSS4 than donor-derived cornea cells. On the contrary, only 2.3% of cells in the SEAM culture expressed TMPRSS4, but expression was enriched in the ocular surface ectoderm, with 7.1% of the cornea clusters and 22.1% of the limbus clusters expressing TMPRSS4. TMPRSS4 is preferentially expressed in secretory cells (Singh et al., 2020), and our data are consistent with other published scRNA-seq datasets showing elevated TMPRSS4 expression in limbal/conjunctival cells compared with central cornea (Collin et al., 2021). Further studies are required to properly assess the involvement of TMPRSS4 in ocular infection, but TMPRSS4 has been implicated in SARS-CoV-2 infection of small intestinal enterocytes and could perhaps be more important than TMPRSS2 in infection of the limbus (Zang et al., 2020). scRNA-seq analysis of different human organs in the context of virus infection suggests that ACE2 is induced by IFN-I/-III (Ziegler et al., 2020) and it is possible that the cytokine signaling between cell types can modulate the permissivity to infection, but we were unable to detect any changes in response to infection of ACE2 and TMPRSS2 in our transcriptional data (Table S1).
IFN-I/-III serves as the first line of defense against viral infections in all mammals, but successful viruses also have the capacity to antagonize the IFN-I/-III response (tenOever, 2016; García-Sastre, 2017). SARS-CoV-2 encodes multiple accessory proteins capable of muting the cellular antiviral response, including nsp1, which interferes with the translation of host mRNA including IFNβ (Narayanan et al., 2008; Thoms et al., 2020) and ORF6, which blocks STAT1 nuclear translocation (Miorin et al., 2020; Frieman et al., 2007). This seems to cause a delay in induction and a dampening of IFN-I/-III in tissue culture, animal models, and patient data while leaving NF-κB-mediated transcription unchecked (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Hoagland et al., 2021), culminating in the improper recruitment of inflammatory monocyte and macrophage populations that lead to severe COVID-19 (Merad and Martin, 2020). Our scRNA-seq analysis of the infected SEAM culture shows the induction of ISGs in all cells in the infected SEAM but to a lesser extent in the cells that support active viral replication. This suggests that the induction of IFN-I/-III may not be effectively inhibited in infected cells or that debris caused by virus-induced cell death results in IFN-I/-III production in neighboring cells that is subsequently released into the medium, where it signals in a paracrine manner (Hoagland et al., 2021; Karki et al., 2021). However, the lower levels of ISGs in infected cells compared with bystander cells indicate that the virus is able to antagonize the induction of ISGs within the cell. On the contrary, NF-κB-signaling is induced in both infected and uninfected bystander cells but is more highly expressed in the infected cells, which suggests that some aspect of SARS-CoV-2 biology results in NF-κB-activation in addition to autocrine cytokine signaling. This is further supported by our mRNA-seq analysis, in which most of the genes that increase in gene expression in response to infection are products of NF-κB-signaling. The IFN-I/-III response can also be suppressed by NF-κB-signaling itself, suggesting that there is a tightly controlled orchestration of inflammatory signaling upon SARS-CoV-2 infection (Cheng et al., 2011). The health of the cell likely contributes to how effective SARS-CoV-2 is in antagonizing ISGs. Although there are few whole-transcriptome RNA-seq reports currently exploring the response of SARS-CoV-2 infection to cells of the eye, non-infected ocular datasets suggest that robust inflammatory responses are present. RNA-seq and ATAC-seq of the same samples report open chromatin and transcriptional expression of inflammatory regulators, including TNF, IFN-I/-III, and components of NF-κB signaling (Collin et al., 2021).
In conclusion, the data presented show that cells of the human ocular surface are susceptible to infection by SARS-CoV-2, and thus the eye should be considered a potential entry site warranting protection. The limbus seems to be the most at risk because of the expression of ACE2, TMPRSS2, and other SARS-CoV-2-associated genes, as well as a higher infection propensity compared with other ocular cell types. The SEAM eye model is useful to study SARS-CoV-2 infection dynamics and for screening compounds for infection prevention. SARS-CoV-2 causes a transcriptional signature in cells of the eye, which is consistent with its effect in other tissues of the body. Future studies are warranted to better understand how infection in the eye may lead to transmission to other regions of the body.
