Significance
Resurrection genomics is an alternative to ancient DNA approaches in studying the genetics and evolution of past and possibly extinct populations. By reviving biological material such as germinating ancient seeds from archaeological and paleontological sites, or historical collections, one can study genomes of lost populations. We applied this approach by sequencing the genomes of seven Judean date palms (Phoenix dactylifera) that were germinated from ∼2,000 y old seeds recovered in the Southern Levant. Using this genomic data, we were able to document that introgressive hybridization of the wild Cretan palm Phoenix theophrasti into date palms had occurred in the Eastern Mediterranean by ∼2,200 y ago and examine the evolution of date palm populations in this pivotal region two millennia ago.
Keywords: domestication, ancient DNA, introgression, crop evolution, hybridization
Abstract
Seven date palm seeds (Phoenix dactylifera L.), radiocarbon dated from the fourth century BCE to the second century CE, were recovered from archaeological sites in the Southern Levant and germinated to yield viable plants. We conducted whole-genome sequencing of these germinated ancient samples and used single-nucleotide polymorphism data to examine the genetics of these previously extinct Judean date palms. We find that the oldest seeds from the fourth to first century BCE are related to modern West Asian date varieties, but later material from the second century BCE to second century CE showed increasing genetic affinities to present-day North African date palms. Population genomic analysis reveals that by ∼2,400 to 2,000 y ago, the P. dactylifera gene pool in the Eastern Mediterranean already contained introgressed segments from the Cretan palm Phoenix theophrasti, a crucial genetic feature of the modern North African date palm populations. The P. theophrasti introgression fraction content is generally higher in the later samples, while introgression tracts are longer in these ancient germinated date palms compared to modern North African varieties. These results provide insights into crop evolution arising from an analysis of plants originating from ancient germinated seeds and demonstrate what can be accomplished with the application of a resurrection genomics approach.
Genome sequencing of ancient samples has provided an unprecedented window into the evolutionary history and biology of past populations and even extinct species. Data from ancient genomes has given us glimpses into human evolution, including extinct hominin species such as Neanderthals (1), and also helped us understand the evolutionary history of species as divergent as mammoths (2), horses (3), grapes (4), and maize (5). Sequencing ancient DNA, however, comes with numerous challenges (6, 7). DNA from fossilized samples is degraded—the molecules are short and chemically modified—and the amount of recovered endogenous DNA is small, even under hospitable preservation conditions (8). Plants present particular challenges (9); in general, plant materials do not have the protective bone tissue found in vertebrates that helps in preserving ancient DNA, and the limited size of archaeobotanical remains often yields only small quantities of endogenous DNA (10). Nevertheless, studies of ancient plant DNA have been conducted successfully, particularly in maize (5), emmer wheat (11), barley (12), and rice (13) as well as other crop species (14, 15). Most of these studies center on archaeobotanical samples found in arid (12), heavily waterlogged (16), or even mineralized environments (17) that have helped to preserve DNA material.
An alternative approach to ancient DNA studies in plants is to germinate ancient seeds from archaeological sites (18), permafrost (19), or historical collections (11) and study the genomics of these revived individuals—an approach we refer to as resurrection genomics. Seeds ranging in age from ∼1,300 y old in the sacred lotus Nelumnbo nucifera (20) to ∼30,000 y old for Silene stenophylla (19) have been germinated successfully, providing the ability to obtain living, intact biological material from otherwise ancient samples. Access to such material may lead to information on earlier phenotypes and (for genomic analysis) would circumvent sequencing errors that arise from degraded ancient DNA, overcome the low amount of endogenous DNA, and allow sequencing at deep coverage.
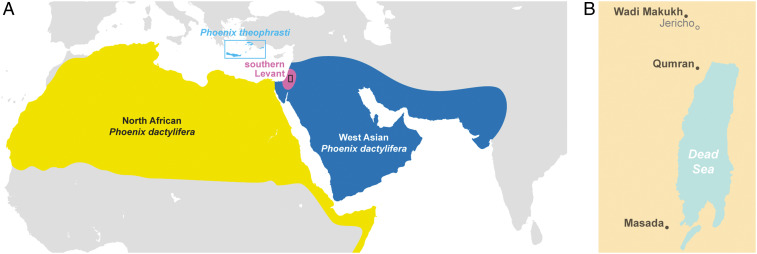
We applied this resurrection genomics approach to date palms (Phoenix dactylifera L.), which represent one of the best examples of successful germination of ancient seeds (21, 22). Date palms are a domesticated fruit crop species that are the iconic perennial plant of the arid lands of West Asia and North Africa (Fig. 1A). This species is believed to have been domesticated ∼7,000 y ago in the region around the Arabian Gulf (23, 24). From there, dates presumably spread westward and were widely cultivated in Egypt from at least the mid-second millennium BCE and further west to the Maghreb at least by the first millennium BCE (23, 25–27).
Fig. 1.
Map of date palms. (A) Geographical distribution of P. dactylifera and P. theophrasti. The Southern Levant is indicated, and the box in this area indicates where the archaeological samples were collected (see next panel). (B) Location of archaeological sites where the germinated Judean date palm seeds were collected.
The Southern Levant (modern-day Israel, Palestine, and Jordan) (Fig. 1B) is situated at the crossroads of Asia, Africa, and Europe, and during the 11th century BCE, this area saw the rise of the ancient geopolitical entity of Judea. Date palms grown in antiquity around Jericho and along the Dead Sea in Judea were referred to as Judean date palms, although it is unclear whether this referred to a distinct genetic population. These Judean date palms were discussed by classical writers such as Josephus and Pliny, who described them as producing superior fruit (28). Indeed, analysis from archaeological excavations in the region do show that the sizes of ancient seeds from these palms were significantly larger than those from modern date varieties (22).
Judean date cultivation is thought to have continued sporadically through the Byzantine and Arab periods (fourth to 11th century CE). By the 19th century, however, no trace of the Judean date palms remained, as date palm agriculture was extinguished in the area (28). Although Judean date palms are believed to be extinct (21, 22), it is unclear if some of their descendants still exist among modern date palm varieties.
Given their presence in a pivotal geographic location at the edge of West Asia and North Africa, the study of Judean date palm genomes could provide insights into the nature and timing of the spread of domesticated dates across the region. Studying the genomes of Judean date palms, as well as reviving this hitherto extinct population, became possible in 2008 when we reported the germination of a ∼2,000 y old date seed recovered from the historic archaeological site of Masada overlooking the Dead Sea (21). Six additional ancient date palm seeds from archaeological sites in Masada, Qumran, and Wadi Makukh in the Judean Desert and dated from the same time period were subsequently germinated (22) (Fig. 1B).
The genetic analyses of these germinated Judean date palms using microsatellite markers established that they represent a mix of North African and West Asian ancestry (22). Modern date palm varieties are genetically differentiated into West Asian [previously referred to by us as Middle Eastern (29–31)] and North African populations (29, 32–34) (Fig. 1A), the latter resulting in part from introgressive hybridization of the Cretan palm Phoenix theophrasti, or a theophrasti-like population, into P. dactylifera (30). The location and timing of this interspecies hybridization, which may be associated with the rise and spread of North African dates, remains unclear (35). In the Nile Valley, date palms were present as early as the predynastic period ∼5,000 y ago but were not extensively cultivated in Egypt until at least the New Kingdom ∼3,500 y ago (26, 27). Further west, date palm remains are dated to about ∼2,800 to 2,400 y ago in Libya (36) but are not found in the Saharan Maghreb and the sub-Saharan Sahel until much later (30, 37).
