Significance
Small CAG (sCAG) RNAs are neurotoxic, but their role in polyglutamine degeneration remains to be fully elucidated. We observed that cellular expression of sCAGs is sufficient to induce neuronal DNA damage and discovered that the transcript level of NUDT16 was reduced in HD models. The NUDT16 protein has previously been linked to the DNA damage pathway. At the structural level, sCAGs form double-stranded CAG–CUG heteroduplex RNA with NUDT16 transcript which led to its gene silencing. We showed that the bisamidinium-based compound DB213 specifically interacts with duplex CAG RNA; consequently, both NUDT16 expression and DNA damage were rescued in HD mice. Our findings describe a pathogenic pathway that induces DNA damage in polyglutamine degeneration and demonstrate its therapeutic potential.
Keywords: DNA damage, Huntington’s disease, NUDT16, R6/2
Abstract
DNA damage plays a central role in the cellular pathogenesis of polyglutamine (polyQ) diseases, including Huntington’s disease (HD). In this study, we showed that the expression of untranslatable expanded CAG RNA per se induced the cellular DNA damage response pathway. By means of RNA sequencing (RNA-seq), we found that expression of the Nudix hydrolase 16 (NUDT16) gene was down-regulated in mutant CAG RNA-expressing cells. The loss of NUDT16 function results in a misincorporation of damaging nucleotides into DNAs and leads to DNA damage. We showed that small CAG (sCAG) RNAs, species generated from expanded CAG transcripts, hybridize with CUG-containing NUDT16 mRNA and form a CAG-CUG RNA heteroduplex, resulting in gene silencing of NUDT16 and leading to the DNA damage and cellular apoptosis. These results were further validated using expanded CAG RNA-expressing mouse primary neurons and in vivo R6/2 HD transgenic mice. Moreover, we identified a bisamidinium compound, DB213, that interacts specifically with the major groove of the CAG RNA homoduplex and disfavors the CAG-CUG heteroduplex formation. This action subsequently mitigated RNA-induced silencing complex (RISC)-dependent NUDT16 silencing in both in vitro cell and in vivo mouse disease models. After DB213 treatment, DNA damage, apoptosis, and locomotor defects were rescued in HD mice. This work establishes NUDT16 deficiency by CAG repeat RNAs as a pathogenic mechanism of polyQ diseases and as a potential therapeutic direction for HD and other polyQ diseases.
Polyglutamine (PolyQ) diseases are a group of disorders caused by the expansion of CAG triplet repeat sequences in the disease loci. Nine neurodegenerative diseases fall into this group (1). DNA damage has been reported in polyQ diseases (2), such as Huntington’s disease (HD) (3), and higher levels of DNA strand breaks are observed in polyQ patients (4). It has been reported that the polynucleotide kinase 3′-phosphatase, a major DNA strand break repair enzyme, is recruited to polyQ protein aggregates (5) and that its enzymatic activity is inhibited by the expanded mutant polyQ disease protein, resulting in an accumulation of DNA damage (4). In addition, DNA damage response was found to be activated by expanded CAG tracts (6). The expanded polyQ protein can induce neuronal apoptosis through the ataxia telangiectasia-mutated kinase (ATM)-p53 DNA damage response (DDR) pathway (5, 7). The expanded polyQ protein has also been shown to interact with ATM kinase, leading to a delay in the nuclear translocation of ATM (3). This causes an impairment of DDR in HD. Furthermore, DNA double-strand breaks were also detected in HD mice (8). All of the abovementioned observations clearly indicated that the integrity of DDR mechanisms is compromised in polyQ diseases.
In addition to the mutant polyQ proteins, the RNA transcripts that carry an expanded CAG repeat are also neurotoxic (1). Interestingly, the simultaneous transcription of complementary CAG and CTG repeat sequences reduces the repeat stability of DNA and causes cell death through the ATM and Rad3-related (ATR) DNA damage pathway (9). Other than inducing DNA instability, expanded CAG repeat-containing RNA is also known to be the target of the ribonuclease Dicer (10), and the mutant transcripts serve as substrates for small CAG (sCAG) RNA production. The sCAG RNAs per se have been reported to confer toxicity (11) through the induction of cellular gene silencing (10, 11). Both Dicer and Argonaute 2 (AGO2), major components of the RNA interference pathway, have been reported to be involved in sCAG RNA toxicity in polyQ diseases (11).
Nucleoside diphosphate linked moiety X (Nudix) hydrolases (12, 13) are enzymes that are responsible for cleaving a wide range of cellular substrates, including nucleoside triphosphates. As a member of the Nudix hydrolase superfamily, the nucleoside diphosphate linked moiety X-type motif 16 (NUDT16) protein was first identified as a nuclear nucleoside diphosphatase and serves as an RNA decapping enzyme to regulate cellular RNA stability (14). Later, NUDT16 was found to possess deoxyinosine diphosphatase activity (15). This nuclear-localized enzyme hydrolyzes deoxyinosine nucleotides through which unwanted nucleotides are eliminated from the cellular nucleotide pools. The incorporation of nonstandard nucleotides, such as deaminated purine, into DNA strands can cause genomic mutations (16). In line with this function of NUDT16, knockdown of NUDT16 expression causes DNA strand breaks in cells (16).
Although the transcription of expanded CAG RNAs has been reported to cause DNA damage and induce apoptosis (9), how mutant CAG transcripts are involved in these processes is not entirely clear. In the current study, we found that the expression level of NUDT16 is down-regulated in polyQ disease models, which promotes DNA damage and apoptosis. Mechanistically, NUDT16 transcripts, which bear naturally occurring CUG sequences in their messenger RNA (mRNA), form the RNA heteroduplex with neurotoxic sCAG RNAs, thus causing NUDT16 silencing. Anticipating that disrupting the CUG-CAG heteroduplex formation could be a therapeutic approach for suppressing CAG RNA-induced DNA damage and apoptosis, we discovered a bisamidinium-based small molecule, DB213 (17), which is capable of promoting the formation of a CAG-CAG RNA homoduplex, thus disfavoring CUG-CAG heteroduplex formation and preventing CUG sequence-containing NUDT16 transcripts from being degraded through the RNA-induced silencing complex (RISC)-dependent gene silencing mechanism. Using NMR spectroscopy, we showed that DB213 interacts with the major groove in the CAG RNA homoduplex. Distinct from other published compounds (e.g., refs. 18 and 19) that mainly target A·A mismatches of the CAG RNA duplex, DB213 recognizes the overall major groove structure of duplex CAG RNA, which is believed to confer a more specific inhibition on CAG RNA targets. Neuronal synaptic pathology has been reported in HD mice (20). We showed that DB213 treatment could rescue CAG repeat RNA-induced synapse loss in mouse primary neurons. Most importantly, DB213 could directly reach the rodent brain through brain-targeted intranasal administration without unnecessary systemic exposure (21–23). After DB213 treatment, DNA damage, apoptosis, and locomotor defects that we observed in a mouse model of polyQ diseases were mitigated. Our study unveiled a CAG RNA-mediated DNA damage-inducing pathway that involves the silencing of NUDT16 and opens a therapeutic strategy to target CAG RNA toxicity.
Results
Expanded CAG RNA Triggers DNA Damage and Apoptosis.