Limitations of study
Although we find compelling evidence that ocular cells can be readily infected with SARS-CoV-2, the study has some clear limitations. First, we obtained only three to seven genetically distinct donors per ocular cell type. Second, whether the ocular cells can be directly infected by aerosol droplets, or whether the infection observed in post-mortem donors originated as an airway infection and spread to the ocular surface systemically, is yet unknown. With regard to the SEAM organoid model, we recognize that this is a model based on stem cells and that the cells in the SEAM cultures are not fully mature ocular cells, seen for instance by the low expression of KRT3 and KRT12. It should also be mentioned that there will be cells at different developmental stages in the SEAM organoid, which might affect the infectivity of the cells in the SEAM, as we see a higher infectivity in the limbus, which is known to harbor a stem cell population. It would be interesting to perform scRNA-seq on the infected cadaver donor ocular cells to investigate whether there is a specific cell niche more prone to infection than other and whether such a niche exhibits stem cell properties.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-SARS-CoV-2 Spike (S) | Center for Therapeutic Antibody Discovery at the Icahn School of Medicine at Mount Sinai | Clone 2B3E5; RRID: AB_431451 |
| Goat Anti-ACE2 antibody | Abcam | Cat# ab15348; Lot GR3333640-8; RRID: AB_301861 |
| Human/Mouse/Rat/Hamster ACE-2 Antibody | R&D systems | Cat# AF933; Lot HOK0320061; RRID: AB_355722 |
| F(ab’)2-Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A11017; Lot 2108802; RRID: AB_143160 |
| F(ab’)2-Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A21246; Lot 2069609; RRID: AB_1500778 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Flour 488 | Invitrogen | Cat# A11055; Lot 1463163; RRID: AB_2534102 |
| F(ab’)2-Goat anti-Rabbit IgG (H+L)Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Invitrogen | Cat# A11071; Lot 1789904; RRID: AB_1500774 |
| F(ab’)2-Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A21237; Lot 1597026; RRID: AB_1500743 |
| Anti-Cytokeratin3/CK-3 antibody [AE5] | abcam | Cat# ab68260; Lot GR3329790-1; RRID: AB_1140695 |
| Recombinant Anti-Keratin 12/K12 antibody [EPR17882] | abcam | Cat# ab185627 Lot GR224164-15; RRID: AB_2889825 |
| Cytokeratin 15 Antibody (ST04-05) | Novus | Cat# NBP2-67525; Lot HM-701; RRID: AB_2889827 |
| Anti-STRO1 antibody [7i35] | abcam | Cat# ab102969; Lot GR8271-1; RRID: AB_10710681 |
| Human Pax6 Antibody | R&D systems | Cat# AF8150; RRID: AB_2827378 |
| Purified anti-Pax-6 Antibody | Biolegend | Cat# PRB-278P; Lot E11EF01096; RRID: AB_291612 |
| Anti-alpha smooth muscle Actin antibody [1A4] | abcam | Cat# ab7817; Lot GR3257713-4; RRID: AB_262054 |
| Anti-MiTF antibody [D5] | abcam | Cat# ab3201; Lot GR3356560-1; RRID: AB_303601 |
| Recombinant Anti-Calreticulin antibody [EPR3924] | abcam | Cat# ab92516 Lot GR3185160-1; RRID: AB_10562796 |
| Purified anti-Tubulin β 3 (TUBB3) antibody | Biolegend | Cat# 801213; RRID: AB_2728521 |
| Sheep anti Human chx 10 (Visual system homeobox 2) (CT) | Exalpha | Cat# X1179P; Lot 13703; RRID: AB_2889828 |
| E-Cadherin (24E10) Rabbit mAb | Cell Signaling | Cat# 3195; Lot 13; RRID: AB_2291471 |
| Recombinant Anti-TMPRSS2 antibody [EPR3862] | abcam | Cat# ab242384; Lot GR3340286-1; RRID: AB_2889829 |
| SARS-CoV-2 (COVID-19) nucleocapsid antibody | GeneTex | Cat# GTX135357; RRID: AB_2868464 |
| p63 antibody | Santa Cruz | Cat# SC-8431; RRID: AB_628091 |
| Bacterial and virus strains | ||
| SARS-CoV-2, isolate USA-WA1/2020 | BEI Resources | Cat# NR-52281 |
| Biological samples | ||
| Human eye globes | Eye-Bank for Sight Restoration | N/A |
| SARS-CoV-2/USA-WA1/2020 isolate | GenBank | Accession number MN985325.1 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase | Worthington | Cat# LS004176 |
| Ambion DNase I Solution | Invitrogen | Cat# AM2222 |
| Thiazovivin | Stem Cell Technologies | Cat# 100-0247 |
| Synthemax II | Corning | Cat# 3535 |
| CryoStor CS2 Freeze Media | Sigma-Aldrich | Cat# C3124 |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |
| Matrigel | Corning | Cat# 354230 |
| Critical commercial assays | ||
| Chromium Single Cell 3′ Library and Gel Bead Kit v3.0 | 10x Genomics | Cat# 1000078 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | Cat# 1000127 |
| Chromium Next GEM Single Cell 5′ Library and Gel Bead Kit v1.1 | 10x Genomics | Cat# 1000167 |
| Chromium Single Cell 5′ Library Construction Kit | 10x Genomics | Cat# 1000020 |
| 10X Genomics Chromium controller v3.16 | 10x Genomics | N/A |
