Significance
Bacterial cell walls contain a protective exoskeleton, peptidoglycan, which is a target of several clinically important antimicrobials. In gram-negative bacteria, peptidoglycan is covered by an additional lipid layer, outer membrane, that serves as a permeability barrier against the entry of toxic molecules. In some bacteria, an abundant lipoprotein, Lpp, staples the outer membrane to peptidoglycan to maintain the structural integrity of the cell envelope. In this study, we identify a previously unknown hydrolytic enzyme that cleaves Lpp from the peptidoglycan and show how the outer membrane–peptidoglycan linkages are modulated in Escherichia coli. Overall, this study helps in understanding the fundamental bacterial cell wall biology and in the identification of alternate drug targets for the development of new antimicrobials.
Keywords: bacterial cell wall, peptidoglycan, Lpp, LdtF
Abstract
The gram‐negative bacterial cell envelope is made up of an outer membrane (OM), an inner membrane (IM) that surrounds the cytoplasm, and a periplasmic space between the two membranes containing peptidoglycan (PG or murein). PG is an elastic polymer that forms a mesh-like sacculus around the IM, protecting cells from turgor and environmental stress conditions. In several bacteria, including Escherichia coli, the OM is tethered to PG by an abundant OM lipoprotein, Lpp (or Braun’s lipoprotein), that functions to maintain the structural and functional integrity of the cell envelope. Since its discovery, Lpp has been studied extensively, and although l,d-transpeptidases, the enzymes that catalyze the formation of PG−Lpp linkages, have been earlier identified, it is not known how these linkages are modulated. Here, using genetic and biochemical approaches, we show that LdtF (formerly yafK), a newly identified paralog of l,d-transpeptidases in E. coli, is a murein hydrolytic enzyme that catalyzes cleavage of Lpp from the PG sacculus. LdtF also exhibits glycine-specific carboxypeptidase activity on muropeptides containing a terminal glycine residue. LdtF was earlier presumed to be an l,d-transpeptidase; however, our results show that it is indeed an l,d-endopeptidase that hydrolyzes the products generated by the l,d-transpeptidases. To summarize, this study describes the discovery of a murein endopeptidase with a hitherto unknown catalytic specificity that removes the PG−Lpp cross-links, suggesting a role for LdtF in the regulation of PG–OM linkages to maintain the structural integrity of the bacterial cell envelope.
The gram-negative bacterial cell envelope is made up of an outer membrane (OM), an asymmetric bilayered lipid membrane which is surface exposed, and an inner membrane (IM) consisting of a phospholipid bilayer surrounding the cytoplasm. In between these two membranes is the periplasmic space in which a sac-like molecule, the peptidoglycan (PG or murein), is located (1). PG is an elastic heteropolymer that protects bacterial cells from lysis by internal osmotic pressure and from external stress conditions. It is a single, large macromolecule made up of multiple linear glycan strands that are interconnected with each other by short peptide chains, forming a net-like sacculus around the cytoplasmic membrane (Fig. 1). The glycan strands are polymers of alternating β-1,4–linked N-acetylglucosamine and N-acetylmuramic acid (MurNAc) disaccharide units, in which the d-lactoyl moiety of each MurNAc residue is covalently attached to the first amino acid of the stem peptide. Normally, the peptide chains are of two to five amino acids with a pentapeptide made up of l-alanine (l-ala1)−d-glutamic acid (d-glu2)−mesodiaminopimelic acid (mDAP3)−d-ala4−d-ala5 residues. In Escherichia coli, ∼40% of the neighboring peptide chains are linked to each other, either between the d-ala4 and mDAP3 (d-ala−mDAP or 4−3) or two mDAP3 residues (mDAP−mDAP or 3−3). Of these, the 4−3 cross-links are more prevalent and are formed by d,d-transpeptidases, whereas 3−3 cross-links are much less abundant and are catalyzed by l,d-transpeptidases (LDTs) LdtD and LdtE (2−4).
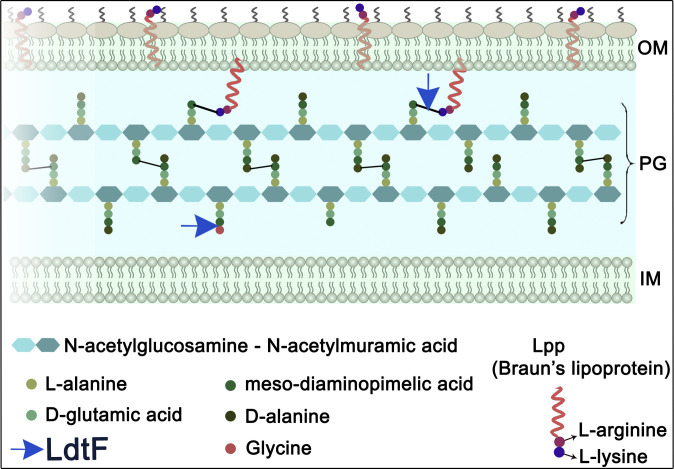
Fig. 1.
Diagrammatic representation of the cell envelope of E. coli. The cell envelope consists of three layers: OM, PG, and IM. PG is stapled to the OM by Lpp or Braun’s lipoprotein (red helix), which exists in either bound or free form (5−9). In the bound form, the N-terminal end of Lpp is anchored to the OM, whereas the C-terminal lysine residue (purple circle) is covalently attached to an mDAP residue (dark green) of the PG stem peptides. The free form of Lpp spans the OM and is exposed to the surface (11). LdtF is identified in this study as an endopeptidase that cleaves the PG−Lpp cross-links and as a glycine-specific carboxypeptidase.
