Significance
Coral reefs are in catastrophic decline worldwide, in part due to increasingly warm surface waters that cause mass coral bleaching and mortality. However, corals in the northern Red Sea and Gulf of Aqaba have shown no sign of bleaching, despite local seawater temperature rising faster than the global average. We show that the exceptional heat tolerance of the common symbiotic reef-building coral Stylophora pistillata from the Gulf of Aqaba is based on a rapid gene expression response and recovery pattern when exposed to heat stress up to 32 °C. Such temperatures are not anticipated to occur in the region within this century, giving real hope for the preservation of at least one major coral reef ecosystem for future generations.
Keywords: coral bleaching, microbiome, heat stress, gene expression profiling, metaorganism
Abstract
Corals from the northern Red Sea and Gulf of Aqaba exhibit extreme thermal tolerance. To examine the underlying gene expression dynamics, we exposed Stylophora pistillata from the Gulf of Aqaba to short-term (hours) and long-term (weeks) heat stress with peak seawater temperatures ranging from their maximum monthly mean of 27 °C (baseline) to 29.5 °C, 32 °C, and 34.5 °C. Corals were sampled at the end of the heat stress as well as after a recovery period at baseline temperature. Changes in coral host and symbiotic algal gene expression were determined via RNA-sequencing (RNA-Seq). Shifts in coral microbiome composition were detected by complementary DNA (cDNA)-based 16S ribosomal RNA (rRNA) gene sequencing. In all experiments up to 32 °C, RNA-Seq revealed fast and pervasive changes in gene expression, primarily in the coral host, followed by a return to baseline gene expression for the majority of coral (>94%) and algal (>71%) genes during recovery. At 34.5 °C, large differences in gene expression were observed with minimal recovery, high coral mortality, and a microbiome dominated by opportunistic bacteria (including Vibrio species), indicating that a lethal temperature threshold had been crossed. Our results show that the S. pistillata holobiont can mount a rapid and pervasive gene expression response contingent on the amplitude and duration of the thermal stress. We propose that the transcriptomic resilience and transcriptomic acclimation observed are key to the extraordinary thermal tolerance of this holobiont and, by inference, of other northern Red Sea coral holobionts, up to seawater temperatures of at least 32 °C, that is, 5 °C above their current maximum monthly mean.
In recent decades, global climate change has tested the resilience and acclimation ability of a multitude of organisms in the tropics, where species live close to their thermal thresholds. In the marine environment, increasingly frequent periods of anomalously high sea surface temperatures (SSTs) have pushed reef-building corals to and beyond their thermal limits (1–3). The result of such disturbance can be coral bleaching (i.e., loss of the endosymbiotic photosynthesizing algae Symbiodiniaceae from the coral host tissue) (4). In highly oligotrophic waters, severe bleaching eventually causes the sessile coral organisms to starve to death on a timescale of weeks, because photosynthates produced by the Symbiodiniaceae are no longer sufficient to cover their nutrient requirements (1, 5–8). When corals die on a massive scale, entire coral reef ecosystems can collapse (1–3, 9). At this rate, by 2050—even under the most environmentally favorable climate change scenarios—most of the world’s coral reef ecosystems will have been greatly diminished, massively transformed, or entirely eliminated (10). However, in some localities, reef-building corals exhibit higher than average tolerance to heat stress (11–15), and it is of vital importance to understand the mechanistic underpinnings of this phenomenon.
Characterization of differentially expressed genes (DEGs) compared with control conditions (i.e., transcriptomics) offers an effective way to assess response and recovery patterns of a coral holobiont during and after thermal stress (16–18). The capability of an organism to rapidly mount a gene expression response when exposed to heat stress and bring the majority of these genes back to baseline expression levels after the heat stress is removed, is referred to as transcriptomic resilience and has been shown to play a key role in coral holobiont survival (17, 19–23). Adding complexity to the heat stress response, corals are also host to a bacterial microbiome that is thought to contribute to the physiology of the holobiont and in particular its thermal tolerance (24–28).
Here, we expand upon previous studies with a time series design across a range of temperatures to investigate the rate and extent of heat stress response and recovery patterns in three major holobiont compartments (i.e., the coral host, the symbiotic algae, and the bacterial microbiome) in the common coral Stylophora pistillata from the Gulf of Aqaba. These corals in the Gulf of Aqaba constitute arguably one of the most thermally tolerant populations in the world with respect to water temperatures above their natural maximum monthly mean (MMM) (11, 29), but the underlying gene expression regulations responsible for this tolerance are still poorly described (30).
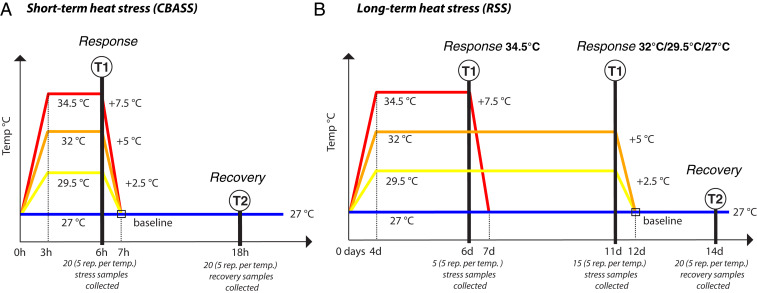
In order to simulate a representative range of heat stress exposures for the Gulf of Aqaba within and beyond this century, corals were exposed to water temperature increases from a baseline of 27 °C to 29.5 °C, 32 °C, and 34.5 °C in short-term (i.e., 3 h ramp-up, 3 h of heating hold, and 1 h ramp-down) and long-term (i.e., 4 d ramp-up, 7 d of heating hold, and 1 d ramp-down) experiments, with a subsequent recovery period at baseline temperatures (Fig. 1). RNA-Sequencing (RNA-Seq) was performed to profile changes in gene expression in both the coral host and dinoflagellate algae during heat stress and after the recovery period. In addition, complementary DNA (cDNA)-based 16S ribosomal RNA (rRNA) gene sequencing was carried out to determine changes in the transcriptionally active bacterial microbiome during and after heat exposures (Fig. 1).
Fig. 1.
Experimental design. Colonies/genotypes (n = 5 genets/treatment) of S. pistillata holobionts (i.e., Stylophora pistillata, Symbiodinium microadriaticum A1, bacterial microbiome) were exposed to two different thermal stress experiments. Note that, for reasons of visual clarity, the x-axes are not linear. (A) A short-term thermal stress experiment was carried out with the CBASS. These thermal treatments included constant temperature at 27 °C (baseline), ramping up to 29.5 °C, 32 °C, or 34.5 °C in 3 h, a 3 h hold at target temperatures (“response” sampling point T1), ramping down to 27 °C over 1 h, and a “recovery” sampling point (T2) 11 h later. (B) The long-term thermal stress experiment using the RSS included constant temperature at 27 °C (baseline), ramping up to 29.5 °C, 32 °C, or 34.5 °C over 4 d, a 7 d temperature hold (“response” sampling point T1; except for the 34.5 °C treatment in which sampling was done after 2 d due to high levels of bleaching), ramping down to 27 °C in 1 d, and a recovery sampling point (T2) 2 d later. Note that the long-term heat stress experiments correspond to 2.8 (29.5 °C), 6.3 (32 °C), and 4.7 (34.5 °C) degree heating weeks (DHW) (77). Ramets of the same five genotypes were sampled for each treatment at each time point. The color coding used throughout the paper is: blue: 27 °C (control), yellow: 29.5 °C, orange: 32 °C, and red: 34.5 °C.
