Abstract
As the most abundant protein with a variety of physiological functions, albumin has been used extensively for the delivery of therapeutic molecules. Thiolactone chemistry provides a powerful tool to prepare spin-labeled albumin-based multimodal imaging probes and therapeutic agents. We report the synthesis of a tamoxifen homocysteine thiolactone derivative and its use in thiol-‘click’ chemistry to prepare multi-functionalized serum albumin. The released sulfhydryl group of the homocysteine functional handle was labeled with a nitroxide reagent to prepare a spin-labeled albumin–tamoxifen conjugate confirmed by MALDI-TOF-MS, EPR spectroscopy, UV-vis and fluorescent emission spectra. This is the basis for a novel multimodal tamoxifen–albumin theranostic with a significant (dose-dependent) inhibitory effect on the proliferation of malignant cells. The response of human glioblastoma multiforme T98G cells and breast cancer MCF-7 cells to tamoxifen and its albumin conjugates was different in tumor cells with different expression level of ERα in our experiments. These results provide further impetus to develop a serum protein for delivery of tamoxifen to cancer cells.
A tamoxifen–homocysteine conjugate provides a tool to prepare a multifunctionalized albumin theranostic with a significant cytotoxicity on the malignant cells.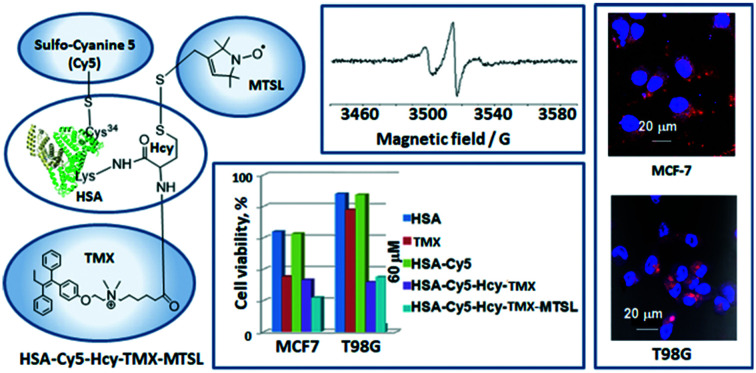
Introduction
The long circulatory half-life of approximately 19 days of human serum albumin (HSA) is facilitated by engagement with the cellular recycling neonatal Fc receptor (FcRn).1–4 Together with its anti-immunogenic properties, HSA is attractive as the sole carrier or as a component of a carrier system for many therapeutic and diagnostic agents.5–15 Different HSA conjugates have proved to be highly biocompatible, with some FDA approved and in current clinical use.5–7 The nanoplatforms with imaging function is crucial for tumor diagnosis and treatment, which could be used for imaging-guided therapy or monitoring the corresponding therapeutic efficacy.
To link different applications, it is desirable to use one-and-the-same imaging agent, functionalized with multiple diagnostic labels. A potential application for such multimodal compounds could be an integrated use preoperative diagnostics and surgical planning (radionuclide-based and magnetic-resonance-based), with intraoperative surgical image guidance (fluorescence-based) to the predefined lesion location. There are two classes of imaging agents that us albumin as the carrier: 99mTc-aggregated albumins for single photon emission computed tomography (SPECT), such as Pulmolite and Nanocoll; and albumin-bound gadolinium chelates for magnetic resonance imaging (MRI), such as gadofosveset (Ablavar). SPECT modality has sensitivity in the nanomolar range, but suffer from limited spatial resolution, not mentioning the disadvantage of ionizing radiation. A reasonable radiofrequency penetration depth in tissues makes MRI and low-field electron paramagnetic resonance (EPR)-based techniques the most appropriate approaches for noninvasive assessment of the chemical parameters tumor microenvironment.16 MRI relies largely on imaging of water protons and is widely used in clinical settings providing anatomical resolution while lacking functional sensitivity.17 EPR imaging is particularly useful in monitoring hypoxic zones in tumors which are highly resistant to radiation and chemotherapeutic treatment.16,18 The recent progress in EPR instrumentation designed to be used in humans19 and the development of biocompatible paramagnetic probes eventually will make possible the translation of EPR techniques for multifunctional profiling of tumor microenvironment into clinical settings. Delicate tumor therapy should take full advantage of the specific tumor microenvironment (TME) including mild acidity, overproduced hydrogen peroxide (H2O2) and glutathione (GSH). Therefore, the value of HSA as a drug carrier may significantly increase if spin-labeled protein will be used for targeting, because this could allow for EPRI/MRI/OMRI methods to follow the drug distribution making spin-labeled HSA a real theranostic tool. However, the chemistry for site-directed spin labeling of HSA conjugates is quite limited.
There are amine and sulfhydryl groups in the albumin that could be used for HSA modification.6,7 To start with, amide coupling based on lysine residue is the most classical method for in vitro covalent albumin conjugation. At present, the functional moieties often contain p-isothiocyanate (p-SCN) or NHS ester (N-hydroxysuccinimide) that can be obtained by in situ activation.6 NHS esters react with diverse nucleophiles on protein surfaces, most importantly the amine group of lysine residues. However, there are as many as 59 lysine residues in albumin, which indicate 59 amine groups as potential modification sites. Given the abundance of surface Lys on protein, the reaction typically results in a mixture of products modified at different protein positions.6,7,20,21 For example, a spin count of between 9–10 spins/molecule of albumin was obtained for active esters of the spin labels.20 This in turn makes functional characterization of modified proteins difficult, because one is dealing with a heterogeneous protein sample rather than a single, homogenous molecule. Therefore, reactions that enable chemists to selectively install modifications on protein surface at pre-defined positions would facilitate both the chemical characterization of these modifications and investigation of their exact biological function.
Site-directed spin labeling (SDSL) of albumin can be achieved if the single free thiol group, present on the cysteine at position 34, is the target site for the conjugation.22–24 Note, that the sulfhydryl group being a target easily and specificity reacts with methanethiosulfonate agent24 that will make possible development of biocompatible disulfide paramagnetic probes for noninvasive in vivo glutathione and redox assessment. It is known, the small paramagnetic disulfide probes can be paramagnetic versions of Ellman's reagent.25 The thiol-specific paramagnetic analogs of Ellman's reagent, disulfide nitroxide biradicals, were for the first time applied for intratumoral GSH assessment in living mice.26 The glutathione (GSH) redox status in vivo, characterized by both the GSH concentration and the GSH/glutathione disulfide (GSSG) ratio, is considered as an important biochemical indicator of oxidative stress and a potentially relevant predictive biomarker of tumor response to therapy in the clinic. In normal conditions, reduced GSH highly predominates over the oxidized form.27 Metabolic alterations affect the tumor tissue redox and result in accumulation of reducing equivalents,28 including a significant increase of GSH, key component of intracellular redox buffer.29 CW (continuous wave) imaging of the spin-labeled disulfide albumin conjugate can be useful in monitoring hypoxic zones in tumors which are highly resistant to radiation and chemotherapeutic treatment. Upon reaction with reduced thiols via the thiol-disulfide exchange reaction, the EPR spectrum will undergoes a drastic change from the complex spectrum of a spin-labeling protein conjugate to the typical triplet pattern of mononitroxides. Consequently, the spin-labeled disulfide protein may permit characterization of the catabolic processes in vivo, which could not be achieved with conventional radiolabels and fluorescent labels. However, the chemistry for SDSL of albumin conjugates is quite limited.
