Significance
The entomopathogenic fungus Beauveria bassiana can overcome insecticide resistance and represents a promising tool for controlling mosquitoes and other insect pests. Improvement of fungal efficacy requires better understanding of fungus–mosquito interactions. Here, we report on a new strategy of insect defense against fungal infection that employs translocation of miRNAs to silence virulence genes. Fungal infection activates production of Anopheles let-7 and miR-100 microRNAs (miRNAs). These miRNAs translocate into fungal hyphae to specifically silence the sec2p and C6TF fungal genes. Both genes are essential for fungal invasive growth and pathogenicity. Notably, virulence of a B. bassiana strain expressing both anti–let-7 and anti–miR-100 “sponge” RNAs is dramatically increased. This study may lead to new strategies for improved fungal-based vector control efficacy.
Keywords: insect immunity, entomopathogenic fungus, host–microbe interactions, microRNA, cross-kingdom RNAi
Abstract
Chemical insecticides remain the main strategy to combat mosquito-borne diseases, but the growing threat of insecticide resistance prompts the urgent need to develop alternative, ecofriendly, and sustainable vector control tools. Entomopathogenic fungi can overcome insecticide resistance and represent promising biocontrol tools for the control of mosquitoes. However, insects have evolved robust defense mechanisms against infection. Better understanding of mosquito defenses against fungal infection is critical for improvement of fungal efficacy. Here, we show that as the pathogenic fungus Beauveria bassiana penetrates into the host hemocoel, mosquitoes increase expression of the let-7 and miR-100 microRNAs (miRNAs). Both miRNAs translocate into fungal hyphae to specifically silence the virulence-related genes sec2p and C6TF, encoding a Rab guanine nucleotide exchange factor and a Zn(II)2Cys6 transcription factor, respectively. Inversely, expression of a let-7 sponge (anti–let-7) or a miR-100 sponge (anti–miR-100) in the fungus efficiently sequesters the corresponding translocated host miRNA. Notably, B. bassiana strains expressing anti–let-7 and anti–miR-100 are markedly more virulent to mosquitoes. Our findings reveal an insect defense strategy that employs miRNAs to induce cross-kingdom silencing of pathogen virulence-related genes, conferring resistance to infection.
Insects are the most diverse and widespread animals on earth. Mosquitoes are vectors of numerous deadly infectious diseases such as malaria, Zika, dengue, chikungunya, and yellow fever (1). Vector control using chemical insecticides remains the most effective weapon for the control of mosquito-borne diseases (2). However, progress in disease control is under threat by widespread and escalating mosquito insecticide resistance (3–5), emphasizing the urgent need to develop alternative, eco-friendly, and sustainable vector control tools that can overcome insecticide resistance (6).
Entomopathogenic fungi are the most common insect disease-causing organisms. The fungi infect insect hosts by penetrating their exoskeleton and are especially suitable for controlling sap-sucking pests and blood-sucking mosquitoes (7, 8). The insecticidal fungi, particularly those belonging to the genera Beauveria and Metarhizium, are equally effective at killing insecticide-resistant and insecticide-susceptible mosquitoes and are considered to be the next-generation mosquito control agents (9–13). However, fungal insecticides act slower than chemical insecticides (14), which restricts their widespread use (15), and insects have evolved a variety of defense mechanisms that reduce infection (16). Improvement of fungal efficacy requires better understanding of fungal-induced host immune defenses.
Fungal infection is initiated by the attachment of spores to the insect cuticle, followed by germination, cuticle penetration, and development within the host’s hemocoel (17). Upon fungal infection, insects can sense and activate both cellular and humoral immune responses (16). These include phagocytosis, encapsulation, nodulation, melanization, and expression of antimicrobial peptides. However, successful pathogens have developed means to overcome host immunity (17–19). This circular battle drives an evolutionary arms race between fungal pathogens and insect hosts.
MicroRNAs (miRNAs) are short regulatory noncoding small RNAs (sRNAs) that regulate expression of target genes at transcriptional and posttranscriptional levels (20), contributing to various biological processes such as development, immunity, and host–microorganism interactions (21–24). Recently, we found that during infection, Beauveria bassiana (B. bassiana) transports a miRNA-like RNA (bba-milR1) into mosquito cells, silencing host defense genes (25). Conversely, recent studies have shown that host-encoded sRNAs can also be transferred to pathogens. Sickle cell erythrocytes can deliver miRNAs into the malaria parasite Plasmodium falciparum to inhibit parasite translation and growth (26). miRNAs secreted by human and mouse intestinal epithelial cells can enter gut bacteria to influence their gene expression (27). In addition, plants also can transport their sRNAs into fungal pathogens to suppress infection (22, 28). However, to our knowledge, transkingdom miRNAs transfer from arthropod hosts to their pathogen cells has not been described.
Here, we report that, after infection with the insect pathogenic fungus B. bassiana, host insects produce miRNAs that translocate into the fungal cells and silence virulence-related genes to reduce fungal pathogenicity. These findings suggest that insects have developed a cross-kingdom RNA interference (RNAi)-mediated defense strategy in an evolutionary arms race to combat fungal pathogens.
Results
Insect miRNAs Are Induced in Response to Fungal Infection and Translocated into Fungal Hyphae.