Results
Corals were subjected to a short-term heat stress using the Coral Bleaching Automated Stress System (CBASS) (31, 32). Long-term heat stress experiments were carried out in the Red Sea Simulator (RSS) (33). The same five coral genotypes were exposed to the full set of experimental conditions and sampled for transcriptomic analysis plus bacterial community composition at two different time points: at the end of the thermal exposure (T1) and after a period of time in which the water temperature had been brought back to baseline temperature (T2) (cf. Fig. 1 and Materials and Methods for more detail). We refer to samples collected at T1 as the “response” of the holobiont to the heat stress and samples collected at T2 as holobionts in “recovery” from the heat stress (Fig. 1).
Sequencing Overview.
Sequencing of the 80 transcriptomes of five genotypes, exposed to four temperatures, sampled at two time points, in two different heat stress experiments yielded ∼2 billion paired reads (PE-150) with 22,263,882 ± 7,159,826 paired reads/sample (mean ± SD). We pseudoaligned reads (Kallisto 0.44.0) (34) to a mixed reference of the predicted genes of S. pistillata (35) and Symbiodinium microadriaticum (clade A1) (36), downloaded from reefgenomics.org/ (37); the S. pistillata colonies used in this study hosted only S. microadriaticum (SI Appendix, Figs. S1 and S2). These alignments resulted in averages of 5,799,280 ± 2,465,759 and 5,309,168 ± 3,421,935 pseudoaligned reads per sample for S. pistillata and S. microadriaticum, respectively (mean ± SD). The cDNA-based 16S rRNA gene sequencing produced a total of 10,337,830 paired reads (PE-301) obtained from 68 samples out of 80; 12 samples in recovery (i.e., at T2) from long-term heat stress (29.5 °C, 32 °C, and 34.5 °C) did not have sufficient RNA for cDNA generation for bacterial community analysis. After denoising, merging read pairs, and removing chimeric sequences, 2,719,742 sequences remained with 39,996 ± 16,596 sequences per sample (mean ± SD). A total of 8,660 amplicon sequence variants (ASVs) were inferred with an average of 771 ± 163 ASVs per sample (mean ± SD).
Holobiont Expression Patterns Driven by Temperature and Coral Genotype.
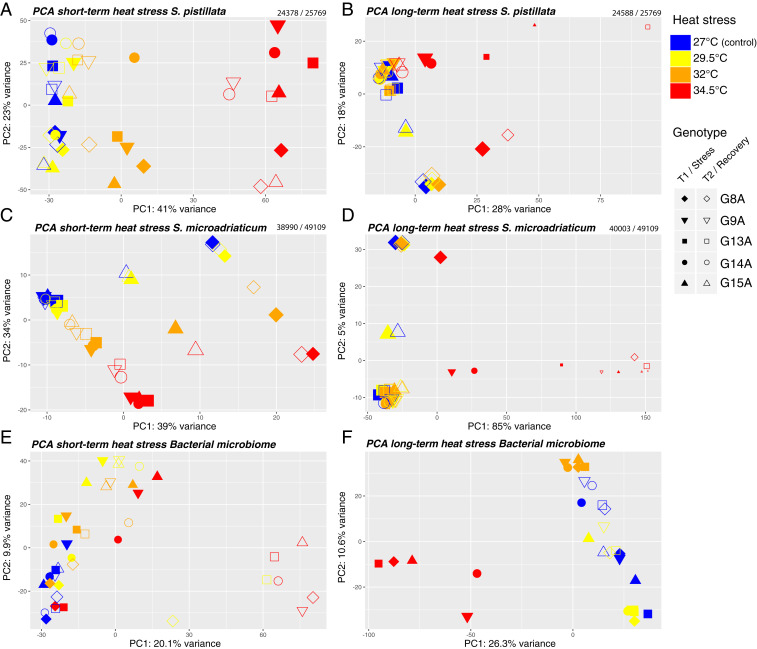
Principal component analysis (PCA) using the PCA function from the DESeq2 (38) software package was carried out for the coral and algal partners in both experiments (Fig. 2), revealing general patterns of whole transcriptome change in relation to temperature, sampling time point, and genotype. Temperature was found to be the major factor (PC1) explaining S. pistillata and S. microadriaticum gene expression at both sampling time points in all experiments (Fig. 2 A–D, permutational multivariate analysis of variance: PERMANOVA, P = 0.001). A conserved genotype effect was apparent with individual genotypes separated mostly along PC2 (Fig. 2 A, B, and D) and slightly along PC1 (Fig. 2C) for both corals and algal symbionts in the two experiments (PERMANOVA for genotype, P = 0.001); one particular genotype (G15A) deviated from this pattern, but single nucleotide polymorphism (SNP) analysis and comparison of sample photos (SI Appendix, Fig. S3) confirmed that these replicates actually came from two individual, but fused, colonies. Interactions between sampling time points (T1, T2) and temperatures were significant for S. pistillata in the short-term stress experiment and for S. microadriaticum in both the short- and long-term experiments (PERMANOVA, P = 0.001), indicating that the response over time operated differently at different temperatures.
Fig. 2.
Ordination plots of coral and algal symbiont gene expression and bacterial ASV composition. PCA of transcriptome-wide gene expression of (A) S. pistillata under short-term heat stress, (B) S. pistillata under long-term heat stress, (C) S. microadriaticum under short-term heat stress, and (D) S. microadriaticum under long-term heat stress. Numbers above each plot represent the number of genes (with at least 1 read) used to build the PCAs and the total number of genes in the genome. PCA based on clr-transformed ASV abundances during (E) short-term and (F) long-term thermal heat stress. Colors as defined in Fig. 1. Symbol shape represents the five colonies (genotypes) used as biological replicates. Filled symbols represent T1, empty symbols represent T2. Symbol size indicates the relative number of reads mapped to their respective reference transcriptome for each sample.
In the short-term experiments at 29.5 °C and 32 °C, the transcriptomes for both coral and algae generally recovered between T1 and T2 in the direction of control levels (Fig. 2 A and C). At 34.5 °C, both coral and algal transcriptomes had begun to recover when comparing T1 and T2 but were still far from baseline gene expression levels (27 °C).
In the long-term experiment at 29.5 °C and 32 °C, gene expression of corals and algae were indistinguishable from the baseline at T1 (P = 0.99). Only at 34.5 °C (P = 0.009) did major changes in gene expression (compared with the baseline) in coral and algae appear at the T1 sampling point, with subsequent uneven (but generally minimal) recovery or death among genotypes (Fig. 2 B and D). Note that the corals experiencing long-term treatment temperatures of 34.5 °C were sampled after 4 d of temperature ramp-up and 2 d at constant heat stress, as they showed clear signs of bleaching. These corals thus had a longer recovery phase, lasting 7 d (Fig. 1). After this 2 d exposure to 34 °C and 7 d recovery at 27 °C, only genotypes G9A, G14A, and G15A seemed to return to the gene expression profiles of control samples, but genotypes G13A and G8A did not return to baseline gene expression. This illustrates that transcriptional behavior is highly uneven among genotypes.
