Significance
We report somatic copy-number alterations (SCNAs) that contribute to an immune microenvironment switch during human papillomavirus-negative head and neck cancer (HNSC) development. Specific and nonspecific SCNA levels were examined in a large prospective oral precancer (188 patients) cohort, 2 HNSC (343, 196 patients) cohorts, and 32 cell lines. Chromosome 9p21.3 loss in precursor lesions, the genomic driver of malignant transition, was enhanced by cumulative 9p-arm gene-dosage decreases, cell-intrinsic senescence suppression, and extrinsic decreases in chemokine, cytokine, and NF-κB pathways. Furthermore, 9p-arm loss and JAK2-PD-L1 codeletion were associated with PD-1 inhibitor resistance. These data reveal an oncogenic immune paradox, aneuploid checkpoint to neoplastic transformation, and immune interception and therapeutic strategies for HPV− HNSC and possibly other CN-driven tumors or diseases.
Keywords: head and neck cancer, genomic copy number variation, immunotherapy, premalignancy, aneuploidy
Abstract
An aneuploid-immune paradox encompasses somatic copy-number alterations (SCNAs), unleashing a cytotoxic response in experimental precancer systems, while conversely being associated with immune suppression and cytotoxic-cell depletion in human tumors, especially head and neck cancer (HNSC). We present evidence from patient samples and cell lines that alterations in chromosome dosage contribute to an immune hot-to-cold switch during human papillomavirus-negative (HPV−) head and neck tumorigenesis. Overall SCNA (aneuploidy) level was associated with increased CD3+ and CD8+ T cell microenvironments in precancer (mostly CD3+, linked to trisomy and aneuploidy), but with T cell-deficient tumors. Early lesions with 9p21.3 loss were associated with depletion of cytotoxic T cell infiltration in TP53 mutant tumors; and with aneuploidy were associated with increased NK-cell infiltration. The strongest driver of cytotoxic T cell and Immune Score depletion in oral cancer was 9p-arm level loss, promoting profound decreases of pivotal IFN-γ-related chemokines (e.g., CXCL9) and pathway genes. Chromosome 9p21.3 deletion contributed mainly to cell-intrinsic senescence suppression, but deletion of the entire arm was necessary to diminish levels of cytokine, JAK-STAT, and Hallmark NF-κB pathways. Finally, 9p arm-level loss and JAK2-PD-L1 codeletion (at 9p24) were predictive markers of poor survival in recurrent HPV− HNSC after anti–PD-1 therapy; likely amplified by independent aneuploidy-induced immune-cold microenvironments observed here. We hypothesize that 9p21.3 arm-loss expansion and epistatic interactions allow oral precancer cells to acquire properties to overcome a proimmunogenic aneuploid checkpoint, transform and invade. These findings enable distinct HNSC interception and precision-therapeutic approaches, concepts that may apply to other CN-driven neoplastic, immune or aneuploid diseases, and immunotherapies.
The genetic bases for predisposition, and neoplastic transformation, to cancer have been increasingly well described. However, it remains less clear how early, precancer cells employ these genetic alterations to acquire the characteristic features and properties (1) of malignant disease. For example, studies of the immune landscape led to breakthrough trials of programmed death-1 (PD-1) inhibitors for recurrent, metastatic head and neck squamous cell carcinoma (HNSC) therapy (2–4). This underscores the importance of immune modulation in these tumors, despite a still suboptimal overall response rate of less than 20% in advanced cancers. Immune response within tumors has been observed to be strongest at the earliest neoplastic stages, as reported recently in lung adenocarcinoma precursors (5). As such, new, immune-based strategies could be developed to reduce the high global burden of HNSC, by intercepting the most common precursor of the most common HNSC presentation: HPV− oral squamous cell carcinomas (6–8).
Studies of chromosome somatic copy-number (CN) alteration (SCNA) profiles have reported the impact of 3p14, 9p21, or 17p13 loss in molecular models of HNSC progression (9) and risk (10–15). Early studies reported that patients with oral precancers harboring 9p21 and/or 3p14 loss were at significantly greater cancer risk than those with retention at these loci (10, 16). A comprehensive, prospective validation study examined the relative contribution of six candidate chromosome-arm regions. 9p21 loss had the greatest influence on cancer risk (13). The mechanism underlying the association between CN and malignant transformation of precancers, however, is unclear (17–20). Studies of CN-altered neoplastic cells have shown that SCNAs can trigger a cytotoxic response in experimental precancer systems (21, 22) but, paradoxically, were associated with immune evasion (23) and suppression (24) in computational studies of naturally occurring human cancers. The latter, in melanoma, found that nonresponders to PD-1 and CTLA-4 blockade had higher CN alteration and loss burdens, which correlated with immunologically cold tumors, characterized by cytotoxic-cell, marker, and metric reductions, and suppressive microenvironment cell, network, and signal increases (23–26). This SCNA-cold association was particularly strong in our previous, pan-The Cancer Genome Atlas (TCGA) computational study in HNSC (23). These data point to a putative in vivo switch from immune hot-to-cold in the precancer–cancer transition, and raise the hypothesis that SCNAs in precursor lesions contribute to malignant transformation through genomic events and mechanisms that enable the acquisition of immune-suppressive, evasive properties. To test this hypothesis, we evaluated CN influence on immune profiles and outcomes in a large prospective oral precancer patient cohort, and HPV− HNSC (tissue specimens and cell lines) and anti–PD-1–treated recurrent-disease cohorts.
Results
Oral Precancer Prospective Cohort.
Demographic, CN, and immune studies.
We assembled a clinical cohort of 188 HPV− oral precancer patients (median age 56-y-old, range 23 to 82 y) who were systematically followed and rigorously annotated until reaching the protocol-specified primary endpoint of invasive cancer (SI Appendix, Table S1A). The 5-y oral cancer-free survival (OCFS) for this population was 71.8%, with a median follow-up time for censored observations of 90.8 mo. Multiplex immunofluorescence was used to assess the amount of infiltrating CD3+, CD8+, and CD68+ cells. This investigation was focused on the association of immune-cell infiltrate with three SCNA metrics: 1) reported major oral-cancer risk SCNAs, CN loss at 3p14, 9p21.3, and 17p13.1 (10, 13, 15); 2) chromosome-7 (chr7) gain [extra copies of this, or other single-chromosome gains, have similar precancer phenotypes, and are thus used to mark lesion trisomy or tetrasomy (11)]; and 3) overall SCNA or aneuploidy level. The latter comprised all CN events, including CN loss at major and minor risk regions on seven chromosome arms plus chr7 gain, in our precancer molecular profiling platform (Methods). CN loss at 3p14, 9p21.3, or 17p13.1, or chr7 gain tended to co-occur, particularly 3p14 loss and 9p21.3 loss with a P = 1.29E-07 (event frequency and co-occurrence rates) (SI Appendix, Table S1B).
In precancer, SCNA level, especially chromosome trisomy/tetrasomy, was associated with increased CD3+ and CD8+ T cell infiltrates.
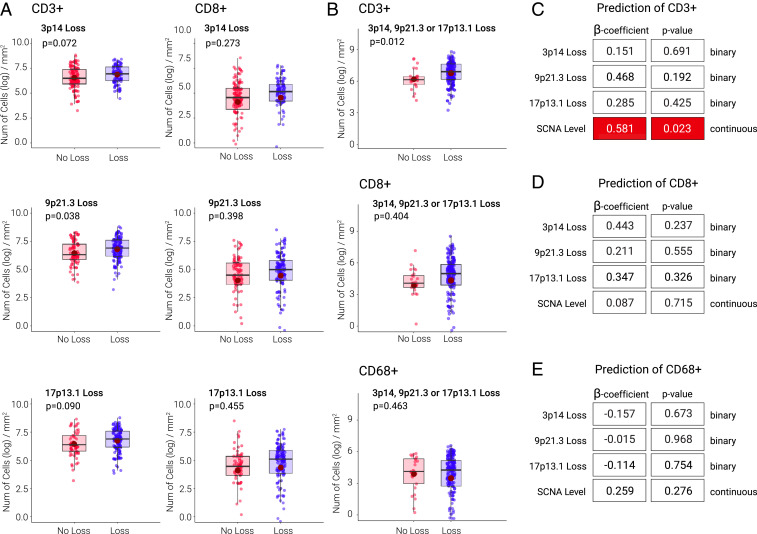
We first correlated major-risk SCNAs (i.e., 3p14, 9p21, and 17p13 loss) with immune-cell infiltrate (Fig. 1 and SI Appendix, Table S2). When considered individually (in univariate analysis), 9p21.3 loss was associated with a near twofold increase in CD3+ cell density (P = 0.038) (Fig. 1A and SI Appendix, Table S2A); 3p14 loss (P = 0.072) and 17p13.1 loss (P = 0.090) were associated with increased CD3+ cell trends. Overall SCNA level was associated with increased CD3+ (P = 0.023), but not CD8+ (P = not significant), cell infiltrate. Chr7 trisomy and tetrasomy was strongly associated with immune-hot precancer lesions: CD3+ (P = 0.0004) and CD8+ (P = 0.015) T cell infiltration in univariate, 35% mixed-effect models (confirmed with median, 50%, cutoff) (SI Appendix, Table S2C). Since SCNAs tended to co-occur (SI Appendix, Table S1B), we performed multivariable logistic regression to assess the contribution of each individual CN alteration and overall SCNA level to cell infiltrates, independent of the others (Fig. 1 C–E and SI Appendix, Table S2). Chromosome gain (trisomy, tetrasomy) was the dominant driver of immune-cell infiltration in precancer lesions in multivariable analysis (CD3+, P = 0.008; CD8+, P = 0.058; CD68+, P = 0.051) (see right side of SI Appendix, Table S2B), contributing to the overall SCNA level-hot precancer association. When chr7 gain or SCNA level were taken into account, 3p14, 9p21.3, or 17p13.1 did not correlate with CD3+, CD8+, or CD68+ cell levels (Fig. 1 C–E and SI Appendix, Table S2B). In every precancer analysis—regardless of CN or immune metric, cutoff (and whether univariate or multivariate)—SCNAs were never associated with a decreased T cell count in the microenvironment.
Fig. 1.
