Abstract
Insects are among the most diverse and successful groups of animals and exhibit great morphological diversity and complexity. The innovation of wings and metamorphosis are some examples of the fascinating biological evolution of insects. Most microRNAs (miRNAs) contribute to canalization by conferring robustness to gene networks and thus increase the heritability of important phenotypes. Though previous studies have demonstrated how miRNAs regulate important phenotypes, little is still known about miRNA evolution in insects. Here, we used both small RNA-seq data and homology searching methods to annotate the miRNA repertoires of 152 arthropod species, including 135 insects and 17 noninsect arthropods. We identified 16,212 miRNA genes, and classified them into highly conserved (62), insect-conserved (90), and lineage-specific (354) miRNA families. The phylogenetic relationship of miRNA binary presence/absence dynamics implies that homoplastic loss of conserved miRNA families tends to occur in far-related morphologically simplified taxa, including scale insects (Coccoidea) and twisted-wing insects (Strepsiptera), leading to inconsistent phylogenetic tree reconstruction. The common ancestor of Insecta shares 62 conserved miRNA families, of which five were rapidly gained in the early winged-insects (Pterygota). We also detected extensive miRNA losses in Paraneoptera that are correlated with morphological reduction, and miRNA gains in early Endopterygota around the time holometabolous metamorphosis appeared. This was followed by abundant miRNA gains in Hymenoptera and Lepidoptera. In summary, we provide a comprehensive data set and a detailed evolutionary analysis of miRNAs in insects. These data will be important for future studies on miRNA functions associated with insect morphological innovation and trait biodiversity.
Keywords: miRNA, annotation, evolution, pterygota, endopterygota, gain and loss
Introduction
Insects are among the most diverse clades of eukaryotes, in terms of morphology, biomass, species numbers, and ecological niches (Grimaldi and Engel 2005; Mayhew 2007). After diverging from their crustacean ancestors, insects went through at least two morphological innovations. The emergence of wings was the first of these innovations and occurred during the early evolution of Pterygota. This innovation enabled insects to conquer the sky and improved their ability to disperse and exploit new environments. The second innovation was the emergence of holometabolan metamorphosis in Endopterygota (Truman and Riddiford 1999). This allowed insects to form an adult body phenotype in the pupa stage. The separation of adults and larvae reduced the competition for food and habitat between the two stages. Both of these morphological innovations significantly contributed to the success of insects, since modern Endopterygota insects comprise >60% of all described metazoan species (Kristensen 1999; McKenna et al. 2019; Truman 2019).
MicroRNAs (miRNAs) are approximately 22 nucleotide (nt) small RNAs that posttranscriptionally repress messenger RNA (mRNA) targets (Bartel 2018). These very small noncoding RNAs play pivotal roles in development, apoptosis, cell differentiation, reproduction, behaviors and physiology in distinct groups of eukaryotes, including plants and animals (Ambros 2004; Lucas and Raikhel 2013; Shenoy and Blelloch 2014; Moran et al. 2017). During evolution, when new miRNAs are integrated into gene regulatory networks, they stabilize the expression of protein-coding genes (PCGs) (Berezikov 2011). This canalization process allows more genes and their respective traits to be susceptible to the action of natural selection, and thus may drive the morphological complexity of animals (Peterson et al. 2009; Wu et al. 2009). However, the manner in which miRNA evolution influenced the emergence of novel phenotypes in insects remains largely unknown.
The process of loss or gain of miRNAs is concomitant with morphological evolution in animals (Sempere et al. 2006; Prochnik et al. 2007; Heimberg et al. 2008; Dai et al. 2009; Wheeler et al. 2009; Hertel and Stadler 2015; Deline et al. 2018). In vertebrates, miRNA families have undergone at least two rounds of expansions. The first expansion occurred at the basal vertebrate clade. The second occurred at the stem of Eutherian mammals (Heimberg et al. 2008). Both expansions were concordant with an increase in vertebrate morphological complexity. In contrast, the extensive miRNA losses in parasitic flatworms indicate that miRNA loss might be related to morphological simplification or parasitic lifestyle (Fromm et al. 2013; Bai et al. 2014). Amounting evidence has indicated that miRNAs are involved in the regulation of insect metamorphosis and wing development (Biryukova et al. 2009; Gomez-Orte and Belles 2009; Bejarano et al. 2010; Lozano et al. 2015; Belles 2017). Within the species-rich and well-studied phylogeny of Lepidoptera, a burst of miRNA innovation occurred early in the clade (Quah et al. 2015). Moreover, a phylogenetic study on insect miRNAs showed no significant correlation between miRNA diversity and innovative holometabolan metamorphosis (Ylla et al. 2016).
The recent availability of insect genomes over the past years makes it possible to obtain a relatively complete miRNA annotation in a high number of insect species (Dannemann et al. 2012; Fromm et al. 2015). Here, we assembled the largest curated data set of miRNAomes, comprising 16,212 miRNA genes sampled from 152 arthropod species. We used these data to build an insect phylogeny with both miRNA and PCGs, and to reconstruct the evolution of miRNAs in insects.
Results
MiRNA Annotation and Family Analysis in Insects
To reflect the evolution of different insect clades, we analyzed 152 arthropod species, including 134 insects and 17 noninsect arthropods (supplementary data set S1, Supplementary Material online). The miRNA annotation of Drosophila melanogaster was downloaded from MirGeneDB (Fromm et al. 2020). For the other 151 species, we annotated 36 species with public available small RNA-seq data (supplementary data set S2, Supplementary Material online) using both evidence-based (miRDeep2) and homology searching-based methods (MapMi and BLAST). For the remaining species, for which only the genome data were available, we annotated using the homology searching-based method (see Methods). We annotated a repertoire of 16,212 miRNA genes from the 152 arthropod species (supplementary table S1 and data set S3, Supplementary Material online). The lack of substantial small RNA-seq data limited our approach and we were only able to identify homologous miRNAs in the 115 species. Although this slightly impacts analysis on miRNA gains, this is still the most comprehensive data set presently available, and sufficiently reliable for large scale evolution analysis, especially the loss of conserved miRNAs.
In this study, we defined miRNA family as a true evolutionary homologous group of miRNA descendants from one common ancestor. Generally, one seed family represents one true family if its members share highly conserved mature (De Wit et al. 2009; Wheeler et al. 2009; Bartel 2018). To assign miRNAs into families, we first grouped miRNAs with identical seed regions (nucleotides 2–8) into “seed families.” Nevertheless, a small portion of “seed families,” for example, MIR-989 and MIR-BgeN1, their mature sequences are highly divergent. If these miRNAs with homologous flanking regions, we still assigned them into the same family. The miRNAs without such evidence were divided into different families in case the mature identity was <0.7 or if they were not locate in a monophyletic gene group (contradictory to insect phylogeny). For the miRNAs belonging to the same family but having different names in miRBase or MirGeneDB, we used the earliest named ID as the family name (supplementary table S2, Supplementary Material online). For example, miR-956 in Diptera and miR-3850 in Tribolium share the same seed sequence and homologous flanking region and were thus grouped into MIR-956 (supplementary fig. S1, Supplementary Material online). Even though some miRNA families shared evidence of homology, they nevertheless contained different seeds (e.g., MIR-10 family) and were regarded as different miRNA families. In total, we obtained 513 miRNA families, including 263 newly described families (supplementary table S3, Supplementary Material online).
We divided the insect miRNA families into four categories based on sequence conservation. The families presenting only in one species or genus were assigned as lineage-specific miRNAs (354). Those conserved in both insects and other arthropods were assigned as highly conserved miRNAs (62). The families conserved only in insects or crustaceans were assigned to insect-conserved miRNAs (90) and crustacean-conserved miRNAs (7), respectively. Because lineage-specific miRNAs can only be identified by the expression-evidence method, the number of these miRNAs mainly depends on the quality of the small-RNA libraries (Tarver et al. 2018). Hence, we focused on conserved miRNAs for the phylogenetic and gain/loss analyses.