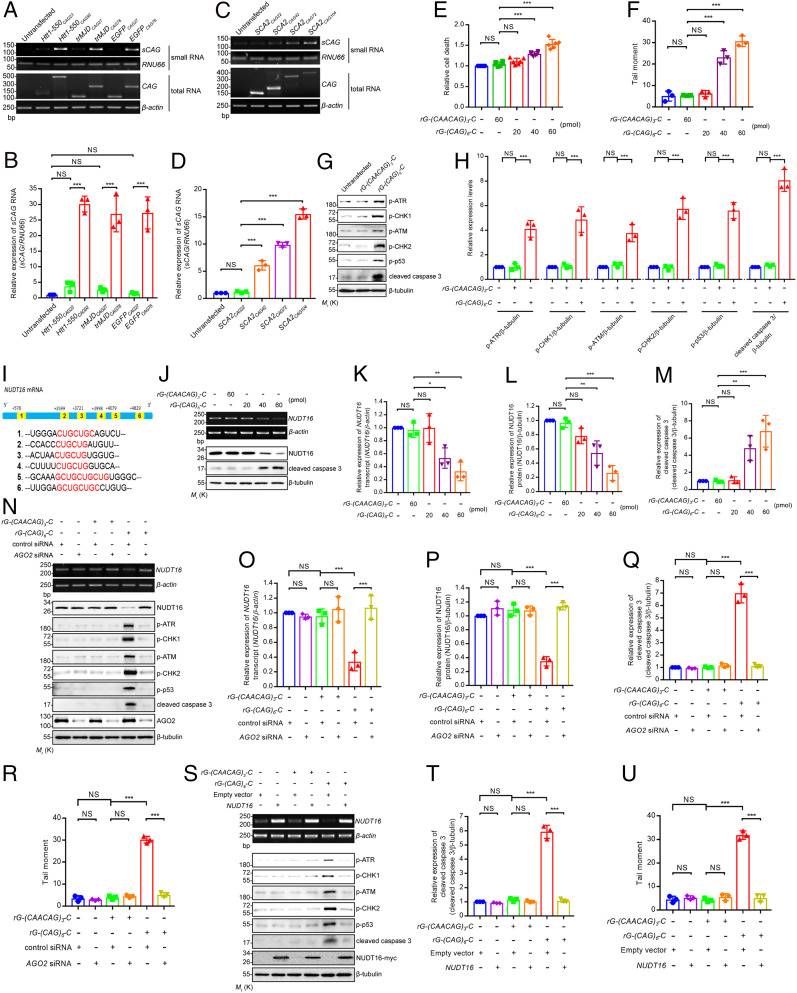
DNA damage has been reported in polyQ diseases (2). We conducted single-cell electrophoresis, also referred to as the “comet assay,” to examine whether simply expressing non-translatable expanded CAG transcripts would induce DNA damage in SK-N-MC neuroblastoma cells. The CAG repeat sequences were placed within the untranslated region of the expression construct to avoid polyQ protein translation. Our data showed that the cells that expressed such non-translatable expanded CAG RNA (EGFPCAG78) induced higher levels of DNA damage than those that expressed the EGFPCAG27 unexpanded control and untransfected cells (Fig. 1 A and B). By means of quantitative real-time PCR, we showed that the expression level of EGFPCAG27 and EGFPCAG78 CAG RNA was comparable in transfected SK-N-MC cells (Fig. 1C). We further performed Western blotting (Fig. 1D) and immunostaining (Fig. 1E) to determine whether the CAG repeats of EGFPCAG27/78 constructs were translated in our cell model. Our data showed that these CAG repeats, which appeared in the 3′-untranslated region of the EGFP constructs, were not translated (Fig. 1 D and E). In contrast, polyQ proteins were detected in cells transfected with the EGFP Htt1-550CAG23/89 constructs whose CAG repeats were located in the opening reading frame (Fig. 1 D and E). We next determined whether the cellular DNA damage triggered by CAG RNA would lead to the activation of DDR pathways and found that expanded CAG RNA induced the phosphorylation of all ATR, ATM, and checkpoint kinases (CHKs) 1 and 2 (Fig. 1 F and G). The total level of these proteins remained unaltered (SI Appendix, Fig. S1A). These protein kinases are involved in both single-strand (ATR and CHK1) and double-strand (ATM and CHK2) DDR pathways (24). We further detected the phosphorylation of p53 and an enhanced level of caspase 3 cleavage (Fig. 1 F and G) in cells transfected with expanded EGFPCAG78 RNA. Consistent with previous findings (11, 25), we observed enhanced cell death, using the lactate dehydrogenase (LDH) assay, in EGFPCAG78-expressing SK-N-MC cells when compared with the unexpanded EGFPCAG27-expressing cells and the untransfected control (Fig. 1H).
Fig. 1.
Expanded CAG repeat RNA induces DNA damage via down-regulating the expression levels of NUDT16. (A) DNA damage was observed in the SK-N-MC cells transfected with EGFPCAG78 construct. The red arrows indicate cells with DNA strand breaks. (B) Quantification of A (n = 3). (C) Real-time PCR showed that expression levels of CAG RNA in SK-N-MC cells transfected with EGFPCAG27/78 constructs were comparable (n = 3; NS indicates not significant, two-tailed unpaired Student’s t test). No translated polyQ protein was detected in EGFPCAG27/78-expressing SK-N-MC cells by Western blotting (D) and immunostaining (E) (n = 3). (Scale bars, 10 μm.) (F) Increased levels of phosphorylated ATR (p-ATR), CHK1 (p-CHK1), ATM (p-ATM), CHK2 (p-CHK2), p53 (p-p53), and cleaved caspase 3 proteins were detected in EGFPCAG78-expressing SK-N-MC cells. (G) Quantification of F) (n = 3). (H) LDH assay demonstrates cell death was induced in the SK-N-MC cells transfected with EGFPCAG78 construct (n = 6). (I) Heat map showing the log2 (fold change) values, with reference to the untransfected control, of 35 DEGs identified from RNA-seq analysis (n = 2). (J) Comet assay showed NUDT16 knockdown induced DNA damage in SK-N-MC cells (n = 3; ***P < 0.001, two-tailed unpaired Student’s t test). (K) Knockdown of NUDT16 expression in SK-N-MC cells induced the cleavage of caspase 3 in a dose-dependent manner. (L) Quantification of K (n = 3). (M) EGFPCAG78-induced DNA damage could be rescued by NUDT16 overexpression in SK-N-MC cells (n = 3). (N) Overexpression of NUDT16 reduced the cleavage of caspase 3 in EGFPCAG78-expressing SK-N-MC cells. (O) Quantification of N (n = 3). Error bars represent ± SD. Unless otherwise specified, statistical analysis was performed using one-way ANOVA. NS indicates not significant, **P < 0.01, ***P < 0.001. Beta-actin or beta-tubulin was used as loading control. Only representative images, gels, and blots are shown.
The NUDT16 Gene Is Involved in CAG RNA-Induced DNA Damage and Apoptosis.
