Significance
The plasma membrane protects cell content from the outer environment. Overcoming this obstacle and delivering drugs efficiently into cells still remain a major hurdle for modern pharmacology and medicine. We developed and demonstrated a simple nanomaterial platform—a dimer of small-diameter carbon nanotube porins (CNTPs)—which functions as a potent membrane fusogen. Molecular simulations revealed a distinct fusion mechanism. CNTP-studded vesicles loaded with a chemotherapeutic agent, doxorubicin, delivered the drug to cancer cells, killing a majority of them. Our work provides new opportunities for understanding membrane fusion mechanisms, designing synthetic fusogens, and developing simple and efficient drug carriers for delivery of therapeutics and vaccines.
Keywords: carbon nanotube porins, membrane fusion, drug delivery, liposomes
Abstract
Drug delivery mitigates toxic side effects and poor pharmacokinetics of life-saving therapeutics and enhances treatment efficacy. However, direct cytoplasmic delivery of drugs and vaccines into cells has remained out of reach. We find that liposomes studded with 0.8-nm-wide carbon nanotube porins (CNTPs) function as efficient vehicles for direct cytoplasmic drug delivery by facilitating fusion of lipid membranes and complete mixing of the membrane material and vesicle interior content. Fusion kinetics data and coarse-grained molecular dynamics simulations reveal an unusual mechanism where CNTP dimers tether the vesicles, pull the membranes into proximity, and then fuse their outer and inner leaflets. Liposomes containing CNTPs in their membranes and loaded with an anticancer drug, doxorubicin, were effective in delivering the drug to cancer cells, killing up to 90% of them. Our results open an avenue for designing efficient drug delivery carriers compatible with a wide range of therapeutics.
Modern medicine relies on an extensive arsenal of drugs to combat deadly diseases such as pneumonia, tuberculosis, HIV and AIDS, and malaria (1). Chemotherapy agents have prolonged lives for millions of cancer patients and in some cases cured the disease or turned it into a chronic condition (2). Yet, the safe and efficient delivery of drugs to target tissues remains a major challenge. Drugs are often poorly soluble, strongly toxic to other tissues, or face rapid degradation in the different chemical environments inside an organism (3). They can accumulate in nontarget tissues and bind to other cellular components or may not internalize efficiently into the target cells (4). Liposomal delivery systems aim to mitigate these problems by encapsulating drugs in external carriers that circulate through the bloodstream (5–7). However, these strategies involve a trade-off between enhancing liposomal stability on the way to the target and easing payload release into the cytosol of the target cell (5). Most current liposomal delivery strategies rely on the endosomal pathway for cell entry, which is inherently inefficient and often results in degradation of the cargo (8). Commonly used cationic lipids, which enhance liposomal fusion with the target membrane and thus enhance endosomal escape, proved to be toxic (9, 10). Another method attempting direct delivery via the plasma membrane required the placement of SNARE-like peptides on the target membrane, which severely limits clinical applications (11).
An alternative approach would facilitate direct payload delivery from liposomes through the plasma membrane into the cell interior by facilitating direct fusion of the carrier membrane with the cell. Our previous molecular dynamics (MD) simulations (12) indicated that carbon nanotube porins (CNTPs)—short pieces of carbon nanotubes inserted into lipid membranes (13)—could potentially facilitate fusion of lipid membranes. That theoretical study raised the intriguing possibility that 10-nm-long and 1.5-nm-wide CNTPs, which resemble hydrophobic stalks of influenza hemagglutinin and HIV-1 Env (14–17), can insert into opposite membranes, promote the formation of a short-lived hourglass-shaped fusion intermediate where the inner leaflets are still intact, and then drive a transition to full fusion (12). However, the simulation study left open many questions, such as whether CNTP-mediated fusion was even practically possible and whether the concept was applicable for drug delivery. Moreover, how additional parameters such as thickness or aggregation of CNTPs might affect the fusion kinetics in a real-world setting remains largely unclear. In this study we explore the hypothesis that small-diameter CNTPs can serve as generic minimal synthetic analogs of viral fusion machines. We show that 0.8-nm-diameter CNTPs indeed facilitate efficient membrane fusion. Moreover, our in vitro experiments demonstrate that CNTP-studded liposomes loaded with an anticancer drug enable efficient payload delivery into target cells. Our studies also reveal a surprising mechanism of fusion, where CNTP dimers show strong fusogenic activity.
Results and Discussion
CNTP-Mediated Vesicle Fusion.