Association between MiRNA and Genome Completeness
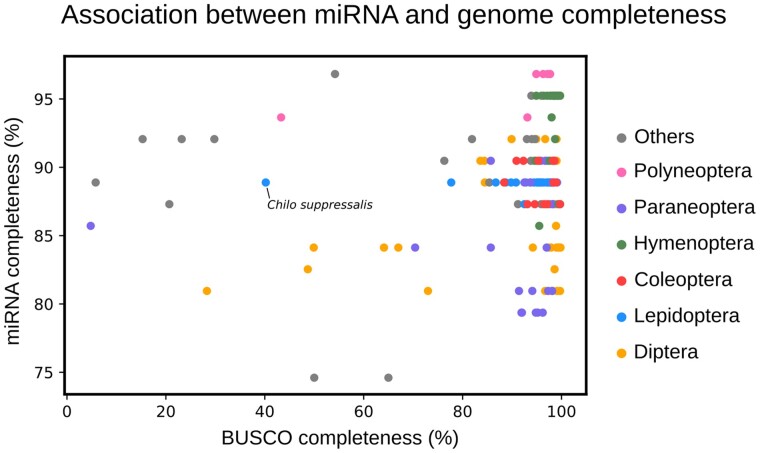
Before using miRNAs for phylogenetic analysis, we first tested whether genome completeness affects the integrity of the miRNA repository. Here, we defined miRNA completeness as the proportion of highly conserved miRNAs that are present in each species. We compared this number with the genome completeness as measured by the Benchmarking Universal Single-Copy Orthologs (BUSCO). The results showed no correlation exists between miRNA and genome completeness (Pearson's r = 0.152) (fig. 1 and supplementary data set S4, Supplementary Material online). Instead, despite the considerable variation in genome completeness for clades like Lepidoptera, we observed no such differences in the miRNA completeness of different species. For example, the genome of Chilo suppressalis had a low BUSCO completeness (40.2%) but showed a high miRNA completeness (88.8%), similar to most Lepidoptera species with a more complete genome assembly, such as Bombyx mori. This suggests that our data are unbiased and suitable for evolutionary analysis.
Fig. 1.
Association between miRNA and genome completeness. The data suggest no correlation exists between miRNA and genome completeness (Pearson's r = 0.152).
Discrepancy between Insect Phylogenetic Trees Constructed by PCGs and MiRNAs
To study the evolution of miRNA gain and loss, we first reconstructed a phylogenetic tree of 152 arthropods based on single-copy PCGs using a maximum likelihood (ML) method. The PCG phylogenetic tree was, in general, concordant with that obtained by Misof et al. (2014) (fig. 2 and supplementary data set S5 and S6, Supplementary Material online). Interestingly, the Phthiraptera and Thysanoptera clades form a sister group with Hemiptera rather than Endopterygota, and the Diplura taxa is closer to Collembola than Insecta.
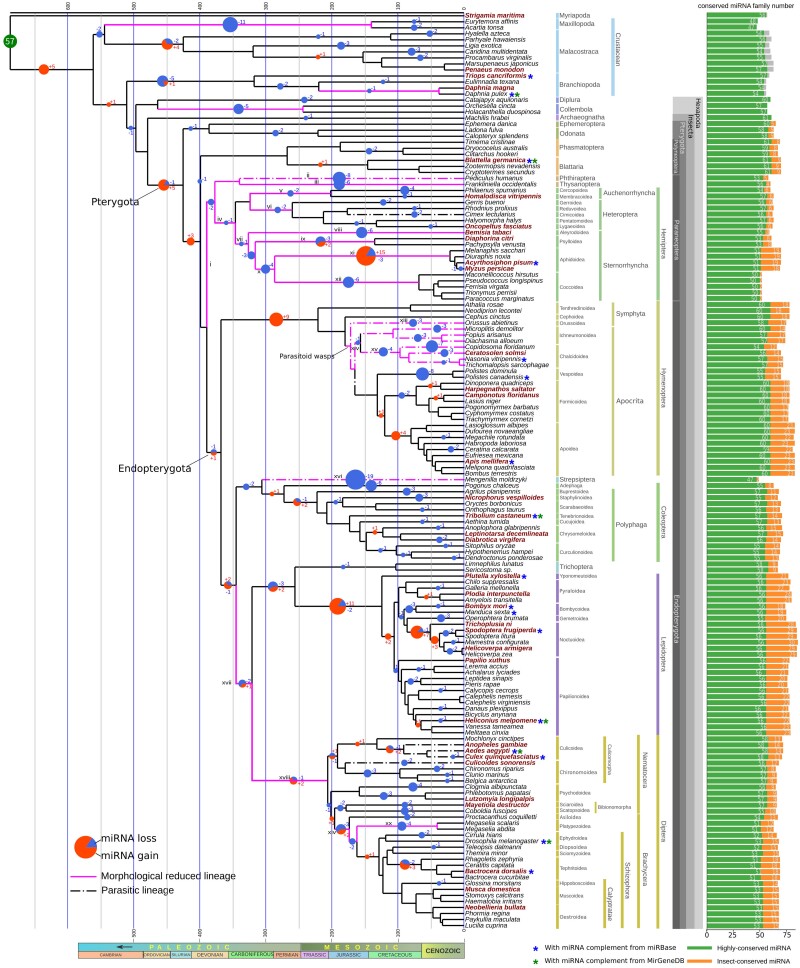
Fig. 2.
The evolution of miRNA families in insects. We constructed a phylogenomic tree representing the phylogeny of all 152 arthropod species. Gains and losses of miRNA families were reconstructed on the tree using the Dollo parsimony model. The size of the pie chart on the branches represents the total number of events at the corresponding branch. The red fraction denotes miRNA gain events and the blue denotes loss events. The species annotated with both publicly available small RNA-seq data and homologous searching are marked in bold and dark red. Lineages with morphology reduction are marked in pink. The important reduced characters with miRNA loss are listed in supplementary table S5, Supplementary Material online and identified with Roman numerals. Lineages leading to parasitic lifestyle are marked with a dotted line. The bars on the right denote the number of conserved miRNA families in each species.
The miRNA binary data have been widely used to construct the phylogeny of Diptera, Tardigrada and basal Hexapoda (Campbell et al. 2011; Wiegmann et al. 2011; Liu et al. 2020). Hence, we conducted a phylogenetic analysis using presence/absence binary data for 159 conserved miRNAs across 74 insect species under the stochastic Dollo model (supplementary data set S7, Supplementary Material online). The miRNA tree topology is generally consistent with the PCG phylogeny (fig. 3A). Nevertheless, in miRNA tree, the extremely morphologically reduced endoparasite Strepsiptera, belonging to the Endopterygota group and considered a sister group to Coleoptera, was grouped with a high confidence (0.94) inside Hemiptera (Paraneoptera) and as a sister group to Coccoidea and Aphidoidea. Moreover, the miRNA tree failed to support the monophyly of Paraneoptera.
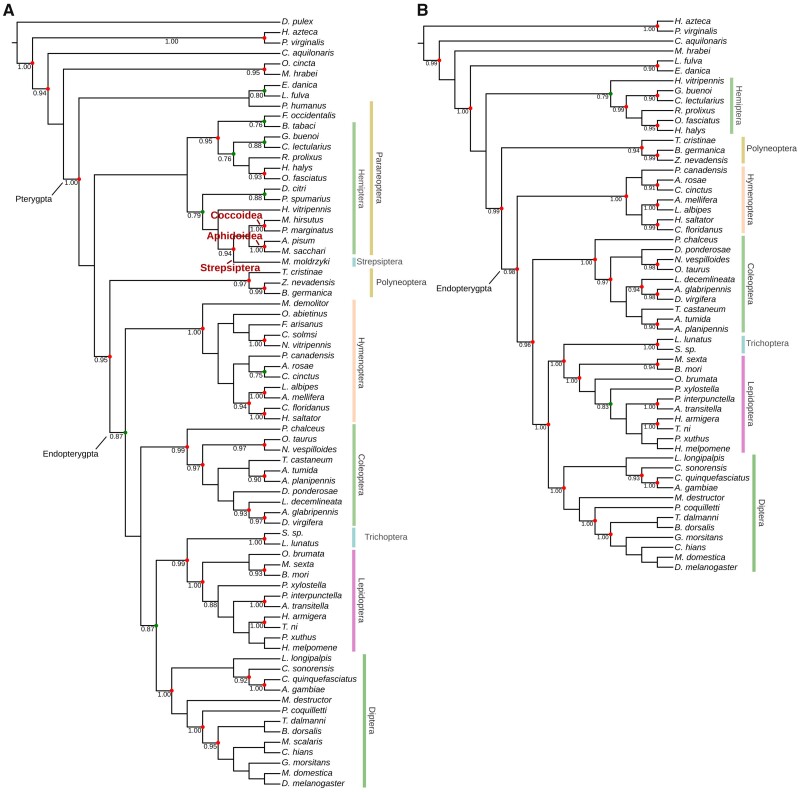
Fig. 3.