We next conducted RNA sequencing (RNA-seq) on our SK-N-MC CAG RNA toxicity-only cell model to determine the genes that are involved in CAG RNA-associated cellular DNA damage and identified 207 differentially expressed genes (DEGs) between the normal (CAG27) and disease (CAG78) conditions (SI Appendix, Fig. S1 D and E). We further validated 35 DEGs, among which 18 were up-regulated and 17 were down-regulated in CAG78 (Fig. 1I). We examined whether any of the 18 up-regulated DEGs were involved in CAG RNA-mediated cellular DNA damage using a small interfering RNA (siRNA) approach. In our EGFPCAG78-expressing SK-N-MC cell model (Fig. 1A), none of the DEGs whose expression levels were knocked down could lead to a satisfactory rescue of DNA damage (SI Appendix, Fig. S1 F and G). We thus considered that none of these DEGs play a major role in CAG RNA-induced cellular DNA damage. For the 17 down-regulated DEGs, we individually knocked down their expression in SK-N-MC cells and found that only the silencing of NUDT16 expression caused a striking DNA damage phenotype (a 6.6-fold increase when compared to the control siRNA treatment; Fig. 1J and SI Appendix, Fig. S1 H and I). Our findings are consistent with those in the literature suggesting that NUDT16 functional impairment causes DNA breakage (16). We further demonstrated that NUDT16 knockdown is capable of inducing caspase 3 cleavage in an siRNA dose-dependent manner (Fig. 1 K and L). In addition, the overexpression of NUDT16 in EGFPCAG78-expressing SK-N-MC cells was sufficient to inhibit CAG RNA-induced DNA damage (Fig. 1M) and caspase 3 cleavage (Fig. 1 N and O). Our findings clearly indicate the involvement of NUDT16 in CAG RNA toxicity.
To confirm the down-regulation of NUDT16 in polyQ degeneration, we examined its transcript and protein levels in the R6/2 HD transgenic mouse model (26). Consistent with our RNA-seq analysis (Fig. 1I) and validation in EGFPCAG78-expressing SK-N-MC cells (SI Appendix, Fig. S2 A–C), NUDT16 expression was reduced at both the RNA and protein levels in R6/2 mouse brains (Fig. 2 A–C). The expression of NUDT16 has previously been reported in the human brain (16). In adult mice, Nudt16 expression was detected in numerous brain regions, including the striatum and hippocampus (SI Appendix, Fig. S2D) (27). This corresponds well with the brain atrophy in the cortex, striatum, and hippocampus reported in R6/2 mice (28). Because R6/2 mice produce both mutant CAG RNA transcripts and the polyQ-Htt protein, we next determined the effect of toxic CAG RNA on NUDT16 expression in wild-type mouse primary cortical neurons. When neurons were transfected with the non-translatable expanded CAG expression construct EGFPCAG78, NUDT16 expression was reduced compared with that in the unexpanded EGFPCAG27 and untransfected controls (Fig. 2 D–F). We further observed that neurons expressing the EGFPCAG78 transcripts displayed a dramatically reduced puncta number of neuronal synaptic markers Synapsin I (Fig. 2 G and H), Bassoon (Fig. 2 I and J), and Homer1 (Fig. 2 K and L). Synapsin I is a neuronal-specific phosphoprotein which mediates trafficking of synaptic vesicles and regulates neurotransmitter release (29). Bassoon and Homer1 are specific structural components of the pre- and postsynaptic machineries, respectively. Reduced expression of these markers (Fig. 2 G–L) suggested that EGFPCAG78 RNA compromised synaptic activities on both pre- and postsynaptic sides. We further examined neurite morphologies. The expression of EGFPCAG78 transcripts caused extensive neurite loss, including all primary (Fig. 2M), secondary (Fig. 2N), and tertiary neurites (Fig. 2O). Taken together, CAG RNA reduces NUDT16 level and causes synaptic defects in neurons.
Fig. 2.
Expanded CAG repeat RNA induces neurites and synapse loss via down-regulating the expression levels of NUDT16 in mouse primary neurons. Both RNA and the protein levels of NUDT16 were decreased in (A) R6/2 mouse brains of 11-wk-old and (D) EGFPCAG78-transfected mouse primary cortical neurons. (B and C) Quantification of A (n = 6, ***P < 0.001, two-tailed unpaired Student’s t test). (E and F) Quantification of D (n = 3). (G) Representative images of Synapsin I staining of mouse primary neurons transfected with EGFPCAG constructs. (H) Quantification of Synapsin I puncta per 100 μm of MAP2- or EGFP-positive neurites. Synapsin I puncta were significantly decreased in EGFPCAG78-expressing neurons (n = 3). (I) Representative images of Bassoon staining of mouse primary cortical neurons. (J) Quantification of Bassoon puncta per 100 μm of MAP2- or EGFP-positive neurites. EGFPCAG78 expression dramatically reduced Bassoon puncta in neurons (n = 3). (K) Representative images of Homer1 staining of mouse primary cortical neurons. (L) Quantification of Homer1 puncta per 100 μm of MAP2- or EGFP-positive neurites. Homer1 expression was significantly decreased upon EGFPCAG78 transfection of neurons (n = 3). Both primary (M), secondary (N), and tertiary (O) neurites in EGFPCAG78-transfected neurons were diminished (n = 3). For the quantification, 10 transfected neurons per replicate were used. (Scale bars, 100 μm.) Error bars represent ± SD. Unless otherwise specified, statistical analysis was performed using one-way ANOVA. NS indicates not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Beta-actin or beta-tubulin was used as loading control. Only representative images, gels, and blots are shown.
Small CAG RNAs Induce Gene Silencing by Targeting CUG Motifs in NUDT16 Transcripts.
We then aimed to determine how NUDT16 expression is silenced in polyQ diseases. Transcripts carrying expanded CAG repeats are known to be cleavage targets of the ribonuclease Dicer (10), and the resultant sCAG RNA products have been reported to confer neurotoxicity and induce apoptosis (11). We detected increased levels of sCAG RNA production in SK-N-MC cells transfected with various polyQ disease mutant constructs (HD and Machado–Joseph disease/spinocerebellar ataxia type 3 [MJD/SCA3]; Fig. 3 A and B). The production of sCAG RNAs was also detected in SK-N-MC cells transfected with the generic non-translatable expanded EGFPCAG78 construct (Fig. 3 A and B). In contrast, no significant sCAG RNA production could be detected in cells transfected with the respective control unexpanded CAG constructs (Fig. 3 A and B). We also conducted a small RNA-seq analysis on these cells and detected higher levels of sCAG RNA species in samples collected from EGFPCAG78-expressing SK-N-MC cells (SI Appendix, Fig. S2E). From cells transfected with a series of SCA type 2 (SCA2) mutant constructs of different CAG repeats, we further showed that the levels of sCAG RNAs produced positively correlated with the CAG repeat lengths of the SCA2 mutant constructs (Fig. 3 C and D).
Fig. 3.