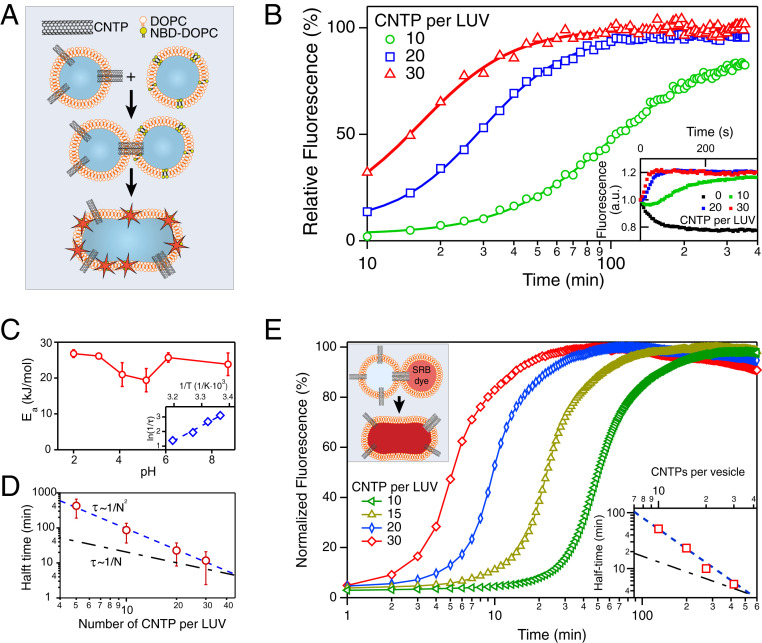
We first tested our hypothesis that CNTPs can promote vesicle fusion by adopting a widely used dequenching lipid mixing assay (18). We mixed large unilamellar 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) vesicles containing 0.8-nm-diameter CNTPs in the lipid bilayer (CNTP-LUVs) with another population of pure DOPC vesicles containing lipids labeled with an NBD dye (NBD-LUVs) at a concentration just above the self-quenching threshold (19). As the lipids from different vesicles mixed during fusion, the NBD dye dequenched (Fig. 1 A and B), allowing us to quantify this process by monitoring the change in the dye fluorescence. Indeed, after mixing of these two vesicle populations the fluorescence signal rose on the timescale of tens of minutes before eventually plateauing. By contrast, fluorescence kinetics recorded in control experiments where CNTP-LUVs were replaced by pure DOPC vesicles (LUVs) did not show dequenching (Fig. 1B, Inset), indicating that the presence of CNTPs in the vesicle shell was critical to inducing fusion. As the CNTP concentration in vesicles increased, the dequenching signal reached the plateau faster (Fig. 1B), again indicating that CNTPs were responsible for the fusion events. Surprisingly, the fusion rate, calculated as , with the fusion half-time (see Materials and Methods for details), did not scale linearly with the CNTP concentration. Instead, it followed an inverse quadratic dependence (Fig. 1D), suggesting that the key fusion step was mediated by a CNTP dimer, formed by CNTPs associating in the membrane. Docking-leveled experiments, designed to separate vesicle docking kinetics from the fusion kinetics (20), indicate that docking kinetics may also contribute to the overall kinetics at higher CNTP concentrations (SI Appendix, Fig. S1).
Fig. 1.
CNTPs facilitate membrane fusion. (A) Schematics of the vesicle fusion assay. CNTP-LUVs fuse with the vesicles containing DOPC lipid labeled with NBD dye in self-quenching concentration, dequench the dye, and increase its fluorescence signal. (B) Kinetics of the vesicle fusion. The fluorescence intensity was recorded as NBD-LUVs were mixed with CNTP-LUVs with different average numbers of CNTPs per vesicle (as indicated on the graph). Solid lines represent best fits to the Hill equation. (C) Activation energy, , for vesicle fusion plotted as a function of pH. (Inset) A representative Arrhenius plot used to obtain the values (n = 3). (D) Plot of the fusion half-time as a function of the average number of CNTPs per vesicle (n = 3 for 10, 20, and 30 CNTP/LUV and n = 2 for 5 CNTP/LUV). The blue dashed line represents a fit to the second-order kinetics. The dashed-dotted black line, which corresponds to the first-order kinetics, is provided as a guide to the eye. (E) Content-mixing assay showing fluorescence signal kinetics recorded as CNTP-LUVs were exposed to LUVs encapsulating SRB dye (each curve is an average of two runs; see SI Appendix, Fig. S2 for raw traces). (Inset) The plot of the fusion half-time as a function of the average number of CNTPs per vesicle.
To verify that our system proceeded to full fusion and complete mixing of the vesicular compartments we set up a different dye dequenching assay, in which the target DOPC vesicles were filled with a solution of sulforhodamin B (SRB) dye in a concentration above its self-quenching threshold. After these vesicles were mixed with the CNTP-LUVs, we observed gradual dequenching of the SRB dye fluorescence (Fig. 1E; see also SI Appendix, Fig. S2), indicating full vesicle content mixing. Content-mixing kinetics proceeded in a CNTP-concentration-dependent manner and followed the same inverse quadratic dependence observed in the membrane mixing assay (Fig. 1E, Inset). We also confirmed vesicle fusion, full content mixing, and the absence of content leakage in similar CNTP-mediated experiments with smaller, 100-nm-diameter DOPC vesicles with 30% cholesterol (SI Appendix, Fig. S3).
As a function of temperature, the fusion rate followed an Arrhenius dependence (Fig. 1C, Inset) with an activation energy, , of ca. 25 kJ/mol or 10 (Fig. 1C). This value is significantly smaller than the activation energy of 30 that was recently reported for the spontaneous fusion of small vesicles (21), indicating that CNTPs indeed lower the energy barrier for membrane fusion. Interestingly, the activation energy showed a weak dependence on pH (Fig. 1C), with the barrier dropping by an additional 2 at pH values between 4 and 5 and recovering back to the original value of 10 at pH values below 4. DOPC remains in a zwitterionic charge state over the whole range of pH used in our measurements. Therefore, we believe that this behavior must originate in the charge state of the CNTP ends, which start to become protonated (22) at pH 4 to 5. We, therefore, speculate that COOH/ interactions stabilize the CNTP dimers that facilitate fusion. This finding indicates that end-group functionalization may be exploited to further tune the selectivity and efficiency of CNTP-mediated fusion.
MD Simulations of Vesicle Fusion.