Phylogenetic tree constructed using the binary data of presence/absence of conserved miRNAs. (A) miRNA phylogenetic tree of 74 arthropod species using the Bayes stochastic Dollo model. The red dots denote branches supported by a posterior value >90%, and the green dots represent posterior values between 75% and 90%. (B) Modified miRNA phylogenetic tree constructed after removing morphologically reduced taxa and parasitoid wasps.
The miRNA tree also failed to support internal relationships in Hymenoptera, especially for parasitoid wasps. However, after removing the clades containing morphologically simplified and parasitoid taxa, we were able to reconstruct a modified miRNA tree that supports the known relationships between the four major orders in Endopterygota (fig. 3B). Interestingly, Endopterygota in both the miRNA trees display a closer relationship to Polyneoptera than Paraneoptera. This may be caused by extensive miRNA losses in Paraneoptera.
Rapid Gain of MiRNAs in the Early Evolution of Winged Insects
To elucidate miRNA gain and loss during insect evolution, we used the PCG tree as a phylogenetic background (fig. 2). We found 57 miRNA families that are conserved since the common ancestor of Myriapoda and Pancrustacea. At the basal branches in Pancrustacea, the miRNA family number increased to 62 in basal Hexapoda. However, no miRNA gain or loss events were observed in the branch leading from Hexapoda to Insecta. Taken all these factors into consideration, we suggested that the common ancestor of insects shared 62 conserved miRNA families (supplementary table S4, Supplementary Material online).
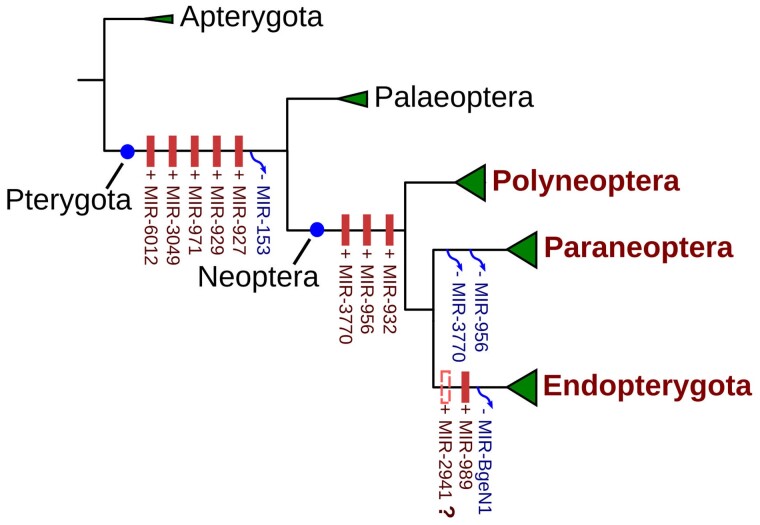
A rapid gain of miRNAs occurred at the appearance of winged insects (Pterygota) in the Early Devonian (∼412 Ma). A total of five miRNA families were gained in this period (MIR-927, MIR-929, MIR-971, MIR-3049, and MIR-6012), at the branch leading from Apterygota to Pterygota (fig. 4). These miRNAs are conserved in both Neoptera and Paleoptera. MIR-927, MIR-929, and MIR-971 are extremely conserved across all insects, whereas MIR-3049 and MIR-6012 show a secondary loss in Panorpida (Lepidoptera and Diptera). MIR-153, another highly conserved miRNA, was lost in Pterygota but is highly conserved across metazoan animals (supplementary fig. S2, Supplementary Material online).
Fig. 4.
MiRNA evolution in Exopterygota.
To gain insights into the putative functions of the miRNAs gained in Pterygota, we searched for their target genes a in seven insect species from Exopterygota to Endopterygota (supplementary data set S8, Supplementary Material online). Gene Ontology analysis showed these target genes were enriched in biological regulation (GO: 0065007), the regulation of biological process (GO: 0050789), response to stimulus (GO: 0050896), localization (GO: 0051179), signaling (GO: 0023052), locomotion (GO: 0040011), and growth (GO: 0040011) in all seven species. In contrast, KEGG pathway analysis showed an enrichment in the MAPK and FoxO signaling pathways (supplementary fig. S3, Supplementary Material online). The MAPK signaling pathway is crucial for insect wing development (Marenda 2006), whereas the FoxO signaling pathway plays a crucial role in stress response and the control of insect behaviors, including dormancy and diapause (Sim and Denlinger 2008; Mattila et al. 2009; Sim et al. 2015). These results imply that the miRNAs originating in Pterygota may contribute to wing formation and stress responses that enabled winged insects to explore new terrestrial ecosystems during their early evolution.
We found that three miRNA families (MIR-932, MIR-956, and MIR-3770) were gained at the branch leading from Paleoptera to Neoptera (fig. 4 and supplementary table S4, Supplementary Material online). MIR-932 is extremely conserved across insect lineages but the other two miRNAs showed secondary losses in some insect lineages. MIR-956 was lost in Paraneoptera and Lepidoptera, whereas MIR-3770 can only be found in Polyneoptera and Hymenoptera insects. In contrast, the branch leading to Polyneoptera and Paraneoptera showed a reduction of miRNA acquisition, although each group contains species with small RNA-seq data of high quality. Only one miRNA (MIR-BgeN4) was found in Polyneoptera and was gained at the branch leading to Blattaria.
Extensive MiRNA Losses in Paraneopteran with Apparent Trait Loss
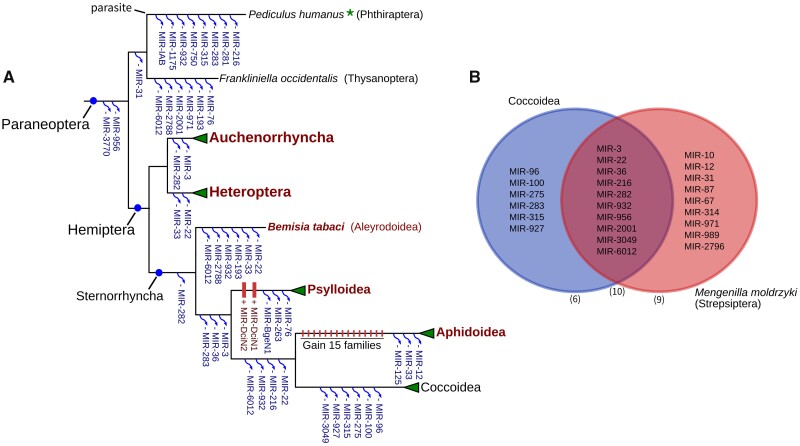
The common ancestor of Paraneoptera shared a comprehensive set of miRNAs with 67 conserved families. Only one miRNA was lost at the stem of Paraneoptera, and the common ancestor of Sternorrhyncha contains 65 conserved families (supplementary table S4, Supplementary Material online). However, extensive miRNA losses occurred in various Paraneopteran clades (fig. 5A), including eight families in Phthiraptera and six in Thysanoptera. Most major Sternorrhynchan clades included in our analysis are supported by small RNA-seq data, suggesting these losses were not result from the limitation in homologous searching.
Fig. 5.
Extensive miRNA loss during Paraneoptera evolution. (A) MiRNA loss events in Paraneoptera Clades, the species whose miRNA annotation is supported by small RNA-seq reads are marked in bold red. (B) Comparison of miRNA family losses in Coccoidea and Mengenilla moldrzyki (Sterpsiptera).
In Hemiptera, only two miRNAs were lost, in Auchenorrhyncha (leafhoppers and froghoppers) and Heteroptera (bugs), which are among the least specialized Hemiptera groups. Sternorrhyncha show more severe miRNA losses, with a total of 26 independent losses along each major branch (fig. 5A and supplementary table S5, Supplementary Material online).