Bacterial communities from short-term heat stress showed significant differences at different temperatures (PERMANOVA, P = 0.001, R2 = 0.220), time points (PERMANOVA, P = 0.002, R2 = 0.063), and genotypes (PERMANOVA, P = 0.001, R2 = 0.233) (Fig. 2E). While host genotype had a strong effect on the bacterial community composition (23.3% of the variance explained), we also observed shifts in the microbiome community structure in response to thermal-stress (i.e., at T1) and absence of recovery toward baseline temperatures at T2. This was particularly clear after exposure to 34.5 °C and recovery (T2), when bacterial communities of all coral genotypes had converged into a unique microbiome distinct from other time points and temperatures (Fig. 2E). In the long-term heat stress experiment, temperature explained the largest fraction of the variation in bacterial community composition (PERMANOVA, P = 0.001, R2 = 0.376), evidenced by a clear separation between samples under heat stress (T1) at 34.5 °C in the PCA (Fig. 2F). For complete PERMANOVA results, see Dataset S1.
Coral Holobiont Response and Recovery: Short-Term Heat Stress.
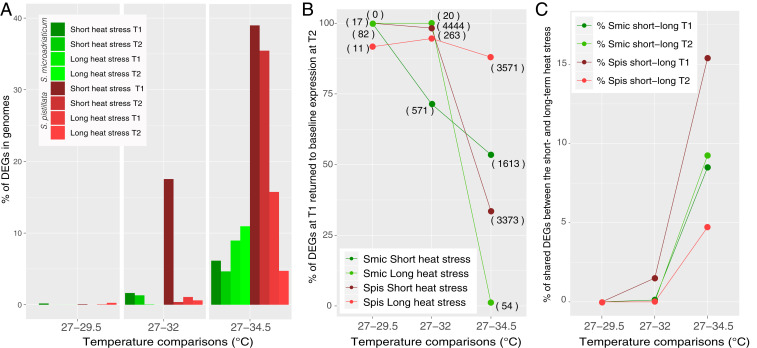
In the short-term heat stress experiments at T1 (cf. Fig. 1A), the number of DEGs in S. pistillata increased with increasing temperature relative to the baseline (27 °C), from 17 DEGs (0.07% of transcriptome) at 29.5 °C, to 4,519 (17.5%) at 32 °C, and to 10,045 (39.0%) at 34.5 °C (Fig. 3A, SI Appendix, Fig. S4, and Dataset S2). Following recovery (i.e., at T2) after exposure to 32 °C, the vast majority (4,444 or 98.3%) of these DEGs had returned to baseline expression levels (i.e., were no longer significantly different from the 27 °C treatment; Fig. 3B). These genes were part of 282 (up) and 285 (down) significantly enriched gene ontology (GO) terms (Dataset S2). In samples exposed to short-term heat stress at 34.5 °C, the percentage of DEGs returning to baseline expression at T2 dropped to 33.6% (Fig. 3B). The genes that remained differentially expressed at T2 (i.e., 66.4%) were part of a large number of GO categories, with most of these DEGs significantly upregulated over baseline expression (378 GOs/446 GOs), including genes involved in the unfolded protein response (UPR, adjusted P < 0.001) (Dataset S2). In the short-term 34.5 °C experiment, 2,464 DEGs were unique to T2 (SI Appendix, Fig. S5).
Fig. 3.
(A) Percentage of DEGs in genomes and (B) percentages of DEGs at T1 that returned to baseline expression at T2 in short- and long-term heat stress for S. pistillata and S. microadriaticum, with number of genes in brackets. (C) Percentage of shared DEGs between the short- and long-term heat stress experiments for coral host (Spis) and algal symbionts (Smic).
In the associated algal symbionts (S. microadriaticum), a relatively small number of genes were differentially expressed relative to the baseline (27 °C) in the short-term experiment at T1 compared to the coral host, with 82 DEGs (0.17% of transcriptome) at 29.5 °C, 800 (1.6%) at 32 °C, and 3,015 (6.1%) at 34.5 °C (Fig. 3 A and C, SI Appendix, Fig. S4, and Dataset S3). At T2, 100%, 71.4%, and 53.5% of these DEGs had returned to baseline expression levels for the 29.5 °C, 32 °C, and 34.5 °C treatments, respectively (Fig. 3 A and B, SI Appendix, Fig. S1, and Dataset S3). A large fraction of DEGs (414) appeared only at T2 in S. microadriaticum after exposure to 32 °C. Importantly, these genes were linked to the down-regulation of photosynthesis (adjusted P < 0.001).
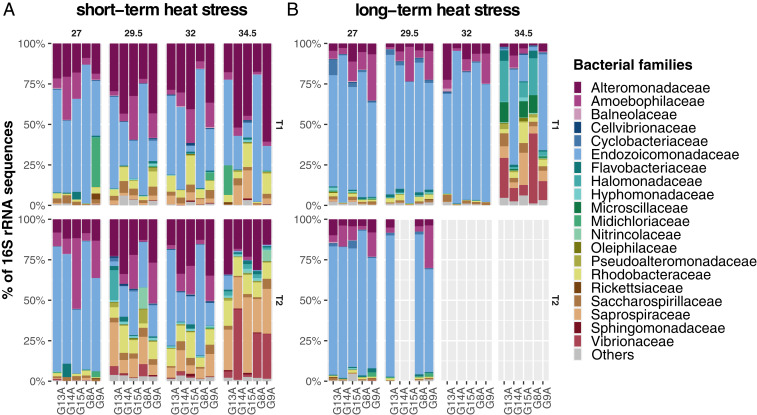
With regard to the bacterial microbiome, in the short-term experiment at T1, we identified 231, 240, and 294 differentially abundant ASVs (DAAs) (Dataset S4) at 29.5 °C, 32 °C, and 34.5 °C, respectively, and 456, 348, and 1,103 DAAs at T2. Of these, 72, 83, and 180 DAAs were significantly more abundant at both T1 and T2 in all heat stress experiments. Members of the genus Endozoicomonas were consistently depleted at all heat stress temperatures and represented the majority of DAAs (17.34%). Members of the Alteromonadaceae (24.42%), Rhodobacteraceae (17.67%), and Saprospiraceae (7.99%) represented the largest fraction of DAAs consistently enriched during heat stress, while DAAs (n = 89) affiliated to Vibrionaceae were mostly enriched at 34.5 °C with fold changes up to 9.7 (ASV0177) (Dataset S4). Of note, ASVs classified as Aureispira sp. (Saprospiraceae) (ASV0015, ASV0034, ASV0035), and Phaeobacter sp. (Rhodobacteraceae) (ASV0077) were consistently more abundant at T1 and T2 across all temperatures, while ASVs classified as Endozoicomonas (Endozoicomonadaceae) (ASV0592, ASV1080) and an unclassified member of the Gammaproteobacteria (ASV1939) were consistently less abundant at T1 and T2 across all temperatures. Furthermore, 56 ASVs classified as Vibrio sp. (Vibrionaceae) were significantly more abundant at T2 and 34.5 °C. In agreement with these shifts in ASV relative abundance, changes in the bacterial community were also reflected at the family level as shown by a prominent relative decrease of members of the Endozoicomonadaceae and an increase in relative abundances of the Vibrionaceae and Saprospiraceae with increasing temperatures (Fig. 4A). ASV raw counts per treatment, as well as the taxonomic information and sequences, can be found in Dataset S5.
Fig. 4.
Relative abundance of the 20 most abundant bacterial families of S. pistillata determined by cDNA-based 16S rRNA gene sequencing during (A) short-term and (B) long-term heat stress. ASV counts were aggregated into bacterial families. Only the most abundant 20 families are shown; less abundant families were aggregated into the “Others” category. Upper panels represent the bacterial community at T1 and lower panels at T2. Each bar represents one of the five coral genotypes. Twelve samples are missing at T2 in the long-term heat stress experiment due to insufficient RNA for cDNA generation for bacterial community analysis.
