Significance
Hypoxia (reduced oxygen availability) is a common finding in the tumor microenvironment and plays a critical role in stimulating the metastasis of breast cancer cells from the primary tumor to distant organs, which is closely related to patient mortality. Critical transcriptional responses to reduced O2 availability are mediated by hypoxia-inducible factors (HIFs). In this study, we demonstrate that hypoxia induces HIF-dependent expression of a disintegrin and metalloproteinase 12 (ADAM12), which clips off the extracellular domain of the membrane-bound heparin-binding epidermal growth factor-like growth factor (HB-EGF). The liberated extracellular domain of HB-EGF binds to the epidermal growth factor receptor, triggering a signal transduction pathway that endows breast cancer cells with increased capability for cell migration and invasion, leading to distant metastasis.
Keywords: tumor microenvironment, cell motility, migration, invasion, metastasis
Abstract
Breast cancer patients with increased expression of hypoxia-inducible factors (HIFs) in primary tumor biopsies are at increased risk of metastasis, which is the major cause of breast cancer-related mortality. The mechanisms by which intratumoral hypoxia and HIFs regulate metastasis are not fully elucidated. In this paper, we report that exposure of human breast cancer cells to hypoxia activates epidermal growth factor receptor (EGFR) signaling that is mediated by the HIF-dependent expression of a disintegrin and metalloprotease 12 (ADAM12), which mediates increased ectodomain shedding of heparin-binding EGF-like growth factor, an EGFR ligand, leading to EGFR-dependent phosphorylation of focal adhesion kinase. Inhibition of ADAM12 expression or activity decreased hypoxia-induced breast cancer cell migration and invasion in vitro, and dramatically impaired lung metastasis after orthotopic implantation of MDA-MB-231 human breast cancer cells into the mammary fat pad of immunodeficient mice.
Breast cancer is the most frequently occurring malignancy in women, with approximately two million new cases diagnosed annually worldwide. Breast cancer is also the leading cause of cancer-related deaths among women, accounting for 15% of all cancer-related deaths (1). The acquisition of invasive properties allows tumor cells to spread into surrounding tissues and ultimately to disseminate and form distant metastases in secondary organs. The clinical outcome worsens significantly for patients who develop distant metastasis: the overall 5-y relative survival rate is 99% for patients with localized disease, compared to 27% for patients with distant metastasis (2).
Intratumoral hypoxia is commonly observed in solid tumors, including breast cancer, and a growing body of data indicates that hypoxia is a powerful driving force for cancer progression (3–5). Hypoxia is a stimulus for angiogenesis, cancer stem cell maintenance, metabolic reprogramming, immune evasion, and other survival responses, as well as invasion and metastasis (3–9). The response of cancer cells to reduced O2 availability is mediated by hypoxia-inducible factors (HIFs), which are heterodimeric proteins composed of an O2-regulated HIF-α subunit (HIF-1α, HIF-2α, or HIF-3α), and a constitutively expressed HIF-1β subunit (8). In both estrogen receptor-positive (ER+) and ER− breast cancers, immunohistochemistry studies have revealed that overexpression of HIF-1α in the primary tumor biopsy is associated with an increased risk of metastasis, relapse, and patient mortality (10–13).
The metastatic process consists of a series of discrete steps beginning with acquisition of a motile phenotype by tumor cells. It has been increasingly recognized that hypoxia induces the HIF-dependent expression of genes that contribute to each step of metastasis, from local tissue invasion, intravasation, and extravasation, to secondary organ colonization, indicating that HIFs function as master regulators of the metastatic process (4, 7, 8). Despite this progress, it is apparent that these complex biological processes require the coordinated induction of large batteries of genes, many of which remain to be identified.
A disintegrin and metalloprotease (ADAM) refers to a large family of integral membrane or secreted glycoproteins that are comprised of several distinct domains, with important roles in regulating cell adhesion and migration (14, 15). ADAMs mediate proteolytic processing of the ectodomains of diverse cell surface receptors and signaling molecules, including Notch receptors and their ligands, epidermal growth factor receptor (EGFR) ligands, and tumor necrosis factor and its receptors (14, 15). Several previous studies have highlighted the role of ADAM12 in regulating multiple molecular pathways, which promote cancer progression (16). Similar to the effect of HIFs (17), increased ADAM12 expression was found to contribute to the cancer stem cell phenotype of claudin-low triple-negative breast cancer (TNBC) cells, as well as resistance to neoadjuvant chemotherapy in ER− breast cancer (18, 19). ADAM12 expression is up-regulated by transforming growth factor-β in activated hepatic stellate cells, and is associated with liver cancer progression (20). ADAM12 is also involved in heparin-binding epidermal growth factor-like growth factor (HB-EGF) ectodomain shedding to potentiate invadopodia formation in lung and pancreatic cancer cells under hypoxic conditions (21).
In the present study, we have investigated the mechanisms and consequences of ADAM12 expression in breast cancer. Our studies have revealed that exposure of human breast cancer cells to hypoxia is sufficient to increase the levels of ADAM12 in an HIF-dependent manner, leading to increased ectodomain shedding of HB-EGF, activation of EGFR signaling to focal adhesion kinase (FAK), and increased cell motility and basement membrane invasion, thereby establishing hypoxia as a major physiological stimulus in the tumor microenvironment that directly connects regulation of ADAM12 expression to the invasive phenotype of breast cancer. Most remarkably, in an orthotopic transplantation model, we find that ADAM12 expression is required for the spontaneous metastasis of breast cancer cells to the lungs.
Results
ADAM12 Expression Is Correlated with the HIF Signature in Breast Cancers and Induced by Hypoxia.
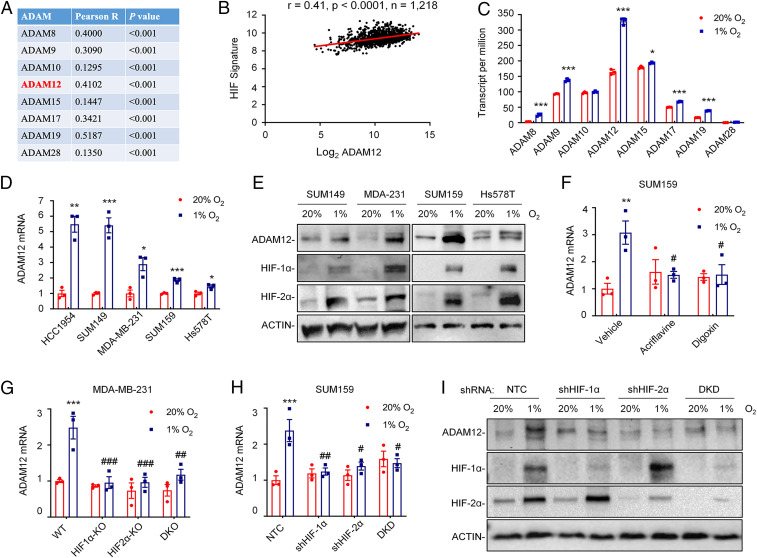
Several ADAM family members are aberrantly expressed in human cancers and involved in cancer progression, metastasis, and therapy resistance (14, 15, 22–24). Among 12 catalytically active human ADAMs, 8 are ubiquitously expressed (15). To test whether ADAM expression is regulated by intratumoral hypoxia, we first analyzed whether the expression of mRNAs encoding ADAMs is correlated with the expression of an HIF metagene signature (25) consisting of 10 HIF-regulated genes that play key roles in breast cancer metastasis (CXCR3, L1CAM, LOX, P4HA1, P4HA2, PDGFB, PLOD1, PLOD2, SLC2A1, and VEGFA). Analysis of mRNA expression data from 1,218 breast cancer specimens in the Cancer Genome Atlas (TCGA) database by Pearson’s test revealed a significant correlation of several ADAM mRNAs with the HIF signature. Among the eight ADAMs analyzed, ADAM19 showed the highest correlation coefficient (R = 0.52) (Fig. 1A). ADAM12 was also highly correlated (R = 0.41) (Fig. 1B).
Fig. 1.