In several gram-negative bacteria, including E. coli, the OM is stapled to PG by a lipoprotein, Lpp or Braun’s lipoprotein. Lpp is the first OM lipoprotein to be discovered almost five decades ago and has been studied extensively (5−9). It is a small protein of 58 amino acids, which is numerically the most abundant (∼106 molecules per cell), and is known to exist in two conformations, each occupying a distinct subcellular location in the cell envelope (9−11). One-third of total Lpp is in the periplasm covalently attached to the mDAP3 residues of the PG peptides (bound form), whereas the other two-thirds spans the OM and is surface exposed (free form) (Fig. 1). The OM–PG tethering by Lpp has been shown to determine the width of the periplasm by controlling the IM–OM distance and to contribute to the stiffness of the cell envelope (12, 13). Although Lpp is not essential for the viability of E. coli, mutants that lack Lpp show several pleiotropic defects, such as increased sensitivity to hydrophobic agents, leakage of periplasmic contents, OM blebbing, vesiculation, cell separation defects, and deficiency in virulence, highlighting the role of Lpp in the maintenance of envelope integrity (8, 9, 14).
Three redundant LDTs, LdtA, LdtB, and LdtC, catalyze the formation of PG−Lpp cross-links by covalently attaching the extreme C-terminal residue of Lpp, lysine to the mDAP3 residue of a tetrapeptide in the mature PG sacculus (15). In this reaction, the terminal d-ala residue of the tetrapeptide is lost, leading to the formation of a tripeptide–Lpp cross-link (Fig. 1). About 10% of the peptides in a PG sacculus are attached to Lpp, and this frequency is presumed to vary during conditions of stress (2, 5, 16−18).
E. coli encodes six LDTs, LdtA to LdtF, belonging to the YkuD family of proteins (3, 18–20). Of these, LdtA, LdtB, and LdtC catalyze the covalent attachment of Lpp to the PG; though these are redundant, LdtB is shown to be physiologically relevant because deleting ldtB alone abrogates the attachment of Lpp to a significant extent (15). Ribosome profiling (10) has also shown the abundance of LdtB to be much higher (∼5,000 copies per cell) than LdtA and LdtC (∼50 and 500, respectively). On the other hand, the 3−3 cross-link formation in the PG sacculus is catalyzed by LdtD and LdtE (21).
Apart from their ability to form cross-links, LdtA through LdtE catalyze an amino acid exchange reaction in the periplasm, wherein the terminal d-ala4/5 residue of the stem peptides is substituted with either glycine or a variety of noncanonical d-amino acids (NCDAA), such as d-tryptophan, d-methionine, or d-aspartate (2, 21, 22). The significance of this exchange in E. coli is not clear, although it is believed that these substituted muropeptides do not participate in further steps of PG polymerization. LDTs are presumed to have a larger role in the maintenance of structural integrity of PG because of their ability to form cross-links de novo in a mature PG sacculus independent of active PG precursor synthesis (18).
LdtF is a recently identified paralog of LDTs and is implicated in facilitating the formation of 3−3 cross-links; however, its precise function remains unclear (18–20). Here, we show that LdtF (encoded by yafK) is a murein l,d-endopeptidase that cleaves Lpp from the PG sacculus. We initially identified ldtF because of its genetic interaction with mepS, a gene encoding a major PG elongation–specific d,d-endopeptidase (23). Further genetic and biochemical analysis demonstrated the role of LdtF in hydrolyzing the products generated by the activity of other LDTs. LdtF cleaves Lpp, which is bound to the PG sacculus, and, in addition, cleaves the terminal glycine residue that is incorporated into the stem peptides due to the periplasmic exchange reaction of LDTs. However, LdtF was not able to cleave the terminal NCDAA residues from the muropeptides. To summarize, this study identifies a murein endopeptidase with a previously unknown catalytic specificity having an ability to modulate the Lpp-mediated OM–PG linkages.
Results
Absence of PG–Lpp Linkages Confer Growth Advantage to an E. coli Mutant Lacking an Elongation-Specific d,d-Endopeptidase, mepS.
