Significance
Structural maintenance of chromosomes (SMC) complexes are essential for chromosome organization and functions. Within this family of complexes, the Smc5/6 complex has unique roles in regulating DNA replication and repair; however, our understanding of its structural organization is limited. Our integrative study presented here reveals several distinct features of the Smc5/6 complex. Unlike other SMCs, the long filamentous region of Smc5/6 does not fold back; rather, it associates with unique, coevolved subunits of the complex, including the Nse2 SUMO ligase and the Nse5/6 subcomplex. We also present a cryoelectron microscopy structure of the Nse5/6 subcomplex and evidence for its involvement in Nse2-mediated sumoylation. Our findings thus provide insights into Smc5/6 structural anatomy and functions.
Keywords: Smc5/6 complex, Nse5, Nse6, sumoylation, structural maintenance of chromosomes
Abstract
Structural maintenance of chromosomes (SMC) complexes are critical chromatin modulators. In eukaryotes, the cohesin and condensin SMC complexes organize chromatin, while the Smc5/6 complex directly regulates DNA replication and repair. The molecular basis for the distinct functions of Smc5/6 is poorly understood. Here, we report an integrative structural study of the budding yeast Smc5/6 holo-complex using electron microscopy, cross-linking mass spectrometry, and computational modeling. We show that the Smc5/6 complex possesses several unique features, while sharing some architectural characteristics with other SMC complexes. In contrast to arm-folded structures of cohesin and condensin, Smc5 and Smc6 arm regions do not fold back on themselves. Instead, these long filamentous regions interact with subunits uniquely acquired by the Smc5/6 complex, namely the Nse2 SUMO ligase and the Nse5/Nse6 subcomplex, with the latter also serving as a linchpin connecting distal parts of the complex. Our 3.0-Å resolution cryoelectron microscopy structure of the Nse5/Nse6 core further reveals a clasped-hand topology and a dimeric interface important for cell growth. Finally, we provide evidence that Nse5/Nse6 uses its SUMO-binding motifs to contribute to Nse2-mediated sumoylation. Collectively, our integrative study identifies distinct structural features of the Smc5/6 complex and functional cooperation among its coevolved unique subunits.
Structural maintenance of chromosomes (SMC) complexes regulate genome organization and maintenance in both prokaryotic and eukaryotic cells. Each complex contains a pair of SMC subunits and a set of non-SMC subunits (1). Studies of several SMC proteins reveal that they form tripartite filamentous structures. An SMC subunit folds back on itself at its middle “hinge” region, enabling its N- and C-terminal ATPase domains to associate forming a “head” region, and its two long coiled-coil regions located in between the hinge and the head to pair in an antiparallel manner, forming an “arm” region (SI Appendix, Fig. S1A) (1). The two SMC subunits of each complex form its backbone and can associate with each other at hinge, head, and arm regions (1).
Much of the molecular understanding of SMC complexes has come from studies of those acting as DNA organization and separation factors, such as prokaryotic Smc-ScpAB and MukBEF complexes and eukaryotic cohesin and condensin. These complexes can entrap and loop DNA, resulting in DNA tethering and folding (2–4). One emerging feature of these complexes is that their long arm regions bend sharply at so-called “elbow” sites (SI Appendix, Fig. S1A). Elbow bending causes the hinge to contact the head-proximal coiled-coil or head-bound non-SMC proteins, a conformation thought to facilitate DNA loop extrusion (5–8). In these SMCs, the head and hinge regions that associate with other proteins and DNA are conserved, while the arm regions are not and act mainly as connecting elements (9).
Differing from cohesin (containing Smc1/3) and condensin (containing Smc2/4), the third eukaryotic SMC complex, containing Smc5 and Smc6, does not appear to affect chromatid intertwining or mitotic chromosome structures (10–12). Rather, the Smc5/6 complex directly regulates DNA replication and recombinational repair (13–15). These unique functions correlate with the acquisition of a special set of six subunits, namely non-SMC elements (Nse)1 to 6. Three Nse subunits, Nse2, Nse5, and Nse6, are not found in any other SMC complexes in either prokaryotes or eukaryotes. Nse2 (also known as Mms21) is a SUMO ligase that promotes the sumoylation of more than a dozen genome maintenance factors (16–19). Nse5 and Nse6 are thought to act distinctly from Nse2 by forming a subcomplex that recruits the Smc5/6 complex to DNA damage sites (13–15, 20–22). The acquisition of the Nse2, Nse5, and Nse6 subunits is one of the most unique features that sets the Smc5/6 complex apart from other SMC complexes.
Our understanding of how the Smc5/6 complex gained unique functions among the SMC family of complexes is hindered by the limited structural information of its holo-complex. Studies of subunits and their fragments or subcomplexes have provided insights into potential intersubunit interactions (23–25). However, these data may not reflect structures and interactions within the entire complex. Thus, it is imperative to determine, in the context of the holo-complex, whether Smc5 and Smc6 adopt distinct conformations relative to other SMCs, how they associate with Nse subunits, and what the functional relationships are among the complex-specific Nse2, -5, and -6 subunits. Here, we provide an integrative structural analysis of the Smc5/6 holo-complex isolated from budding yeast that addresses these challenges. We use negative-staining electron microscopy (EM), cross-linking mass spectrometry (CL-MS), single-particle cryo-EM, structural modeling, and functional analyses to identify several unique features of the Smc5/6 complex that distinguish it from the other SMC complexes. We also provide evidence that the coevolved Nse2, -5, and -6 subunits are connected at both structural and functional levels.
Results
Negative-stain EM Analysis of the Smc5/6 Complex Reveal a Rod-Shaped Structure without Elbow-Bending.
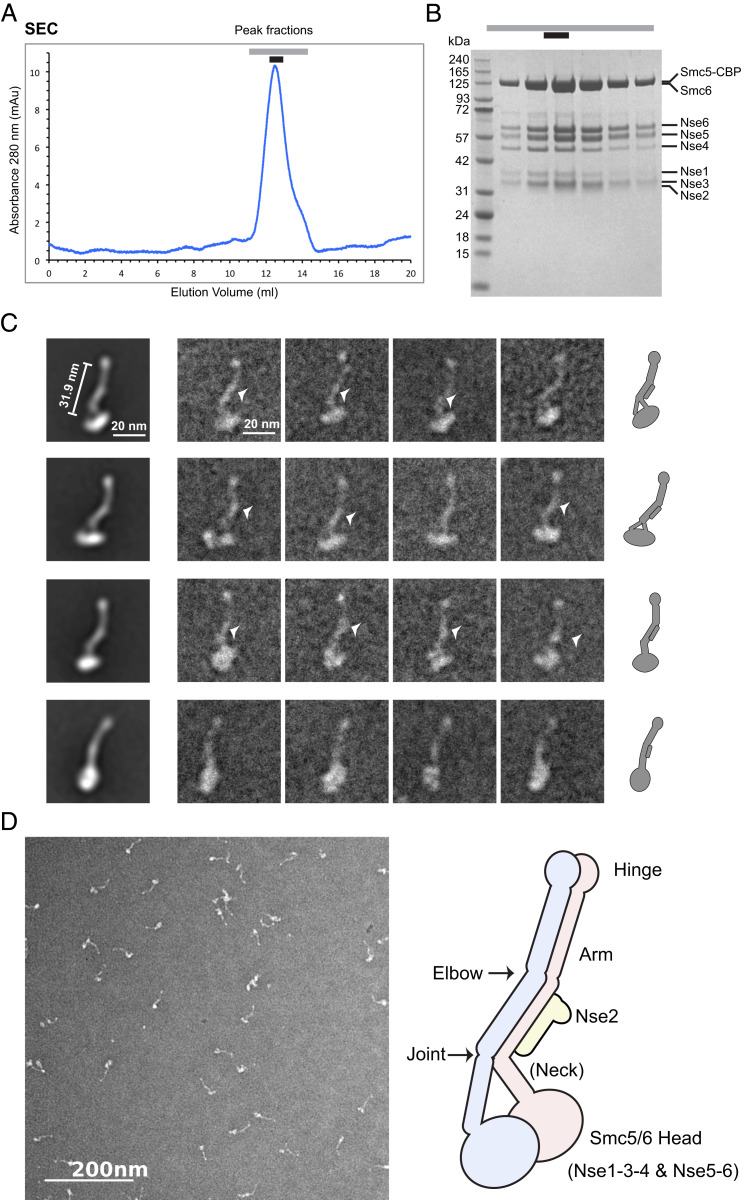
To gain structural insight into the Smc5/6 holo-complex (holo-Smc5/6), we coexpressed all eight subunits of the budding yeast complex in its native host cells (Materials and Methods). The complex was prepared by affinity purification followed by size-exclusion chromatography, which showed that the entire complex eluted in a single peak (Fig. 1A). The presence of all eight subunits in the peak fraction was confirmed by SDS/PAGE and by MS analyses (Fig. 1B and SI Appendix, Fig. S1B).