ADAM12 expression in breast cancer is correlated with HIF target gene expression and is induced by hypoxia in an HIF-dependent manner. (A and B) The mRNA expression of eight ADAM family members was compared with a 10-gene HIF signature in 1,218 human breast cancers from TCGA database by Pearson’s correlation test. For each comparison, the Pearson product-moment correlation coefficient (R = −1 to 1) and statistical significance (P) are shown (A). Individual data points for ADAM12 are shown (B). (C) RNA-seq data for the expression of eight ADAM family genes in SUM159 cells following exposure to 20% or 1% O2 for 24 h are shown (mean ± SEM; n = 3). *P < 0.05, ***P < 0.001 versus 20% O2 (Student’s t test). (D) RT-qPCR was performed to quantify ADAM12 mRNA levels in five human breast cancer cell lines following exposure to 20% or 1% O2 for 24 h. For each cell line, the expression of ADAM12 mRNA was quantified relative to 18S rRNA and then normalized to the result obtained from cells at 20% O2 (mean ± SEM; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 versus 20% O2 (Student’s t test). (E) Immunoblot assays were performed to determine ADAM12, HIF-1α, and HIF-2α protein levels in breast cancer cell lines following exposure to 20% or 1% O2 for 48 h. ACTIN was analyzed here (and elsewhere) as a loading control. (F) SUM159 cells were exposed to 20% or 1% O2 for 24 h in the presence of vehicle, digoxin (100 nM), or acriflavine (5 μM), and expression of ADAM12 mRNA was assayed by RT-qPCR. **P < 0.01 versus vehicle at 20% O2; #P < 0.05 versus vehicle at 1% O2 (two-way ANOVA with Tukey’s posttest). (G and H) RT-qPCR was performed to quantify ADAM12 mRNA levels in MDA-MB-231 (G) and SUM159 (H) subclones exposed to 20% or 1% O2 for 24 h. Data were normalized to WT (G) or NTC (H) cells at 20% O2 (mean ± SEM; n = 3). ***P < 0.001 versus WT or NTC at 20% O2; #P < 0.05, ##P < 0.01, ###P < 0.001 versus WT or NTC at 1% O2 (two-way ANOVA with Tukey’s posttest). (I) Immunoblot assays were performed to determine ADAM12, HIF-1α, and HIF-2α protein levels in whole cell lysates prepared from SUM159 subclones exposed to 20% or 1% O2 for 48 h. DKD, double knockdown; DKO, double knockout.
To provide more direct evidence for hypoxia-induced expression of ADAM family members in breast cancer, we analyzed RNA-sequencing (RNA-seq) data from SUM159 cells exposed to 20% or 1% O2 for 24 h, which revealed that expression of six ADAM family members (ADAM8, -9, -12, -15, -17, and -19) was induced by hypoxia, with ADAM12 showing the greatest expression at 1% O2 (Fig. 1C). To extend the results from SUM159 cells, we exposed a panel of human breast cancer cell lines, including ER+ MCF-7 and T47D cells, HER2+ HCC1954 cells, and triple-negative SUM149, MDA-MB-231, SUM159, and Hs578T cells to 20% or 1% O2 for 24 h. Reverse-transcription and quantitative real-time PCR (RT-qPCR) analysis of total RNA isolated from the cells showed that ADAM12 mRNA levels were increased significantly under hypoxic conditions in most of the breast cancer cell lines (Fig. 1D). When normalized to the expression level in HCC1954 cells, we found that ADAM12 was very highly expressed in TNBC cell lines as compared to ER+ or HER2+ lines (SI Appendix, Fig. S1A).
We next analyzed the effect of hypoxia on ADAM12 protein expression by immunoblot assay and found that ADAM12 protein levels were also increased, along with HIF-1α and HIF-2α proteins, under hypoxic conditions (Fig. 1E and SI Appendix, Fig. S1B). Taken together, these data indicate that hypoxia induces ADAM12 mRNA and protein expression in human breast cancer cell lines.
Hypoxia-Induced ADAM12 Expression Is HIF-Dependent.
To investigate whether hypoxia induces ADAM12 expression in an HIF-dependent manner, we first took a pharmacologic approach to inhibit HIF activity. Digoxin and acriflavine are known HIF inhibitors, which act by distinct molecular mechanisms: Digoxin inhibits the accumulation of HIF-1α protein in hypoxic cells (26), whereas acriflavine blocks the dimerization of HIF-1α or HIF-2α with HIF-1β, which subsequently leads to degradation of the proteins (27). Treatment of SUM159 and SUM149 cells with either digoxin or acriflavine blocked hypoxia-induced ADAM12 mRNA expression as determined by RT-qPCR (Fig. 1F and SI Appendix, Fig. S1C).
To determine the individual and joint contributions of HIF-1α and HIF-2α to the regulation of ADAM12 expression under hypoxic conditions, we utilized MDA-MB-231 subclones with knockout of HIF-1α (HIF1α-KO), HIF-2α (HIF2α-KO), or both (double knockout), which were previously established using the CRISPR/Cas9 technique (28). SUM159 and SUM149 subclones stably transduced with expression vectors encoding a nontargeting control (NTC) short-hairpin RNA (shRNA), or vectors encoding shRNA targeting HIF-1α (shHIF1α), HIF-2α (shHIF2α), or both (double knockdown) (29) were also established. RT-qPCR revealed that both HIF-1α and HIF-2α knockdown/knockout abrogated the induction of ADAM12 mRNA expression in cells exposed to hypoxia (Fig. 1 G and H and SI Appendix, Fig. S1D). Furthermore, hypoxic induction of ADAM12 protein expression was also blocked by expression of shRNA targeting HIF-1α or HIF-2α or both, as determined by immunoblot assays of whole cell lysates (Fig. 1I). In summary, these data indicate that ADAM12 is induced in response to hypoxia in a HIF-1– and HIF-2–dependent manner in human breast cancer cells.
ADAM12 Is a Direct HIF Target Gene.
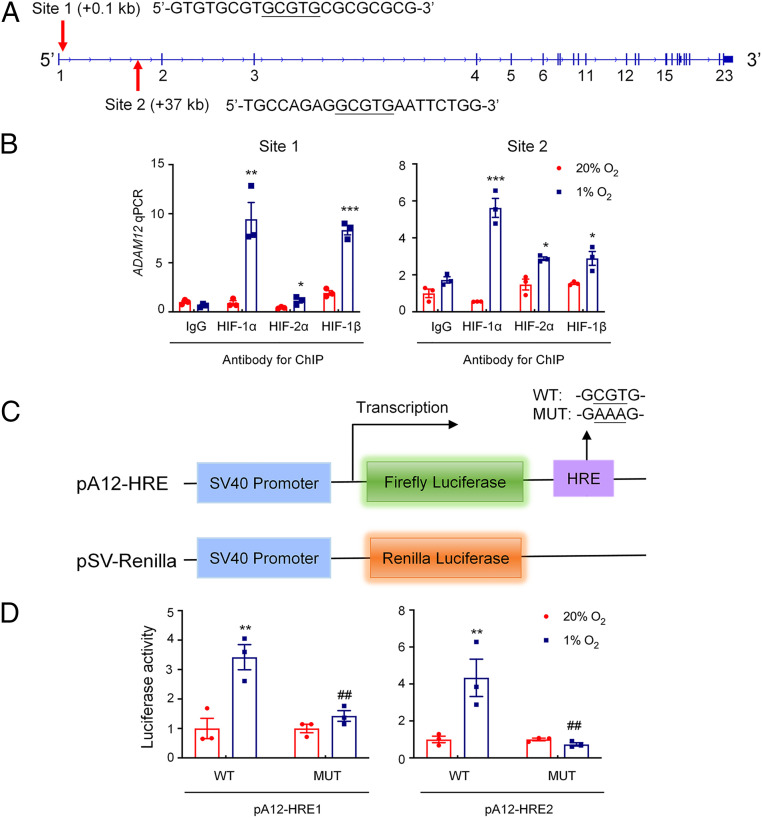
The human ADAM12 gene sequence was searched for matches to the consensus HIF binding site sequence 5′-(A/G)CGTG-3′ (30) that were located within DNase I hypersensitive chromatin domains. Several candidate HIF binding sites in the ADAM12 gene were interrogated by chromatin immunoprecipitation (ChIP) assays of SUM159, MDA-MB-231, and SUM149 cells exposed to 20% or 1% O2 for 16 h. Two HIF binding sites, which were located at +0.1 kb (site 1) and +37 kb (site 2) relative to the ADAM12 transcription start site, respectively, were identified (Fig. 2A). Chromatin fragments containing either of the two DNA sequences were immunoprecipitated with antibodies against HIF-1α, HIF-2α, or HIF-1β in a hypoxia-inducible manner from SUM159 cells (Fig. 2B), as well as MDA-MB-231 and SUM149 cells (SI Appendix, Fig. S2 A and B), indicating that hypoxia induces direct binding of HIF-1 and HIF-2 to the ADAM12 gene.
Fig. 2.