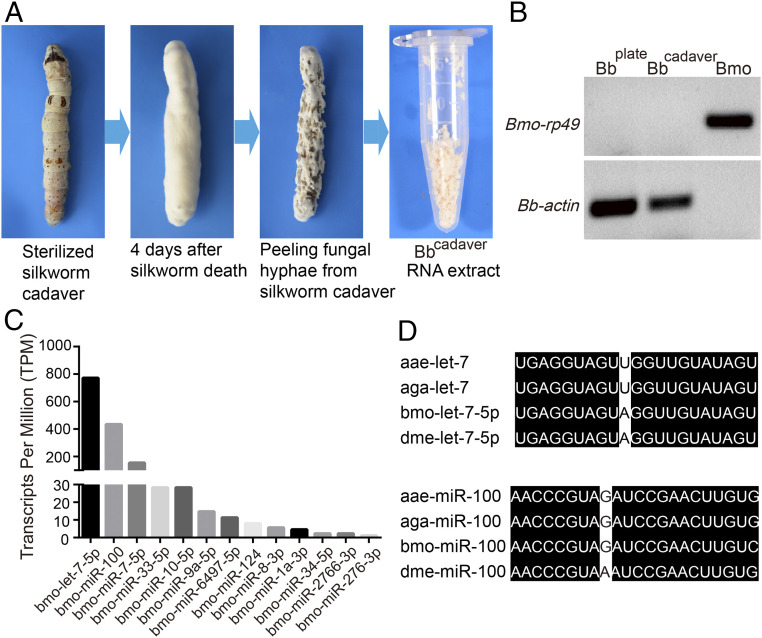
To investigate possible roles of insect miRNAs in defense against fungal infection, we searched for host miRNAs in infecting fungal hyphae. Silkworm (Bombyx mori) larvae were infected with B. bassiana ARSEF252 and fungal mycelia were recovered from the surface of the infected larva cadavers (designated as Bbcadaver) (Fig. 1A). Silkworm larvae have a large body size that facilitates recovery of mycelia from the cadaver surface while avoiding contamination with host tissues. Total Bbcadaver sRNAs were deep-sequenced and the abundant B. mori rp49 transcript was not detected, indicating that fungal RNA was not contaminated by host RNAs (Fig. 1B). We identified 13 small RNAs whose sequences cannot be mapped to the B. bassiana genome and whose sequences are identical to the host insect miRNAs (Fig. 1C). These findings suggest that host insect miRNAs are transferred into fungal hyphae. Among them, let-7 and miR-100 are the most abundant and were selected for further characterization (Fig. 1C). Both, let-7 and miR-100, are highly conserved across insects, including the silkworm B. mori, mosquitoes (Anopheles, Aedes), and Drosophila melanogaster (Fig. 1D). Moreover, RT-PCR analysis confirmed that let-7 and miR-100 are present in fungal hyphae recovered from infected silkworm cadavers and Anopheles stephensi cadavers (Bbcadaver), but not in cultured B. bassiana hyphae (Bbplate) (Fig. 2A and SI Appendix, Fig. S1). This finding prompted us to use Anopheles mosquitoes as the host insect to investigate the role of let-7 and miR-100 in mosquito–fungal interactions.
Fig. 1.
Deep sequencing identification of host miRNAs in fungal hyphae recovered from infected insect cadavers. (A) Schematic representation of the study design. Silkworms killed by B. bassiana were surface sterilized with 1% bleach and placed in petri dishes containing wet filter papers to facilitate fungal emergence. Four days later, fungal hyphae grown out of the Bbcadavers were recovered carefully for RNA isolation. (B) RT-PCR detection confirmed that fungal mycelia recovered from Bbcadavers were not contaminated with silkworm tissues. B. mori ribosomal protein 49 gene (Bmo-rp49) was used to detect the B. mori nucleic acids. B. bassiana actin (Bb-actin) was used to detect the B. bassiana nucleic acid. Bbplate, fungal mycelia grown on plates; Bmo, noninfected B. mori silkworm. (C) Host miRNAs identified in fungal hyphae recovered from silkworm cadavers. (D) Mature let-7 and miR-100 sequences are highly conserved across insect species. aae, Aedes aegypti; aga, Anopheles gambiae; bmo, B. mori; and dme, Drosophila melanogaster. All miRNA sequences were obtained from the miRNA database.
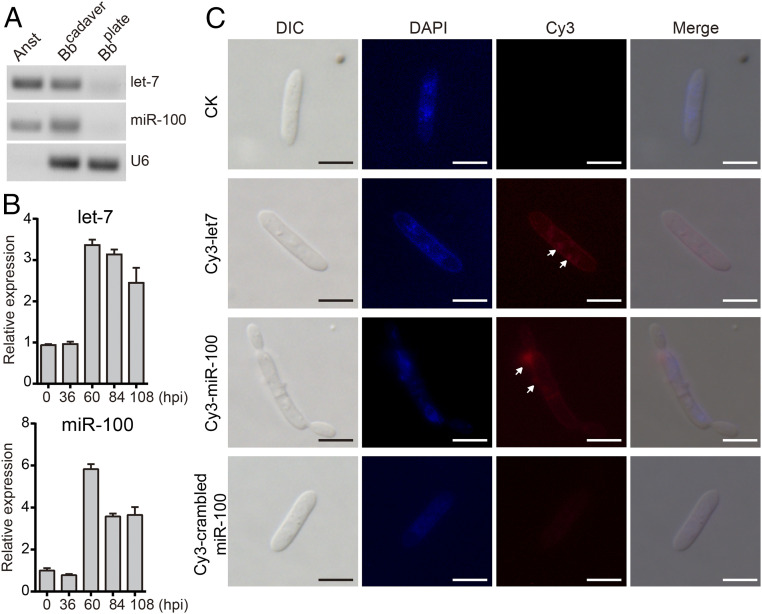
Fig. 2.