Precancer SCNA and immune-cell infiltrate associations. (A) Distributions of CD3+ and CD8+ cells by 3p14, 9p21.3, and 17p13.1 loss and P values for comparing the two groups for each covariate using the linear mixed-effect models on log-transformed data. Note that throughout the report, and in all main and supplemental figures and tables, “9p21.3” (3p14 or 17p13.1) loss means either focal or arm loss (i.e., deletion at 9p21.3 region could derive from arm or focal events), unless specifically qualified as “focal only.” (B) Distributions of CD3+, CD8+, and CD68+ cells by any loss of 3p14, 9p21.3, 17p13.1, and P values for comparing the two groups for each covariate using the linear mixed-effect models on log-transformed data. (C–E) Coefficients of estimates (SE) and P values from multivariable logistic regression models of CD3+, CD8+, and CD68+ cells dichotomized based on top and bottom 35% of the distribution. Each model includes 3p14, 9p21.3, or 17p13.1 loss and SCNA level as independent variables. A positive coefficient indicates that patients with the SCNA event were more likely to have higher CD3+, CD8+, and CD68+ cell levels than those without the event; statistically significant positive associations as in C are indicated by dark red box. Findings were confirmed by dichotomizing the parameters based on median, in addition to top and bottom 35%, of the distribution.
9p21.3 loss (followed by dysplasia) was the strongest, independent prognostic marker of poor OCFS.
The presence of dysplasia in oral precancer has been previously associated with cancer risk (6–8). As expected (12), dysplastic lesions harbored more SCNAs overall and 9p21.3 loss. We observed a disproportionately stronger SCNA–dysplasia association with CD3+ (vs. CD8+) cell levels, possibly reflecting increased CD3+/CD4+ Tregs in histologic high-grade lesions (Fig. 1 A–D and SI Appendix, Table S2 A–C). Compared to nondysplastic lesions, dysplasia was associated with an increase in CD3+ cells (P < 0.01) (SI Appendix, Table S2A). To determine the contributions of each clinical, histological, genomic, and immune marker in predicting outcomes, we performed a univariate analysis of OCFS. Four parameters predicted poor OCFS: histology (dysplasia versus hyperplasia; β = 0.693, P = 0.009, hazard ratio [HR] = 2.000, 95% CI = 1.180 to 3.370), 9p21.3 loss (β = 0.659, P = 0.011, HR = 1.932, 95% CI = 1.160 to 3.200), chr7 gain (β = 0.601, P = 0.010, HR = 1.824, 95% CI = 1.151 to 2.889), and overall SCNA level (β = 0.372, P = 0.010, HR = 1.451, 95% CI = 1.092 to 1.927) (SI Appendix, Table S2D).
Next, we built a multivariable Cox-proportional hazard model—including histology, three major risk SCNAs, chr7 gain and overall SCNA—and demonstrated that only dysplasia and 9p21.3 loss were individually associated with poor OCFS (SI Appendix, Fig. S1 and Table S2E). Patients with both dysplasia and 9p21.3 loss had the lowest OCFS (P = 0.0001) (SI Appendix, Fig. S1B), statistically significantly lower OCFS than patients with dysplasia or 9p21-loss alone (P = 0.014 or P = 0.012, log-rank test, respectively). Including age, gender, smoking status, alcohol use, T cell infiltrates, major CN-loss loci, overall SCNA level, or chr7 gain in eight distinct multivariable prediction models revealed 9p21.3 loss as having the most consistent, significant, negative association with OCFS, followed by, and independent of, histology (SI Appendix, Fig. S1C and Table S2E).
Oral Cancer–HPV− HNSC (TCGA).
In TCGA HPV− HNSC, SCNAs were associated with decreased T cell infiltrate: Pivotal role of 9p loss.
The comprehensive genomic and transcriptomic data from TCGA was leveraged to investigate in 343 HPV− HNSC samples the associations of 3p14, 9p21.3, and 17p13.1 loss and SCNA level with tumor-cell infiltrates and the previously described immune score (IS) (23). The IS was based on the expression level of cytotoxic T cell markers, consistent with metrics utilized in other reports (24–29). We used RNA expression levels of CD3D, CD8A, or CD68 as a proxy to recapitulate in HNSC the analyses performed on oral precancer biopsy samples for CD3+, CD8+, and CD68+ levels (SI Appendix). Expression levels of CD3+ and CD8+, but not CD68+ (Pearson’s r = 0.34), strongly correlated (Pearson’s r = 0.91 and 0.93 for CD3D and CD8A, respectively) with IS (SI Appendix, Fig. S2).
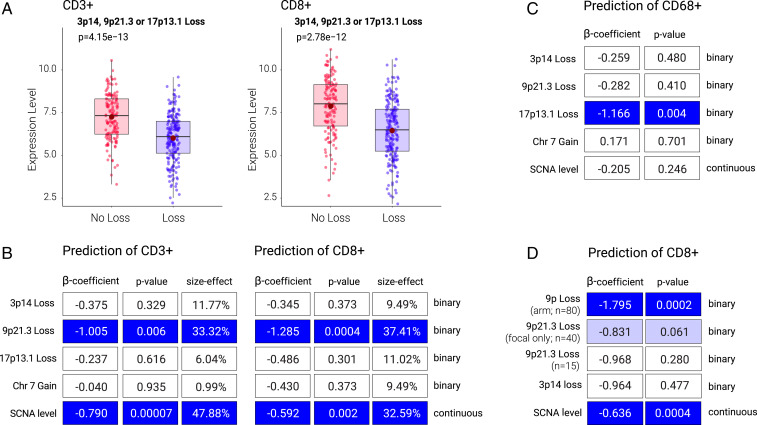
To define loss at 3p14, 9p21.3, or 17p13.1 loci, we considered purity-adjusted CN measured in the same genomic location used to probe loss in precancers (using pathology- and ABSOLUTE-based purity estimates) (Methods). Unless otherwise specified, loss of a genomic region (for example “9p21.3 loss”), refers to a loss at this region irrespective of the length of the deletion (e.g., whether focal or arm). In all cases, analyses were performed considering both specific-risk CN (e.g., 9p21.3 loss) as a binary variable (presence or absence of SCNA) and CN as a continuous variable (log-transformed CN level). 3p14, 9p21.3, and 17p13.1 loss were most associated with SCNA level, and CN arm losses (versus gains) (SI Appendix, Fig. S4). Chr7 gain, 9p loss and SCNA level (using cancer and precancer metrics) were associated with reduced CD3+/CD8+ cell numbers, activation markers (granzyme B [GZMB], interferon-γ [IFNG]), and IS (Fig. 2 and SI Appendix, Fig. S3).
Fig. 2.
HPV− HNSC SCNA and immune-cell infiltrate associations. (A) Relationship between expression level of CD3+ (CD3D gene) and CD8+ (CD8A gene) with CN loss (arm or focal) at 9p21.3, 3p14, or 17p13.1. P value is from Wilcoxon test. (B) Multivariable logistic regression model for the prediction of CD3+ and CD8+ cell levels. Variable importance (size effect) is also shown. (C) Multivariable logistic regression model for the prediction of CD68+ cells. (D) Multivariable logistic regression similar to B but considering 9p (arm) loss, 9p21.3 (focal) loss, and 9p21.3 (arm and focal) loss as separate variables. Note that in B–D the dark blue boxes mark significantly decreased (immune cold) associations of the CN metric with immune cell indicated; light blue boxes mark marginally statistically significant immune-cold associations (e.g., panel D; p=0.061) for 9p21.3 focal-loss association with CD8+.
Since individual SCNAs tended to co-occur (e.g., 3p14 loss and 9p21.3 loss, P = 1.73E-26) (SI Appendix, Table S3F) and were associated with overall SCNA level, we determined the individual contribution of each major CN event in predicting immune marker and infiltrate parameter. Multivariable logistic regression was performed to predict the level of infiltrating CD3+ and CD8+ T cells, dividing the tumors into those containing high or low levels using predefined 35% and median cutoffs (SI Appendix, Table S3 A–E and G–L). As predictive elements, CN metrics included those used in the precancer cohort: 3p14, 9p21.3, or 17p13.1 loss, chr7 gain, and overall SCNA level. According to this model, 3p14 or 17p13.1 losses were not statistically significantly associated with CD3+ or CD8+ T cell infiltration (Fig. 2B and SI Appendix, Table S3A), IS, or GZMB/IFNG (SI Appendix, Table S3A and Fig. S3). In contrast, 9p21.3 loss had a strong negative association with CD3+ and CD8+ T cell infiltrates (β = −1.005, P = 0.006, odds ratio [OR] = 0.366, 95% CI = 0.178 to 0.744 for CD3+; and β = −1.285, P = 0.0004, OR = 0.277, 95% CI = 0.133 to 0.562 for CD8+) (Fig. 2B and SI Appendix, Table S3A). Importantly, this robust association was independent of overall SCNA level, which was also statistically significantly negatively associated with CD3+ and CD8+ T cell infiltrates (Fig. 2B). Although immune-cold in univariate analysis (SI Appendix, Fig. S3C), the chr7-tumor microenvironment signal was lost in multivariable analyses, which included SCNA level (Fig. 2B and SI Appendix, Table S3A). Neither 9p21.3 loss nor overall SCNA level was associated with CD68+ cells (Fig. 2C and SI Appendix, Table S3 A and B). Analyses using CN metric for major or minor loci loss and overall (precancer- and HNSC-defined) SCNA levels as a continuous variable (in addition to binary loss/gain classification based on a CN threshold) and with purity correction (using ABSOLUTE) gave similar results. 9p21.3 loss and SCNA level were identified as the two strongest predictors of T cell depletion (SI Appendix, Table S3 C and G). Size-effect analysis showed that 9p21.3 loss could explain 33.32% of the variance for CD3+ and 37.41% for CD8+ T cells (Fig. 2B). These results were similar when using fivefold cross-validation and adding other covariates, such as age, gender, TP53, and stage (SI Appendix, Table S3K).