ADAM12 is a direct HIF target gene. (A) Two matches to the HIF consensus binding site (5′-RCGTG-3′) were identified in the human ADAM12 gene at +0.1 kb (site 1) and +37 kb (site 2), relative to the transcription start site. (B) SUM159 cells were incubated at 20% or 1% O2 for 16 h, and ChIP assays were performed using antibodies against HIF-1α, HIF-2α, or HIF-1β. Primers flanking site 1 or site 2 were used for qPCR, and results were normalized to IgG at 20% O2 (mean ± SEM; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 versus 20% O2 (Student’s t test). (C) The following reporter plasmids were generated: pA12-HRE, containing a 55-bp candidate hypoxia response element (HRE1 or HRE2, encompassing site 1 or 2, respectively), which was either WT or mutant (MUT), downstream of an SV40 promoter and firefly luciferase coding sequences (Upper); and pSV-Renilla, a control plasmid containing Renilla luciferase coding sequences downstream of the SV40 promoter (Lower). (D) SUM159 cells were cotransfected with pSV-Renilla and firefly luciferase reporter pA12-HRE1 or pA12-HRE2 (WT or MUT), and exposed to 20% or 1% O2 for 24 h. Fluc:Rluc ratio was determined and normalized to WT at 20% O2 (mean ± SEM; n = 3). **P < 0.01 versus WT at 20% O2; ##P < 0.01, versus WT at 1% O2 (two-way ANOVA with Tukey’s posttest).
Next, we investigated whether DNA fragments encompassing these two HIF binding sites function as hypoxia response elements (HREs). We generated two reporter plasmids, pA12-HRE1 and pA12-HRE2, by inserting a 55-bp oligonucleotide, encompassing either site 1 or site 2, downstream of SV40 promoter and firefly luciferase (Fluc) coding sequences in the pGL2-promoter reporter plasmid (Fig. 2 C, Upper). Plasmids containing HRE1 or HRE2 with a 5′-CGT-3′ to 5′-AAA-3′ mutation that abrogates HIF binding (30) were also constructed. The pSV-Renilla plasmid, encoding Renilla luciferase (Rluc) driven by the SV40 promoter alone (Fig. 2 C, Lower), was cotransfected. The Fluc:Rluc ratio is a measure of HRE activity. Exposure to hypoxia resulted in a significant increase in the Fluc:Rluc ratio in SUM159 cells that were cotransfected with either wild-type pA12-HRE1 or pA12-HRE2 and pSV-Renilla, whereas mutation of the HIF binding sequence within HRE1 or HRE2 completely abolished hypoxia-induced Fluc activity (Fig. 2D), demonstrating that each of the 55-bp oligonucleotides functions as an HRE. Taken together, the data presented in Fig. 2 demonstrate that both HIF-1 and HIF-2 bind directly to the ADAM12 gene to activate transcription in hypoxic breast cancer cells, which is consistent with our previous finding that both HIF-1α and HIF-2α are required for hypoxic induction of endogenous ADAM12 expression (Fig. 1 H and I).
ADAM12 Knockdown Impairs Hypoxia-Induced Breast Cancer Cell Motility.
Cell motility is a necessary prerequisite for tissue invasion and metastasis (31–33). As previously reported, hypoxia mediates increased cell motility in a HIF-dependent manner and promotes distant metastasis (34). To investigate the role of ADAM12 expression in hypoxia-induced cell motility, we generated shRNA-mediated ADAM12 knockdown subclones from MDA-MB-231 and SUM159 cells by stably transducing lentiviral vectors encoding one of five different shRNAs against ADAM12, or a vector expressing NTC shRNA. Decreased ADAM12 mRNA and protein expression by the subclones was confirmed by RT-qPCR and immunoblot assay, respectively (SI Appendix, Fig. S3).
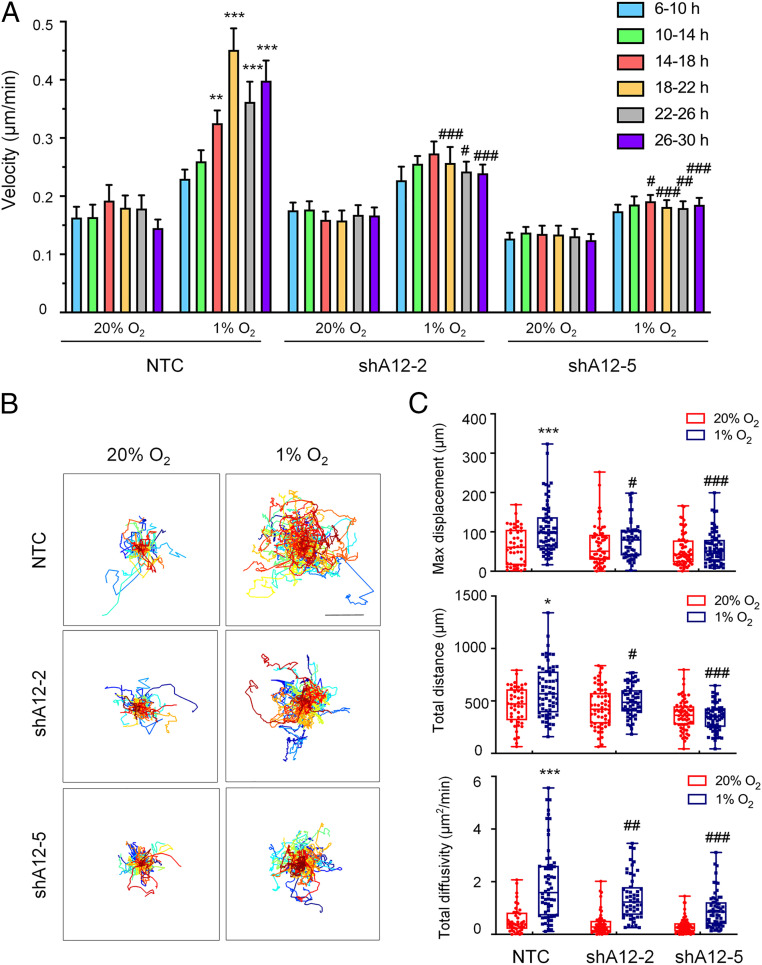
To determine whether ADAM12 expression regulates breast cancer cell motility, we dynamically monitored the random motility of MDA-MB-231 NTC and ADAM12 knockdown subclones exposed to 20% or 1% O2 for 30 h using time-lapse photomicroscopy. Mean cell velocity, which was determined at 4-h intervals, revealed increased velocity starting at 14 h of exposure to 1% O2 in NTC subclones, but not in ADAM12 knockdown subclones (Fig. 3A). Individual (x, y) coordinates were determined for each cell every 10 min to construct cell trajectory maps (Fig. 3B). The total distance traversed and the maximum displacement of cells relative to the origin were also calculated based on their position coordinates. Consistently, the increased movement of cells induced by hypoxia was significantly impaired by ADAM12 knockdown in MDA-MB-231 cells (Fig. 3C). Velocity (S) and persistence time (P; mean time between significant changes in the direction of movement) were calculated by fitting cell trajectories to an anisotropic persistent random walk model of cell motility. Total diffusivity (D) describes overall cell movement within a specific time frame and is calculated according to the formula D = S2P/4 (35). Cell diffusivity was increased at 1% O2 compared to 20% O2 in NTC subclones, as previously reported (36), but subclones with decreased ADAM12 expression showed significantly impaired cell movement under hypoxic conditions (Fig. 3C).
Fig. 3.
ADAM12 knockdown impairs MDA-MB-231 cell motility. (A) Cell velocity over 4-h intervals was determined for subclones, which were stably transfected with a NTC shRNA, or shRNA targeting ADAM12 (shA12-2, shA12-5) and exposed to 20% or 1% O2 for 30 h. Data are shown as mean ± SEM (n = 44 to 50 cells). **P < 0.01, ***P < 0.001 versus NTC cells at 20% O2 at the corresponding time point; #P < 0.05, ##P < 0.01, ###P < 0.001 versus NTC cells at 1% O2 at the corresponding time point (two-way ANOVA with Tukey’s posttest). (B) Cell trajectories (n = 44 to 50) are plotted using (x, y) coordinates obtained at 10-min intervals over a 30-h time course. (Scale bar, 50 μm.) (C) The maximum displacement, total distance, and total diffusivity of subclones were calculated and data are shown as mean ± SEM (n = 44 to 50 cells). *P < 0.05, ***P < 0.001 versus NTC cells at 20% O2; #P < 0.05, ##P < 0.01, ###P < 0.001 versus NTC cells at 1% O2 (two-way ANOVA with Tukey’s posttest for all comparisons).