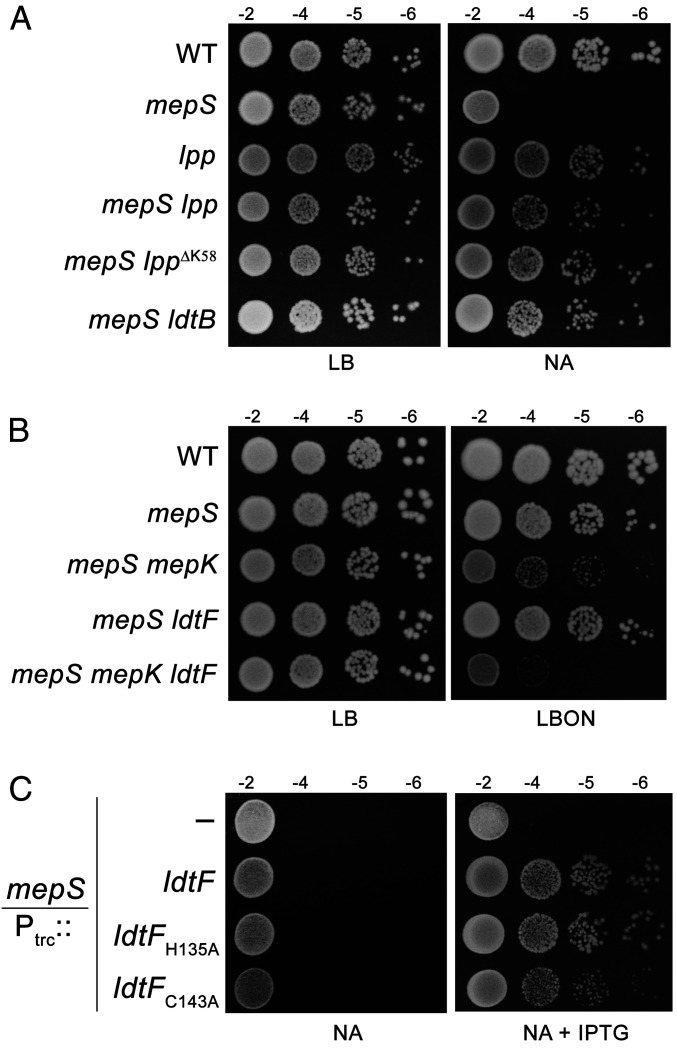
We showed earlier that the absence of 3−3 cross-link–forming LDTs (ldtD and ldtE) confers a growth advantage to a mutant lacking an elongation-specific 4−3 cross-link–cleaving d,d-endopeptidase, MepS, signifying the importance of cleavage of both 4−3 and 3−3 cross-links to make space for incorporation of new PG material during cell elongation (23, 24). To examine whether the tethering of OM to PG by Lpp also affects the process of PG expansion, we introduced a deletion of Lpp into a mutant lacking MepS and examined the growth phenotypes of the double mutant on nutrient agar (NA) because the mepS deletion mutant is unable to grow on NA at high temperatures (25). Fig. 2A shows that the absence of Lpp restores a moderate amount of growth to the mepS deletion mutant on NA. In addition, a mutant lpp allele that lacks the C-terminal lysine residue and hence is unable to bind PG (LppΔK58; ref. 11) confers suppression like that of Lpp deletion. Likewise, deletion of LdtB, which catalyzes the formation of mDAP−Lpp linkages, conferred growth to the mepS deletion mutant (Fig. 2A and SI Appendix, Fig. S1A and S2). However, deletion of LdtA or LdtC, which also link Lpp to mDAP, did not have any effect on mepS growth (SI Appendix, Fig. S1A). Surprisingly, deletion of LdtF, the newly identified paralog of LDTs, conferred a very small yet consistent growth defect to the mepS single mutant (SI Appendix, Fig. S1B), which was further exacerbated in an mepS mutant lacking mepK, a gene encoding the 3−3 cross-link–cleaving PG hydrolase (Fig. 2B).
Fig. 2.
Genetic interactions of mepS with LDTs. (A) WT (MG1655) and its mutant derivatives carrying either deletion of lpp, ldtB, or lpp∆K58, an allele lacking C-terminal lysine residue, were tested for viability on NA at 37 °C. (B) Indicated strains were grown and viability was tested on LBON agar plates at 37 °C. (C) An mepS deletion mutant carrying either vector pTrc99a (Ptrc::), pRB1 (Ptrc::ldtF), pRB2 (Ptrc::ldtFH135A), or pRB3 (Ptrc::ldtFC143A) were tested for viability on NA at 37 °C with or without IPTG (0.25 mM).
As these results intrigued us, we further investigated the role of LdtF by introducing the plasmids encoding each of the LDTs (26) into the mepS mutant. As shown in SI Appendix, Fig. S3, plasmids encoding ldtA, ldtB, ldtC, ldtD, and ldtE did not confer growth to the mepS mutant, whereas a plasmid encoding ldtF alone was able to moderately suppress the growth defects of mepS mutant. Another plasmid derivative carrying cloned ldtF downstream to an IPTG-dependent promoter (Ptrc::ldtF) also suppressed the growth defects of the mepS mutant on NA (Fig. 2C), indicating that LdtF may have a distinct function compared to that of other LDTs. LdtF belongs to the YkuD family of proteins, and members of this family contain the LDT domain with an invariant cysteine residue at the active site (27). To further validate the role of LdtF, we constructed a mutant derivative with an alanine residue substituted for cysteine (LdtF-C143A) and examined its ability to suppress the mepS mutation. Fig. 2C shows that a plasmid encoding LdtF-C143A poorly suppressed the mepS deletion mutant, whereas another variant coding for LdtF-H135A behaved like that of wild type (WT). However, deletion of ldtF alone in a WT strain did not confer any discernible phenotype when grown on Luria–Bertani (LB), LB broth without NaCl (LBON), or NA plates at 30, 37, or 42 °C, except for a slight reduction in the doubling time during the exponential phase of growth (SI Appendix, Fig. S4A). In addition, the ldtF deletion mutant did not exhibit significant sensitivity to any of the cell wall antibiotics, such as cephalexin, cefsulodin, mecillinam, or vancomycin (SI Appendix, Fig. S4B).
LdtF Modulates PG−Lpp Linkages In Vivo.