Coral Holobiont Response and Recovery: Long-Term Heat Stress.
In general, there were substantially fewer DEGs in S. pistillata in the long-term heat stress experiment at both time points compared with the short-term heat stress experiment. At T1, there were 12 DEGs (0.05% of transcriptome) at 29 °C, 278 DEGs (1.08%) at 32 °C, and 4,056 DEGs (15.7%) at 34.5 °C (Fig. 3A, SI Appendix, Fig. S4, and Dataset S2), of which 92%, 95%, and 88% of DEGs returned to baseline expression at T2 (Fig. 3B). At 34.5 °C, among the DEGs up-regulated at T1 that returned to baseline expression at T2 were genes involved in protein folding (adjusted P < 0.01) and reactive oxygen species metabolic processes (adjusted P < 0.05). DEGs down-regulated at T1 that returned to baseline expression at T2 included genes involved in biomineral tissue development (adjusted P < 0.001). At T2 in the 34.5 °C treatment, 731 new DEGs appeared (SI Appendix, Fig. S5), 397 of them down-regulated (Dataset S2).
In the S. microadriaticum transcriptome at T1, there were no DEGs at 29 °C (Dataset S3) and only 20 DEGs at 32 °C, all of which returned to baseline expression at T2 (Fig. 3B) (i.e., complete recovery). In the 34.5 °C treatment at T1, DEGs represented 9% of the S. microadriaticum genes and included up-regulation of 11 tubulin genes (alpha, beta, beta-4, beta-5). Only 1.2% of these genes returned to baseline expression at T2 (Fig. 3B), including previously up-regulated tubulin genes implicated in a broad suite of metabolic processes (Dataset S3). At T2, DEGs represented 11% of the S. microadriaticum transcriptome in the 34.5 °C treatment. However, at this temperature, the overall number of reads of S. microadriaticum was low, and we did not consider gene expression based on such low numbers to be very meaningful.
For the bacterial microbiome, at T1 in the long-term heat stress, 613, 1,082 and 1,666 DAAs were identified at 29.5 °C, 32 °C, and 34.5 °C, respectively, in comparison to the 27 °C baseline temperature (Dataset S4). The majority of DAAs were consistently depleted during heat stress across all temperatures and were affiliated to the bacterial families Rhodobacteraceae (10.0%), Endozoicomonadaceae (9.20%), Alteromonadaceae (6.33%), and Flavobacteraceae (4.36%). Members of the Rhodobacteraceae (14.29%), Flavobacteraceae (10.88%), Cyclobacteraceae (9.18%), and Saprospiraceae (9.18%) were consistently enriched at all temperatures, while members of the Vibrionaceae (n = 19) were only enriched at 34.5 °C with fold change increases of up to 13.7 (Dataset S4). These substantial changes in ASV abundances observed during long-term heat stress (T1) at 34.5 °C were also evident at the family level (Fig. 4B). Comparisons between T1 and T2 were not possible for 32 °C and 34.5 °C due to unavailability of samples at T2.
Holobiont Response Similarity between Short- and Long-Term Heat Stress.
Short-term and long-term heat stress revealed a generally low degree of similarity in the gene expression response (i.e., the percentage of shared DEGs between the experiments for coral host and algae symbionts) at T1 and T2 up to 32 °C (Fig. 3C); indeed, response similarity was nearly zero at 29 °C and 32 °C (Fig. 3C). However, in coral holobionts brought to 34.5 °C, a clear response similarity was observed between short- and long-term heat stress, with more than 15% DEG similarity at T1 (Fig. 3C), distributed over 62 up-regulated and 92 down-regulated GO terms. Of these up-regulated genes, many were implicated in the response to a stimulus/stress (adjusted P < 0.001), protein catabolic processes, cell redox homeostasis, or reactive oxygen species metabolic processes. Among the down-regulated genes, many were implicated in developmental processes (adjusted P < 0.001) (Dataset S6). At T2, fewer genes (∼5%) exhibited similar responses between both experiments in the coral host (Fig. 3C), but those that did included up-regulation of alpha amino acid catabolic processes (adjusted P = 0.003). In the symbiont algae, gene expression similarity was around 8 to 9% in the 34.5 °C treatments at both T1 and T2; 99% of these genes were down-regulated (578 out of 580 at T1 and 646 out of 648 at T2) (Dataset S6). In the bacterial community, 25% of the DAAs under short- and long-term heat stress were affiliated to Rhodobacteraceae, 10% to Alteromonadaceae, and 10% to Sphingomonadaceae. Moreover, 10 DAAs were differentially abundant only during heat stress (T1) and common in the short and long-term heat stress. These ASVs were affiliated to the families Cyclobacteriaceae (ASV0218, ASV0416, and ASV2144), Cryomorphaceae (ASV0275), Flavobacteriaceae (ASV1814), Saprospiraceae (ASV0357), Vibrionaceae (ASV0090), unclassified Gammproteobacteria (ASV1128, ASV4729), and unclassified bacteria (ASV0563).
Discussion
The capability of a coral holobiont to rapidly increase the number of DEGs in response to environmental stress and subsequently relax this response poststress has been termed transcriptomic resilience (20, 23). The main finding from our study is that S. pistillata holobionts from the Gulf of Aqaba can mount an extraordinarily fast and pervasive gene expression response and show extremely strong transcriptional resilience when exposed to thermal stress up to 32 °C, which is ∼5 °C above their maximum monthly mean summer temperatures (MMM) (11, 29). Heat exposure to 5 °C above the natural MMM should (in analogy with coral holobionts from almost any other reef locality) cause high levels of stress and bleaching—if not almost instant death. Yet, S. pistillata from the Gulf of Aqaba demonstrates extreme tolerance to these elevated seawater temperatures (11, 29). Transcriptomic resilience of the coral host in particular appears key to this exceptional thermal tolerance.
Coral and algae transcriptomes were substantially modified and displayed low transcriptomic resilience when subjected to temperatures reaching 34.5 °C, even during short-term exposure, after which only about 34% of coral and 53% of algae DEGs at T1 returned to baseline expression levels at T2 (Fig. 3 A and B). At 34.5 °C, the bacterial microbiome showed no resilience but exhibited a shift to an opportunistic bacterial community, even during recovery. Specifically, long-term exposure to 34.5 °C resulted in symbiont bacteria loss, dominance of Vibrio species, commonly regarded as opportunistic pathogens, and coral death within a few days.
The gene expression in coral host and symbiont algae showed little similarity between short- and long-term heat stress exposure at 32 °C (Fig. 3C). Yet, modifications to the coral and algal transcriptomes began to exhibit higher levels of similarity in both short- and long-term thermal stress exposures at 34.5 °C, providing the first transcriptomic snapshot of the consequences of exceeding the upper critical thermal threshold temperature for these corals that, based on physiological observations, appears to be between 33.8 °C and 34.2 °C (32).
The rate at which the coral transcriptome can recover to baseline expression levels following thermal stress exposure has previously been linked to thermal tolerance. In the present study, S. pistillata exposed to 32 °C was able to re-establish its baseline gene expression levels in just 11 h following short-term stress, and in just 2 d after long-term stress (Figs. 1 and 3 A and B), revealing exceptionally fast transcriptomic resilience for this coral population. In a study of two species of Acropora in the Ofu Island back reef in American Samoa, which underwent a period of more than a month of natural heat stress at ∼1 °C above the regional bleaching threshold (29 °C), Acropora gemmifera colonies went from 3,504 DEGs immediately following heat stress to 12 DEGs (i.e., nearly complete recovery) 4 mo later, with no significant mortality observed. In the same time interval, nearby Acropora hyacinthus maintained roughly half (1,063) of its DEGs and suffered 85% mortality (22).