HSA has 35 cysteine residues; 34 are paired in 17 disulfide bonds leaving only Cys34 available for site-specific chemical modification.22–24,30–33 The utility of cysteine in protein modification cannot be overstated. Its versatile reactivity has enabled access to a range of modified proteins that allowed insight into complex biological problems. Moreover, this utility has driven chemical methodology to develop mild, selective reaction for cysteine and cysteine derivatives. However, DTNB (5,5′-dithio-bis(2-nitrobenzoic acid)) titrations indicate the sulfhydryl titer for most commercial plasma HSA preparations is approximately 30–40%.34,35 In healthy subjects, Cys34 is mainly present in a reduced form (about 70%) (HSAred) while 25–30% is reversibly oxidized as a mixed disulfide, mainly with cysteine or in minor amounts with cysteinyl glycine, homocysteine and glutathione (HSAox).35 Very small amounts (3–4%) of irreversibly oxidized forms, e.g., sulfinate and sulfonate, are also found.36 Pathologic conditions, like kidney or liver diseases, may increase the level of oxidized HSA to 70%.36,37 The heterogeneity of HSA is central to its physiological role and presents a complication for protein modification, but that is the biological reality. Consequently, novel SDSL strategies that are directly applicable to native proteins, that avoid the need for genetic engineering, and that do not rely on the relatively rare free sulfhydryl group of cysteine remain highly desirable.
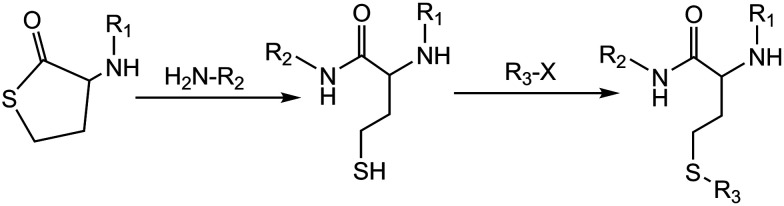
The simultaneous requirements of chemoselectivity, speed, stability and low toxicity of the labelling reagents makes development of selective chemical modification methods for proteins a significant challenge for chemists and biologists and hampers the application of many chemistry techniques. The decoration of biopolymers with reactive functional handles and their subsequent modification, via ‘click’ chemistry is an attractive approach, potentially leading to a plethora of synthetic biomaterials for many applications areas (bio-conjugation,38–42 labeling,43 medicine,44etc.). Conjugation and modification reactions involving thiols (thiol-‘click’ reactions45,46) are valuable metal-free alternatives to the ‘click’ benchmark: the widely exploited copper assisted azide–alkyne cyclo-addition (CuAAC).47,48 We previously reported using the reactivity of a homocysteine thiolactone (a cyclic thioester) as a latent thiol functionality in thiol-‘click’ chemistry for the synthesis of an HSA-based theranostic agent.14,21 The thiols were released by nucleophilic ring-opening (aminolysis) by amino groups on the HSA and subsequently reacted with a thiol ‘scavenger’ (a maleimide of the drug) according to the scheme in Fig. 1.
Fig. 1. A thiolactone entity as a latent thiol functionality: the thiol is released by nucleophilic ring-opening of the cyclic thioester for a subsequent thiol click reaction to incorporate the R3 residue. 19F-MR imaging (R1) – fluorinated compounds (perfluorotoluene residues). Drug carrier (R2) – HSA. Chemotherapeutic agent (R3) – 5-trifluoromethyl-2′-deoxyuridine 5′-monophosphate (pTFT).13,40.
Homocysteine thiolactone (HTL) is a quite valuable synthetic building block to prepare different multifunctional agents.14,21,31,49–53 Owing to its dual aminoacyl-thioester character, HTL undergoes facile electrophilic and nucleophilic reactions at its amino and activated carboxyl groups, respectively. Amidation reactions are most frequently used for the synthesis of different HTL derivatives.31,49,52,53 Surprisingly, N-substituted HTL appears to be a suitable starting material for medicinal chemistry.14,21,31,49–51 For example, an important HTL derivative is N-substituted HTL, N-acetyl HTL, (2-acetamido-4-mercaptobutyric acid γ-thiolactone, medically known as citiolone), a commercial compound that was introduced as thiolating agent for proteins.49–51 Under N-homocysteinylation of HSA by HTL in vivo54 and HTL derivatives in vitro,21,31 only three of the 59 protein lysine residues appeared to be modified. Moreover, no atoms are wasted as thiolactone chemistry results in 100% atom-efficient conjugation reactions. For example, although the reactivity of S-ethyl trifluorothioacetate and NHS esters might be higher in an aminolysis reaction, these have the intrinsic disadvantage of releasing the corresponding thioethanol and NHS as the side products.20,55
Here, we suggest taking advantage of natural posttranslational modification of albumin – its N-homocysteinylation54 – to use the amino group of the homocysteine (Hcy) residue as a site of anticancer drug (tamoxifen) attachment to the protein. The procedure of spin labeling will involve modifying the released sulfhydryl group of a homocysteine functional handle in the albumin structure by a nitroxide reagent. The presented “click chemistry” approach will expedite the parallel development of new disulfide paramagnetic probes this facilitate of the selection of the optimal probe and technique for in vitro and in vivo studies.