We note that 3p14 loss was statistically significantly associated with immune-cold tumors in only one of the models tested here (continuous-variable model, using pathology-based estimate for purity correction). 3p14 loss associated with lower CD8+ (β = 0.583, P = 0.007, OR = 1.791, 95% CI = 1.174 to 2.765) and CD3+ (β = 0.585, P = 0.003, OR = 1.796, 95% CI = 1.231 to 2.662) T cells and IS (β = 0.601, P = 0.005, OR = 1.824, 95% CI = 1.208 to 2.801) (SI Appendix, Table S3B). 3p14 loss association with immune markers (β-coefficient) was weaker than that of 9p21.3 loss in every analysis conducted here. The analysis of arm- versus focal-level events in the continuous-data model showed a 3p-arm loss association with low CD3+ (P = 0.030) (SI Appendix, Table S3H), and a trend for CD8+ (P = 0.091) (SI Appendix, Table S3D) cell infiltrates (using standardized CN values). Furthermore, 3p14 focal-only loss also showed a trend in the continuous-data model for CD3+ (P = 0.078) (SI Appendix, Table S3H) but not for CD8+ (P = 0.15) (SI Appendix, Table S3D) cells. Neither 17p-arm nor 17p13.1-focal loss were associated with CD3+ and CD8+ T cells (SI Appendix, Table S3 E and I).
To match the oral-precancer data by HNSC anatomic subsite, we repeated the analyses of CD8+, CD3+, and CD68+ cells, IS, and arm/focal analysis conducted for all HPV− HNSC (SI Appendix, Table S3), for the oral-cavity cancer-only subset in TCGA HPV− HNSC samples. The latter primary tumor samples were obtained from the alveolar ridge, buccal mucosa, floor of mouth, hard palate, lip, oral tongue, or oral cavity (not otherwise specified). After this filter, the sample number decreased from 343 to 232, and the associations remained similar to that observed by considering all TCGA HPV− HNSC samples (see SI Appendix, Table S4 for oral-cavity only). Taken together, these data indicate that 9p loss in HPV− HNSC is linked to immune-exclusion, cold microenvironments. The 9p linkage, unlike 3p or 17p, was strong, specific, and consistent in every cancer-association analysis, whether it be univariate or multivariable, independent of threshold/cutoff and type of parameter (binary or continuous) or anatomic subsite. The linkage was confirmed in two purity-correction methods.
The association between 9p21.3 loss and immune-cold HPV− HNSC: Driven mainly by entire 9p-arm loss.
Loss at 9p21.3 can occur due to loss of the entire 9p arm, focal region at 9p21.3, or both (where arm and focal events are present in different chromosome copies). In all of the analyses described so far, SCNA was assessed at a specific genomic locus (e.g., 9p21.3) without information on deletion size. In precancer, estimates of CN level at 9p21.3 (by PCR) did not allow a distinction between arm or focal loss. However, in the context of HNSC data, this distinction was possible. Thus, we set out to determine the contribution of arm- versus focal-level loss to the association between 9p21.3 loss and immune infiltrates. Specifically, we assessed 9p21.3 loss coming from arm-level events (9p arm loss) and 9p21.3 loss derived from focal-level only events. A profound effect of 9p arm-level loss was observed in predicting low CD3+ (β = −1.568, P = 0.0008, OR = 0.209, 95% CI = 0.080 to 0.508) (SI Appendix, Table S3G) and CD8+ (β = −1.795, P = 0.0002, OR = 0.166, 95% CI = 0.061 to 0.413) (SI Appendix, Table S3C) T cell infiltrate, while the 9p21.3 focal-level event showed nonstatistically significant decreases of CD8+ and CD3+ T cell infiltration (Fig. 2D and SI Appendix, Tables S3 C and G).
Similar results were obtained when considering deletion of specific candidate genes on 9p—specifically CDKN2A, IFNA, JAK2, and CD274—instead of the entire 9p21 or 9p24 regions. In all cases, arm-level deletion was a stronger predictor (in terms of β-coefficient and P value) of CD3+ and CD8+ cells (SI Appendix, Table S3L), than focal deletion of these specific genes. Furthermore, in the Lasso classification method, 9p loss was the top scoring parameter selected to predict IS (P < 0.001) across all possible arm-level losses or gains (SI Appendix, Table S5). In this analysis, neither 3p nor 17p loss were found to be statistically significantly associated with CD3+ or CD8+ cell infiltrates. In analyses of whole-chromosome associations, chromosome-9 loss was the only event significantly associated with low CD3+/CD8+ T cell infiltrates and IS (P = 0.008). This whole-chromosome association was seemingly due to the strong negative influence of 9p-arm loss on T cell infiltrates, as the association of 9q loss with T cell infiltrate was not statistically significant (SI Appendix, Table S5). The phenotype is likely due to the cumulative effect of loss of many interacting or cooperating 9p genes outside of 9p21.3 (SI Appendix, Fig. S5).
The association between 9p loss and immune infiltrate: Influenced by stage and TP53 mutation.
To test whether tumor microenvironment association was influenced at the early-invasive transition and by disease extent, we stratified tumors by stage and found that the negative association of 9p21.3 loss, 9p arm-level loss, and SCNA level with CD3+/CD8+ levels was no longer statistically significant in early HNSC (SI Appendix, Figs. S6 A and B, S7 A and B, S8 A and B and Table S6A). Since in dysplastic precancer, SCNA level was associated with a predominant CD3+ cell increase, we inferred that specific immune-cell types may be associated with SCNA level in early-stage HNSC (SI Appendix, Table S2A). The CIBERSORT algorithm (30) was used to estimate the fraction of different cell types in the tumor microenvironment by CN metric. Natural killer (NK)-cell levels were increased in SCNA-high tumors, specifically in early-stage primary disease (P = 0.007) (SI Appendix, Fig. S7D). Chr7 gain did not show any stage-specific differences in NK or CD8+ cells. CIBERSORT analysis of CD8+ T cell levels, however, revealed a stage-specific pattern opposite to that of NK cells; that is, CD8+ levels were negatively associated with SCNA level (and 9p21 loss) in advanced (vs. early) stage HNSC samples, a finding confirmed by logistic regression, purity-controlled analysis (SI Appendix, Fig. S7C and Table S6A). The association of 9p21.3 or 9p loss with CD3+ and CD8+ T cells was stronger in TP53 mutant than wild-type tumors, especially in early-stage lesions (SI Appendix, Figs. S6 C and D, S8 C and D, and Table S6 B–D).
Effect of SCNAs on immune infiltrate in HPV+ HNSC.
We investigated SCNA frequencies and immune-cell associations in all 36 HPV+ HNSCs available from TCGA. Compared to HPV− tumors, HPV+ tumors showed a similar frequency of 17p13.1 loss but lower frequencies of both 3p14 and 9p21.3 loss (SI Appendix, Table S7A). Logistic regression for the prediction of CD3+, CD8+, and CD68+ cells showed that none of the CN parameters or metrics studied here were statistically significantly associated with any of these three cell types in HPV+ tumors (SI Appendix, Table S7 B and C).
HPV− HNSC Cell Lines.
9p loss in HPV− HNSC cell lines was associated with decreased SASP, JAK-STAT, and cytokine pathways.
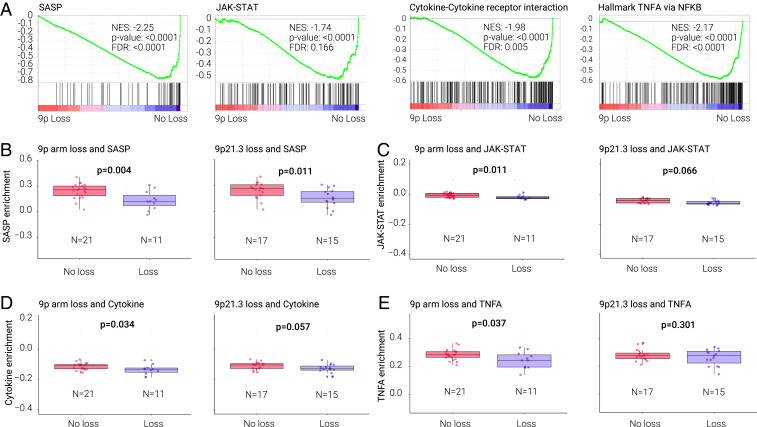
We next studied gene-expression profiles in HNSC cell lines (SI Appendix, Table S8), both containing or not containing CN alterations associated with T cell or IS suppression in the TCGA HPV− HNSC analyses above. Although in vitro cell-line studies do not allow analyses of interactions with immune cells, they can identify cell-autonomous mechanisms that may mediate tumor-immune interactions. We performed gene-set enrichment analysis (GSEA) on transcriptomes of a set of 32 HPV− HNSC cell lines (SI Appendix, Tables S9–S14), comparing cell lines with or without 9p-arm or 9p21.3 loss. Similar analyses were also performed considering 3p-arm or 3p14 loss. Among the top 10 pathways depleted in cell lines containing 9p-arm loss, 7 were associated with immune processes or interactions. Three of the most significantly depleted pathways in cell lines containing 9p-arm loss were the senescence-associated secretory pathway (SASP) (false-discovery rate [FDR] P < 0.0001), cytokine–cytokine–receptor interaction (FDR = 0.005), and JAK-STAT signaling pathways (FDR = 0.166) (Fig. 3A and SI Appendix, Table S14). These pathways represent interrelated gene sets that encode for mostly secreted molecules that promote tumor-immune infiltration. A similar gene-set analysis showed that the top depleted Hallmark pathway was TNFA Signaling via NF-κB (FDR < 0.0001) (Fig. 3A). In contrast, 3p14 or 3p loss were not associated with depletion of these or other pathways related to immune processes, but instead, associated with enrichment of cytokine–cytokine–receptor interaction and JAK-STAT Signaling (FDR = 0.084 and 0.231, respectively) pathways, and proinflammatory molecule expression (SI Appendix, Table S11).
Fig. 3.
SASP, JAK/STAT, cytokine receptor, tumor necrosis factor-α (TNFA) signaling via NF-κB pathways in relationship with 9p or 9p21.3 loss in HPV− HNSC cell lines (A) Pathway depletion from GSEA analysis of HPV− HNSC cell lines comparing lines with and without 9p loss. GSEA plots, NES (normalized enrichment score), P value, and FDR are shown for and SASP, JAK/STAT, cytokine–cytokine receptor, and TNFA signaling via NF-κB pathways. (B–E) Relationship between gene expression of the SASP (B), JAK/STAT (C), cytokine–cytokine receptor pathway (D), and TNFA signaling via NF-κB (E) (ssGSEA analysis) and 9p arm or 9p21.3 loss. P value is from Wilcoxon test.