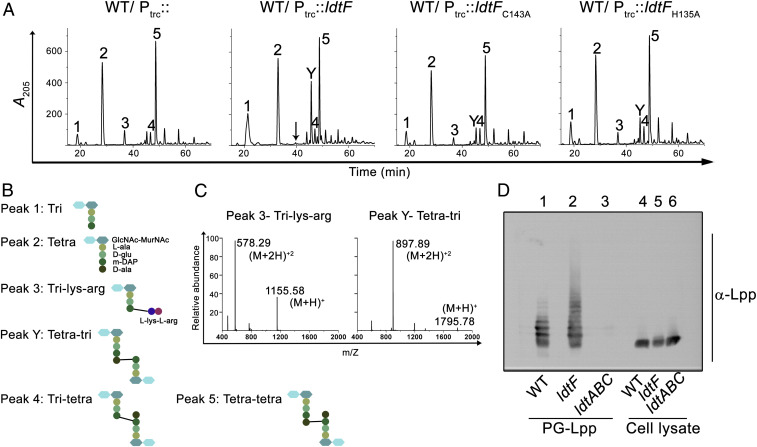
To understand the basis of LdtF’s function, we examined the composition of PG in strains either having a deletion or multiple copies of ldtF. PG sacculi from these strains were prepared and digested with a muramidase (mutanolysin), followed by separation of soluble muropeptides by reverse-phase-high-performance liquid chromatography (RP-HPLC) and identification of the peaks by mass spectrometry (MS) or MS–MS analysis. No major difference was observed in the muropeptide profile of the ldtF deletion derivative compared to that of WT (SI Appendix, Fig. S5). In contrast, the PG sacculi of cells carrying additional copies of ldtF had considerable alterations (Fig. 3A and SI Appendix, Table S3), the most significant being the absence of peak 3 (tri-lys-arg), which is a disaccharide tripeptide attached to a lys-arg dipeptide (Fig. 3 B and C). Tri-lys-arg muropeptides are generated because of the proteolytic activity of pronase, which is used during preparation of PG sacculi to remove bound Lpp. Pronase cleaves Lpp at the 56th position, leaving the extreme C-terminal lys-arg dipeptide attached to the mDAP residue of the stem peptides, resulting in the generation of several species of muropeptides bound to the lys-arg dipeptide (2). In addition to the absence of peak 3, a muropeptide peak eluting at 46 min (labeled “Y”) was significantly elevated in the PG sacculi of cells carrying additional copies of LdtF (Fig. 3A). MS–MS analysis indicated this peak to be a tetra-tri dimer linked by a 4−3 cross-bridge with a molecular mass of 1,794 Da (Fig. 3 B and C and SI Appendix, Fig. S6). The absence of peak 3 with a concomitant increase in peak 1 (a monomer of tri) allowed us to speculate that LdtF may have an ability to modulate the mDAP−Lpp linkages. Though the source of peak Y is not clear, it was not detected in a strain deleted for Lpp, suggesting it may have originated by the activity of LdtF on PG−Lpp cross-links (SI Appendix, Fig. S7A). Additionally, the incidence of peak Y was not dependent on the presence of functional LdtD and LdtE (SI Appendix, Fig. S7B). Furthermore, all other alterations observed due to overexpression of LdtF disappeared in a strain lacking Lpp, reinforcing the suggestion that LdtF functions downstream of Lpp (SI Appendix, Fig. S7A). As shown in Fig. 3A, an analysis of PG in strains carrying plasmids encoding either LdtF-C143A or LdtF-H135A indicated a direct role for LdtF in modulation of mDAP−Lpp linkages (SI Appendix, Table S3).
Fig. 3.
LdtF modulates PG−Lpp linkages. (A) HPLC chromatograms of PG sacculi of WT carrying either vector (Ptrc::), pRB1(Ptrc::ldtF), pRB2 (Ptrc::ldtFH135A), or pRB3 (Ptrc::ldtFC143A). Cultures were grown to an A600 of ∼1 in LB containing 0.2 mM IPTG for PG isolation. The black arrow indicates the absence of tri-lys-arg peak. (B) Structures of muropeptides. (C) Mass spectra of peaks 3 and Y, showing molecular mass (M+H)+ of 1,155.58 and 1,795.78 Da. (D) Determination of PG−Lpp linkages in the WT and ldtF mutant was done by treating the intact sacculi with mutanolysin, followed by electrophoresis of the soluble muropeptides. Lpp-containing muropeptides were visualized by Western blot using anti-Lpp antibody. PG from the ldtABC mutant was used as negative control. Cell lysates of WT, ldtF, and ldtABC were used as controls (lanes 4, 5, and 6).
Because the above results implicated LdtF in the regulation of PG−Lpp linkages, we examined the extent of these cross-links in cells lacking LdtF. To perform this experiment, Lpp-bound PG sacculi were prepared from WT and ldtF deletion mutant. Sacculi from both strains were digested with mutanolysin, soluble muropeptides were collected, and normalized amounts (SI Appendix, Fig. S8) were electrophoresed using SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) followed by Western blotting and detection with anti-Lpp antibody. Fig. 3D shows that the PG sacculi derived from the LdtF deletion mutant indeed contain a greater abundance of high-molecular-weight Lpp-bound muropeptides compared to that of WT; however, the level of low-molecular-weight Lpp-bound muropeptides were unchanged. This observation suggested an interesting possibility of LdtF specifically modulating the larger oligomeric Lpp−cross-linked muropeptides of the PG sacculus, and the implications of this result are further discussed below.
LdtF Is a Murein Endopeptidase that Cleaves PG−Lpp Linkages.
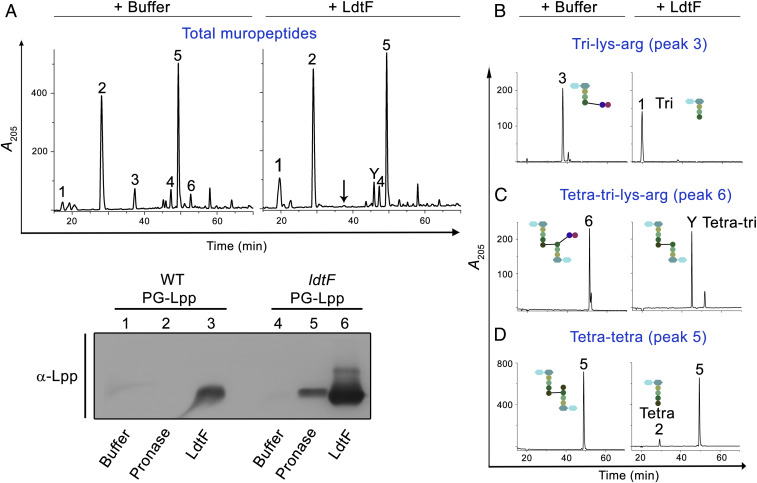
To examine the enzymatic activity of LdtF, a signal-less, hexa-histidine–tagged LdtF (LdtF20–246-His6) was overexpressed and purified. Treatment of soluble muropeptides derived from the PG sacculi of WT E. coli with purified LdtF yielded muropeptide fraction that totally lacked tri-lys-arg (peak 3) and tetra-tri-lys-arg (peak 6) with a concomitant increase in tri- and tetra-tri muropeptides (Fig. 4A). Cleavage of tri-lys-arg and tetra-tri-lys-arg into tri- or tetra-tri muropeptides was also confirmed using purified fractions (Fig. 4 B–D).