Among the different Sternorrhyncha clades, Coccoidea underwent several episodes of miRNA loss throughout the evolution of Sternorrhyncha, making it one of the groups with the least miRNA families in Paraneoptera (52 families). All these miRNAs are absent in five species in Pseudococcidae, suggesting these loss events occurred before the radiation of this clade. Such massive loss can also be found in Strepsiptera in Endopterygota. Since only one species was analyzed in Strepsiptera, it is difficult to determine the origin of these loss events to a specific period. A total of ten miRNAs were lost in the same family in both Coccoidea and Strepsiptera (fig. 5B), which explained the erroneous placement of these two clades in our miRNA tree. Since both Coccoidea and Strepsiptera share a similar trend of morphological simplification (Beutel et al. 2014), we propose a connection with homoplastic miRNA loss.
We then explored the function of lost miRNAs by searching for their target genes in the red flour beetle Tribolium castaneum (Strepsiptera) and the true bug Halyomorpha halys (Coccoidea). We found the targets of these lost miRNAs are enriched in the Wnt and MAPK signaling pathways and dorso-ventral axis formation in both species (supplementary data set S9, Supplementary Material online). This observation corroborates possible miRNA implication in morphogenesis.
Interestingly, the Psylloidea and Aphidoidea clades gained novel miRNAs, an observation that was supported by small RNA-seq data. In Aphidoidea, 14 of these newly gained miRNAs are conserved across the four Aphid species. Importantly, these new miRNAs may be connected with specific phenotypical changes in these groups, including polymorphism.
Although we found evidence for parallel loss of some miRNA families in distantly related lineages, the majority of miRNA losses in Paraneoptera occurred independently in different lineages. For example, MIR-33 was independently lost in Heteroptera, Aleyrodoide and Aphidoidea. These results suggest homoplastic loss often occurs in parallel across morphologically reduced lineages.
No Apparent MiRNA Innovations in the Appearance of Holometabolous Endopterygota
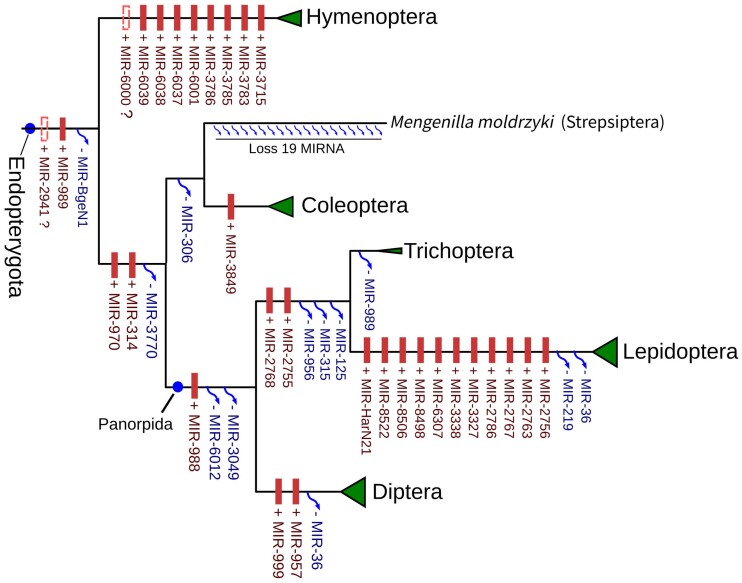
The development of complete metamorphosis is a process that emerged during the evolution of Endopterygota and which contributes significantly to the biodiversity of insect life (Truman and Riddiford 1999; Mayhew 2007). The important changes in development patterns and the emergence of holometabolan metamorphosis were accompanied by only up to 2 miRNA gains at the transition from Exopterygota to Endopterygota (fig. 6).
Fig. 6.
Gain and loss of miRNA families in major Endopterygota lineages.
Ylla et al. (2016) previously suggested that two additional miRNAs, MIR-1006 and MIR-1007, were gained during the transition from Exopterygota to Endopterygota, but they analyzed only four Endopterygota species. We revisited the history of these two miRNAs by including more taxa and suggest they do not have an Endopterygota origin. While Ylla et al. claimed Mir-1006 was present in Apismellifera and D. melanogaster and was thus of Endopterygota origin, we could not find any Mir-1006 homologue in other Hymenopteran insects. The structure of the A. mellifera Mir-1006 precursor in miRBase only forms a hairpin-like structure with a minimum free energy (MFE) of –7.10 kcal/mol, and the extension of this precursor leads to the loss of the hairpin-like structure (supplementary fig. S4A, Supplementary Material online). Moreover, no reads matched to the A. mellifera Mir-1006 when using both the miRBase records and the small RNA-seq data. The only work including Mir-1006 in the Apis genus was reported by Chen et al. (2010), but that study was based on a small amount of reads mapping to the 3′ arm, and had no reads from 5′ arm. These observations, together with our analysis suggest Mir-1006 was spuriously annotated in A. mellifera.
Mir-1007 is another interesting case, and its seeds are identical in Drosophila and Hymenoptera, with nearly all pairwise identities between respective miRNAs above 0.7 (our threshold for miRNA family assignment) (supplementary fig. S4C, Supplementary Material online). However, the phylogenetic assignment of these two miRNAs is complex. We were only able to find MIR-1007 in Drosophila, and the miRNA is missing in any other Ditpera, Amphiesmenoptera and Coleoptera species. Hence, in case MIR-1007 is indeed homologous to MIR-6037, at least 8 independent miRNA losses occurred (supplementary fig. S4B, Supplementary Material online). This number seems unreasonably high. Moreover, the precursor of these two miRNAs showed great disparity (supplementary fig. S4D, Supplementary Material online), whereby it is likely they arose from two independent events and were thus assigned to different families. MIR-1007 and MIR-6037 provide a good example of homoplasy in miRNAs.
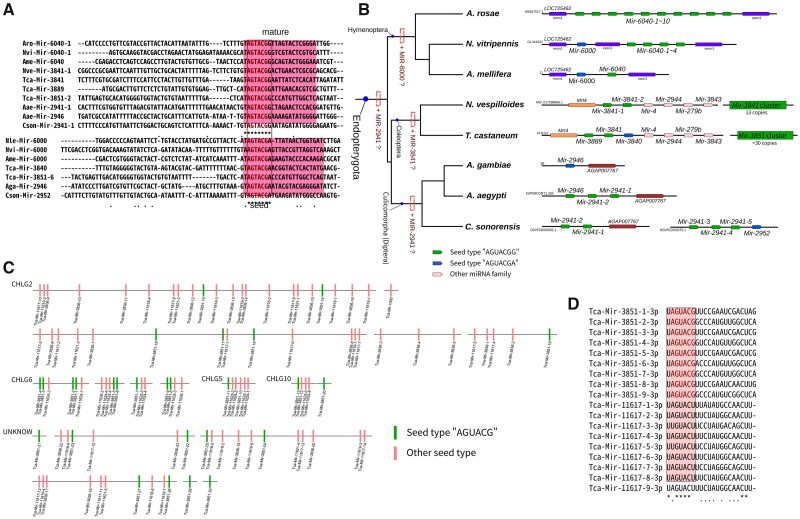
There are another three enigmatic miRNAs that may have originated in the common ancestor of Endopterygota, namely MIR-2941 in mosquito, MIR-3841 in Coleoptera and MIR-6000 in Hymenoptera. These three miRNAs share the same seeds but are otherwise highly divergent (fig. 7A). In miRBase, these miRNAs were assigned into three different families. In MirGeneDB, MIR-6000 was not included, whereas MIR-2941 and MIR-3841 belong to different families. These three miRNAs share no homologous synteny (fig. 7B) but were featured as tandem clusters in many species. In beetles Nicrophorusvespilloides and sawflies Andrenarosae, MIR-3841 and MIR-6000 both formed tandem duplication clusters with more than ten units. In Tribolium, miR-3841 share the same seeds as a variety of other members of MIR-3851 (fig. 7C and D). As a specific fast-evolving miRNA, MIR-3851 forms large tandem duplication clusters in the Tribolium genome (Ninova et al. 2016), implying that these clustered miRNAs probably belong to a fast evolving miRNA family in Endopterygota.
Fig. 7.
MIR-6000, MIR-3841, and MIR-2941 in Endopterygota insects (A) Sequence alignments of MIR-6000, MIR-3841, and MIR-2941. The two seed types (AGUACGG and AGUACGA) were aligned separately. (B) Flanking genomic regions of MIR-6000, MIR-3841, and MIR-2941 in different Endopterygota lineages. (C) MIR-3851 clusters in Tribolium castaneum. Green stripes denote the members sharing the same seed sequence with MIR-2941. (D) Alignment of mature sequences of MIR-3851 members in T. castaneum.