Host miRNAs are induced in response to fungal infection and are transferred into fungal hyphae. (A) Insect miRNAs let-7 and miR-100 are detected in fungal hyphae recovered from infected An. stephensi mosquito cadavers (Bbcadaver) but not in cultured B. bassiana hyphae (Bbplate). The uninfected mosquito (Anst) was used as a positive control for host miRNA detection. The B. bassiana snRNA U6 was used for fungal detection. (B) qRT-PCR analysis of let-7 and miR-100 expression in mosquitoes at 36, 60, 84, and 108 h postinfection (hpi) with B. bassiana. RNA was extracted from fungus-infected mosquitoes and mosquito ribosomal protein S7 mRNA was used as an internal reference. The expression values are normalized to uninfected mosquitoes (time 0). Data represent three biological repeats, each with three technical replicates. Error bars indicate SEM (n = 3). (C) let-7 and miR-100 localization in B. bassiana hyphal bodies. At 60 h after topical infection with B. bassiana (108 conidia/mL), let-7, miR-100 and scrambled miR-100 control RNAs (all Cy3-labeled) and phosphate-buffered saline negative control (CK) were injected into the hemocoel of An. stephensi female mosquitoes. 24 h later, the resulting hyphal bodies were recovered from the hemolymph, washed with PBS, fixed in 4% paraformaldehyde, and stained with 4′,6-diamidino-2-phenylindole (DAPI). DIC, differential interference contrast microscopy; blue, DAPI; red, Cy3. (Scale bars, 10 μm.)
To examine host miRNA expression profile during fungal infection, we extracted RNA from B. bassiana-infected An. stephensi mosquitoes at 0, 36, 60, 84, and 108 h post-fungal topical infection (hpi) and quantified let-7 and miR-100 expression by quantitative RT-PCR (qRT-PCR). The expression of both let-7 and miR-100 were markedly induced by the fungus, starting at 60 hpi (Fig. 2B). Similar results were obtained for the fat body of B. mori (SI Appendix, Fig. S2). These results indicate that host insects increase the production of let-7 and miR-100, and both miRNAs are transferred to fungal hyphae after B. bassiana infection.
Next, we investigated whether externally provided miRNAs are able to translocate into the fungus. We injected fluorescently labeled let-7, miR-100, or scrambled miR-100 control sRNAs into the hemocoel of adult An. stephensi mosquitoes at 60 h post-topical infection with B. bassiana and examined fungal hyphal bodies by fluorescence microscopy. We found that Cy3-labeled let-7 and miR-100 bba-milR1 were loaded in vesicles visible either on the surface or in the cytoplasm of the fungal hyphal bodies (Fig. 2C). These results suggest that the fungus may import host miRNAs from host insect hemolymph.
Host let-7 and miR-100 miRNAs Confer Resistance to Fungal Infection.
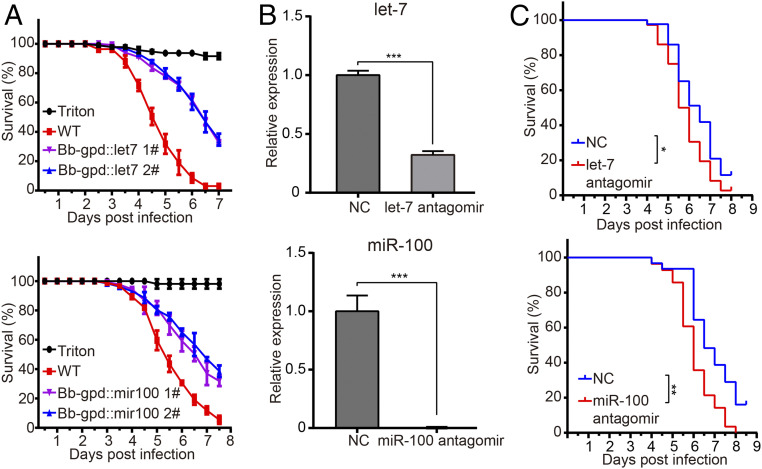
To test whether host insect miRNAs confer resistance to the fungal pathogen, we generated transgenic B. bassiana strains that express the host let-7 or miR-100 (SI Appendix, Fig. S3A). Expression of both host miRNAs in transgenic B. bassiana strains was validated by qRT-PCR (SI Appendix, Fig. S3B). No obvious growth defects were observed in the transgenic strains (SI Appendix, Fig. S3C). Importantly, virulence of the fungal strains expressing either let-7 or miR-100 against An. stephensi mosquitoes was largely compromised (Fig. 3A). Similarly, the transgenic B. bassiana strains expressing host miRNAs let-7 or miR-100 exhibited decreased virulence to B. mori larvae (SI Appendix, Fig. S3D). To further confirm the role of let-7 and miR-100 in host defense against fungal infection, we silenced let-7 and miR-100 expression in mosquitoes by systemic injection of let-7 or miR-100 antagomirs followed by B. bassiana infection. Antagomirs are microRNA inhibitors whose sequences are complementary to the mature miRNAs. Antagomir injection markedly decreased let-7 or miR-100 levels (Fig. 3B) and rendered mosquitoes more susceptible to B. bassiana infection (Fig. 3C). Taken together, these results indicate that the host miRNAs let-7 and miR-100 confer resistance to fungal infection.
Fig. 3.
The effect of host miRNAs on fungal virulence. (A) Survival of adult female An. stephensi mosquitoes following topical application of a suspension of 107 conidia/mL of the WT and transgenic B. bassiana strains, as indicated. Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as a negative control (Triton). Expression of let-7 and miR-100 significantly reduced fungal virulence to mosquitoes compared with WT (P < 0.001, log-rank test). The experiments were repeated three times with similar results. Error bars indicate the SEM of three technical repeats. (B) Silencing efficiency of let-7 and miR-100 in mosquitoes injected with antagomir-let-7 or antagomir-miR-100 was measured by qRT-PCR. The mosquito ribosomal protein S7 mRNA was used as an internal reference. Error bars indicate the SEM (n = 3). ***P < 0.001 (Student’s t test). (C) Effects of let-7 antagomir (200 nM) or miR-100 antagomir (200 nM) injection on survival of mosquitoes following topical application of a suspension of B. bassiana (107 conidia/mL). Mosquitoes injected with negative control antagomir (NC) served as controls. The experiments were repeated three times with similar results. *P < 0.05 and **P < 0.01. P value < 0.05 was regarded as statistically significant (log-rank test).
Host miRNAs Target Fungal Genes sec2p and C6TF.