Fig. 1.
Negative-stain EM analysis of the Smc5/6 holo-complex. (A) SEC profile of the Smc5/6 holo-complex. Peak fractions are indicated by lines above the elution profile from a Superose 6 increase column. (B) Analysis of Smc5/6 holo-complex peak fractions by SDS/PAGE. A picture of the Coomassie-stained gel is shown and the protein band corresponding to each subunit is indicated. The peak fraction marked by a black bar above the picture was used for negative-stain EM analyses. (C) Negative-stain EM analysis of the Smc5/6 holo-complex prepared in the presence of ATPγS. (Left) Two-dimensional class averages using a circular mask of 700 Å. (Center) Raw images of representative particles, with thickened part of the arm marked by arrowheads. (Right) Cartoons depicting the overall shape of holo-Smc5/6 seen in 2D class averages. Nse2 (bar) is presumably positioned along the midsection of the arm and accounts for the thickening of this section compared with other parts of the arm. (D) A typical field view of the negative stain EM of the Smc5/6 holo-complex (Left) and a cartoon model of the overall structure of the complex (Right), with its main structural features labeled. We suspect that Nse subunits (not drawn), except Nse2, are located close to the head regions (see text).
Since ATP-bound state is critical for SMC functions, we set out to assess the Smc5/6 holo-complex in the presence of ATPγS, a nonhydrolysable ATP analog (2–4). Negative-stain EM data revealed that most particles of the Smc5/6 complex formed tripartite filaments, containing hinge, arm, and head regions, as seen for other SMC complexes (Fig. 1 C and D). Instead of adopting an open ring shape, as speculated previously, Smc5 and Smc6 appeared to align with each other throughout most of their length. Class averages generated by reference-free, two-dimensional (2D) image classification suggested that the Smc5/6 holo-complex contained a small lobed hinge region, a larger head region, and an essentially linearly aligned rod-shaped arm connecting the head and hinge regions (Fig. 1 C and D). Both raw and 2D classification images revealed differently shaped head regions, which might reflect their distinct orientations, among other possibilities (Fig. 1C). In addition, the head-proximal arm segment, often referred to as the “neck,” appeared as two separated threads in some images, suggesting the possibility of local separation (Fig. 1C, second row). In many images, the arm showed a notable thickening in the middle, likely stemming from Nse2 binding to the middle of Smc5 coiled-coil region, as shown previously (25) (Fig. 1 C and D). In addition, two kinks could be seen at defined positions of the arm (Fig. 1C). We suspect that these kinks occur at the two discontinuities of the coiled-coil arm regions noted previously for SMCs and are referred to as the elbow and the “joint” (Fig. 1D).
Strikingly, an elbow-bent feature seen for cohesin and condensin, wherein roughly one-third of the coiled-coil regions, proximal to the hinge, bends back sharply (SI Appendix, Fig. S1A) (5–8), was not seen for holo-Smc5/6 prepared in the presence of ATPγS. We explored this distinct finding further by comparing the arm length of the Smc5/6 complex with those of cohesin and condensin. Our measurement estimated the Smc5/6 complex arm length to be ∼31.9 nm (Fig. 1C). This is 25 to 34% longer than the arm length of elbow-bent yeast condensin (25.5 nm) and cohesin (24.0 nm), respectively (5, 8). Because the coiled-coil sequences of Smc5 and Smc6 are about three-quarters in length of those of cohesin and condensin (8), an elbow-bent structure of Smc5/6 would predict an arm length shorter than those of cohesin and condensin, which was not the case. We thus concluded that the arm region of Smc5/6 holo-complex examined in the presence of ATPγS does not adopt an elbow-bent configuration. It may be noted that our data do not exclude the possibility that other conformations may exist in the absence of nucleotides or during the Smc5/6 ATP hydrolysis cycle and DNA binding events (Discussion).
CL-MS Data Suggest No Elbow-Bending of the Arm Region in the Smc5/6 Holo-Complex.
Next, we employed CL-MS as an orthogonal approach to examine the Smc5/6 complex structure. Purified Smc5/6 holo-complex mixed with ATPγS was cross-linked with disuccinimidyl sulfoxide (DSSO) or N,N′-carbonyldiimidazole (CDI) (SI Appendix, Fig. S1C), and analyzed by liquid chromatography nanoelectrospray ionization high-resolution MS in three biological replicates. In total, we observed 337 unique cross-linked sites involving all 8 subunits (SI Appendix, Fig. S1D and Dataset S1). Among these, 214 were intramolecular and 123 were intermolecular cross-links. We examined these results alongside the only known structure of the budding yeast Smc5/6 complex that includes Nse2 and its associated Smc5 coiled-coil region (25). Mapping all 12 unique amino acid cross-links involving these regions onto the Nse2-Smc5 structure showed that they were within the expected distance constraints for each cross-linker (i.e., less than 30 Å and 20 Å for DSSO and CDI, respectively) (SI Appendix, Fig. S1E). This is consistent with the accuracy of CL-MS observed in prior studies (26), and indicates high confidence in detected interactions.
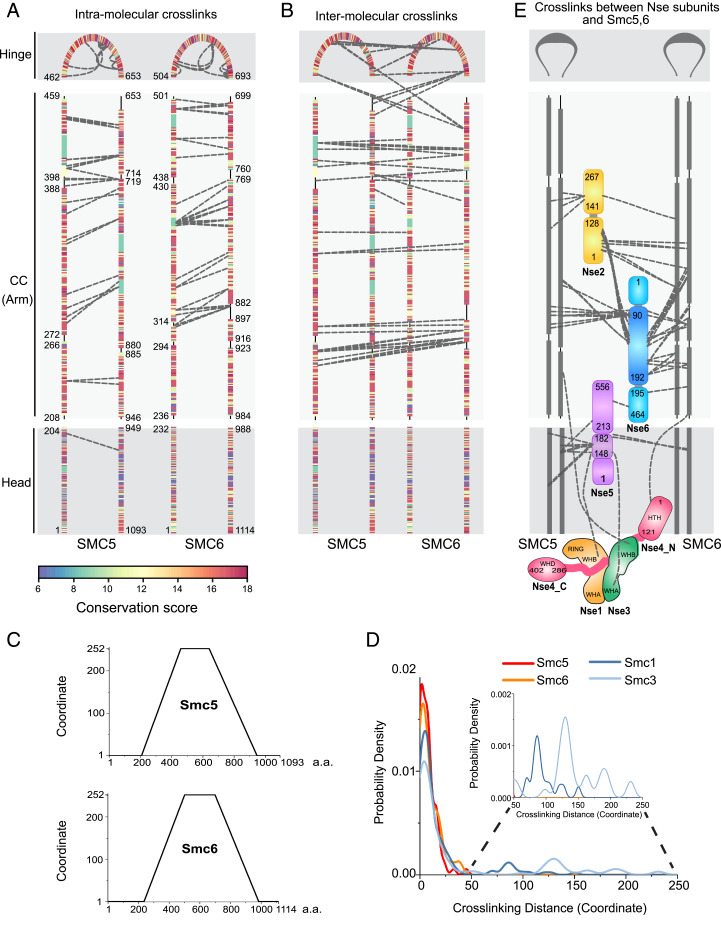
A substantial fraction of the detected cross-links involved Smc5 and Smc6, with 87 intra-Smc5, 80 intra-Smc6, and 44 Smc5-Smc6 intermolecular cross-links (Dataset S1). These cross-links spanned most of the Smc5 and -6 sequences, with an enrichment in their arm regions (Fig. 2 A and B and SI Appendix, Fig. S1D). The abundant Smc5 and -6 cross-links allowed us to derive features of their topology in solution. First, and as expected from antiparallel folding of both proteins, numerous intra-Smc5 and intra-Smc6 cross-links showed association of their N- and C-terminal coiled-coil segments (Fig. 2A). Second, 28 cross-links connected the coiled-coil regions of Smc5 with those of Smc6 in a manner consistent with a zipped-up arm configuration (Fig. 2B). The apparent absence of cross-links between some parts of Smc5 and Smc6 arm regions, particularly at the neck regions, may suggest local separation within this segment (Fig. 2B). Third, the two hinge regions formed multiple intermolecular cross-links, consistent with their dimerization (Fig. 2B) (20, 24). The fewer intra- or interhead cross-links suggest that the two heads may be shielded from cross-linking due to association with Nse subunits or not being closely associated (Fig. 2 A and B, SI Appendix, Fig. S1D, and Dataset S1). Overall, our CL-MS data are in agreement with the negative-stain EM data regarding the overall shape of Smc5/6, and provide greater resolution as to the manner of Smc5 and -6 association within the complex.