Overexpression of NUDT16 suppresses the sCAG RNA-induced DNA damage and apoptosis. (A) Upon transfection of Htt1-550CAG92, trMJDCAG78, and EGFPCAG78 constructs, higher levels of sCAG (of ∼21 nucleotides) were observed. (B) Quantification of A (n = 3). (C) The level of sCAG detected positively correlated with the CAG repeats of the SCA2 constructs. (D) Quantification of C (n = 3). (E) Synthetic rG-(CAG)6-C RNA induced cell death dose dependently (n = 6). (F) DNA damage in rG-(CAG)6-C-transfected SK-N-MC cells was observed (n = 3). (G) Increased p-ATR, p-CHK1, p-ATM, p-CHK2, p-p53, and cleaved caspase 3 proteins were detected in rG-(CAG)6-C-transfected cells. (H) Quantification of G (n = 3). (I) The six CUG-rich regions of NUDT16 mRNA are highlighted in yellow, and the CUG sequences are marked in red. (J) rG-(CAG)6-C transfection decreased NUDT16 mRNA and protein levels and induced caspase 3 cleavage dose dependently. (K–M) Quantification of J (n = 3). (N) Knockdown of AGO2 expression restored rG-(CAG)6-C-induced down-regulation of NUDT16 and reduced the phosphorylation of ATR, CHK1, ATM, CHK2, and p53 and the cleavage of caspase 3 and rescued the rG-(CAG)6-C–induced DNA damage (R) (n = 3). (O–Q) Quantification of N (n = 3). (S) Overexpression of NUDT16 suppressed rG-(CAG)6-C–induced cleavage of caspase 3 and the phosphorylation of ATR, CHK1, ATM, and CHK2. (T) Quantification of S (n = 3). (U) Synthetic rG-(CAG)6-C–induced DNA damage was rescued upon the overexpression of NUDT16 (n = 3). All the experiments were performed in SK-N-MC cells. Error bars represent ± SD. Statistical analysis was performed using one-way ANOVA. NS indicates not significant, *P < 0.05, **P < 0.01, ***P < 0.001. RNU66, beta-actin, or beta-tubulin was used as loading control. Only representative gels and blots are shown.
To examine the cytotoxic effect of sCAG RNAs, we transfected cells with different concentrations of the synthetic sCAG RNA, rG-(CAG)6-C, and observed that they induced cell death in a dose-dependent manner (Fig. 3E). Synthetic rG-(CAACAG)3-C RNA, which carried interrupted CAG repeats, did not elicit cytotoxicity. Notably, CAA also codes for glutamine. This indicated that not any small RNA sequence confers cytotoxicity. We next performed a comet assay to determine whether sCAG RNAs could induce cellular DNA damage and found that cells transfected with sCAG RNAs showed higher levels of DNA damage than the untransfected control cells and cells transfected with the interrupted sCAG RNA control (Fig. 3F) and that the extent of cellular DNA damage was positively correlated with the amount of sCAG RNA introduced. We further showed that the synthetic sCAG RNA induced the phosphorylation of ATR, ATM, CHK1, and CHK2 (Fig. 3 G and H and SI Appendix, Fig. S2F) in the DDR pathways (24). We also detected the phosphorylation of p53 (Fig. 3 G and H) and an enhanced caspase 3 cleavage (Fig. 3 G and H) in cells transfected with the synthetic sCAG RNAs rG-(CAG)6-C. Our findings highlight that sCAG RNAs specifically confer cytotoxicity by triggering cellular DNA damage.
We identified several naturally occurring CTG repeats in the NUDT16 gene sequence, among which each of six such regions consist of at least six nucleotides of CTG repeats and are located within the transcribed region (Fig. 3I). Besides, we also detected distinct CTG-rich regions in the mouse Nudt16 gene (SI Appendix, Fig. S2G). When transcribed, these CUG sequences could be targets of sCAG RNAs generated from expanded mutant CAG transcripts (Fig. 3 A–D), which subsequently lead to NUDT16 gene silencing. As expected, the NUDT16 transcript level was reduced in cells transfected with synthetic sCAG RNA (Fig. 3 J–L), and the silencing effect was positively correlated with the amount of sCAG RNA used in the transfection as well as proportional to the extent of caspase 3 activation (Fig. 3 J and M). No such effects were observed in cells transfected with noncontinuous sCAG RNA rG-(CAACAG)3-C. Our findings thus indicated that synthetic sCAG RNA transfection per se is capable of inducing a specific down-regulation of NUDT16 and caspase activation.
Disruption of the Gene Silencing Complex Is Sufficient to Rescue sCAG/NUDT16 Cellular Toxicity.
Argonaute 2 (AGO2) plays a key role in sCAG RNA-mediated gene silencing (11). We observed that both the transcript and the protein levels of NUDT16 were restored after AGO2 knockdown (Fig. 3 N–P). This indicated that the down-regulation of NUDT16 expression is mediated through the RISC mechanism. In addition, the phosphorylation of DNA damage kinases (Fig. 3N), p53 (Fig. 3N), cellular DNA damage (Fig. 3R), and caspase 3 cleavage (Fig. 3 N and Q) were all restored in sCAG RNA-transfected cells after AGO2 knockdown. The total protein level of DNA damage kinases and p53 remained unaltered (SI Appendix, Fig. S2H). This clearly demonstrated that sCAG RNA-directed RISC gene silencing contributes to the down-regulation of NUDT16.
Next, we aimed to determine whether heterologous NUDT16 overexpression could rescue the sCAG RNA-induced hyperphosphorylation of DNA damage kinases (Fig. 3 G and H). Our results showed that the phosphorylation levels of the ATR, ATM, CHK1, and CHK2 kinases were uniformly restored to baseline when NUDT16 was heterologously overexpressed in sCAG RNA-expressing cells (Fig. 3S), as was cellular DNA damage (Fig. 3U) and caspase activation (Fig. 3 S and T). Overexpression of NUDT16 did not alter the total protein level of ATR, ATM, CHK1, CHK2, and p53 (SI Appendix, Fig. S2I).
DB213, a Bisamidinium Compound that Binds to the Major Groove of Homoduplex CAG RNA.
Our results indicated that targeting the RISC mechanism led to the rescue of sCAG RNA-mediated cellular DNA damage (Fig. 3 N–R). This prompted us to explore the possibility of developing a therapeutic approach to target NUDT16 silencing in CAG RNA toxicity. We previously reported a series of bisamidinium-based RNA groove binders as potential therapeutic agents for the treatment of myotonic dystrophy type I, a neuromuscular condition caused by expanded CUG RNA (30–32). We showed that DB213 (Fig. 4A and SI Appendix, Figs. S3 and S4), a weak CUG RNA binder (17), interacts with sCAG RNA at a KD = 3.8 µM (Fig. 4A). Under the same experimental conditions, no interaction could be detected between DB213 and the noncontinuous CAG control small RNA in which the CAG sequence is interrupted by CAA (Fig. 4A), which suggested that this compound is in favor of binding to continuous CAG RNA sequences. We have also observed an increase in the melting temperature of the sCAG RNA by ca. 7 °C in the presence of one equivalent DB213 (SI Appendix, Table S1), which indicates a higher thermodynamic stability of the sCAG RNA upon binding with DB213.
Fig. 4.