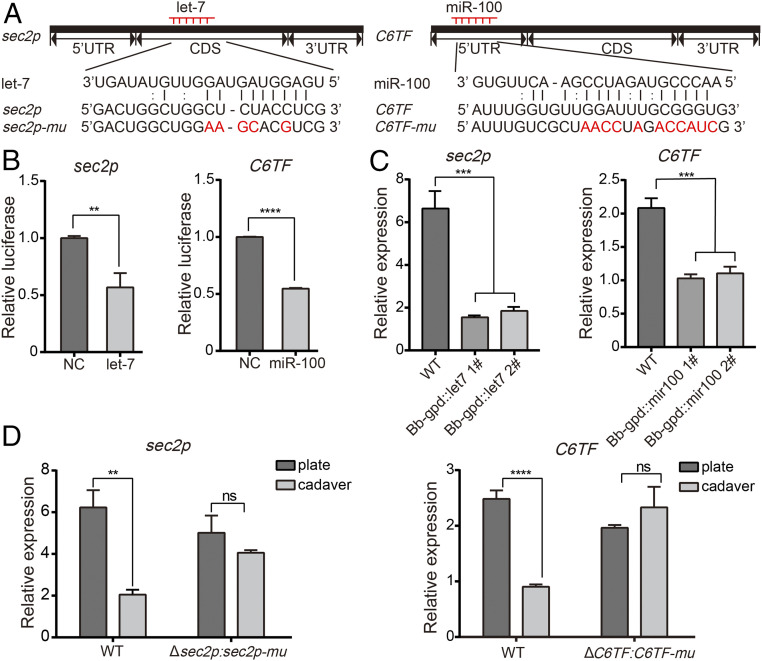
We used three stringent miRNA target prediction softwares (miRanda, microTar, and Probability of Interaction by Target Accessibility) to predict potential targets of let-7 or miR-100 in the 3′ untranslated region (UTR), 5′ UTR and coding sequences of B. bassiana transcriptome. We found that let-7 and miR-100 could base pair with some genes involved in pathogenicity (29) (Fig. 4A and SI Appendix, Tables S1 and S2). To validate the predicted interactions, we first conduced target verification assay ex vivo using luciferase reporter assays. DNA fragments surrounding predicted target sites were PCR-amplified and separately cloned into the plasmid of siRNA silencing (psiCHECK-2, Premega) downstream of the Renilla luciferase stop codon. Dual luciferase reporter assays revealed that let-7 significantly silences the gene encoding a Rab guanine nucleotide exchange factor sec2p (BBA_07763), and miR-100 suppresses a gene encoding the fungal-specific transcription factor belonging to the Zn(II)2Cys6 family (30) (BBA_03208, designated C6TF) (Fig. 4B). We also performed in vivo assays to validate the fungal genes targeted by host miRNAs. As expected, the transcript levels of sec2p and C6TF were markedly suppressed in the transgenic strains expressing let-7 (Bb-gpd::let7) and miR-100 (Bb-gpd::mir100), respectively (Fig. 4C). To further examine whether the fungal gene expression could be specifically affected by the transferred host miRNAs, we generated miRNA-resistant complemented strains Δsec2p:sec2p-mu and ΔC6TF:C6TF-mu, carrying a let-7–resistant form of sec2p (sec2p-mu) or a miR-100–resistant form of C6TF (C6TF-mu) mutated at the miRNA binding sites, respectively (Fig. 4A). As predicted, the transcript levels of sec2p and C6TF were markedly decreased in fungal hyphae recovered from the wild type (WT)-infected cadavers but not suppressed in hyphae recovered from the miRNA-resistant complemented strain-infected cadavers, compared with their hyphae grown on plates (Fig. 4D). These results suggest that the fungal genes sec2p and C6TF are targeted by the host miRNAs let-7 and miR-100, respectively. Ex vivo and in vivo assays for other predicted target genes were not conclusive (SI Appendix, Figs. S4 and S5).
Fig. 4.
Host miRNAs target fungal genes sec2p and C6TF. (A) Sequence alignments of let-7 and miR-100 with fungal target genes (sec2p and C6TF) and the miRNA-resistant mutated version at the predicted binding sites (sec2p-mu and C6TF-mu). Mutated nucleotides are in red. (B) let-7 and miR-100 suppress the expression of sec2p and C6TF, respectively. The interactions between miRNAs (let-7 and miR-100) and the target sites of sec2p and C6TF were determined by dual luciferase reporter assays in HEK293T cells that were cotransfected with let-7 or miR-100 overexpression vectors and psiCHECK-2 vectors containing target sequences of sec2p or C6TF. Cotransfection of empty vector (NC) with psiCHECK-2 vectors containing target gene sequences was used as controls. (C) Expression of target genes sec2p and C6TF is suppressed in transgenic B. bassiana strains overexpressing let-7 (Bb-gpd::let7) or miR-100 (Bb-gpd::mir100). Gene expression was quantified by qRT-PCR. B. bassiana actin was used as an internal reference. The experiments were repeated three times with similar results. (D) qRT-PCR analysis of target genes sec2p and C6TF expression in fungal mycelia recovered from silkworm cadavers infected with B. bassiana WT or with miRNA-resistant complemented strains (Δsec2p:sec2p-mu and ΔC6TF:C6TF-mu) and their cultured hyphae on plates. The experiments were repeated three times with similar results. B. bassiana actin was used as an internal reference. Values are mean ± SEM (n = 3). **P < 0.01 and ***P < 0.001; ****P < 0.0001; ns, not significant (Student’s t test).
Fungal Genes sec2p and C6TF Are Required for Pathogenicity.