| DNA-free DNA removal kit | Invitrogen | Cat# AM1906 |
| KAPA SYBR FAST qPCR Master Mix Kit Universal | Kapa Biosystems | Cat# KK4601 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat# 20020594 |
| Deposited data | ||
| Mouse Gene Atlas | BioGPS | http://biogps.org/downloads/ |
| Single cell RNA sequencing of SEAM organoid and SEAM organoid infected with SARS-CoV-2 | NCBI GEO | GSE165477 |
| Response to SARS-CoV-2 infection in cornea, limbus and sclera from human donors | NCBI GEO | GSE164073 |
| Additional methods and sequencing data deposited at Mendelay Data | https://data.mendeley.com/datasets/jgw2mcgb67/1 | 10.17632/jgw2mcgb67.1 |
| Experimental models: Cell lines | ||
| Human: Passage 35-40 H9 ES cells | WiCell | N/A |
| Adult human RPE cells, Passage 1-3 | This paper | N/A |
| Adult human Cornea cells, Passage 1-3 | This paper | N/A |
| Adult human Limbus cells, Passage 1-3 | This paper | N/A |
| Adult human choroid cells, Passage 1-3 | This paper | N/A |
| Adult human Iris cells, Passage 1-3 | This paper | N/A |
| Adult human Iris cells, Passage 1-3 | This paper | N/A |
| Vero E6 | ATCC | Cat# CRL-1586; RRID: CVCL_0574 |
| Oligonucleotides | ||
| Primer: Sars-CoV-2 N sgRNA Forward: CTCTTGTAGATCTGTTCTCTAAACGAAC | Blanco-Melo et al., 2020 | Blanco-Melo et al., 2020 |
| Primer: Sars-CoV-2 N sgRNA Reverse: GGTCCACCAAACGTAATGCG | Blanco-Melo et al., 2020 | N/A |
| Primer: TUBA1A Forward: GCCTGGACCACAAGTTTGAC | Blanco-Melo et al., 2020 | N/A |
| Primer: TUBA1A Reverse: TGAAATTCTGGGAGCATGAC | Blanco-Melo et al., 2020 | N/A |
| Primer: ACE2 Forward: CGAGTGGCTAATTTGAAACCAAGAA | Zhang et al., 2020 | N/A |
| Primer: ACE2 Reverse: ATTGATACGGCTCCGGGACA | Zhang et al., 2020 | N/A |
| Primer: TMPRSS2 Forward: GTCCCCACTGTCTACGAGGT | Vidal et al., 2015 | N/A |
| Primer: TMPRSS2 Reverse: ATTGATACGGCTCCGGGACA | Vidal et al., 2015 | N/A |
| Software and algorithms | ||
| Prism 8 | GraphPad | https://graphpad.com/ |
| Cell Ranger Single-Cell Software Suite (v3.1) | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Seurat R package | Satija et al., 2015 | Stuart et al., 2019 |
| Jensen TISSUES text | Jensen Lab | https://tissues.jensenlab.org/Search |
| ImageJ | Schneider et al., 2012 | Schneider et al., 2012 |
| bowtie2 R package | Langmead and Salzberg, 2012 | Langmead and Salzberg, 2012 |
| ggplot2 R package | Wickham, 2016 | https://ggplot2.tidyverse.org |
| R | R Foundation for Statistical Computing, | https://www.R-project.org/ |
| BaseSpace | Illumina | https://login.illumina.com/platform-services-manager/?rURL=https://basespace.illumina.com&clientId=basespace&clientVars=aHR0cHM6Ly9iYXNlc3BhY2UuaWxsdW1pbmEuY29tL2Rhc2hib2FyZA&redirectMethod=GET#/ |
| DEseq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| RNA-Seq Alignment App v.2.0.2 | Illumina | https://login.illumina.com/platform-services-manager/?rURL=https://basespace.illumina.com&clientId=basespace&clientVars=aHR0cHM6Ly9iYXNlc3BhY2UuaWxsdW1pbmEuY29tL2Rhc2hib2FyZA&redirectMethod=GET#/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Timothy A. Blenkinsop (timothy.blenkinsop@mssm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The datasets generated during this study are available at NCBI GEO: GSE165477, GSE164073.
Experimental model and subject details
Adult human eye post-mortem donor material
Globes from human donors aged between 36 and 90 years were obtained within 24h of death from the Eye-Bank for Sight Restoration (EBSR), Inc., New York, NY, USA, and Miracles in Sight, Winston-Salem, NC, USA. Informed consent was obtained from all subjects and de-identified at EBSR. All experiments were conducted under an Institutional Review Board of the Mount Sinai School of Medicine protocol review, in accordance with Mount Sinai’s Federal Wide Assurances (FWA#00005656, FWA#00005651) to the Department of Health and Human Services and followed guidelines set by HIPAA.
Human embryonic stem cells culture and maintenance
Human embryonic stem cell (hESC) line H9 was obtained from WiCell. hESCs were cultured and maintained on irradiated MEFs in Dulbecco’s Modified Eagle Medium: Nutrient Mix F-12 (DMEM/F12, Life Technologies), containing 20% knockout serum replacement (KSR, Life Technologies) and supplemented with 1X L-Glutamine (Life Technologies), 1X MEM Non-Essential Amino Acids Solution (Life Technologies), 1X Penicillin-Streptomycin (10,000 U/mL, Life Technologies), 0.1 mM 2-mercaptoethanol (Sigma). 10 ng/ml FGF-basic (GIBCO) was freshly added to the medium before use. Prior to differentiation, hESCs were seeded onto Matrigel-coated dishes (Corning) in mTeSR1 (STEMCELL Technologies) media for at least three passages.