Tamoxifen, (TMX) trans-1-(4-b-dimethylamino ethoxyphenyl)-1,2-diphenylbut-1-ene (ICI 46 474), is the most commonly-used drug for the treatment of estrogen receptor positive breast cancer and has been saving lives worldwide for the past four decades.56 In addition to inhibition of estrogen receptor, tamoxifen can also inhibit other pathways, including protein kinase C.57–61 Some trials investigating the use of tamoxifen to treat malignant glioma produced encouraging results.61–66 Based on randomized trials and retrospective studies that tested the efficacy of tamoxifen in the treatment of glioblastoma multiforme, there were recommendations for further research on the role of tamoxifen in the treatment of glioblastoma.67 However, TMX suffers from low solubility and poor selectivity, and thus long-term use of the drug exposes patients to increased risk of uterine malignancies.58 Efforts to circumvent the selectivity-uptake challenge and other problems, such as solubility, stability, and toxicity, have been the focus for tamoxifen-based compounds in medicinal chemistry.68,69 The progress made in the last few years in tamoxifen research has been reviewed.56
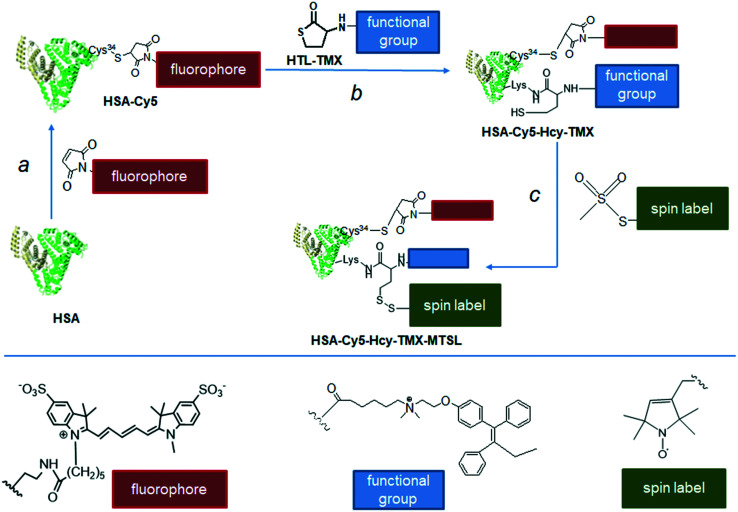
We present novel multimodal tamoxifen conjugates based on a biodegradable and biocompatible protein: the tamoxifen–HSA conjugates HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL (Fig. 2), for targeted delivery of tamoxifen in order to achieve the full potential of this important anticancer drug. The accumulated knowledge on the chemical reactivity and selectivity of thiolactones and subsequent thiol-‘click’ reactions, provides particularly interesting routes to spin-labeled drug delivery systems based on HSA. We have synthesized and characterized a novel conjugate of homocysteine thiolactone (HTL) with tamoxifen (HTL–TMX), attached HTL–TMX to the amino group of HSA, and spin labeled this tamoxifen–HSA conjugate to produce disulfide HSA-Cy5-Hcy-TMX-MTSL (Fig. 2).
Fig. 2. Synthetic route to the multifunctionalized HSA architectures – HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL. Drug carrier (shown schematically as a heart-like bundle of helices) – HSA. Effector – tamoxifen: chemotherapeutic agent (functional group, blue rectangle). Note that a homocysteine (Hcy-TMX) is used as a functional handle. The procedure of spin labeling modifies the released sulfhydryl group of the homocysteine functional handle by a nitroxide reagent. Spin label shown schematically as a green rectangle. Optical imaging fluorophore (red rectangle) – fluorescent dye Cy5 was conjugated with Cys34.
The spin-labeling procedures, spectroscopic and EPR characteristics of the obtained conjugates HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL are reported, together with preliminary examination of the biological behavior in vitro, such as their internalization by tumor cells and cytotoxicity.
Results and discussion
Synthesis of N-substituted homocysteine thiolactone HTL–TMX
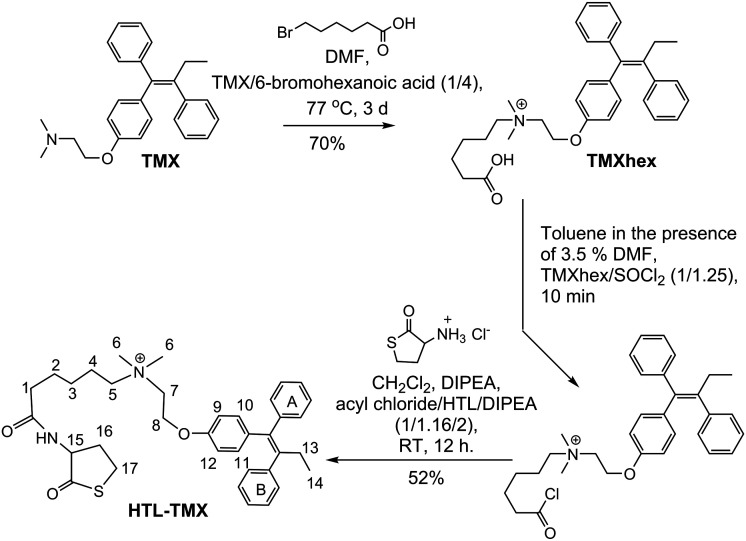
HTL is the key component in design of the modified protein.14,31,49,53,70 The fundamental requirements are to convert the amino group of thiolactone into a functional handle and, most importantly, to conjugate it to the reactive system of interest. The functionalization of the amino group of thiolactone must maintain the integrity of the thiolactone ring. Because of its dual aminoacyl-thioester character, homocysteine thiolactone is susceptible to both nucleophilic and electrophilic attack. Self-condensation of two thiolactones occurs during attempts to obtain the free amino-group; while mutual aminolysis can form the corresponding diketopiperazine derivative (3,6-bis(2-mercaptoethyl)piperazine-2,5-dione).31,71–73 We used N-acylating agents to develop robust HTL derivatives. The intrinsic instability of the neutral HTL requires an efficient reaction between the amino group and an electrophile. Amidation reactions are most frequently used: conjugation with acyl halides,74 (in situ) activated carboxylic acids,75 and anhydrides.70 We chose to use a carboxylic acyl chloride, which is a stronger acylating agent than thioesters. We synthesized the thiolactone derivative: (Z)-N-(2-(4-(1,2-diphenylbut-1-enyl)phenoxy)ethyl)-N,N-dimethyl-6-oxo-6-(2-oxotetrahydro thiophen-3-ylamino)hexan-1-aminium (conjugate HTL–TMX, Scheme 1) and then used its thiolactone moiety for conjugation to HSA.
Scheme 1. Synthesis of (Z)-N-(2-(4-(1,2-diphenylbut-1-enyl)phenoxy)ethyl)-N,N-dimethyl-6-oxo-6-(2-oxotetra hydrothiophen-3-ylamino)hexan-1-aminium (HTL–TMX).
The linker between the tamoxifen and the thiolactone residue is a flexible hexanoic acid. This linker serves several purposes; the principal two being to avoid steric clashes between the drug and HSA, and to facilitate conjugation via electrostatic attraction between HTL–TMX and HSA which are oppositely charged at neutral pH.