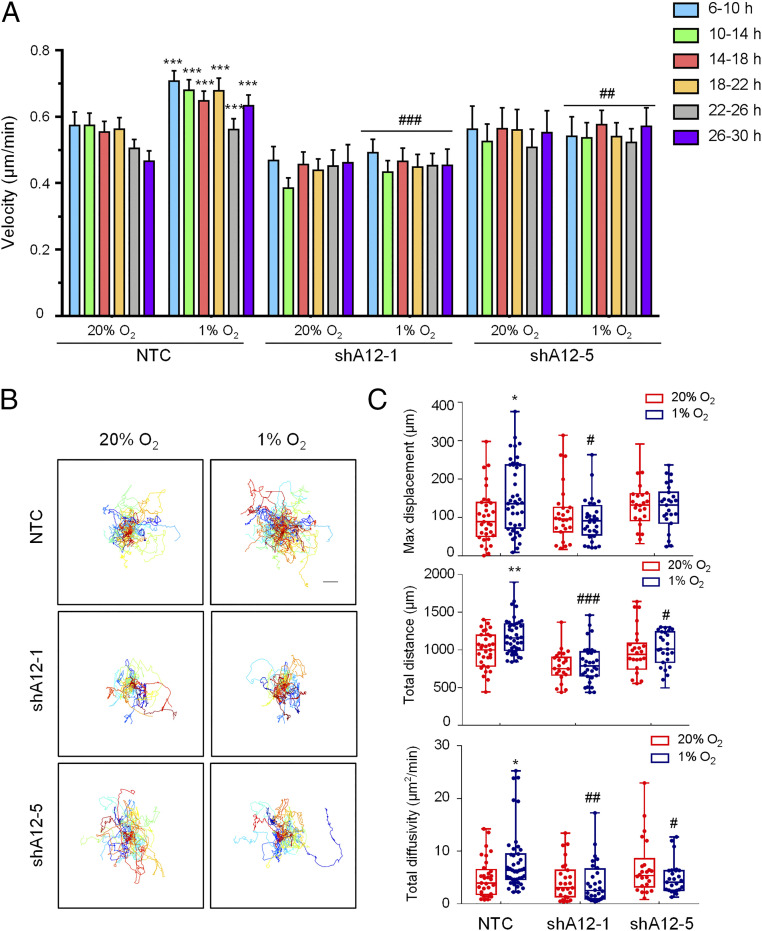
We extended these observations by analyzing the motility of SUM159 cells on collagen-coated plates over the course of 30 h. Hypoxia increased the velocity, maximal displacement, total distance migrated, and total diffusivity of the NTC subclone; in contrast to the movement of MDA-MB-231 cells on plastic, the effect of hypoxia on SUM159 NTC subclone cells was already maximal at the first time point analyzed (Fig. 4). However, as in the case of MDA-MB-231 cells on plastic, hypoxia had no significant effect on the motility of two ADAM12 knockdown subclones of SUM159 on collagen. Thus, motility is stimulated in an ADAM12-dependent manner when human breast cancer cells are exposed to hypoxia.
Fig. 4.
ADAM12 knockdown impairs SUM159 cell motility. (A) Cell velocity over 4-h intervals was determined for subclones, which were stably transfected with a NTC shRNA, or shRNA targeting ADAM12 (shA12-1, shA12-5) and exposed to 20% or 1% O2 for 30 h. Data are shown as mean ± SEM (n = 26 to 44 cells). ***P < 0.001 versus NTC cells at 20% O2, independent of time. ##P < 0.01, ###P < 0.001 versus NTC cells at 1% O2 independent of time (two-way ANOVA with Tukey’s posttest). (B) Cell trajectories are plotted using (x, y) coordinates obtained at 10-min intervals over a 30-h time course. (Scale bar, 50 μm.) (C) The maximum displacement, total distance, and total diffusivity of subclones were calculated and data are shown as mean ± SEM (n = 26 to 44 cells). *P < 0.05, **P < 0.01 versus NTC cells at 20% O2; #P < 0.05, ##P < 0.01, ###P < 0.001 versus NTC cells at 1% O2 (two-way ANOVA with Tukey’s posttest for all comparisons).
ADAM12 Increases Migration and Invasion of Hypoxic Breast Cancer Cells.
We next analyzed the effect of ADAM12 knockdown in MDA-MB-231 and SUM159 cells on directional cell migration and invasion in Boyden chamber assays. The results demonstrated that hypoxia significantly increased migration of NTC cells across the Transwell membrane approximately twofold, whereas hypoxia-induced migration was significantly impaired by ADAM12 knockdown (Fig. 5 A–D). Similarly, the number of cells that invaded through Matrigel, a tumor-derived basement membrane/extracellular matrix preparation layered on top of a Transwell insert, was also analyzed. Compared with NTC cells, ADAM12 knockdown subclones had decreased invasive properties and the stimulatory effect of hypoxia on invasion was abolished by ADAM12 knockdown (Fig. 5 E–H). Based on these data, we conclude that ADAM12 expression is required for the increased migration and invasion of hypoxic breast cancer cells in vitro.
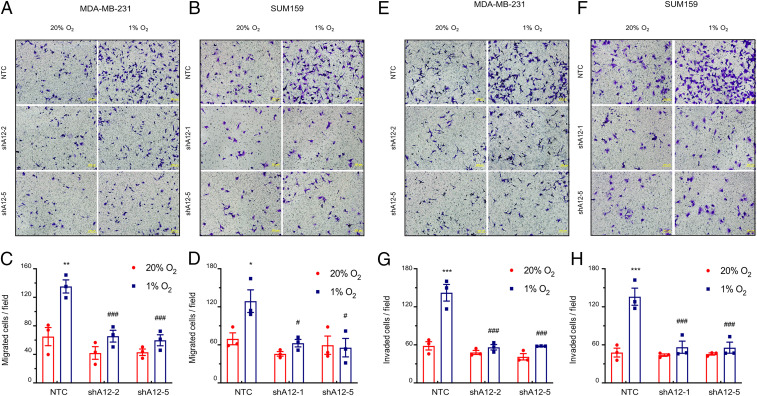
Fig. 5.
ADAM12 knockdown impairs breast cancer cell migration and invasion. (A–D) The migration of NTC and ADAM12 knockdown subclones exposed to 20% or 1% O2 was analyzed. MDA-MB-231 (A) and SUM159 (B) cells that migrated through the filter to the underside were stained with Crystal violet and representative images are shown. (Scale bars, 200 μm.) The number of migrated MDA-MB-231 (C) and SUM159 (D) cells per field were determined and data are presented as mean ± SEM (n = 3). *P < 0.05, **P < 0.01 versus NTC at 20% O2; #P < 0.05, ###P < 0.001 versus NTC at 1% O2 (two-way ANOVA with Tukey’s posttest). (E–H) The invasion of NTC and ADAM12 knockdown subclones exposed to 20% or 1% O2 was analyzed. MDA-MB-231 (E) and SUM159 (F) cells that invaded through the Matrigel-coated Boyden chamber inserts were stained with Crystal violet and representative images are shown. (Scale bars, 200 μm.) The number of invaded MDA-MB-231 (G) and SUM159 (H) cells per field were determined and data are presented as mean ± SEM (n = 3). ***P < 0.001 versus NTC at 20% O2; ###P < 0.001 versus NTC at 1% O2 (two-way ANOVA with Tukey’s posttest).
Hypoxia Induces Breast Cancer Cell Invasion through ADAM12-EGFR-FAK Signaling.
ADAM12 is a major ectodomain sheddase on the cell membrane, overexpression of which usually leads to release of mature growth factor ligands and activation of EGFR signaling. FAK is an intracellular protein tyrosine kinase that regulates the cycle of focal adhesion contact formation and disassembly required for regulation of cell shape, adhesion, and motility (37). EGFR-FAK pathway activation was detected by the phosphorylation of specific tyrosine (Y) residues using immunoblot assays, which revealed that the phosphorylation of both EGFR (at Y1173) and FAK (at Y397) was increased in response to hypoxic exposure of MDA-MB-231 and SUM159 cells, although total EGFR and FAK protein levels were not altered by hypoxia (Fig. 6A), indicating that the EGFR-FAK pathway was specifically activated in these breast cancer cells under hypoxic conditions.
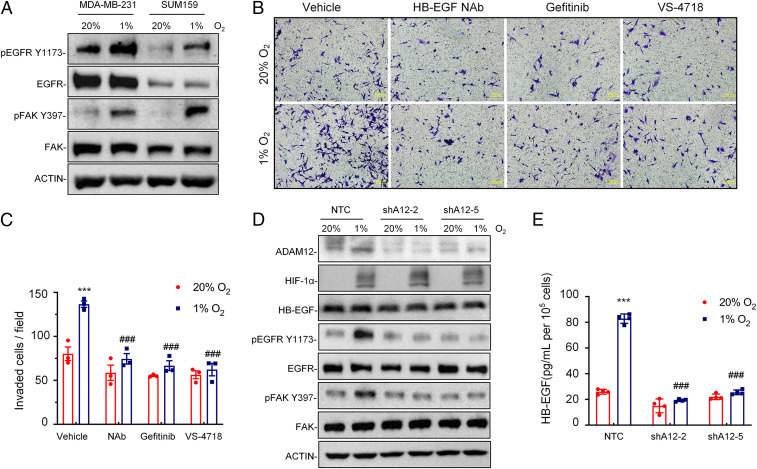
Fig. 6.