We do not know if these early examples of date palm cultivation in Africa were already impacted by genetic contributions from P. theophrasti, and determining when this interspecific hybridization occurred could tell us whether this took place during antiquity or much later in the evolution of this domesticated fruit species. An analysis of the full genome sequences of the Judean date palms germinated from ∼2,000 y old seeds could also advance our understanding of the role that interspecies hybridization has played in the evolution of this fruit crop species. Here, we report whole-genome sequencing of the seven germinated ancient Judean date palm samples, which provides an opportunity to study the change in genomic composition of date palms in the Southern Levant two millennia ago. We demonstrate that hybridization between P. dactylifera and P. theophrasti took place at least by the second century BCE, and we show increasing levels of Cretan palm introgression in the Southern Levant date palm populations across a period that spanned the fourth century BCE to mid-second century CE. We also use genome data to examine genes associated with fruit color and sugar composition, providing information on genetic characteristics of previously extinct (but now resurrected) Judean dates from ∼2,000 y ago.
Results
Genome Resequencing of Ancient Germinated Judean Date Palms.
A total of 35 well-preserved P. dactylifera seeds from archaeological remains were initially planted of which seven were germinated successfully (21, 22). The germinated seeds were excavated from three archaeological sites in the Judean desert (present-day Israel) (Fig. 1B) and radiocarbon dated to ∼2,000 y before present (B.P.). We generated whole-genome resequencing data from these seven Judean date palms, designated as Methuselah, Hannah, Adam, Judith, Boaz, Jonah, and Uriel (Table 1) (38). The two oldest (Methuselah and Hannah) date to between the fourth and first century BCE and the youngest (Uriel and Jonah) from the first to second centuries CE (21, 22). These germinated samples were analyzed alongside previously published whole-genome resequencing data from modern date palm (P. dactylifera) and wild Phoenix species (29–31) (SI Appendix, Table S1). Modern accessions are represented by 25 and 46 cultivated date palms from North Africa and West Asia, respectively, as well as three wild date palms from Oman. Between one and 18 samples of five wild Phoenix species were also included—18 P. theophrasti, eight P. sylvestris, three P. atlantica, six P. canariensis, and one P. reclinata.
Table 1.
Description of the seven germinated Judean date palms
| Accession | Origin | Radiocarbon date* | Sequencing coverage (x) |
| Methuselah | Masada | 351 BCE to 21 CE | 18.1 |
| Hannah | Wadi Makukh | 355 BCE to 56 BCE | 40.2 |
| Adam | Masada | 202 to 2 BCE | 35.8 |
| Judith | Qumran | 165 BCE to 5 CE | 35.9 |
| Boaz | Qumran | 44 BCE to 63 CE | 41.1 |
| Jonah | Qumran | 25 CE to 134 CE | 40.6 |
| Uriel | Qumran | 25 CE to 134 CE | 42.3 |
From Sallon et al. (22).
The Judean date palms were sequenced using 2 × 100 base pair (bp) paired-end Illumina sequencing. Reads from both Judean date palms and present-day Phoenix samples were mapped to the Barhee BC4 reference genome assembly (39), resulting in deep coverage for the Judean date palms (36.3× on average), while the other Phoenix samples were sequenced to an average depth of 17.6× (Table 1 and SI Appendix, Table S2). We identified 5,332,805 single-nucleotide polymorphisms (SNPs) across the 117 Phoenix accessions after quality filtering, which we used in subsequent analyses.
Seed storage over long periods prior to germination is potentially associated with accumulation of DNA damage, including single and double strand breaks, along with increased levels of base loss and modification (40). Nevertheless, DNA repair mechanisms are active during germination, allowing seeds to remain viable (40). Modification of guanine to 8-oxoguanine is one of the most common DNA lesions (41), leading to G-to-T (and C-to-A) substitutions (42). To explore whether Judean date palm genomes are enriched in G-to-T/C-to-A substitutions compared to modern date palms, we compiled the singleton mutational profile for each accession. We found no difference in singleton SNP profiles between ancient germinated and modern date palms (Wilcoxon rank sum tests, P > 0.05; SI Appendix, Fig. S1A and Dataset S1). During seed storage, DNA strand breaks also occur as DNA damage, and their repair leads to short insertion/deletion (indel) mutations at the break junctions (43). Here, we did not observe an elevated number of short singleton indels in ancient samples compared to modern date palms (multivariate Wilcoxon test, W = 236, P = 0.71; SI Appendix, Fig. S1B and Dataset S2).
Genetic Relationships between Ancient Judean and Modern Date Palms.
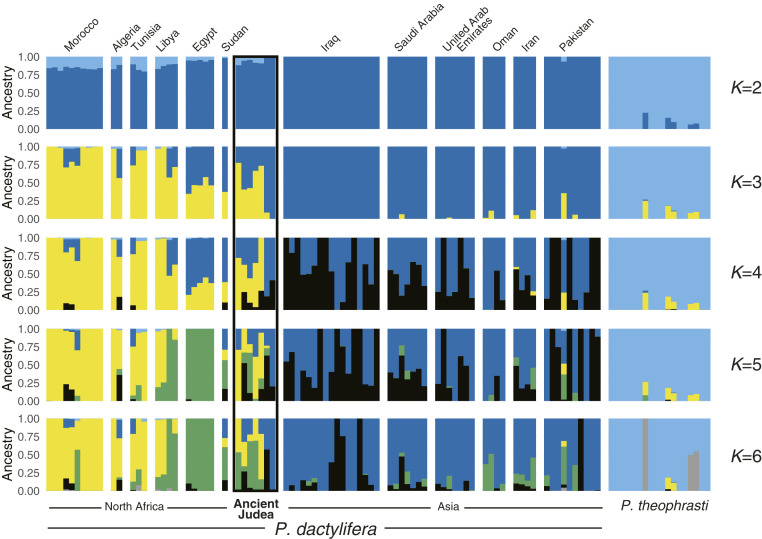
To examine population structuring and the genetic relationships of the seven Judean date palms with present-day Phoenix spp., we performed model-based genetic clustering using ADMIXTURE (44). Analyses with all Phoenix spp. accessions (SI Appendix, Figs. S2 and S3A) or including only date palms (P. dactylifera) and P. theophrasti (Fig. 2 and SI Appendix, Figs. S3B and S4) showed that the germinated ancient seedlings have a genetic makeup resembling that of modern date palms.
Fig. 2.
Ancestry coefficients inferred in 99 date palms and P. theophrasti accessions. The Judean date palms are ordered as follows from left to right: Boaz, Adam, Judith, Uriel, Jonah, Hannah, Methuselah. The number of clusters (K) range from two to six. Additional K values may be found in SI Appendix, Fig. S4. Cross-validation error plot may be found in SI Appendix, Fig. S3B.
At K = 2, Methuselah and Hannah resemble modern West Asian date palms, while the five others show mixed ancestry from the West Asian cluster and the Cretan palm P. theophrasti, similar to what is observed in modern North African date palms (Fig. 2). The proportion of ADMIXTURE genomic ancestry shared by the Judean date palms and P. theophrasti falls between 4.51 and 11.8%, in the same range seen in modern North African P. dactylifera (∼1.90 to 20.6%).
