Significance
The functional role of the N-terminal tail (NT) of galectin-3 (Gal-3) with its numerous proline residues (27 out of 113 in human Gal-3) has remained enigmatic since its discovery. Our study enlightens the galectin field on the role of specific Gal-3 NT prolines and opens a window into understanding the mechanisms of Gal-3 proline polymorphisms (e.g., at P64) in various pathological disorders. In addition, we show that Gal-3 oligomerization, triggered by binding to glycoconjugates either in solution or on the plasma membrane and disrupted endomembranes, is a dynamic process analogous to liquid–liquid phase separation (LLPS). This dynamic behavior of Gal-3 illuminates a mechanism underlying the formation, regulation, and function of clusters of glycosylated cell surface receptors.
Keywords: galectin-3, proline-rich protein, phase separation, oligomerization, glycan
Abstract
Galectin-3 (Gal-3) has a long, aperiodic, and dynamic proline-rich N-terminal tail (NT). The functional role of the NT with its numerous prolines has remained enigmatic since its discovery. To provide some resolution to this puzzle, we individually mutated all 14 NT prolines over the first 68 residues and assessed their effects on various Gal-3–mediated functions. Our findings show that mutation of any single proline (especially P37A, P55A, P60A, P64A/H, and P67A) dramatically and differentially inhibits Gal-3–mediated cellular activities (i.e., cell migration, activation, endocytosis, and hemagglutination). For mechanistic insight, we investigated the role of prolines in mediating Gal-3 oligomerization, a fundamental process required for these cell activities. We showed that Gal-3 oligomerization triggered by binding to glycoproteins is a dynamic process analogous to liquid–liquid phase separation (LLPS). The composition of these heterooligomers is dependent on the concentration of Gal-3 as well as on the concentration and type of glycoprotein. LLPS-like Gal-3 oligomerization/condensation was also observed on the plasma membrane and disrupted endomembranes. Molecular- and cell-based assays indicate that glycan binding–triggered Gal-3 LLPS (or LLPS-like) is driven mainly by dynamic intermolecular interactions between the Gal-3 NT and the carbohydrate recognition domain (CRD) F-face, although NT–NT interactions appear to contribute to a lesser extent. Mutation of each proline within the NT differentially controls NT–CRD interactions, consequently affecting glycan binding, LLPS, and cellular activities. Our results unveil the role of proline polymorphisms (e.g., at P64) associated with many diseases and suggest that the function of glycosylated cell surface receptors is dynamically regulated by Gal-3.
Galectin-3 (Gal-3) is expressed in a variety of tissues and cell types and is involved in diverse physiological and pathological processes (1–3). As the only chimeric member of the galectin family, Gal-3 consists of a long N-terminal tail (NT) and a highly conserved carbohydrate recognition domain (CRD) at the C terminus. The CRD amino acid sequence folds as a β-sandwich with two β-sheet faces (the sugar-binding S-face and opposing F-face) (4). Whereas the CRD S-face presents the carbohydrate binding site, the aperiodic and highly dynamic NT is not integral to the folded structure (5), has questionable involvement in the proposed pentamer formation (6), and has little apparent effect on carbohydrate binding to the CRD canonical site (5), yet it is crucial to the functional activity of this unique galectin (7). A mysterious feature of the NT is the high incidence of prolines. Human Gal-3 has 250 amino acid residues, 113 of which belong to the NT, with a total of 27 prolines that account for ∼25% of the NT or approximately four times that found in an average protein sequence. Moreover, the positions of prolines within the N-terminal part of the NT of Gal-3 are highly conserved across species, yet their presence and functional value have long puzzled researchers.
Similar to other galectins, Gal-3 is secreted from cells to the extracellular milieu via a nonclassical secretory pathway (8, 9) and binds to glycoconjugates on the cell surface and in the extracellular matrix (ECM) (10). Gal-3 mediates a spectrum of cell processes, including cell surface receptor clustering and lattice formation (11–13), the formation of lipid rafts/microdomains (14, 15), the biogenesis of clathrin-independent carriers (CLIC) (15), the adhesion of cells to the ECM (16), and cell agglutination (17, 18). Gal-3 is monomeric in solution with only one carbohydrate recognition site, yet it remains unknown how Gal-3 achieves the multivalence prerequisite for receptor cross-linking. This longstanding query has been subjected to multiple investigations that yielded no conclusion. It has been proposed that Gal-3 can self-associate into oligomers upon binding to multivalent ligands. It was also proposed that Gal-3 may associate as a pentamer through NT–NT interactions (6). However, other studies claim that Gal-3 forms dimers (19) and higher-order structures (20). Recently, CRD–CRD interactions have been detected on the cell surface, resulting in the termination of Gal-3 function (20, 21). Furthermore, we have shown by NMR studies that the NT interacts transiently with the CRD F-face (5).
Recently, a distinct mode of molecular interaction (liquid–liquid phase separation, LLPS) has emerged as an important modulator for some biological events. In the cytoplasm and nucleoplasm, LLPS contributes to the formation of condensates, such as stress granules, nucleolus, and Cajal bodies (22, 23), and is involved in the regulation of DNA transcription (24), cell cycle control (25, 26), autophagy degradation (27), cancer suppression (28), and nuclear pore passage (29, 30). This emerging field has been reviewed extensively (31). With the expansion of LLPS studies, researchers have extended the recognition of LLPS to the cell membrane. The roles of phase separation in cell–cell junctions (32, 33), signal transmission between neurons (34), and regulation of transmembrane signaling (35) have been reported.
Given that Gal-3 has an intrinsically disordered and dynamic NT leading to unusual oligomerization, it is reasonable to examine whether Gal-3 forms an extended assembly like LLPS, rather than a stationary oligomer, since high concentrations of Gal-3 can form fuzzy structures as determined by NMR and small-angle X-ray scattering (36), and Gal-3 forms liquid droplets at high salt concentrations (37). Although these studies are informative, the results were obtained under nonphysiological conditions. Whether phase separation occurs on the cell membranes and how it is initiated, elongated, and/or regulated remain unknown. Thus, this study addresses the issue of Gal-3 oligomerization/phase separation on cell membranes, with a particular focus on the mode of action of the Gal-3 NT and its enigmatic proline residues in these processes.
Results
Prolines within the Gal-3 NT Differentially Direct Cell Activity.
The NT of human Gal-3 has 113 residues, with the initial 68 residues and 14 highly conserved prolines being crucial to Gal-3 biological function (38, 39). Because of this, we produced a series of 14 alanine-substituted mutants at these NT prolines (SI Appendix, Fig. S1A) to assess how each one affects cell-based activities. In addition, we investigated Gal-3 mutant P64H, a natural polymorphism associated with many diseases (40–43). The addition of exogenous Gal-3 elicits various functional effects on cell migration, activation, adhesion/agglutination, and on the endocytosis and biogenesis of CLIC. In each cell-based assay, we assessed the role of Gal-3 NT prolines by comparing effects from wild-type (WT) Gal-3 and these 14 NT proline mutants.