Hypoxia induces breast cancer cell invasion through ADAM12-EGFR-FAK signaling. (A) Immunoblot assays were performed to analyze phosphorylated EGFR (pEGFR Y1173), total EGFR, phosphorylated FAK (pFAK Y397), and total FAK protein in MDA-MB-231 and SUM159 subclone cells following exposure to 20% or 1% O2 for 24 h. (B and C) MDA-MB-231 cells were treated with vehicle, HB-EGF NAb (10 μg/mL), Gefitinib (5 μM), or VS-4718 (1 μM) and exposed to 20% or 1% O2 for 24 h. Cells that invaded through Matrigel-coated Boyden chamber inserts were stained with Crystal violet (B) and quantified by densitometry (C; mean ± SEM, n = 3). ***P < 0.001 versus cells exposed to vehicle at 20% O2; ###P < 0.001 versus cells exposed to vehicle at 1% O2 (two-way ANOVA with Tukey’s posttest). (D) MDA-MB-231 subclones stably transduced with NTC or ADAM12 shRNA vector (shA12-2 or shA12-5) were exposed to 20% or 1% O2 for 24 h, and whole-cell lysates were subjected to immunoblot assays. (E) MDA-MB-231 subclones stably transduced with NTC or ADAM12 shRNA vectors were exposed to 20% or 1% O2 for 72 h, and ELISA was performed to quantify HB-EGF in cell supernatant. Data shown are mean ± SEM, n = 3. ***P < 0.001 versus NTC cells at 20% O2; ###P < 0.001 versus NTC cells at 1% O2 (two-way ANOVA with Tukey’s posttest).
To determine the molecular mechanism by which ADAM12 promotes cell invasion, we first treated MDA-MB-231 cells with neutralizing antibody (NAb) against the HB-EGF ectodomain, which is an EGFR ligand that is generated by ADAM12. HB-EGF NAb blocked hypoxia-induced cell invasion in the Boyden chamber assay (Fig. 6 B and C). Administration of the EGFR tyrosine kinase inhibitor gefitinib or the FAK inhibitor VS-4718 also blocked hypoxia-induced invasion of MDA-MB-231 cells through Matrigel (Fig. 6 B and C). Hypoxia increased the phosphorylation of EGFR (at Y1173) and FAK (at Y397) in NTC but not in ADAM12 knockdown sh2 and sh5 subclones (Fig. 6D). Total EGFR, FAK, and HB-EGF levels in whole cell lysates were not altered by hypoxia or ADAM12 knockdown (Fig. 6D). However, ELISA revealed that the soluble form of HB-EGF in cell culture conditioned media increased in response to hypoxic exposure of NTC but not ADAM12 knockdown cells (Fig. 6E). Taken together, the data presented in Fig. 6 indicate that ADAM12 → HB-EGF → EGFR → FAK signaling is required for hypoxia-induced breast cancer cell migration and invasion.
ADAM12 Deficiency Impairs Spontaneous Metastasis of Breast Cancer Cells to the Lungs.
As migration and invasion are two critical steps in the process of metastasis, we analyzed the effect of ADAM12 knockdown on cancer progression in vivo. NTC or ADAM12 knockdown subclones of MDA-MB-231 cells were implanted into the mammary fat pad of female severe combined immunodeficiency (SCID) mice, and tumor growth was measured every 3 or 4 d. When the largest tumor volume reached 1,200 mm3, the mice were euthanized and primary tumor and lungs were collected. Primary tumor growth curves were not significantly different between the NTC and ADAM12 knockdown subclones (Fig. 7A), although the final weight of shA12-2 subclone tumors was modestly but significantly decreased (Fig. 7B).
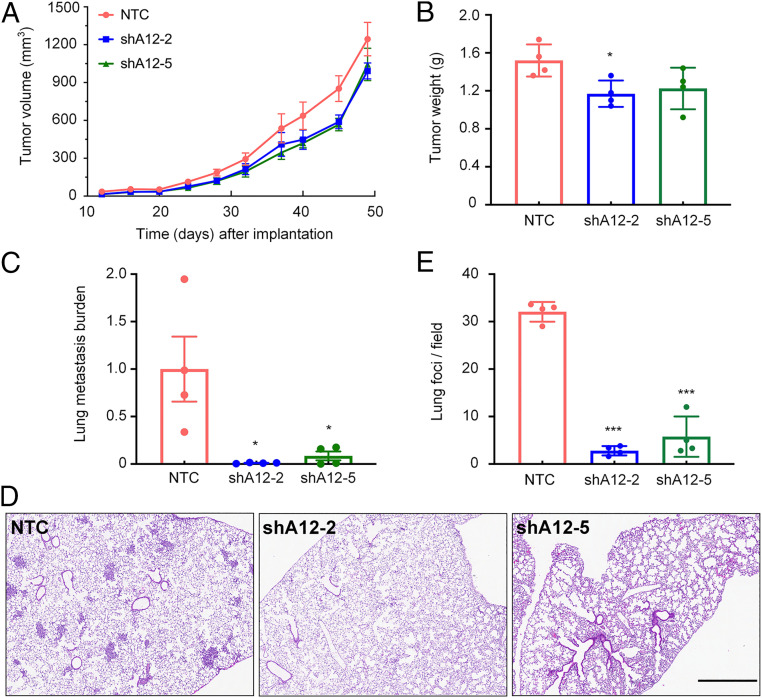
Fig. 7.
ADAM12 knockdown impairs the spontaneous metastasis of breast cancer cells to the lungs. (A and B) MDA-MB-231 subclones (2 × 106 cells) were implanted into the mammary fat pad of 6-wk-old female SCID mice. Primary tumor volume (A) was determined starting at day 12. On day 49, the primary tumor was harvested and weighed (B). (C) To analyze lung metastasis, genomic DNA was purified from one lung for qPCR using human-specific HK2 primers and the results were normalized to the NTC group. (D and E) The other lung was fixed under inflation, paraffin-embedded, and sections were stained with hematoxylin and eosin (D) to count the number of metastatic foci per field (E). For all graphs, mean ± SEM (n = 4) are shown; *P < 0.05, ***P < 0.001 versus NTC (one-way ANOVA). (Scale bar, 0.5 mm.)
To analyze the effect of ADAM12 knockdown in MDA-MB-231 cells on lung metastasis, we processed one mouse lung for histology and isolated genomic DNA from the other lung. The presence of human DNA in the mouse lungs was quantified by qPCR using human-specific primers. This assay revealed a dramatic reduction in metastatic burden in lungs from mice bearing ADAM12 knockdown tumors as compared with mice bearing NTC tumors (Fig. 7C). The contralateral lungs were fixed under inflation, paraffin-embedded, and sections were stained with hematoxylin and eosin. Histological analysis confirmed that ADAM12 knockdown significantly decreased the number of metastatic foci in the lungs (Fig. 7 D and E).
We also injected NTC and ADAM12 knockdown subclones of SUM159 cells into the mammary fat pad of immunodeficient mice. ADAM12 knockdown was associated with a modest increase in primary tumor growth that was not statistically significant (SI Appendix, Fig. S4). Five weeks after tumor cell implantation in the mammary fat pad, we excised the primary tumor, waited another 3 wk, and then analyzed lymph node, lungs, liver, and brain tissue for metastatic cells using human-specific qPCR. No evidence of metastasis was detected in any tissue of any tumor-bearing mouse. Taken together, the data presented in Fig. 7 and SI Appendix, Fig. S4 demonstrate that ADAM12 knockdown has no consistent effect on primary tumor growth but is absolutely required for the spontaneous metastasis of MDA-MB-231 breast cancer cells from the mammary fat pad to the lungs of SCID mice.
ADAM12 Is Highly Expressed in Human Breast Cancers and Predicts Poor Clinical Outcome.