Fig. 4.
Endopeptidase activity of LdtF. Soluble muropeptides of WT PG sacculi (A), purified tri-lys-arg (B), purified tetra-tri-lys-arg (C), or purified tetra-tetra dimer (D) were incubated either with buffer or LdtF (4 μM) for 16 h and separated by RP-HPLC. LdtF cleaved peak 3 (tri-lys-arg) to yield tri (peak 1) and peak 6 (tetra-tri-lys-arg) to yield tetra-tri (peak Y). LdtF showed an extremely weak activity on tetra-tetra dimer (peak 5). (E) Cleavage of Lpp from the PG sacculi (containing bound Lpp) of WT or ldtF mutant was tested by incubating the PG sacculi either with buffer, pronase (0.2 mg/mL), or LdtF (4 µM) for 16 h at 30 °C. Pronase, a nonspecific protease, is used as a positive control.
We next examined the ability of LdtF to cleave the bound Lpp from the intact PG sacculi. To perform this experiment, Lpp-bound PG sacculi from WT and ldtF mutant were isolated, and equal amounts of each were treated with purified LdtF. The soluble fraction was electrophoresed on SDS-PAGE, and Lpp was detected by Western blot analysis using anti-Lpp antibody. As a positive control, PG sacculi treated with pronase were used. Fig. 4E shows that both LdtF and pronase cleave Lpp from the PG sacculi and that the amount of Lpp released from the sacculi of LdtF deletion mutant was considerably higher (approximately fivefold) than that of the WT (compare lanes 2 and 3 with 5 and 6). The remaining insoluble PG fraction was further analyzed by RP-HPLC, and, as expected, the lys-arg muropeptides were not detected in LdtF-treated PG, whereas pronase-treated PG contained the lys-arg muropeptides (SI Appendix, Fig. S9). Overall, these results demonstrate the catalytic specificity of LdtF on PG−Lpp or PG−lys-arg substrates.
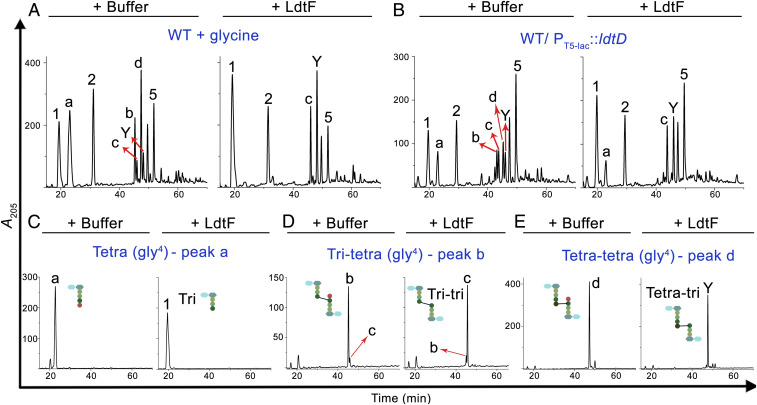
LdtF Is a Glycine-Specific Carboxypeptidase that Cleaves Terminal Glycine Residue from the Stem Peptides.
The above experiments clearly demonstrated hydrolytic activity of LdtF on PG−Lpp linkages formed by LdtA, LdtB, and LdtC. LDTs also exchange the terminal d-ala of stem peptides with a glycine residue. To examine whether LdtF has any activity on glycine-substituted muropeptides, soluble muropeptides of a WT strain grown with glycine supplementation were used as substrates for LdtF. As expected, growth of WT E. coli with exogenously added glycine resulted in the accumulation of a large number of muropeptides with glycine at position four, whose identity is determined by MS analysis (SI Appendix, Fig. S10). Fig. 5A shows that LdtF effectively removes glycine from a variety of glycine-containing muropeptides. LdtF also cleaved several glycine-containing muropeptides prepared from PG sacculi of a strain overexpressing LdtD (Fig. 5B). Of these, three distinct types of glycine-containing muropeptides were purified to homogeneity, and Fig. 5 C–E show that LdtF removes glycine residue from all these muropeptides. However, the abundance of glycine-containing muropeptides remained the same in both WT and ldtF deletion mutant when grown with glycine supplementation, suggesting the existence of alternate carboxypeptidases that cleave the terminal glycine residue. In support of this idea, the ldtF deletion mutant was not sensitive to the addition of glycine and behaved just like that of the WT strain.
Fig. 5.
LdtF is a glycine-specific carboxypeptidase. Soluble muropeptides generated from WT cells grown in minimal A medium (36) supplemented with 50 mM glycine (A), soluble muropeptides of WT/P-ldtD (B), purified tetra (gly4) (C), tri-tetra (gly4) (D), or tetra-tetra (gly4) (E) were incubated either with buffer or LdtF (4 μM) and processed as described above. LdtF cleaved the terminal glycine residue completely from peak “a” (tetra-gly4), “b” (tri-tetra-gly4), or “d” (tetra-tetra-gly4) to yield peak 1 (tri), “c” (tri-tri), or Y (tetra-tri).