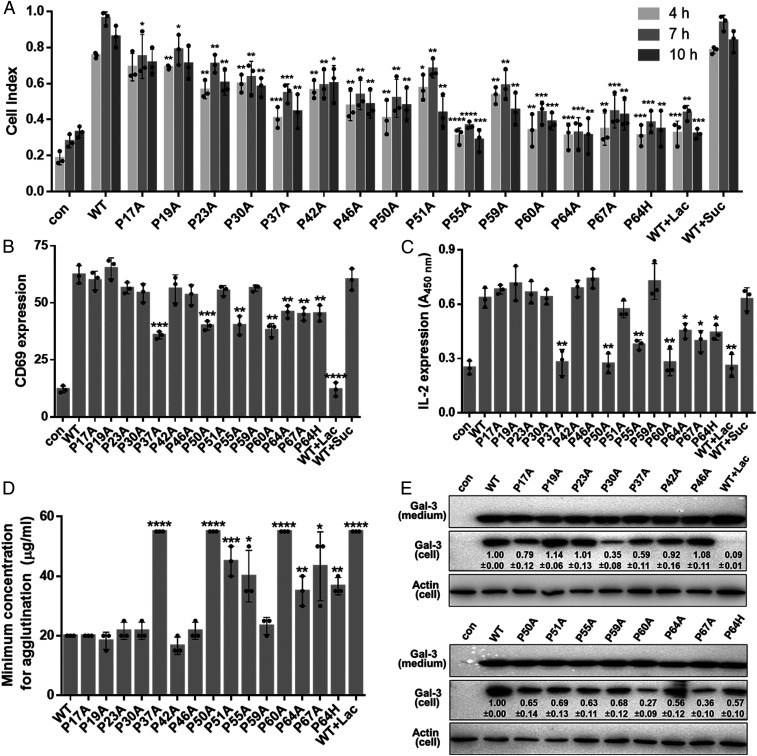
WT Gal-3 induces human microvascular endothelial cell (HMEC-1) migration by interacting with CD146 (44). Fig. 1A and SI Appendix, Fig. S2 A and B show the effects of WT Gal-3 and its NT proline mutants on HMEC-1 cell migration. Compared to WT Gal-3, HMEC-1 migration is attenuated by ∼10 to 15% with P17A and P19A; by ∼20 to 40% with P23A, P30A, P42A, P46A, P50A, P51A, and P59A; by ∼40 to 50% with P37A, P60A, and P67A; and by >60% with P55A and P64A. P64H has a similar effect to P64A (Fig. 1A and SI Appendix, Fig. S2B). Moreover, most of these NT proline mutants elicit their peak effects at times less than the ∼7-h interval observed for WT Gal-3, with the effect being dependent on glycan binding because it is inhibited by lactose (Lac) (Fig. 1A and SI Appendix, Fig. S2B).
Fig. 1.
The function of NT prolines in various cell processes. (A) Proline substitutions inhibit Gal-3–induced HMEC-1 migration. HMEC-1 migration induced by 0.7 µM WT Gal-3 (with or without Lac or sucrose [Suc]) or Gal-3 NT proline mutants was monitored in real time by using xCELLigence real-time cell analyzer (SI Appendix, Fig. S2 A and B). Quantitative data are shown here. Con means without attractant. (B and C) Proline substitutions inhibit Gal-3–induced T cell activation. Jurkat cells were treated with 1.0 µM WT Gal-3 (with or without Lac or Suc) or its NT proline mutants, followed by the detection of CD69 expression by flow cytometry (B) or of IL-2 secretion by ELISA (C). (D) Quantitative data showing chicken erythrocyte agglutination in the presence of increasing concentrations of WT Gal-3 (with or without Lac) or its NT proline mutants. (E) Effect of proline mutants on endocytosis. Values in each lane are relative intensities of the bands of Gal-3 mutants, shown as the mean ± SEM (n = 3). In A–D, WT Gal-3 and each NT proline mutant are compared, and data are shown as the mean ± SD (n = 3). P values were determined by using Student’s two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We examined the effects of WT Gal-3 and its NT proline mutants on T cell activation with two readouts: CD69 expression on the cell surface by flow cytometry analysis and IL-2 cell release by enzyme-linked immunosorbent assay (ELISA). Although most NT proline mutants showed no apparent effects on CD69 expression relative to WT Gal-3, six of them (P37A, P50A, P55A, P60A, P64A, and P67A) did significantly inhibit CD69 expression, with P37A and P50A having the greatest effect (Fig. 1B). Similar trends were observed in the IL-2 assay, with P37A, P50A, P55A, P60A, P64A, and P67A significantly suppressing the release of IL-2 (Fig. 1C). Again, P64H and P64A displayed similar effects in both the CD69 expression and IL-2 release assays (Fig. 1 B and C). Thus, we find that a subset of NT prolines is crucial to T cell activation, and, because Lac inhibits this activity, its function is correlated to glycan binding.
Hemagglutination is commonly used to assess galectin function as a model of cell–cell adhesion. Using chicken erythrocytes [that are highly sensitive to Gal-3 (45)], we observe that agglutination is most attenuated by P37A, P50A, and P60A, with less significant effects from P51A, P55A, P64A, and P67A (Fig. 1D and SI Appendix, Fig. S3F). P64H and P64A showed similar effects. With human erythrocytes (AB, A, B, and O types), the same set of prolines renders essentially the same trend, albeit less pronounced (SI Appendix, Fig. S3 B–E), in all red blood cell types, but type O was the least sensitive to NT proline substitutions. Although it is unclear as to why this is, we hypothesize that the answer lies in blood group glycan structures themselves. Type O has a linear glycan chain, and types A and B have branched glycan chains with an additional sugar residue (SI Appendix, Fig. S3A).
The effect of NT proline mutations on Gal-3–triggered CLIC formation and endocytosis was investigated by treating HMEC-1 cells with these mutants and examining their intracellular content (Fig. 1E). Endocytosis with NT mutants P30A, P60A, and P67A was drastically reduced compared to that of WT Gal-3. P64A and P64H displayed similar effects. Interestingly, this set of prolines involved in endocytosis was not the same as that affecting cell migration (Fig. 1A), even though both assays were performed using the same cell line. Overall, our cell-based assays demonstrate that subsets of NT prolines are crucial to Gal-3–mediated activities that are mediated by Gal-3 binding to cell surface glycans.
Glycoprotein–Gal-3 Binding Induces Oligomerization.
The varied and differential functions of prolines within the Gal-3 NT imply that these residues are involved in a common process governing Gal-3–mediated function. Gal-3 oligomerization is fundamentally important to cellular responses (12). Thus, we focused on Gal-3 oligomerization for mechanistic insight into the biological role of NT prolines. To achieve this goal, we employed the model of LLPS, a distinct mode of molecular interaction that has recently emerged as an important modulator of some biological events. LLPS is usually performed with proteins at high salt concentrations (46), conditions that are not truly biologically relevant. Because extracellular Gal-3 exerts most of its function by binding to glycoconjugates, we questioned whether glycoconjugates can mediate LLPS. Here, we explored the possibility of glycoproteins (GPs) CD146, CD45, CD71, and CD7 triggering Gal-3–based phase separation, monitored by microscopic imaging, solution turbidity, and droplet size measured by dynamic light scattering (DLS).
For comparison with previously published reports, we initially performed LLPS experiments with Gal-3 in high salt and found that droplet formation solely relies on the NT, with residues 13 to 68 being most essential. The CRD is not required, and NT–NT hydrophobic interactions apparently provide the driving force. Detailed information is presented in SI Appendix, Supplementary Results and Fig. S4.