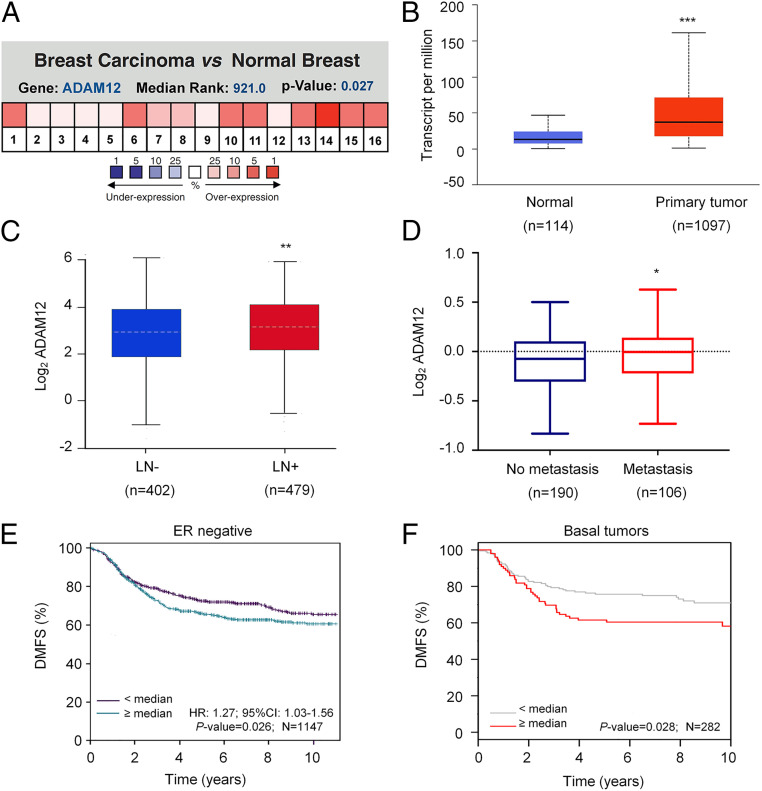
A metaanalysis of 16 datasets in the Oncomine database revealed that ADAM12 is one of the most highly differentially expressed genes in breast cancer as compared to normal breast tissue (Fig. 8A). Analysis of breast cancer gene-expression data from TCGA also revealed that ADAM12 mRNA was significantly overexpressed in breast cancer as compared with adjacent normal tissue (Fig. 8B). ADAM12 mRNA expression was significantly increased in patients with lymph node metastasis as compared to patients without metastasis (Fig. 8C). ADAM12 expression was also correlated with distant metastasis: analysis of the van de Vijver et al. microarray dataset (38) revealed that ADAM12 mRNA levels were higher in primary tumors from patients with distant metastasis compared to those without distant metastasis (Fig. 8D). Kaplan–Meier analysis of TCGA data using the bcGenExMiner online platform (39) revealed that high levels of ADAM12 expression were associated with poor distant metastasis-free survival of patients with ER− breast cancer. Analysis of the gene-expression–based outcome (GOBO) dataset (40) also revealed a significant correlation (Fig. 8 E and F). Taken together, the data presented in Fig. 8 demonstrate that ADAM12 is highly expressed in human breast cancers and that ADAM12 gene expression is associated with distant metastasis and mortality in patients with breast cancer.
Fig. 8.
ADAM12 is highly expressed in human breast cancers and is correlated with patient mortality. (A) Analysis of ADAM12 gene expression in breast cancer and normal breast tissue using the Oncomine database. Metaanalysis of 16 datasets from 6 different microarray studies shows ADAM12 is one of the most highly differentially expressed genes in breast cancer versus normal tissue. Color scale represents over- or underexpression rank by percentile. (B) ADAM12 mRNA expression in primary breast cancer samples (n = 1,097) relative to adjacent normal tissue (n = 114) from TCGA database are shown. ***P < 0.001 versus normal, by Student’s t test. (C) ADAM12 mRNA expression, in primary breast cancer from patients with (LN+) or without (LN−) lymph node metastasis (TCGA database) was analyzed. **P < 0.01 versus LN−, by Student’s t test. (D) ADAM12 mRNA expression in 296 primary breast cancers stratified according to distant metastasis status (38) were analyzed. *P < 0.05 versus no metastasis, by Student’s t test. Horizontal line, median; box, 75th to 25th percentile; whiskers, maximum and minimum values. (E and F) Kaplan–Meier analysis of distant metastasis-free survival (DMFS) of breast cancer patients, stratified by ADAM12 mRNA expression above or below the median, was performed using data obtained from two independent databases, bcGenExMiner V4.4 (n = 1,147) and GOBO (n = 282). Statistical analysis was performed using log-rank tests; HR, hazard ratio.
Discussion
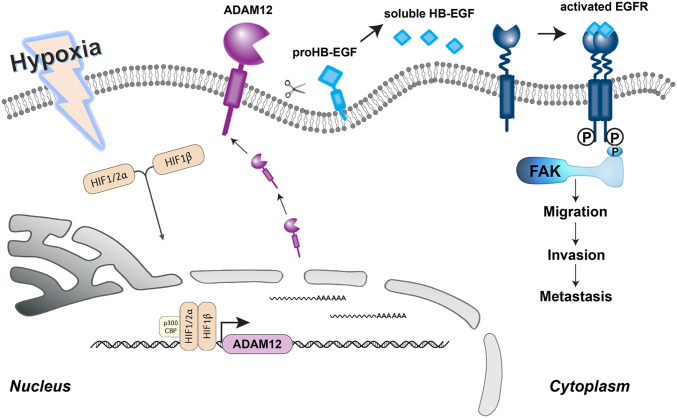
Hypoxia is a common phenomenon found in a wide range of solid tumors and is often related to poor prognosis (4, 41). Hypoxia promotes metabolic reprograming, angiogenesis, cancer stem cell specification, as well as invasion and metastasis, by activating the expression of a large battery of genes that are subject to transcriptional activation by hypoxia-inducible factors (3–9). However, within each cancer, the specific target genes activated and the pathogenic consequences of their activation are unique. In the present study, we have demonstrated the critical role of ADAM12, a direct HIF target gene, in promoting breast cancer metastasis. Hypoxia induces ADAM12-mediated shedding of HB-EGF in an HIF-dependent manner, leading to cell migration, invasion, and distant metastasis, through activation of EGFR-FAK signaling (Fig. 9). FAK, a tyrosine kinase that localizes to focal adhesions, integrates signals from multiple intracellular pathways to control cell motility and invasion (37, 42). Hypoxia-induced invasion was abrogated by the knockdown of ADAM12 or by administration of Gefitinib or VS-4718 to inhibit EGFR or FAK activation, respectively. Taken together, our findings delineate a molecular mechanism by which ADAM12 activates a critical signal transduction pathway that triggers breast cancer cell motility, invasion, and metastasis.
Fig. 9.
HIF-mediated ADAM12 expression promotes hypoxia-induced migration, invasion, and metastasis. Schematic diagram of hypoxia-induced and HIF-dependent expression of ADAM12 mRNA and protein, which cleaves membrane-bound HB-EGF, thereby increasing EGFR-to-FAK signaling, cell migration, invasion, and distant metastasis.
ADAM12 is comprised of extracellular metalloprotease, disintegrin-like, cysteine-rich, and EGF-like domains, followed by a transmembrane domain and a cytoplasmic domain (43). In pituitary adenoma, ADAM12 was shown to induce epithelial to mesenchymal transition and promote cell migration and invasion (44). In esophageal cancer, ADAM12 overexpression forms a positive feedback loop with FAK to promote metastasis (45). In pancreatic cancer, ADAM12 is a circulating marker for stromal activation and predicts response to chemotherapy (46). The ADAM12 metalloprotease domain can cleave a range of transmembrane substrate proteins (16, 21, 43). Here, we report that ADAM12 expression is induced in hypoxic breast cancer cells, which increases HB-EGF ectodomain shedding and thereby augments EGFR signaling and downstream FAK activation.
We have previously shown that HIFs mediate enhanced motility of breast cancer cells by hypoxia-induced transactivation of RHOA, ROCK1, and ITGA5 gene expression (34, 36). In the present study, we dynamically monitored the random motility of cells for 30 h to evaluate the role of hypoxia and ADAM12 in breast cancer cell motility. No significant change in cell motility was observed during the first 14 h of exposure to hypoxia, which is consistent with the requirement for induction of ADAM12 mRNA and protein expression. Cell motility is a complex biological process involving coordinated changes in the interaction between cells and their extracellular environment. Many studies have highlighted that ADAM12 may mediate cellular and cell–matrix interaction through its disintegrin-like and cysteine-rich domains (47). ADAM12 knockdown has been shown to influence several β1-integrin–dependent functions, causing reorganization of the actin cytoskeleton and reduced cell attachment to extracellular matrix components, thereby regulating integrin-dependent cell migration (48, 49). Our data indicate that HIF-dependent ADAM12 signaling is required to induce not only cell motility, but also invasion through the extracellular matrix under hypoxic conditions. Knockdown of ADAM12 did not completely abrogate hypoxia-induced migration and invasion (Fig. 5), which is consistent with the independent effects of HIF-dependent RHOA/ROCK/ITGA5 (34, 36) and LOX/LOXL2/LOXL4 (50–52) gene expression on motility and invasion, respectively. The role of ADAM12 in promoting cell motility and invasion under hypoxia complements prior studies showing that ADAM12 contributes to the regulation of invadopodia and focal adhesions to promote metastasis (21, 53). Hypoxia-induced ADAM12 → HB-EGF → EGFR (Fig. 9) and RHOA → ROCK1 (34) signaling both converge on FAK and both signals appear to be required for efficient FAK activation in hypoxic breast cancer cells. Whereas signal transduction pathways can be activated in cancer cells by somatic mutations, these studies demonstrate that similar effects can also be triggered by intratumoral hypoxia.