Fig. 2.
CL-MS analysis of Smc5/6 holo-complex. (A) Intra-Smc5 and intra-Smc6 cross-links. Three Smc5 and Smc6 regions, namely head, coiled-coil (CC, or arm), and hinge, are marked and residues bordering each region are labeled. Cross-links connecting the N- and C-terminal portions of each region are represented by dashed lines between the corresponding amino acid pairs. Cross-links connecting adjacent sequences listed in Dataset S1 are omitted to highlight the antiparallel nature of the coil-coiled pairing of Smc5 and Smc6. Coiled-coil regions are interrupted by noncoiled-coil segments and residues bordering each predicted coiled-coil segment are indicated. Amino acids are colored based on conservation scores as shown in SI Appendix, Fig. S1F and Dataset S3. (B) Intermolecular cross-links between Smc5 and Smc6. The graph is presented as in A. (C) Transformed amino acid coordinates for Smc5 and Smc6 (for details, see Materials and Methods). (D) Probability density of cross-linking distance based on transformed coordinates for Smc5 and Smc6 in the Smc5/6 complex, and for Smc1 and Smc3 in cohesin. CL-MS data for Smc1 and 3 are based on Bürmann et al. (8) (for details, see Materials and Methods and Dataset S2). (E) Intersubunits cross-links among Nse1-6 subunits, and between them and Smc5 and -6. Smc5 and Smc6 are presented similarly as in A and B. Domain structures of Nse subunits are not drawn to scale to highlight the cross-links. As shown previously, Nse1 contains WHA, WHB, and RING domains, while Nse3 contains WHA and WHB domains (23, 60), the Nse4 N-terminal region (Nse4_N) containing a helix-turn-helix (HTH) domain and its C-terminal region (Nse4_C) containing a WHD domain are connected by a largely unstructured region.
Cross-linking distance analyses of cohesin have shown that 4 to 20% of Smc1 and Smc3 cross-links at their coiled-coil regions are separated by more than 100 amino acids (8). Such long-distance cross-links reflect contacts between hinge- and head-proximal coiled-coils caused by elbow bending and are consistent with EM images (8). In contrast, all intraprotein cross-links that fall onto the coiled-coil regions of Smc5 and -6 showed a distance of less than 45 amino acids (Fig. 2 C and D and Dataset S2). In addition, no head-hinge cross-links were observed for holo-Smc5/6 in the presence of ATPγS (Dataset S1). Both of these observations are incompatible with a bent elbow model; rather, they support the conclusion from our EM analyses that holo-Smc5/6 prepared in the presence of ATPγS adopts a straight, rod-shaped configuration.
Association of Unique Nse Subunits with Smc5/6 Arm and Head Regions.
Among the six Nse subunits, Nse5 and Nse6, which form a subcomplex (Nse5/6), and Nse2 are unique to the Smc5/6 complex, whereas Nse1 and Nse3 resemble the KITE subunits of bacterial SMC complexes, and Nse4 is similar to the kleisin subunit seen in most SMC complexes (13–15, 27). Nse1, Nse3, and Nse4 form a subcomplex (Nse1/3/4) that binds to the Smc5/6 head regions (20, 28). Accordingly, we found that Nse1, Nse3, and Nse4 cross-linked near the Smc5/6 head regions (Fig. 2E, SI Appendix, Fig. S1D, and Dataset S1). Interestingly, Nse2 and Nse5/6 showed numerous cross-links with residues across two-thirds of the Smc5 and -6 arm regions (Fig. 2E, SI Appendix, Fig. S1D, and Dataset S1). Consistent with earlier structural data, Nse2 cross-linked to the middle section of the Smc5 coiled-coils and the corresponding region of Smc6 (Fig. 2E) (25). A region spanning 100 aa at the Nse6 N terminus (90 to 192 aa) showed multiple cross-links with Smc5 and -6 arm regions adjacent to the Nse2-binding site (Fig. 2E). We refer to this Nse6 region as the coiled-coil–associated or proximal domain (CAD). Consistent with the observation that Nse2 and Nse6-CAD are cross-linked to adjacent arm regions, cross-links between these Nse subunits were also observed (Fig. 2E). Thus, the unique Nse2 and -6 subunits are also physically close to each other within the Smc5/6 complex.
Association of SMC midarm regions with non-SMC subunits has not been seen in other SMCs (2–4), indicating a highly specific feature of Smc5/6. In line with this conclusion, the arm sequences of Smc5 and Smc6, unlike those in Smc1/3 (cohesin) and Smc2/4 (condensin), are conserved (Fig. 2A and SI Appendix, Fig. S1F) (9). In fact, conservation scores of Smc5 and Smc6 coiled-coil sequences are comparable to or higher than those of their head and hinge sequences (SI Appendix, Fig. S1F and Dataset S3). This is consistent with the observation that the Smc5/6 arm regions uniquely engage in Nse interactions. Overall, our analyses suggest that the arm sequences of Smc5/6 complex are distinctively conserved and associate with Nse subunits not found in other SMC complexes. We envision that these interactions could limit the flexibility of the arm region as well as influence the behavior of the entire Smc5/6 complex.
While the N-terminal CAD of Nse6 is in close proximity with Smc5/6 arm regions, its C-terminal region cross-linked with Nse5, consistent with them forming a subcomplex (Fig. 2E) (20, 29). However, unlike Nse6, Nse5 cross-linked to the Smc5 and Smc6 head regions (Fig. 2E). The Nse5 residues involved in these linkages were mainly restricted to a small segment in the middle of the protein (148 to 182 aa), referred to as the head-associated or proximal domain (HAD) hereafter. In line with Nse5 being close to the Smc5 and Smc6 head regions, cross-links were found between Nse5 and the Nse1/3/4 subcomplex, which is also situated close to the head regions (Fig. 2E, SI Appendix, Fig. S1D, and Dataset S1). We did not observe cross-links between Nse1/3/4 and the more hinge-proximally located Nse2 and Nse6 subunits (Fig. 2E and Dataset S1). In summary, our CL-MS data enable us to place the six Nse subunits relative to each other and to Smc5 and -6, and further suggest connection of Nse5/6 to distal parts of the complex. Given that the SMC arm regions can affect the conformations and functions of the entire complex, the distinct features of the Smc5/6 arm regions observed in the presence of ATPγS, including no elbow-folding and association with unique Nse subunits, are likely an underlying principle supporting some or all of its rather specialized functions (Discussion).
Structure of the Nse5/6 Complex Reveals Distinct Domains Used for an Interaction Network.