Relatively fast transcriptomic resilience was also observed in A. hyacinthus corals from the Ofu back reef that were experimentally exposed to 6 °C above the 29 °C regional bleaching threshold for 1 h (20), with 8,913 DEGs (27% of the transcriptome) observed 1 h after heat exposure, dropping to 3,831 DEGs (a 57% drop) 15 h into recovery at 29 °C. Under natural conditions, Ruiz-Jones and Palumbi (39) observed strong and fast transcriptomic resilience in A. hyacinthus corals 2 d after a natural temperature spike at ∼31 °C (i.e., 2 °C above the regional bleaching threshold). However, this gene expression response involved only 177 coexpressed genes (∼0.5% of the A. hyacinthus transcriptome), far from the large plasticity observed in S. pistillata in this study. On the other hand, the gene expression observed in these A. hyacinthus corals pointed to the “unfolded protein” response as a first line of defense of corals against thermal stress, consistent with the findings in Acropora sp. from Ofu Island (American Samoa) (40) and in S. pistillata from the Gulf of Aqaba (30) (this study) (Dataset S2). In our study, the number of DEGs observed in S. pistillata exclusively at the recovery time point (T2) increased with increasing experimental temperature (SI Appendix, Fig. S5). At 34.5 °C, the transcriptomic pattern in S. pistillata was not only characterized by a low percentage of gene recovery but also by a large increase in new DEGs at T2 (SI Appendix, Fig. S5), consistent with the pattern observed in the less heat-tolerant A. hyacinthus (22). Thus, low ability to recover normal gene expression and a tendency for new DEGs to appear during recovery seems to characterize a coral that has exceeded its thermal tolerance limit.
The transcriptomic resilience of S. microadriaticum was strong and similar to that of its coral host up to 32 °C (Fig. 3B) in both the short-term and long-term experiments. It did, however, decrease at 34.5 °C, especially in the long-term experiments (Fig. 3B), indicating a sharp transition in resilience of the symbiont population, similar to the coral host, between 32 °C and 34.5 °C (41). In addition, 414 DEGs appeared in the 32 °C treatment at T2 in the short-term experiment (Dataset S3), mainly as a down-regulation of genes involved in photosynthesis, indicating that reduced photosynthetic activity might be beneficial to the symbionts immediately after thermal stress, possibly to reduce the abundance of reactive oxygen species.
By analogy to the concept of transcriptomic resilience, we define the ability of the bacterial microbiome to dynamically change its composition in response to heat stress and return to the initial composition during the recovery phase as “microbiome resilience,” which further adds to the notion of microbiome flexibility (28). In contrast to the strong transcriptomic resilience of the coral and algal symbionts, the bacterial microbiome composition showed no resilience during short-term heat stress experiments, even at temperatures of 29.5 °C and 32 °C (Fig. 4). Rather, microbiome composition shifted significantly during recovery (i.e., at T2) (Fig. 4A), indicating that different dynamics are at play compared to coral host and algal gene expression. Importantly, in contrast to the set of all genes present in the genomes of coral host and symbiotic algae, which is a fixed entity, the microbiome is “fluid” (42) and an open entity (i.e., there is a dynamic exchange of external and holobiont-associated microbes). This was demonstrated by the appearance of opportunistic bacteria, such as Saprospiraceae and Vibrionaceae, in the short-term experiment at 34.5 °C (Figs. 2E and 4A). Notably, bacteria of the genus Aureispira (Saprospiraceae) increased in abundance at all experimental temperatures above 27 °C and remained highly abundant in the recovery time point. Members of the Aureispira are assumed to be primarily seawater residents (43) but have also been found in tissues of Acropora muricata affected by White Syndrome, suggesting their involvement in opportunistic infections (44). There is also evidence that members of the Aureispira prey on Vibrio sp. in a process linked with the availability of calcium ions (45) and thus may help to counter an increasing abundance of opportunistic bacteria (46).
Together, our observations indicate that in short-term heat stress events up to 32 °C, the S. pistillata holobiont can achieve near-complete gene expression recovery (98.3% of the genes recovered), but that substantial shifts in the bacterial microbiome composition are seen, consistent with previous work (26). During long-term heat stress, the bacterial microbiome composition was stable up to 32 °C, with a dominance of the tissue-associated Endozoicomonadaceae (Fig. 4B), commonly present in S. pistillata (47, 48) and assumed to be abundant in healthy corals (49). As such, their decrease in relative abundance at 34.5° is consistent with the notion that these bacteria are important for holobiont functioning (48, 50, 51).
Acclimation, defined as a return to a normal state of gene expression during a long-term thermal stress, is an important factor in coral thermal tolerance (21). In our study, accepting the assumption that the long-term experiments can be considered a continuation of the short-term experiments, we also investigated the capability of the S. pistillata holobiont to acclimate to elevated temperatures. To do so, we compared the number of DEGs at T1 in the short-term (T1 = 6 h) and long-term (T1 = 11 d) heat stress experiments (Fig. 3A). Both the coral host and algal symbiont host showed signs of acclimation at stress temperatures of 29.5 °C and 32 °C, with a much smaller fraction of DEGs at T1 in the long-term heat stress compared with T1 in the short-term heat stress (Fig. 3A). Similarly, the bacterial microbiome showed signs of acclimation up to 32 °C (Fig. 4B), with a more stable community composition among genotypes and between temperatures (27 °C, 29.5 °C, and 32 °C) in the long-term heat stress experiment (Figs. 2F and 4B). Signs of acclimation in the microbiome disappeared at the experimental temperature of 34.5 °C, along with the appearance of opportunistic bacteria.
Conclusion
The northern Red Sea and Gulf of Aqaba constitute a coral reef refugium from global warming (11, 29, 52, 53), which is thought to stem from selection for thermal tolerance during northbound migration through the warmer waters of the southern Red Sea following the last ice age (11, 29). Our study has shown that S. pistillata from the northern Red Sea and Gulf of Aqaba exhibits an exceptional transcriptomic resilience and is capable of producing a fast and pervasive gene expression response to both short-term (hours) and long-term (days and weeks) thermal stress. Indeed, we observed only negligible transcriptomic change(s) following an 11 d exposure to temperatures >2 °C above the local MMM, which is thought to induce bleaching in most other coral populations. Even at 5 °C above their MMM, clear signs of acclimation were observed. Only at 7.5 °C above the local MMM did these corals exhibit irreversible transcriptomic change(s), loss of symbiont algae, an increase of opportunistic (and potentially pathogenic) bacteria in their microbiome, and high mortality. Importantly, our multitemperature/multitimepoint approach indicates a link between the short-term transcriptomic resilience and longer-term acclimation capacity. Overall, our study represents a significant step toward understanding the complex transcriptomic basis of coral thermal tolerance. Assuming that our results for S. pistillata are representative of the general coral population in the Gulf of Aqaba and the northern Red Sea, these corals represent the best chance for humanity to preserve a major, highly biodiverse coral reef ecosystem that, to date, has been unaffected by ocean warming, in contrast with most other reefs that will continue to be decimated by the combined effects of local and global anthropogenic stressors (2).
Materials and Methods
Samples Collection.