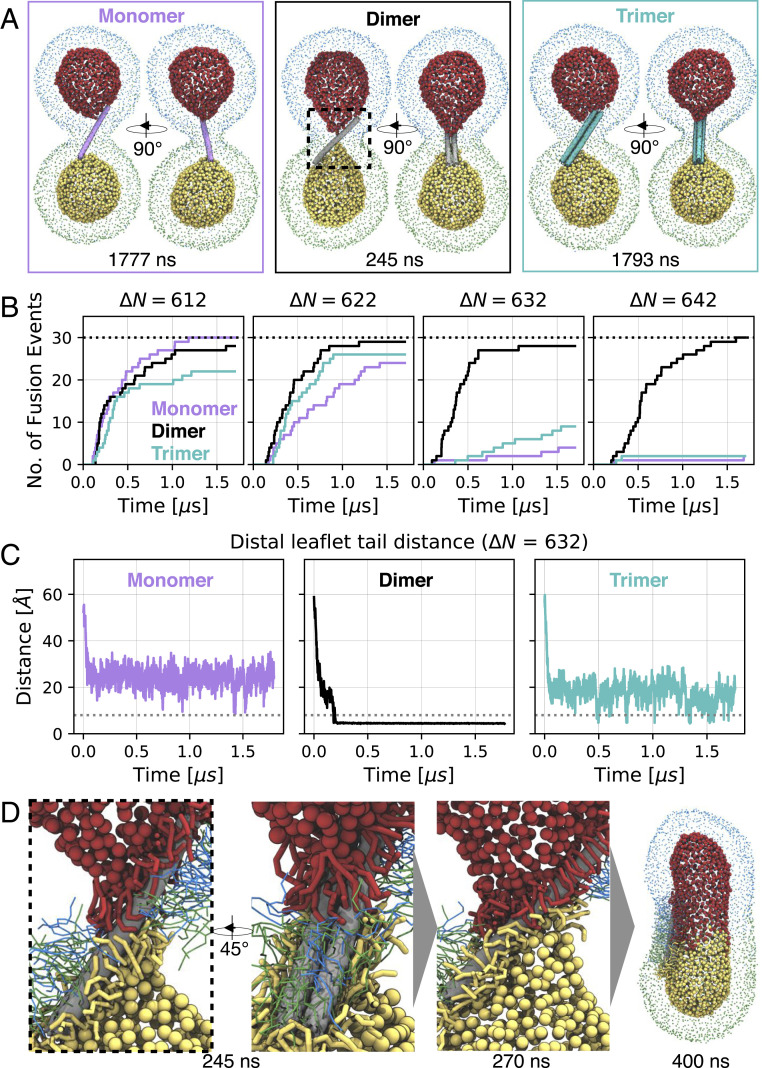
We confirmed the enhanced fusogenic properties of CNTP dimers and elucidated the underlying fusion mechanism using coarse-grained MD simulations. The simulation systems contained two 15-nm DOPC vesicles with a bridging 0.8-nm CNTP monomer, dimer, or trimer inserted in their membranes (Fig. 2A, SI Appendix, Fig. S4 and Table S1 and Movie S1). To control the driving force and kinetic rate of vesicle fusion, we varied the asymmetry in the number of lipids in the outer and inner leaflets of the two vesicles, defined as . By increasing the number asymmetry we lowered the bilayer strain and fusion propensity of vesicles, allowing us to differentiate more clearly between the fusogenic characteristics of CNTP monomers, dimers, and trimers.
Fig. 2.
Coarse-grained MD simulations of CNTP-mediated vesicle fusion. (A) Snapshots of simulated systems of a CNTP monomer (magenta), dimer (gray), and trimer (cyan) (). Lipid phosphate groups are colored uniquely for each leaflet and vesicle (outer leaflets: blue/green for top/bottom vesicle; inner leaflets: red/yellow for top/bottom vesicle). Inner leaflet phosphate groups are drawn larger for clarity. Times of snapshots are indicated (see also Movie S1). (B) Cumulative number of CNTP-mediated vesicle fusion events as a function of time at different number asymmetries N. Monomer (magenta), dimer (black), and trimer (cyan) simulations are compared. A total of 30 simulations were performed for each starting configuration (indicated as black dashed line). Simulations were 1.7 s long. (C) Minimal distance of C5A/B tail beads of the opposing inner leaflet lipids at . Exemplary trace shown for monomer, dimer, and trimer, respectively. All traces for all systems are shown in SI Appendix, Fig. S6. The dashed line at 8 Å indicates contact of the opposing inner leaflets. (D) Zoom-in on CNTP dimer-mediated fusion. Time points of snapshots are indicated. Lipids within 8 Å of the CNTP are shown. Color scheme as in A. Inner leaflet lipids are drawn thicker for clarity. Outer leaflet phosphate groups are omitted for clarity (see also Movie S2).
Surprisingly, CNTP dimers rapidly fused vesicles without any significant changes in fusion time over the full range of tested asymmetries. By contrast, CNTP monomers and trimers fused only vesicles with low number asymmetry, i.e., only in the presence of significant bilayer strain (Fig. 2B). At high number asymmetry, monomer and trimer fusion, but not dimer fusion, slowed down dramatically, with only a few fusion events observed during the simulation time.
We gained a more detailed insight into the fusion mechanism by monitoring the minimal distance between any two lipid tail groups in the inner leaflets of the two vesicles. In all simulations, CNTP dimers achieved initial inner-leaflet contact, which is a prerequisite for fusion, more rapidly than CNTP monomers (Fig. 2C and SI Appendix, Fig. S6). Consistently, in all relaxed prefusion systems, CNTP dimers distorted inner leaflets to a higher degree than monomers and trimers, leading to rapid inner-leaflet contact and subsequent fusion (Fig. 2C, SI Appendix, Fig. S6, and Movie S1).
The asymmetric shape of the CNTP dimer explains its distinct fusogenic properties. The energetic drive to cover the hydrophobic surface of the wide faces by lipid tails (Fig. 2D and Movie S2) pulls the two vesicles together and causes significant distortion of the distal leaflets. The narrow edges of the CNTP dimer facilitate tail–tail contacts between the opposing inner leaflets. By contrast, single thin CNTPs are not coated as densely with lipid tails and consequently do not distort the distal leaflets significantly (Fig. 2 A and C and SI Appendix, Fig. S6), impeding fusion. Trimers are too thick to establish sufficient tail–tail interactions (Fig. 2 A and C and SI Appendix, Fig. S6) and thus fail to induce fusion on the MD simulations timescale. We emphasize that this mechanism, where the distinct geometry of the hydrophobic CNTP dimer surface facilitates lipid migration and subsequent fusion, is distinct from the common viral fusion mechanism, which relies on structural rearrangements of the fusion peptide stalks to bring the interacting membranes together (14–17). Our results may also point to the simplest structure of a membrane fusogen.