As shown in Scheme 1, reaction of tamoxifen with commercially available 6-bromohexanoic acid without carboxyl group protection afforded the (Z)-6-((2-(4-(1,2-diphenylbut-1-en-1-yl)phenoxy)ethyl)dimethyl ammonio)-hexanoic acid (TMXhex) with satisfactory yield.
Analytical HPLC found the retention time for TMXhex to be 48 min, but 92 min for tamoxifen (Fig. S1 in ESI†). Then, the TMXhex was reacted with thionyl chloride in the presence of 3.5% DMF to give the acyl chloride of TMXhex, which was further conjugated with thiolactone to produce HTL–TMX at 52% yield. Detailed synthetic procedures and spectroscopic data of synthesized compound are given in ESI.† ESI MS (m/z): calculated for HTL–TMX C36H45N2O3S 585.31, observed 585.3.
Synthesis of albumin-drug conjugate HSA-Cy5-Hcy-TMX
With HTL–TMX in hand, we embarked on the synthesis of a theranostic agent. The construction of HSA-Cy5-Hcy-TMX is illustrated in Fig. 2, step b. In order to visualize the cellular uptake, HSA was labeled with the fluorophore Cy5, whose strong fluorescence is readily detected when it is internalized in a cell.31 The Cy5 was coupled to the surface-exposed Cys34 of HSA via a Michael addition76 (Fig. 2, step a). The synthetic procedure was adapted from Chubarov et al.31 The conjugation reaction of HTL–TMX to HSA-Cy5 was carried out in PBS buffer (pH 7.4) at 37 °C. Low molecular weight homocysteine derivatives were removed from the HSA conjugates by centrifugal filtration with Centricon concentrators having a molecular weight cut-off of 3000 Da.
Spin-labeling of HSA-Cy5-Hcy-TMX
Nitroxide radicals have been the most extensively used and most reliable spin labels, and have been used for interspin distance measurements in various doubly-labeled biomolecules.77 One of the most widely used compounds for SDSL of cysteine is 3-maleimido-proxyl.22 However, it can produce significant alkylation of the terminal amino group of lysine and, to a lesser extent, alkylation of the imidazole group of histidine.78,79 In contrast, the methanethiosulfonate group is widely thought to label free sulfhydryl groups with essentially absolute specificity.80,81 It is the basis for the nitroxide spin label
MTSL, which is frequently used in paramagnetic relaxation enhancement nuclear magnetic resonance (PRE-NMR) studies of proteins because it is readily attached via a disulfide bond to cysteine residues (despite possible disulfide exchange reactions), and is also relatively small, inexpensive, and commercially available. Therefore, we used the methanethiosulfonate label nitroxide. The assembly of HSA-Cy5-Hcy-TMX-MTSL is shown in Fig. 2, step c. The conjugation reaction of MTSL to homocystamide HSA-Cy5-Hcy-TMX was carried out in PBS (pH 7.4) at 37 °C. Ellman's test showed that no free sulfhydryl groups remaining after overnight incubation.
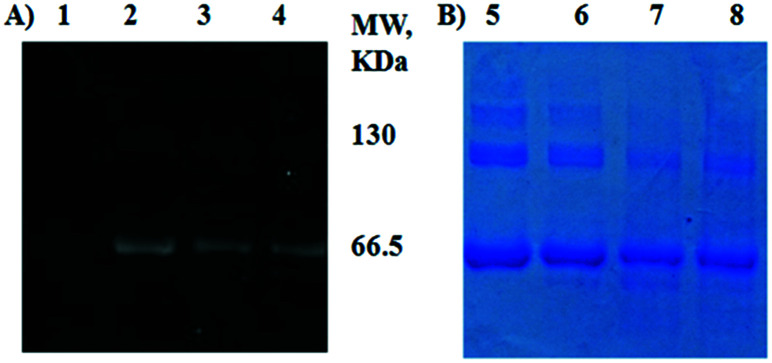
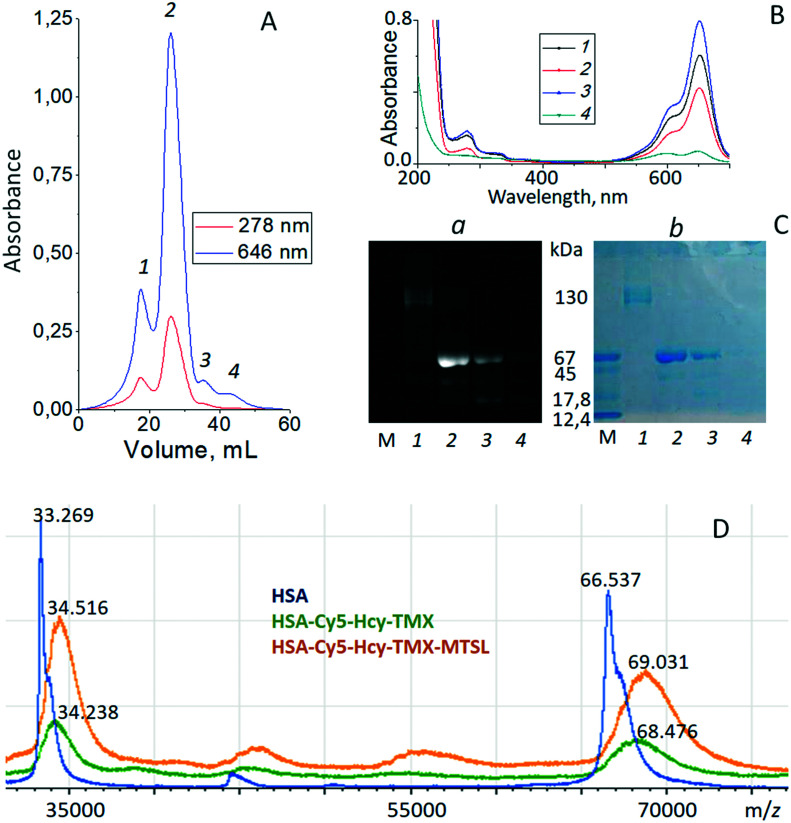
All HSA samples were assessed by SDS-PAGE and Coomassie blue staining. All showed major bands at approximately 65 kDa, similar to the published molecular weight for HSA of 66.5 kDa (ref. 82) (Fig. 3). Gels of fluorophore-equipped HSA and its homocystamide conjugates HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL were also scanned for fluorescence (Fig. 3A). Our starting protein, HSA A3782, is 62% monomeric with MW ∼ 66.5 kDa and 24% dimeric with only 14% of higher aggregates as shown by SDS-PAGE (Fig. 3B, lane 5, and ESI† Table S1). N-Homocysteinylation of a lysine residue produces a new thiol group in a protein, which affects the structure and biochemical properties of the modified protein. The N-Hcy-HSA is more susceptible to oxidation than is HSA and the amount of aggregates increases.54 However, the total amount of oligomers decreased to 23–33% in the conjugates from 38% in the starting HSA (Fig. 3B, lanes 6–8, and ESI† Table S1). At the same time, a new band appeared in the HSA conjugates with MW ∼ 46.5 kDa by MALDI-TOF. This lower MW band suggests partial cleavage of the initial protein oligomers. In addition to its ligand binding ability, HSA displays remarkable enzymatic properties,83 including esterase-like activity, a serine protease activity,84 and a new intrinsic proteolytic activity under reducing conditions.85 These are not major reactions under the experimental conditions for spin labeling the homocysteinylated HSA, but they could be in some situations.