Eip93F is a Target of MIR-989 in Endopterygota
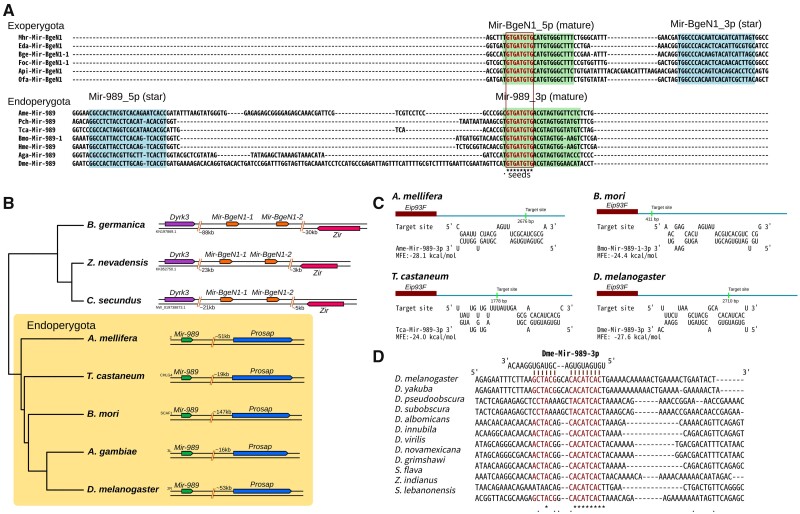
Specifically, MIR-989 was gained at the branch leading from Exopterygota to Endopterygota, together with the loss of MIR-BgeN1 (which corresponds to BGE-NOVEL-1 in the MirGeneDB and to miR-bg5 in a previous analysis [Ylla et al. 2016]). Even though is derives from a different arm, the mature MIR-BgeN1 (5′-arm in Exopterygota) and MIR-989 (3′-arm in Endopterygota) share an identical seed (fig. 8A). This opens the possibility that MIR-989 in Endopterygota originated from a hairpin-shifting event of MIR-BgeN1 in Exopterygota. Instead, it might have arisen independently as a result of convergent evolution. We searched the flanking region of both miRNAs and found they share no homologous synteny. In Endopterygota, MIR-989 is located upstream of the gene Prosap, whereas in Exopterygota no deep conserved synteny relationship exists. However, in cockroach and termites (Blattaria), MIR-BgeN1 is linked with the genes Dyrk3 and Zir (fig. 8B). Hence, MIR-989 more likely arose independently due to convergent evolution in Endopterygota, whereby we placed the two miRNAs in two independent families.
Fig. 8.
MIR-989 in Endopterygota insects. (A) Precursor sequence alignments of MIR-BgeN1 in Exopterygota and MIR-989 in Endopterygota. (B) Flanking genomic regions of MIR-BgeN1 in Blattaria and MIR-989 in Endopterygota. (C) Target sites of MIR-989 on the 3′-UTR of Eip93F in four Endopterygota insects. (D) The MIR-989 target site on Eip93F in Drosophila was conserved at both the seed and 3′-complimentary regions.
To further investigate the role of the MIR-989 family in insects, we implemented a search for its targets among four Endopterygota insects (A. mellifera, T. castaneum, B. mori, D. melanogaster), and performed a similar approach for the MIR-BgeN1 family in three Exopterygota species (Acyrthosiphonpisum, H. halys, and Zootermopsisnevadensis) (supplementary data sets S10 and S11, Supplementary Material online). We found that the transcription factor Eip93F (E93), which is related to metamorphosis, is targeted by MIR-989 in all four Endopterygota insects (fig. 8C). The inclusion of more Endopterygota taxa allowed to unveil this relationship between MIR-989 and Eip93F and confirms it is widely distributed in Endopterygota (supplementary data set S12, Supplementary Material online). Moreover, the target site was conserved at both the seed and the 3′-complementary regions across in Drosophila and Apis (fig. 8D and supplementary fig. S5, Supplementary Material online). Nevertheless, we failed to find any target-site relationship between MIR-BgeN1 and Eip93F in the three analyzed Exopterygota species. This implies that the relationship between Eip93F and miRNAs is unique to Endopterygota, and perhaps explains the origin of this group. In both Exopterygota and Endopterygota, Eip93F plays key roles in promoting adult metamorphosis and is essential for the transition between nymphal (pupal) and adult states (Mou et al. 2012; Urena et al. 2014). Hence, the origin of MIR-989 with its target Eip93F in Endopterygota might have played a very important role in the development of metamorphosis in this clade.
MiRNA Explosion and Collapse within Endopterygota
Despite a very limited number of miRNA gains during early Endopterygota evolution, we estimated a total of 27 gain events occurred at the major branches after Endopterygota diversification (fig. 6 and supplementary table S4, Supplementary Material online). A total of nine families were gained on the branch leading to Hymenoptera, making this clade second in terms of highest number of miRNA gains in Endopterygota insects. These miRNAs are shared by sawfly A. rosae and other Hymenoptera insects but do not exist in other species. In Hymenoptera, parallel miRNA losses occurred across the clades of parasitoid wasps. As observed in Sternorrhyncha, the ancestor of parasitoid wasps also retained a relatively complete miRNA complement and several loss events only occurred in parallel after the radiation of parasitoid wasps (supplementary fig. S6A, Supplementary Material online). For example, two subfamilies of Braconidae, namely Microgatrinae and Opiinae, lost three miRNAs in different miRNA families, indicating these loss events happened very recently during their evolution.
A further two miRNAs, MIR-314 and MIR-970, were obtained in the common ancestors of Coleopterodae and Panorpida, after the split from Hymenoptera. Strepsiptera showed an extensive loss of 19 miRNA families (supplementary table S4, Supplementary Material online), one of which (MIR-3849) was gained in Coleoptera and is shared by both Pogonus chalceus (Adephaga) and Polyphaga. Moreover, miR-988 first appeared in the common ancestor of Panorpida and is shared by Diptera, Trichoptera, and Lepidoptera. As for MIR-2755 and MIR-2768, both are shared by Lepidoptera and Trichoptera and were gained at the root of Amphiesmenoptera. We also observed a significant miRNA burst in Lepidoptera, with a total of 11 families gaines. This is in agreement with a previous study (Quah et al. 2015) and makes Ditrysia Lepidoptera the clade with the highest miRNA gains in Endopterygota.
In Diptera, the miRNA families MIR-999 and MIR-957 are shared by both mosquitos and flies and first emerged at the root of this group. We also observed a series of miRNA gains and losses from basal Diptera to crown Schizophora (supplementary fig. S6B, Supplementary Material online), which provides support to the paraphyly of the old taxon Nematocera. The MIR-994 family is shared by Mayetiola destruotor (Bibionomorpha) and Brachycera, suggesting a closer relationship between these groups. Furthermore, MIR-284 and MIR-958 are shared by Proctacanthus coquilletti (Asiloidea) and upper Brachycera and were gained at the root of Brachycera. This branch also witnessed the loss of three conserved miRNAs, which was followed by another two loss events at the stem of Cyclorrhapha. The gain of MIR-987 occurred after the split of Platypezoidea at during basal Schizophora evolution, and this phylogenetic relationship is consistent with a previous analysis of Diptera miRNA (Wiegmann et al. 2011).
Discussion
Multicellular animals developed a complex system of genomic innovations, including the expansion of key signaling pathways, transcription factors and regulatory DNA and RNA classes (Sogabe et al. 2019). High degrees of noncoding DNA elements and a large number of expressed noncoding RNAs are believed to confer organisms the ability to coordinate the biological complexity of morphology (Taft and Mattick 2003). However, the miRNA repertories of a limited number of insect taxa have so far failed to provide convincing evidence to explain the evolution of miRNAs in detail. In this study, we extend the miRNA annotation to 152 arthropod species and thus provide the most comprehensive data set of miRNA complements to date. This allowed us to reconstruct the evolutionary relationship between miRNAs in different insect species. The completeness of the miRNA annotation shows no correlation to genome completeness, suggesting our pipeline works equally well even in the cases of low genome quality. By including trace data for searching, short-length miRNAs are easier to retrieve in fragmented genomes compared with PCGs. Notably, only homologous miRNAs could be found in the 115 species lacking small RNA library data. Hence, we needed to be more stringent in analyzing the gain of novel miRNAs in some lineages because we might unintentionally exclude some newly evolved miRNAs due to the lack of small RNA library data (Guerra-Assunção and Enright 2012; Tarver et al. 2018). Despite this, we present the first large-scale analysis of miRNA evolution across more than one hundred insect species, and our data provide the most reliable analysis of miRNA gain/loss in insects at present.