SEAM ocular cultures
The differentiation of hESCs was performed as outlined in Figure 4A. First, hESCs were seeded as single cells onto Matrigel-coated dishes at 500cells cm-2 (5,000 cells in one well of a 6-well plate), after which they were cultured in mTeSR1 medium (StemCell Technologies, Inc.) for 10 days to achieve round small-to-medium-sized separate colonies. Half of the culture medium was then changed to SEAM differentiation medium; GMEM (Life Technologies), 10% knockout serum replacement (KSR, Life Technologies) and supplemented with 1X L-Glutamine (Life Technologies), 1X MEM Non-Essential Amino Acids Solution (Life Technologies), 1X Penicillin-Streptomycin (10,000 U/mL, Life Technologies), Sodium Pyruvate (100mM, Life Technologies) and 55 μM 2-mercaptoethanol (Sigma). Medium changes were performed three times per week throughout the differentiation, in which half of the medium was changed for the first week of differentiation, after which full medium changes were made. Between four to seven weeks of differentiation, SEAM structures were mature and pigmented. At seven weeks they were analyzed and/or harvested.
Cell lines and primary cell culture
Vero E6 cells (kidney epithelial cells from female African green monkey, ATCC, CRL-1586) were maintained in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO) supplemented with 10% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Millipore Sigma) at 37°C in a humidified atmosphere with 5% CO2. Globes were placed in preservation media within 12 hours of death and obtained from the Eye-Bank for Sight Restoration, Inc., New York, NY. The donors died of cardiac, uterine cancer, smoke inhalation, obesity, metabolic acidosis, respiratory or renal failure, myocardial infarction with no ocular disease history. The anterior segment was separated from the rest of the eye by performing a circumferential incision 6mm posterior from the ora serrata. The iris was then removed by manual manipulation using forceps. Cornea cells were isolated from the central part of the cornea leaving approximately 1mm distance from the edge of the transparent section of the anterior segment of the eye. This corneal tissue was then further cut into 1mm2 pieces and digested with collagenase 1mg/ml. The full thickness was used including corneal epithelium, corneal stroma and corneal endothelium. The limbal region was dissected from the remaining tissue after the cornea was removed. The limbus sample included the full thickness of 1mm of transparent cornea out to 1mm of opaque tissue. The sclera sample included the full thickness of greater than 2mm away from the beginning of opaque region of the anterior segment to 6mm of opaque region. The ciliary body and iris were pulled away from the cornea/limbus/sclera tissue using #5 Dumont forceps. The iris was then gently pulled away from the ciliary body. These separated tissues were then further cut into 1mm2 pieces and digested with collagenase 1mg/ml with 3ug/ml DNase Solution (STEMCELL Technologies) in Earle’s balanced salt solution for 3 hours. Following digestion, tissue pieces were triturated for 2 minutes using a 10ml pipette. The tissue pieces are allowed to settle at the bottom of the conical tube and the supernatant is collected, centrifuged and resuspended and frozen using CS2 medium (Cryostore).
Taking the posterior segment, the vitreous and retina was encouraged with the use of angled forceps to separate from the back of the eyecup to the point where the retina was only still attached to the eye at the location of the optic disk. Using micro scissors, the retina was cut from the optic disk. The vitreous was then separated from the retina using forceps and the whole retina was cut into approximately 1mm2 pieces and digested with hyaluronidase (6mg/ml) in Earle’s balanced salt solution for 3 hours. The tissue pieces are allowed to settle at the bottom of the conical tube and the supernatant is collected, centrifuged, resuspended and frozen using CS2 medium (Cryostore).
The posterior eyecup was placed in a cup with the optic nerve facing down. The eyecup is filled with 0.25% trypsin with 3ug/ml DNase Solution and incubated at 37°C for 50 minutes. RPE were then brushed off the Bruch’s Membrane, collected and frozen using CS2 medium. After the RPE are removed, choriocapillaris/Bruch’s membrane is cut into 1mm2 pieces of tissue and placed into 0.25% collagenase 2.5mg/ml with 3ug/ml DNase Solution. Afterward, tissue pieces are triturated for 2 minutes using a 10ml pipette. The tissue pieces are allowed to settle at the bottom of the conical tube and the supernatant is collected, centrifuged and resuspended and plated in Synthemax II coated tissue culture plates (Corning, NY) or frozen using CS2 medium (Biolife Solutions, WA). Cells were cultured using DMEM/F12 with 10% FBS medium containing Thiazovivin (2 μM), Glutamax, NaPyruvate, NEAA, Nicotinamide, N1 and Pen/Strep at 1% each. Cells were tested bi-weekly for the presence of mycoplasma using MycoAlert Mycoplasma Detection Kit (Lonza). None of the cell lines were authenticated by an external service but Vero E6 cells were obtained directly from ATCC and primary cell cultures were validated through expression of characteristic markers by immunofluorescence (Figure 2A).