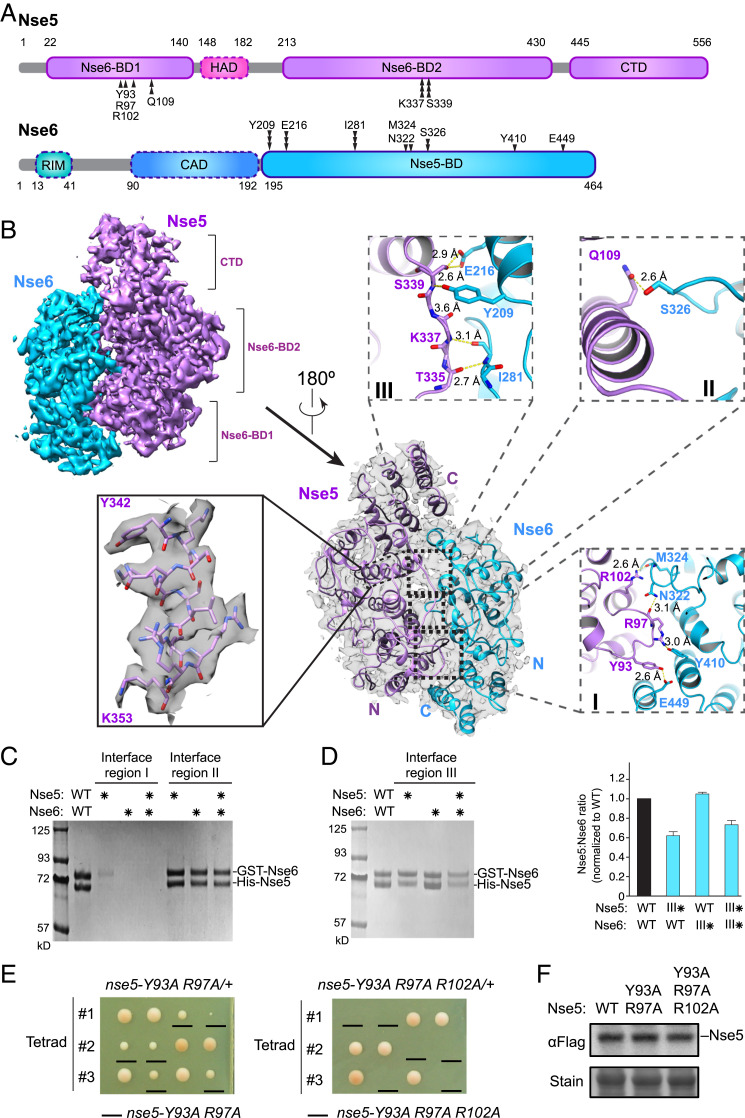
Although the Nse5/6 subcomplex presents one of the most unique features of the Smc5/6 complex and serves as a linchpin connecting other subunits together, its structure has not been solved. Addressing this, we examined the Nse5/6 structure using cryo-EM. The heterodimer Nse5/6 complex eluted as a single peak in size-exclusion chromatography (SI Appendix, Fig. S2A) and the high purity of the complex was confirmed by SDS/PAGE analyses (SI Appendix, Fig. S2B). Two-dimensional class averages of cryo-EM data revealed the overall clasped-hand shape of the complex (SI Appendix, Fig. S2C). Three-dimensional (3D) classification, 3D autorefine, and a subsequent gold-standard refinement procedure (SI Appendix, Fig. S2 C and D) resulted in a reconstruction with an overall resolution of 3.2 Å (SI Appendix, Fig. S2 D and E and Table S1). Analyzing local resolution indicated that the Nse5 C-terminal domain (CTD; 445 to 556 aa) contributed to low-resolution density (SI Appendix, Fig. S2F and Fig. 3A). In addition, the N-terminal region of Nse6 (1 to 194 aa) was invisible in the density map, presumably due to structural flexibility. This region of Nse6 contains both the CAD element identified in this study for coiled-coil association and the Rtt107-interacting motif (RIM) reported previously (Fig. 3A) (21).
Fig. 3.
Cryo-EM structure of the Nse5/Nse6 complex. (A) Domain organization of Nse5 and Nse6 proteins. Residues involved in Nse5 and Nse6 interaction at regions I, II, and III are marked by single, double, and triple vertical arrowheads, respectively. The domains seen in the cryo-EM structure are outlined in solid lines while dotted outlines indicate the other domains. RIM, Rtt107-interacting motif. (B) Cryo-EM structure of the Nse5/6 complex. Two views of the structure are shown with 180° rotation, one with a transparent surface containing ribbon model (Center) and one with surface rendering (Upper Left). An example of secondary structure elements with amino acid side chains is shown (Left). Inlets depict detailed interactions between Nse5 and Nse6 at their binding regions I, II, and III. Yellow dashed lines indicate the distances between atoms involved in hydrogen bond formation. (C) Mutating Nse5 and Nse6 interaction regions I and II has different effects on the Nse5/Nse6 complex levels. A representative SDS/PAGE gel picture of in vitro two-step pull-down results for wild-type or mutant Nse5 and Nse6 proteins. Asterisks indicate mutated form of the proteins. Region I mutations include Nse5-Y93, R97A, and Nse6-N322, Y410, E449A. Region II mutations include Nse5-Q109A and Nse6-S326W. (D) Mutating Nse5 residues at the interaction region III reduces its binding to Nse6. Data are presented as in C, except Nse5-K337, S339W, and Nse6-Y209A, I281W mutant proteins were examined. The graph on the right depicts the relative ratios of Nse5:Nse6 in the pull-down assays based on quantification from two experiments with average and SDs shown. (E) Mutating Nse5 residues at the interaction region I in cells causes slow growth or cell lethality. Diploid cells with indicated genotype were dissected and three representative tetrads for each diploid are shown. (F) Mutating Nse5 residues at the interaction region I does not impair protein expression levels. Flag-tagged Nse5 proteins extracted from cells were examined by immunoblotting. Loading is shown by Ponceau S stain (stain).
The resolution of the core of the Nse5/6 complex was ∼3.0 Å (SI Appendix, Fig. S2F) and, hence, we were able to build a de novo atomic model (Fig. 3B). Based on this model, Nse5 and Nse6 associate with each other largely in an antiparallel fashion (Fig. 3B). Nse6 uses its C-terminal half, referred to as Nse5-binding domain (Nse5-BD, 195 to 464 aa), to bind to two regions of Nse5: the Nse6-binding domain 1 (Nse6-BD1) located at its N terminus (22 to 140 aa) and domain 2 (Nse6-BD2) that is more C-terminally situated (213 to 430 aa) (Fig. 3A). These two domains of Nse5 flank the HAD domain associating with the head (Fig. 3A). Collectively, our cryo-EM and CL-MS data suggest that Nse5 and Nse6 use several distinct domains to engage in a network of interactions, including binding to each other, anchoring to arm regions, contacting the head regions, and binding to Rtt107.
Nse5 and Nse6 Differ from the HEAT Repeat Subunits in Cohesin and Condensin.
It is a matter of debate whether Nse5 and Nse6, enriched in α-helices, are similar to the HEAT repeat subunits of cohesin and condensin; this is partly due to the difficulty in predicting HEAT repeat structures based on sequence analysis (1, 27). Most HEAT repeat proteins, including several cohesin and condensin subunits, contain more than 15 tandemly repeated modules of two antiparallel α-helices linked by a short loop (30). The cohesin and condensin HEAT repeat subunits can bind to DNA and form flexible horse-shoe like structures capable of stretching and scrunching; both features are critical for entrapping and extruding DNA (30).
Our structure shows that Nse5 and Nse6 are made up of α-helices interrupted by loops and are devoid of β-sheet elements. However, distinct from cohesin and condensin HEAT repeat subunits, vast majorities of the Nse5 and -6 helices are not HEAT repeat units (Fig. 3B). Although four Nse6 helices (residues 387 to 464) resemble a pair of HEAT repeats, the Nse5/6 complex as a whole does not have the characteristics of HEAT repeat proteins. Our DALI search (31) and structural alignment analysis indeed showed no homologous structures related to Nse5, Nse6, or their complex in the Protein Data Bank to date, suggesting that Nse5/6 adopts a novel structure.
We next addressed whether the Nse5/6 subcomplex can associate with DNA. We found that Nse5/6 did not interact with either single-stranded or double-stranded DNA (SI Appendix, Fig. S3 A and B). As a comparison, we examined the Nse1/3/4 subcomplex, as the homologous subcomplex in human and fission yeast associated with DNA (32). Indeed, we found that purified budding yeast Nse1/3/4 subcomplex interacted with both single-stranded DNA and double-stranded DNA (SI Appendix, Fig. S3 A and B), suggesting that Nse1/3/4 DNA binding is a highly conserved feature and is likely mainly responsible for Nse-mediated DNA binding in the Smc5/6 complex.
In summary, our data suggest that Nse5 and Nse6 are distinct from the HEAT repeat subunits of cohesin and condensin at both structural and functional levels, and that Nse5/6 engages in multiple protein–protein, rather than protein–DNA, interactions.