Reconsidering the Use of MiRNAs in Phylogenetic Analysis
The use of miRNAs as phylogenetic markers to infer species relationships was successfully implemented in many eukaryotes, including metazoan (Tarver et al. 2013), Tardigrada (Campbell et al. 2011), arthropods (Rota-Stabelli et al. 2011), and basal Hexapoda (Liu et al. 2020). Here, we compared insect phylogenetic trees using two independent data sets comprising 1) PCGs or 2) miRNAs. In comparison to the PCG tree, the miRNA tree performed poorly at the nodes of Paraneoptera and parasitic clades and erroneously placed Strepsiptera into Sternorrhyncha with a high support (fig. 2). This discordance provides a good example that homoplastic miRNA loss can occur due to convergent morphological simplification in distantly related lineages. Therefore, the phylogenetic analysis using miRNAs needs to be considered carefully, especially when species of morphological reduction or parasitic taxa are included.
The richness of the selected taxa is another important aspect to achieve a confident phylogenetic analysis with miRNA data. Ylla et al. (2016) only used seven insect species, and the inclusion of a single spurious or miss annotation (Mir-1006) led to incorrect conclusions. This risk can be mitigated by the inclusion of more closely related species and comparisons between annotation results.
Another important aspect is how to properly define miRNA families. To date, most studies (including ours) adopted seed or sequence similarity to define miRNA families (Guerra-Assunção and Enright 2010; Meunier et al. 2013; Ylla et al. 2016). A gene family represents as a set of homologous genes derived from one common ancestral gene (Krebs 2018). In our case, some Endopterygota miRNAs evidenced rapid evolution and shared limited similarities in nonseed regions (MIR-2941, MIR-3841 and MIR-6000; MIR-956 and MIR-3850). In contrast, we provide some evidence for convergent evolution during miRNA evolution, as demonstrated by similar mature miRNAs in distantly related taxa (e.g., MIR-989 and MIR-BgeN1; MIR-1007 and MIR-6037). The classification of these miRNA into single or miRNA separated families can affect the conclusions, whereby it is necessary to take phylogeny into account during miRNA family assignment.
The quality of the analyzed source data source should also be carefully examined, as publicly available data may contain spurious annotations (due to sampling error). Previous studies asserted the pervasive secondary loss of miRNAs in different lineages (Thomson et al. 2014; Hertel and Stadler 2015), but intensively querying miRNAs led to opposite conclusions (Tarver et al. 2018). Here, we obtained the most complete annotation of insect miRNAs to date. The miRNA phylogenetic relationships were partially supported by the PCG phylogeny after removing the morphologically reduced species. Since miRNA and its flanking sequence served as a sort of phylogenetic marker (Kenny et al. 2015), we suggest that miRNAomes have the potential to be considered auxiliary evidence in further phylogenomic analysis.
The Gain and Loss of MiRNAs in Insects
Morphological innovations, such as the appearance of insect wings and holometabolan metamorphosis, significantly contributed to the success of insects (Grimaldi and Engel 2005). Here, we found five miRNAs that were gained in basal Pterygota, at the origin of winged insects. These newly gained miRNAs may thus have played crucial roles in the development of Pterygota morphological innovations, in particular wing vein formation, as well as stress resistance. These likely provided winged insects with the ability to explore previously unexplored terrestrial niches. In contrast, we found extensive miRNA losses in Paraneoptera and Parasitoid wasps. The common ancestor of Paraneoptera still had a complete miRNA compliment, and miRNA loss events occurred after the radiation of the clade. We suggest these miRNA losses may be connected to the independent morphological reduction observed in each clade. Paraneoptera insects are good examples to understand how miRNA loss drives morphological reduction (or vice versa). The loss of miRNAs in Parasitoid wasps likely resulted from parasitic lifestyle. Considering the conservation of miRNA sequences across distantly related taxa, parasitoid wasps may be able to exploit the miRNAs present in the host by using parasitism factors (like virus). In fact, it has been demonstrated that parasitic wasps can adopt miRNAs from virus to modulate host development (Wang et al. 2018).
In Endopterygota, only few miRNAs were gained at the stem compared with numerous miRNAs that emerged after the radiation of the major Endopterygotan clades, especially in the branch leading to Hymenoptera and Lepidoptera. This result supports more of the pronymph theory that both larvae and pupa in Endopterygota derived from already exiting life stages in Exopterygota (pronymph and nymph). Hence, the increment in morphological complexity is not as high as expected. In Neuropterida and some basal Coleoptera, the larvae (Oligopod larvae) and pupa type (Decticous pupa) are considered more primitive and similar to the adult stage than Endopterygota groups like Hymenoptera and Lepidoptera. Since the primary development of organ systems in these taxa occurs during the larval stage, the metamorphosis has likely progressed slowly (Gillott 2005; Belles 2020). Hymenoptera is placed as the most basal linage of Endopterygota in modern phylogenetic relationships. If the degree of metamorphosis in ancestral Endopterygota was still as rudimentary as observed in modern Megaloptera and Neuroptera, and advanced metamorphosis accumulated independently after radiation, it is possible to expect a concomitant accumulation of genes during this evolutionary process. Extensive analysis with PCGs or other functional elements and more Endopterygotan taxa, such as Megaloptera and Neuroptera, might confirm this possibility.
Our work provides a more well-curated miRNA data set and a robust gain and loss phylogenetic analysis of insect miRNA families. Due to the shortage of 3′UTR sequences, it is still hard to accurately predict miRNA targets. This hampers a deeper understanding on the function of many of the gained and lost miRNAs. To fully elucidate the relationship between miRNA evolution and organism complexity and trait diversity, it is necessary to develop more high-quality small RNA sequencing and genome data that improve the accuracy and coverage of miRNA prediction. Moreover, the biological functions of newly evolved miRNAs should be experimentally verified to confirm their putative roles in contributing to organismal complexity. The establishment of novel miRNA-target relationships may contribute to establish the time of appearance of certain important developmental features (Moran et al. 2017; Kawahara et al. 2019), which is very important for fully uncovering the roles played by miRNAs during insect evolution.
Materials and Methods
Data Sources and Preprocessing
The masked and unmasked genomes of 152 selected arthropod species were retrieved from the NCBI Assembly Database, Ensembl Metazoa, and BCM-HGSC (supplementary data set S1, Supplementary Material online). Genome completeness was assessed using BUSCO v4.1.4 with the library arthropoda_odb10 (Simão et al. 2015). The small RNA-seq libraries used for annotation were downloaded from the NCBI Sequence Read Archive (SRA) database (supplementary data set S2, Supplementary Material online). SRA files were decompressed by the fastq-dump program in the SRA toolkit package. The raw reads were filtered using Trimmomatic v0.38 (Bolger et al. 2014). The adaptors were removed by Cutadapter (Martin 2011). The Kraken package was used to remove low complexity reads and collapse redundant reads (Davis et al. 2013). The reads that mapped to Rfam (Kalvari et al. 2018) (except miRNA) and Repbase (Jurka et al. 2005) were also removed, and were those with <18 nt or >24 nt length. All samples in one species were combined as clean reads before proceeding with the miRNA annotation.
The miRNA sequences used for homology searches were downloaded from the miRBase v22 (Kozomara and Griffiths-Jones 2014) and the MirGeneDB 2.0 (Fromm et al. 2020) databases. For the latter, we selected the miRNA repertories of seven arthropods (D. melanogaster, Aedes aegypti, Heliconius melpomene, T.castaneum, Blattella germanica, Daphnia pulex, and Ixodes scapularis), totaling 1,184 miRNAs. The miRNA repertories of 12 arthropods were selected from miRBase, specifically: A. aegypti, Anopheles gambiae, A.mellifera, A.pisum, Bactrocera dorsalis, B.mori, Culex quinquefasciatus, H.melpomene, Plutella xylostella, Spodoptera frugiperda, Triops cancriformis, and T. castaneum. Considering that the miRNAs present in the miRBase reportedly contain numerous false positives, we used a homemade python script (available at https://gitee.com/mamading/my_mi-rna_work) to filter these miRNAs (Fromm et al. 2020). These criteria include: 1) Reads with a size of 20–26 nt map to each of the miRNA hairpin precursor arms; 2) The 5′ end of >90% of the reads from the same arm are located at the same position; 3) The 5p and 3p of mature miRNAs show 2 nt overhang; 4) The loop length is equal to or longer than 8 nt. We set the maximum length of the loop to infinite given most insects contain two or more Dicer proteins (Fromm et al. 2015). This allowed us to obtain 1,087 high-confident miRNAs. The miRNAs from both sources were then combined and used as query for further BLAST analysis. The miRNA annotation of D. melanogaster was downloaded directly from MirGeneDB (Fromm et al. 2015).