Fig. 3. SDS-PAGE of thiolactone-derived homocystamide conjugates of the HSA under Laemmli condition with subsequent fluorescence detection (A) and Coomassie blue staining (B). HSA homocystamide (lanes 1 and 5), HSA-Cy5 (lanes 2 and 6), HSA-Cy5-Hcy-TMX (lanes 3 and 7), HSA-Cy5-Hcy-TMX-MTSL (lanes 4 and 8).
Purification and characterization of HSA theranostic conjugates
Multifunctional theranostic agents HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL were purified by size exclusion chromatography (SEC) on Sephadex-G 150 and then characterized by SDS-PAGE, UV-vis, MALDI-TOF mass spectrometry, and fluorescent emission spectroscopy (Fig. 4 and ESI† Fig. S2). Detailed purification procedures and spectroscopic data of synthesized conjugates are given in ESI.† The SEC showed the main product, peak 2, eluting at approximately 60–150 minutes post-injection for all samples (Fig. 4A and ESI† Fig. S2A).
Fig. 4. Panel A. representative chromatogram of HSA-Cy5-Hcy-TMX-MTSL products obtained by SEC utilizing a Sephadex-G 150 superfine column (300 × 12 mm column) at a flow rate of 0.18 ml min−1 (0.1 m sodium chloride). Protein elution was monitored at 278 and 646 nm. Panel B. UV-vis spectra of HSA homocystamides in PBS buffer, pH 7.4. Products from SEC: peak 1 − black; peak 2 − red; peak 3 − blue; peak 4 − green. Panel C. SDS-PAGE of homocystamide conjugates of the HSA under Laemmle condition with subsequent fluorescence detection (a) and Coomassie blue staining (b). SDS-PAGE had been performed with a molecular weight marker (m), lane 1 – products from SEC peak 1, lane 2 – products from SEC peak 2, lane 3 – products from SEC peak 3, lane 4 – products from SEC peak 4. Panel D. MALDI-TOF spectra of HSA (blue line), HSA-Cy5-Hcy-TMX (green line) and HSA-Cy5-Hcy-TMX-MTSL (orange line). The family of molecular ions is compatible with the structures shown in the Fig. 2.
Changes in the molecular masses of the HSA conjugates were monitored with MALDI-TOF mass spectrometry. HSA in native form is a monomer of 585 amino acid residues.82 However, plasma-derived HSA generally exhibits a broad range of post-translationally modified forms due to glycation, truncation, oxidation or genetic variants.34–37 The TOF mass analyzers used in these experiments did not measure the m/z values for [M + H]+ ions in MALDI spectra with great accuracy for masses over 60 kDa. MALDI mass spectra for each sample were recorded in four replicates with m/z values that differed by 20–100 Da. Additionally, the different post-translationally modified forms caused spectral overlap. The molecular mass of our HSA A3782 (Sigma-Aldrich) in our mass experiments averaged 66.537 kDa while HSA-Cy5, using the same system, gave 67.303 kDa. The MALDI data indicated one Cy5 dye molecule per protein.
MALDI-TOF spectra of unmodified HSA and its homocysteinylated derivatives are reported in Fig. 4D. The HSA-Cy5-Hcy-TMX conjugate had a calculated molecular mass of 68.476 kDa and 34.238 kDa for the double charged protein (Fig. 4D, green line). The difference between the homocysteinylated and native species is 1939 Da, which corresponds to two N-homocysteinyl moieties, N-linked by amide linkages to two Lys of HSA (N-Lys-Hcy-TMX; 586.31 Da) plus one dye moiety on Cys34 (S-Cys-Cy5; 766.4 Da). The N-homocysteinylated residues were identified by MALDI-TOF/TOF mass spectrometry of peptide mixtures formed via trypsinolysis of modified HSA. The N-homocysteinylation of HSA-Cy5 by HTL–TMX can modify three of the 59 lysine residues. According to MALDI-TOF MS data (ESI† Table S2), the Hcy-TMX residue was attached to Lys-199/Lys-174/Lys524 residues of HSA.
The proposed binding site for TMX in HSA overlaps the seventh fatty acid binding site (FA7).86 TMX forms a salt bridge with Glu153 (in 3-IB) in the hydrophobic pocket of Sudlow site I close to Trp residue (3.42 nm). Sudlow site I of HSA is a large binding site with its opening defined by three helices (the long h10 of domain I and the h3 and h6 of domain II). Charged residues, Lys-195, Arg-218, and Glu-292, are located at the opening of Sudlow site I, while the catalytic residue Lys-199 is located at the middle of site I and defines one wall of the binding site. The reactivity of Lys-199 can be explained by the unusually low pKa of its ε-amino group (pKa ≈ 8),87 which may be due to the close proximity of positively charged residues, Lys-195, Arg-281, Arg-222, which would disfavor protonation of Lys-199.88 The lower pKa should favor reaction of the amino group with tamoxifen homocysteine thiolactone derivative, since the reactive amino group must be uncharged for this chemical modification to occur. The other sites with high reactivity towards HTL–TMX were Lys-174 or Lys-524. All of these residues are located on the surface of HSA where they are available to solvent (ESI† Fig. S3).
When HSA-Cy5-Hcy-TMX was treated with MTSL, the MALDI-TOF spectrum of the HSA-Cy5-Hcy-TMX-MTSL product showed an increase in the mass of ∼555 Da relative to unreacted HSA-Cy5-Hcy-TMX (Fig. 4D, orange line). Based on the mass spectrometry analysis, there are approximately three MTSL labels conjugated to one HSA (elemental composition of nitroxide radical is C9H15NOS, so the mass of a single spin label is 185.1 amu).