The Nse5 and Nse6 Dimeric Interface.
The dimeric interface of Nse5 and Nse6 is extensive, covering 1,942-Å2 area and consisting of three regions (Fig. 3B). Region I is the largest among the three, with side chains and main chains of three Nse5 residues (Tyr93, Arg97, and Arg102) forming multiple polar interactions with side chains of three Nse6 residues (Asn322, Tyr410, and Glu449) and main chain of M324 from Nse6 (Fig. 3B). Region II is the least extensive, with a single side chain from Gln109 of Nse5 contacting the side chain of Ser326 of Nse6 via polar interactions (Fig. 3B). Interaction region III showed intermediate levels of contact compared with regions I and II. Here, polar interactions were formed between the main chains of Thr335, Lys337, and Ser339 in Nse5 and main chain of Ile281 and side chain of Tyr209 in Nse6, as well as between the side chain of Ser339 in Nse5 and side chain of Glu216 in Nse6 (Fig. 3B).
We performed mutational analyses to validate our structure of Nse5 and Nse6, and to assess the importance of the aforementioned three interfacial regions. Key residues supporting interactions in each region were mutated, and Nse5 and Nse6 mutations were examined individually or in combination. Coexpressed His6-tagged Nse5 and GST-tagged Nse6 were examined for complex formation in a two-step in vitro pull-down experiment. To this end, His6-Nse5 was first enriched by binding to Ni-NTA beads and the eluate was then bound to glutathione beads to assess the Nse5/Nse6 complex level. Compared with the recovered wild-type Nse5/6 complex, mutants showed different effects on the levels of complex formation (Fig. 3 C and D and SI Appendix, Fig. S3C). Correlating with their different interfacial structural features, mutations affecting region I caused the strongest disruption of Nse5 and Nse6 interaction, whereas those at region II had no detectable effects (Fig. 3C). Mutating region III residues led to a moderate reduction in Nse5/6 complex formation (Fig. 3D). The different levels of importance among the three Nse5 and -6 interfacial regions were consistent with their different degrees of sequence conservation. While region I residues show the strongest conservation, those in regions II and III are less well conserved (SI Appendix, Fig. S3 D and E). These analyses suggest that Nse5 and Nse6 form a stable dimeric complex and region I is the most critical in supporting their binding.
Evidence for Maintaining the Nse5/6 Structure in the Smc5/6 Complex.
Next, we addressed whether the Nse5/6 structure is maintained in the Smc5/6 holo-complex. To this end, we examined the structure against the CL-MS data obtained from the holo-complex. If the Nse5/6 structure is maintained in the holo-complex, we can expect the holo-complex’s cross-link data to fit with the structure model. Indeed, all seven cross-links (six intramolecular links and one intermolecular link) mapped to the Nse5/6 structure are within the constraints imposed by the cross-linkers (SI Appendix, Fig. S3F). This finding supports the view that the Nse5/6 structure can be maintained in the Smc5/6 complex.
We also performed cellular studies to examine the residues predicted to be critical for the Nse5 and Nse6 interaction based on our structure model. Mutating two Nse5 residues important for region I interaction (Y93A, R97A) severely impaired cell growth (Fig. 3E). Further mutating a third Nse5 residue involved in region I interaction (R102) led to lethality (Fig. 3E). Both mutant proteins expressed at wild-type levels, thus their phenotype was not caused by reduced protein levels (Fig. 3F). As Nse5-Y93A, R97A greatly impaired the Nse5/6 complex formation in vitro, we infer from these findings that the Nse5 and Nse6 interaction predicted by our structure model is critical for Smc5/6 complex functions in cells. These in vivo findings strengthen the idea that the Nse5/6 structure is maintained in the Smc5/6 complex. We note that our data do not exclude the possibility of Nse5/6 structural alterations upon holo-Smc5/6 interaction with DNA and other proteins.
Integrative Structure Modeling of a Five-Subunit Smc5/6 Complex.
The structures of Nse5/6 from this study and Nse2-bound Smc5 arm region from our previous work (25), in conjunction with our CL-MS data, made it feasible to compute structural models. We omitted Nse-1/3/4 due to insufficient data to determine their juxtaposition in the complex. We used an integrative approach (33–37) to generate a structure of the five-subunit complex composed of Smc5, -6, and Nse2, -5, and -6 (designated Smc5/6–Nse2/5/6) (Fig. 4A). The integrative structure model was computed by combining comparative models of Smc5/6 head and hinge regions, parametrically designed backbone models of their coiled-coil regions, the crystal structure of Nse2, our cryo-EM derived Nse5/6 structure, and our CL-MS data (Materials and Methods and SI Appendix, Fig. S4A).
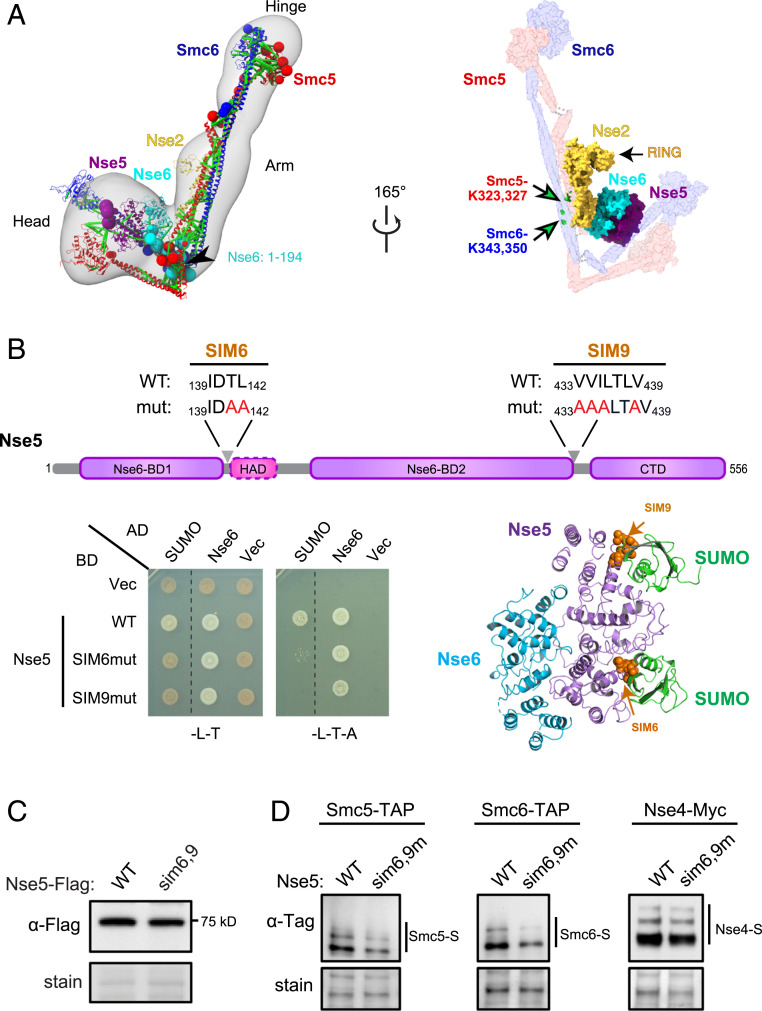
Fig. 4.
Three-dimensional model of the Smc5/6 complex and Nse5 affecting Smc5 and -6 sumoylation. (A) Three-dimensional model of the five-subunit Smc5/6 complex. (Left ) Model representing the centroid of the ensemble of good scoring models obtained through Monte Carlo sampling (PDB-Dev code PDBDEV_00000081). Regions with available atomic structures were modeled as rigid bodies and shown in ribbon representation, while regions lacking atomic structure were modeled as coarse-grained flexible beads and are shown with spheres whose radii are proportional to the average size of the corresponding modeled fragment. The structural ensemble is demonstrated as a 3D localization probability density (61) whose surface is rendered transparent for visual clarity. Satisfied cross-links are shown in green while violated links are colored light brown. (Right) Surface presentation of the model highlights the proteins with available atomistic structure. Smc5 and Smc6 sumoylation sites are marked on our 3D model as green balls and indicated by arrows. (B) Nse5 SIM sequences contribute to its interaction with SUMO. (Upper) Two SIM sequences are indicated in the Nse5 domain structure diagram. (Lower Left) Mutations in Nse5-SIM6 or SIM9 abolish the interactions with SUMO in yeast-two-hybrid assay. AD, Gal4 active domain; BD, Gal4 DNA binding domain; vec, vector. SC-Leu-Trp media (-L-T) selects for BD and AD plasmids, while SC-Leu-Trp-Ade media (L-T-A) reports for positive interactions. (Lower Right) Nse5 SIM6 and SIM9 are located on the surface of the protein and accessible for SUMO binding based on structural modeling using the SUMO-SIM complex structure [PDB ID code 3V62 (62)]. (C) The nse5-sim6,9 mutant does not affect its protein level. (D) The sumoylation levels of Smc5 and Smc6, but not Nse4, were reduced in nse5-sim6,9 mutant cells. Sumoylated proteins were enriched by Ni-NTA resins that bind to His8-tagged SUMO and examined by immunoblotting using antibodies recognizing the tag fused to the endogenous proteins. Loading is shown by Ponceau S stain (stain). The dashed lines indicate removal of superfluous rows in B.