Ramets of five S. pistillata colonies (genotypes) all harboring S. microadriaticum (clade A1, see below) were collected from the nursery at the Interuniversity Institute (IUI) for Marine Science in Eilat, Israel (29° 30′ 06.5″ N, 34° 55′ 04.6″ E). Per colony/genotypes, 16 ramets (total of 80 ramets) were collected as replicates for the two experiments. The samples were collected mid-January 2019 with a water temperature of 22 °C at a depth of 8 m and were placed after collection in acclimatization tanks.
Short- and Long-Term Heat Stress Experimental Designs.
Two experiments (Fig. 1) were conducted to simulate short- and long-term thermal stress events. In each of these systems, the holobionts were stressed with four temperatures: 27 (ambient/control), 29.5, 32, and 34.5 °C (Fig. 1). The temperatures were selected to illicit a full thermal stress response in these corals based on preliminary experiments and previous studies (11, 29, 32).
CBASS (Fig. 1A) (31, 32) was used to generate short-term heat stresses of 18 h between February 27 and 28, 2019. Two replicates of each of the five collected genotypes were placed in four 10 L tanks (40 ramets). These four tanks corresponded to the four temperatures. The number of tanks and samples were double for physiological measurements (32). For RNA extraction and library preparation, we consistently took the first replicate of the two tanks. The temperature profile was constantly monitored, and only the tanks that had the expected temperature profile (following Fig. 1A) were used.
The long-term heat stress experiment was performed using the RSS (Fig. 1B) (33) for 2 wk from January 18 to February 2, 2019. This system is a flow-through, mesocosm system with constant and automated monitoring of the temperature. Similar to the CBASS, four 40 L tanks contained two replicates of each of the five genotypes (40 ramets). Ramets were held at max temperatures for 6 d and were collected after this period for all treatments except for the highest temperature (34.5 °C). For this temperature, the ramets were collected after 4 d of ramping and 2 d of hold, because these corals were already showing signs of tissue disintegration.
For both experiments, a total of 80 coral fragments were collected, 40 at the end of each thermal stress (T1) and 40 after a time of recovery (T2). These samples were immediately snap-frozen in liquid nitrogen and then stored at −80 °C. CBASS and RSS are referred to as short-term and long-term heat stress, respectively, in the following sections.
RNA Extraction and RNA-Seq Library Preparation and Sequencing.
Samples frozen in liquid nitrogen were rapidly placed in a new and sterile ziplock bag and 1,000 μL of sterile and filtered seawater was added. The coral tissue was then blasted off in less than 3 min with an air pump connected to a sterile 1,000 μL pipette tip. An aliquot of the slurry was directly transferred into the RNeasy Mini Kit (QIAGEN) buffer and frozen in liquid nitrogen until extraction. The 80 aliquots were extracted for total RNA using the RNeasy Mini Kit following manufacturer guidelines. RNA quality was verified a first time with NanoDrop 2000 (ThermoFisher Scientific Inc.). The RNA was then transported to the sequencing facility using a GenTegra-RNA plate. After recovery of the RNA, all samples were measured with a Qubit Flurometer (ThermoFisher Scientific Inc.), and a subset of samples were checked for integrity with a 2100 Bioanalyzer mRNA Nano assay (Agilent Technologies, Inc.).
Eighty libraries were constructed with 1 µg total RNA each following the Illumina reference guide of the 96 sample TruSeq Stranded mRNA protocol (Illumina). Seven RNA samples were less concentrated than the minimum input recommendation, and the total volume of the RNA aliquot was used for the construction of these libraries. The SuperScript II Reverse Transcriptase kit was used to generate cDNA and Agencourt AMPure XP beads were used to clean cDNA. Each library’s quality was assessed with the high sensitivity DNA assay on the 2100 Bioanalyzer (Agilent Technologies, Inc.). Libraries that showed signs of primer dimer contamination were cleaned with a repeat round of AMPpure beads with ratio change. The libraries were then multiplexed in sets of 10 across eight lanes of 150 base pair (bp) paired-end sequencing on an Illumina HiSeq4000.
RNA-Seq Analysis.
Raw reads were demultiplexed by the sequencing facility. The quality of the demultiplexed reads was checked with FastQC (54) before and after read trimming. For a better visualization and comparison of the qualities of all libraries, MultiQC (55) was used to concatenate the results of FastQC. The reads were trimmed with Trimmomatic (version 0.36) (56) to remove Illumina adapters, low quality reads, and reads smaller than 40 bp. For initial analysis, a composite transcriptome was built combining the predicted gene transcripts of S. pistillata (35), S. microadriaticum (36), and Cladocopium goreaui (clade C) (57). Indeed, S. pistillata of the Red Sea associates with both algal symbiont species. S. microadriaticum is more common in shallow water colonies, while Cladcopium spp. is more prevalent in deeper water (58). Samples were collected in the shallow; thus only S. microadriaticum reads were expected. In order to confirm this, filtered reads of the 80 libraries were pseudoaligned to this reference composite transcriptome with Kallisto (0.44.0) (34). Once the absence of Cladocopium spp. reads was confirmed, filtered reads of the 80 libraries were pseudoaligned to a composite transcriptome made only of S. pistillata and S. microadriaticum. Tximport (59) was used to import the pseudoaligned counts into the R environment with RStudio (R Core Team, 2018, R version 3.5.2, RStudio Team (2016), RStudio version 1.1.463) for transcript-level count estimation from Kallisto for gene expression analysis.
The differential gene expression analysis was performed with the package DESeq2 (version 1.22.2) (38) in R. The design used in DESeq2 included temperature and genotype. The analysis was separated by experiment (short-term and long-term), and in each of these experiments differential expression was analyzed separately for S. pistillata and S. microadriaticum. All genes that had a minimum mean of five reads across five genotypes in at least seven out of the eight treatments assayed (4 temperatures × 2 time points) for each experiment were kept for downstream analysis. This retained 18,757 and 20,276 (short-term and long-term heat stress) out of 25,769 genes for the coral and 30,843 and 23,775 (short-term and long-term heat stress) out of 49,109 genes for the algal symbiont, respectively. Normalization for sequencing depth was applied through the DESeq dispersion function. Wald testing for significance difference of coefficients with a negative binomial general linear model (GLM) was applied in the DESeq function.
To investigate the difference in expression in each experiment as well as for the coral and algal symbiont, differential expression analysis contrasts were applied for a “standard” temperature increase, with the following temperature comparisons: 27 °C versus 29.5 °C, 27 °C versus 32 °C and 27 °C versus 34.5 °C for the stress time point (T1) and similarly for the recovery time point (T2). DESeq2 P values were corrected using Benjamini–Hochberg (BH) at a default false discovery rate (FDR) of 0.1. Only the genes ending up with an adjusted P value below 0.05 were considered differently expressed. The number of differentially expressed genes reported for the different experiments and species was compared to the total number of genes in their respective species transcriptome in order to scale the effect of the different treatments as a percentage of transcriptome change. The PCA representations for the two experiments and the coral and algal symbionts were plotted with vst normalized gene expression on all genes except nonexpressed genes. A filter for nonexpressed genes was added to remove genes that did not have at least one read across all treatments. PCA data were replotted with the package ggplot2 (60). Significance of temperature, genotype, and time-point were tested by PERMANOVA using the adonis function from the vegan package (Package version 2.5–6) (61). All gene lists of DEGs were then used to detect genes that are differentially expressed in the temperature comparison during heat stress (T1) but not differentially expressed in the temperature comparison during recovery (T2). A second gene list included the genes that were found differentially expressed in both time points (T1 and T2). The third gene list included the newly differentially expressed genes during recovery (T2). Venn diagrams were built using the R package VennDiagram (62).