We also probed whether such a dimer-based fusion mechanism is unique to the 0.8-nm CNTPs by testing for fusion in simulations of CNTP monomers and dimers with larger diameters of 1.2 and 1.5 nm. We found that at larger diameters the difference in fusion performance between the dimers and monomers vanished (SI Appendix, Figs. S7–S11) and monomers could also catalyze fusion. This is not surprising, as CNTP monomers with a larger diameter provide sufficient hydrophobic surface to pull the lipid tails from the opposite bilayers into proximity. Conversely, the diameter is still small enough to allow contact between tails of the inner leaflets. Our previous simulations of fusion with CNTPs of 1.5-nm diameter already showed that they could catalyze membrane fusion (12). Even though CNTP dimers with larger diameters also catalyzed fusion, they did not further increase the already fast fusion rates. However, we cannot rule out that the differential dimer effect might reemerge for the larger-diameter vesicles where the bilayer stress is significantly reduced, such as those typically used in the experiments. These findings align with our mechanistic model and show that the key structural features of a fusogen can be realized in multiple ways. We also note that only the dimer-based fusion mechanism is relevant for our experiments, which used a narrow diameter distribution of the CNTPs (0.81 0.14 nm) (23).
We observed that not all CNTP fusion simulations proceeded directly to fusion pore formation. In several replicas across parameter sets we noticed the formation of an intermediate with a 25-Å minimum headgroup distance (SI Appendix, Fig. S12). Visual inspection revealed a hemifusion diaphragm, where the lipids of the inner leaflets form a bilayer-like structure (SI Appendix Fig. S12, Top), which was in some instances stable for long times. In this intermediate state that followed the stalk state, the outer leaflet lipids could equilibrate while the vesicle content remained separated. The hemifusion diaphragm spontaneously opened in a distinct step, typically away from the CNTP, forming a fusion pore that completed the fusion process. The formation of hemifusion diaphragms was particularly pronounced in systems with low asymmetry (i.e., higher lipid density), where the diaphragm accommodated excess lipids of the inner leaflet. In systems with larger diameters, i.e., 1.2 and 1.5 nm, where fusion was observed over the full range of asymmetries, high-asymmetry systems fused faster because the formation of hemifusion diaphragms was disfavored.
Drug Delivery with CNTPs.
To demonstrate drug delivery to cancer cells using CNTP-mediated membrane fusion (Fig. 3A), we encapsulated a widely used first-line-of-defense chemotherapeutic agent, doxorubicin (DOX), in CNTP-LUVs. Systemic administration of DOX is complicated by its significant cardiac toxicity, which is often mitigated by encapsulating high amounts of the drug into PEGylated liposomal carriers (used widely in cancer treatment in commercial formulation as Doxil or Caelyx) (24). Doxil liposomes have a very slow background DOX release profile (25) that protects healthy tissues during circulation and allows the carriers to accumulate in tumors passively and then enter the tumor cells via endocytosis (26). Replacing endocytosis with a more direct fusion-based entry pathway could significantly improve the delivery efficiency for the liposome-encapsulated drugs.
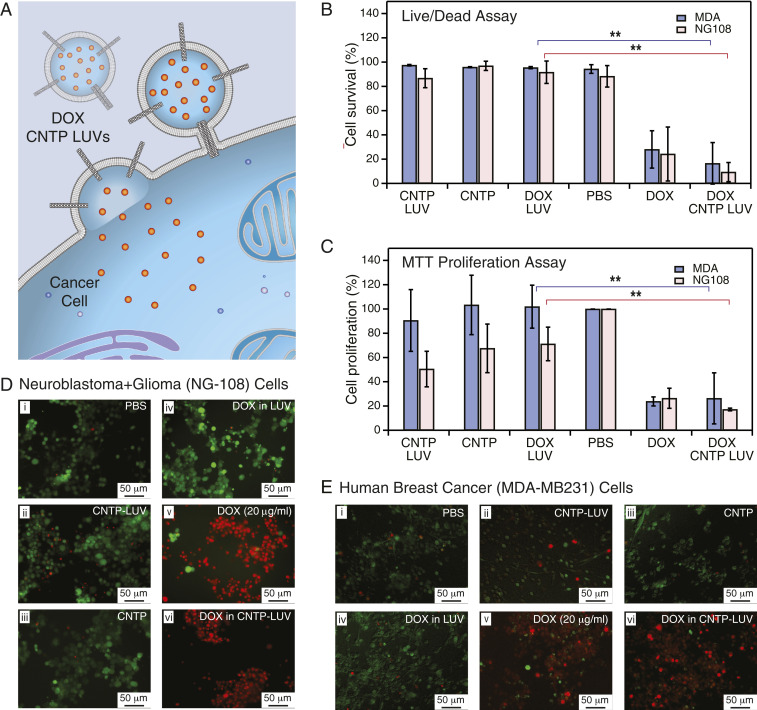
Fig. 3.