Fig. 4A shows a representative model of the ensemble of structures obtained through structural sampling (SI Appendix, Supplemental Methods), for the five-subunit complex that sufficiently satisfies the input information. The structural ensemble has excellent fit with our CL-MS data, with 97.6% of the crosslinks satisfied by at least one model in the ensemble (SI Appendix, Fig. S4B). The structural precision of the ensemble, which refers to the average Cα RMSD of all models in the ensemble from the representative centroid model, is 3.9 nm, and is 12% of the dimension of the Smc5/6 complex measured from the EM averages (31.9 nm) (Figs. 1D and 4A). The model depicts an overall shape of the complex largely in agreement with our negative-stain EM data. Specifically, Smc5 and -6 are paired at their hinge and aligned throughout coiled-coil regions without elbow bending (Fig. 4A). The middle section of the arms of the complex is thickened due to Nse2 binding (Fig. 4A). A noticeable difference from the EM images is a sharp turn of the head-proximal coiled-coil regions (neck) at an angle larger than those seen in the EM data. This difference may reflect flexibility of the neck region and the model lacking Nse1/3/4, which bind to neck and head regions, thus potentially constraining these regions. In the model, the Nse5/6 core is placed above the Smc5 and Smc6 head regions, which are separated. The relative separation of the heads raised the question whether or not the complex is in ATP-bound state. As our EM data have insufficient resolution, clarification of this point will require future work. On the opposite side of the Nse5, the Nse6’s CAD (90 to 192 aa) is in close proximity to Smc5 and -6 arm regions and Nse2 (Fig. 4A). The spatial closeness between Nse2 and Nse5/6 raises the possibility of their functional coupling.
Nse5 SUMO-Interacting Motifs Affect Smc5 and -6 Sumoylation.
To test the possibility that the coevolved Nse2 and Nse5/6 have functional connections, we considered the unique sumoylation function of the Smc5/6 complex that is not shared by other SMCs (16–19). Nse2 sumoylates Smc5, Smc6, and Nse4, among other substrates, and the sumoylation of Smc5 and -6 contributes to DNA replication and repair (16). However, the functional mechanisms of Nse2 as a SUMO ligase remain to be understood. Activities of other SUMO ligases require binding to the SUMO E2 through their RING domain and binding to the donor SUMO via SUMO interaction domains, and the latter interaction supports a productive conformation required for SUMO transfer (38). While Nse2 contains a RING domain, it has not been reported to interact with SUMO (25, 39), raising the possibility that Nse2 may require cofactors for optimal sumoylation efficiency for specific substrates.
We and others have found that Nse5 can associate with SUMO in yeast two-hybrid assays (40), raising the possibility that it may assist Nse2-mediated sumoylation. To test this idea, we first located several 4-aa sequences resembling SUMO-interacting motifs (SIMs) on Nse5 (SI Appendix, Fig. S5A). SIMs harbor two to four hydrophobic residues and may contain an acidic residue (41–43). Taking advantage of our Nse5/6 structure, we assessed these sequences for accessibility for SUMO binding and found that only three SIMs (SIMs 1, 6, and 9) fit this criterion (SI Appendix, Fig. S5A). These and a few other SIMs were mutated and examined for SUMO and Nse6 association in yeast two-hybrid assays (SI Appendix, Fig. S5A). We found that mutating Nse5-SIM6 located between Nse6-BD1 and HAD, as well as SIM9 between Nse6-BD2 and CTD, abolished SUMO interaction without affecting Nse6 association (Fig. 4B). In contrast, mutating SIM1 or other sequences reduced Nse6 interaction (SI Appendix, Fig. S5B). We concluded that SIM6, which has good sequence conservation (SI Appendix, Fig. S5C), and SIM9 of Nse5 primarily mediate SUMO interaction.
We proceeded to ask whether the SUMO binding of Nse5 contributes to Nse2-dependent sumoylation. MS-based mapping showed that Smc5 and -6 are sumoylated at their coiled-coil regions (44). In our 3D model, these sites are located close to Nse2 and the Nse5/6 complex (Fig. 4A), implicating their feasibility to be influenced by both Nse2 and Nse5/6. Indeed, we found that the nse5-sim6,9 mutant, which expressed at wild-type levels (Fig. 4C), was defective for the sumoylation of both Smc5 and Smc6 (Fig. 4D). This effect was specific because the sumoylation of Nse4 or several other Nse2 substrates was unaffected (SI Appendix, Fig. S5D). Taken together, our in vivo data suggest that Nse5 SIMs favor the sumoylation state of the Smc5 and Smc6 proteins, providing a functional connection between the coevolved Nse2 and Nse5/6 proteins.
Discussion
The SMC family of proteins are primordial DNA modulators postulated to have evolved prior to histones (45). They are essential for maintaining the structures and functions of the genome. Extensive studies focusing on a subgroup of the SMC complexes, such as cohesin and condensin, have revealed their abilities to extrude DNA loops and cohese sister chromatids to sculpt chromatin (2–4). While the predominant view is that all SMC complexes behave similarly, several lines of evidence suggest that this perspective may not apply to the Smc5/6 complex. Despite being essential for cell growth, the Smc5/6 complex does not appear to affect chromatin organization or topologically entrap chromatin (10–12). Rather, the Smc5/6 complex can directly regulate DNA replication and repair (13–15). The structural and molecular basis for the uniqueness of the Smc5/6 complex has been unclear. Our integrative, multiscale structural and functional analyses of the yeast Smc5/6 complex have begun to address and fill in this knowledge gap, thereby unveiling several properties of this complex that are not shared by other SMC complexes.
Our negative-stain EM and CL-MS data, and integrative models corroborate each other and suggest that the arm regions of the Smc5/6 holo-complex examined in the presence of ATPγS have four distinct and interrelated features. Our EM images and CL-MS analyses show a linear alignment of the arm regions of Smc5 and Smc6, with thickening and two kinks in the middle, but no elbow bending as seen for cohesin and condensin. Our measurement of arm length from EM data and integrative modeling are consistent with the much-shortened arm sequences for Smc5/6 compared with cohesin and condensin, as noted previously (8). Also, different from cohesin and condensin, Smc5/6 arm regions show interactions with its unique subunits, Nse2 and Nse6. In fact, our data suggest that only the hinge-proximal arm regions of Smc5/6, approximately one-third of the arm, may be free of Nse association. Such extensive arm–Nse connection may provide a partial explanation for the lack of elbow bending. Furthermore, Smc5/6 arm sequences have a high level of conservation, notably absent in cohesin and condensin. Altogether, these features allow us to propose that the arm regions of the Smc5/6 complex are engaged in multiple protein–protein interactions, in addition to linking the head and hinge regions. As the Smc5 arm region also binds to the Mph1 DNA helicase (46), the Smc5/6 arm could also serve as a platform for associating with additional genomic maintenance factors.
One of the most unique features of the Smc5/6 complex is its three coevolved Nse subunits, namely Nse2 and the Nse5/6 complex. Our cryo-EM structure revealed a clasped-hand shaped Nse5/6 with a rather rigid appearance. Such a structure is distinct from those of HEAT repeat subunits of cohesin and condensin that are built with intrinsic elasticity required for their functions. Also, unlike these HEAT repeat proteins, we found that Nse5/6 lacked DNA binding ability. These features are a significant departure from the HEAT repeat subunits common to cohesin and condensin, which employ their DNA binding and flexibility to support multistep DNA trapping and extrusion processes (30). The absence of these features in Nse5/6 suggests that the Smc5/6 complex is unlikely to be involved in chromatin organization in a manner seen for cohesin and condensin.