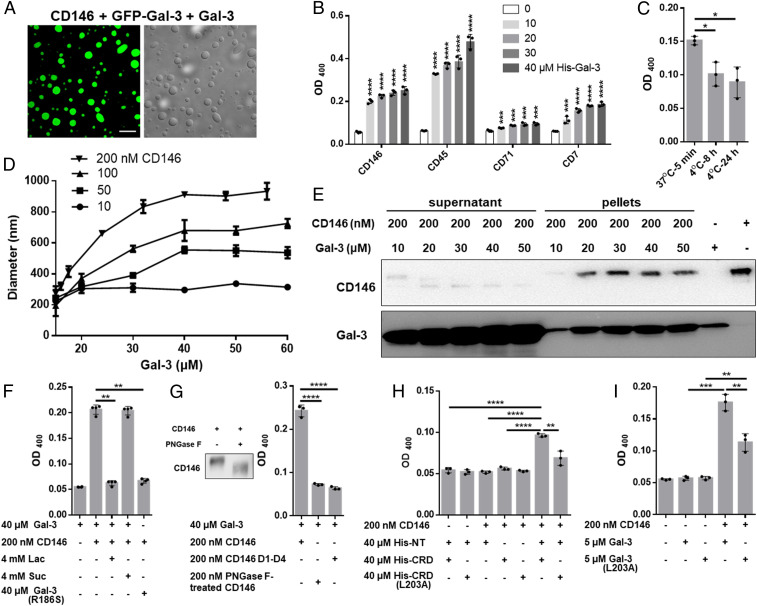
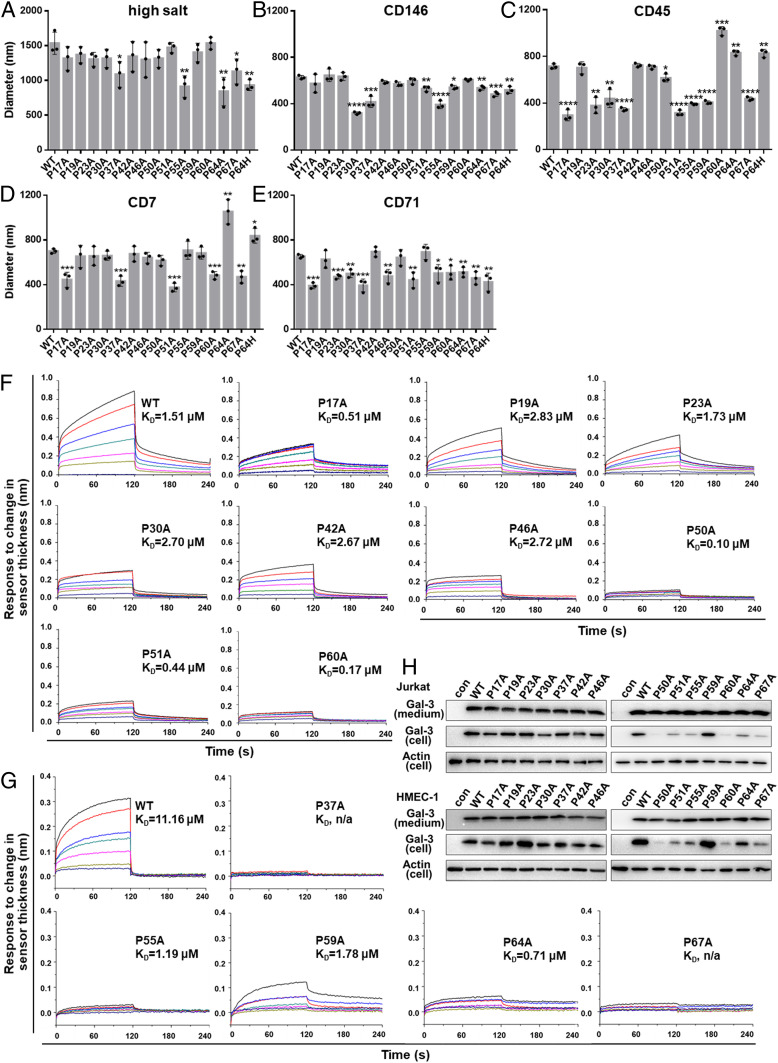
Next, we found that Gal-3–based droplets form with GPs CD146, CD45, CD71, and CD7 in the absence of high salt (Fig. 2 A and B). Under the same conditions, neither Gal-1 nor Gal-2 induces this effect (SI Appendix, Fig. S5A). Unlike high salt-induced LLPS, GP-induced droplet formation is only partially dependent on temperature (Fig. 2C), suggesting that both hydrophobic and hydrophilic interactions provide the driving force in this instance. In addition, droplet size is dependent on the concentrations of both Gal-3 and any of these GPs (i.e., CD146, CD45, CD7, and CD71), with droplet size increasing with Gal-3 concentration and leveling off at diameters of 250, 500, 700, and 900 nm when the CD146 concentration is 10, 50, 100, and 200 nM, respectively (Fig. 2D and SI Appendix, Fig. S5B). Gal-3:CD146 droplet stoichiometry was assessed in sedimentation experiments, in which protein content was analyzed in pellets and supernatant (Fig. 2E). The Gal-3:CD146 stoichiometry was calculated to be 26:1, 46:1, 61:1, 80:1, and 82:1 at 10, 20, 30, 40, and 50 μM Gal-3, respectively (SI Appendix, Table S1), showing that droplet compositions are Gal-3 dependent.
Fig. 2.
Gal-3 oligomerization/LLPS is induced by GPs. (A) Confocal and differential interference contrast images showing Gal-3 droplets in the presence of CD146 (4 μM GFP–Gal-3 + 250 μM Gal-3 + 400 nM CD146). (Scale bar, 10 μm.) (B) Turbidity of Gal-3 in the presence of 200 nM each GP. (C) Effect of temperature on droplet stability after 8 and 24 h standing at 4 °C. (D) Size of droplets at different concentrations of Gal-3 and CD146 measured by DLS. The Gal-3 concentrations were 15, 16, 17.5, 24, 32, 40, 48, and 56 μM for the 200 nM CD146 curve and 15, 20, 30, 40, 50, and 60 μM for the other curves. (E) Sedimentation analysis. CD146 was mixed with Gal-3 to allow phase separation followed by sedimentation. Gal-3 and CD146 levels in the supernatant and pellets were detected by Western blotting. The CD146 sample (200 nM fixed concentration) used here consists mostly of highly glycosylated species (major gel band at 120 kDa) with which Gal-3 primarily interacts. (F) Turbidity of Gal-3 or Gal-3(R186S) in the presence of CD146 with or without Lac and Suc. (G) Turbidity of Gal-3 in the presence of CD146 D1 to D4 or PNGase F-treated CD146. (H) Turbidity of Gal-3 variants or their combinations with or without 200 nM CD146. (I) Turbidity of 5 μM WT Gal-3 or mutant Gal-3(L203A) in the presence of 200 nM CD146. Turbidity data are shown as the mean ± SD (n ≥ 3). P values were determined by using Student’s two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Droplet formation depends on glycan binding because of the following: 1) Lac, but not Suc, inhibits the assembly (Fig. 2F); 2) Gal-3(R186S), a mutant that strongly reduces glycan affinity (47), barely induces droplet formation (Fig. 2F); 3) CD146 D1 to D4 [a truncated form of CD146 absent highly glycosylated domain 5 (44)], or PNGase F-treated CD146 that depletes bound N-glycans, attenuates droplet formation (Fig. 2G); and 4) the NT alone does not induce droplet formation (SI Appendix, Fig. S5C). The predominantly hydrophobic NT and hydrophilic CRD are both required for CD146-mediated droplet formation (Fig. 2H), with NT residues 13 to 68 being the most crucial (SI Appendix, Fig. S5C). Nevertheless, the CRD and NT need not be covalently linked because simply mixing free truncated CRD and free NT peptide is sufficient (Fig. 2H). In addition, the mutation of Gal-3 CRD F-face residue L203 to alanine (L203A) in truncated CRD or in full-length Gal-3 significantly reduces droplet formation (Fig. 2 H and I), an observation that is consistent with interactions between the NT and CRD F-face (5).
The best model that appears to explain these data is one in which the Gal-3 canonical CRD S-face binds to glycans on GPs, allowing the free CRD F-face to interact with NTs from additional Gal-3 molecules, thus establishing a polymerization-like network of Gal-3/CD146 molecules. Confocal microscopy shows that the spherical droplets formed by these associations are solid structures filled throughout with this lattice network as opposed to buffer-filled vesicles with an outer membrane layer formed by the Gal-3/CD146 network (Fig. 2A).