DB213 specifically interacts with sCAG RNA and suppresses DNA damage and cell death via releasing the CUG-rich NUDT16 transcripts from sCAG. (A) Isothermal titration calorimetric analysis shows DB213 specifically interacted with rGG-(CAG)6-CC but not rGG-(CAACAG)3-CC RNA (n = 3). (B) An overview of the superimposed five structures of DB213-sCAG complex (PDB ID: 7D12). (C) A working model for DB213 action. DB213 releases NUDT16 transcripts from the sCAG/NUDT16 heteroduplex, and this prevents NUDT16 degradation. (D, Top) NMR spectra showing the Watson–Crick imino H1/H3 signals of 0.1 µmol of sCAG homoduplex. (Middle) Mixture of 0.09 µmol of CUG and 0.1 µmol of sCAG, which formed 0.09 µmol sCAG/CUG heteroduplex and a tiny population of sCAG homoduplex. (Bottom) Titration of 1 µmol of DB213 to the mixture; peak intensities of H1/H3 signals from the sCAG/CUG heteroduplex decreased by ∼22% (green) (n = 3). The sCAG and CUG sequences were 5′-rGCAGCAGCAGC-3′ and 5′-CAAAG CUGCUGCUG-3′, respectively. (E) DB213 treatment dose-dependently restored NUDT16 expression in rG-(CAG)6-C–transfected cells. (F and G) Quantification of E (n = 3). (H) DB213 treatment suppressed rG-(CAG)6-C–induced cell death (n = 6). (I) The rG-(CAG)6-C–induced phosphorylation of ATR, CHK1, ATM, CHK2, and p53 and the cleavage of caspase 3 were diminished upon DB213 treatment. (J and K) Quantification of I (n = 3). (L) DNA damage induced by rG-(CAG)6-C was suppressed upon DB213 treatment (n = 3). DB213 treatment restored rG-(CAG)6-C–induced down-regulation of LTBP1 (M), MOGS (N), NDUFV1 (O), and QARS1 (P) transcriptional levels (n = 3). All cell-based experiments were carried out in SK-N-MC cells. Error bars represent ± SD. Statistical analysis was performed using one-way ANOVA. NS indicates not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Beta-actin or beta-tubulin was used as loading control. Only representative gels and blots are shown.
The binding mechanism of DB213 to the sCAG RNAs was studied in more details using high-resolution NMR spectroscopy (SI Appendix, Fig. S4 A–D). The NMR solution structures of the complex formed between DB213 and an sCAG hairpin which contains a UUCG loop and two CAG repeats in the stem were refined using Amber 18 based on 85 RNA intramolecular and 120 RNA-DB213 intermolecular proton–proton distance restraints derived from nuclear Overhauser effect (NOE) peak intensities (SI Appendix, Tables S2 and S3). The purpose of introducing an ultra-stable UUCG loop to the RNA is to facilitate the formation of an intramolecular hairpin of which the stem contains two consecutive CAG repeats. The stem region of our apo-form sCAG hairpin is comparable to the published intermolecular CAG homoduplex (Protein Data Bank [PDB] ID: 5VH7 (33); SI Appendix, Fig. S4A). Among the 100 refined sCAG-DB213 complex structures, five with the lowest total energies were selected as the final representative ensemble (Fig. 4B; PDB ID: 7D12), and their refinement statistics are shown in SI Appendix, Table S4. The superimposed five structures revealed that DB213 localizes at the major groove of the sCAG RNA and specifically interacts with A3·A18 and A6·A15 mismatches in a nearly symmetrical manner (Fig. 4B). The large chemical shift perturbations observed for A3, A6, A15, and A18 and their flanking residues upon DB213 binding, and NOEs between the methyl/methylene protons of DB213 and H8 proton of A3/A15, are also consistent with DB213 as a major groove binder (SI Appendix, Fig. S4 B–D). Hydrogen bonds formed between the imino protons H161/H464 of DB213 and A3 N7/A15 N7 of sCAG RNA at, respectively, a distance of 1.9 ± 0.1Å and 2.0 ± 0.1Å, and also between the amino protons H1/H7 of DB213 and G4 O6/G16 O6 of sCAG RNA, were identified (SI Appendix, Fig. S4E).
We next investigated whether the stabilization of sCAG by DB213 could inhibit the undesired hybridizations between the sCAG and CUG-containing NUDT16 mRNA and therefore impact NUDT16 gene silencing (Fig. 4C). Using 1H NMR spectroscopy, we observed that the addition of DB213 clearly reduced the guanine imino signals from C-G Watson–Crick base pairs (13.95 to 12.90 ppm) in the sCAG-CUG heteroduplex. A corresponding increase in the intensity of the G7 (12.47 ppm) signal from the sCAG homoduplex was observed, suggesting that DB213 has the ability to reduce the sCAG-CUG RNA heteroduplex by stabilizing the sCAG homoduplex (Fig. 4D). In line with this finding, our isothermal titration calorimetry (ITC) results showed that DB213 had a higher binding affinity to rGG-(CAG)6-CC (Kd = 3.8 µM) than rGG-(CAG)4-(CUG)2-CC (Kd = 19.2 µM) (SI Appendix, Fig. S4 F and G), suggesting that DB213 formed a stronger complex with the rCAG homoduplex over the rCAG/CUG heteroduplex.
In synthetic sCAG RNA-transfected SK-N-MC cells, DB213 treatment restored cellular NUDT16 expression, an effect that was observed to be DB213 dose dependent (Fig. 4 E–G). In addition, both cell death (Fig. 4H) and caspase 3 cleavage (Fig. 4 I and K) were suppressed in the DB213-treated cells. We further showed that DB213 treatment is capable of restoring the phosphorylation status of DNA damage kinases (Fig. 4I) and cellular DNA damage (Fig. 4L). The total protein level of DNA damage kinases and p53 remained unchanged (SI Appendix, Fig. S5A) upon the treatment of DB213.
In addition to NUDT16, we performed quantitative real-time PCR on the remaining 16 down-regulated DEGs (Fig. 1I) to determine whether they also demonstrated similar down-regulation in sCAG RNA-transfected SK-N-MC cells (SI Appendix, Fig. S5 C–N). Our results showed that the expression level of LTBP1 (Fig. 4M), MOGS (Fig. 4N), NDUFV1 (Fig. 4O), and QARS1 (Fig. 4P) was reduced. We further found that these four genes all harbor CUG-rich regions in their transcripts, and each of these regions constitutes at least six nucleotides of continuous CUG sequences (SI Appendix, Fig. S5B). The above observations suggested that sCAG RNA forms a heteroduplex with CUG sequences in general. Importantly, we discovered that DB213 treatment could restore the expression of all these four genes (Fig. 4 M–P).
To further investigate whether DB213 could affect the expression of endogenous CAG repeat-containing genes, we measured the transcript level of the spinocerebellar ataxia 17 (SCA17) CAG repeat disease gene TATA box binding protein (TBP) (34). Our quantitative real-time PCR data showed that DB213 treatment did not alter the mRNA level of TBP (SI Appendix, Fig. S6A). We also investigated the effect of DB213 on SK-N-MC cells transfected with non-translatable expanded CAG construct EGFPCAG78 (Fig. 1 A–H). This model produces sCAG RNAs from expanded CAG transcripts (Fig. 3 A and B). First, our quantitative real-time PCR analysis showed that the expression levels of CAG repeats were not affected by DB213 in EGFPCAG27/78-transfected SK-N-MC cells (SI Appendix, Fig. S6B). We next found that DB213 treatment did not alter sCAG RNA production in EGFPCAG78-transfected cells, even though the transcriptional and translational levels of NUDT16 were restored, and caspase 3 activation was suppressed after DB213 treatment (SI Appendix, Fig. S6 C–G). This demonstrated that the compound rescues NUDT16 gene silencing downstream to sCAG RNA production. DB213 treatment also suppressed DNA damage (SI Appendix, Fig. S6H) and cell death (SI Appendix, Fig. S6I) induced by EGFPCAG78 expression in these cells.