MiRNA Annotation
To obtain a curated miRNA data set, we established a pipeline for miRNA annotation. First, we annotated miRNA repertories of 36 species with publicly available small RNA-seq data using miRDeep2 (Friedlander et al. 2012) and MapMi (Guerra-Assunção and Enright 2010). In contrast, we only used MapMi for the remaining 115 species. For both two programs, we used masked genome data to build the indexes and unmasked genomes for sequence extraction. For miRdeep2, we set a cut-off score >5 and a Significant Randfold P-value = yes. For MapMi, we set a cut-off score ≥ 25 and MFE < –18 kcal/mol. The miRDeep2 predicted miRNAs that are currently not included in available databases were considered novel miRNAs if passing the criteria described by Fromm et al. (2015).
After this, and in order to reduce the possibility that real miRNAs might be neglected by the annotation programs or not covered by the genome assemblies, we also searched the missing miRNAs in genomic data. If one miRNA was missing in a certain species but present in another closely related species, we assumed this miRNA exists but corresponded to a false negative in the annotation programs. We then manually searched for its homologous sequence (BlastN, e-value <1e-5) and conserved genomic region. The putative sequence (if present) was filtered according to structural criteria (stem-loop or stem-apical bulge structure & MFE < –18 kcal/mol). In case the sequence was still missing, we assumed the miRNA exists but was not covered by the genome assembly. We searched raw genome sequencing data in the NCBI SRA database by online BlastN (e-value <1e-5). The hits were downloaded and assembled by Cap3 (Huang and Madan 1999). If no hit was found, we considered the miRNA as truly absent. We then removed redundant miRNAs. Finally, the miRNA annotations for all species were collected for further analysis.
MiRNA Family Classification
We first grouped miRNAs with identical seed (site 2–8 in mature sequence) into “seed families.” For each seed family, we checked the sequence identity of mature miRNAs and the corresponding flanking genomic regions. The miRNAs that shared the same seed and an homologous flanking sequence, independently of the discrepancy between their mature sequences, were assigned to the same miRNA family. For those miRNAs with no homologous flanking sequence, we used the globalxx function in the BioPython package (http://www.biopython.org) to align each pair of mature miRNA sequences in each seed family. The sequences showing an identity <0.7 or not locating in a monophyletic gene group (contradictory to insect phylogeny, like Mir-1007 and Mir-6037) were divided into different miRNA families. The family names of known miRNAs followed the nomenclature of MirGeneDB. For novel families predicted by miRDeep2, and in case they existed in MirGeneDB, we renamed them by abbreviating the species name, adding an “N” to represent “novel” and an index number. For example, the novel 1 family in B. germanica (termed BGE-NOVEL-1 in MirGeneDB) was renamed MIR-BgeN1 in this study.
Construction of Phylogenomic Trees
To place miRNA family gain and loss events in context, we constructed a phylogenomic tree for the 152 selected arthropod species using orthologous single-copy PCGs. To assign all genes into orthologous groups, we downloaded the protein gene annotations from 107 species with OGS data (supplementary data set S5, Supplementary Material online), and used the software Diamond to conduct all-versus-all BLASTp for their peptides (Buchfink et al. 2015). We then used OrthoMCL to assign genes into orthologous groups (Li et al. 2003) and obtained 2,380 single-copied orthologous genes. The groups that were present in single-copy in >95% of the species were selected as single-copy orthologous genes. Next, we identified the orthologous region of these selected genes in the 45 species for which no protein annotations were available. The protein sequences of orthologous groups obtained from closely related species were used as query, and the TBlastN program was applied to perform the search. The matched regions were extracted from the genome and confirmed by reciprocal BLASTp to the protein sets from the other species. The obtained sequences were selected as corresponding to single-copy orthologous genes in these species.
For each orthologous group, we aligned their respective amino acid sequences using mafft with L-INS-I algorithms (Katoh and Standley 2013). Low quality alignments and poly conserved sites were removed with Gblocks (we set -b1 = 0 and -b2 = 0) (Castresana 2000). The alignment of all the genes was then concatenated into a supermatrix using an in-house Python script. The obtained supermatrix contained a total of 365,877 amino acids (aa). This matrix was used to construct a ML phylogenetic tree using IQ-tree (Nguyen et al. 2015). We set the replacement model to LG+I+G and used the Ultrafast bootstrap method with a bootstrap value of 1,000. Next, we selected 18 nodes using the divergence time inferred from Misof et al. as calibration (supplementary table S6, Supplementary Material online) and the r8s to infer the divergence time of the whole tree (Sanderson 2003; Misof et al. 2014).
Phylogenetic Analysis Using MiRNAs
We reconstructed two binary matrices using the presence/absence of conserved miRNA families in 74 arthropod species. We removed the species from Sternorrhyncha, Thysanoptera, Phthiraptera, Strepsiptera and Phoridae in Diptera, and parasitoid wasps in Hymenoptera, as these species show some degree of morphological simplification. A conserved miRNA family was defined as a miRNA family that was present in more than one species but not in the same genus. The software BEAST v1.8.4 was used to perform the phylogenetic analysis under a Bayes stochastic Dollo model (Suchard et al. 2018). The clock model was set as an uncorrelated relaxed clock with Gamma relaxed distribution. The tree prior was set to Birth–Death process. The length of the chain was set to 10,000,000, with samples taken every 1,000 iterations. The final tree was summarized by TreeAnnotator, available in the BEAST package.
Gain and Loss Analysis
To investigate the miRNA evolution in insects, we analyzed the gain and loss events of miRNA families. We assumed that miRNA families evolved under a Dollo parsimony model (Rogozin et al. 2005). This model allows for each gene family to be gained only once in the course of evolution but poses no limitation on the secondary loss of each family after it is gained. We used this model by implementing the Dollop program in the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html). The presence/absence of each family was used as input data and the constructed phylogenomic tree was used as a background tree. We manually mapped the gain and loss of each miRNA family to the corresponding branches of the tree. The obtained tree showing the gain and loss events on each branch was then used to explore the evolution of miRNAs in insects.
Target Searching
To search for miRNA targets in the species of interested, we first extracted the 3′-UTR sequences of this species with a homemade python script. For each PCG, we selected the region extending from the stop codon to the 3′-end of the transcripts as 3′-UTR sequences. The mature miRNAs of interest were used as query. We used three different miRNA target prediction programs for target searching, namely Miranda (Enright et al. 2003), RNAhybrid (Kruger and Rehmsmeier 2006), and Pita (Kertesz et al. 2007). In Miranda, we set a cut-off score of <120 and a max energy < –18; in Pita, we set a cut-off value score < 8; in RNAhybrid, we set the cut-off value to MFE < –18 kcal/mol. The targets that were predicted in more than two programs were selected as putative targets. The function of the target genes was annotated by comparing to homologous genes in D. melanogaster using BLASTp. Finally, we conducted KEGG and Gene Ontology enrichment analyses of target genes using the clusterProfiler package available in R (Yu et al. 2012).
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 31972354, 31772238, 31701785), the National Science & Technology Fundamental Resources Investigation Program of China (2019FY100400), and the Fundamental Research Funds for the Central Universities (2020QNA6024).
Author Contributions
F.L. conceived the project and designed the research; X.M. annotated the miRNAs; X.M. and F.L. analyzed the data; X.M., K.H., Z.S., M.L., X.C., and F.L. interpreted and discussed the results; X.M., K.H., and F.L. wrote the manuscript.
Data availability
The source data are available as supplementary files. All data used for miRNA annotation, phylogenetic tree construction, and target analysis can also be accessed at http://insect-genome.com/miRNAomes/.