Virus
SARS-related coronavirus 2 (SARS-CoV-2), isolate USA-WA1/2020 (NR-52281) was deposited by the Center for Disease Control and Prevention (Atlanta, GA, USA) and obtained through BEI Resources. The virus was propagated in Vero E6 cells in DMEM (GIBCO) supplemented with 2% FBS (Atlanta Biologicals), 4.5g/L D-glucose (GIBCO, 4mM L-glutamine (GIBCO), 10mM non-essential amino acids (GIBCO), 1mM sodium pyruvate (GIBCO), and 10mM HEPES (Fisher Scientific). Virus stock was washed twice in 15ml propagation medium to dilute out cytokines secreted into the culture medium during propagation by filtration through an Amicon Ultra-15 Centrifugal filter unit (100kDa, Millipore Sigma). Virus stock was resuspended in propagation medium. All infections were performed with the same passage 3 virus stock.
Method details
Adult human eye tissue dissection and dissociation
The eye tissues were separated from the eye globe using forceps and scissors then cut into 1mm2 pieces and placed into 0.25% collagenase (Worthington, NJ, USA), 3μg/ml DNase I Solution (STEMCELL Technologies) and 2μM Thiazovivin ROCK Inhibitor for a minimum of 2 hours. Cells were isolated from the tissue pieces by trituration, then pelleted by centrifugation at 356 x g for 5 min and plated into tissue culture treated plates coated with Synthemax II (Corning, AZ, USA) with Dulbecco’s Modified Eagle Medium: Nutrient Mix F-12 (DMEM/F12, Life Technologies), containing 10% heat-inactivated fetal bovine serum (FBS, Sigma), and 2μM Thiazovivin ROCK inhibitor. Cells were frozen down prior to single-cell RNA sequencing using CryoStor CS2 Freeze Media (Millipore Sigma) in Mr. Frosty containers.
Immunohistochemistry of adult human eye tissue
Tissue samples consistent of sclera, limbus and cornea were obtained from the Eye Bank for Sight Restoration and fixed in 4% paraformaldehyde (PFA, FisherScientific) for 24h. The tissue was then dehydrated in 30% sucrose for 24h before cryopreserving in Fisher Healthcare Tissue-Plus O.C.T. Sections of 14 μm were cut and transferred onto glass slides. Sections were re-hydrated by washing 3 × 5 min with PBS before blocking and permeabilizing the cells with 5% normal donkey serum (NDS), 1% BSA and 0.3% Triton X-100 in PBS for 1h at room temperature. Primary antibodies were added diluted in 5% NDS, 1% BSA in PBS and incubated overnight at 4°C. Sections were then washed 3 × 5min with 1% BSA in PBS before adding secondary antibodies and DAPI diluted in 5% NDS, 1% BSA in PBS and incubated for 1h at room temperature in the dark. Lastly the sections were washed 3 × 5 min with 1% BSA in PBS and mounted with ProLong Gold antifade mountant (ThermoFisher). Images were obtained using a Leica DM5500 upright microscope. Tile-scans were taken with 20x and 40x objectives using 10% overlap in both x- and y-directions, before automatically stitching the images in the Leica Micro Systems imaging software. Image processing was performed in FiJi (Schindelin et al., 2012).
Immunofluorescence staining
Cells were fixed with 4% PFA for 24 h, rinsed 3 times with phosphate buffered saline (PBS), and incubated with 0.3% Triton X-100 (Fisher), 1% BSA and 5% Normal Goat Serum (Jackson ImmunoResearch Laboratories, Inc.) for 1h to permeabilize cells and block non-specific reaction. Primary antibodies (see Key Resources Table) were resuspended in 1% BSA in PBS and added for overnight incubation at 4 °C. Cells were rinsed 3 times and incubated with the appropriate fluorochrome-conjugated secondary antibodies and DAPI (see Key Resources Table) at room temperature for 45 min. Finally, cells were rinsed 3 times and PBS was added. Images were taken using a Leica DMI6000 inverted microscopy.
SEAM ocular culture dissociation
Differentiated SEAM cultures were dissociated for 1 hour at 37 °C, 20% O2, 5% CO2 using 2.5 mg/ml Collagenase 2 (Worthington Biochemical Corporation) in Hanks’ Balanced Salt Solution with 3mg/ml DNase I Solution (StemCell Technologies, Inc.) and 2 μM Thiazovivin ROCK Inhibitor. The harvested cells were washed with Dulbecco’s Modified Eagle Medium: Nutrient Mix F-12 (DMEM/F12, Life Technologies), containing 0.1% BSA, 3 μg/ml DNase I Solution and 2 μM Thiazovivin and filtered through a 40μm cell strainer (Corning). Cells were frozen down prior to single-cell RNA sequencing in CS2 medium (Biolife Solutions, WA).