At K = 3, most North African date palms form a separate cluster, and the rest, especially the Egyptian samples, appear admixed between this cluster and the West Asian date palms. Likewise, the Judean date palms have mixed ancestry from the North African and West Asian date palms, except Methuselah, which has 100% ancestry from West Asia. At large K values, the Judean date palms still do not form a unique genetic cluster but instead display ancestry from both western (Morocco, Algeria, Tunisia, and Libya) and eastern (Egypt and Sudan) North Africa and from West Asia (Fig. 2 and SI Appendix, Fig. S4). We should note that at K > 2, the contribution of P. theophrasti to the Judean and North African date palm genomes is less apparent, and the clustering that is observed appears to represent population structuring.
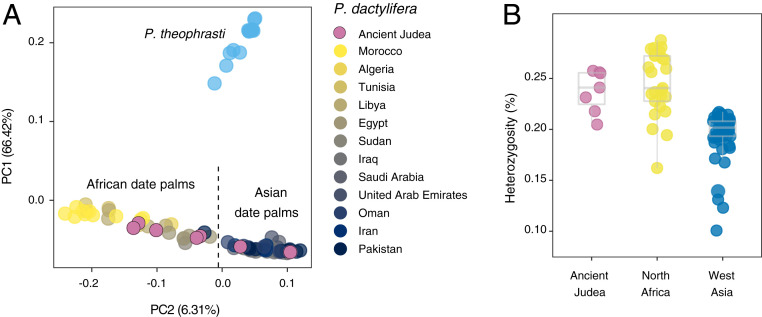
We then applied a principal component analysis (PCA) to the SNP diversity of date palms and P. theophrasti accessions. We found that the first principal component (PC1) explains 66.42% of the variance in SNP diversity and separates P. dactylifera from P. theophrasti, while PC2 explains 6.31% of SNP diversity and reveals an east (Asia) and west (North Africa) axis of genetic diversity (Fig. 3A and SI Appendix, Fig. S5). In the PCA analysis, the ancient Judean date palms do not form a distinct genetic cluster. Instead, Methuselah and Hannah are found within the cluster of West Asian date palms, while the five others show closer affinity with North African populations. Judith and Adam, which have intermediate PC2 scores, cluster with Egyptian and Sudanese varieties, while Boaz, Uriel, and Jonah have more negative PC2 values and are closely associated with modern western North African varieties from Morocco, Algeria, and Tunisia. Additional principal components together only explain an additional 2.61% of the genetic variation (SI Appendix, Fig. S5B). This pattern, where two ancient Judean palms cluster with present-day West Asian while the five others cluster with modern North African varieties, is also supported by a phylogenetic analysis (SI Appendix, Fig. S6) and corroborates the microsatellite-based genetic study (22).
Fig. 3.
Structure and diversity of date palms. (A) PCA of 99 Phoenix accessions including the seven Judean date palms genotyped for 24,689 SNPs. The variance explained by each principal component (PC) is given in parentheses. Plots showing additional PCs may be found in SI Appendix, Fig. S5A. (B) Heterozygosity in each date palm accession.
Genetic Diversity of Ancient Judean Date Palms.
We calculated the heterozygosity of each date palm accession (Fig. 3B and SI Appendix, Table S3) and confirmed that, on average, North African date palms (mean heterozygosity = 0.245 ± 0.00635%) are more diverse than the West Asian date palms (0.196 ± 0.00341%) (one-sided Wilcoxon rank sum test, W = 1,097, P = 7.66 × 10−11) as previously reported (29–31). The Judean date palms have a mean heterozygosity similar to North African P. dactylifera (0.238 ± 0.00781%; Wilcoxon rank sum test, W = 68, P = 0.40). This can be attributed to the five samples previously shown to cluster with North African date palms and that show an elevated proportion of heterozygous sites compared to West Asian date palms (one-sided Wilcoxon rank sum test, W = 240, P = 3.49 × 10−7). In contrast, the heterozygosity of Methuselah and Hannah, which cluster with West Asian date palms in previous analyses, is comparable to other samples in this cluster (Wilcoxon rank sum test, W = 75, P = 0.22). Furthermore, we find that only 0.7% of the total date palm polymorphisms is unique to the ancient germinated samples (SI Appendix, Fig. S7), indicating that most of the diversity found in the ancient Judean date palms is shared with modern P. dactylifera.
Introgression from P. theophrasti into 2,000 y Old Date Palms.
Oasis agriculture and the cultivation of the date palm is first apparent in southern Mesopotamia during the fifth millennium BCE and was developed across the Gulf region from around 3,000 BCE (23). Phoenix dactylifera was presumably domesticated in this region, supported by the discovery of wild date populations in Oman (31, 35). In North Africa, date palm cultivation may not have begun in Egypt until ∼3,500 y ago (26, 27) and further west in the Maghreb, by the first millennium BCE (25, 36). Modern North African date palms are both strongly differentiated from and have higher levels of nucleotide diversity than West Asian P. dactylifera (29–31). It was shown recently that this pattern can be explained in part by interspecific hybridization of wild P. theophrasti or a theophrasti-like population into West Asian P. dactylifera (30). Phoenix theophrasti is found on the island of Crete and in isolated populations on other Aegean islands and southwest Turkey (45) but may have had a broader geographic range in the Eastern Mediterranean in the past (46). The timing and localization of the introgression event(s) remain largely unknown (30, 35), and genome analysis of the germinated Judean date palms provides an opportunity to characterize this introgression in ∼2,000 y old samples originating from a region at the geographic juncture of North Africa and West Asia.
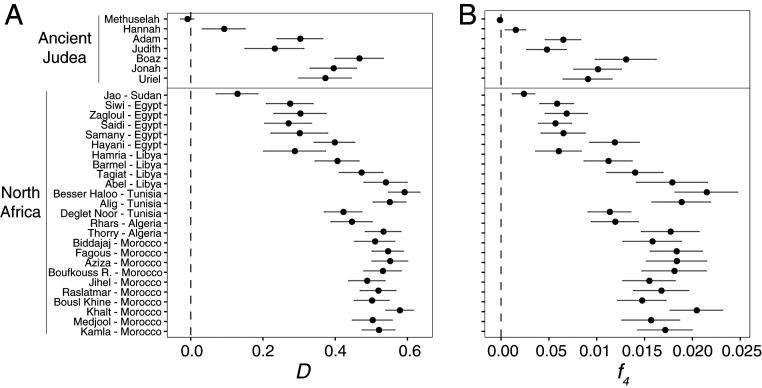
To assess whether ancient Judean date palms show evidence of introgression from P. theophrasti, we calculated the D-statistic (47, 48) using the following population tree: (((W, X), Y), Z), where W are individual African or Judean date palms, X are the West Asian date palms, Y are P. theophrasti, and Z an outgroup species. With P. reclinata set as an outgroup, an analysis of individual genomes found that Methuselah (D = −0.0094 ± 0.0092, |z-score| < 2) has a D-statistic that overlaps with zero (Fig. 4A and SI Appendix, Table S4). For the other six ancient date palms, we find positive D values whose CIs do not overlap with zero (range of D = 0.0915 ± 0.0296 for Hannah to D = 0.468 ± 0.032 for Boaz, |z-score| > 2), which is associated with an excess of shared derived alleles with P. theophrasti and consistent with hybridization with this species. The use of P. canariensis as an outgroup, instead of P. reclinata, does not lead to significant differences in the resulting D-statistics (SI Appendix, Fig. S8A; mean of difference in D-statistics = 0.00125).
Fig. 4.
Admixture between ancient Judean/modern North African date palms and P. theophrasti. (A) Admixture D-statistic and (B) admixture f4-statistic, both testing whether ancient Judean and modern North African date palms show an excess of shared alleles with P. theophrasti, in which case they are significantly positive. We estimated the D-statistics and f4-statistics using the following tree: (((test sample, West Asian date palms), P. theophrasti), P. reclinata). The same analysis with P. canariensis as the outgroup is in SI Appendix, Fig. S8.