Literature Cited
- Ambros V. 2004. The functions of animal microRNAs. Nature 431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- Bai Y, et al. 2014. Genome-wide sequencing of small RNAs reveals a tissue-specific loss of conserved microRNA families in Echinococcus granulosus. BMC Genomics 15(1):736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2018. Metazoan microRNAs. Cell 173(1):20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Smibert P, Lai EC.. 2010. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 338(1):63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles X. 2017. MicroRNAs and the evolution of insect metamorphosis. Annu Rev Entomol. 62:111–125. [DOI] [PubMed] [Google Scholar]
- Belles X, editor. 2020. Insect metamorphosis: from natural history to regulation of development and evolution. 1st ed. San Diego (CA): Academic Press is an Imprint of Elsevier. [Google Scholar]
- Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 12(12):846–860. [DOI] [PubMed] [Google Scholar]
- Beutel RG, Friedrich F, Yang XK, Ge SQ.. 2014. Insect morphology and phylogeny. Berlin, Boston: De Gruyter. [Google Scholar]
- Biryukova I, Asmar J, Abdesselem H, Heitzler P.. 2009. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 327(2):487–496. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Campbell LI, et al. 2011. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc Natl Acad Sci USA. 108(38):15920–15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. 2010. Next-generation small RNA sequencing for microRNAs profiling in the honey bee Apis mellifera: honey bee microRNAs profiling by deep sequencing. Insect Mol Biol. 19(6):799–805. [DOI] [PubMed] [Google Scholar]
- Dai ZH, et al. 2009. Characterization of microRNAs in cephalochordates reveals a correlation between microRNA repertoire homology and morphological similarity in chordate evolution. Evol Dev. 11(1):41–49. [DOI] [PubMed] [Google Scholar]
- Dannemann M, Nickel B, Lizano E, Burbano HA, Kelso J.. 2012. Annotation of primate miRNAs by high throughput sequencing of small RNA libraries. BMC Genomics 13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ.. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63(1):41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E, Linsen SEV, Cuppen E, Berezikov E.. 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19(11):2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deline B, et al. 2018. Evolution of metazoan morphological disparity. Proc Natl Acad Sci USA. 115(38):E8909–E8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, et al. 2003. MicroRNA targets in Drosophila. Genome Biol. 5(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N.. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, et al. 2015. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 49:213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, et al. 2020. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 48(D1):D1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Worren MM, Hahn C, Hovig E, Bachmann L.. 2013. Substantial loss of conserved and gain of novel microRNA families in flatworms. Mol Biol Evol. 30(12):2619–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott C. 2005. Entomology. Netherlands: Springer Science & Business Media. [Google Scholar]
- Gomez-Orte E, Belles X.. 2009. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc Natl Acad Sci USA. 106(51):21678–21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS.. 2005. Evolution of the insects. New York, Cambridge University Press. [Google Scholar]
- Guerra-Assunção JA, Enright AJ.. 2010. MapMi: automated mapping of microRNA loci. BMC Bioinformatics 11:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Assunção JA, Enright AJ.. 2012. Large-scale analysis of microRNA evolution. BMC Genomics 13:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PCJ, Peterson KJ.. 2008. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci USA. 105(8):2946–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Stadler PF.. 2015. The expansion of animal microRNA families revisited. Life (Basel) 5(1):905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Madan A.. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9(9):868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1–4):462–467. [DOI] [PubMed] [Google Scholar]
- Kalvari I, et al. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46(D1):D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara AY, et al. 2019. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc Natl Acad Sci USA. 116(45):22657–22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, et al. 2015. The phylogenetic utility and functional constraint of microRNA flanking sequences. P Roy Soc B-Biol Sci. 282(1803):20142983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat Genet. 39(10):1278–1284. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S.. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(D1):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Goldstein ES, Kilpatrick ST. 2018. Lewin’s genes XII. Burlington: Jones & Bartlett Learning. [Google Scholar]
- Kristensen NP. 1999. Phylogeny of endopterygote insects, the most successful lineage of living organisms. Eur J Entomol. 96(3):237–253. [Google Scholar]
- Kruger J, Rehmsmeier M.. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34(Web Server issue):W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AM, et al. 2020. MicroRNA evolution provides new evidence for a close relationship of Diplura to Insecta. Syst Entomol. 45(2):365–377. [Google Scholar]
- Lozano J, Montanez R, Belles X.. 2015. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc Natl Acad Sci USA. 112(12):3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas K, Raikhel AS.. 2013. Insect MicroRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 43(1):24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenda DR. 2006. MAP kinase subcellular localization controls both pattern and proliferation in the developing Drosophila wing. Development 133(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J, Bremer A, Ahonen L, Kostiainen R, Puig O.. 2009. Drosophila FoxO regulates organism size and stress resistance through an adenylate cyclase. Mol Cell Biol. 29(19):5357–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Mayhew PJ. 2007. Why are there so many insect species? Perspectives from fossils and phylogenies. Biol Rev Camb Philos Soc. 82(3):425–454. [DOI] [PubMed] [Google Scholar]
- McKenna DD, et al. 2019. The evolution and genomic basis of beetle diversity. Proc Natl Acad Sci USA. 116(49):24729–24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, et al. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767. [DOI] [PubMed] [Google Scholar]
- Moran Y, Agron M, Praher D, Technau U.. 2017. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 1(3):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou XC, Duncan DM, Baehrecke EH, Duncan I.. 2012. Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc Natl Acad Sci USA. 109(8):2949–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S.. 2016. MicroRNA evolution, expression, and function during short germband development in Tribolium castaneum. Genome Res. 26(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA.. 2009. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 31(7):736–747. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA.. 2007. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 217(1):73–77. [DOI] [PubMed] [Google Scholar]
- Quah S, Hui JH, Holland PW.. 2015. A burst of miRNA innovation in the early evolution of butterflies and moths. Mol Biol Evol. 32(5):1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Babenko VN, Koonin EV.. 2005. Dollo parsimony and the reconstruction of genome evolution. Parsimony, phylogeny, and genomics 2005, 190: 200. [Google Scholar]
- Rota-Stabelli O, et al. 2011. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. P Roy Soc B-Biol Sci. 278(1703):298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ.. 2006. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B. 306(6):575–588. [DOI] [PubMed] [Google Scholar]
- Sim C, Denlinger DL.. 2008. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 105(18):6777–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Kang DS, Kim S, Bai X, Denlinger DL.. 2015. Identification of FOXO targets that generate diverse features of the diapause phenotype in the mosquito Culex pipiens. Proc Natl Acad Sci USA. 112(12):3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Shenoy A, Blelloch RH.. 2014. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 15(9):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogabe S, et al. 2019. Pluripotency and the origin of animal multicellularity. Nature 570(7762):519–522. [DOI] [PubMed] [Google Scholar]
- Suchard MA, et al. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4(1):vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Mattick JS.. 2003. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biol. 5(1):P1. [Google Scholar]
- Tarver JE, et al. 2013. miRNAs: small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 30(11):2369–2382. [DOI] [PubMed] [Google Scholar]
- Tarver JE, et al. 2018. Well-annotated microRNAomes do not evidence pervasive miRNA loss. Genome Biol Evol. 10(6):1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson RC, Plachetzki DC, Mahler DL, Moore BR.. 2014. A critical appraisal of the use of microRNA data in phylogenetics. Proc Natl Acad Sci USA. 111(35):E3659–E3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW. 2019. The evolution of insect metamorphosis. Curr Biol. 29(23):R1252–R1268. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM.. 1999. The origins of insect metamorphosis. Nature 401(6752):447–452. [DOI] [PubMed] [Google Scholar]
- Urena E, Manjon C, Franch-Marro X, Martin D.. 2014. Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc Natl Acad Sci USA. 111(19):7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZZ, et al. 2018. Parasitic insect-derived miRNAs modulate host development. Nat Commun. 9(1):2205–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BM, et al. 2009. The deep evolution of metazoan microRNAs. Evol Dev. 11(1):50–68. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, et al. 2011. Episodic radiations in the fly tree of life. Proc Natl Acad Sci USA. 108(14):5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T.. 2009. Evolution under canalization and the dual roles of microRNAs-A hypothesis. Genome Res. 19(5):734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylla G, Fromm B, Piulachs M-D, Belles X.. 2016. The microRNA toolkit of insects. Sci Rep. 6:srep37736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GC, Wang LG, Han YY, He QY.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data are available as supplementary files. All data used for miRNA annotation, phylogenetic tree construction, and target analysis can also be accessed at http://insect-genome.com/miRNAomes/.