Single-cell RNA sequencing
Viability of single cells was assessed using Trypan Blue staining, and debris-free suspensions of > 80% viability were deemed suitable for single cell RNA Seq. Uninfected SEAM cultures were processed using the 10X Genomics Chromium controller v3.16 and the Chromium Single Cell 3′ Library and Gel Bead Kit v3.0 and the infected SEAM culture was processed using the Chromium Single Cell 5′ Library and Gel Bead Kit v1.1 using an input of ~10,000 cells. Gel beads were prepared according to standard manufacturer’s protocols. Oil partitions of a single-cell and oligo coated gel beads (GEMs) were captured and reverse transcription was performed, resulting in cDNA tagged with a cell barcode and unique molecular index (UMI). Next, GEMs were broken, and cDNA was amplified and quantified using Agilent Bioanalyzer High Sensitivity chip (Agilent Technologies) and QuBit analysis (ThermoFisher). The samples were sequenced on the Novaseq 6000 Illumina sequencer with S4 flow cell, (100/paired end reads) targeting a depth of 50,000-100,000 reads per cell using v3 chemistry at the genomics core facility at Mount Sinai or on a Nextseq 500 High Capacity flow cell in-house. Fastq files were generated using Cell Ranger Single-Cell Software Suite (v3.1) and were aligned to the grch38 reference genome. Downstream analyses and graph visualizations were performed in the Seurat R package (v. 3.1.2). Briefly, we removed cells with unique gene counts greater than 7,000 (potential doublets) and less than 200. The remaining cells were normalized by a global-scaling normalization method (LogNormalize) with the default scale factor (10,000). Linear dimensional reduction was performed by PCA, following which clustering was performed. The results were visualized using Uniform Manifold Approximation and Projection (UMAP) plots for dimension reduction. Violin and individual gene UMAP plots were generated using the Seurat R package. Heatmaps were generated from the top 20 differentially expressed genes per cluster. Cluster annotation was guided by manual gene expression notation, which was complemented by Enrichr gene set enrichment analysis, using the top 100 differentially expressed genes for each cluster. Jensen TISSUES text mining provided the association between genes and human tissues, the Mouse Gene Atlas from BioGPS was used for cell type specifications and gene ontologies were generated using the GO biological process 2018 terms.
For integrated analysis (Healthy versus Infected) a similar protocol was followed, initially removing counts greater than 7,000 (potential doublets) and less than 200 for both datasets and normalizing the individual datasets with NormalizaData. The two datasets were anchored to each other and integrated. The combined data was then scaled and Linear dimensional reduction was performed by PCA and clustering was performed with a resolution of 0.75. The results were visualized using UMAP, and cluster annotation was performed as described above.
Quantitative real-time PCR analysis
Cells were lysed in TRIzol Reagent (Invitrogen) and separated into an aqueous and an organic layer by centrifugation following the addition of chloroform. RNA was precipitated from the aqueous layer with isopropanol, washed in 75% ethanol and resuspended in nuclease free ddH2O. Prior to reverse transcription, samples were DNase treated with DNA-free DNA removal kit (Invitrogen). cDNA was generated using oligo d(T) primers (Invitrogen) and SuperScript II Reverse Transcriptase (Thermo Fisher). Quantitative real-time PCR was performed on a LightCycler 480 Instrument II (Roche) using KAPA SYBR FAST qPCR Master Mix Kit (Kapa Biosystems) and primers specific for SARS-CoV-2 N subgenomic RNA and TUBA1A, ACE2, and TMPRSS2 spanning at least one intron (see Key Resources Table). Delta-delta-cycle threshold (ΔΔCt) was determined relative to mock-infected samples. Viral RNA levels were normalized to TUBA1A and depicted as fold change over mock-infected samples.
RNA sequencing of viral infections
1μg of total RNA was enriched for polyadenylated RNA species and prepared for next-generation sequencing using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. Sequencing libraries were sequenced on an Illumina NextSeq 500 platform.
Bioinformatic analyses
Raw reads were aligned to the human genome (hg19) using the RNA express app on Basespace (Illumina) and differential gene expression was determined using the DEseq2 protocol (Love et al., 2014). Raw reads were aligned to the SARS-CoV-2 genome (SARS-CoV-2/USA-WA1/2020 isolate, GenBank accession number MN985325.1) using bowtie2 (Langmead and Salzberg, 2012) and read coverage was visualized using ggplot2 (Wickham, 2016).
Virus infections
All experiments that involved live SARS-CoV-2 virus were carried out in a CDC/USDA-approved biosafety-level 3 facility at the Icahn School of Medicine at Mount Sinai (New York, USA) in accordance with institutional biosafety requirements. Virus stock was diluted in the appropriate growth medium for the specific cell type or SEAM culture with serum reduced to 2%. Cells were inoculated in a minimal volume for 1h at 37°C with agitation every 10 minutes to facilitate attachment. Inoculum was then removed, cells washed once in 1 x PBS and supplied with fresh appropriate culture medium with 2% serum. All infected samples were inactivated by a validated method before removal from the facility in accordance with institutional policies.