Glycan-Triggered Gal-3 Oligomerization on Cell Membranes.
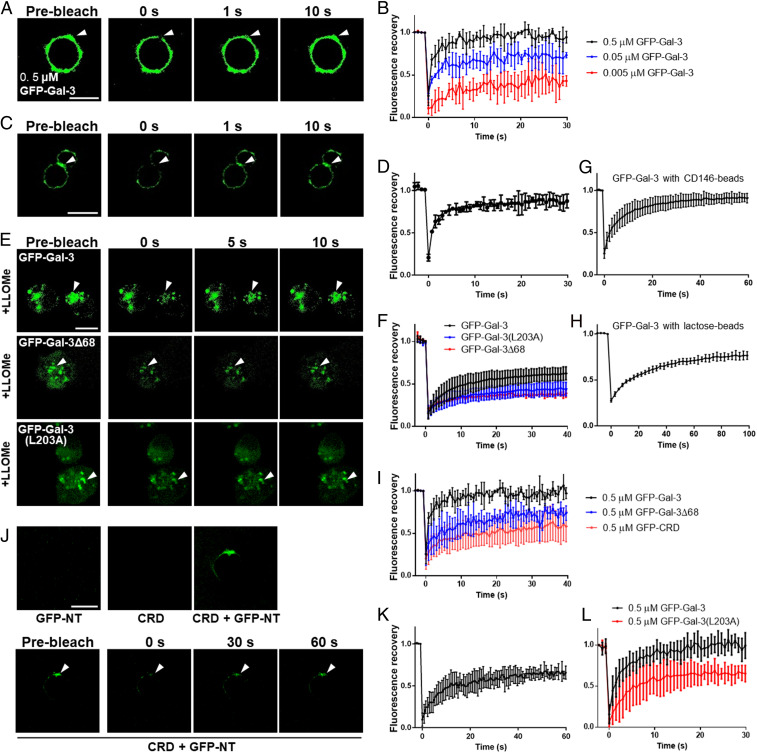
Three cell-based assays model three distinct Gal-3 oligomerization states. First, we assessed Gal-3 clustering on the cell surface by incubating fixed T cells (to avoid receptor internalization and lateral movement) with GFP–Gal-3, photobleaching the mixture, and measuring fluorescence recovery. GFP–Gal-3 cell binding was found to be glycan dependent (SI Appendix, Fig. S6A), with fluorescence quickly recovering after photobleaching (Fig. 3 A and B and SI Appendix, Fig. S6B). Next, we assessed Gal-3 accumulation between cells in the Gal-3–mediated adhesion/agglutination of human erythrocytes of type AB and discovered that fluorescence bridging the cells recovered quickly after photobleaching (Fig. 3 C and D). Finally, we expressed GFP–Gal-3 (as well as GFP–Gal-3(R186S), GFP–Gal-3(L203A), and GFP–Gal-3Δ68) in human embryonic kidney (HEK) 293T cells and assessed recruitment of cytosolic Gal-3 to damaged endomembranes, a glycan-dependent process related to cell stress and microbial infection (48–50). For this purpose, we used a model of lysosomal membrane damage induced by Leu-Leu methyl ester hydrobromide (LLOMe). As shown in SI Appendix, Fig. S6C, GFP–Gal-3 was expressed in some cells and evenly distributed in the cytoplasm but accumulated in puncta upon LLOMe treatment, verifying the accumulation of cytosolic GFP–Gal-3 on the damaged membranes. After photobleaching, fluorescence recovered quickly (Fig. 3 E and F), with a maximum recovery of ∼50%. These data indicate that membrane-bound Gal-3 exchanges rapidly with Gal-3 in the solution or in the cytosol, analogous to GP-mediated Gal-3 LLPS. Gal-3 exchange was also observed using CD146-coated latex beads (Fig. 3G and SI Appendix, Fig. S6D) or Lac-coated latex beads (Fig. 3H and SI Appendix, Fig. S6E), supporting the conclusion that glycan binding initiates this fast exchange process. Fluorescence recovery was significantly attenuated when the first 68 NT residues (GFP–Gal-3Δ68) were deleted (Fig. 3 E, F, and I) and more so when the entire NT (GFP–CRD) was deleted (Fig. 3I), consistent with the full NT sequence being required for maximal effect. The data also suggest that CRD–CRD interactions may also contribute to Gal-3 condensation on cell membranes.
Fig. 3.
GP-triggered Gal-3 oligomerization on cell membranes. (A) Confocal images before and after photobleaching of Jurkat cells with GFP–Gal-3. (B) Quantitative FRAP data of A. (C) Confocal images before and after photobleaching of human erythrocytes type AB with GFP–Gal-3. (D) Quantitative FRAP data of C. (E) Confocal images before and after photobleaching of HEK 293T cells expressing GFP–Gal-3, GFP–Gal-3(L203A), or GFP–Gal-3Δ68 treated with LLOMe. (F) Quantitative FRAP data of E. (G) FRAP data from CD146-coated beads with GFP–Gal-3. (H) FRAP data from Lac-coated beads with GFP–Gal-3. (I) FRAP data from Jurkat cells with GFP–Gal-3, GFP–Gal-3Δ68, or GFP-CRD. (J) Assessment of NT–CRD interactions. Cells were treated with CRD, GFP-NT, or combinations thereof. (K) Quantitative FRAP data of J. (L) FRAP data from Jurkat cells with GFP–Gal-3 or GFP–Gal-3(L203A). FRAP data are shown as the mean ± SD (n ≥ 5). (Scale bar, 10 μm.)
To assess the involvement of intermolecular NT–CRD interactions, we incubated fixed T cells with GFP-NT, unlabeled Gal-3 CRD, and combinations thereof. However, only cells incubated with the combination fluoresced (Fig. 3J), indicating that the CRD promotes recruitment of GFP-NT. Furthermore, photobleaching of these cells allows >50% fluorescence recovery, suggesting that NT–CRD interactions drive NT condensation on the cell surface (Fig. 3K). In support of this, we observed that Gal-3 mutant L203A [that significantly attenuates NT–CRD interactions (5)] displays reduced florescence recovery compared to WT GFP–Gal-3 (Fig. 3L). When expressed in HEK 293T cells, cytosolic GFP–Gal-3(L203A) shows reduced accumulation on LLOMe treatment and reduced florescence recovery following photobleaching compared to WT Gal-3 (Fig. 3 E and F and SI Appendix, Fig. S6C). Interestingly, GFP–Gal-3(L203A) condensation/accumulation was not completely lost, possibly because of CRD–CRD or NT–NT interactions.