Functional Analysis.
The GOseq package (1.34.1) (63) was used for GO enrichment analysis for all gene lists. GOseq applies a correction based on the gene length that was shown to be important for RNAseq based data. Significantly enriched categories after P value adjustment with the BH method were plotted by category, Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) and with a -log10 of the adjusted P value.
Symbiont and Coral Genetic Structure and Genotypes.
To identify the type of algal symbionts in our samples, two approaches were taken. First, the previously detailed pseudoalignment of the trimmed reads against the composite transcriptome of S. pistillata, S. microadriaticum, and Cladocopium goreaui was used to detect reads belonging to S. microadriaticum or Cladocopium spp. or both. At the same time, reads were pseudoaligned with Kallisto (34) to the SymPortal ITS2 database (64) to identify the origin of the ITS2 sequence present in our samples.
In order to confirm the clonality of ramets replicates and to understand expression variation associated with genotypes, SNPs for the different samples were retrieved for both the coral and the algal symbiont. Raw RNA-Seq reads were mapped with STAR (2.5.3a) (65) with a single pass. BAM files were used for calling SNPs with the bcftools option call -v -m from SAMtools (1.8) (66). Low quality SNPs (<30 mapping quality) were filtered out. A single vcf file including all shared SNPs across all samples was created. Plink was used to modify the vcf format (1.90) (67) in order to be used by the adegenet package (68) in the R environment for PCA genetic variation plotting. For this analysis, four S. pistillata and nine S. microadriaticum samples mostly from bleached samples at 34.5 °C in both CBASS and RSS experiments were not included due to low number of sequenced reads (SI Appendix, Fig. S3 A and B). All bioinformatic analysis were performed on the Wally cluster of the Scientific Computing and Research Support Unit of the University of Lausanne, and all statistical analyses were performed in R with RStudio (RStudio Team, 2016).
Bacterial Community 16S rRNA Gene Library Preparation and Sequencing.
16S rRNA amplicons were generated from cDNA as template in order to assess the composition of the transcriptionally active bacterial community. Residual DNA contamination was removed from RNA samples using the RQ1 RNase-Free DNase (Promega) according to the manufacturer's protocol, with the exception that incubation times for digestion and termination were reduced by half to avoid RNA degradation. Synthesis of cDNA was done using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher) according to manufacturer's instructions. Amplification of the hyper variable regions V5-V6 of the 16S rRNA gene transcript (cDNA) was done using primers 781F (5′ AGGATTAGATACCCTGGTA 3′) and 1061R (5′ CRRCACGAGCTGACGAC 3′) (69) attached to Illumina universal adaptors. The Nextera XT DNA library prep kit was used for indexing according to the manufacturer’s instructions, and libraries were normalized using the SequalPrep Normalization Plate Kit (Thermo Fisher Scientific) before pooling. Pooled libraries were sequenced on the Illumina MiSeq platform using 2 × 301 bp paired end reads (Illumina Reagent Kit v3).
Bacterial Community Analysis.
Distinct 16S rRNA amplicon sequence variants (ASVs) were inferred using DADA2 (70). First, forward and reverse reads were truncated at the 3′ end at 220 and 190 base pairs, respectively, based on quality profiles. Reads were truncated at the first instance of a quality score ≤10. After truncation, reads with an expected error > 2 or with the presence of ambiguous bases were discarded. ASVs were inferred from each read pair independently using the “pool=T” option to increase the resolution of singletons and doubletons. ASV pairs were merged and checked for chimeras. After chimeric ASVs were removed, final ASVs were taxonomically annotated using the SILVA database (version 138) (71). ASV raw counts per treatment and taxonomic information can be found in Dataset S5.
ASV counts were transformed using the centered-log ratio (clr) transformation described by Aitchison (72) to better handle comparisons of compositional data (73, 74). PCA analyses were calculated from Euclidean distances of clr-transformed counts using Phyloseq (75), and differences between bacterial communities were determined using PERMANOVA implemented in Vegan (61). Differential abundance analysis of 16S rRNA amplicon ASVs was done using the R package Analysis of Composition of Microbiomes with Bias Correction (ANCOMBC) (v.1.0.2), an algorithm for compositional analysis that uses a linear regression framework to estimate the unknown sampling fractions from counts (76). Unlike standard approaches based on relative abundances, ANCOMBC estimates absolute abundances and identifies differentially abundant ASVs relative to the geometric mean abundance of the reference (27 °C in all comparisons) and relative to the geometric mean abundance of bacterial taxa in the respective sample under consideration. ANCOMBC function was applied on ASV counts to compare heat stress samples (29, 32, or 34 °C) to baseline temperature (27 °C). To correct for multiple testing, the FDR method was used to adjust P values. Differentially abundant ASVs (DAAs) were declared if adjusted P values were <0.05 between treatment comparisons. Finally, relative abundances of the most abundant 20 families were generated using ggplot2 (60).
Supplementary Material
Acknowledgments
R.S. was supported by a fellowship from the AXA foundation for “Ocean, marine biodiversity and conflict prevention” and funding provided by Ecole Polytechnique Fédérale de Lausanne. Experimental work was supported by US-Israeli Bi-National Science Foundation Grant #2016403 (D.J.B. and M.F.) and US National Science Foundation Grant #1833201 (D.J.B.). C.R.V. was supported through the Deutsche Forschungsgemeinschaft (German Research Foundation) Project Number 433042944. A.M. was supported by Swiss National Science Foundation through Grant Number 200021_179092. Corals used in this study were collected under permit 2019/42143 from Israel’s Nature and Parks Authority.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023298118/-/DCSupplemental.
Data Availability
The raw RNA-Seq and bacterial 16S rRNA amplicon sequencing data are available at the National Center for Biotechnology Information (NCBI) under BioProject PRJNA674053. Data (DEGs, GOs, ASVs) can be found in supplementary datasets (Datasets S2, S3, S5, and S6), and scripts for gene expression abundance and gene expression analysis can be accessed at GitHub, https://github.com/rsavary/Fast-and-pervasive-transcriptomic-resilience. Bacterial community scripts can be accessed at GitHub, https://github.com/ajcardenasb/RSS_CBASS_16S.