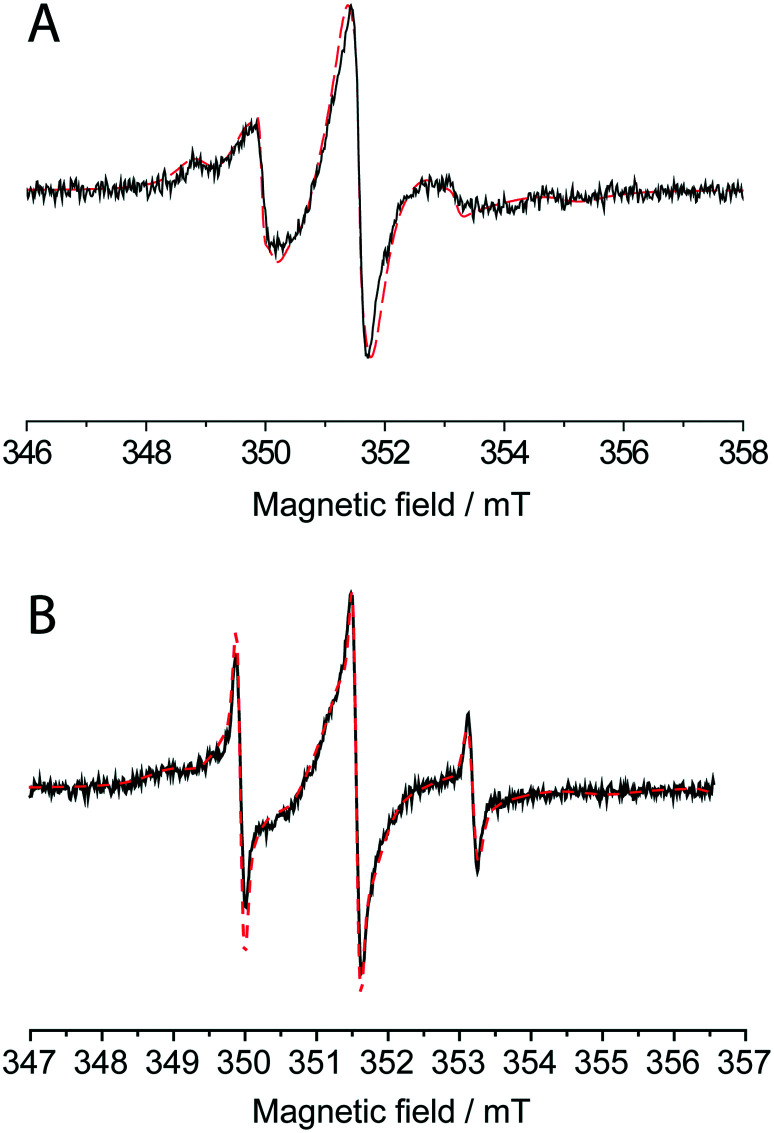
The attachment of MTSL to HSA was proven by the room-temperature CW EPR spectra (Fig. 5A). The spin label concentration measured by EPR is 3.3 times the protein concentration measured by UV-vis spectroscopy. In solution, the rotation correlation time of the spin label significantly decreases upon macromolecule binding,23 which strongly affects the shape of the CW EPR spectrum. The spectrum of HSA-Cy5-Hcy-TMX-MTSL can only be simulated assuming a mixture of two partially immobilized labels with correlation time τc = 2.2 ns (60%) and τc = 10 ns (38%) that confirms successful attachment of the spin-label. The remaining 2% of the EPR spectra corresponds to the incompletely washed-out free spin-label (τc ∼ 0.5 ns).
Fig. 5. X-band CW EPR spectrum of HSA-Cy5-Hcy-TMX-MTSL conjugate at 300 k (A) and spectrum after partial release of nitroxide label from albumin conjugate (B). The red dash line shows the simulated spectrum for a mixture of two types of protein-bound spin-labels with correlation times τc = 2.2 ns (60% in A, 42% in B) and τc = 10 ns (38% in A, 51% in B) and of free spin-label with τc = 0.5 ns (2% in A and 7% in B).
Continuous wave EPR spectroscopy and MALDI-TOF analysis of the HSA-Cy5-Hcy-TMX-MTSL conjugate both indicate an average of three nitroxide radicals per HSA. This is rather surprising because there should only be two free sulfhydryl groups available for spin labeling: one from each of the two adducted thiolactones. The MTSL reagent is widely regarded by the spin-labeling community to react with free sulfhydryl groups with essentially absolute specificity.80,81 However, amines do react with compounds similar to MTSL to produce sulfenamides.89 The number of spins in HSA-Cy5-Hcy-TMX-MTSL is greater than the number of free SH-groups and may indicate reaction with a single activated group such as a lysine. This additional spin label will improve the EPRI/MRI/OMRI aspects of this theranostic agent even if that label was attached by an unexpected reaction.
We previously reported using of a N-substituted HTL as a latent thiol functionality in thiol-‘click’ chemistry for the synthesis of an anti-cancer albumin–trifluorothymidine conjugate with disulfide bond for on-demand delivery of the chemotherapeutic agent 5-trifluoromethyl-2′-deoxyuridine 5′ monophosphate (pTFT) in response to redox trigger.14,21 Upon addition of glutathione, which is relatively abundant in tumor cells, was cleavage of the disulfide bond occur resulting in the active pTFT drug release. We present here, a novel anticancer HSA-Cy5-Hcy-TMX-MTSL conjugate that can be the thiol-specific paramagnetic analog of Ellman's reagent. Upon reaction with reduced thiol via the thiol-disulfide exchange reaction, the EPR spectrum of albumin conjugate undergoes a change, in particular, a fraction with three narrow lines appears, which corresponds to the spectrum of a free (detached) nitroxide (Fig. 5B).
In vitro studies
The cytotoxicity of tamoxifen involves more than one pathway.57–59,63 Tamoxifen acts as a breast cancer drug/chemoprevention agent by antagonizing the action of estradiol, via binding to the ligand binding domain of ERα and provoking a conformational state of the protein that is incapable of binding estrogen.56 There may be an additional estrogen receptor-independent pathway. Previous studies47–52 show that high doses of tamoxifen can also inhibit the proliferation of malignant ER-negative glioma cells. Such high doses of tamoxifen have limited efficacy in a majority of the patients. Nonetheless, there continues to be interest in tamoxifen and other selective estrogen receptor modulators for the treatment of gliomas, even though these tumors are estrogen receptor negative. In several ER-negative tumors, tamoxifen increased the levels of transforming growth factor-b1 (TGF-b1), a pleiotropic cytokine that regulates the proliferation and functional activity of a wide range of cell types.90 Therefore, we examined how the tamoxifen derivatives conjugated to HSA accumulated in cells and their subsequent cytotoxicity both to the human breast cancer MCF-7 cell line (ER-positive cells) and to the human glioblastoma multiforme T98G cell line (ER-negative cells).