Second, we tested for admixture between Judean date palms and P. theophrasti with f4-statistics (47) and using the same population tree. Similarly, we find significant positive f4 for all North African and Judean date palms except for Methuselah (Fig. 4B and SI Appendix, Table S5), again consistent with gene flow from P. theophrasti. The results for f4-tests do not change if we use P. canariensis or P. reclinata as outgroups (SI Appendix, Fig. S8B; mean of difference in f4-statistics = 0.00024). We also conducted a related f3-test for admixture, and in this analysis, only four Judean date palms (Jonah, Boaz, Adam, and Hannah) had significantly negative f3-statistics that signal gene flow from P. theophrasti (SI Appendix, Fig. S9 and Table S6). The other three resurrected date palms do not show a significantly negative f3-statistic, which for this test is uninformative (49) but not inconsistent with admixture.
We also calculated those three statistics, namely D, f4, and f3, in order to identify whether the Judean date palms show evidence of admixture with the wild relative P. sylvestris and found no significant evidence of hybridization (SI Appendix, Tables S4–S6).
P. theophrasti Introgression Fraction Increases Over Time in Ancient Date Palms.
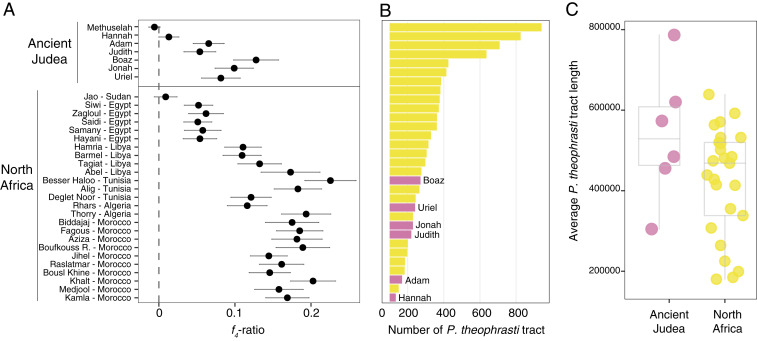
The D- and f4-statistics indicate that most Judean date palms, like modern North African P. dactylifera, have mixed ancestry due to introgressive hybridization. Using the f4-ratio statistics (47), we estimated the percentage of P. theophrasti ancestry in North African and ancient Judean genomes (Fig. 5A and SI Appendix, Table S7). With P. reclinata set as an outgroup, we find the present-day North African date palms have between 0.58 and 20.28% (mean = 12.58%) of their ancestry shared with P. theophrasti, which is qualitatively similar to our previous report (30). Among Judean date palms, Methuselah has no significant evidence of introgression from P. theophrasti. However, the six other germinated date palms have varying levels of P. theophrasti ancestry, ranging from 1.3% (Hannah) to 12.8% (Boaz). There are no significant differences in estimated levels of P. theophrasti introgression whether we use P. reclinata or P. canariensis as the outgroup (SI Appendix, Fig. S10; mean of difference in f4-ratio statistics = 0.0023). We observe that the two oldest samples, from the fourth to first century BCE (Methuselah and Hannah), have no or very little P. theophrasti ancestry. With the two other samples dated from the second and first century BCE (Judith and Adam), they have on average 3.31 ± 1.57% of P. theophrasti ancestry, while the three youngest samples from after this date (Boaz, Uriel, and Jonah) have a significantly higher P. theophrasti fraction in their genome (mean 10.28 ± 1.35%, one-sided Wilcoxon rank sum test, W = 9, P = 0.05).
Fig. 5.
Admixture proportion and introgressive tracts in ancient Judean and modern North African date palms. (A) f4-ratio statistic, indicating the fraction of P. theophrasti genomes found in the test sample and calculated as f4(A,O; X,C)/f4(A,O; B,C), where X is the test sample; A is a sister species, namely P. sylvestris; (B and C) are the mixing populations, namely West Asian date palms and P. theophrasti, respectively; and O is the outgroup, that is, P. reclinata. The same analysis with Phoenix canariensis as the outgroup is in SI Appendix, Fig. S10. (B) Introgressive tract count in each North African (yellow) and ancient Judean (pink) date palms. (C) Average introgression tract length in North African and ancient Judean date palms.
Following admixture, the length of introgressed tracts is expected to decrease with time as recombination breaks down these segments at each generation (50). In the absence of continuing gene flow from P. theophrasti after ∼2,000 y ago, the Judean date palms would be expected to have longer P. theophrasti tracts compared to modern North African date palms. We tested this hypothesis by performing local ancestry analyses in both North African and Judean date palms, excluding Methuselah, which did not show evidence of admixture. We used RFMix (51) to identify genomic segments inherited from either West Asian date palms or P. theophrasti. This revealed that Judean date palms display significantly fewer P. theophrasti tracts (34 to 171 tracts, with an average of 110.7 ± 8.4) than North African varieties (50 to 841 tracts, with an average of 274.6 ± 8.0; one-sided Wilcoxon rank sum test, W = 25.5, P = 0.0071) (Fig. 5B). Nonetheless, the introgressed segments of P. theophrasti are on average 49.7% longer in the Judean date palms (530.40 ± 1.0 Mb) compared to the modern North African genomes (354.4 ± 0.08 Mb), and this difference is significant (one-tailed Student’s t test, t = −9.5741, degrees of freedom (df) = 1506.9, P < 10−15) (Fig. 5C).
Genes for Fruit Color and Sugar Content in Judean Date Palms.
Date palms are characterized by substantial phenotypic variation, and there has been recent success in identifying genes associated with key fruit traits such as color and sugar content using genome-wide association studies (GWAS) (39). The knowledge of functional alleles associated with these traits offers a glimpse into the features of fruits harvested 2,000 y ago.
Date palm fruits vary in color from deep red to pale yellow, and this is controlled by the VIRESCENS (VIR) gene, which encodes an R2R3-myb transcription factor (29, 39). Analysis shows that red coloration is associated with a wild-type VIR+ allele, while yellow fruits have either the dominant VIRIM or are homozygous for the recessive virsaf allele (39). Genotype analysis at the VIR locus indicates that all three alleles are present in Judean date palms (SI Appendix, Table S8). This reveals that ∼2,000 y ago, both the yellow and the less common red fruits were produced in ancient Judea as suggested by the Babylonian Talmud, a text compiled during late antiquity (28).
Sugar content is another key trait of date fruits and contributes to the flavor profile of individual varieties. Most date fruits have high fructose/glucose content, but some varieties have high sucrose content (39, 52). GWAS analysis indicates this is largely controlled by a 1.1 Mb genomic region that contains multiple copies of cell wall (CWINV1 to 3) and alkaline/neutral (A/N-INV1) invertase genes (39). High sucrose fruits are characterized by an ∼40 kb deletion that removes portions of the CWINV1-3 genes and/or a separate ∼5 kb deletion that takes out the 5′ portion of A/N-INV1. In the current study, we do not detect these deletions in any of the seven Judean date palms (SI Appendix, Table S8). This suggests these 2,000 y old palms would likely be associated with higher fructose and glucose levels (and therefore lower sucrose levels) in fruits.