As with all HIF target genes, we are left to answer the question: Is it more effective to target an individual HIF target gene product such as ADAM12 or to target HIF activity? Many metalloproteases of the ADAM and matrix metalloprotease families mediate invasion and many are encoded by HIF target genes (7). Indeed, our preliminary analysis suggests that ADAM8, -9, -17, and -19 are likely to be HIF-regulated in human breast cancers (Fig. 1A). Targeting HIF activity will block hypoxia-induced expression of hundreds of genes. Although the effect of HIF inhibition is broad, it is not as deep as targeted therapies, as HIF inhibition will not affect gene expression that is controlled by other transcriptional regulatory mechanisms. For example, NOTCH and TWIST1 (21, 53), as well as DNA hypomethylation (54), contribute to ADAM12 gene transcriptional regulation. On the other hand, there is considerable evidence that targeting individual metalloproteases is not an effective therapeutic strategy, which may be due to expression of multiple enzymes with overlapping specificities (55). Combining HIF inhibition with individualized targeted therapies may provide a more effective strategy for the prevention or control of metastatic disease.
Materials and Methods
Cell Culture and Reagents.
Breast cancer cell lines MCF-7, MDA-MB-231, and Hs578T were maintained in high-glucose (4.5 mg/mL) DMEM. SUM159 cells were maintained in DMEM/F12 (50:50) medium. SUM-149 cells were maintained in Ham’s F-12 media supplemented with hydrocortisone and insulin. HCC1954 and T47D cells were maintained in RPMI-1640 medium. All culture media were supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin. Cells were maintained at 37 °C in a 5% CO2, 95% air incubator (20% O2). Hypoxic cells were maintained at 37 °C in a modular incubator chamber (Billups–Rothenberg) flushed with a gas mixture containing 1% O2, 5% CO2, and 94% N2. Gefitinib (Sigma-Aldrich) and VS-4718 (TOCRIS) were dissolved in DMSO at 1,000× final concentration.
RT-qPCR Assays.
Total RNA was extracted using TRIzol (Invitrogen) and cDNA synthesis was performed using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s instructions. qPCR analysis was performed on the CFX96 Real-Time PCR detection system using SYBR Green qPCR master mix (Bio-Rad). The expression of each target mRNA relative to 18S rRNA was calculated based on the cycle threshold (Ct) as 2−Δ(ΔCt), in which ΔCt = Ct (target mRNA) – Ct (18S rRNA), and Δ(ΔCt) = ΔCt (treatment) − ΔCt (control). PCR primer sequences are shown in SI Appendix, Table S1.
Immunoblot Assays.
Whole-cell lysates were prepared in modified RIPA buffer (Millipore) supplemented with 1% protease inhibitor mixture and PMSF (Roche). Equal amounts of protein extract were separated by SDS/PAGE, electroblotted onto nitrocellulose membranes, and probed with primary antibodies (SI Appendix, Table S2). HRP-conjugated secondary antibodies were used and chemiluminescent signals were detected using ECL Plus (GE Healthcare).
ChIP-qPCR Assays.
As previously described (56), breast cancer cells were incubated at 20% or 1% O2 for 16 h, cross-linked in 3.7% formaldehyde for 15 min, quenched in 0.125 M glycine for 5 min, and lysed with SDS lysis buffer. Chromatin was sheared by sonication, and lysates were precleared with salmon sperm DNA/protein A agarose slurry (Millipore) and incubated with antibodies against HIF-1α, HIF-2α, or HIF-1β (Novus Biologicals) in the presence of protein A-agarose beads overnight. After serial washes of the agarose beads with low salt, high salt, and Tris-EDTA buffers, DNA was eluted in 1% SDS with 0.1 M NaHCO3, and cross-links were reversed by addition of 0.2 M NaCI. DNA was purified by phenol–chloroform extraction and ethanol precipitation, and candidate HIF binding sites were analyzed by qPCR (see SI Appendix, Table S3 for primer sequences).
Construction of HRE Reporter Plasmids.
Complementary oligonucleotides (see SI Appendix, Table S4 for sequences) were annealed and inserted into pGL2-Promoter (Promega), which contains a basal SV40 promoter upstream of firefly luciferase coding sequences.
Lentiviral Vectors and Transduction.
Lentiviral vectors encoding shRNA targeting HIF-1α and HIF-2α were described previously (29). pLKO.1-puro lentiviral vectors encoding shRNA targeting ADAM12 mRNA were purchased from Sigma-Aldrich, and shRNA sequences are shown in SI Appendix, Table S5. As previously described (56), lentiviral vectors were transfected into 293T cells for packaging. Viral supernatant was collected after 48 h and used for transfection as described previously (29). Puromycin (Sigma-Aldrich) was added to the medium of cells transduced with lentivirus for selection of stably transfected clones. Each subclone represents a pool of stably transfected cells that survived puromycin selection.
Cell Motility Measurements.
MDA-MB-231 cells were seeded on polystyrene tissue culture plates. SUM159 cells were seeded on collagen-coated six-well plates, which contained 1 mL of a 1-mg/mL solution of rat type I collagen (Corning, cat. no. 354236) per well. Cell movement over time was imaged using a Lionheart FX Automated Microscope (BioTek Instruments) maintained at 37 °C with controlled humidity. The microscope was maintained inside a customized sealed chamber (Coy Lab Products) equilibrated to 20% or 1% O2. Images were taken at 10× with an Olympus UPLFLN 10XPh phase objective every 10 min for 30 h in phase contrast. Cells in the time-lapse movies were tracked using MetaMorph software (Molecular Devices) to calculate x and y coordinates at each time interval. Cell displacement, , was calculated using the x and y coordinates of each cell (in micrometers) for each measurement recorded. Anisotropic persistent random walk model analysis was performed using MATLAB (MathWorks) as described previously (35). Cell trajectory data were used to statistically profile cell migration using the mean squared displacement.
Boyden Chamber Migration and Invasion Assays.
Transwell chambers with 8-μm pore membrane inserts, which were either uncoated or coated with Matrigel (Corning), were used to assay migration and invasion, respectively. A total number of 5 × 104 (migration) or 1 × 105 (invasion) cells were resuspended in serum-free medium, seeded onto the upper membrane of a Transwell insert (migration) or a Matrigel-coated Transwell insert (invasion) in the presence of cell culture medium that was supplemented with 10% FBS in the bottom chamber, and exposed to 20% or 1% O2 for 16 h (migration) or 24 h (invasion). Cells on the bottom side of the membrane were fixed with paraformaldehyde, permeabilized with methanol, stained with 1% Crystal violet at room temperature, and counted in five randomly selected fields under bright field microscopy. To examine the effect of HB-EGF on cell invasion, a NAb to HB-EGF (10 μg/mL; R&D Systems) was added to the medium.
ELISA.
Cells were exposed to 20% or 1% O2 for 72 h and shed HB-EGF protein in the conditioned media was measured using an ELISA kit (R&D Systems) according to the manufacturer’s instructions.
Animal Studies.
Animal protocols were approved by the Johns Hopkins University Animal Care and Use Committee, and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (57). Two million MDA-MB-231 or SUM159 subclone cells were injected into the mammary fat pad of 5- to 7-wk-old female SCID mice in a 1:1 (vol:vol) suspension of Matrigel (BD Biosciences) and PBS. Primary tumors were measured for length (a) and width (b), and the volume (V) was calculated according to the following formula: V = ab2 × 0.52. Tumors and lungs were harvested at the end of the experiment. One lung was inflated for formalin fixation, paraffin embedding, and hematoxylin and eosin staining; the other lung was harvested to isolate genomic DNA by digestion with proteinase K, phenol–chloroform extraction, and isopropanol precipitation. A 200-ng aliquot of genomic DNA was used for qPCR with human-specific HK2 gene primers and mouse 18S rRNA gene primers.
Bioinformatics and Statistics.