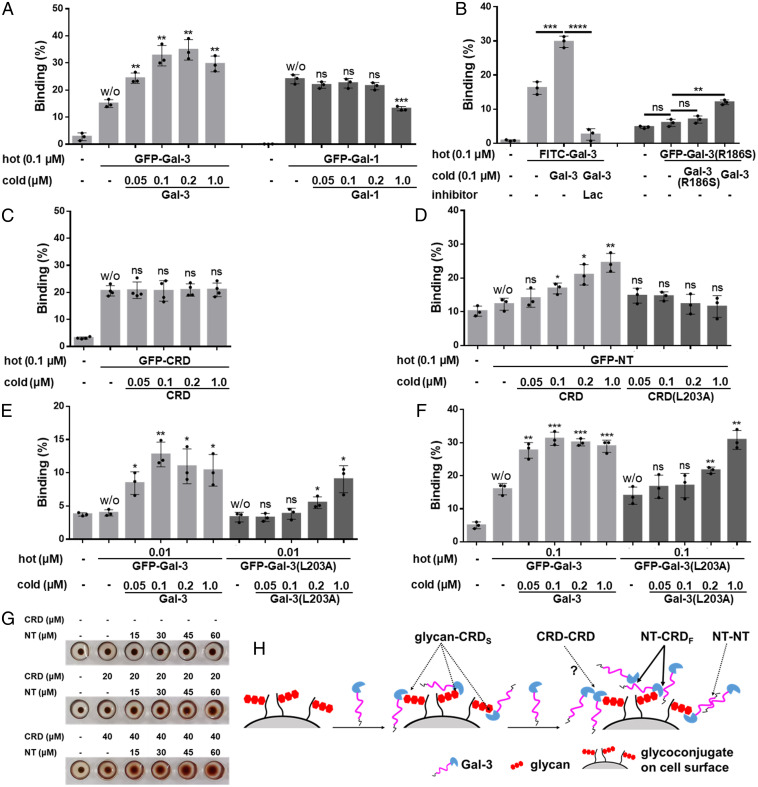
In addition, we used a T cell surface binding assay with fluorescently labeled Gal-3 (hot Gal-3) and unlabeled Gal-3 (cold Gal-3). Classically, cell surface binding of hot Gal-3 should be decreased upon the addition of cold Gal-3 because of competition for cell surface receptors. Surprisingly, the binding of hot Gal-3 (0.1 μM GFP–Gal-3 or FITC–Gal-3) was not attenuated upon the addition of cold Gal-3, but, rather, it was enhanced in contrast to the use of Gal-1 (Fig. 4 A and B and SI Appendix, Fig. S7 A and B). This cannot be explained with a simple model of cell surface binding (i.e., one ligand:one receptor) but rather fits well with an LLPS/condensation model. Lac totally inhibits Gal-3 binding and condensation (Fig. 4B and SI Appendix, Fig. S7B). Gal-3 mutant R186S shows minimal binding to cells, although its binding is significantly enhanced by the presence of cold WT Gal-3 but not by cold Gal-3(R186S) (Fig. 4B and SI Appendix, Fig. S7B). These data indicate that Gal-3 oligomerization/condensation on the cell surface is initiated by glycan binding. To investigate which interactions drive this process, we assessed CRD–CRD interactions and found that this type of interaction is relatively weak, if at all present, under the experimental conditions used (Fig. 4C and SI Appendix, Fig. S7C).
Fig. 4.
Cell surface Gal-3 condensation. (A–F) The fixed Jurkat cells were incubated with hot (fluorescently labeled) Gal-1 and Gal-3 variants with and without cold (nonfluorescently labeled) Gal-1 and Gal-3 variants and inhibitor as indicated. The binding of the hot was analyzed by flow cytometry and is shown as the mean ± SD (n ≥ 3). Representative images are shown in SI Appendix, Fig. S7 A–F. In A and C–F, the binding in the presence of cold was compared to the binding in the absence of cold (marked w/o for without cold on top of the columns) in each group. (G) The agglutination of human erythrocytes type AB was performed in the presence of CRD, NT, or their combination. (H) Proposed interaction model on the cell surface. CRDS, CRD S-face. CRDF, CRD F-face. P values were determined by Student’s two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns, not significant.
Thereafter, we concentrated our attention on NT–CRD and NT–NT interactions. WT Gal-3 possesses both NT–CRD and NT–NT interaction potentials, whereas mutant Gal-3(L203A) has attenuated NT–CRD interactions (Fig. 4D and SI Appendix, Fig. S7D) but maintains NT–NT interaction potential (SI Appendix, Fig. S4F). As shown in Fig. 4 E and F and SI Appendix, Fig. S7 E and F, the binding of hot Gal-3(L203A) is not promoted by lower concentrations (≤0.1 μM) of cold Gal-3(L203A) but is promoted by higher concentrations (≥0.2 μM) of cold Gal-3(L203A) compared to WT Gal-3 that promotes effects at both lower and higher concentrations. These data suggest that at lower concentrations (≤0.1 μM Gal-3), NT–CRD interactions provide the primary driving force, with NT–NT interactions being negligible. However, at higher concentrations (≥0.2 μM), NT–NT interactions have a significant impact, with both NT–CRD and NT–NT interactions playing roles. Supporting the presence of NT–CRD interactions, we observed that hot NT (GFP-NT) could be recruited to cells by the presence of cold CRD but not by cold CRD(L203A) (Fig. 4D and SI Appendix, Fig. S7D).
Similar results were obtained using endothelial cells (HMEC-1) and His-tagged hot full-length Gal-3. The binding of 0.4 μM hot His–Gal-3 was increased dramatically upon the addition of 0.8 and 8 μM cold Gal-3 and Gal-3 CRD but not by free NT (SI Appendix, Fig. S7G). Increased binding was inhibited by Lac (SI Appendix, Fig. S7H), and cold CRD promoted hot NT binding to these cells but not the other way around (SI Appendix, Fig. S7I). Furthermore, intermolecular CRD–NT–CRD interactions are demonstrated in the hemagglutination assay, in that the CRD alone could not (or could only minimally) mediate cell–cell interactions, whereas the addition of NT fragments increased agglutination (Fig. 4G) with the NT bridging cells (SI Appendix, Fig. S7J). Collectively, our data support the interaction model discussed above and depicted in Fig. 4H.
NT Prolines Are Crucial to Gal-3 Oligomerization/Phase Separation.
Using full-length Gal-3 mutants (SI Appendix, Fig. S1A), we found that salt-induced LLPS was significantly decreased by using mutants P37A, P55A, P64A/H, and P67A compared to WT Gal-3 (Fig. 5A). On the other hand, GP-mediated Gal-3 LLPS was significantly reduced by different GPs and NT proline subsets. Inhibition was greatest with P30A, P37A, P51A, P55A, P64A/H, and P67A using CD146 (Fig. 5B); with P17A, P23A, P30A, P37A, P51A, P55A, P59A, and P67A using CD45 (Fig. 5C); with P17A, P37A, P51A, P60A, and P67A using CD7 (Fig. 5D); and with P17A, P23A, P30A, P37A, P46A, P51A, P64A/H, and P67A using CD71 (Fig. 5E). P37A, P51A, and P67A were common to all of the above. Interestingly, P64A/H had an inhibitory effect with CD146 and CD71 but promoted LLPS with CD7 and CD45 (along with P60A).
Fig. 5.
Multiple prolines participate in Gal-3 oligomerization/phase separation. (A) Droplet size of 40 μM WT Gal-3 or its mutants in the presence of 2.5 M NaCl was measured by using DLS. (B–E) Droplet size of 25 µM WT Gal-3 or its mutants in the presence of 200 nM CD146 (B), 100 nM CD45 (C), 250 nM CD7 (D), and 400 nM CD71 (E) was measured by using DLS. (F and G) Gal-3 condensation on CD146-coated Ni-NTA (F) or SA (G) sensors assessed by BLI. WT Gal-3 and its mutants were tested at 0, 500, 1,000, 1,500, 2,000, 2,500, 3,000, and 3,500 nM. Partial sensorgrams for association (0 to 120 s) and dissociation (120 to 240 s) are presented. Full sensorgrams are shown in SI Appendix, Fig. S8 C and D. KD values are presented in each sensorgram. n/a, not applicable. (H) Association of WT Gal-3 and its mutants with Jurkat cells or HMEC-1 cells. Proteins associated with cells and in culture medium were detected by Western blotting. The concentration of Gal-3 mutants in culture medium was determined to control for equal amounts of protein. In A–E, comparisons were made between WT Gal-3 and each mutant, and the data are shown as the mean ± SD (n = 3). P values were determined by Student’s two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
GP-triggered Gal-3 condensation on a solid surface was also examined using CD146-coated biosensors with biolayer interferometry (BLI) (44). Partial sensorgrams for the association and dissociation steps are presented in Fig. 5 F and G, with full sensorgrams shown in SI Appendix, Fig. S8 C and D. Compared to WT Gal-3, all NT proline mutants display reduced sensor thickness upon association, especially for Gal-3 mutants P37A, P50A, P55A, P60A, P64A, and P67A. The most affected mutants displayed lower dissociation constant (KD) values (i.e., greater affinity/avidity) for CD146 than WT Gal-3. This may be explained by our model in which the binding of inner layer Gal-3 to GP glycans is much stronger than the binding of outer layer Gal-3 to the inner layer Gal-3. The observation of reduced sensor thickness upon association of any Gal-3 mutant (especially with P37A, P50A, P55A, P60A, P64A, and P67A), compared to WT Gal-3, implies that the outer layer is much thinner with fewer bound Gal-3 molecules. The larger KD values (weaker affinity/avidity) for WT Gal-3 binding reflect the dissociation of outer layer Gal-3 molecules first, leading to the observed faster dissociation rates that result in larger KD values. SI Appendix, Fig. S8E supports our finding that KD values are dependent on the concentration of Gal-3.