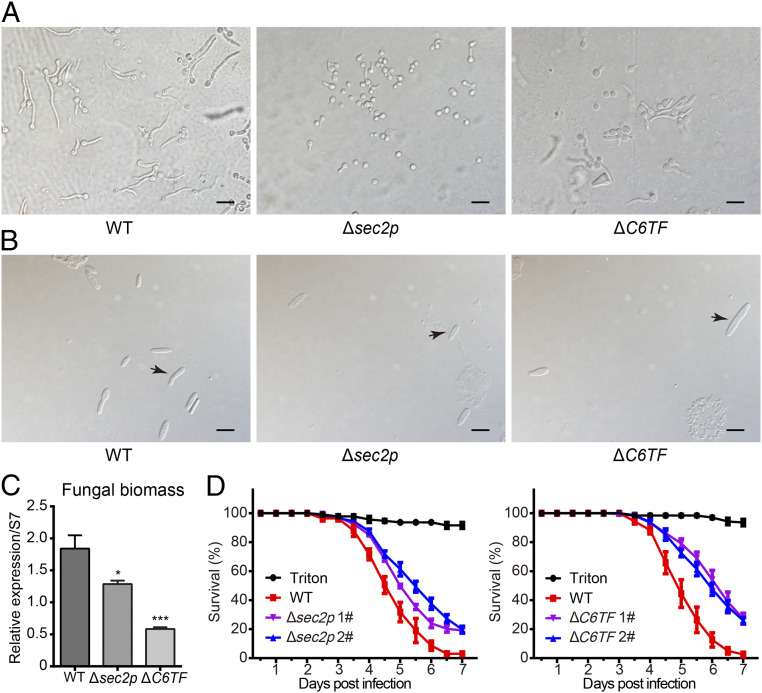
To investigate the biological functions of sec2p and C6TF in B. bassiana, we generated gene deletion mutants by homologous recombination (SI Appendix, Fig. S6 A and B) and analyzed fungal growth and pathogenicity. Deletion of the sec2p resulted in a significant reduction in vegetative growth on Sabouraud dextrose agar plus yeast extract (SDAY) compared with the WT strain (SI Appendix, Fig. S6C). Moreover, the germination rate of the sec2p and C6TF deletion mutants on cicada hindwings was markedly reduced (Fig. 5A). Furthermore, the formation of hyphal bodies in the insect hemocoel, a proxy for fungal invasion efficiency, was significantly decreased at 60 h after topical infection with Δsec2p or ΔC6TF compared to WT (Fig. 5B). The fungal biomass in mosquitoes infected with either Δsec2p or ΔC6TF was lower than that of WT (Fig. 5C). Pathogenicity assays revealed that deletion of sec2p or C6TF markedly reduced fungal virulence in both An. stephensi mosquitoes (Fig. 5D) and B. mori larvae (SI Appendix, Fig. S7) compared with WT. These results indicate that the fungal genes sec2p and C6TF are involved in fungal invasive growth and pathogenicity to insects. To further verify the contribution of sec2p and C6TF to fungal virulence, we performed bioassays with the miRNA-resistant complemented strains. The defects in growth phenotype in Δsec2p were restored in the let-7–resistant and sec2p-complemented strain Δsec2p:sec2p-mu (SI Appendix, Fig. S6C). The miR-100–resistant and C6TF-complemented strain ΔC6TF:C6TF-mu also showed growth phenotype similar to WT (SI Appendix, Fig. S6C). Importantly, the miRNA-resistant complemented strains Δsec2p:sec2p-mu and ΔC6TF:C6TF-mu exhibited stronger virulence to mosquitoes than WT (SI Appendix, Fig. S8). These results show that both sec2p and C6TF are involved in fungal pathogenicity.
Fig. 5.
sec2p and C6TF are required for invasive growth and fungal pathogenicity. (A) Conidial germination of WT, sec2p, and C6TF deletion mutants (Δsec2p and ΔC6TF) at 12 h postincubation on cicada hind wings. (Scale bars, 10 μm.) (B) Microscopic observation of fungal hyphal body formation in the silkworm hemocoel at 60 h after topical infection with 5 × 106 conidia/mL of the WT, Δsec2p, and ΔC6TF strains. Arrows point to hyphal bodies. (Scale bars, 10 μm.) (C) qPCR-based quantification of fungal load in mosquitoes infected with WT or mutants at 60 hpi. Fungal mass was estimated by measuring fungal 18S rRNA abundance relative to that of An. stephensi ribosomal protein S7 (AsS7) DNA. Values are mean ± SEM (n = 3). *P < 0.05 and ***P < 0.001. P value < 0.05 is regarded as statistically significant (Student’s t test). (D) Survival of adult female mosquitoes infected with WT and mutants (Δsec2p and ΔC6TF), following topical application of a spore suspension (107 conidia/mL). Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as a negative control (Triton). Δsec2p and ΔC6TF mutants exhibited reduced virulence compared to WT (P < 0.01, log-rank test). The experiments were repeated three times with similar results. Error bars indicate the SEM of three technical repeats.
Expression of Anti–let-7 or Anti–miR-100 Enhances Fungal Virulence.