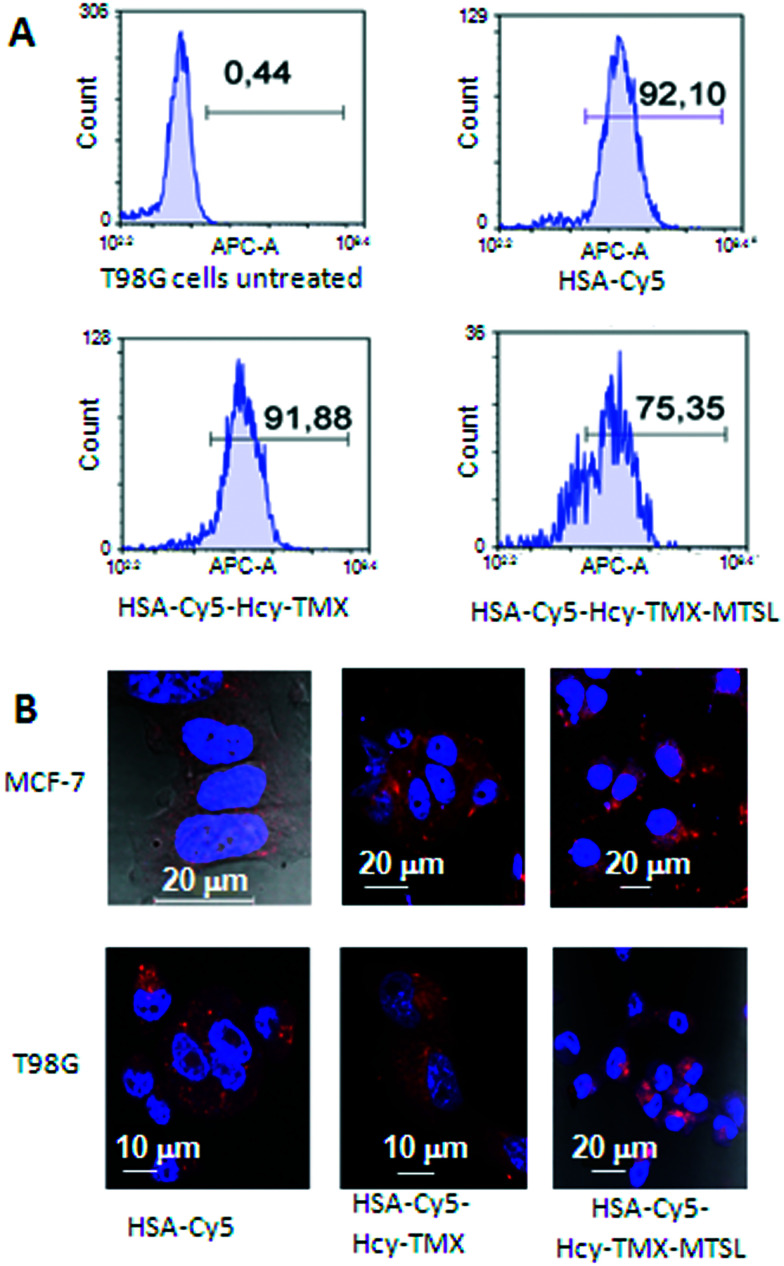
Fluorescence microscopy clearly shows that both HSA conjugates enter the cells (Fig. 6B and ESI† Fig. S4). Although HSA-Cy5-Hcy-TMX and HSA Cy5-Hcy-TMX-MTSL are comparable in size, they have different uptake profiles.
Fig. 6. Uptake of HSA-Cy5, HSA Cy5-Hcy-TMX and HSA Cy5-Hcy-TMX-MTSL conjugates in cancer cells. A. In vitro cellular uptake of HSA-Cy5, HSA Cy5-Hcy-TMX and HSA Cy5-Hcy-TMX-MTSL by human glioblastoma multiforme T98G cells measured by flow cytometry. Blue colour: FACS analysis; numbers show percent cellular uptake of HSA conjugates. The data were normalized to non-treated cells (control). B. Representative fluorescence micrographs showing HSA conjugates in cells (DAPI nuclear stain, blue; Cy5-fluor 510 HSA labeled conjugates, red). Scale bar: 10/20 μm.
The presence of tamoxifen on HSA-Cy5-Hcy-TMX does not decrease uptake in T98G cells (Fig. 6A), but the presence of spin labels on the HSA-Cy5-Hcy-TMX-MTSL does decrease uptake efficiency by 16% in T98G cells (Fig. 6A). The influence of spin labeling on uptake needs further investigation, but does not seem to be caused by differences in cell viability because cells exposed to HSA-Cy5-Hcy-TMX and HSA-Cy5-Hcy-TMX-MTSL conjugates showed comparable viability (Fig. 7).
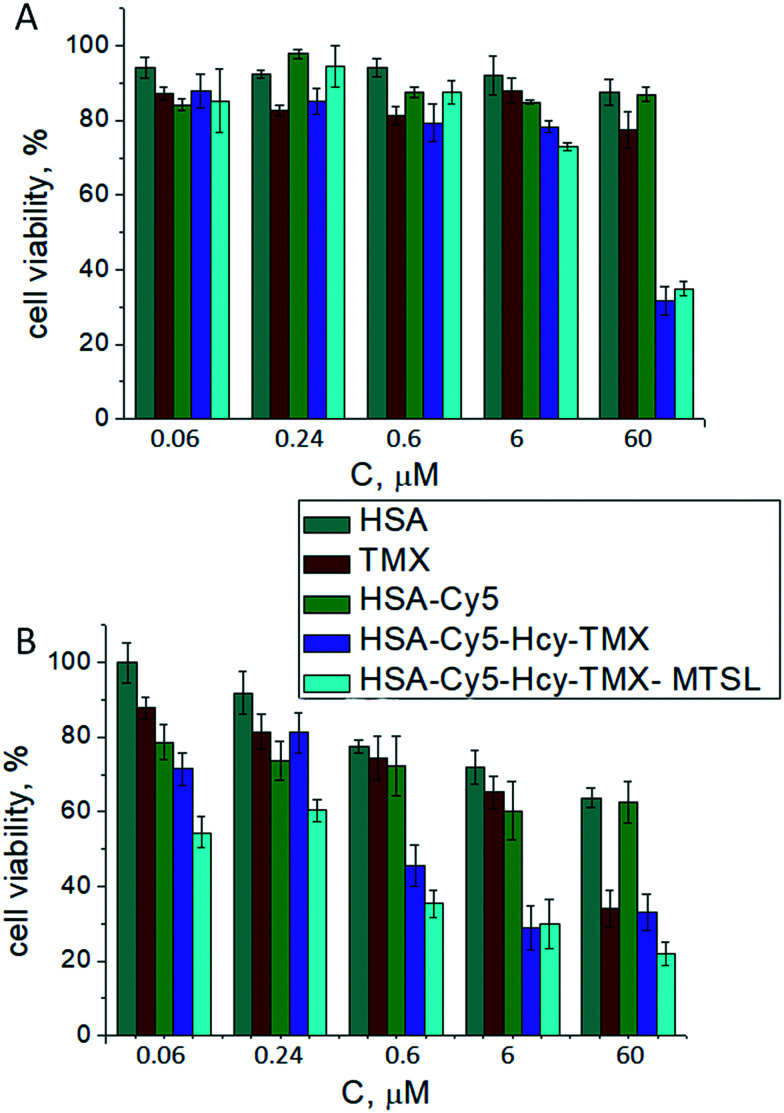
Fig. 7. In vitro cytotoxicity studies in T98G cell (A) and MCF-7 lines (B) for the drug delivery system HSA-TMX conjugates. Cell viability was normalized to PBS treated control 100%.