The effect of NT proline mutations on cell surface Gal-3 oligomerization is evidenced by the following binding assay. WT Gal-3 or mutants were allowed to bind to the surface of Jurkat or HMEC-1 cells at 4 °C (to avoid internalization), and bound proteins were visualized by Western blotting (Fig. 5H). Compared to WT Gal-3, NT mutants P50A, P51A, P55A, P60A, P64A, and P67A showed significantly reduced binding in both cell types, whereas Gal-3 mutant P23A displayed increased binding on HMEC-1 cells but not on Jurkat cells.
Gal-3 NT–CRD Exchange Dynamics Are Mediated by NT Prolines.
Next, we investigated why and how multiple prolines within the NT modulate Gal-3 oligomerization/phase separation. For this purpose, we employed NMR spectroscopy to assess NT–CRD exchange dynamics because NT–CRD interactions are the primary driving force for glycan-triggered Gal-3 oligomerization/phase separation. Previously, we reported that the Gal-3 NT transiently interacts with the F-face of the CRD (5). For this, we compared 15N-HSQC spectra of WT Gal-3 and Gal-3 NT proline mutants as exemplified in SI Appendix, Fig. S9 with NT mutant P46A. Analyses of chemical shift changes (Δδ) and resonance broadening (ΔINT) of all NT proline mutants indicate significant changes throughout the lectin SI Appendix, Table S2. Most importantly, we find that NT proline mutants appear to modulate CRD S-face residue dynamics (SI Appendix, Fig. S9E). In this regard, NT–CRD interactions and exchange dynamics vary somewhat among mutants, primarily because some NT mutants show regions of increased or decreased resonance broadening suggestive of stronger or weaker interactions, respectively, with the CRD F-face. Detailed information is presented in SI Appendix, Supplementary Results and Fig. S9.
NT Octapeptides and NT Proline Mutants Shed Light on Proline-Mediated CRD–NT Binding.
For additional insight into the roles of NT prolines, we synthesized a series of overlapping octapeptides that run through NT residues 21 to 69 (SI Appendix, Fig. S10A). Microscale thermophoresis (MST) with CRD or NT peptides alone or in combination (SI Appendix, Fig. S10B) show that only NT–CRD interactions are relatively strong (KD between 21 nM [with *NT] and 1.5 μM [with *CRD]), and CRD F-face mutant L203A nearly abolished NT–CRD interactions. MST also demonstrated that all NT octapeptides bind to the WT Gal-3 CRD, with KD values for the best binders ranging from about 2 (pep21/40) to 11 (pep28) or 35 mM (pep35), and weaker interactions with F-face CRD mutant L203A (SI Appendix, Fig. S10C). Some NT octapeptides can compete with full-length NT and inhibit WT Gal-3 function (SI Appendix, Fig. S11). In addition, because NT octapeptide affinities are ∼1,000 times less than that for the full-length NT, it appears that the WT Gal-3 NT may bind to more than one CRD simultaneously. We also prepared several full-length Gal-3 NT fragment mutants (SI Appendix, Fig. S1B) and found that CRD binding affinities of mutants P17A, P30A, and P51A are similar to (or greater than) those of their WT counterparts, whereas affinities of mutants P46A, P50A, P55A, and P67A are markedly less than those of their native counterparts (SI Appendix, Fig. S10D). Thus, prolines within the NT can regulate NT–CRD interactions in line with our NMR results.
Gal-3 Binding to Glycans Is Modulated by NT Prolines.
Our NMR observation that NT proline mutants modulate CRD S-face residue dynamics (SI Appendix, Fig. S9E) suggests a potential link to glycan binding. To assess this possibility, we assessed the affinity/avidity of WT Gal-3 and proline mutants (and F-face mutant L203A) for three well-known glycan ligands, Lac, LacNAc, and (LacNAc)3, using isothermal titration calorimetry (ITC). Table 1 and SI Appendix, Fig. S12 show the following: 1) binding affinity/avidity ranks as ordered (LacNAc)3 > LacNAc > Lac; 2) mutations alter binding strength, with P19A binding more strongly and others binding more weakly than WT Gal-3; 3) mutants P17A, P37A, P50A, P51A, P59A, P60A, P64A/H, and P67A impact binding the most; 4) any given mutation may have different effects on the binding of different glycans; and 5) Gal-3(L203A) behaves similarly to P46A in terms of affinity for these glycans.
Table 1.
The affinity/avidity (KD, μM) of WT Gal-3 and mutants for Lac, LacNAc, and (LacNAc)3 measured by using ITC approach
| Mutants | Lac | LacNAc | (LacNAc)3 |
| WT | 374 | 129 | 10 |
| P17A | — | — | 210 |
| P19A | 150 | 50 | 9.8 |
| P23A | — | >1,000 | 25 |
| P30A | — | 231 | 22 |
| P37A | — | — | 171 |
| P42A | — | — | 53 |
| P46A | 642 | 199 | 15 |
| P50A | — | — | 149 |
| P51A | — | — | 117 |
| P55A | — | — | 36 |
| P59A | — | — | 265 |
| P60A | — | — | 309 |
| P64A | — | — | 72 |
| P67A | — | — | 110 |
| P64H | — | — | 78 |
| L203A | 779 | 165 | 15 |
—, KD was not obtained because of weak interactions.
We also used an N-glycan array assay to further assess the effects of proline mutations on glycan binding (SI Appendix, Fig. S13). These data show that mutation of some prolines alters the binding profile for P23A, P30A, P37A, and P60A and affinities/avidities for P42A, P46A, P50A, P51A, P55A, P64A/H, and P67A. Overall, these data indicate that Gal-3 CRD glycan binding is dependent on the positions of specific prolines within the NT. In general, mutations of prolines modulate NT–CRD exchange dynamics, thereby inducing allosteric effects through the CRD β-sandwich that, in turn, increase internal motions of CRD S-face residues and modify the glycan-binding footprint and/or affinity/avidity.
Discussion
The functional role of the highly conserved, proline-rich Gal-3 NT has remained enigmatic since its initial reporting. Our present investigation has provided some pieces to this puzzle. Most importantly, we find that prolines within the NT modulate Gal-3 function by differentially affecting EC migration, T cell activation, hemagglutination, and endocytosis. In this regard, alanine substitution of any one NT proline has a significant inhibitory effect, especially P37, P55, P60, P64, and P67 (SI Appendix, Table S3). Interestingly, these prolines are sequentially closer to the CRD and possibly elicit a greater effect on transient NT folding onto the CRD F-face compared to the other mutated NT proline residues.