The expression of interferon (IFN)-α molecules was lower in tumors (TCGA) containing 9p21.3 loss, but not in cell lines harboring 9p21.3 or 9p loss, consistent with an extrinsic microenvironment effect (SI Appendix, Fig. S10). JAK2 was also decreased in TCGA samples (but not cell lines) harboring 9p loss. Single-sample GSEA (ssGSEA) (31) analysis confirmed that 9p-arm loss was associated with suppressed SASP, JAK-STAT, cytokine–cytokine–receptor interaction, and TNFA signaling via NF-κB pathways (Fig. 3 B–E). Since the TCGA analysis above suggested that 9p21.3 loss contributes to immune-cold tumors promoted by 9p-arm deletion, we examined this association in the cell lines. GSEA analysis comparing cell lines with or without loss at 9p21.3 showed profound depletion of SASP (FDR < 0.0001), and a trend toward decreased JAK-STAT and cytokine pathways (Fig. 3 B–E and SI Appendix, Table S10). These cell-line results suggest that 9p21.3 deletion contributes mainly to the cell-intrinsic SASP suppression, while deletion of the entire 9p arm is necessary for cell-line suppression of cytokines and other molecules related to cytokine, JAK-STAT, and NF-κB pathways. Compared to SASP, 9p-loss associated down-regulation of the cytokine–cytokine–receptor interaction, JAK-STAT, and NF-κB pathways were stronger in the tumor samples (SI Appendix, Fig. S9) than cell lines (Fig. 3). Immune-regulatory genes (belonging to one or more of the four pathways) down-regulated in cell lines and tumors containing 9p loss, were: CCL2, CCL24, CSF3R, KDR, IER5, IRF1, IL2RG, LTB, MCL1, SAT1, TNFSF10, and TNFRSF6B (SI Appendix, Fig. S11). Molecules specifically decreased in tumors but not cell lines (SI Appendix, Fig. S11) notably included CCL19 (log2 fold-change [log2FC] = −1.347, FDR = 1.74E-06), CCL21 (log2FC = −0.894, FDR = 0.001), CXCL9 (log2FC = −2.040, FDR = 1.46E-15), and CXCL10 (log2FC = −2.513, FDR = 1.44E-20) (Dataset S1).
We performed logistic regression model analysis for the prediction of IS, taking into account SCNA level (as a covariate), to define potential candidate genes on 9p. Based on the logistic model, 84 genes were associated with IS (FDR < 0.001) and among these, 42 showed significance (FDR < 0.001) in at least two of the following parameters: correlation between RNA level and IS in tumors, between RNA level and DNA CN in tumors, or correlation between RNA and DNA CN in cell lines (Dataset S2). These genes—which contained JAK2 and CDKN2A (32)—included 21 genes in 9p13, 5 in 9p21, 6 on 9p22, and 10 genes on 9p24. New candidate genes identified in cell lines—such as RANBP6, IL33, and the SUMO E3 ligase TOPORS—can decrease proinflammatory and proimmunogenic molecules, and thus promote immune escape (FDR < 0.0001) (SI Appendix, Table S24). In sum, these data indicate that in cancer-cell lines, 9p-arm deletion of key immunoregulatory genes can lead to T cell depletion observed in tumors. Deletion of individual regions located on 9p, such as 9p21.3 or 9p24, were not sufficient to recapitulate the effects observed in cell lines harboring deletion of the entire arm, suggesting a cumulative effect of sets of genes located on this chromosome arm.
Real-World Evidence Cohort.
Chromosome-9p loss predicted survival of HPV− HNSC patients after immunotherapy.
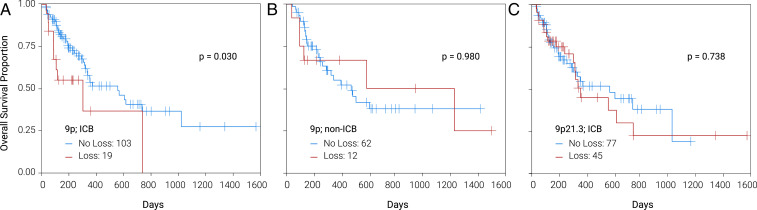
Based on the strong association between 9p loss and immune depletion and IS in TCGA and cell-line analyses, respectively, we examined whether there was a correlation between 9p loss and patient survival after immunotherapy in a real-world evidence (RWE) cohort. The independent deidentified RWE dataset contained genomic profiles annotated with clinical outcomes data (SI Appendix, Fig. S12). Briefly, this cohort included 196 HPV− HNSC patients who received first- or second-line anti–PD-1 checkpoint therapy (pembrolizumab, nivolumab) or chemotherapy (with no prior or subsequent immunotherapy). Kaplan–Meier survival plots in this patient cohort with 9p loss treated with anti–PD-1 therapy, or chemotherapy alone, are shown in Fig. 4; 9p21, 9p13, and 9p24 chromosomal loss or gene-deletion survival analyses are shown in SI Appendix, Table S25. Within the context of the above PD-1 inhibitor therapy, median survival was longer in patients without 9p loss (HR = 0.468, 95% CI = 0.232 to 0.944, log-rank P = 0.03) (Fig. 4A). To assess whether this survival difference reflected a generalized prognostic association (unrelated to PD-1 blockade), we analyzed survival in patients treated with chemotherapy only (i.e., not treated with PD-1, checkpoint, inhibitors) and observed no difference between 9p loss vs. no-loss groups (P = 0.98) (Fig. 4B). These results indicated that 9p loss was a strong, specific predictive marker of clinical benefit from PD-1 inhibitors. In contrast, 3p, 17p, or 9p21.3 loss were not predictive (of anti–PD-1 therapy efficacy) (Fig. 4C), or prognostic in patients who were not treated with immune-checkpoint blockade (ICB) (SI Appendix, Table S25).
Fig. 4.
Chromosome 9p loss in (A) ICB and (B) non-ICB-treated HPV− HNSC; focal 9p21 loss in ICB-treated patients (C).
PD-L1 and PD-L2 expression in tumor cells have been associated with benefits from nivolumab and pembrolizumab in HNSC patients (2–4, 26, 33). To assess whether the predictive effect of 9p-arm loss could be entirely attributed to loss of the PD-L1 (or PD-L2) gene, we evaluated survival in anti–PD-1–treated patients according to the presence of PD-L1/-L2 gene deletion. Borderline predictive associations of PD-L1, -L2, and JAK2 deletion (P = 0.071–0.081) (at 9p24) were observed; while combined deletion of all three genes or 9p24.1 locus associations with ICB survival were P = 0.018 and P = 0.028, respectively (SI Appendix, Table S25). In regard to the strongest CN ICB-predictive marker in this study, patients with PD-L1-JAK2 codeletion, had an inferior median survival vs. those without this codeletion (6 vs. 19 mo, P = 0.007). As with 9p loss, deletions of PD-L1, PD-L2, JAK2, and 9p24, as well as PD-L1-JAK2 codeletion, were not prognostic in non-ICB–treated patients (SI Appendix, Table S25). Tumor-cell PD-L1 IHC was associated with 9p loss—negative (<1%) PD-L1 was more common in patients with (58%) versus without (21%; P = 0.013) 9p loss. Survival analysis of ICB-treated patients according to PD-L1 found a HR of 0.83 (95% CI 0.34 to 2.03) for the PD-L1 positive versus negative group (P = 0.682). Tumor mutational burden (TMB; threshold of 10 mutations per megabase) was not associated with 9p loss or survival differences after PD-1 blockade (HR = 0.762; 95% CI 0.423 to 1.372; P = 0.364). None of our samples displayed microsatellite-instability high.
Discussion
Aneuploid Checkpoint and Immune Microenvironment Switch in Precancer–Cancer Transition.
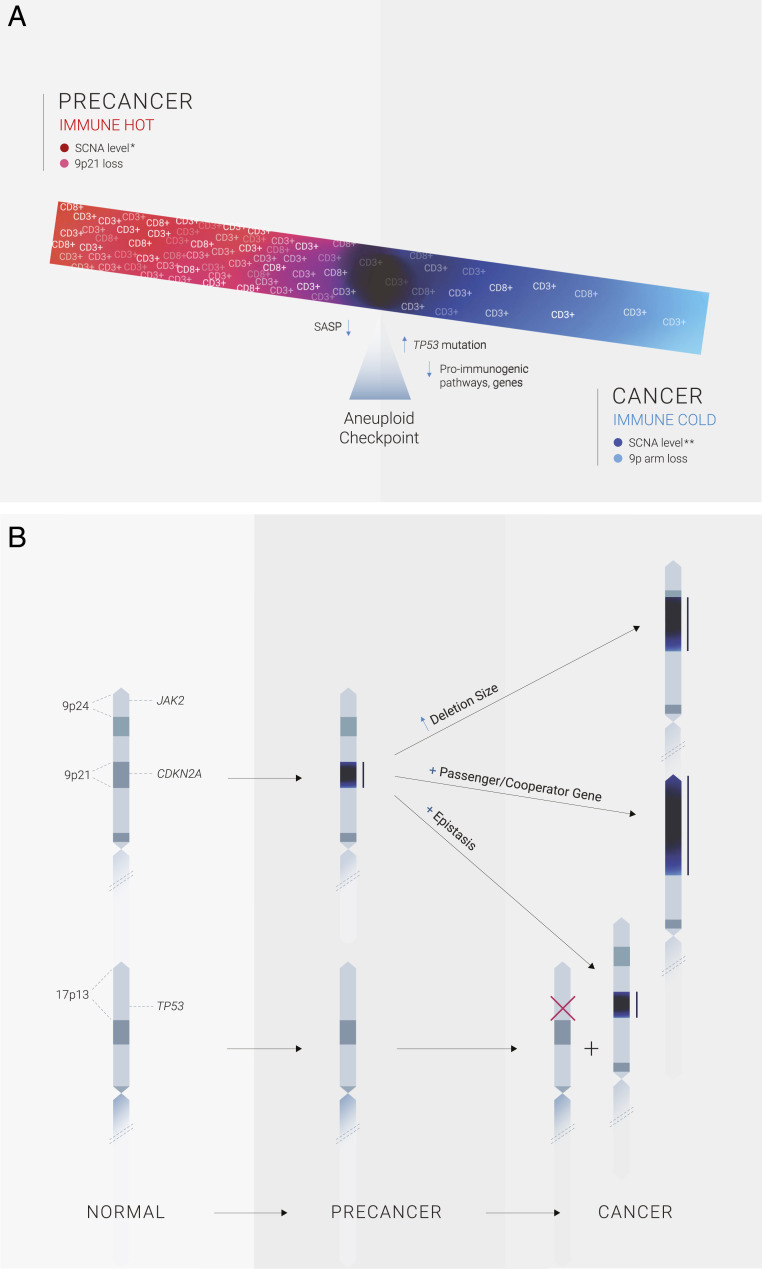
We report here that chromosome-dosage imbalance contributes to an immune T cell switch during human HPV− head and neck tumorigenesis (Fig. 5A). Experimental cell transfer of single chromosomes, or genome-engineered focal alterations, trigger cytotoxic responses, regardless of the cell system or identity of the extra chromosome (24, 34, 35). Supporting these preclinical experiments, we found that in clinical oral precancer lesions single-chromosome trisomy/tetrasomy correlated with increased CD3+ and CD8+ T cell infiltrates, which contributed to overall SCNA level-hot associations. This raises the possibility that CN-altered preneoplastic cells elicit host recognition. The opposite SCNA-cold phenotype was observed in oral cancers, mostly with 9p-arm loss, marked by profound suppression of tumor-infiltrating cytotoxic T cell number and activation markers (GZMB, IFNG, IS) (Fig. 2 B and D and SI Appendix, Fig. S3 and Table S3 A–C). This posits that during tumorigenesis, CN-generated immunogenic precancer cells acquire cold states, properties that allow escape of an aneuploid checkpoint immune-surveillance, -response, and eventual invasion and metastatic spread (24, 36, 37). The latter is consistent with recent studies of metastatic colorectal and renal cancer, which found that metastases that persisted or progressed were the least immunogenic (38), some harboring large-scale CN aberrations (notably 9p loss) (39), and other mechanisms of immune escape. Tumor evolution and selection of cells with further, complex, karyotypic imbalance and pivotal mutations (TP53) likely augment this process. These mechanisms have been observed under conditions of prolonged in vitro culture stress (24, 36) and in vivo immune pressure (37). These data uncover a striking context-dependent switch of SCNA influence on neoplastic microenvironments during oral precancer-to-cancer transition, progression, and metastasis.