Plaque assay
Infectious particles were quantified by plaque assay on Vero E6 cells (ATCC #CRL-1586). 3.5x105 cells were plated on 12-well plates two days before infection. 10-fold serial dilutions were prepared of the virus containing samples diluted in DMEM (GIBCO) + 2% FBS in triplicate. Cells were inoculated with 200μl virus dilution for 1 hour at room temperature with agitation every 10 minutes. After 1 hour, the inoculum was aspirated and replaced with an overlay of MEM (GIBCO) with 4mM L-glutamine, 0.2% BSA, 10mM HEPES, 0.12% NaHCO3, 2% FBS and 0.7% Oxoid agar and incubated at 37°C for 48 hours. Cells were then fixed in 5% formaldehyde in PBS for 24 hours at room temperature. Plaques were stained with crystal violet solution (0.2% crystal violet w/v, 20% ethanol v/v). Supernatants were all frozen once at −80°C before quantification.
Quantification and statistical analysis
Statistical significance was determined by one-way ANOVA and the p-value was corrected for multiple comparisons using Dunnett’s test. Statistical analyses were performed with Prism 8 (Graphpad) software. For all data analysis of RNA sequencing, statistical significance was determined using the DEseq2 protocol apart from the quantification of viral reads in Figure 3B in which statistics were calculated using a one-way ANOVA. Statistical analysis of single cell RNA-seq data was performed using the Wilcox analysis in the Seurat package.
For all data, a p-value or an adjusted p-value < 0.05 was considered to be statistically significant. Details about statistical analyses for each experiment are outlined in the figure legends.
Acknowledgments
We would like to thank the patients and families who generously donated for the research conducted. We would like to thank the Eye Bank for Sight Restoration for their continual support and hard work at procuring donations. We would like to thank Dr. Barbara Corneo for manuscript review. This work was supported in part by a Challenge Grant award from Research to Prevent Blindness, the National Eye Institute (NEI), extramural grant 1R21EY030215-01, and the Icahn School of Medicine at Mount Sinai. We would like to thank the Carlsberg Foundation and the Lundbeck Foundation for supporting this work.
Author contributions
Conceptualization, T.A.B. and B.R.t.; methodology, B.M., A.Z.E., R.M., S.A.U., T.A.B., and B.R.t.; software, T.A.B. and B.R.t.; investigation, B.M., A.Z.E., and R.M.; resources, T.A.B.; writing, T.A.B., B.M., A.Z.E., R.M., and B.R.t.; visualization, B.M., A.Z.E., and R.M.; supervision, T.A.B. and B.R.t.; project administration, T.A.B. and B.R.t.; funding, T.A.B. and B.R.t.
Declaration of interests
The authors declare no competing interests.
Published: May 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2021.04.028.
Supplemental information
Differentially expressed genes in postmortem donor cells after infection with SARS-CoV2 at an MOI = 1 for 24h, identified by RNA-seq, divided by cell type: Limbus, cornea and Sclera.
Categorical and functional analysis of limbus genes using databases: BioPlanet2019, GO Biological Processes 2017, 2018, MGI Mammalian Phenotype, COVID-19 Regulated Gene Sets, Jensen Compartments and MSigDB Hallmark 2020. All databases accessed through Enrichr.
References
- Aiello F., Gallo Afflitto G., Mancino R., Li J.O., Cesareo M., Giannini C., Nucci C. Coronavirus disease 2019 (SARS-CoV-2) and colonization of ocular tissues and secretions: a systematic review. Eye. 2020;34:1206–1211. doi: 10.1038/s41433-020-0926-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bchetnia M., Girard C., Duchaine C., Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J. Infect. Public Health. 2020;13:1601–1610. doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O’Donoghue S.I., Schneider R., Jensen L.J. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014;2014:bau012. doi: 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappy P., Candotti D., Sauvage V., Lucas Q., Boizeau L., Gomez J., Enouf V., Chabli L., Pillonel J., Tiberghien P., et al. No evidence of SARS-CoV-2 transfusion transmission despite RNA detection in blood donors showing symptoms after donation. Blood. 2020;136:1888–1891. doi: 10.1182/blood.2020008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu M., Zhang Z., Qiao K., Huang T., Chen M., Xin N., Huang Z., Liu L., Zhang G., Wang J. Ocular manifestations of a hospitalised patient with confirmed 2019 novel coronavirus disease. Br. J. Ophthalmol. 2020;104:748–751. doi: 10.1136/bjophthalmol-2020-316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.S., Feldman K.E., Lee J., Verma S., Huang D.B., Huynh K., Chang M., Ponomarenko J.V., Sun S.C., Benedict C.A., et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal. 2011;4:ra11. doi: 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schunemann H.J., COVID-19 Systematic Urgent Review Group Effort (SURGE) Study Authors Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin J., Queen R., Zerti D., Dorgau B., Georgiou M., Djidrovski I., Hussain R., Coxhead J.M., Joseph A., Rooney P., et al. Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul. Surf. 2021;19:190–200. doi: 10.1016/j.jtos.