Discussion
Our previous work shows that ancient P. dactylifera seeds recovered from archaeological digs and dated to ∼2,000 y ago can be successfully germinated (21, 22). Despite the prolonged age of the seed prior to their germination, they do not appear to have substantially increased mutation levels. The levels of heterozygosity in the ancient date palms are within range of contemporary samples, and there is no excess of G-to-A/C-to-T singletons associated with substantial increase in mutational damage (40). We also do not find an excess of short indel singletons that may arise from elevated levels of DNA strand break repair (43). The main drivers of DNA damages are humidity, oxygen, and ionizing radiations (40, 53), and the absence of any significant increase in new mutations associated with seed aging in the ancient samples may be due to the unique environmental conditions around the Dead Sea where the seeds were discovered (22). Moreover, active genomic maintenance mechanisms associated with longer seed life spans may have repaired any accumulated lesions (40). Finally, it should be noted that longer seed lifespans are believed to have evolved as an adaptation to arid environments where seeds need to survive prolonged dry periods (54).
Our ancient date palm samples were recovered in the Levant, an area between Asia, Africa, and Europe that played a critical role throughout history, from early human dispersal (55) to the origins of agriculture (56). It notably served as a waystation for the dispersal of crops such as emmer wheat (57) and barley (12) as they moved south and west from the Fertile Crescent. In this region, the oldest archaeobotanical records of date palms stretch to the fifth millennium BCE, but it is unclear whether they derive from wild or cultivated stands and if they are of local origin (25, 30, 58, 59). The availability of whole-genome sequences of date palm plants from ∼2,000 y ago allowed us to examine the dynamics of date palm evolution in the Southern Levant at a crucial time, when this area was influenced or even controlled by several great empires from both east and west, including Babylon, Persia, Egypt, and Rome.
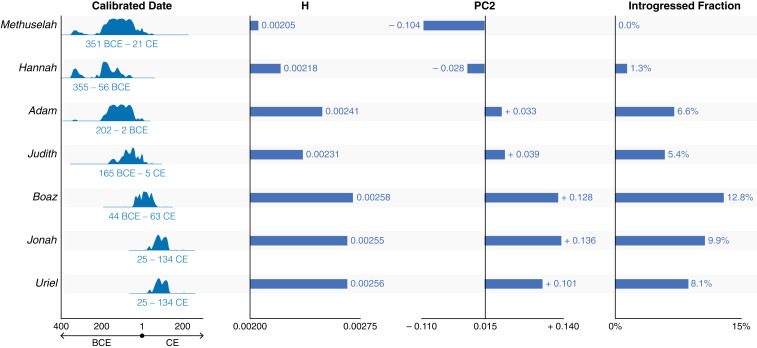
As a group, the seven Judean date palms germinated from ancient seeds do not form a distinct genetic cluster but instead show shifting genetic affinities over time (Figs. 3A and 6); in this respect, the term Judean date palm may represent less a discrete genetic population but rather a designation of origin. Methuselah and Hannah, which are the oldest samples and radiocarbon dated from between approximately fourth to first centuries BCE, are found within the genetic cluster of West Asian date palms. The five other Judean date palms, however, show closer affinities with the North African population (Fig. 3A). Judith and Adam, which are of intermediate age among the samples (approximately first to second centuries BCE), group with eastern North African accessions comprised of Egyptian and Sudanese varieties. The most recent samples (Boaz, Uriel, and Jonah), which are dated to between the first century BCE to second century CE, are closely associated with the more western North African accessions of Morocco, Tunisia, and Algeria (Figs. 3A and 6).
Fig. 6.
Shift of ancestry and genetic attributes through time in the Judean date palms. The calibrated dates are derived from Sallon et al. (22). H refers to the nucleotide heterozygosity of the sample (Fig. 3B), PC2 is the coordinate in the PCA (Fig. 3A), and the values of introgression fraction is the f4-statistics derived for each sample using P. reclinata as the outgroup (Fig. 5A).
One cannot tell whether the genetic relationships between the ancient samples and present-day traditional cultivars indicate contributions of external sources into the Judean date palm gene pool or whether these contemporary cultivars are descendants from this Levantine population. Given that some of our ancient date palms show genetic similarities to West Asian while others to different North African cultivars, it is more likely that the genetic affinities with contemporary cultivars represent ancient dispersal of western and eastern genotypes into the Southern Levant. Interestingly, the changing genetic clustering of our ancient samples over time coincides with transitions in imperial control in the region, from the eastern Neo-Babylonian (early seventh century BCE) and Persian Empires (539 to 331 BCE) to the later more western Hellenistic Roman Empire (331 BCE to 476 CE).
We previously showed that the cultivated date palms of North Africa evolved as a hybrid between the domesticated palms of West Asia and the wild P. theophrasti, although when and where this hybridization took place remains unclear (30, 35). From our analyses of the Judean date palms, it is apparent that by ∼2,400 to 2,000 y ago, the hybridization event(s) that introduced P. theophrasti genomic segments into the P. dactylifera cultivated gene pool had already occurred. This is also consistent with sequencing of an ancient date palm leaf from an archaeological excavation of a temple complex of the animal necropolis in Saqqara, Egypt, that dates to ∼2,100 y ago and which likewise shows P. theophrasti introgression (60).
We observe an influx of date palms with increasing P. theophrasti ancestry over the ∼550 y period represented by our samples (Fig. 6). This changeover is also seen in Egypt, where archaeological remains of date palm seeds show an increase of western morphotypes, which may reflect growing control of the Roman Empire in the area with accompanying movement of date palm varieties from the west (61). Another possible explanation for the increase in introgression fraction with time, however, is the onset or continuing gene flow with P. theophrasti, which may have grown in the Levant in the past (49, 62). This is less likely, however, given the small introgression fraction in the Judean date palm genomes; if indeed the interspecific hybridization took place in the Levant, then we would expect to see early on a large amount of P. theophrasti introgression in the area. The movement of date palms from the west may also explain the trend of increasing nucleotide heterozygosity over time across greater than five centuries (Fig. 6). If the original hybridization of P. dactylifera and P. theophrasti occurred outside this region (possibly in North Africa/Egypt), then the inflow of interspecific hybrids from the west would indeed lead to greater genetic diversity in date palms of the Southern Levant during the Roman period.
We surmised that the Judean date palms, being ∼2,000 y older than modern cultivars, should be closer in time to the interspecific crossing event(s) of P. dactylifera × P. theophrasti that gave rise to the hybrid genomes of North African date palms. We would expect to observe longer P. theophrasti introgression tracts in the date palm genome soon after this hybridization event, with subsequent recombination with backcrossing to P. dactlyifera resulting in shorter tract lengths over time. We indeed find a pattern consistent with the age of these Judean date palm samples. Specifically, these ancient date palms have significantly longer introgression tracts when compared to modern North African varieties, which we attribute to less recombination between the introduced P. theophrasti genomic regions and P. dactylifera genome sequences in the ∼2,000 y old palms when compared to modern North African varieties (Fig. 5C).
This analysis has provided key insights into the evolution of domesticated date palm, and highlights once again the impact that interspecific hybridization plays in the evolution of domesticated crop species (30). It also demonstrates the role that geopolitical transformations play in crop evolution; in this case, we observe changes in the genetic composition of date palm plants that likely arose as different empires and peoples took control of the Southern Levant, possibly facilitating the movement of crops and other organisms. What we observe in this region may inform our understanding of the dynamics of the post-domestication geographic spread of crop species and the roles that human migration and colonial/imperial expansion play in shaping evolutionary trajectories of species.