DOX delivery with CNTPs. (A) Schematic showing CNTP-LUV loaded with the DOX payload fusing to a cancer cell and delivering DOX to the cell interior. (B) Cell survival in live/dead assay after 48-h exposure of neuroblastoma-glioma (NG-108) and human breast cancer (MDA) cell cultures to DOX-CNTP-LUVs, CNTP-LUVs, free CNTPs, DOX-LUVs, free DOX, and PBS buffer (n = 9). **P 0.01. (C) Results of MTT cell proliferation assay after 48-h exposure of NG-108 and MDA cell cultures to DOX-CNTP-LUVs, CNTP-LUVs, free CNTPs, DOX-LUVs, free DOX, and PBS buffer. (NG-108 cells: n = 9; MDA cells: n = 15). **P 0.01. (D) Fluorescence microscopy images of NG108 cell culture with live and dead cells stained with green and red dye, respectively. Prior to imaging the cells were exposed for 48 h to (i) PBS buffer, (ii) CNTP-LUVs without the drug payload, (iii) CNTP solution, (iv) LUVs encapsulating DOX, (v) 20 g/mL of DOX, or (vi) CNTP-LUVs with encapsulated DOX. (E) Fluorescence microscopy images of MDA cell culture with live and dead cells stained with green and red dye, respectively. Prior to imaging the cells were exposed for 48 h to (i) PBS buffer, (ii) CNTP-LUVs without the drug payload, (iii) CNTP solution, (iv) LUVs encapsulating DOX, (v) 20 g/mL of DOX, or (vi) CNTP-LUVs with encapsulated DOX.
To evaluate this hypothesis, we tested DOX-CNTP-LUV performance in a series of cell viability assays on two different cell lines: NG108-15 (mouse neuroblastoma and rat glioma hybrid cells) and MDA MB-231 (human breast cancer cells) (Fig. 3). For these experiments we loaded CNTP-LUVs with a relatively low encapsulated DOX concentration of 10 g/mL. DOX molecule size is larger than the CNTP pore size, excluding the possibility that DOX would leak through the nanotubes. Control experiments (SI Appendix, Fig. S13) also confirm the absence of long-term drug leakage from CNTP-LUVs. To mimic some of the current liposomal delivery strategies, we used smaller, 100-nm-diameter liposomes with lipid composition of 70% DOPC and 30% of cholesterol. This vesicle composition also showed high fusion efficiency with an average of less than 1 h, similar to what we observed for the pure DOPC vesicles at a similar size (SI Appendix, Fig. S3 A and B), indicating the cholesterol presence did not interfere with the fusion mechanism.
After 48-h exposure to DOX-loaded CNTP-LUVs, cell viability decreased significantly compared to the phosphate-buffered saline (PBS) control, with only 9% of NG108 cells and 16% of MDA cells surviving the treatment (Fig. 3 B; D, iv; and E, iv). The efficiency of the CNTP-LUVs loaded with 10 g/mL of DOX (Fig. 3 B and C; D, v; and E, v) was mostly comparable to administering 20 g/mL of free DOX (Fig. 3 B and C; D, v; and E, v). This observation is significant because DOX-CNTP-LUVs used in our experiments contain a much smaller overall amount of drug (10 g/mL) compared to what is used in commercial Doxil formulation (2 mg/mL) (27). Thus, CNTP-LUV carriers could potentially exhibit dramatically lower systemic toxicity and still would maintain the high efficiency of drug release.
By contrast, control experiments (Fig. 3 B; D, i–iii; and E, i–iii) with cells exposed to CNTP-LUVs and free CNTPs showed very low cytotoxicity, with typically over 85% of the cells remaining alive after the same 48 h of exposure. These viability numbers were on par with those measured after exposure to pure PBS buffer (88% and 94% for NG108 and MDA cells, respectively). Interestingly, when pure LUVs were loaded with 20 g/mL of DOX their cytotoxicity was also on par with control experiments (Fig. 3 B; D, i; and E, i), showing little to no efficiency without the presence of a viable delivery mechanism.
Cell proliferation (MTT) assays results (Fig. 3C) tracked the trends obtained in the cell viability (live/dead) assay across all samples that we tested. Exposure of both cell lines to DOX-loaded CNTP-LUVs led to a significant decrease in the cells’ proliferation ability. Control experiments where we exposed cells to CNTPs and CNTP-LUVs in the presence of free DOX in solution did not show a statistically significant cell viability decrease (SI Appendix, Figs. S14 and S15), indicating that CNTP-mediated fusion was indeed the main pathway for the drug entry into the cancer cells, and that the drug did not enter through defects on the cell membranes created by free CNTPs or CNTP-LUVs. Additional experiments (SI Appendix, Fig. S16) also showed a dose-dependent cell response to CNTP-DOX-LUV treatment, with higher doses resulting in progressively lower cell survival probabilities. Additional corroboration of the proposed delivery mechanism comes from experiments in which we exposed MDA cells to CNTP-LUVs loaded with a self-quenched concentration of SRB dye and observed gradual dequenching of the dye as it entered the cells (SI Appendix, Fig. S17).
We also noticed that NG108 cells exposed to free CNTPs and CNTP-LUVs showed a small decrease in the cell proliferation percentage relative to the PBS buffer control. Visual observations of the NG108 cell morphology in the images of those samples showed that cells in the cultures exposed to CNTP and CNTP-LUV were still alive and building neural networks (SI Appendix, Fig. S18). Literature reports show that neural cell hybrids, such as NG108, can differentiate under certain stresses (28). Similar to the images of differentiated NG108 cells in the literature, our cultures started to form abundant neurites and varicosities after incubating them with free CNTPs and CNTP-LUVs. By contrast, the control populations of cells exposed only to the PBS buffer looked more flat and circular and had significantly fewer neurite formations (SI Appendix, Fig. S18). Thus, we hypothesize that the cells incubated with CNTP-containing samples started to differentiate instead of growing.