LLPS (or LLPS-like) was used here to model Gal-3 oligomerization in situ. Previously, Chiu et al. (37) investigated high salt-induced Gal-3 LLPS. For greater biological relevance, we focused the present study on GP-mediated Gal-3 oligomerization/LLPS on cell membranes. Unlike high salt-induced Gal-3 LLPS, GP-mediated Gal-3 LLPS is highly correlated with Gal-3 concentration, with NT–CRD interactions dominating at lower concentrations (≤0.1 μM) and both NT–CRD and NT–NT interactions contributing at higher concentrations (≥0.2 μM) (Fig. 4 E and F). This finding explains some inconsistencies with different assays [i.e., our MST assay with 25 nM CRD or NT was used (SI Appendix, Fig. S10B) vis-à-vis GP-induced LLPS when Gal-3 was in the micromolar range (Fig. 2I)]. This observation also clarifies previous reports that use micromolar concentrations of Gal-3 and conclude that NT–NT interactions dominate (6, 36, 37). Because physiological concentrations of Gal-3 are normally lower than 0.1 μM, we propose that the primary driving force for Gal-3 oligomerization/condensation on cell membranes in situ is provided by NT–CRD interactions. It is worth noting that because intracellular CRD–NT interactions (5) are attenuated in Gal-3 F-face mutant L203A, the NT is more solvent exposed and free to promote greater NT–NT interactions vis-à-vis WT Gal-3. Thus, NT–NT interactions are likely overestimated with Gal-3(L203A) (Fig. 4 E and F).
Here, we demonstrated that GP-mediated Gal-3 LLPS is dependent on prolines within the NT. The substitution of specific NT prolines with alanine modulates Gal-3 NT–CRD exchange dynamics, usually leading to attenuation of GP-mediated Gal-3 macromolecular associations. In a previous study, Chiu et al. characterized the salt-induced self-association of three Gal-3 NT constructs (37). However, in their investigation, all tyrosine, all tryptophan, or all tyrosine and tryptophan residues within the NT were replaced with glycine residues, and even though doing this produced significant effects on LLPS, effects from NT aromatic residue substitutions may in fact be indirect. In formulating the present study, we noted that most tyrosine and tryptophan residues within the Gal-3 NT are positioned at the N-terminal side of prolines that modulates cis-trans proline isomerization by stabilizing cis proline (51). We propose here that proline residues within the Gal-3 NT control transient conformations of the Gal-3 NT and NT–CRD interactions, and this in turn mediates LLPS. In this regard, our present work, in which one proline residue at a time is replaced by alanine, better demonstrates how the NT mediates LLPS and GP-mediated effects.
The present study also underscores differential effects with different GPs (i.e., CD146, CD45, CD7, and CD71), an effect that is likely related to the type and number of glycan chains present on each CD. Indeed, our glycan array and ITC data demonstrate that different NT proline mutants have different glycan-binding footprints and different binding affinities/avidities. Therefore, the removal of some NT prolines modifies NT–CRD F-face dynamic interactions that, in turn, allosterically modulate internal motions/dynamics of residues on the S-face affecting S-face sugar binding.
GP-mediated Gal-3 LLPS is a complex process of protein–protein and protein–glycan associations. Given the present data, we now know that the CRD S-face of Gal-3 molecules binds to glycans on GPs, whereas their F-faces (and possibly NTs) bind to the NTs and CRD F-faces of other Gal-3 molecules, inducing a polymerization-like cascade that leads to phase separation and droplet formation. The Gal-3:GP droplet stoichiometry varies proportionally to the Gal-3 concentration (Fig. 2E). Moreover, because the Gal-3 NT has multiple epitopes for intermolecular CRD F-face binding, the NT likely can bind multiple Gal-3 molecules, resulting in a highly cross-linked GP–Gal-3 network that produces solid-like droplets with both GP and Gal-3 randomly arranged, unlike organized structures found in bilaminar vesicles. We summarized Gal-3 oligomerization/phase separation in a model illustrated in SI Appendix, Fig. S14A, with effects from NT proline mutations and NT truncations summarized in SI Appendix, Fig. S14B and Table S3.
We also showed in this investigation that glycan-triggered LLPS of cytosolic Gal-3 occurs on damaged endomembranes. Whereas glycans are generally displayed on the cell surface or confined within the lumen of organelles, they can become exposed to the cytosolic milieu upon disruption of organelle membranes by stress or exposure to pathogens (50). Gal-3, Gal-8, and Gal-9 are recruited to vesicles that contain intracellular bacteria (e.g., Salmonella typhimurium, Shigella flexneri, and Listeria monocytogenes) and are ruptured by these bacteria as they escape from the vesicles (52, 53). The recruitment and accumulation of galectins on ruptured vesicles depend on the binding of galectins to host glycans (48, 49, 53). Glycan-dependent recruitment of galectins (Gal-1, Gal-3, Gal-8, and Gal-9) to sterile endomembranes damaged by various agents has also been reported (53–57). By recognizing glycans on damaged endomembranes, cytosolic galectins “sense” the danger of pathogenic invasion and other stresses, thereby mediating appropriate responses. Interestingly, these galectins display variations in their puncta composition, location, and function in response to a given stimuli (57). Although Gal-8 and Gal-3 both accumulate on vesicles with L. monocytogenes, the functional consequence differs, with Gal-3 down-regulating antibacterial autophagy and Gal-8 promoting autophagy (50). Although these opposing responses are not fully understood, our findings of Gal-3–mediated LLPS in the cytoplasm provides an explanation.
Glycan-triggered Gal-3 condensation (LLPS-like) on the cell surface occurs in a similar fashion. Because these macromolecular assemblies are dependent on the concentrations of both Gal-3 and GP-linked glycans, it is likely that oligomeric states in situ are dependent on the cellular microenvironment, including variations in the glycans on different GPs. An important role of galectins on the cell surface is to cross-link and reorganize glycosylated receptors (58). The dynamic behavior of Gal-3 provides an additional layer of control and may represent a new mechanism underlying the formation, regulation, and function of clusters of cell surface receptors. Many receptors elicit cellular behavior consistent with phase separation, such as formation of nanometer-to-micrometer scale structures (59). Although phase separation of membrane proteins has been largely reported to occur intracellularly (60), here, we have demonstrated extracellular phase separation of these proteins.
Overall, it was surprising that the replacement of a single proline within the structurally aperiodic, highly dynamic NT can have such dramatic and differential effects on cellular activities. In fact, mutation of a single Gal-3 NT residue, P64, presents a real-life example because of a common polymorphism, rs4644, that codes for P64 or H64 of Gal-3. The P64H mutation is associated with the risk of prostate cancer, phenotypic variations in Chagas disease, ovarian carcinoma, breast cancer, and chronic kidney disease (40–43). The P64H mutant influences the susceptibility of Gal-3 to cleavage by matrix metalloproteinase (41), possibly resulting from a reduced binding of the Gal-3 NT to its CRD F-face and release into solution, as we observed with the P64A mutant by NMR. Here, we found that P64A and P64H both attenuate LLPS and inhibit Gal-3–CLIC formation and endocytosis, EC migration, and T cell activation. Currently, many disease states have been correlated with LLPS, such as neurodegenerative diseases and tumor formation (28, 41, 61). For example, amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that affects nerve cells in the brain and spinal cord. ALS-linked proline mutations affect the phase separation of a proline-rich protein (proteasome shuttle factor UBQLN2), leading to the formation of inclusion bodies that are pathologically characteristic of ALS (62). Our present discovery also provides a better understanding into the mechanisms of proline polymorphisms in pathological disorders.