Cancer datasets were downloaded from Oncomine (Compendia Bioscience, https://www.oncomine.com/). The rank and P value for ADAM12 differential mRNA expression between breast carcinoma and normal breast tissue across the 16 datasets from 6 independent studies (58–63) were obtained directly from the Oncomine website. A comprehensive gene-expression microarray dataset of 296 primary breast tumors (38) was used to compare ADAM12 mRNA levels in metastatic versus nonmetastatic breast cancer. TCGA breast cancer data were obtained from an online data portal (https://xenabrowser.net/). The Pearson correlation test was used to compare expression of ADAM family genes with the HIF signature, based on mRNA levels from TCGA BRCA dataset (64). Prognostic significance of ADAM12 expression in breast cancer was performed using patient data obtained from the bcGenExMiner (39) and GOBO (40) online databases. Survival plots were created using Kaplan–Meier methods and the log rank test was performed. All data are presented as mean ± SEM unless otherwise noted. Differences between two groups were analyzed by Student’s t test, whereas differences between multiple groups were analyzed by ANOVA. For all statistical tests, P < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Weibo Luo (University of Texas Southwestern Medical Center, Dallas) for generously providing CRISPR knockout subclones of MDA-MB-231 cells; Samantha Garcia and Rachel Geisler (Novus Biologicals) for generously providing antibodies against HIF-1α, HIF-2α, HIF-1β, phospho-EGFR, and HB-EGF; and Byung Hyun Yi for assistance with cell motility measurements. G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at the Johns Hopkins University School of Medicine. This work was supported by grants from the American Cancer Society, Armstrong Family Foundation, and the Cindy Rosencrans Fund for Triple-Negative Breast Cancer (to G.L.S.), and the China Scholarship Council (to R.W.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020490118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Bray F., et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Harris A. L., Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Semenza G. L., The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim. Biophys. Acta 1863, 382–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araos J., Sleeman J. P., Garvalov B. K., The role of hypoxic signalling in metastasis: Towards translating knowledge of basic biology into novel anti-tumour strategies. Clin. Exp. Metastasis 35, 563–599 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Lu X., Kang Y., Hypoxia and hypoxia-inducible factors: Master regulators of metastasis. Clin. Cancer Res. 16, 5928–5935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schito L., Semenza G. L., Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer 2, 758–770 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Yuen V. W., Wong C. C., Hypoxia and innate immunity in liver cancer. J. Clin. Invest. 130, 5052–5062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Heer E. C., Jalving M., Harris A. L., HIFs, angiogenesis, and metabolism: Elusive enemies in breast cancer. J. Clin. Invest. 130, 5074–5087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schindl M.et al.; Austrian Breast and Colorectal Cancer Study Group , Overexpression of hypoxia-inducible factor 1α is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin. Cancer Res. 8, 1831–1837 (2002). [PubMed] [Google Scholar]
- 11.Bos R., et al., Levels of hypoxia-inducible factor-1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer 97, 1573–1581 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Dales J. P., et al., Overexpression of hypoxia-inducible factor HIF-1α predicts early relapse in breast cancer: Retrospective study in a series of 745 patients. Int. J. Cancer 116, 734–739 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Generali D., et al., Hypoxia-inducible factor-1α expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin. Cancer Res. 12, 4562–4568 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki S., Okada Y., ADAMs in cancer cell proliferation and progression. Cancer Sci. 98, 621–628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards D. R., Handsley M. M., Pennington C. J., The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loechel F., Gilpin B. J., Engvall E., Albrechtsen R., Wewer U. M., Human ADAM 12 (meltrin α) is an active metalloprotease. J. Biol. Chem. 273, 16993–16997 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Xiang L., Semenza G. L., Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv. Cancer Res. 141, 175–212 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Duhachek-Muggy S., et al., Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol. Cancer 16, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Duhachek-Muggy S., Dubnicka S., Zolkiewska A., Metalloproteinase-disintegrin ADAM12 is associated with a breast tumor-initiating cell phenotype. Breast Cancer Res. Treat. 139, 691–703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Pabic H., et al., ADAM12 in human liver cancers: TGF-β-regulated expression in stellate cells is associated with matrix remodeling. Hepatology 37, 1056–1066 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Díaz B., Yuen A., Iizuka S., Higashiyama S., Courtneidge S. A., Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J. Cell Biol. 201, 279–292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romagnoli M., et al., ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis. EMBO Mol. Med. 6, 278–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlomann U., et al., ADAM8 as a drug target in pancreatic cancer. Nat. Commun. 6, 6175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szalad A., Katakowski M., Zheng X., Jiang F., Chopp M., Transcription factor Sp1 induces ADAM17 and contributes to tumor cell invasiveness under hypoxia. J. Exp. Clin. Cancer Res. 28, 129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H.et al., Chemotherapy-induced Ca2+ release stimulates breast cancer stem cell enrichment. Cell Rep. 18, 1946–1957 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., et al., Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc. Natl. Acad. Sci. U.S.A. 105, 19579–19586 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K., et al., Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U.S.A. 106, 17910–17915 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Chen Y., et al., ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 128, 1937–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., et al., HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene 31, 1757–1770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Semenza G. L., et al., Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Palmer T. D., Ashby W. J., Lewis J. D., Zijlstra A., Targeting tumor cell motility to prevent metastasis. Adv. Drug Deliv. Rev. 63, 568–581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells A., Grahovac J., Wheeler S., Ma B., Lauffenburger D., Targeting tumor cell motility as a strategy against invasion and metastasis. Trends Pharmacol. Sci. 34, 283–289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trepat X., Chen Z., Jacobson K., Cell migration. Compr. Physiol. 2, 2369–2392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilkes D. M., et al., Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 111, E384–E393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Wu P.-H., Giri A., Wirtz D., Statistical analysis of cell migration in 3D using the anisotropic persistent random walk model. Nat. Protoc. 10, 517–527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ju J. A., et al., Hypoxia selectively enhances Integrin α5β1 receptor expression in breast cancer to promote metastasis. Mol. Cancer Res. 15, 723–734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra S. K., Hanson D. A., Schlaepfer D. D., Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 (2005). [DOI] [PubMed] [Google Scholar]
- 38.van de Vijver M. J., et al., A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Jézéquel P., et al., bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 131, 765–775 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Ringnér M., Fredlund E., Häkkinen J., Borg Å., Staaf J., GOBO: Gene expression-based outcome for breast cancer online. PloS One 6, e17911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buffa F. M., Harris A. L., West C. M., Miller C. J., Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer 102, 428–435 (200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlaepfer D. D., Mitra S. K., Ilic D., Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim. Biophys. Acta 1692, 77–102 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Kveiborg M., Albrechtsen R., Couchman J. R., Wewer U. M., Cellular roles of ADAM12 in health and disease. Int. J. Biochem. Cell Biol. 40, 1685–1702 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Wang J., et al., ADAM12 induces EMT and promotes cell migration, invasion and proliferation in pituitary adenomas via EGFR/ERK signaling pathway. Biomed. Pharmacother. 97, 1066–1077 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Luo M. L., et al., An ADAM12 and FAK positive feedback loop amplifies the interaction signal of tumor cells with extracellular matrix to promote esophageal cancer metastasis. Cancer Lett. 422, 118–128 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Veenstra V. L., et al., ADAM12 is a circulating marker for stromal activation in pancreatic cancer and predicts response to chemotherapy. Oncogenesis 7, 87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bridges L. C., Bowditch R. D., ADAM-integrin interactions: Potential integrin regulated ectodomain shedding activity. Curr. Pharm. Des. 11, 837–847 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi N., et al., ADAM12 induces actin cytoskeleton and extracellular matrix reorganization during early adipocyte differentiation by regulating β1 integrin function. J. Cell Sci. 116, 3893–3904 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Huang J., Bridges L. C., White J. M., Selective modulation of integrin-mediated cell migration by distinct ADAM family members. Mol. Biol. Cell 16, 4982–4991 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erler J. T., et al., Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 (2006). Retracted in: Nature579, 456 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Wong C. C., et al., Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc. Natl. Acad. Sci. U.S.A. 108, 16369–16374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong C. C., et al., Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. (Berl.) 90, 803–815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eckert M. A., et al., ADAM12 induction by Twist1 promotes tumor invasion and metastasis via regulation of invadopodia and focal adhesions. J. Cell Sci. 130, 2036–2048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendaza S., et al., ADAM12 is a potential therapeutic target regulated by hypomethylation in triple-negative breast cancer. Int. J. Mol. Sci. 21, 903–919 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kessenbrock K., Plaks V., Werb Z., Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 141, 52–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang T., et al., Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. U.S.A. 111, E3234–E3242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 58.Radvanyi L., et al., The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 11005–11010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turashvili G., et al., Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 7, 55 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma X. J., Dahiya S., Richardson E., Erlander M., Sgroi D. C., Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 11, R7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gluck S., et al., TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabije ± trastuzumab. Breast Cancer Res. Treat. 132, 7871–791 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Finak G., et al., Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14, 518–527 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Curtis C.et al.; METABRIC Group , The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Network , Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.