References
- 1.Hoegh-Guldberg O., Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshwater Res. 50, 839–866 (1999). [Google Scholar]
- 2.Hughes T. P., et al., Coral reefs in the anthropocene. Nature 546, 82–90 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Hughes T. P., et al., Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83 (2018). [DOI] [PubMed] [Google Scholar]
- 4.LaJeunesse T. C., et al., Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Morris L. A., Voolstra C. R., Quigley K. M., Bourne D. G., Bay L. K., Nutrient availability and metabolism affect the stability of coral-symbiodiniaceae symbioses. Trends Microbiol. 27, 678–689 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Davy S. K., Allemand D., Weis V. M., Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopp C., et al., Subcellular investigation of photosynthesis-driven carbon assimilation in the symbiotic reef coral Pocillopora damicornis. mBio 6, e02299-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rädecker N., et al., Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. U.S.A. 118, e2022653118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glynn P. W., D’Croz L., Experimental evidence for high temperature stress as the cause of El Nifio-coincident coral mortality. Coral Reefs 8, 181–191 (1990). [Google Scholar]
- 10.Hoegh-Guldberg O., Poloczanska E. S., Skirving W., Dove S., Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 4, 158 (2017). [Google Scholar]
- 11.Fine M., Gildor H., Genin A., A coral reef refuge in the Red Sea. Glob. Change Biol. 19, 3640–3647 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Gardner S. G., et al., Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol. Evol. 9, 938–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hume B. C. C., et al., Ancestral genetic diversity associated with the rapid spread of stress-tolerant coral symbionts in response to Holocene climate change. Proc. Natl. Acad. Sci. U.S.A. 113, 4416–4421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howells E. J., et al., Corals in the hottest reefs in the world exhibit symbiont fidelity not flexibility. Mol. Ecol. 29, 899–911 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Osman E. O., et al., Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 8, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSalvo M. K., et al., Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Barshis D. J., et al., Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M., Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113 (2010). [Google Scholar]
- 19.Bellantuono A. J., Granados-Cifuentes C., Miller D. J., Hoegh-Guldberg O., Rodriguez-Lanetty M., Coral thermal tolerance: Tuning gene expression to resist thermal stress. PLoS One 7, e50685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seneca F. O., Palumbi S. R., The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol. 24, 1467–1484 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Bay R. A., Palumbi S. R., Rapid acclimation ability mediated by transcriptome changes in reef-building corals. Genome Biol. Evol. 7, 1602–1612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas L., López E. H., Morikawa M. K., Palumbi S. R., Transcriptomic resilience, symbiont shuffling, and vulnerability to recurrent bleaching in reef-building corals. Mol. Ecol. 28, 3371–3382 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Franssen S. U., et al., Transcriptomic resilience to global warming in the seagrass Zostera marina, a marine foundation species. Proc. Natl. Acad. Sci. U.S.A. 108, 19276–19281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peixoto R. S., Rosado P. M., Leite D. C. d. A., Rosado A. S., Bourne D. G., Beneficial Microorganisms for Corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8, 341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torda G., et al., Rapid adaptive responses to climate change in corals. Nat. Clim. Chang. 7, 627–636 (2017). [Google Scholar]
- 26.Ziegler M., Seneca F. O., Yum L. K., Palumbi S. R., Voolstra C. R., Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler M., et al., Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 3092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voolstra C. R., Ziegler M., Adapting with microbial help: Microbiome flexibility facilitates rapid responses to environmental change. BioEssays 42, e2000004 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Krueger T., et al., Common reef-building coral in the Northern Red Sea resistant to elevated temperature and acidification. R. Soc. Open Sci. 4, 170038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maor-Landaw K., et al., Gene expression profiles during short-term heat stress in the red sea coral Stylophora pistillata. Glob. Change Biol. 20, 3026–3035 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Voolstra C. R., et al., Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Change Biol. 26, 4328–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Evensen N. R., Fine M., Perna G., Voolstra C. R., Barshis D. J., Remarkably high and consistent tolerance of a Red Sea coral to acute and chronic thermal stress exposures. Limnol. Oceanogr. 9999, 1–12 (2021). [Google Scholar]
- 33.Bellworthy J., Fine M., The Red Sea simulator: A high-precision climate change mesocosm with automated monitoring for the long-term study of coral reef organisms: A large future ocean mesocosm in the Gulf of Aqaba. Limnol. Oceanogr. Methods 16, 367–375 (2018). [Google Scholar]
- 34.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Voolstra C. R., et al., Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci. Rep. 7, 17583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aranda M., et al., Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liew Y. J., Aranda M., Voolstra C. R., Reefgenomics.Org–A repository for marine genomics data. Database (Oxford) 2016, baw152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Jones L. J., Palumbi S. R., Tidal heat pulses on a reef trigger a fine-tuned transcriptional response in corals to maintain homeostasis. Sci. Adv. 3, e1601298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas L., et al., Mechanisms of thermal tolerance in reef-building corals across a fine-grained environmental mosaic: Lessons from Ofu, American Samoa. Front. Mar. Sci. 4, 434 (2018). [Google Scholar]
- 41.Voolstra C. R., et al., Contrasting heat stress response patterns of coral holobionts across the Red Sea suggest distinct mechanisms of thermal tolerance. Research Square, 10.21203/rs.3.rs-117181/v1 (2020). [DOI] [PubMed]
- 42.Jaspers C.et al.; Consortium of Australian Academy of Science Boden Research Conference Participants , Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Zoology (Jena) 133, 81–87 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Yang S.-H., et al., Long-term survey is necessary to reveal various shifts of microbial composition in corals. Front. Microbiol. 8, 1094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweet M., Bythell J., White syndrome in Acropora muricata: Nonspecific bacterial infection and ciliate histophagy. Mol. Ecol. 24, 1150–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furusawa G., Hartzell P. L., Navaratnam V., Calcium is required for ixotrophy of Aureispira sp. CCB-QB1. Microbiology (Reading) 161, 1933–1941 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Welsh R. M., et al., Bacterial predation in a marine host-associated microbiome. ISME J. 10, 1540–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayer T., et al., The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neave M. J., et al., Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roder C., Bayer T., Aranda M., Kruse M., Voolstra C. R., Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol. Ecol. 24, 3501–3511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neave M. J., Apprill A., Ferrier-Pagès C., Voolstra C. R., Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neave M. J., Michell C. T., Apprill A., Voolstra C. R., Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7, 40579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grottoli A. G., Tchernov D., Winters G., Physiological and biogeochemical responses of super-corals to thermal stress from the Northern Gulf of Aqaba, Red Sea. Front. Mar. Sci. 4, 215 (2017). [Google Scholar]
- 53.Osman E. O., et al., Thermal refugia against coral bleaching throughout the northern Red Sea. Glob. Change Biol. 24, e474–e484 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Andrews S., Fast QC: A quality control tool for high throughput sequence data. Version 0.11.9 (2010). https://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 13 April 2021.
- 55.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., et al., Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1, 95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byler K. A., Carmi-Veal M., Fine M., Goulet T. L., Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS ONE 8, e59596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2016). [Google Scholar]
- 61.Oksanen J., et al., Community Ecology Package. R package version 2 (2013). http://sortie-admin.readyhosting.com/lme/R%20Packages/vegan.pdf. Accessed 13 April 2021.
- 62.Chen H., Boutros P. C., VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young M. D., Wakefield M. J., Smyth G. K., Oshlack A., Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hume B. C. C., et al., SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell S., et al., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jombart T., Ahmed I., adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersson A. F., et al., Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3, e2836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Callahan B. J., et al., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quast C., et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aitchison J., The Statistical Analysis of Compositional Data (Royal Statistical Society, 2nd Ed., 1982), 44, pp. 139–177. [Google Scholar]
- 73.Knight R., et al., Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J., Microbiome Datasets are compositional: And this is not optional. Front. Microbiol. 8, 2224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMurdie P. J., Holmes S., phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin H., Peddada S. D., Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu G., et al., Reef-scale thermal stress monitoring of coral ecosystems: New 5-km global products from NOAA coral reef watch. Remote Sens. 6, 11579–11606 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-Seq and bacterial 16S rRNA amplicon sequencing data are available at the National Center for Biotechnology Information (NCBI) under BioProject PRJNA674053. Data (DEGs, GOs, ASVs) can be found in supplementary datasets (Datasets S2, S3, S5, and S6), and scripts for gene expression abundance and gene expression analysis can be accessed at GitHub, https://github.com/rsavary/Fast-and-pervasive-transcriptomic-resilience. Bacterial community scripts can be accessed at GitHub, https://github.com/ajcardenasb/RSS_CBASS_16S.