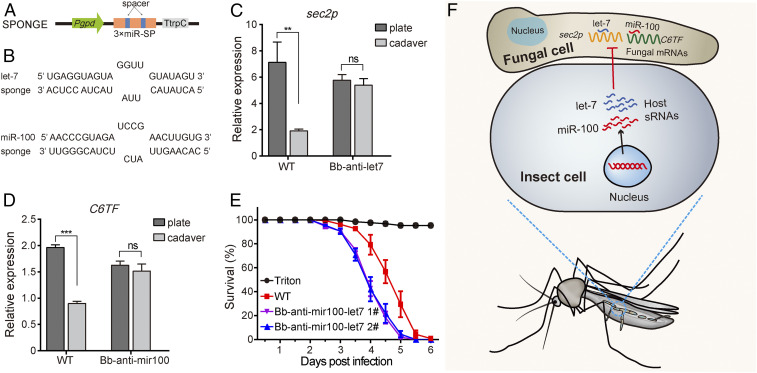
miRNA sponges have been used to inhibit the function of endogenous miRNA (31). To sequester the transferred host miRNAs let-7 or miR-100, we generated transgenic B. bassiana strains expressing let-7 sponge or miR-100 sponge (Bb-anti–let7 and Bb-anti–mir100) for inhibition of host let-7 or miR-100, respectively (Fig. 6 A and B). We found that the transcript abundance of sec2p and C6TF was significantly decreased in fungal mycelia recovered from insect cadavers infected with WT fungus, but not influenced in the mycelia recovered from the cadavers infected with transgenic strains expressing let-7 sponge or miR-100 sponge, compared to mycelia grown on plates (Fig. 6 C and D). These results indicate that transcriptional reduction of fungal genes is induced by the transferred host miRNAs, and that expression of miRNA sponges in the fungus inhibits this effect. To enhance fungal virulence, we generated Bb-anti–mir100-let7 strain that expresses both let-7 and miR-100 sponges and found that this strain exhibits drastically enhanced virulence to mosquitoes (Fig. 6E).
Fig. 6.
Expression of anti–let-7 and anti–miR-100 enhances fungal virulence against mosquitoes. (A) Schematic diagram of the miRNA sponge expression cassette. Pgpd, Aspergillus nidulans constitutively active gpdA promoter; TrpC, A. nidulans trpC terminator. (B) Sequence alignments of let-7 and miR-100 with sponge sequences. let-7 sponge and miR-100 sponge sequences separately target the seed region of let-7 and miR-100, and base pairs with each miRNA mature sequence imperfectly with central bulges. (C and D) Expression of sec2p (C) and C6TF (D) is suppressed in fungal hyphae recovered from WT-infected silkworm cadavers but not suppressed in the hyphae recovered from the cadavers infected with strains that carried anti–let-7 sponge or anti–miR-100 sponge. Gene expression was quantified by qRT-PCR. The experiments were repeated three times with similar results. B. bassiana actin was used as an internal reference. Values are mean ± SEM (n = 3). **P < 0.01 and ***P < 0.001; ns, not significant (Student’s t test). (E) Survival of adult female mosquitoes infected with WT and Bb-anti–mir100-let7 strain following topical application of a spore suspension (5 × 107 conidia/mL). Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as a negative control (Triton). Bb-anti–mir100-let7 strain exhibited stronger virulence compared to WT (P < 0.0001, log-rank test). Error bars indicate the SEM of three technical repeats. (F) Model of host insect employing miRNAs to inhibit virulence gene expression in an entomopathogenic fungus. Upon infection with the B. bassiana pathogen, mosquitoes increase let-7 and miR-100 expression, and both miRNAs translocate into fungal cells, silencing the fungal virulence genes sec2p and C6TF and causing decrease of fungal pathogenicity.
Discussion
B. bassiana has received considerable attention as a promising environmentally friendly alternative to chemical insecticides for the control of mosquitoes and agricultural pests. In addition to insecticidal activity, fungal infection leads to sublethal effects, including reduction of mosquito fecundity, host location ability, and feeding propensity, which further reduces vector competence (9, 32). To combat fungal infection, insects have evolved potent defense strategies. Here, we report that insects have evolved an unusual miRNA-based antifungal host defense by employing miRNAs to silence fungal genes involved in pathogenicity, revealing a new cross-kingdom defense strategy (Fig. 6F).
Insects employ a range of defense mechanisms to combat microbial infection. As insecticidal fungi penetrate the integument, insects can sense and activate innate immune responses via recognition of pathogen-associated molecular patterns and pattern recognition receptors. This results in the activation of prophenoloxidase-mediated melanization and production of antifungal peptides via the Toll immune signaling pathway (16, 33). As a counter response, fungal pathogens have also evolved the ability to suppress or avoid insect’s immune responses (18). We recently discovered that B. bassiana deploys a miRNA-like RNA (bba-milR1) to mosquito cells during fungal penetration of the mosquito integument, which induces cross-kingdom RNAi to silence the host’s Toll receptor Spätzle 4 gene (25).
Once B. bassiana invades the insect hemocoel, the invading fungus switches from filamentous hyphal growth to yeast-like hyphal bodies that possess fewer carbohydrate epitopes, helping fungi to avoid detection by host hemocytes (17). How insects defend against fungal infection during the later stages of infection in the hemocoel is largely unknown. Successful launch of host immune defenses requires rapid and fine-tuning regulation of gene expression in response to infection (34). miRNAs are key regulators of gene expression reprograming and play an important role in insect immune responses, pathogen virulence, and communication between hosts and pathogens (25, 35). In this study, we found that as B. bassiana penetrates the hemocoel, insects increase production of the let-7 and miR-100 miRNAs and transfer both miRNAs into the fungal hyphae for specific silencing. In many insects, let-7 and miR-100 are cotranscribed in an evolutionarily conserved locus, and play roles in regulating development, metabolism, longevity, or innate immunity (36–38). The present study demonstrates that the translocated host miRNAs let-7 and miR-100 selectively suppress the fungal genes sec2p and C6TF, respectively, both of which are essential for fungal virulence. These findings provide an example of miRNA trafficking from arthropod hosts to pathogen cells leading to cross-kingdom regulatory interactions.
Small RNA trafficking between interacting organisms has previously been observed (22–27), but mechanisms by which sRNAs are transferred from hosts to interacting pathogens remains largely unknown. A recent study reported extracellular vesicles (EVs) as vehicles of cross-kingdom sRNA trafficking between the Arabidopsis plant and the fungal pathogen Botrytis cinerea (28). In mammals, EVs were shown to play a role in systemic sRNA transport among cells within the organism (39). In this study, we found that Cy3-labeled let-7 and miR-100 are loaded into vesicles visible either on the surface or in the cytoplasm of the fungal hyphal bodies (Fig. 2C), suggesting that EVs may also be used by insects to transfer miRNAs to the fungal pathogen.