Conclusions and Outlook
Our results establish CNTPs as potent fusogens that exploit the unique structure and geometry of CNTP dimers to facilitate membrane fusion. Our MD simulations revealed the mechanism of CNTP-mediated fusion and explain the observed fusion kinetics: CNTP dimers pull the membranes together with their flat faces and bring the inner leaflets into contact across their narrow faces. Researchers can apply the same principles that enable CNTP-mediated membrane fusion to design other synthetic fusogens for even more efficient and targeted delivery to specific cell types. Computational screening by molecular simulations could be used to guide the systematic design of novel nanomaterial-based fusogens and to improve the properties of the accompanying liposomes.
Our experiments demonstrate that CNTP-studded liposomes can provide the basis for constructing the long-desired, but so far elusive, inert versatile carrier for direct and highly efficient delivery of drugs and DNA and RNA vaccines (29) across the plasma membrane. This strategy could bypass the endocytotic pathway entirely and thus avoid some of the problems encountered by the previous delivery strategies.
Finally, the use of carbon nanomaterials for drug delivery raises some understandable safety concerns. We note, however, that recent studies of the in vivo biocompatibility of short small-diameter CNTs reported their efficient renal clearance in mice (30, 31) and nonhuman primates (32), pointing to the feasibility of using this material for therapeutics development. Further research on the long-term fate and clearance mechanisms of ultrashort carbon nanotubes in the tissues should clarify these important questions.
Materials and Methods
Materials and Equipment.
All of the lipids (DOPC, 1-oleoyl-2-6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl-sn-glycero-3-phosphocholine [NBD-PC], and cholesterol) were obtained from Avanti. All of the other chemicals were purchased from Sigma-Aldrich and used as received, unless specified. Live/dead assay and MTT cellular assay kits were obtained from Abcam. The size-exclusion columns for LUV separation used Sepharose CL-6B (Sigma-Aldrich). The ultrashort CNTPs were synthesized by sonication-assisted cutting of 0.8-nm single-walled carbon nanotubes according to the previously published procedure (33). Previous studies have confirmed that this procedure produces CNTPs with an extremely tight diameter distribution of 0.81 0.14 nm as measured by transmission electron microscopy (23). Some CNTP batches were chemically coupled to the 6-aminofluorescein (6-AF) dye using a 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) coupling procedure based on a published protocol (34). All fluorescence and absorbance spectra were measured with the Spectramax iD3 Microplate Reader (Molecular Devices) and Cytation 5 (Biotek). Vesicle size was measured using a dynamic light scattering (DLS) setup (Malvern Analytical).
Large Unilamellar Vesicles Formation.
The LUVs and CNTP-LUVs were formed and characterized using previously described protocols (33). Briefly, the CNTPs were added to the lipid mixtures prior to rehydration and extrusion. The average number of CNTPs per vesicle was quantified using assays described in the same protocol (33). We note that this procedure is based on a calibrated proton permeability of an individual CNTP and thus could introduce a small systematic error. LUV and CNTP-LUVs loaded with DOX were prepared using the same protocol, but the sonication time was extended to 10 min from 2 min. To form NBD-LUVs we used 85% of DOPC and 15% of NBD-DOPC, and for SRB-LUVs we added 28 mM SRB to the solution before sonication. All LUVs went through 10 freeze–thaw cycles to remove multilamellar vesicles. LUVs used for drug delivery experiments followed the same protocol, except that the lipid composition was 70% DOPC and 30% cholesterol, 300 mM solution of ammonium sulphate was used instead of buffered KCl solution, and the vesicles were extruded through a 100-nm membrane filter. In the final step the vesicles were purified on a column conditioned with PBS at pH 7.4. The size of LUVs was determined using DLS. The drug encapsulation efficiency was 10%, as determined by literature protocols (35). To quantify DOX leakage from DOX-loaded LUVs and CNTP-LUVs we monitored fluorescence (480 excitation/590 emission) for 18 h at 37 C (SI Appendix, Fig. S13).
Lipid-Mixing and Content-Mixing Assays.
To obtain a self-quenched concentration of NBD dye in LUVs we used a 15% NBD-PC and 85% DOPC mixture, as determined from calibration experiments. Lipid fusion assays were performed at different pH (2, 3.15, 4.11, 5.15, 6.11, and 8.7) with buffer pH adjusted with 1 M HCl. In each fusion assay CNTP-LUVs and NBD-LUVs were mixed at 1:1 vol ratio and the fluorescence kinetics (474 excitation/530 emission) was recorded for at least 3 h at a preset temperature maintained by the plate reader. Each assay was repeated at least three times. For content-mixing assays, CNTP-LUVs were mixed with SRB-LUVs at 1:1 ratio in the presence of tetramethylrhodamine polyclonal antibody from Thermo Fisher to quench the signal from any leaked SRB dye. The amount of antibody used was calculated to quench at least 80% of all SRB dye contained in the sample LUV. The fluorescence kinetics (550 excitation/595 emission) was monitored for at least 18 h at 24 C. All content fusion assays were repeated at least two times.
To extract fusion half-times from the fusion kinetics data we fitted the fluorescence traces to the Hill-like equation (36):
| [1] |
where F is normalized fluorescence signal, is the fusion half-time, is the time, and is the Hill coefficient. The fusion rate was then calculated as . The values of the fit parameter typically varied between 2 and 3.
DOX Delivery to NG108-15 and MDA-MB231 Cells.
NG108-15 (mouse neuroblastoma × rat glioma hybrid) and MDA-MB231 cell lines were obtained from ATCC. The NG108 cells were cultured in growth media (Dulbecco’s modified Eagle’s medium, 1% penicillin–streptomycin, and hypoxanthine-aminopterin-thymidine medium 1× with 10% fetal bovine serum, from Gibco) at 37 C in 5% CO2. The MDA-MB-231 cells were cultured in DMEM/F-12, GlutaMAX supplement (Thermo Fisher) with 10% fetal bovine serum (Gibco) at 37 C in 5% CO2. The cells were seeded in 96-well plates at 5,000 cells per well and cultured for 2 d before the experiment. Each well was treated with growth media and sample at 1:1 vol ratio for 48 h. Drug delivery experiments were conducted using different batches of cells purchased and cultured at different times and the results were averaged between batches wherever possible.