Fig. 5.
Immune hot-to-cold aneuploid switch model of oral precancer-to-cancer transition. (A) In precancer, the dominant CN driver of immune-hot lesions was chromosome gain (trisomy, tetrasomy) and 9p21 loss (Left). Immunogenic precancers with SCNAs must acquire other genomic alterations (see B) in order to overcome an aneuploid checkpoint, or barrier (Center), that then leads to immune evasion (CD3+/CD8+ T cell depletion) (Right) during the precancer-to-cancer transition. Tumors with complex karyotypes and other key, associated genomic events—notably TP53 mutation or loss (associated with aneuploidy in human tumors) or downstream pathway inactivation—could mask SASP, cGAS–STING, or other cell-intrinsic responses triggered by aneuploid cells (Center, left of aneuploid checkpoint), and shed light on the pivotal influence of CXCL9 and related chemokines on cytotoxic T cell recruitment, and other immune regulatory pathways, during oral precancer progression to invasive disease, the latter associated with profound reductions in CXCL9/10 levels in tumors with 9p loss (see Discussion). (B) Possible genetic mechanisms for the aneuploid switch involving early loss of chromosome 9p21 and augmentation or expansion of deletion size in later stages of HPV− head and neck tumorigenesis. (Right, Top) Increased deletion size in cancer. (Right, Center) expanded 9p-arm loss to encompass and simultaneously affect hundreds of neighboring, interacting CN-altered genes could possibly support the complex transformational (and metastatic) cascade by altering many functional, e.g., immune, phenotypes. The 9p genes involved depend on whether the deletion expansion is restricted to 9p21 (and includes CDKN2A), is telomeric (in this illustration, could include JAK2 on 9p24) or centromeric (and so could include SASP-related cytokine-cytokine pathway genes in 9p13) (Dataset S2). (Right Bottom) Epistatic interactions between 9p21 loss and events elsewhere in the cancer genome (in this illustration, inactivating mutation or loss of the TP53 gene on 17p13). Chromosome loss regions are indicated in dark blue and delineated by adjacent solid black lines. Mutation is indicated with a red “X.”
Role of 9p Loss in Immune Evasion.
Our data, consistent with a previous study (13), indicate that 9p21 loss in oral precancer is the major genomic driver of cancer risk. Contributing to its pivotal function in tumorigenesis and malignant transformation, we documented an integral role for 9p loss in immune escape (Fig. 2 B and D). In HPV− HNSC, a profound influence was observed of 9p loss, but not 3p or 17p, on immune-exclusive, cold microenvironments. These results also suggest that the previously reported association of 3p loss with decreased T cell infiltrate and activation in HNSC specimens (40) may be due to marked co-occurrence of 3p14 loss and 9p21.3 loss (P = 1.73E-26 in HPV− HNSC) (SI Appendix, Table S3F). Furthermore, in cell lines not only was 3p loss not associated with immune depletion, but surprisingly in contrast, was associated with enrichment of immune-response and -pathway markers and metrics (SI Appendix, Table S11), consistent with experimental 3p deletion in lung cells (24).
The findings clearly suggest that a microenvironment switch is activated by arm-level 9p loss, with genes on 9p21.3 contributing, but not being sufficient, to drive immune evasion (Figs. 2D and 3 and SI Appendix, Fig. S9 and Table S20A). Specific 9p loci relevant to immune response and escape include IFN-signaling pathway components, namely the IFN-α gene cluster (on 9p21.3) as well as the key IFN-γ pathway gene JAK2 (on 9p24) (26) (SI Appendix, Fig. S10). In our experiments the IFN-α gene cluster loss, postulated to promote tumor cell-intrinsic evasion in melanoma (32), does not appear to be operative in immune-cold HNSC with 9p loss. Our prediction model implicates a cumulative effect of up to 40 potential candidate genes contributing to the role of 9p deletion in immune evasion, including a prominent 21-gene cluster deletion in 9p13 (Dataset S2). Notable among the new candidates were RANBP6 and IL33, which can influence central processes involved in tumor microenvironment-cell recruitment and checkpoint-blockade efficacy by silencing STAT3 activity (41) and attracting immune cells to sites of tissue injury (42), respectively. A third possible culprit, TOPORS, can promote NF-κB activity (43), potentially contributing to NF-κB pathway suppression, T cell depletion, and escape after 9p loss.
From a mechanistic standpoint (Fig. 5), it is possible that chromosome loss needed for immune exclusion is larger in size or discontinuous, a concept first suggested in a cross-sectional 3p-mapping study of human lung squamous precancer progression (44), later extended to other chromosome-region losses (5q, 9p, 13q, 17p) (45), precancer types, and model systems. The underlying basis for greater effects of larger chromosomal losses on neoplastic- and immune-transition in precancer is unclear. The influence of aneuploidy level in cancer precursor lesions on invasive cancer risk has been best studied in Barrett esophagus patients who progressed to esophageal adenocarcinoma (46), where some genomic features are known to be early drivers of tumorigenesis (e.g., TP53), but few other features have any clearly associated cancer-related activity (Fig. 5). Targets within 9p altered regions identified here may be augmented in effect by cooperating genes and regulatory elements elsewhere on the arm whose dosage titrates the effect of primary target genes (47, 48). It is also possible that 9p (the top arm-level event to predict low IS) (SI Appendix, Table S5) targets are epistatic with genomic events on other chromosomes, thereby creating synthetic physiological effects that exacerbate phenotype severity (49, 50) (Fig. 5B).
A well-established limitation of SCNA–microenvironment association studies involves tumor purity. Lower purity has been correlated with CN loss, microenvironment profile, and checkpoint blockade efficacy. We therefore rigorously controlled for this important potential confounder in all genome–microenvironment association analyses conducted here. For each genomic site analyzed as a continuous and binary variable, pathology- and ABSOLUTE-based purity correction gave similar results, suggesting that our HNSC SCNA/genome-immune association findings were not confounded by purity issues. Furthermore, in our extensive statistical analyses, some reported associations were limited by marginal statistical significance within the context of multiple testing. Nevertheless, such results highlighted here were rigorously controlled for multiple testing (e.g., FDRs < 0.001), consistent through various analyses, cutoffs, and thresholds.
9p Loss Inhibits Proimmunogenic Pathways.
SCNA-pathway analyses provide insight into the mechanistic basis of 9p-loss–driven immune evasion. We found four interrelated pathways (SASP, cytokine–cytokine–receptor interaction, JAK-STAT signaling, and TNFA signaling via NF-κB) statistically significantly decreased in primary tumors and cell lines harboring 9p loss. A key mutational difference between oral precancer and cancer is the substantially greater TP53 mutation frequency in the latter (9, 14, 51), which may remodel SCNA-generated immune-regulatory networks. TP53 plays a fundamental role in inducing SASP and senescence (52), and the TP53 target p21 (together with interleukin-1, IL-1) is among the top genes up-regulated after induction of chromosome missegregation and aneuploidy in TP53 wild-type cells (53). Chromosome instability and segregation errors, that can lead to aneuploidy, are known to activate cGAS-STING pathways (32) and in turn NF-κB. Differential activation of the cGAS-STING pathway in CN altered cells may also account for SCNA-induced contrasting effects in precancer and cancer contexts. As with SASP, cell-intrinsic loss of cGAS or STING have been shown to have opposing roles in tumorigenesis. Early in precancer with focal 9p loss and limited SCNAs, acute cGAS-STING can lead to SCNA-detection and tumor-suppressive defense of transformation, then evolve immune-suppressive, tumor-promoting effects in later neoplastic stages characterized by progressive 9p-arm loss and karyotype complexity, and consequently chronic cGAS-STING signaling, which could inhibit or redirect downstream pathway promotion of immune evasion (21, 32, 37).