Our data collectively show that Nse5/6 contains distinct domains that engage in a network of protein–protein interactions. Besides the domains that form extensive interfaces in the Nse5/6 heterodimer, Nse5-HAD and Nse6-CAD are in close proximity with the head and arm regions, respectively. Yeast two-hybrid and protein binding assays have suggested that the human and fission yeast Nse6 homologs interact with the Smc5/6 arm fragments, indicating that this interaction is evolutionarily conserved (28, 47). Although Nse5/6 was reported to associate with the fragments of Smc5 and -6 hinge in vitro (20), their cross-link in the holo-Smc5/6 was not seen here. While this interaction involving protein fragments may not reflect physiological association, we do not rule out other possibilities, such as the interaction may be involved in intercomplex association, and further clarification will be needed in the future. We have previously shown that Nse6 uses another domain for binding to Rtt107 that in turn recruits the Smc5/6 complex to damaged chromatin (21). Collectively, our data suggest that Nse5/6 plays a critical role by building a protein–protein interaction network that supports Smc5/6 complex structure and function.
Our integrative model points to a spatial proximity among the three coevolved Nse subunits (Nse2, -5, and -6) that is unique to the Smc5/6 complex. Our in vivo data show that Nse5 contains SIM sequences that mediate its SUMO interaction. Furthermore, Nse5-SIM mutations specifically reduce Nse2-dependent Smc5 and Smc6 sumoylation that occurs at their arm regions located close to Nse2 and Nse5/6. These data suggest a functional collaboration between Nse2 and Nse5/6. We consider at least two nonmutually exclusive mechanisms for the roles of Nse5-SIM in Smc5 and -6 sumoylation. Given that other SUMO ligases bind to SUMO to support SUMO transfer to substrates, the apparent lack of SUMO binding by Nse2 suggests that Nse5 may substitute for it during specific sumoylation events. Interestingly, we recently found that the Esc2 protein acts as another cofactor that helps Nse2-mediated sumoylation of the STR complex and Pol2 (48). It is thus possible that Nse2 may collaborate with different cofactors to efficiently sumoylate distinct substrates, and that the Smc5/6 complex may act as a “composite SUMO ligase.” This model does not exclude another possibility wherein Nse5 simply helps to increase local SUMO concentration via binding to SUMO. Testing these and other potential models in the future will shed light into how the Smc5/6 complex acts as a SUMO ligase and uses sumoylation to regulate DNA transactions. As the sumoylation function of Nse2 is not essential for growth, Nse5/6 likely carry out additional roles (17, 19). Our computational model suggests that Nse5/6 is positioned above the head domains; this may resemble bacterial MukE that can localize above the head domain of MukB in the asymmetric complex (49). Such a location could suggest a role for Nse5/6 in regulating head functions, such as binding to DNA and Nse1/3/4 as well as ATP hydrolysis cycle.
During the course of our work, negative-stain EM and CL-MS data of a human and a budding yeast Smc5/6 complex were reported; only the latter study suggested an elbow-bent configuration of the complex (50, 51). The EM images from this latter study (51) resemble ours, including a similar arm length of the complex, which suggests the absence of elbow-bending, as discussed in Results. Additionally, our integrative model supports a straight-arm configuration of Smc5 and Smc6. We note that two recent preprints reporting on the budding yeast holo-Smc5/6 also concluded no elbow-bending (52, 53). One of these studies also reports an X-ray–based Nse5/6 core structure highly similar to ours (52). While our work describes an effect of Nse5 on sumoylation, these studies report Nse5/6 inhibition of the Smc5/6 ATPase activity (52, 53). Together, these findings reveal a multifunctional nature of the Nse5/6, which has been implicated in associating Smc5/6 with chromatin in multiple organisms (21, 22, 54, 55).
Given that the studies conducted so far prepared and examined Smc5/6 complex in somewhat different conditions, it remains possible that the complex can adopt other configurations during its ATPase cycle or upon DNA interaction. Future studies to define structural changes in Smc5/6 during each stage of its reaction cycle will provide additional insights into its DNA-linking and sumoylation activities observed previously (16, 50, 51, 56).
Overall, our integrative structural analyses of the Smc5/6 complex provide insights into both the similarities shared with other SMC complexes, as well as multiple distinct features. One distinct feature entails a straight Smc5/6 arm region that binds to three unique and coevolved Nse subunits, which themselves exhibit physical proximity and functional coupling. We propose that these Nse proteins and their interacting arm regions can both regulate Smc5/6 conformational changes and serve as platforms for binding and collaborating with DNA replication and repair factors. It will be interesting to elucidate how they collaborate with the DNA-binding parts of the complex, including the Nse1/3/4 subcomplex and the Smc5/6 head and hinge regions (24, 32, 57). In conjunction with previous findings, we highlight that Nse5/6 can regulate sumoylation status and recognize chromatin markers for targeting the Smc5/6 complex to replication and lesion sites, among other potential roles. Future work to elucidate how different sets of Nse subunits collaborate among themselves and with Smc5 and -6 to enable the complex to carry out its described roles in DNA replication and repair will further our understanding of this multifunctional genome maintenance complex.
Materials and Methods
Yeast Strains and Genetic Methods and Plasmids.
All yeast strains are derivatives of W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535). For other in vivo assays, at least two strains per genotype were examined in each experiment, and only one is listed in SI Appendix, Table S2. Plasmids used are listed in SI Appendix, Table S3. Detailed strain and mutation construction and standard yeast two-hybrid assay procedure are described in the SI Appendix, Supplemental Methods.
Purification of Smc5/6 Holo-Complex, Nse5/6, and Nse1/3/4 Subcomplexes, and Biochemical Assays.
The Smc5/6 holo-complex was purified from yeast cells while the two subcomplexes were purified in bacteria. Details of protein purification, in vitro pulldown tests, and DNA binding assays are described in SI Appendix, Supplemental Methods.
Negative-Stain EM of the Smc5/6 Holo-Complex and Cryo-EM Analyses of the Nse5/6 Complex.
Purified SMC5/6 holo-complex were examined by a JEOL JEM-1230 electron microscope and images were recorded at a calibrated magnification of 60,000×, yielding the pixel size of 4.0 Å at specimen level. The purified Nse5/6 complex was applied onto glow-discharged UltrAuFoil and images were collected on a FEI Titan Krios electron microscope with a Gatan K3 camera with a 1.064-Å pixel size. Image processing was performed by RELION 3.0 and a total of 657,200 particles were selected for 3D classification. Details are outlined in SI Appendix, Supplemental Methods.
Cross-Linking MS and Data Analyses.
The Smc5/6 complex was cross-linked with DSSO (Thermo) or CDI (Sigma Aldrich), and purified before digestion with LysC followed by Trypsin. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (58) partner repository with the dataset identifier PXD023164. Details are in SI Appendix, Supplemental Methods.
Integrative Structure Determination of Smc5/6–Nse2/5/6 Complex.
Integrative structure modeling proceeded through the standard four-stage protocol (33–37), which was scripted using the Python Modeling Interface package, a library for modeling macromolecular complexes based on the Integrative Modeling Platform software (35), version develop-255ae6c (https://integrativemodeling.org). Details are listed in SI Appendix, Supplemental Methods; files containing input data, scripts and output results are available at https://github.com/integrativemodeling/smc56_nse256.
Detection of In Vivo Protein Sumoylation.
For all proteins, except Pol2, examined here, SUMOylated proteins were pulled and probed for specific substrates by immune-blotting (59). For Pol2, the protein was immuno-precipitation before immune-blotting for its sumoylated forms. Details are listed in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Shelly Lim and Jake Bonner for assisting in yeast strain construction in the early stage of this work; Dirk Remus and his laboratory members for helping in protein expression; Prabha Sarangi, Shengliu Wang, and Dirk Remus for comments on the manuscript; and Yi Ren, Richard Hite, De La Cruz Michael Jason V, and Misha Kopylov for advice and consultation on electron microscopy studies. Negative-stain EM images were collected in the New York Structural Biology Center supported by grants from the Simons Foundation (Grant SF349247), NYSTAR, and the National Institute of General Medical Sciences (Grant GM103310). Z.S. is supported by an A*STAR scholarship. This work is supported by National Institute of General Medical Science Grants R01GM080670 and R01GM131058 (to X.Z.), and R01GM083960 and P41GM109824 (to A.S.); National Cancer Institute Grant R01 CA214812 (to A.K.); the Maloris Foundation (D.J.P.); a Beene Cancer Center grant (jointly to X.Z. and D.J.P.); and Memorial Sloan-Kettering Cancer Center Core Grant P30-CA016086.