Materials and Methods
Reagents, protein expression and purification, cell culture, endothelial cell migration, T cell activation assays, hemagglutination assays, endocytosis assays, turbidity assays, DLS measurements, confocal imaging, fluorescence recovery after photobleaching (FRAP), Western blotting, sample preparation for phase separation, phase separation induced by high salt and glycan in solution, assessment of cell surface Gal-3 oligomerization/condensation by FRAP, flow cytometry, assessment of Gal-3 oligomerization/condensation on the endomembranes by FRAP, glycan array studies, BLI assay, MST, ITC, sedimentation assay, NMR spectroscopy, and statistical analyses are described in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 31870796 and 31770852), the Scientific and Technologic Foundation of Jilin Province (Grant 20190304082YY), and the National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program,” China (Grant 2019ZX09735001). NMR instrumentation was provided with funds from the NSF (BIR-961477), the University of Minnesota Medical School, and the Minnesota Medical Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021074118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Raz A., et al., Evidence for the role of 34-kDa galactoside-binding lectin in transformation and metastasis. Int. J. Cancer 46, 871–877 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Johannes L., Jacob R., Leffler H., Galectins at a glance. J. Cell Sci. 131, jcs208884 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Braeuer R. R., et al., Galectin-3 contributes to melanoma growth and metastasis via regulation of NFAT1 and autotaxin. Cancer Res. 72, 5757–5766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seetharaman J., et al., X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution. J. Biol. Chem. 273, 13047–13052 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Ippel H., et al., Intra- and intermolecular interactions of human galectin-3: Assessment by full-assignment-based NMR. Glycobiology 26, 888–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad N., et al., Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J. Biol. Chem. 279, 10841–10847 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Dumic J., Dabelic S., Flögel M., Galectin-3: An open-ended story. Biochim. Biophys. Acta 1760, 616–635 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Sato S., Hughes R. C., Regulation of secretion and surface expression of Mac-2, a galactoside-binding protein of macrophages. J. Biol. Chem. 269, 4424–4430 (1994). [PubMed] [Google Scholar]
- 9.Popa S. J., Stewart S. E., Moreau K., Unconventional secretion of annexins and galectins. Semin. Cell Dev. Biol. 83, 42–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thijssen V. L. J. L., Poirier F., Baum L. G., Griffioen A. W., Galectins in the tumor endothelium: Opportunities for combined cancer therapy. Blood 110, 2819–2827 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Nieminen J., Kuno A., Hirabayashi J., Sato S., Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J. Biol. Chem. 282, 1374–1383 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Nabi I. R., Shankar J., Dennis J. W., The galectin lattice at a glance. J. Cell Sci. 128, 2213–2219 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Thiemann S., Baum L. G., Galectins and immune responses-just how do they do those things they do? Annu. Rev. Immunol. 34, 243–264 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hsu D. K., et al., Endogenous galectin-3 is localized in membrane lipid rafts and regulates migration of dendritic cells. J. Invest. Dermatol. 129, 573–583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakshminarayan R., et al., Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat. Cell Biol. 16, 595–606 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Zhao W., et al., Galectin-3 mediates tumor cell-stroma interactions by activating pancreatic stellate cells to produce cytokines via integrin signaling. Gastroenterology 154, 1524–1537.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Sano H., et al., Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J. Immunol. 165, 2156–2164 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Glinsky V. V., et al., Intravascular metastatic cancer cell homotypic aggregation at the sites of primary attachment to the endothelium. Cancer Res. 63, 3805–3811 (2003). [PubMed] [Google Scholar]
- 19.Yang R. Y., Hill P. N., Hsu D. K., Liu F. T., Role of the carboxyl-terminal lectin domain in self-association of galectin-3. Biochemistry 37, 4086–4092 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Lepur A., Salomonsson E., Nilsson U. J., Leffler H., Ligand induced galectin-3 protein self-association. J. Biol. Chem. 287, 21751–21756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundqvist M., et al., Galectin-3 type-C self-association on neutrophil surfaces; the carbohydrate recognition domain regulates cell function. J. Leukoc. Biol. 103, 341–353 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y.-H., Forman-Kay J. D., Chan H. S., Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry 57, 2499–2508 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Hofweber M., et al., Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Woodruff J. B., et al., The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Rai A. K., Chen J.-X., Selbach M., Pelkmans L., Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Zhang G., Wang Z., Du Z., Zhang H., mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506.e22 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Bouchard J. J., et al., Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey S., et al., Surface properties determining passage rates of proteins through nuclear pores. Cell 174, 202–217.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Milles S., et al., Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 163, 734–745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeynaems S., et al., Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwayer C., et al., Mechanosensation of tight junctions depends on ZO-1 phase separation and flow. Cell 179, 937–952.e18 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Beutel O., Maraspini R., Pombo-García K., Martin-Lemaitre C., Honigmann A., Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Wu X., et al., RIM and RIM-BP form presynaptic active-zone-like condensates via phase separation. Mol. Cell 73, 971–984.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Su X., et al., Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y.-H., et al., The intrinsically disordered N-terminal domain of galectin-3 dynamically mediates multisite self-association of the protein through fuzzy interactions. J. Biol. Chem. 292, 17845–17856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu Y.-P., et al., Liquid-liquid phase separation and extracellular multivalent interactions in the tale of galectin-3. Nat. Commun. 11, 1229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue H., et al., Selective effects of ginseng pectins on galectin-3-mediated T cell activation and apoptosis. Carbohydr. Polym. 219, 121–129 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Gao X., et al., The two endocytic pathways mediated by the carbohydrate recognition domain and regulated by the collagen-like domain of galectin-3 in vascular endothelial cells. PLoS One 7, e52430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L., et al., Galectin-3 in cancer. Clin. Chim. Acta 431, 185–191 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Balan V., et al., Racial disparity in breast cancer and functional germ line mutation in galectin-3 (rs4644): A pilot study. Cancer Res. 68, 10045–10050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer A., et al., Apoptosis gene polymorphisms and risk of prostate cancer: A hospital-based study of German patients treated with brachytherapy. Urol. Oncol. 31, 74–81 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Rebholz C. M., et al., Plasma galectin-3 levels are associated with the risk of incident chronic kidney disease. Kidney Int. 93, 252–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Zheng Y., Wang H., Zhou Y., Tai G., CD146 interacts with galectin-3 to mediate endothelial cell migration. FEBS Lett. 592, 1817–1828 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Gao X., et al., The inhibitory effects of a rhamnogalacturonan I (RG-I) domain from ginseng pectin on galectin-3 and its structure-activity relationship. J. Biol. Chem. 288, 33953–33965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salomonsson E., et al., Mutational tuning of galectin-3 specificity and biological function. J. Biol. Chem. 285, 35079–35091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paz I., et al., Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 12, 530–544 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Feeley E. M., et al., Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc. Natl. Acad. Sci. U.S.A. 114, E1698–E1706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weng I.-C., et al., Cytosolic galectin-3 and -8 regulate antibacterial autophagy through differential recognition of host glycans on damaged phagosomes. Glycobiology 28, 392–405 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Wu W. J., Raleigh D. P., Local control of peptide conformation: Stabilization of cis proline peptide bonds by aromatic proline interactions. Biopolymers 45, 381–394 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Dupont N., et al., Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6, 137–149 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Thurston T. L. M., Wandel M. P., von Muhlinen N., Foeglein A., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maejima I., et al., Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X., et al., Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J. Biol. Chem. 289, 11162–11174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aits S., et al., Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia J., et al., Galectins control mTOR in response to endomembrane damage. Mol. Cell 70, 120–135.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boscher C., Dennis J. W., Nabi I. R., Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 23, 383–392 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Zeng M., et al., Phase separation-mediated TARP/MAGUK complex condensation and AMPA receptor synaptic transmission. Neuron 104, 529–543.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Case L. B., Ditlev J. A., Rosen M. K., Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel A., et al., A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Dao T. P., et al., ALS-linked mutations affect UBQLN2 oligomerization and phase separation in a position- and amino acid-dependent manner. Structure 27, 937–951.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.