Resurrection carries two meanings—to bring back from the dead and to bring back something into existence that had disappeared or ended. The use of the term resurrection in the study of revived biological material derived from ancient sources is not new and has been discussed over the last few years in both contexts. First, the possibility of reviving extinct species, or de-extinction, using genetic material from long-dead individuals has generated interest as a way to bring back lost taxa (63). Second, the resurrection approach has been described as a method to examine biological questions in the context of contemporary and/or experimental evolution and ecology (64). In the latter case, the term resurrection denotes reviving dormant biological material (seeds, microbes, etc.), and it is in this context that we propose the term resurrection genomics as an approach to studying the genomes of organisms that have lived hundreds to thousands of years in the past.
Aside from these 2,000 y old date palms, we have already noted studies of germination of 1,300 y old lotus seeds (20) and even ∼30,000 y old Silene sp. from the Siberian permafrost (19). Investigators have also reported germinating 151 y old acacia seeds (65) and 680 y old peatland Sphagnum spores (66) as well as obtaining live callus tissue from 1,600 y old Anagyris foetida seeds (67). Reviving organisms need not be limited to plants, as there have been sediments with reported revivals of 100 y old diatoms (68), 150 y old dinoflagellates (69), and 700 y old Daphnia pulicaria (70). As in ancient DNA studies, it will be important to ensure that the samples have good provenance and are properly dated to ensure authenticity. Moreover, there is a possibility that a bias in which samples can be revived may lead to a skew in the resulting data, and this should be considered when drawing conclusions from these studies. Nevertheless, the prospect of investigating the genomes of individuals that have lived 102 to 104 y ago provides unprecedented opportunities to directly examine the biology of life from the past.
Materials and Methods
Sampling, Library Preparation, and Genome Resequencing.
The seven ancient Judean date palm accessions originate from the germination of date palm seeds collected during archaeological excavations of three sites in the Judean desert (21, 22). The genome sequences of the ancient Judean date palms were compared to those of 110 contemporary Phoenix dactylifera varieties and other wild Phoenix species (SI Appendix, Table S1) (29–31). Genomic DNA was extracted from leaf tissue as explained in Sallon et al., (22). Libraries (2 × 100 paired end) were constructed with Nextera library preparation protocols and sequenced on an Illumina HiSeq 2500 system according to the manufacturer’s protocols. More details on this and subsequent analyses are found in SI Appendix, Supplementary Information Text.
Read Processing and Genome Alignment.
Reads were demultiplexed and those passing Illumina quality control filters were processed with Trimmomatic (71) v. 0.39 to remove contaminating adapter sequences. Only read pairs where both reads were 76 bp or longer following trimming were retained for subsequent steps. We aligned reads to the reference genome from Hazzouri et al., (39). Processed reads were aligned to this unmasked genome using bwa mem (72) v. 0.7.15-r1140. We used MarkDuplicates (Picard tools) to flag duplicate read pairs. Further details of read processing as well as all subsequent analyses can be found in SI Appendix.
Variant Calling and SNP Filtering.
SNP calling and genotyping were performed with the Genome Analysis Toolkit v. 3.5-0-g36282e4 HaplotypeCaller (73) run in Genome Variant Call Format (GVCF) mode followed by joint genotyping with GenotypeGVCFs. Reads were filtered from the HaplotypeCaller step to exclude those with a mapping quality <10 and to exclude those marked as secondary alignments. We excluded SNPs in repetitive regions identified during the annotation of the genome assembly (39). We also excluded multiallelic SNPs and SNPs within 6 bps of indel polymorphisms. For further filtering, thresholds were determined following Flowers et al. (30), from which most of our data come from. Statistical analyses were conducted with the R Statistical programming language (74).
Identifying DNA Lesions Associated with Prolonged Period of Seed Storage.
Using a custom Perl script, we identified singleton mutations in each accession, computed the number in each substitution class (G-to-C, A-to-C, etc.), and normalized by the total number of singletons observed in the accession. We used Wilcoxon rank sum tests to examine differences among ancient germinated and present-day genomes in each of the six mutation type fractions (wilcox.test function in R). We also used a custom Perl script to identify singleton indels. We restricted our analysis to indels having the following features: quality (QUAL) ≥ 30, depth (DP) ≥5, and maximum missing data of 15%. We tested whether there is a significant difference in small indel numbers using a multivariate Wilcoxon test implemented in the mWilcoxonTest function from the R package DepthProc v.2.1.3 (https://www.rdocumentation.org/packages/DepthProc).
Population Clustering and Relatedness among Samples.
We estimated individual ancestries on 1) all accessions and 2) date palms and P. theophrasti only using ADMIXTURE (44) v. 1.3.0. We used 53,845 randomly sampled SNPs and default parameters. The optimization method was a block relaxation algorithm, and the termination criterion was to stop when the log-likelihood increases by less than ε = 10−4 between iterations. We ran the PCA on date palms and P. theophrasti accessions using genotype calls with the pcadapt (75–77) R package v. 4.3.3 using a subset of randomly sampled SNPs and further filtered out SNPs with minor allele frequencies below 5%, resulting in 24,689 SNPs.
We generated maximum likelihood trees of Phoenix spp. accessions using RAXML-NG (78) v. 0.9.0 and a randomly downsampled SNP set including 1% of our full SNP set (53,845 SNPs). We performed 20 maximum likelihood tree searches using 10 random and 10 parsimony-based starting trees. We picked the best scoring topology and checked the robustness of the tree by performing 100 bootstrap replicates.
Individual and Population Statistics.
We calculated for each date palm and P. theophrasti accessions the level of heterozygosity using Variant Call Format (VCF) tools v. 0.1.14 (79) in nonoverlapping sliding windows of 5 kb. The level of polymorphism was calculated in each population or species using a custom Perl script calculating the number of polymorphic sites in each group. We then identified private polymorphisms that count each SNP segregating for an allele (“private allele”) that is restricted in its distribution to a particular focal population or species in our analysis. For this analysis, we removed admixed samples in both date palms and P. theophrasti.
Admixture Tests and Ancestry Proportions.
We tested for the presence and extent of admixture between Phoenix populations/species using the R package admixr (80) v.0.9.1.9000, which provides an implementation of ADMIXTOOLS (47). We used a script available online to generate the input in EIGENSTRAT format from our VCF (https://github.com/joanam/scripts/blob/master/convertVCFtoEigenstrat.sh). We ran the tool using the full SNP set after filtering out regions associated with sex determination (see above).
We calculated Patterson’s D (47, 48), also known as the ABBA-BABA test, in admixr. We tested for gene flow between both North African and Judean date palms individually (test sample) and two wild relative species, namely P. theophrasti and P. sylvestris. We used both P. reclinata and P. canariensis as outgroup species following our tree reconstruction (SI Appendix, Fig. S6B). In this and subsequent introgression analyses, significance was tested with the block jackknife with each linkage group and unplaced contig treated as a separate block. The SE was used to calculate a standard score (z-score = statistics/SE), and we used an absolute z-score >2 to assess statistically significant results. The f3-statistic was used to test whether sample C is a mixture of two populations A and B (47), with significance tested as above. To infer the proportion of ancestry in the North African and ancient Judean date palms from West Asian date palms and P. theophrasti, we calculated the f4-ratio statistics as described in Patterson et al. (47). Significance was tested as above.
Chromosome Painting and Introgression Tract Lengths.