Additionally, the 9p21.3 region contains CDKN2A, which plays a central role in cell-cycle inhibition and SASP promotion in the presence of DNA damage and other cellular stresses, including aneuploidy (54, 55). 9p21.3 loss appeared important (and probably necessary) for depletion of SASP-related immune-regulatory molecules. In cell lines, 9p21.3 loss had a strong cell-intrinsic negative effect on SASP-pathway genes (Figs. 3 and 5 and SI Appendix, Fig. S9 and Table S20A). In fact, it was the one pathway decreased in cell lines with only focal 9p21.3 loss (SI Appendix, Tables S20 and S22). Decreased SASP proinflammatory molecule production can impair T cell recruitment (56). When TP53 is mutant, CDKN2A loss through 9p21.3 deletion may lead to decreased SASP, resulting in cold tumors. Consistent with these results, TP53-mutant, early-invasive disease with 9p21 deletion had a greater magnitude of CD3+/CD8+ T cell depletion compared to TP53 wild-type, 9p-deleted counterparts. The association with 9p loss and mutant TP53 suppression of CD8+ and CD3+ T cells was stronger with 9p21 than 9p arm loss (SI Appendix, Figs. S6 C and D and S8 C and D and Table S6 B–D).
A profound decrease in chemokines CXCL9 and CXCL10 (FDR = 1.46E-15 and = 1.44E-20) was found to be associated with 9p loss (57, 58). These IFN-γ-inducible chemokines, which attract CXCR3 T cells requisite for cytolytic activity and further IFN-γ production, are known to be secreted by tumor- and microenvironment-derived cells, such as dendritic cDC1 subset (59). IFN-γ plays a prominent role in HNSC, and an IFN-γ signature was found to correlate with IS and anti–PD-1 benefit in HNSC (29). Secretion of CXCR3 ligands greatly enhances tumor-antigen cross-presentation essential for T cell priming and activation (60). Additionally, we found that 9p loss in TCGA (but not cell lines) was associated with a paucity of this chemokine family, including very low levels of CCL19 and CCL21, and the latter can lead to a severe decrement in CD8+ effector T cells infiltrating the tumor site, and, thus, a promising IFN-γ-mechanism-based intervention (61) (NCT03546361) in this context (62, 63). A lack of chemokine decrease in cell lines suggests the centrality of extracellular signals that limit chemokine production. These data suggest that 9p loss diminishes tumor-site effector T cell recruitment, infiltration, and capacity for effective cell-mediated antitumor immunity. 9p loss orchestrates a chemokine-profile deficit, thus promoting a cold microenvironment, inhospitable to lymphocyte infiltration of the tumor. Since the pathways controlling immune-cell recruitment to the tumors are likely not cell-intrinsic, possibly involving secreted molecules, the presumably larger fraction of cells in precancer without arm deletions may promote cell recruitment. This may partially mask the effect of 9p loss on cytokine and chemokine decrements.
CN in Early Lesions and Immune Surveillance.
Although SCNAs are central events in most cancers, the relationship between SCNAs in precancers and A) malignant transformation, or B) immune response, has not been well studied. Study of early-stage tumors, typically harboring one or a few arm/chromosome trisomies, may inform precancer karyotypic states, and shed light on the SCNA/immune-cell balance occurring during the pivotal invasive transition (36). In stage-specific analyses, the negative associations of 9p21.3, 9p arm-level loss, and SCNA level with CD3+ and CD8+ T cell levels were weaker, and no longer statistically significant, in patients with early-stage HNSC in contrast to more advanced disease (SI Appendix, Figs. S6 A and B, S7 A–C, and S8 A and B). Interestingly, SCNA level was associated with increased tumor-infiltrating NK-cell levels in stage I HNSC samples (P = 0.007) (SI Appendix, Fig. S7D). This is consistent with a previous preclinical study showing that aneuploidy in normal human cells can provoke NK-cell activation (21, 64). These stage-specific, cytotoxic-cell and TP53-mutation results could infer an aneuploid-immune switch emerging during an invasive transition inflection point, which strengthens with tumor evolution and progression. CN-altered lesions, therefore, must evolve properties and mechanisms to silence these immune responses and evade recognition by NK and other cytotoxic cells. Although CN burden in precursor lesions confers a high risk of malignant transformation, these SCNA-defined early lesions retained, to some extent, T cell (Fig. 1 A–D and SI Appendix, Table S2 A–C) and NK cell (SI Appendix, Fig. S7D) infiltration, possibly holding CN-altered preinvasive lesions in check—a model by which invasive cancer transition may involve an altered relationship of neoplastic CN with immune microenvironment (65, 66). Augmenting a residual microenvironment immune response in the oral precancer setting with anti–PD-1/PD-L1–centered prevention, however, may not have a high therapeutic yield, since PD-L1–independent evasion is expected to emerge as lesions with 9p loss obviate an aneuploid checkpoint. We suggest, therefore, considering other immune therapies and combinations to intercept this high-risk precancer, notably chemokine enrichment strategies or Treg-targeted/CTLA4 inhibitor-based strategies and CD40 agonists. The latter, which can activate NK cells (67), was recently shown to prevent oral tumor development in mice (68).
The aneuploid-checkpoint concept proposed here may apply to cancers of sites other than the oral cavity, most notably lung squamous precancers/cancers. The latter track experimentally (24) and computationally with HPV− HNSC in pan-cancer genomic-SCNA association studies, possibly reflecting shared coevolution of immune evasion and neoplastic invasion (23, 24, 28, 69, 70). This was corroborated in a cross-sectional study of host detection in low-grade lung squamous precursors, through activation of resident immune cells, and escape through suppressive cells, networks, and signals, including ILs and checkpoints, operative in high-grade disease (24, 71, 72). Furthermore, longitudinal studies of this precancer found that persistent, progressive lesions were associated with suppression of IFN-signaling, pathway gene expression, and depletion of innate and adaptive cells (45). In these precancer studies, regressive high-grade lesions harbored more infiltrating immune cells than those that progressed to cancer (71). Consistent with lung squamous-cell carcinoma (LUSC) precursor studies, adaptive escape was already evident in lung adenocarcinoma precursors (5). Overall, lung carcinoma in situ—the direct, high-grade precursor to LUSC—although preinvasive, had the full CN alteration profile displayed in invasive cancer, a finding mirrored in preinvasive studies of high-grade, progressive esophageal, pancreatic, oral, breast, and lung adenocarcinoma in situ (20, 66, 73, 74). The most striking, consistent driver of SCNA-induced LUSC, esophageal adenocarcinoma and HPV− HNSC was TP53 mutation (18, 46). Virtually all lung carcinoma in situ that progressed to invasive cancer were TP53 mutant and T cell cold (71). The latter is consistent with our observation of greater influence of mutant TP53 on 9p-loss induced suppression of T cell infiltration in early-stage disease, and suggest potential of interception, targeting this pivotal event (75). In addition to lung cancer precursors (18), acquiring specific (e.g., large-scale 9p21.3, 17p, TP53) allelic losses and SCNA increases have been linked to immune-exclusive (cytotoxic-cell depleted) cold, and -suppressive (emergence of CD3+/CD4+ Tregs) (Fig. 1 A–D and SI Appendix, Table S2 A–C) microenvironments in corresponding high-grade (and early invasive) progressive esophageal, colorectal, breast and pancreatic lesions (19, 66, 74, 76, 77). The latter (and HNSC) were the two strongest aneuploid/immune cold signals observed in our pan-TCGA study (23).
Prediction of Survival after Cancer Immunotherapy.
Because of the 9p associations with lower cytotoxic T cell abundance, we hypothesized that 9p-arm loss could represent a novel biomarker that, in addition to (and potentially in lieu of) PD-L1 expression, could more accurately predict clinical benefit from PD-1–targeted agents. ICB of PD-1 receptor interaction with both PD-L1 and PD-L2 (on 9p24.1) (33) would be expected to have limited benefit in this setting. We identified 9p loss (Fig. 4A) as a highly specific predictive marker of anti–PD-1 clinical benefit in HPV− HNSC (P = 0.03). Specific 9p alterations relevant to response and resistance to immunotherapy include deletion or impaired IFN-signaling pathway components, namely loss of IFN-α genes (32) and the IFN-γ pathway gene JAK2. The latter (e.g., JAK2 loss-of-function mutations) has been associated with anti–PD-1 and -CTLA4 resistance in melanoma (26). Although we found consistent, marginally statistically significant associations of PD-L1, -L2 or JAK2 deletion (P = 0.071–0.081) with anti–PD-1 resistance in our HNSC RWE, our data strongly support a dual-hit scenario. JAK2-PD-L1 codeletion was associated with striking anti–PD-1 resistance, more than threefold difference in median survival, P = 0.007, a critical clinical impact that could be even further amplified by broader SCNA- (aneuploidy) induced cold tumors.
In stark contrast, 9p loss and PD-L1-JAK2 codeletion in non-ICB chemotherapy-only–treated patients lack any prognostic impact, with HRs and P values of near 1.0. Although CN gain was not associated with clinical benefit in ICB-treated melanoma patients, recurrent 9p loss was a statistically significant event in the no-clinical benefit subgroup (25). Consistent with our PD-L1-JAK2 codeletion resistance findings, JAK2, PD-L1 (and PD-L2) (9p24.1) amplification, or 9p CN gain, have been associated with opposite effects, namely anti–PD-1 benefit, primarily in metastatic melanoma (78, 79). Loss of 9p24 (and to a lesser degree 9p13) was associated with reduced survival trends after nivolumab or pembrolizumab (SI Appendix, Table S25). Loss of 9p21 (IFN-α gene set, CDKN2A deletion), 3p, and 17p were not predictive or prognostic in this cohort. Although TMB has been associated with anti–PD-1 benefit in limited HNSC studies (80, 81), we were not able to detect such an effect in this 196-patient HNSC cohort. Furthermore, TMB did not correlate with CN loss in our cohort, consistent with an earlier HNSC report (82). Direct therapeutic implications for patients with HPV− HNSC include deprioritizing single-agent PD-1 blockade and prioritizing novel immune agents and strategies that could elicit immune response, especially therapy associated with a microenvironment enriched by IFN-γ-pathway CXCL9 and related chemokines [associated with response to pembrolizumab when upregulated in HNSC (83)]. Some chemokines, notably CCL2, decreased in our cell line and TCGA studies of 9p loss, can have striking context-dependent CCR2–CCL2 axis effects—either tumor-suppressing or -promoting, via cell-autonomous and/or extrinsic pathways (17, 84). SCNAs tend to differ by tissue type (22). However, these 9p loss and JAK2-PD-L1 codeletion findings in anti–PD-1–treated HPV− HNSC may apply to other tumors/sites and therapies, especially given the pivotal, broad role of JAK2 in cancer cell sensitivity to IFN-γ, impaired antigen presentation, T cell sensitivity, and evasion (26).