Footnotes
Competing interest statement: A.K. is a consultant for Novartis and Rgenta.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026844118/-/DCSupplemental.
Data Availability
Cross-linking mass spectrometry data have been deposited in the ProteomeXchange Consortium (accession no. PXD023164). The atomic coordinates of the Nse5/6 subcomplex have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank with the code 7LTO. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession code EMD-23517. The integrative model of the pentameric Smc5/6 complex has been deposited in the PDB-Dev database with accession code PDBDEV_00000081.
References
- 1.Uhlmann F., SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gruber S., Shaping chromosomes by DNA capture and release: Gating the SMC rings. Curr. Opin. Cell Biol. 46, 87–93 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Hassler M., Shaltiel I. A., Haering C. H., Towards a unified model of SMC complex function. Curr. Biol. 28, R1266–R1281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatskevich S., Rhodes J., Nasmyth K., Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Lee B.-G., et al., Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat. Struct. Mol. Biol. 27, 743–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong M., et al., Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol. Cell 79, 99–114.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Z., Gao H., Bai X.-C., Yu H., Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368, 1454–1459 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Bürmann F., et al., A folded conformation of MukBEF and cohesin. Nat. Struct. Mol. Biol. 26, 227–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hons M. T., et al., Topology and structure of an engineered human cohesin complex bound to Pds5B. Nat. Commun. 7, 12523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farcas A.-M., Uluocak P., Helmhart W., Nasmyth K., Cohesin’s concatenation of sister DNAs maintains their intertwining. Mol. Cell 44, 97–107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venegas A. B., Natsume T., Kanemaki M., Hickson I. D., Inducible degradation of the human SMC5/6 complex reveals an essential role only during interphase. Cell Rep. 31, 107533 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Ohta S., et al., Proteomics analysis with a nano Random Forest approach reveals novel functional interactions regulated by SMC complexes on mitotic chromosomes. Mol. Cell. Proteomics 15, 2802–2818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palecek J. J., SMC5/6: Multifunctional player in replication. Genes (Basel) 10, E7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aragón L., The Smc5/6 complex: New and old functions of the enigmatic long-distance relative. Annu. Rev. Genet. 52, 89–107 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kegel A., Sjögren C., The Smc5/6 complex: More than repair? Cold Spring Harb. Symp. Quant. Biol. 75, 179–187 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Solé-Soler R., Torres-Rosell J., Smc5/6, an atypical SMC complex with two RING-type subunits. Biochem. Soc. Trans. 48, 2159–2171 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Zhao X., Blobel G., A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potts P. R., Yu H., Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25, 7021–7032 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews E. A., et al., Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25, 185–196 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan X., et al., Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J. Biol. Chem. 284, 8507–8515 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan B., Wu J., Meng X., Lei M., Zhao X., Molecular basis for control of diverse genome stability factors by the multi-BRCT scaffold Rtt107. Mol. Cell 75, 238–251.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oravcová M., et al., Brc1 promotes the focal accumulation and SUMO ligase activity of Smc5-Smc6 during replication stress. Mol. Cell. Biol. 39, e00271-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle J. M., Gao J., Wang J., Yang M., Potts P. R., MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 39, 963–974 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alt A., et al., Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat. Commun. 8, 14011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan X., et al., Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell 35, 657–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ser Z., Cifani P., Kentsis A., Optimized cross-linking mass spectrometry for in situ interaction proteomics. J. Proteome Res. 18, 2545–2558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells J. N., Gligoris T. G., Nasmyth K. A., Marsh J. A., Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr. Biol. 27, R17–R18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palecek J., Vidot S., Feng M., Doherty A. J., Lehmann A. R., The Smc5-Smc6 DNA repair complex. bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J. Biol. Chem. 281, 36952–36959 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R. 3rd, Boddy M. N., The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 26, 1617–1630 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimura S. H., Hirano T., HEAT repeats—Versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 129, 3963–3970 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zabrady K., et al., Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 44, 1064–1079 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rout M. P., Sali A., Principles for integrative structural biology studies. Cell 177, 1384–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S. J., et al., Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russel D., et al., Putting the pieces together: Integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward A. B., Sali A., Wilson I. A., Biochemistry. Integrative structural biology. Science 339, 913–915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saltzberg D. J., et al., Using Integrative Modeling Platform to compute, validate, and archive a model of a protein complex structure. Protein Sci. 30, 250–261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Streich F. C. Jr, Lima C. D., Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536, 304–308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makhnevych T., et al., Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33, 124–135 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Bustard D. E., Ball L. G., Cobb J. A., Non-Smc element 5 (Nse5) of the Smc5/6 complex interacts with SUMO pathway components. Biol. Open 5, 777–785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannich J. T., et al., Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280, 4102–4110 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Hecker C.-M., Rabiller M., Haglund K., Bayer P., Dikic I., Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Zhu J., et al., Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283, 29405–29415 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhagwat N. R., et al., SUMO is a pervasive regulator of meiosis. eLife 10, e57720 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirano T., The ABCs of SMC proteins: Two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399–414 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Xue X., et al., Restriction of replication fork regression activities by a conserved SMC complex. Mol. Cell 56, 436–445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adamus M., et al., Molecular insights into the architecture of the human SMC5/6 complex. J. Mol. Biol. 432, 3820–3837 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Li S., et al., Esc2 orchestrates substrate-specific sumoylation by acting as a SUMO E2 cofactor in genome maintenance. Genes Dev. 35, 261–272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo J.-S., et al., Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136, 85–96 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Serrano D., et al., The Smc5/6 core complex is a structure-specific DNA binding and compacting machine. Mol. Cell 80, 1025–1038.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez-Escribano P., et al., Purified Smc5/6 complex exhibits DNA substrate recognition and compaction. Mol. Cell 80, 1039–1054.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taschner M., et al., Nse5/6 inhibits the Smc5/6 ATPase to facilitate DNA substrate selection. bioRxiv [Preprint] (2021). 10.1101/2021.02.09.430422. (Accessed 1 March 2021). [DOI]
- 53.Hallett S. T., et al., Nse5/6 is a negative regulator of the ATPase activity of the Smc5/6 complex. bioRxiv [Preprint] (2021). 10.1101/2021.02.12.430902. (Accessed 1 March 2021). [DOI]
- 54.Etheridge T., et al., Live-cell single-molecule tracking highlights requirements for stable Smc5/6 chromatin association in vivo. bioRxiv [Preprint] (2020). 10.1101/2020.06.19.148106 (Accessed 13 April 2021). [DOI] [PMC free article] [PubMed]
- 55.Räschle M., et al., DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanno T., Berta D. G., Sjögren C., The Smc5/6 complex is an ATP-dependent intermolecular DNA linker. Cell Rep. 12, 1471–1482 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Jo A., Li S., Shin J. W., Zhao X., Cho Y., Structure basis for shaping the Nse4 protein by the Nse1 and Nse3 dimer within the Smc5/6 complex. J. Mol. Biol. 433, 166910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez-Riverol Y., et al., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ulrich H. D., Davies A. A., In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol. Biol. 497, 81–103 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Pebernard S., Perry J. J. P., Tainer J. A., Boddy M. N., Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol. Biol. Cell 19, 4099–4109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alber F., et al., The molecular architecture of the nuclear pore complex. Nature 450, 695–701 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Armstrong A. A., Mohideen F., Lima C. D., Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cross-linking mass spectrometry data have been deposited in the ProteomeXchange Consortium (accession no. PXD023164). The atomic coordinates of the Nse5/6 subcomplex have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank with the code 7LTO. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession code EMD-23517. The integrative model of the pentameric Smc5/6 complex has been deposited in the PDB-Dev database with accession code PDBDEV_00000081.