We performed local ancestry estimation (chromosome painting) with the software RFMix (51) v2.03-r0, restricting our analysis to nonadmixed samples and to the 18 linkage groups (see above). We first phased the VCF with Beagle (81) v. 4.1 using the genotype field (gt option). We then ran RFMix using default parameters and with query samples being the Judean and North African date palms, while the reference samples were West Asian date palms and P. theophrasti. In the absence of a genetic map, we assumed that 1 cM = 1 Mb. We then identified all P. theophrasti tracts in the query genomes along with their length. Statistical tests to compare Judean and North African date palms were performed using either Student’s tests (t.test in R) if data were normally distributed (as evidenced by Anderson–Darling tests for normality, ad.test) or Wilcoxon rank sum tests (wilcox.test in R).
Genotyping of the Fruit Color and Sugar Composition Loci.
We genotyped the Judean date palms at key loci involved in fruit color and sugar content identified by Hazzouri et al. (29, 39). Fruit color varies between yellow and red at the khalal stage and has been mapped to the VIRESCENS locus identified on linkage group four (39). Three alleles have been proposed (39): VIR+ (wild-type allele), VIRIM, and VIRIM and virsaf. To genotype the VIRIM allele, we manually examined reads that map to the breakpoint between exon 3 and the retrotransposon insertion that characterizes this allele. The virsaf allele is defined by an SNP at position 24,051,180 (39). Sugar composition is associated with a deletion that spans ∼40 kb at ∼2.467 to 2.507 Mb of linkage group 14, a region with multiple invertase genes (39). To genotype this locus, we identified, in each Judean date palm, whether they have a homozygote deletion in the region as evidenced by an absence of mapped reads.
Supplementary Material
Acknowledgments
We thank Dr. Elaine Solowey of the Arava Institute of Environmental Sciences, Kibbutz Ketura, Israel for supplying leaf samples of the seven germinated ancient date palms; Marc Arnoux, Nizar Drou, Michael Dhar, and the New York University Abu Dhabi Bioinformatics Core for assistance with DNA sequencing and bioinformatic analyses; and Joel Malek for introducing the collaborators to each other. We are grateful to Yann Bourgeois, Nathan Wales, Christopher West, Rafal Gutaker, and Jae Young Choi for insightful discussions; to Martin Petr for support in the use of the R package admixr; and to Ramin Rahni for graphical support. This study was supported in parts by grants from the New York University Abu Dhabi Research Institute, the US NSF Plant Genome Research Program, and the Zegar Family Foundation to M.D.P.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025337118/-/DCSupplemental.
Data Availability
Sources for all downloaded data are stated in SI Appendix, Table S1. Sequencing reads for the seven Judean date palms whose genomes were resequenced for this study have been deposited in the Sequence Read Archive under BioProject accession number PRJNA681290. Methuselah: SAMN16948731; Hannah: SAMN16948732; Adam: SAMN16948733; Judith: SAMN16948734; Boaz: SAMN16948735; Jonah: SAMN16948736; Uriel: SAMN16948737. The custom Perl scripts are available in Dryad (https://doi.org/10.5061/dryad.kkwh70s3w).
References
- 1.Sankararaman S., et al., The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller W., et al., Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456, 387–390 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Gaunitz C., et al., Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 360, 111–114 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Ramos-Madrigal J., et al., Palaeogenomic insights into the origins of French grapevine diversity. Nat. Plants 5, 595–603 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kistler L., et al., Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 362, 1309–1313 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Pääbo S., et al., Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Fulton T. L., “Setting up an ancient DNA laboratory” in Ancient DNA: Methods and Protocols, Shapiro B., Hofreiter M., Eds. (Humana Press, NJ, 2012), pp. 1–11. [Google Scholar]
- 8.Carpenter M. L., et al., Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutaker R. M., Burbano H. A., Reinforcing plant evolutionary genomics using ancient DNA. Curr. Opin. Plant Biol. 36, 38–45 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Wales N., et al., The limits and potential of paleogenomic techniques for reconstructing grapevine domestication. J. Archaeol. Sci. 72, 57–70 (2016). [Google Scholar]
- 11.Scott M. F., et al., A 3,000-year-old Egyptian emmer wheat genome reveals dispersal and domestication history. Nat. Plants 5, 1120–1128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mascher M., et al., Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nat. Genet. 48, 1089–1093 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Fan L. J., et al., Ancient DNA sequences of rice from the lower Yangtze reveal significant genotypic divergence. Chin. Sci. Bull. 56, 3108–3113 (2011). [Google Scholar]
- 14.Wales N., et al., Ancient DNA reveals the timing and persistence of organellar genetic bottlenecks over 3,000 years of sunflower domestication and improvement. Evol. Appl. 12, 38–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith C. C. R., et al., Genetics of alternative splicing evolution during sunflower domestication. Proc. Natl. Acad. Sci. U.S.A. 115, 6768–6773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollmann B., Jacomet S., Schlumbaum A., Morphological and genetic studies of waterlogged Prunus species from the Roman vicus Tasgetium (Eschenz, Switzerland). J. Archaeol. Sci. 32, 1471–1480 (2005). [Google Scholar]
- 17.Willerslev E., et al., Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Minnis P. E., Seeds in archaeological sites: Sources and some interpretive problems. Am. Antiq. 46, 143–152 (1981). [Google Scholar]
- 19.Yashina S., et al., Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost. Proc. Natl. Acad. Sci. U.S.A. 109, 4008–4013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen-Miller J., Mudgett M. B., Schopf J. W., Clarke S., Berger R., Exceptional seed longevity and robust growth: Ancient sacred lotus from China. Am. J. Bot. 82, 1367–1380 (1995). [Google Scholar]
- 21.Sallon S., et al., Germination, genetics, and growth of an ancient date seed. Science 320, 1464 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Sallon S., et al., Origins and insights into the historic Judean date palm based on genetic analysis of germinated ancient seeds and morphometric studies. Sci. Adv. 6, eaax0384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tengberg M., Beginnings and early history of date palm garden cultivation in the Middle East. J. Arid Environ. 86, 139–147 (2012). [Google Scholar]
- 24.Gros-Balthazard M., Hazzouri K. M., Flowers J. M., Genomic insights into date palm origins. Genes (Basel) 9, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss E., Beginnings of fruit growing in the Old World – two generations later. Isr. J. Plant Sci. 62, 75–85 (2015). [Google Scholar]
- 26.Tengberg M., Newton C., “Origine et évolution de la phéniciculture au Moyen-Orient et en Égypte” in Des Fruits d’ici et d’ailleurs : Regards Sur l’histoire de Quelques Fruits Consommés En Europe, Ruas M.-P., Ed. (Éditions Omniscience, 2016), pp. 83–105. [Google Scholar]
- 27.Gros-Balthazard M., et al., On the necessity of combining ethnobotany and genetics to assess agrobiodiversity and its evolution in crops: A case study on date palms (Phoenix dactylifera L.) in Siwa Oasis, Egypt. Evol. Appl. 13, 1818–1840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goor A., The history of the date through the ages in the Holy Land. Econ. Bot. 21, 320–340 (1967). [Google Scholar]
- 29.Hazzouri K. M., et al., Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun. 6, 8824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flowers J. M., et al., Cross-species hybridization and the origin of North African date palms. Proc. Natl. Acad. Sci. U.S.A. 116, 1651–1658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gros-Balthazard M., et al., The discovery of wild date palms in oman reveals a complex domestication history involving centers in the Middle East and Africa. Curr. Biol. 27, 2211–2218.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zehdi-Azouzi S., et al., Genetic structure of the date palm (Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Ann. Bot. 116, 101–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arabnezhad H., Bahar M., Mohammadi H. R., Latifian M., Development, characterization and use of microsatellite markers for germplasm analysis in date palm (Phoenix dactylifera L.). Sci. Hortic. (Amsterdam) 134, 150–156 (2012). [Google Scholar]
- 34.Mathew L. S., et al., A Genome-wide survey of date palm cultivars supports two major subpopulations in Phoenix dactylifera. G3 (Bethesda) 5, 1429–1438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gros-Balthazard M., Flowers J. M., A brief history of the origin of domesticated date palms. arXiv [Preprint] 2020. 2012.00281 [q-bio.E] (Accessed 1 December 2020).