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re’em Y., Redfield S., Austin J.P., Akrami A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. medRxiv. 2020 doi: 10.1101/2020.12.24.20248802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Liu J., Liu J., Yu P., et al. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in Rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.03.13.990036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamashima K., Gautam P., Lau K.A., Khiong C.W., Blenkinsop T.A., Li H., Loh Y.H. Potential modes of COVID-19 transmission from human eye revealed by single-cell atlas. bioRxiv. 2020 doi: 10.1101/2020.05.09.085613. [DOI] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R., Ishikawa Y., Sasamoto Y., Katori R., Nomura N., Ichikawa T., Araki S., Soma T., Kawasaki S., Sekiguchi K., et al. Co-ordinated ocular development from human iPS cells and recovery of corneal function. Nature. 2016;531:376–380. doi: 10.1038/nature17000. [DOI] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54:557–570.e5. doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Patel J., Patel B.C. StatPearls. 2020. Ophthalmic manifestations of coronavirus.https://www.ncbi.nlm.nih.gov/books/NBK556093/ [Google Scholar]
- Huang R., Grishagin I., Wang Y., Zhao T., Greene J., Obenauer J.C., Ngan D., Nguyen D.T., Guha R., Jadhav A., et al. The NCATS BioPlanet—an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharmacol. 2019;10:445. doi: 10.3389/fphar.2019.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K., USFQ-COVID19 Consortium SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T.H.T., Tang E.W.H., Chau S.K.Y., Fung K.S.C., Li K.K.W. Stepping up infection control measures in ophthalmology during the novel coronavirus outbreak: an experience from Hong Kong. Graefes Arch. Clin. Exp. Ophthalmol. 2020;258:1049–1055. doi: 10.1007/s00417-020-04641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chan J.F., Li K.K., Tso E.Y., Yip C.C., Sridhar S., Chung T.W., Chiu K.H., Hung D.L., Wu A.K., et al. Detection of SARS-CoV-2 in conjunctival secretions from patients without ocular symptoms. Infection. 2020 doi: 10.1007/s15010-020-01524-2. Published online September 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. Fields Virol. 2013;1:825–858. [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencucci R., Favuzza E., Becatti M., Tani A., Mazzantini C., Vignapiano R., Fiorillo C., Pellegrini-Giampietro D., Manetti M., Marini M., Landucci E. Co-expression of the SARS-CoV-2 entry receptors ACE2 and TMPRSS2 in healthy human conjunctiva. Exp. Eye Res. 2021;205:108527. doi: 10.1016/j.exer.2021.108527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U S A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S., Vargas J., Hoffmann A. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8:227–241. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungmungpuntipantip R., Wiwanitkit V. Ocular manifestation, eye protection, and COVID-19. Graefes Arch. Clin. Exp. Ophthalmol. 2020;258:1339. doi: 10.1007/s00417-020-04662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. Human Cell Atlas Meeting Participants The Human Cell Atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Khanra I., Wang Z., Merugu S.B., Yunus F.U., Mashausi D.S., Li D. Emerging novel coronavirus is a global threat: insight in the biology of COVID-19 and its hijacking process of hosts’ cell. Curr. Pharm. Des. 2020 doi: 10.2174/1381612826666200909141725. Published online September 9, 2020. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Elicieri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S.J., Rodriguez-Bravo V., Quinn S.A., Rodriguez-Barrueco R., Lujambio A., Williams E., Sun X., de la Iglesia-Vicente J., Lee A., Readhead B., et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27:223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Duan F., Luo C., Liu Q., Qu X., Liang L., Wu K. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020;138:575–578. doi: 10.1001/jamaophthalmol.2020.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020;92:589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen X., Chen L., Deng C., Zou X., Liu W., Yu H., Chen B., Sun X. The evidence of SARS-CoV-2 infection on ocular surface. Ocul. Surf. 2020;18:360–362. doi: 10.1016/j.jtos.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zeng Y., Tong Y., Chen C. Ophthalmologic evidence against the interpersonal transmission of 2019 novel coronavirus through conjunctiva. medRxiv. 2020 doi: 10.1101/2020.02.11.20021956. [DOI] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. HCA Lung Biological Network. Electronic address. lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes in postmortem donor cells after infection with SARS-CoV2 at an MOI = 1 for 24h, identified by RNA-seq, divided by cell type: Limbus, cornea and Sclera.
Categorical and functional analysis of limbus genes using databases: BioPlanet2019, GO Biological Processes 2017, 2018, MGI Mammalian Phenotype, COVID-19 Regulated Gene Sets, Jensen Compartments and MSigDB Hallmark 2020. All databases accessed through Enrichr.
Data Availability Statement
The datasets generated during this study are available at NCBI GEO: GSE165477, GSE164073.