Cell Viability Quantification Using Live/Dead Assay.
The live/dead dye was diluted in PBS to a final concentration of 5× (5 L in 1 mL of PBS). After exposure to the samples, the media was aspirated from the well and replaced by 100 L of dye solution. The cells were incubated for 15 min. The fluorescent images of cells were recorded using a Leica fluorescence microscope with FITC (494/151 nm) and RHO (528/617 nm) filters to visualize live and dead cells, respectively. The number of live and dead cells was counted using ImageJ and normalized to the total number of counted cells. The experiment was repeated three times, three wells each time, using at least three images per well. Since the distribution of our sample averages was not normally distributed and the samples size was less 50, we used Wilcoxon statistical analysis to test for significant differences. For some of the experiments quantifying cell response to free CNTPs, CNTPs were modified by covalent coupling of a 6-AF dye to the end of the CNTP (see Materials and Methods for details). Control experiments indicated that even though this modification produced slower fusion, modified and unmodified CNTPs produced similar outcomes in the fusion experiments at the 1- to 2-h timescale (SI Appendix, Fig. S3C), which is still much shorter than the 48-h timescale of the cell viability experiments.
Cell Proliferation Quantification Using MTT Assay.
The media with samples were removed from wells and 50 L of MTT reagent and 50 L of PBS was added to each well. The cells were incubated at 37 C for 3 h. After the incubation, 150 L of MTT solvent was added into the well. The plate was incubated overnight at room temperature in a dark box. We recorded the absorbance at 590 nm and used it (after subtracting background from PBS and MTT reagent controls) to determine the number of cells in each sample using a calibration curve established separately for each cell line. The cell proliferation percentage was normalized using PBS-exposed sample as a 100% reference.
Monitoring Liposome Content Fusion with Live Cells.
MDA-MB231 cells were seeded at cells per well in a 96-well plate and cultured for 48 h. Right before the measurement, the growth media was removed from the cells and replaced with 1:1 mixture of cell media and solution of CNTP-SRB-LUVs (20 CNTP/LUV). The fluorescent signal (555 excitation/595 emission) was monitored in a plate reader.
Coarse-Grained MD Simulations.
All MD simulations were set up and run as previously described (12) using the MARTINI (v. 2.2) coarse-grained model (37). Simulations were performed using GROMACS 2018.7 (38) with the recommended new parameter set for MARTINI simulations (39). The Verlet neighbor search algorithm was used to update the neighbor list, with the length and update frequency being automatically determined (nstlist = 25, rlist = 1.259 nm). Lennard-Jones and Coulomb forces were cut off at 1.1 nm with the potential shifted to 0 using the Verlet-shift potential modifier. Pressure was maintained at 1 bar using the Parrinello–Rahman barostat and temperature was maintained at 300 K using the velocity rescaling algorithm with characteristic coupling times of 12 and 1 ps, respectively (40, 41).
CNTPs with 0.8-/1.2-/1.5-nm diameter consisted of 30 rings with 5/8/10 beads each, respectively. The total length of all three types of CNTPs was 11.8 nm. For thin 0.8-nm nanotubes the force constant of improper dihedrals, which maintains stiffness, was increased to 550 kJmol−1rad−2, whereas the 1.2- and 1.5-nm CNTPs used the default values (12, 42). System starting configurations were set up following the protocol for system A of ref. 12, where two 15-nm DOPC vesicles were stapled by a thin CNT monomer, dimer, and trimer, respectively (SI Appendix, Fig. S4). The number asymmetry was varied by removing lipids from the inner leaflets of both vesicles, respectively. All simulated systems are summarized in SI Appendix, Table S1. For each setup, 30 replicates were run with different initial velocities.
Supplementary Material
Acknowledgments
We thank Drs. V. Frolov, A. Shnyrova, and R. Tunuguntla for help with the initial stages of this work, P. Lastrico for designing Fig. 3A, and anonymous reviewers for suggesting a number of additional experiments, including the docking-leveled approach for evaluating fusion kinetics. A.N. and K.V.C.’s work on CNTP synthesis and vesicle fusion was supported by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under award SCW0972-17. N.T.H.’s work on vesicle fusion kinetics was supported by the Division of Materials Research of the National Science Foundation under award 1710211. Drug delivery studies were supported by the Lawrence Livermore National Laboratory (LLNL) Laboratory Directed Research and Development (LDRD) Program under award 18-ERD-011. Work at the LLNL was performed under the auspices of the US Department of Energy under contract DE-AC52-07NA27344. Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract DE-AC02-05CH11231. M.S., R.M.B., J.K., and G.H.’s work on computer simulations was supported by the Max Planck Society. M.S. and G.H. were supported by the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz DynaMem program of the state of Hesse. J.M.H.’s work on CNTP modifications and cell culture assays was supported by Engineering and Physical Sciences Research Council grant EP/R004072/1.
Footnotes
Competing interest statement: Some of the results reported in the paper have been disclosed as part of a provisional patent filing.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016974118/-/DCSupplemental.
Data Availability
Kinetics data and cell viability assay data have been deposited in Figshare (DOI: 10.6084/m9.figshare.12440285) (43).