Concluding Remarks
In summary, we present evidence from patient samples in support of a CN-driven hot-to-cold switch in the precancer-invasion transition, and identify immune regulators and genomic alterations (e.g., TP53 mutation) that shape tumor evolution (Fig. 5). According to this model, CN-defined high-risk oral preinvasive and early invasive lesions are immunogenic, suggesting possible clinical benefit of therapeutically augmenting the (presumably) still preserved immune surveillance in this setting. In parallel competing forces, CN-generated neoplastic evolution and immune escape requires acquisition of intrinsic properties to circumvent selective pressures from host surveillance, through further karyotypic, mutational, and other events, to overcome a proimmunogenic aneuploid checkpoint and fuel tumor formation. Cumulative 9p-arm-level loss was the strongest driver of immune evasion (e.g., T cell, IS) in HPV-negative HNSC—profound, specific and consistent by every metric in every analysis and purity-correction here, across the entire CN loss/gain genome. This microenvironment switch may be enabled through the combination of specific 9p-arm loss events with possible epistatic and other genomic events, such as CDKN2A–TP53 interactions leading to cell-intrinsic evasion through SASP and other mechanisms. We identify a prominent role of 9p-arm deletion (as compared to regional/gene hotspot deletions, or other SCNAs, such as losses at 3p or 17p) in promoting depletion of A) cytotoxic cells (mainly CD8+ T cells), and B) IFN-γ-related pathways [e.g., CXCL9 and JAK2 signaling—both associated with response to immunotherapy in HNSC and other tumor types; (83)], and enriching suppressive cells (e.g., Tregs), molecules, and networks operant in a subset of progressive lesions, and ultimately anti–PD-1–resistant tumors. The influence of 9p loss was independent of SCNA level, which was also significantly associated with decreased cytotoxic activity, likely driven by the cumulative, discontinuous loss of 9p-dosage clusters of established immune-regulatory genes at 9p21.3, 9p24, and possible new candidates on 9p13 and remote interactions (e.g., 17p13) (Fig. 5B). Functional insights gained by studying these specific and broad CN-related phenomena and concepts emerging in neoplasia (in this case on tumor-immune interactions), could also provide important clues for understanding CN contribution to the etiology of a wide array of other common complex human diseases, including neurological and immune disorders (85); and genetic diseases with aneuploid etiology, such as Down syndrome, trisomy 21, phenotypes (e.g., activation of IFN signaling) (36). While further experimental studies will be important to interrogate and characterize underlying molecular features and mechanisms operative in the precancer-to-invasive cancer transition, our data resolve the aneuploid paradox and hot-to-cold switch in human neoplastic transformation in the oral precancer-invasion transition. Furthermore, our HNSC cohort data identified a potentially critical marker of anti–PD-1 resistance involving combined JAK2-PD-L1 codeletion and aneupoidy. These findings support a paradigm of personalized immune-based systemic interception and therapy for CN-driven early neoplastic disease, and enable precision-therapeutic approaches for advanced, recurrent disease, models that may apply to other body-site cancers, aneuploid diseases, and immunotherapy approaches.
Methods
Prospective Oral Precancer Cohort.
Clinical, demographic, immune, and genomic studies in the prospective oral precancer cohort are as follows. The prospective cohort included 188 oral precancer patients, characterized by clinical, pathologic, and SCNA risk factors. After study enrollment, patients were stratified by prior oral cancer, and systematically followed until reaching the protocol-specified primary endpoint of invasive cancer. Patients had clinic visits at months 1, 3, 6, 9, 12, and every 6 mo thereafter following protocol-defined and institutional practice. Additionally, an OCFS sweep was performed prior to database lock for the current analysis. The protocol was registered in ClinicalTrials.gov (NCT00402779) and approved by the MD Anderson Cancer Center Institutional Review Board. Participants provided written informed consent for biospecimens to be tested for genomic alterations and status of other biomarkers of interest reported herein, as well as for collection of demographic, clinical, and outcomes data. Precancer SCNAs included previously established major (3p14, 9p21.3, and 17p13.1) and minor (4q26-28, 4q31.1, 8p22, 8p23, 11q13, 11q22, and 13q21) CN-loss risk loci (13, 86) and chr7 gain. See SI Appendix for additional methods details, including SCNA, genomic and immune profiling, and statistical considerations for this and the other major sections below.
HPV− HNSC in TCGA.
SCNA, mutation, gene expression, HPV status, and clinical parameters for HPV− HNSC in TCGA are as follows. CN data of HNSCs in TCGA cohort were derived from Affymetrix SNP 6.0 arrays and obtained from the GDAC Firehose and GDC Data Portal (GISTIC2 analysis, Level 4). The CN for each chromosomal region (given as log2 CN ratios) were adjusted by tumor purity derived from previously reported pathology-based and ABSOLUTE (23, 24, 26, 87–89) methods. The HNSC SCNA level corresponds to the total number of chromosome arm gains or losses across the genome (except where otherwise specified). To distinguish between arm and focal-level events, we considered a threshold of ≥70% (default value in GISTIC2) of arm length (given in units of the fraction of chromosome arm) to identify the arm-level events, while all the others were considered as focal-level events. All the multivariable regression analyses that included a specific SCNA (e.g., 9p21.3 loss) as binary (loss or no loss) were confirmed using log2-transformed CN ratio as continuous variable [after normalized to z-score (23)]. HPV+ HNSC was based on rigorous viral-read (27) and anatomic subsite data criteria.
HPV− HNSC Cell Lines.
For SCNA and gene-expression data, we studied 32 HPV− HNSC cell lines (SI Appendix, Table S8). Segments, SCNA, and gene-expression data were derived from DepMap (CCLE_segmented_cn.csv, DepMap Public 19Q4; CCLE_gene_cn.csv, DepMap Public 19Q4 and CCLE_expression_full.csv, DepMap Public 19Q4) (90).
Survival Analysis of 9p Loss in ICB-Treated HPV− HNSC.
For the RWE cohort, we utilized a real-world cohort of HPV− HNSC patients treated with PD-1 inhibitors nivolumab or pembrolizumab, or chemotherapy, whose tumors were microdissected then underwent next-generation sequencing of a panel of 592 genes covering all autosomes and the X chromosome. We used this information to infer CN (loss) at 3p, 9p, and 17p (consort diagram in SI Appendix, Fig. S12). The 592-gene panel arm-loss algorithm was CAP/CLIA-validated against standard FISH in 436 patients for 1p/19q codeletion (24) showing sensitivity of 96.6% (95% CI: 82.2 to 99.9) and specificity of 99.5% (95% CI: 98.2 to 99.9) and further validated against whole-exome sequencing on 369 cases for 9p and 9p21 loss, finding a Pearson’s correlation between the two assays of 0.828 for 9p loss and 0.773 for 9p21.3 loss. Tumor-cell PD-L1 protein expression IHC with antibodies 22C3 or 28-8, demonstrated to show comparable analytical performance for assessment of PD-L1 expression on tumor cells (Blueprint), were performed according to manufacturer specifications. TMB in our 592-gene panel was found to be equivalent to the FDA-approved 324-gene companion diagnostic test related to the agnostic use of pembrolizumab for tumors with ≥10 mutations per megabase. The 592-gene MI Tumor Seek panel was used to validate anti–PD-1 therapy efficacy in relation to microsatellite-instability status (SI Appendix, SI Text).
Supplementary Material
Acknowledgments
This research was supported by several grants, including a Cancer Prevention and Research Institute of Texas Grant RP140464 and a Conquer Cancer Foundation Career Development Award, support from the Barbanti Funds for Cancer Research and Instituto Cura (to W.N.W.); Cancer Research UK Grand Challenge, Mark Foundation for Cancer Research (C5470/A27144), R00 CA212621, MRA Young Investigator Award, V Foundation Fellowship (to T.D.); a postdoctoral fellowship from the National Cancer Center (to J.J.B.); Stand Up To Cancer–LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Cancer Research Grant SU2C-AACR-DT-23-17, NIH Grant 5U01CA196408, and National Center for Advancing Translational Sciences Grant UL1TR001881 (to S.M.D.); support for Precancer Genome Atlas (PCGA) work by the Packard Fellowship for Science and Engineering (to L.B.A.); and Stand Up To Cancer–Lustgarten Foundation Pancreatic Cancer Interception Dream Team Translational Cancer Research Grant SU2C-AACR-DT-25-17, NIH Grants P01 CA106451, P50 CA097007, R01DE026644, and P30 CA023100 (to S.M.L.). S.M.L. is Distinguished Chugai Professor and Director of Moores Cancer Center at University of California San Diego.
Footnotes
Competing interest statement: W.N.W. has received honoraria, speaker’s fees, and/or participated in advisory boards from Roche/Genentech, BMS, Eli Lilly, Merck, AstraZeneca, Bayer, Boeringher Ingelheim, and Pfizer. J.P.A. and D.B.S. are employees of Caris Life Sciences. S.M.D. is on the Scientific Advisory Boards of Early Diagnostics, Johnson & Johnson Lung Cancer Initiative, Lung Life AI, Inc., and T-Cure Bioscience, Inc. He has research funding from Johnson & Johnson Lung Cancer Initiative and Novartis. W.C. is on the Board of Directors of Genetron Health LLC and a Founder of Interleukin Combinatorial Therapies, Inc. and InVaMet, Inc. S.M.L. is on the Human Longevity, Inc. (uncompensated) and Biological Dynamics, Inc. Scientific Advisory Boards.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022655118/-/DCSupplemental.
Data Availability
For the analyses of the HNSC samples (TCGA), codes and data used in the report are available in GitHub, https://github.com/breezyzhao/Davoli-Lab_HNSC_immune_infiltrate. All other study data are included in the article and supporting information.