Both CAG RNA and the polyQ protein confer neurotoxicity in polyQ diseases. In addition to targeting CAG RNA toxicity, we aimed to determine whether DB213 exerts any modulatory effect on polyQ protein toxicity. To do this, we transfected SK-N-MC cells with a truncated version of the MJD/SCA3 disease constructs (trMJDCAG27/78), which produce both mutant RNA and protein, along with another MJD/SCA3 disease construct, which confers only protein toxicity. In contrast to trMJDCAG78, the trMJDCAA/G78 construct expressed transcripts that contain both CAG and CAA glutamine codons in its polyQ polymorphic region (35). It was previously reported that a disruption in the CAG continuity in the MJD transcript abolishes its RNA toxicity but that polyQ protein toxicity is retained (36). Because of the lack of RNA toxicity, cells transfected with the trMJDCAA/G78 construct only induced an intermediate level of cell death (SI Appendix, Fig. S6J) and cellular DNA damage (SI Appendix, Fig. S6 K and L) when compared with that of cells transfected with trMJDCAG78 (SI Appendix, Fig. S6M). We further showed that DB213 treatment could only partially rescue cell death induced by trMJDCAG78 expression (SI Appendix, Fig. S6O). The incomplete suppression could be explained by the fact that DB213 only suppresses CAG RNA but not polyQ protein toxicity. Importantly, DB213 treatment was unable to mitigate cell death induced by the trMJDCAA/G78 protein toxicity-only construct. We further found that the expression levels of the polyQ protein were not altered upon the treatment of DB213 in trMJDCAG27/78- and trMJDCAA/G78-transfected SK-N-MC cells (SI Appendix, Fig. S6N). Our results thus indicated that DB213 does not target polyQ protein toxicity.
Furthermore, when we treated mouse primary neurons expressing non-translatable expanded CAG construct with DB213, NUDT16 expression was restored at both RNA and protein levels (Fig. 5 A–C). Furthermore, DB213 treatment also rescued EGFPCAG78-induced loss of Synapsin I (Fig. 5 D and E), Bassoon (Fig. 5 F and G), and Homer1 puncta (Fig. 5 H and I and SI Appendix, Fig. S7). In addition, all the diminished primary (Fig. 5J), secondary (Fig. 5K), and tertiary neurites (Fig. 5L) in EGFPCAG78-transfected neurons were recovered upon the DB213 treatment. This confirmed the suppression effect of this compound in neurons. We also showed that DB213 did not confer an observable cytotoxic effect on rat primary neurons when administered up to micromolar concentration (SI Appendix, Fig. S6P).
Fig. 5.
DB213 rescues CAG repeat RNA-induced neurites and synapse loss via restoring the expression level of NUDT16 in mouse primary neurons. (A) Upon DB213 treatment, both the NUDT16 mRNA and protein levels were restored in EGFPCAG78-transfected mouse primary cortical neurons. (B and C) Quantification of A (n = 3). (D–I) Representative images of Synapsin I (D), Bassoon (F), and Homer1 (H) staining of EGFPCAG78-expressing neurons. Quantification of Synapsin I (E), Bassoon (G), and Homer1 (I) puncta per 100 μm of MAP2- or EGFP-positive neurites. DB213 treatment restored the number of Synapsin I, Bassoon, and Homer1 puncta in EGFPCAG78 neurons (n = 3). (J–L) The diminished primary (J), secondary (K), and tertiary (L) neurites in EGFPCAG78-transfected neurons were restored upon DB213 treatment (n = 3). For the quantification, 10 transfected neurons per replicate were used. (Scale bars, 100 μm.) Error bars represent ± SD. Statistical analysis was performed using one-way ANOVA. NS indicates not significant, **P < 0.01, ***P < 0.001. Beta-actin or beta-tubulin was used as loading control. Only representative images, gels, and blots are shown.
DB213 Mitigates Neurodegeneration and Locomotor Impairment in In Vivo Drosophila Expanded CAG RNA Toxicity Models.
We next employed Drosophila models of CAG RNA toxicity (36) to examine the therapeutic effect of DB213 in vivo. We conducted a deep pseudopupil assay on flies that expressed the non-translatable expanded CAG100 repeat transgene under the control of the gmr-GAL4 driver. We observed severe retinal degeneration, which was indicated by the deterioration of rhabdomere structure in the eyes of CAG100 adult flies (Fig. 6A). DB213 treatment on CAG100 RNA-expressing F1 animals was initiated at the larval stage, and the rescue effect, which peaked at 240 nM, was observed in a dose-dependent manner (Fig. 6 A and B). The expression of another two repeats expansion RNAs, CUG480 (30) and CGG90 (37), in the Drosophila eyes induced external eye degeneration (SI Appendix, Fig. S8 A and B). In contrast to CAG100, treating CUG480- and CGG90-expressing flies with DB213 of up to 480 nM did not yield any observable phenotypic suppression. This suggested that DB213’s rescuing effect exhibits sequence specificity toward CAG repeats in vivo. We next expressed RNA carrying 250 triplet CAG repeats (CAG250) in neuronal tissues using the Elav-GAL4 driver. Adult flies that pan-neurally expressed CAG250 RNA displayed climbing defects resulting from motor neuron damage (Fig. 6C). DB213 treatment initiated at the larval stage partially but statistically significantly rescued adult locomotor impairment (Fig. 6C). DB213 could be detected in adult fly heads (SI Appendix, Fig. S8C) by liquid chromatography tandem mass spectrometry analysis (38). The incomplete suppression effect of DB213 on the expanded CAG RNA phenotypes in our Drosophila model could be explained by the fact that drug administration ceased during the pupal stage because developing pupae are confined to a drug impermeable pupal case, whereas the expanded CAG RNA continued to express and elicit neurotoxicity during pupal development.
Fig. 6.
DB213 treatment restores retinal degeneration and locomotor defects in in vivo Drosophila models. (A) Pseudopupil assay showed that DB213 treatment restored retinal degeneration in DsRedCAG100 flies. (B) DB213 dose-dependently suppressed retinal degeneration in DsRedCAG100 flies (n = 3). (C) Adult fly climbing assay revealed that DB213 treatment mitigated locomotor defects in DsRedCAG250 flies in a dose-dependent manner (n = 3). Flies of genotypes: w; gmr::Gal4 UAS::DsRedCAG0/+; +/+, w; gmr::Gal4/+; UAS::DsRedCAG100/+, elav::Gal4/w; UAS::DsRedCAG0/+; +/+, and elav::Gal4/w; +/+; UAS::DsRedCAG250/+. Error bars represent ± SD. Statistical analysis was performed using one-way ANOVA. NS indicates not significant, **P < 0.01, ***P < 0.001. Only representative images are shown.
DB213 Rescues the Locomotor Defects and sCAG/NUDT16-Induced Apoptosis in an In Vivo HD Mouse Model.
We further examined the effect of DB213 on the R6/2 transgenic HD mouse model (26). The R6/2 mice displayed mild behavioral defects in weeks 7 and 9 in an open-field test, whereas a significantly more severe phenotype was observed at week 11 (Fig. 7A). In a rotarod test on locomotor coordination, the performance of these mice was found to be severely affected at 9 and 11 wk of age (Fig. 7B). The above findings confirmed that R6/2 mice displayed behavioral and locomotor phenotypes under our experimental settings. We observed that the NUDT16 protein level was progressively reduced with age, from 9 to 11 wk, in the R6/2 mouse brains (Fig. 7 C and D). Consistent with our cell-based data, activation of ATR and ATM DNA damage kinases as well as caspase 3 were observed (Fig.7 C and E–G) in R6/2 brains. Our data thus confirmed NUDT16 perturbation and DNA damage in HD mice in vivo. We previously demonstrated that in situ thermosensitive gel-assisted intranasal administration is an effective delivery route for brain-targeted delivery of DB213 in mice (22). The tolerance dose of DB213 was determined empirically (SI Appendix, Table S5). In open-field (Fig. 7 H and I) and rotarod (Fig. 7J) tests, a 4-wk daily treatment of 25 mg/kg DB213 in in situ thermosensitive gel formulation significantly rescued the locomotor defects of the R6/2 mice by week 11 (Fig. 7 I and J). At the molecular level, down-regulated NUDT16 protein expression was restored, and the downstream DNA damage-induced apoptotic pathway was suppressed after DB213 treatment (Fig. 7 K–O). Our findings provide in vivo evidence that DB213 has a positive effect on molecular and motor phenotypes in an HD mouse model.