References
- 1.Katzung B. G.. Basic & Clinical Pharmacology (McGraw-Hill, New York, ed. 14, 2018). [Google Scholar]
- 2.Chabner B. A., Roberts T. G., Chemotherapy and the war on cancer. Nat. Rev. Canc. 5 (1), 65–72 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Timko B. P., et al. , Advances in drug delivery. Annu. Rev. Mater. Res. 41, 1–20 (2011). [Google Scholar]
- 4.Sercombe L., et al. , Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 6, 286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen Y., Gurman P., Elman N., Drug Delivery: An Integrated Clinical and Engineering Approach (CRC Press, 2017). [Google Scholar]
- 6.Peer D., et al. , Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2, 751 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Zylberberg C., Matosevic S., Pharmaceutical liposomal drug delivery: A review of new delivery systems and a look at the regulatory landscape. Drug Deliv. 23, 3319–3329 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Allen T. M., Cullis P. R., Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 65, 36–48 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Knudsen K. B., et al. , In vivo toxicity of cationic micelles and liposomes. Nanomedicine 11, 467–477 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Filion M. C., Phillips N. C., Major limitations in the use of cationic liposomes for DNA delivery. Int. J. Pharm. 162, 159–170 (1998). [Google Scholar]
- 11.Yang J., et al. , Drug delivery via cell membrane fusion using lipopeptide modified liposomes. ACS Cent. Sci. 2, 621–630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaskara R. M., Linker S. M., Vögele M., Köfinger J., Hummer G., Carbon nanotubes mediate fusion of lipid vesicles. ACS Nano 11, 1273–1280 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Geng J., et al. , Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature 514, 612–615 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Harrison S. C., Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epand R. M., Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta Biomembr. 1614, 116–121 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Earp L. J., Delos S. E., Park H. E., White J. M., “The many mechanisms of viral membrane fusion proteins” in Membrane Trafficking in Viral Replication, Marsh M., Ed. (Springer, 2004), pp. 25–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielian M., Rey F. A., Virus membrane-fusion proteins: More than one way to make a haripin. Nat. Rev. Microbiol. 4, 67–76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Struck D. K., Hoekstra D., Pagano R. E., Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20, 4093–4099 (1981). [DOI] [PubMed] [Google Scholar]
- 19.Weber T., et al. , SNAREpins: Minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Hernandez J. M., Kreutzberger A. J. B., Kiessling V., Tamm L. K., Jahn R., Variable cooperativity in snare-mediated membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 12037–12042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.François-Martin C., Rothman J. E., Pincet F., Low energy cost for optimal speed and control of membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 114, 1238–1241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornasiero F., et al. , PH-tunable ion selectivity in carbon nanotube pores. Langmuir, 26, 14848–14853 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Tunuguntla R., Allen F. I., Kim K., Bellivieau A., Noy A., Ultrafast proton transport in sub-1-nm diameter carbon nanotube porins. Nat. Nanotechnol. 11, 639–644 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Barenholz Y. C., Doxil—The first FDA-approved nano-drug: Lessons learned. J. Contr. Release 160, 117–134 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Russell L. M., Hultz M., Searson P. C., Leakage kinetics of the liposomal chemotherapeutic agent doxil: The role of dissolution, protonation, and passive transport, and implications for mechanism of action. J. Contr. Release 269, 171–176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seynhaeve A. L. B., Dicheva B. M., Hoving S., Koning G. A., Ten Hagen T. L. M., Intact doxil is taken up intracellularly and released doxorubicin sequesters in the lysosome: Evaluated by in vitro/in vivo live cell imaging. J. Contr. Release 172, 330–340 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Soundararajan A., Bao A., Phillips W. T., R. Perez, III, Goins B. A., Re-liposomal doxorubicin (Doxil): In vitro stability, pharmacokinetics, imaging and biodistribution in a head and neck squamous cell carcinoma xenograft model. Nucl. Med. Biol. 36, 515–524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tojima T., Yamane Y., Takahashi M., Ito E., Acquisition of neuronal proteins during differentiation of NG108-15 cells. Neurosci. Res. 37, 153–161 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Kowalski P. S., Rudra A., Miao L., Anderson D. G., Delivering the messenger: Advances in technologies for therapeutic mrna delivery. Mol. Ther. 27, 710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggiero A., et al. , Paradoxical glomerular filtration of carbon nanotubes. Proc. Natl. Acad. Sci. U.S.A. 107, 12369–12374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galassi T. V., et al. , Long-term in vivo biocompatibility of single walled carbon nanotubes. PloS One 15, e0226791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alidori S., et al. , Carbon nanotubes exhibit fibrillar pharmacology in primates. PloS One 12, e0183902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunuguntla R. H., Escalada A., Frolov V. A., Noy A., Synthesis, lipid membrane incorporation, and ion permeability testing of carbon nanotube porins. Nat. Protoc. 11, 2029 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Rawson F. J., Garrett D. J., Leech D., Downard A. J., Baronian K. H. R., Electron transfer from proteus vulgaris to a covalently assembled, single walled carbon nanotube electrode functionalised with osmium bipyridine complex: Application to a whole cell biosensor. Biosens. Bioelectron. 26, 2383–2389 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Ong S. G. M., Ming L.-C., Lee K.-S., Yuen K.-H., Influence of the encapsulation efficiency and size of liposome on the oral bioavailability of griseofulvin-loaded liposomes. Pharmaceutics 8 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Focke W. W., Van der Westhuizen I., Musee N., Loots M. T., Kinetic interpretation of log-logistic dose-time response curves. Sci. Rep. 7, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., De Vries A. H., The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Abraham M. J., et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 39.De Jong D. H., Baoukina S., Ingólfsson H. I., Marrink S. J., Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7 (2016). [Google Scholar]
- 40.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 42.Vögele M., Hummer G., Divergent diffusion coefficients in simulations of fluids and lipid membranes. J. Phys. Chem. B 120, 8722–8732 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Ho N. T., et al. , Data from “Membrane fusion and drug delivery with carbon nanotube porins.” Figshare. 10.6084/m9.figshare.12440285. Deposited 23 April 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Kinetics data and cell viability assay data have been deposited in Figshare (DOI: 10.6084/m9.figshare.12440285) (43).