References
- 1.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Burtness B.et al.; KEYNOTE-048 Investigators , Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 394, 1915–1928 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Cohen E. E. W.et al.; KEYNOTE-040 investigators , Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 393, 156–167 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ferris R. L., et al., Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krysan K., et al., The immune contexture associates with the genomic landscape in lung adenomatous premalignancy. Cancer Res. 79, 5022–5033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi A. K., et al., Oral leukoplakia and risk of progression to oral cancer: A population-based cohort study. J. Natl. Cancer Inst. 112, 1047–1054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vokes E. E., Weichselbaum R. R., Lippman S. M., Hong W. K., Head and neck cancer. N. Engl. J. Med. 328, 184–194 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Shield K. D., et al., The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J. Clin. 67, 51–64 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Califano J., et al., Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 56, 2488–2492 (1996). [PubMed] [Google Scholar]
- 10.Mao L., et al., Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat. Med. 2, 682–685 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Hittelman W. N., Genetic instability in epithelial tissues at risk for cancer. Ann. N. Y. Acad. Sci. 952, 1–12 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Rosin M. P., et al., Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin. Cancer Res. 6, 357–362 (2000). [PubMed] [Google Scholar]
- 13.Zhang L., et al., Loss of heterozygosity (LOH) profiles—Validated risk predictors for progression to oral cancer. Cancer Prev. Res. (Phila.) 5, 1081–1089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross A. M., et al., Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat. Genet. 46, 939–943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J. J., et al., Predicting cancer development in oral leukoplakia: Ten years of translational research. Clin. Cancer Res. 6, 1702–1710 (2000). [PubMed] [Google Scholar]
- 16.Lippman S. M., Hong W. K., Molecular markers of the risk of oral cancer. N. Engl. J. Med. 344, 1323–1326 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Krysan K., Tran L. M., Dubinett S. M., Immunosurveillance and regression in the context of squamous pulmonary premalignancy. Cancer Discov. 10, 1442–1444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teixeira V. H., et al., Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spira A., et al., Leveraging premalignant biology for immune-based cancer prevention. Proc. Natl. Acad. Sci. U.S.A. 113, 10750–10758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood H. M., et al., The genomic road to invasion-examining the similarities and differences in the genomes of associated oral pre-cancer and cancer samples. Genome Med. 9, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santaguida S., et al., Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchi J. J., Zhao X., Mays J. C., Davoli T., Not all cancers are created equal: Tissue specificity in cancer genes and pathways. Curr. Opin. Cell Biol. 63, 135–143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davoli T., Uno H., Wooten E. C., Elledge S. J., Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor A. M.et al.; Cancer Genome Atlas Research Network , Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roh W., et al., Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9, eaah3560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenan T. E., Burke K. P., Van Allen E. M., Genomic correlates of response to immune checkpoint blockade. Nat. Med. 25, 389–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney M. S., Shukla S. A., Wu C. J., Getz G., Hacohen N., Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamborero D., et al., A pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin. Cancer Res. 24, 3717–3728 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Ayers M., et al., IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman A. M., et al., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhoum S. F., Cantley L. C., The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yearley J. H., et al., PD-L2 expression in human tumors: Relevance to anti-PD-1 therapy in cancer. Clin. Cancer Res. 23, 3158–3167 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Senovilla L., et al., An immunosurveillance mechanism controls cancer cell ploidy. Science 337, 1678–1684 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Viganó C., et al., Quantitative proteomic and phosphoproteomic comparison of human colon cancer DLD-1 cells differing in ploidy and chromosome stability. Mol. Biol. Cell 29, 1031–1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheltzer J. M., et al., Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31, 240–255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripathi R., Modur V., Senovilla L., Kroemer G., Komurov K., Suppression of tumor antigen presentation during aneuploid tumor evolution contributes to immune evasion. OncoImmunology 8, 1657374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angelova M., et al., Evolution of metastases in space and time under immune selection. Cell 175, 751–765.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Turajlic S., et al., Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandal R., et al., The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 1, e89829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldrini B., et al., EGFR feedback-inhibition by Ran-binding protein 6 is disrupted in cancer. Nat. Commun. 8, 2035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan B. C. L., Lam C. W. K., Tam L. S., Wong C. K., IL33: Roles in allergic inflammation and therapeutic perspectives. Front. Immunol. 10, 364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renner F., Moreno R., Schmitz M. L., SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol. Cell 37, 503–515 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Wistuba I. I., et al., High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 60, 1949–1960 (2000). [PubMed] [Google Scholar]
- 45.Beane J. E., et al., Molecular subtyping reveals immune alterations associated with progression of bronchial premalignant lesions. Nat. Commun. 10, 1856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Killcoyne S., et al., Genomic copy number predicts esophageal cancer years before transformation. Nat. Med. 26, 1726–1732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., et al., Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531, 471–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue W., et al., A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl. Acad. Sci. U.S.A. 109, 8212–8217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller F. L., et al., Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van de Haar J., et al., Identifying epistasis in cancer genomes: A delicate affair. Cell 177, 1375–1383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.William W. N., et al., Genomic and transcriptomic landscape of oral pre-cancers (OPCs) and risk of oral cancer (OC). J. Clin. Oncol. 37, 6009 (2019). [Google Scholar]
- 52.Lujambio A., et al., Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santaguida S., Vasile E., White E., Amon A., Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29, 2010–2021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andriani G. A., et al., Whole chromosome instability induces senescence and promotes SASP. Sci. Rep. 6, 35218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J. Y., et al., Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proc. Natl. Acad. Sci. U.S.A. 116, 2603–2611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue W., et al., Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrott K. M., Wiley C. D., Desprez P. Y., Campisi J., Apigenin suppresses the senescence-associated secretory phenotype and paracrine effects on breast cancer cells. Geroscience 39, 161–173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamane M., et al., Senescence-associated secretory phenotype promotes chronic ocular graft-vs-host disease in mice and humans. FASEB J. 34, 10778–10800 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Spranger S., Dai D., Horton B., Gajewski T. F., Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wculek S. K., et al., Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Lee J. M., et al., Phase I trial of intratumoral Injection of CCL21 gene–modified dendritic cells in lung cancer elicits tumor-specific immune responses and CD8+ T-cell infiltration. Clin. Cancer Res. 23, 4556–4568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hillinger S., et al., EBV-induced molecule 1 ligand chemokine (ELC/CCL19) promotes IFN-γ-dependent antitumor responses in a lung cancer model. J. Immunol. 171, 6457–6465 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Whyte C. E., et al., ACKR4 restrains antitumor immunity by regulating CCL21. J. Exp. Med. 217, e20190634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R. W., Viganò S., Ben-David U., Amon A., Santaguida S., Aneuploid cells activate NF-κB to promote their immune clearance by NK cells. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.06.25.172239v1. Accessed 1 September 2020.
- 65.Foy J. P., et al., Immunological and classical subtypes of oral premalignant lesions. OncoImmunology 7, e1496880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hiraoka N., Onozato K., Kosuge T., Hirohashi S., Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin. Cancer Res. 12, 5423–5434 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Sartorius R., et al., Induction of human NK cell-mediated cytotoxicity by CD40 triggering on antigen presenting cells. Cell. Immunol. 221, 81–88 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Monteiro de Oliveira Novaes J. A., et al., Targeting of CD40 and PD-L1 pathways inhibits progression of oral premalignant lesions in a carcinogen-induced model of oral squamous cell carcinoma. Cancer Prev. Res. (Phila.), 10.1158/1940-6207.Capr-20-0418 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Campbell J. D.et al.; Cancer Genome Atlas Research Network , Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 23, 194–212.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbst R. S., Heymach J. V., Lippman S. M., Lung cancer. N. Engl. J. Med. 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pennycuick A., et al., Immune surveillance in clinical regression of preinvasive squamous cell lung cancer. Cancer Discov. 10, 1489–1499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mascaux C., et al., Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature 571, 570–575 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Qian J., et al., Genomic underpinnings of tumor behavior in in situ and early lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 201, 697–706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abba M. C., et al., A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 75, 3980–3990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rudin C. M., et al., An attenuated adenovirus, ONYX-015, as mouthwash therapy for premalignant oral dysplasia. J. Clin. Oncol. 21, 4546–4552 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spira A., et al., Precancer atlas to drive precision prevention trials. Cancer Res. 77, 1510–1541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kouketsu A., et al., Regulatory T cells and M2-polarized tumour-associated macrophages are associated with the oncogenesis and progression of oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 48, 1279–1288 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Budczies J., Denkert C., Győrffy B., Schirmacher P., Stenzinger A., Chromosome 9p copy number gains involving PD-L1 are associated with a specific proliferation and immune-modulating gene expression program active across major cancer types. BMC Med. Genomics 10, 74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gupta S., et al., JAK2, PD-L1, and PD-L2 (9p24.1) amplification in metastatic mucosal and cutaneous melanomas with durable response to immunotherapy. Hum. Pathol. 88, 87–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cristescu R., et al., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li W., et al., Plasma-based tumor mutational burden (bTMB) as predictor for survival in phase III EAGLE study: Durvalumab (D) ± tremelimumab (T) versus chemotherapy (CT) in recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC) after platinum failure. J. Clin. Oncol. 38, 6511 (2020). [Google Scholar]
- 82.Chen Y. P., et al., Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: Implications for immunotherapy. Ann. Oncol. 30, 68–75 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Litchfield K., et al., Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kubli S. P., Berger T., Araujo D. V., Siu L. L., Mak T. W., Beyond immune checkpoint blockade: emerging immunological strategies. Nat. Rev. Drug Discov. (2021). [DOI] [PubMed] [Google Scholar]
- 85.Shaikh T. H., Copy number variation disorders. Curr. Genet. Med. Rep. 5, 183–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.William W. N. Jr, et al., Erlotinib and the risk of oral cancer: The erlotinib prevention of oral cancer (EPOC) randomized clinical trial. JAMA Oncol. 2, 209–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carter S. L., et al., Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mermel C. H., et al., GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aran D., Sirota M., Butte A. J., Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghandi M., et al., Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the analyses of the HNSC samples (TCGA), codes and data used in the report are available in GitHub, https://github.com/breezyzhao/Davoli-Lab_HNSC_immune_infiltrate. All other study data are included in the article and supporting information.