Sec2p promotes exocytosis (40) and is required for pathogenicity in Fusarium graminearum (41). Our study showed that deletion of sec2p in B. bassiana causes significant reduction in both vegetative and invasive growth, as well as fungal pathogenicity. Deletion of C6TF also markedly reduced B. bassiana virulence and invasive growth. In the rice blast pathogen Magnaporthe oryzae, the C6TF ortholog MoCOD1 is also essential for fungal virulence by modulating appressorium formation and invasive growth (42). Taken together, this study uncovers an insect defense mechanism whereby infection leads to the up-regulation of specific miRNAs that are transferred to the invading fungus and suppress fungal virulence genes, in this way, conferring antifungal resistance. This study opens avenues to improve fungal virulence by expression of host miRNA sponges.
Materials and Methods
Insect Rearing and Fungal Culture.
An. stephensi (Dutch strain) mosquitoes were maintained as previously described (25). The larvae of silkworm, B. mori L. (race: Nistari), were reared on fresh mulberry leaves at 25 °C. The fungus B. bassiana was cultivated in SDAY at 26 °C ± 1 °C. Experimental details can be found in SI Appendix, Materials and Methods.
Fungal Infection.
To conduct fungal infection, adult female An. stephensi were sprayed with fungal conidial suspension (1 × 107−5 × 107 conidia/mL). Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as a negative control. Silkworm larvae were inoculated by topical immersion in fungal spore suspension (5 × 106 conidia/mL) or 0.01% Triton X-100 (control) for 30 s. Experimental details can be found in SI Appendix, Materials and Methods.
sRNA Deep Sequencing.
B. bassiana mycelia harvested from the silkworm larva cadavers were used for total RNA extraction. sRNA reads that did not map to the B. bassiana genome were aligned to the silkworm B. mori genome. miRNAs were identified by matching the reads to the miRNA database (www.mirbase.org). Experimental details can be found in SI Appendix, Materials and Methods.
Detection of miRNA Translocation into Fungal Cell.
To examine the transport and localization of the host miRNA in fungal cells in vivo, we injected the Cy3-conjugated let-7, miR-100, or scrambled miR-100 control (50 μM) into the hemocoel of adult An. stephensi mosquitoes at 60 h post-topical infection with B. bassiana (108 conidia/mL). The resulting hyphal bodies were recovered from the hemolymph after 24 h. Fluorescence signals were visualized by fluorescence microscopy. Experimental details can be found in SI Appendix, Materials and Methods.
qRT-PCR.
To quantify gene expression, total RNA was reverse transcribed to synthesize complementary DNA (cDNA) using a TransScript All-in-One First-Strand cDNA Synthesis SuperMix kit (TransGen). qRT-PCR was performed using Synergy Brands (SYBR) dye technology. To synthesize insect or B. bassiana miRNA cDNAs, 1 μg of total RNA was reverse transcribed using miRcute miRNA First-Strand cDNA Synthesis kit (Tiangen), and qRT-PCR was performed using miRcute miRNA qPCR detection kit (Tiangen) or the AceQ qPCR SYBR Green MasterMix kit (Vazyme). Experimental details can be found in SI Appendix, Materials and Methods.
Dual Luciferase Reporter Assays.
B. mori let-7 and miR-100 precursors were separately cloned into the pmR-mCherry vector (Promega). The ∼400 bp sequences of the predicted let-7 and miR-100 target sites in sec2p and C6TF were separately cloned into the psiCHECK-2 vector (Promega). HEK293T cells were cotransfected with psiCHECK-2-target reporter vector and the miRNA expression vector or empty vector. Experimental details can be found in SI Appendix, Materials and Methods. Primers are shown in SI Appendix, Table S3.
Expression of let-7 or miR-100 in B. bassiana.
To ectopically express let-7 or miR-100 in B. bassiana, the fragments surrounding primary let-7 and primary miR-100 were PCR-amplified from insect genomic DNA and then separately cloned into the binary vector pBarGFP-PgpdA. The resulting miRNA expression vectors were separately transformed into B. bassiana ARSEF252 by Agrobacterium tumefaciens-mediated transformation method. Experimental details can be found in SI Appendix, Materials and Methods. Primers are shown in SI Appendix, Table S3.
Generation of Target Gene Deletion Mutants and miRNA-Resistant Mutated Strains.
The target genes were disrupted in B. bassiana by homologous recombination using A. tumefaciens-mediated transformation. To generate let-7 and miR-100 miRNA-resistant mutated strains, we reverted disruptants Δsec2p and ΔC6TF with miRNA-resistant target site mutated gene sec2p-mu and C6TF-mu that carry the mutated nucleotide sequences in miRNA binding site without changing the amino acid sequences, respectively. Experimental details can be found in SI Appendix, Materials and Methods. Primers are shown in SI Appendix, Table S3.
Generation of B. bassiana Strains Expressing miRNA Sponges.
To sequester the translocated host miRNAs and inhibit their activity in the fungus, we generated Bb-anti–let7, Bb-anti–mir100, and Bb-anti–mir100-let7 strains that express specific miRNA sponge(s). Experimental details can be found in SI Appendix, Materials and Methods.
Determination of Fungal Invasive Growth and Hyphal Body Formation.
The cicada hind wings were used to test fungal invasive growth. To monitor hyphal body differentiation in the insect hemocoel, the hemolymph was collected from the fifth instar larvae of B. mori 60 h post-topical infection (5 × 106 conidia/mL). Hyphal bodies were counted with a hemocytometer under the compound microscope. Experimental details can be found in SI Appendix, Materials and Methods.
miRNA Antagomir Injection.