Fig. 7.
DB213 treatment rescues the locomotor defects in the in vivo R6/2 HD transgenic mouse model. (A) Open-field assay demonstrated progressive spontaneous locomotor defects occurred with age in R6/2 mice (n = 6; error bars represent ± SEM). (B) Locomotor defect of R6/2 mice was also indicated via rotarod test (n = 6; error bars represent ± SEM). (C) NUDT16 protein was progressively decreased with age in the R6/2 mouse brains followed by the activation of p-ATR, p-ATM, and cleaved caspase 3. (D–G) Quantification of C (n = 3; error bars represent ± SD). (H) A brief diagram summary for the DB213 treatment scheme on the R6/2 mouse. Locomotor defects of the R6/2 mice were significantly rescued by DB213 treatment as measured via open-field (I) and rotarod (J) assays (n = 6; error bars represent ± SEM). (K) Upon DB213 treatment, the decreased protein level of NUDT16 was restored in R6/2 mouse brains, and the activation of p-ATR, p-ATM, and cleaved caspase 3 were suppressed. (L–O) Quantification of K (n = 6; error bars represent ± SD). Statistical analysis was performed using one-way ANOVA. NS indicates not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Beta-tubulin was used as loading control. Only representative blots are shown.
Discussion
The Role of NUDT16 in polyQ Diseases.
Previous studies have demonstrated that mutant polyQ proteins affect genome stability by impairing cellular DNA repair mechanisms (39–41). The current study unveiled how cellular CAG RNA species trigger DNA damage in polyQ diseases (SI Appendix, Fig. S9). Expanded mutant CAG transcripts are known targets of Dicer endonuclease for sCAG RNA production (10), and these toxic CAG RNA species are capable of triggering gene silencing (11). The NUDT16 gene was identified from our RNA-seq analysis as a down-regulated DEG in cells expressed with the expanded mutant CAG RNA. It is also the only gene whose function is related to cellular DNA damage and maintenance of genome stability (16). Interestingly, the DNA mismatch repair system protein MLH1 that had previously been reported to be involved in the cell growth arrest phenotype functionally interacts with NUDT16 (42). We showed that heterologous NUDT16 overexpression rescued CAG RNA-induced DNA damage (Fig. 1M). Apart from NUDT16, four other down-regulated DEGs that we recovered from our RNA-seq could also be mediated through the sCAG RNA-mediated RISC gene silencing mechanism (Fig. 4 M–P). It should be noted that heterologous NUDT16 overexpression largely suppressed caspase 3 activation (Fig. 1 N and O), which underlines NUDT16 silencing as the primary target of CAG RNA toxicity in polyQ diseases.
We showed that sCAG production positively correlates with the CAG repeat length (Fig. 3 C and D). Interestingly, the CAG repeat length but not the polyQ stretch protein has been reported to correlate with age at onset in HD (43). Furthermore, genes involved in DNA damage repair, such as MLH1, have been identified to be age-at-onset modifiers of HD (43); therefore, it would be of interest to further examine the sCAG RNA load and NUDT16 transcript levels in polyQ patients with different ages at disease onset and HD patients who have inherited age-at-onset modifiers of the DNA damage repair gene. Such investigations may lead to additional mechanistic insights into RNA toxicity and disease onset modifications.
DB213 as a Drug Candidate to Treat polyQ RNA Toxicity.
The ATM kinase is a treatment target along the DDR pathway. When rat primary neurons that expressed the mutant HD construct were treated with ATM kinase inhibitors, neurotoxicity could be rescued (44). Finding a blood–brain barrier–penetrable hit compound for modulating the DDR pathway would be ideal for counteracting DNA damages in polyQ diseases in general. Our group recently reported a bisamidinium compound, AQAMAN, that is capable of interacting with soluble forms of the polyQ protein and suppresses protein toxicity in both in vitro and in vivo environments (45). Within the past decade, some small molecules have been reported to have affinity toward CAG RNA sequences (46). Some of these CAG RNA-targeting molecules (e.g., refs. 18 and 19) recognize an A·A mismatch of duplex CAG RNA. Our discovery that DB213 is a strong binder to the major groove of sCAG duplex RNA is noteworthy. Although groove binding of bisamidinium compounds to RNA is not unprecedented, addressing complementarity in terms of structure and surface interactions to achieve a strong and specific binding to RNA is never trivial compared with the more common intercalation or hydrogen bonding strategies, not to mention the highly diverse and flexible structures of RNA that add additional challenges to the design of small molecule binders. Our NMR structural studies have revealed that DB213 stretches within the major groove of sCAG RNA and spans across two consecutive A·A mismatches, likely to maximize interactions. We have shown that it is the repeated CAG sequence and hence the specific groove structure of the sCAG RNA that DB213 targets. RNA of different sequences has therefore resulted in much weaker or insignificant binding, which can explain the relatively low cytotoxicity of the small molecule therapeutics. Through intranasal administration, DB213 is capable of targeting the brain (23). This further substantiates its potential as a drug candidate for targeting CAG RNA toxicity in polyQ diseases. An in vivo gene silencing effect has also been reported to be associated with expanded CUG RNA toxicity (47). In addition to rescuing cellular proteins that are recruited to RNA foci, it should be noted that small molecules that target CUG repeat RNAs (30) may mediate phenotypic suppression by promoting the formation of CUG homoduplexes in cells. There are also agents capable of cleaving long repeat RNAs (31, 48). It is of interest to determine whether these compounds would affect the cellular abundance of short repeat RNAs that could trigger detrimental consequences of gene silencing.
An increasing number of small molecule compounds have been reported to target CAG repeat RNAs, some of which can reduce disease protein aggregation (e.g., ref. 19). It would be of interest to determine whether these compounds can also target CAG RNA toxicity. Although it is still challenging to identify a single compound that can target both protein and RNA toxicities, a cotreatment strategy remains an appealing approach. We showed that cotreating R6/2 mice with both anti-RNA and anti-protein toxicity peptides yields better phenotypic suppression (49).
Materials and Methods
Detailed experimental procedures on cell-based assays, Western blotting, quantitative real-time PCR, immunocytochemistry, comet assay, LDH assay, ITC binding assay, NMR experiments, RNA-seq analysis, and in vivo animal studies are provided in SI Appendix.
Statistics and Reproducibility.
Unless otherwise specified, all experiments were independently repeated at least three times. Information about the number of biological replicates and statistical tests are provided in the figure legends.