Considering an earlier report that d,d-carboxypeptidases hydrolyze the terminal glycine from the stem peptides (2, 28), we made a quadruple mutant deleted for major d,d-carboxypeptidases, DacA, DacB, DacC, and DacD, and tested for sensitivity to glycine. As expected, the quadruple mutant formed smaller-sized colonies on glycine-supplemented media (SI Appendix, Fig. S11). The introduction of an ldtF deletion marginally exacerbated the defect of this quadruple mutant, whereas multiple copies of ldtF moderately improved the growth of this mutant on glycine-containing media (SI Appendix, Fig. S11), implicating a role for LdtF in the removal of terminal glycine residue from the stem peptides. In sum, the above results demonstrate that LdtF is a glycine-specific carboxypeptidase.
LdtF Does Not Cleave NCDAAs from the Stem Peptides.
To examine whether LdtF also cleaves the terminal NCDAA residues that are substituted by the exchange reaction of the LDTs, PG sacculi were made from WT E. coli grown in the presence of d-methionine, d-tryptophan, or d-phenylalanine (22). Soluble muropeptides of these PG sacculi were separated, and the peaks containing the NCDAA substitutions were identified by MS analysis (SI Appendix, Fig. S12A) and used as substrates for LdtF (SI Appendix, Fig. S12B). However, LdtF was not able to cleave the terminal NCDAA from any of these muropeptides (SI Appendix, Fig. S12B).
LdtF Removes Lpp-Mediated IM–PG Linkages.
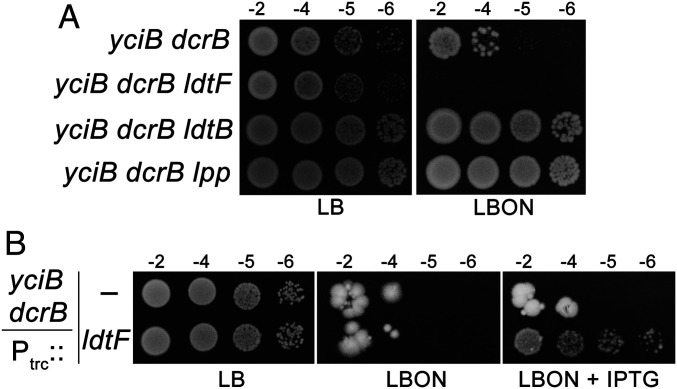
Lpp is transported from the cytosol into the periplasm by Sec-mediated pathway and is eventually translocated to the OM by lipoprotein translocating machinery, LolABCDE (29). However, in certain transport-defective mutants, Lpp is stalled at the periplasmic face of the IM, leading to the formation of IM–PG linkages by LDTs (30, 31). It has been shown recently that the absence of two small cytoplasmic membrane proteins, DcrB and YciB, leads to mislocalization of Lpp at the IM, resulting in lethal IM–PG cross-links, and that this lethality is suppressed by the deletion of either Lpp or LdtB (31). To examine the ability of LdtF to cleave the Lpp bound to IM, we made use of this mutant and observed that a deletion of ldtF exacerbates the growth defect of the yciB dcrB double mutant (Fig. 6A). In addition, the introduction of a multicopy ldtF plasmid (Ptrc::ldtF) partially restored the growth of the yciB dcrB double mutant, suggesting that LdtF may also cleave Lpp bound to the IM (Fig. 6B).
Fig. 6.
LdtF may also cleave Lpp-mediated IM–PG linkages. (A) The yciB dcrB mutant and its ldtF, ldtB, or lpp deletion derivatives were subjected to viability assays on LBON at 30 °C. (B) Indicated strains were grown with or without IPTG (0.1 mM) on LBON at 37 °C.
Discussion
Here, we report the identification of a previously unknown PG hydrolase, LdtF, that cleaves Lpp (or Braun’s lipoprotein), an abundant OM lipoprotein that links OM to the PG sacculus in E. coli. LdtF is also a glycine-specific carboxypeptidase that removes the terminal glycine residue from the PG muropeptides. LdtF is a recently identified member of the YkuD family of proteins in E. coli; the other paralogs of this family comprise LdtA, LdtB, and LdtC, which catalyze the formation of mDAP−Lpp linkages, and LdtD and LdtE, which catalyze the formation of mDAP−mDAP cross-links. This study also represents an instance wherein members of a paralogous family perform contrasting but not overlapping functions. While this manuscript is in review, an independent study by Winkle et al. (32) has also demonstrated identical biochemical activities for LdtF.
Role of LdtF in Maintenance of Envelope Structure and Stability.
We identified LdtF in this study because its absence enhanced growth defects of a mutant lacking two of the PG elongation–specific endopeptidases, MepS and MepK, and, additionally, multiple copies of ldtF rescued the defects of mepS mutant (Fig. 2 B and C). Moreover, we observed that the absence of LdtF increases the PG−Lpp linkages (Fig. 3D), whereas more copies of ldtF decrease the level of PG-bound Lpp (Fig. 3A), suggesting a role for LdtF in modulating the degree of PG−Lpp cross-linkages. Subsequent biochemical analysis confirmed LdtF to be a hydrolase having two distinctive enzymatic functions—an l,d-endopeptidase activity that cleaves mDAP−Lpp cross-links and a carboxypeptidase activity that cleaves mDAP−gly linkages (Figs. 4 and 5). Interestingly, two other YkuD family proteins, Csd6 and Pgp2, function as l,d-carboxypeptidases (33, 34). These hydrolases, including LdtF, have a distinctive YkuD domain with either an alanine or asparagine residue occurring at the second position after the catalytic cysteine, whereas synthases such as LdtA to LdtE contain an arginine residue (32–34).