- 36.Van der Veen M., Westley B., “Paleoeconomic studies” in The Archaeology of Fazzan. Vol. 3: Excavations of C. M. Daniels, Mattingly D. J., Ed. (Society for Libyan Studies, 2010), pp. 489–519. [Google Scholar]
- 37.Fuller D. Q., Pelling R., “Plant economy: Archaeobotanical studies” in Volubilis Après Rome, Fentress E., Limane H., Eds. (Brill, 2019), pp. 349–368. [Google Scholar]
- 38.Gros-Balthazard M., et al., Short read sequencing (illumina) of seven Judean date palms grown from seeds dated to ∼2,000 years. Sequencing Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA681290. Deposited 28 November 2020.
- 39.Hazzouri K. M., et al., Genome-wide association mapping of date palm fruit traits. Nat. Commun. 10, 4680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterworth W. M., Bray C. M., West C. E., Seeds and the art of genome maintenance. Front. Plant Sci. 10, 706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanvah S., et al., Oxidation of DNA: Damage to nucleobases. Acc. Chem. Res. 43, 280–287 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A., 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-—T and A—C substitutions. J. Biol. Chem. 267, 166–172 (1992). [PubMed] [Google Scholar]
- 43.Waterworth W. M., Drury G. E., Bray C. M., West C. E., Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 192, 805–822 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boydak M., A new subspecies of Phoenix theophrasti Greuter (Phoenix theophrasti Greuter subsp. golkoyana Boydak) from Turkey. Forestist 69, 133–144 (2019). [Google Scholar]
- 46.Fuller D. Q., Long and attenuated: Comparative trends in the domestication of tree fruits. Veg. Hist. Archaeobot. 27, 165–176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson N., et al., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green R. E., et al., A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter B. M., Admixture, population structure, and f-statistics. Genetics 202, 1485–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang M., Nielsen R., The lengths of admixture tracts. Genetics 197, 953–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maples B. K., Gravel S., Kenny E. E., Bustamante C. D., RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dowson V. H. W., Aten A., Dates, Handling, Processing and Packing (Food and Agriculture Organization, Rome, 1962). [Google Scholar]
- 53.Alizadeh E., Sanche L., Role of humidity and oxygen level on damage to DNA induced by soft X-rays and low-energy electrons. J. Phys. Chem. C Nanomater. Interfaces 117, 22445–22453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Probert R. J., Daws M. I., Hay F. R., Ecological correlates of ex situ seed longevity: A comparative study on 195 species. Ann. Bot. 104, 57–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frumkin A., Bar-Yosef O., Schwarcz H. P., Possible paleohydrologic and paleoclimatic effects on hominin migration and occupation of the Levantine Middle Paleolithic. J. Hum. Evol. 60, 437–451 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Zeder M. A., The origins of agriculture in the Near East. Curr. Anthropol. 52, S221–S235 (2011). [Google Scholar]
- 57.Nesbitt M., Samuel D., “From staple crop to extinction? The archaeology and history of the hulled wheats” in Proceedings of the First International Workshop on Hulled Wheats: 21-22 July 1995, Padulosi S., Hammer K., Heller J, Eds. (International Plant Genetic Resources Institute (IPGRI), Castelvecchio Pascoli, Tuscany, Italy, 1996), pp. 41–100. [Google Scholar]
- 58.Zohary D., Spiegel-Roy P., Beginnings of fruit growing in the old world. Science 187, 319–327 (1975). [DOI] [PubMed] [Google Scholar]
- 59.Liphschitz N., Bonani G., Wild and cultivated date palm (Phoenix dactylifera) from Qumran Cave 24. Tel Aviv 28, 305–309 (2001). [Google Scholar]
- 60.Perez-Escobar O. A., et al., Archaeogenomics of a ∼2,100-year-old Egyptian leaf provides a new timestamp on date palm domestication. BioRxiv [Preprint] (2020). 10.1101/2020.11.26.400408 (Accessed 1 December 2020). [DOI]
- 61.Terral J. F., et al., Insights into the historical biogeography of the date palm (Phoenix dactylifera L.) using geometric morphometry of modern and ancient seeds. J. Biogeogr. 39, 929–941 (2012). [Google Scholar]
- 62.Kislev M. E., Hartmann A., Galili E., Archaeobotanical and archaeoentomological evidence from a well at Atlit-Yam indicates colder, more humid climate on the Israeli coast during the PPNC period. J. Archaeol. Sci. 31, 1301–1310 (2004). [Google Scholar]
- 63.Abeli T., et al., Ex situ collections and their potential for the restoration of extinct plants. Conserv. Biol. 34, 303–313 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Franks S. J., Hamann E., Weis A. E., Using the resurrection approach to understand contemporary evolution in changing environments. Evol. Appl. 11, 17–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leino M. W., Edqvist J., Germination of 151-year old Acacia spp. seeds. Genet. Resour. Crop Evol. 57, 741–746 (2010). [Google Scholar]
- 66.Bu Z. J., et al., The Methuselah of plant diaspores: Sphagnum spores can survive in nature for centuries. New Phytol. 214, 1398–1402 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Özgen M., et al., Analysis of ancient DNA from in vitro grown tissues of 1600-year-old seeds revealed the species as Anagyris foetida. Seed Sci. Res. 22, 279–286 (2012). [Google Scholar]
- 68.Härnström K., Ellegaard M., Andersen T. J., Godhe A., Hundred years of genetic structure in a sediment revived diatom population. Proc. Natl. Acad. Sci. U.S.A. 108, 4252–4257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delebecq G., Schmidt S., Ehrhold A., Latimier M., Siano R., Revival of ancient marine dinoflagellates using molecular biostimulation. J. Phycol. 56, 1077–1089 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Frisch D., et al., A millennial-scale chronicle of evolutionary responses to cultural eutrophication in Daphnia. Ecol. Lett. 17, 360–368 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). 1303.3997 (Accessed 1 December 2020).
- 73.McKenna A., et al., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2015). [Google Scholar]
- 75.Luu K., Bazin E., Blum M. G. B., pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 17, 67–77 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Duforet-Frebourg N., Bazin E., Blum M. G. B., Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol. Biol. Evol. 31, 2483–2495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duforet-Frebourg N., Luu K., Laval G., Bazin E., Blum M. G. B., Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol. Biol. Evol. 33, 1082–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danecek P.et al.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petr M., Vernot B., Kelso J., admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 35, 3194–3195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sources for all downloaded data are stated in SI Appendix, Table S1. Sequencing reads for the seven Judean date palms whose genomes were resequenced for this study have been deposited in the Sequence Read Archive under BioProject accession number PRJNA681290. Methuselah: SAMN16948731; Hannah: SAMN16948732; Adam: SAMN16948733; Judith: SAMN16948734; Boaz: SAMN16948735; Jonah: SAMN16948736; Uriel: SAMN16948737. The custom Perl scripts are available in Dryad (https://doi.org/10.5061/dryad.kkwh70s3w).