The miRNA antogomir was used to down-regulate the corresponding endogenous miRNA levels. The let-7 antagomir (200 nM) or miR-100 antagomir (200 nM) was microinjected into the thorax of 3-d-old female mosquitoes. Control mosquitoes were injected with negative control antagomir (5′-CAGUACUUUUGUGUAGUACAA-3′) (200 nM). Mosquitoes were allowed to recover for 3 d before conducting fungal infection. Experimental details can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Professor Marcelo Jacobs-Lorena at Johns Hopkins University School of Public Health for comments and proofreading the manuscript. This study was supported by grants from the National Key R&D Program of China (2018YFA0900502, 2020YFC1200100, and 2017YFD0200400), the National Natural Science Foundation of China (grants 31772534, 32021001, 31830086, and 31501703), and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB11010500).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023802118/-/DCSupplemental.
Data Availability
The sRNA sequencing datasets have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession number PRJNA647702. All other study data are included in the article and/or supporting information.
References
- 1.Walker T., Jeffries C. L., Mansfield K. L., Johnson N., Mosquito cell lines: History, isolation, availability and application to assess the threat of arboviral transmission in the United Kingdom. Parasit. Vectors 7, 382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanda E., Ameneshewa B., Bagayoko M., Govere J. M., Macdonald M. B., Harnessing integrated vector management for enhanced disease prevention. Trends Parasitol. 33, 30–41 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Ingham V. A., et al., A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577, 376–380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranson H., Lissenden N., Insecticide resistance in African Anopheles mosquitoes: A worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 32, 187–196 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Moyes C. L., et al., Evaluating insecticide resistance across African districts to aid malaria control decisions. Proc. Natl. Acad. Sci. U.S.A. 117, 22042–22050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai Y., et al., Coordinated regulation of infection-related morphogenesis by the KMT2-Cre1-Hyd4 regulatory pathway to facilitate fungal infection. Sci. Adv. 6, eaaz1659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M. B., Read A. F., Can fungal biopesticides control malaria? Nat. Rev. Microbiol. 5, 377–383 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Peng T. L., Syazwan S. A., Lee S. H., “Soil-borne Entomopathogenic Bacteria and Fungi” in Microbes for Sustainable Insect Pest Management: An Eco-Friendly Approach, Khan M. A., Ahmad W., Eds. (Springer International Publishing, Cham, 2019), vol. 1, pp. 23–41. [Google Scholar]
- 9.Blanford S., et al., Fungal pathogen reduces potential for malaria transmission. Science 308, 1638–1641 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Scholte E. J., et al., An entomopathogenic fungus for control of adult African malaria mosquitoes. Science 308, 1641–1642 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Lovett B., et al., Transgenic Metarhizium rapidly kills mosquitoes in a malaria-endemic region of Burkina Faso. Science 364, 894–897 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Knols B. G., Bukhari T., Farenhorst M., Entomopathogenic fungi as the next-generation control agents against malaria mosquitoes. Future Microbiol. 5, 339–341 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Farenhorst M., et al., Fungal infection counters insecticide resistance in African malaria mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 106, 17443–17447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei G., et al., Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. U.S.A. 114, 5994–5999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimopoulos G., Insect immunity and its implication in mosquito-malaria interactions. Cell. Microbiol. 5, 3–14 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Wang C., Wang S., Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 62, 73–90 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Qu S., Wang S., Interaction of entomopathogenic fungi with the host immune system. Dev. Comp. Immunol. 83, 96–103 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Feng P., Shang Y., Cen K., Wang C., Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. U.S.A. 112, 11365–11370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L., et al., Importance of miRNA stability and alternative primary miRNA isoforms in gene regulation during Drosophila development. eLife 7, e38389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiberg A., et al., Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T., et al., Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Mayoral J. G., et al., Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc. Natl. Acad. Sci. U.S.A. 111, 18721–18726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asgari S., Regulatory role of cellular and viral microRNAs in insect-virus interactions. Curr. Opin. Insect Sci. 8, 104–110 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cui C., et al., A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat. Commun. 10, 4298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaMonte G., et al., Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12, 187–199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S., et al., The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Q., et al., Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valero-Jiménez C. A., Wiegers H., Zwaan B. J., Koenraadt C. J., van Kan J. A., Genes involved in virulence of the entomopathogenic fungus Beauveria bassiana. J. Invertebr. Pathol. 133, 41–49 (2016). [DOI] [PubMed] [Google Scholar]
- 30.MacPherson S., Larochelle M., Turcotte B., A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong S., et al., Broad spectrum immunomodulatory effects of Anopheles gambiae microRNAs and their use for transgenic suppression of Plasmodium. PLoS Pathog. 16, e1008453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanford S., Jenkins N. E., Read A. F., Thomas M. B., Evaluating the lethal and pre-lethal effects of a range of fungi against adult Anopheles stephensi mosquitoes. Malar. J. 11, 365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottar M., et al., Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127, 1425–1437 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M. Y., Lee J. E., Kim L. K., Kim T., Epigenetic memory in gene regulation and immune response. BMB Rep. 52, 127–132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussain M., Asgari S., MicroRNAs as mediators of insect host-pathogen interactions and immunity. J. Insect Physiol. 70, 151–158 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Fullaondo A., Lee S. Y., Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 36, 267–273 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Rubio M., Belles X., Subtle roles of microRNAs let-7, miR-100 and miR-125 on wing morphogenesis in hemimetabolan metamorphosis. J. Insect Physiol. 59, 1089–1094 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Chawla G., Sokol N. S., Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development 139, 1788–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colombo M., Raposo G., Théry C., Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Medkova M., France Y. E., Coleman J., Novick P., The rab exchange factor Sec2p reversibly associates with the exocyst. Mol. Biol. Cell 17, 2757–2769 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H., et al., FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 20, 3378–3392 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Chung H., Choi J., Park S. Y., Jeon J., Lee Y. H., Two conidiation-related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genet. Biol. 61, 133–141 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sRNA sequencing datasets have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession number PRJNA647702. All other study data are included in the article and/or supporting information.