LdtF was earlier identified because a transposon insertion in ldtF (yafK) caused defective biofilm formation in an enteroaggregative E. coli strain (35). LdtF deletion has also been shown to confer additive sickness to a mutant defective in the transport of lipopolysaccharide (18). Although the basis of the above phenotypes is not clear, elevated OM–PG linkages may result in a defective cell envelope, leading to such phenotypes. Excess OM–PG linkages may also alter the plasticity of the cell wall, resulting in decreased fitness and survival of E. coli. Nevertheless, under laboratory conditions, the absence of ldtF did not result in a large effect on the growth of E. coli, except for a small decrease in the doubling time (SI Appendix, Fig. S4).
It is interesting to note that although the abundance of lower-molecular-weight Lpp-bound muropeptides was comparable in both WT and ldtF mutant (Fig. 3D and SI Appendix, Fig. S5), the amount of bound Lpp is considerably higher in the absence of LdtF (Figs. 3D and 4E). The occurrence of larger Lpp-bound oligomeric muropeptides in the ldtF mutant (Fig. 3D) strongly suggests that LdtF may preferentially cleave PG−Lpp cross-links of higher-order structures in the PG sacculus. Lpp-mediated OM–PG cross-linkages resulting in the formation of large oligomers may distort the symmetry or the organization of the cell envelope, and LdtF may perhaps work toward eliminating such linkages. In addition, the effect of ldtF alleles on the growth of mutants in which Lpp is stalled at the IM (Fig. 6), suggests a role for LdtF in the removal of rare IM–PG linkages that may occur during transport of Lpp across the periplasm. LdtF may also facilitate PG turnover as the Lpp-linked muropeptides may not efficiently get recycled unless Lpp is cleaved from the PG sacculi.
Earlier studies implicated LdtF in facilitating the formation of 3−3 cross-links because ectopic expression of LdtF along with coexpression of LdtD or LdtE (in a strain lacking ldtABCDEF) increased the level of 3−3 cross-links (18–20). However, we did not observe increased 3−3 dimers when LdtF was overexpressed in WT; in contrast, we detected increased 4−3 linked dimers (peak Y) whose occurrence was dependent on Lpp but not on LdtD or LdtE (SI Appendix, Fig. S7). The basis of this discrepancy is not clear—it may perhaps be due to the strain background used for these studies and needs to be further investigated.
Regulation of PG−Lpp Linkages.
Lpp is an abundant OM lipoprotein (5, 6, 10) with one-third of it covalently attached to PG, making almost 10% of the peptides linked to Lpp. However, it is not clear how E. coli maintains optimal levels of PG−Lpp linkages. The combined activities of LdtABC and LdtF may control the abundance of PG−Lpp linkages, or, alternatively, structural/conformational constraints of PG sacculi may limit the extent of PG−Lpp cross-link formation. These linkages are reported to be higher during conditions of stress, including the stationary phase of growth and certain mutant conditions (2, 16–18). LdtF-promoter expression is shown to be higher in the stationary phase (18); however, preliminary experiments done in our laboratory to examine the endogenous LdtF expression (using an LdtF–FLAG fusion) show that the protein levels fall during the stationary phase, suggesting a likely basis for the occurrence of a higher amount of PG−Lpp linkages in the stationary phase. It would be worthwhile to further examine how cells achieve a dynamic yet balanced level of PG−Lpp linkages to maintain the structural and functional integrity of the cell envelope.
Role of PG−Lpp Linkages in PG Enlargement.
The absence of PG−Lpp linkages either by deleting Lpp or LdtB or increasing the copy number of LdtF partially rescued the growth defects of an mepS deletion mutant (Fig. 2 A and C); however, the suppression was not very robust (SI Appendix, Figs. S1A and S2). In addition, other phenotypes of mepS, such as sensitivity to β-lactam antibiotic, mecillinam, or its synthetic lethality with deletion of mepM (23), were not suppressed by deletion of Lpp or LdtB, suggesting that the absence of Lpp linkages may not significantly affect the process of PG enlargement. The lack of OM–PG tethering may perhaps alter the mechanical properties of the cell envelope and increase the flexibility of the PG sacculus, consequently resulting in a marginal growth advantage to mepS mutants in low osmolar conditions such as NA.
Materials and Methods
Media, Bacterial Strains, and Plasmids.
LB has 0.5% yeast extract, 1% tryptone, and 1% NaCl (36). LBON is LB without NaCl. NA has 0.5% peptone and 0.3% beef extract. Antibiotics were used at the following concentrations: ampicillin, 50 µg/mL; chloramphenicol, 30 µg/mL; and kanamycin, 50 µg/mL.
Molecular and Genetic Techniques.
Recombinant DNA and plasmid constructions were performed as per the standard methods. MG1655 genomic DNA was used as a template, Phusion high-fidelity DNA polymerase (New England Biolabs) was used for PCR amplifications, and the plasmid clones were confirmed by sequence analysis. P1 phage–mediated transductions and transformations were performed using standard methods (36). All strains are derivatives of MG1655 (Coli Genetic Stock Centre) unless otherwise indicated. Deletion mutations are from the Keio mutant collection (37).
Determination of PG−Lpp Linkages in the PG Sacculi.
Lpp-bound PG sacculi were isolated from cultures of WT and ∆ldtF mutant, treated with mutanolysin, and the soluble fraction was run on 15% SDS-PAGE followed by Western blotting using anti-Lpp antibody.
Determination of Enzymatic Activity.