Supplementary Material
Acknowledgments
This work was supported by the Research Grants Council General Research Fund (Project Nos. 14107118 and 14122019), The Chinese University of Hong Kong Vice-Chancellor’s One-Off Discretionary Fund (Project Nos. 4930713 and VCF2014011), Lui Che Woo Institute of Innovative Medicine Brain Research and Innovative Neuroscience Initiative (Project No. 8303404), Gerald Choa Neuroscience Centre (Project No. 7105306), Faculty of Medicine, The Chinese University of Hong Kong, and the US NIH (R01AR069645). Z.S.C. was supported by a Postdoctoral Fellowship in Clinical Neurosciences program between The Chinese University of Hong Kong and University of Oxford (Nuffield Department of Clinical Neurosciences and Pembroke College). X.L. and T.-F.C. are supported by the Innovation and Technology Commission, Hong Kong Special Administrative Region Government to the State Key Laboratory of Agrobiotechnology (The Chinese University of Hong Kong). Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.T.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022940118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lieberman A. P., Shakkottai V. G., Albin R. L., Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey T. H., Jones L., The central role of DNA damage and repair in CAG repeat diseases. Dis. Model. Mech. 11, dmm031930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlazzo M. L., et al., Mutations of the Huntington’s disease protein impact on the ATM-dependent signaling and repair pathways of the radiation-induced DNA double-strand breaks: Corrective effect of statins and bisphosphonates. Mol. Neurobiol. 49, 1200–1211 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee A., et al., The role of the mammalian DNA end-processing enzyme polynucleotide kinase 3′-phosphatase in spinocerebellar ataxia type 3 pathogenesis. PLoS Genet. 11, e1004749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao R., et al., Inactivation of PNKP by mutant ATXN3 triggers apoptosis by activating the DNA damage-response pathway in SCA3. PLoS Genet. 11, e1004834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahiri M., Gustafson T. L., Majors E. R., Freudenreich C. H., Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol. Cell 15, 287–293 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Gao R., et al., Mutant huntingtin impairs PNKP and ATXN3, disrupting DNA repair and transcription. eLife 8, e42988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasset-Rosa F., et al., Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y., Leng M., Wan M., Wilson J. H., Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol. Cell. Biol. 30, 4435–4451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krol J., et al., Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell 25, 575–586 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bañez-Coronel M., et al., A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 8, e1002481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessman M. J., Frick D. N., O’Handley S. F., The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271, 25059–25062 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Carreras-Puigvert J., et al., A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat. Commun. 8, 1541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peculis B. A., Reynolds K., Cleland M., Metal determines efficiency and substrate specificity of the nuclear NUDIX decapping proteins X29 and H29K (Nudt16). J. Biol. Chem. 282, 24792–24805 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Abolhassani N., et al., NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals. Nucleic Acids Res. 38, 2891–2903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyama T., Abolhassani N., Tsuchimoto D., Nonaka M., Nakabeppu Y., NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res. 38, 4834–4843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcheschi R. J., Tonelli M., Kumar A., Butcher S. E., Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication. ACS Chem. Biol. 6, 857–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S., et al., Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats. Nucleic Acids Res. 47, 10906–10913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan E., Mishra S. K., Mishra R., Mishra A., Kumar A., Discovery of a potent small molecule inhibiting Huntington’s disease (HD) pathogenesis via targeting CAG repeats RNA and Poly Q protein. Sci. Rep. 9, 16872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim R. G.et al.; HD iPSC Consortium , Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci. 20, 648–660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Zhang Y., Wong C. H., Edwin Chan H. Y., Zuo Z., Demonstration of direct nose-to-brain transport of unbound HIV-1 replication inhibitor DB213 via intranasal administration by pharmacokinetic modeling. AAPS J. 20, 23 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wang Q., Wong C. H., Chan H. Y. E., Lee W. Y., Zuo Z., Statistical Design of Experiment (DoE) based development and optimization of DB213 in situ thermosensitive gel for intranasal delivery. Int. J. Pharm. 539, 50–57 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Wang Q., et al., Efficient brain uptake and distribution of an expanded CAG RNA inhibitor DB213 via intranasal administration. Eur. J. Pharm. Sci. 127, 240–251 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Blackford A. N., Jackson S. P., ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Tsoi H., Lau T. C., Tsang S. Y., Lau K. F., Chan H. Y., CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. U.S.A. 109, 13428–13433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangiarini L., et al., Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Lein E. S., et al., Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Cepeda-Prado E., et al., R6/2 Huntington’s disease mice develop early and progressive abnormal brain metabolism and seizures. J. Neurosci. 32, 6456–6467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilfiker S., et al., Synapsins as regulators of neurotransmitter release. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354, 269–279 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong C. H., et al., Targeting toxic RNAs that cause myotonic dystrophy type 1 (DM1) with a bisamidinium inhibitor. J. Am. Chem. Soc. 136, 6355–6361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen L., et al., Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J. Am. Chem. Soc. 137, 14180–14189 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Luu L. M., et al., A potent inhibitor of protein sequestration by expanded triplet (CUG) repeats that shows phenotypic improvements in a Drosophila model of myotonic dystrophy. ChemMedChem 11, 1428–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J. L., VanEtten D. M., Fountain M. A., Yildirim I., Disney M. D., Structure and dynamics of RNA repeat expansions that cause Huntington’s disease and myotonic dystrophy type 1. Biochemistry 56, 3463–3474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura K., et al., SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 10, 1441–1448 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Tsoi H., Lau C. K., Lau K. F., Chan H. Y., Perturbation of U2AF65/NXF1-mediated RNA nuclear export enhances RNA toxicity in polyQ diseases. Hum. Mol. Genet. 20, 3787–3797 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Li L. B., Yu Z., Teng X., Bonini N. M., RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453, 1107–1111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q., et al., A peptidylic inhibitor for neutralizing expanded CAG RNA-induced nucleolar stress in polyglutamine diseases. RNA 24, 486–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q., et al., Pharmacokinetics and brain uptake of HIV-1 replication inhibitor DB213 in Sprague-Dawley rats. J. Pharm. Biomed. Anal. 125, 41–47 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Fujita K., et al., A functional deficiency of TERA/VCP/p97 contributes to impaired DNA repair in multiple polyglutamine diseases. Nat. Commun. 4, 1816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H., et al., A polyglutamine expansion disease protein sequesters PTIP to attenuate DNA repair and increase genomic instability. Hum. Mol. Genet. 21, 4225–4236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enokido Y., et al., Mutant huntingtin impairs Ku70-mediated DNA repair. J. Cell Biol. 189, 425–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneshima Y., et al., Deoxyinosine triphosphate induces MLH1/PMS2- and p53-dependent cell growth arrest and DNA instability in mammalian cells. Sci. Rep. 6, 32849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium . Electronic address: gusella@helix.mgh.harvard.edu; Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium, CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu X. H., et al., Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington’s disease. Sci. Transl. Med. 6, 268ra178 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Hong H., et al., AQAMAN, a bisamidine-based inhibitor of toxic protein inclusions in neurons, ameliorates cytotoxicity in polyglutamine disease models. J. Biol. Chem. 294, 2757–2770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A., et al., Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem. Biol. 7, 496–505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qawasmi L., et al., Expanded CUG repeats trigger disease phenotype and expression changes through the RNAi machinery in C. elegans. J. Mol. Biol. 431, 1711–1728 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Angelbello A. J., et al., Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl. Acad. Sci. U.S.A. 116, 7799–7804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M., et al., Brain-targeting delivery of two peptidylic inhibitors for their combination therapy in transgenic polyglutamine disease mice via intranasal administration. Mol. Pharm. 15, 5781–5792 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.