To examine the activity of LdtF, soluble muropeptides were incubated with either buffer or LdtF (4 µM) at 30 °C for 16 h. Samples were heat inactivated, reduced with sodium borohydride, and separated by RP-HPLC. Lpp cleavage activity was examined by incubating purified LdtF (4 µM) with the Lpp-bound PG sacculi for 16 h in 25 mM Tris⋅HCl (pH 8.0) at 30 °C, followed by electrophoresis of the soluble fraction on SDS-PAGE and Western blotting with anti-Lpp antibody.
Supplementary Material
Acknowledgments
We thank the National BioResource Project, Japan for E. coli Keio collection and ASKA plasmid library; Thomas Silhavy for Lpp-K58 allele and anti-Lpp antibody; Sujata Kumari for initiating the work; V. Krishna Kumari and C. Subbalakshmi for HPLC; B. Raman and Y. Kameshwari for MS; N. Madhusudhana Rao for suggestions; and members of Reddy laboratory for critical comments. This work is supported by funds from the Council of Scientific and Industrial Research (MLP0141) and Department of Biotechnology (Centre of Excellence in Microbial Biology), Government of India (to M.R.). We acknowledge financial support from the Department of Biotechnology (to R.B.) and the University Grants Commission of India (to P.K.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101989118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glauner B., Höltje J. V., Schwarz U., The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095 (1988). [PubMed] [Google Scholar]
- 3.Egan A. J. F., Errington J., Vollmer W., Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 18, 446–460 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Garde S., Chodisetti P. K., Reddy M., Peptidoglycan: Structure, synthesis, and regulation. Ecosal Plus 9, 10.1128/ecosalplus.ESP-0010-2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V., Rehn K., Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10, 426–438 (1969). [DOI] [PubMed] [Google Scholar]
- 6.Braun V., Bosch V., Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur. J. Biochem. 28, 51–69 (1972). [DOI] [PubMed] [Google Scholar]
- 7.Inouye M., Shaw J., Shen C., The assembly of a structural lipoprotein in the envelope of Escherichia coli. J. Biol. Chem. 247, 8154–8159 (1972). [PubMed] [Google Scholar]
- 8.Braun V., Hantke K., Lipoproteins: Structure, function, biosynthesis. Subcell. Biochem. 92, 39–77 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Asmar A. T., Collet J.-F., Lpp, the Braun lipoprotein, turns 50-major achievements and remaining issues. FEMS Microbiol. Lett. 365, fny199 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Li G.-W., Burkhardt D., Gross C., Weissman J. S., Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowles C. E., Li Y., Semmelhack M. F., Cristea I. M., Silhavy T. J., The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79, 1168–1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asmar A. T., et al., Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol. 15, e2004303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathelié-Guinlet M., Asmar A. T., Collet J.-F., Dufrêne Y. F., Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat. Commun. 11, 1789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders A. N., Pavelka M. S., Phenotypic analysis of Escherichia coli mutants lacking L,D-transpeptidases. Microbiology (Reading) 159, 1842–1852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnet S., et al., Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 189, 3927–3931 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pisabarro A. G., de Pedro M. A., Vázquez D., Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J. Bacteriol. 161, 238–242 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwechheimer C., Rodriguez D. L., Kuehn M. J., NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. MicrobiologyOpen 4, 375–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morè N., et al., Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio 10, e02729 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montón Silva A., et al., The fluorescent D-amino acid NADA as a tool to study the conditional activity of transpeptidases in Escherichia coli. Front. Microbiol. 9, 2101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters K., et al., Copper inhibits peptidoglycan LD-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 10786–10791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnet S., Dubost L., Marie A., Arthur M., Gutmann L., Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190, 4782–4785 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caparrós M., Pisabarro A. G., de Pedro M. A., Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol. 174, 5549–5559 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S. K., SaiSree L., Amrutha R. N., Reddy M., Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol. Microbiol. 86, 1036–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Chodisetti P. K., Reddy M., Peptidoglycan hydrolase of an unusual cross-link cleavage specificity contributes to bacterial cell wall synthesis. Proc. Natl. Acad. Sci. U.S.A. 116, 7825–7830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara H., Abe N., Nakakouji M., Nishimura Y., Horiuchi K., Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb. Drug Resist. 2, 63–72 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa M., et al., Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 12, 291–299 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Biarrotte-Sorin S., et al., Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 359, 533–538 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto T., Katane M., Saitoh Y., Sekine M., Homma H., Involvement of penicillin-binding proteins in the metabolism of a bacterial peptidoglycan containing a non-canonical D-amino acid. Amino Acids 52, 487–497 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Szewczyk J., Collet J.-F., The journey of lipoproteins through the cell: One birthplace, multiple destinations. Adv. Microb. Physiol. 69, 1–50 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Yakushi T., Tajima T., Matsuyama S., Tokuda H., Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179, 2857–2862 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mychack A., et al., A synergistic role for two predicted inner membrane proteins of Escherichia coli in cell envelope integrity. Mol. Microbiol. 111, 317–337 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Winkle M., et al., DpaA detaches Braun’s lipoprotein from peptidoglycan bioRxiv [Preprint]. 10.1101/2021.02.21.432140 (21 February 2021). [DOI] [PMC free article] [PubMed]
- 33.Kim H. S., et al., The cell shape-determining Csd6 protein from Helicobacter pylori constitutes a new family of L,D-carboxypeptidase. J. Biol. Chem. 290, 25103–25117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frirdich E., et al., Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J. Biol. Chem. 289, 8007–8018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheikh J., Hicks S., Dall’Agnol M., Phillips A. D., Nataro J. P., Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41, 983–997 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Miller J. H., A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY, 1992). [Google Scholar]
- 37.Baba T., et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.