The effect of tamoxifen and its HSA conjugates on viability of the cancer cells was determined by the standard colorimetric 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay.91 The human glioblastoma multiforme T98G cells and breast cancer MCF-7 cells responded differently to tamoxifen and its HSA conjugates. In MCF-7 cells, the conjugates inhibited cell growth in a dose-dependent manner and with higher cytotoxicity than did free tamoxifen (Fig. 7). Free tamoxifen has minimal toxicity to MCF-7 cells after 72 h incubation at a concentration of 6 μM, whereas HSA-Cy5-Hcy-TMX or HSA Cy5-Hcy-TMX-MTSL caused over 70% toxicity under these conditions, Fig. 7B. When T98G cells were treated with 6 μM HSA-TMX, the cell viability remained high (around 80%). However, upon treatment of the cells with 60 μM HSA-TMX, cell viability decreased to 30%, Fig. 7A. At the same time, the free tamoxifen and drug-free HSA exhibited very low cytotoxic activity (e.g., 80% cell viability at 60 μM after 72 h). The high viability of the cells with HSA shows that the carrier is safe for use. These results are fully consistent with the design expectations and provide experimental support for the suggestion that HSA is an attractive candidate for use in tamoxifen delivery systems. However, more detailed studies with this conjugate involving toxicity, metabolic fate, and immunological consequences are necessary to establish its potential as an in vivo theranostic agent. The investigation of the tamoxifen conjugate as an in vivo theranostic agent is currently in progress along with further its study as the disulfide paramagnetic probe.
Conclusions
This paper describes the authors' preliminary studies directed toward the possibility of the practical implementation of the idea to design efficient antitumor drugs based on hybrid molecules composed of spin-labeled disulfide homocystamide albumin and tamoxifen. We designed and synthesized a tamoxifen homocysteine thiolactone derivative and its used in N-homocysteinylation of the fluorophore-equipped serum albumin. MALDI-TOF-MS analysis indicated that 2 tamoxifen molecules on average were conjugated with one molecule of HSA. The released sulfhydryl group of the homocysteine functional handle was then labeled with a nitroxide reagent to prepare a multi-functionalized serum albumin confirmed by MALDI-TOF-MS, EPR spectroscopy, UV-vis and fluorescent emission spectra. As a result, testing of the cytotoxic activity in vitro for the homocystamide albumin conjugates with the tamoxifen against the human breast cancer MCF-7 cell line (ER-positive cells) and to the human glioblastoma multiforme T98G cell line (ER-negative cells) showed a statistically reliable dose-dependent increase in the number of dead cells, in comparison with the control, that is, cells treated with albumin or tamoxifen alone. These results are fully consistent with the design expectations and provide experimental support for the suggestion that homocystamides HSA are attractive candidates for use in tamoxifen delivery systems. The presented “click chemistry” approach will expedite the parallel development of new disulfide paramagnetic probes this facilitate of the selection of the optimal probe and technique for in vitro and in vivo studies.
Experimental section
Materials and methods
The HSA product used in this study was purchased from Sigma-Aldrich Chem. Co. (St. Louis, MO, USA). S-(1-Oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) was synthesized as described.24d,l-Homocysteine thiolactone hydrochloride, 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB), D2O, and general chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and were used as received, unless otherwise specified. Tamoxifen (TMX) citrate, 6-bromoxeanoic acid were obtained from TCI (Japan). SOCl2 was obtained from Merck (Germany). Cascade blue ethylenediamine tri-sodium salt and MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay kit were purchased from Invitrogen.
MCF7 and T98G cells were obtained from the Russian cell culture collection (Russian Branch of ETCS, St. Petersburg, Russia). Breast cancer cell line MCF-7 and human glioblastoma multiforme T98G cell line were cultured in IMDM medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), penicillin (100 units per mL), and streptomycin (100 μg mL−1) at 37 °C, 5%CO2 in humid atmosphere.
Continuous wave EPR
Samples for EPR were placed in glass capillary tubes (OD 1.5 mm, ID 0.9 mm, with sample volume ca. 10 μL). Continuous wave (CW) EPR experiments were carried out at X-band (9.5 GHz) at 300 K using a commercial Bruker EMX spectrometer. Experimental settings were as follows: sweep width 12 mT; modulation frequency 100 kHz; modulation amplitude 0.1 mT; time constant 81.92 ms; number of points 1024; number of scans 8. The experimental spectra were simulated using EasySpin.92,93 All spectra were simulated using slow-motion regime to describe label mobility.
N-Homocysteinylation of HSA-Cy5 by homocysteine thiolactone derivative
The HSA-Cy5 conjugate is a known HSA conjugate. The synthesis of HSA-Cy5 was adapted from Chubarov et al.31
Preparation of HSA-Cy5-Hcy-TMX theranostic (Fig. 2, step a)
A solution (0.2 mL, 0.565 mM, 0.13 μmol) of the HSA-Cy5 in PBS buffer (pH 7.4) was mixed with HTL–TMX in DMSO (40 μL, 28 mM, 1.12 μmol). The reaction mixture was incubated under constant gently stirring at 37 °C in the dark for 17 h. The protein conjugate was purified by SEC utilizing a Sephadex-G 150 superfine column, then freeze-dried and stored at 4 °C.
The yield of HSA-Cy5-Hcy-TMX was ∼63%. UV-vis (PBS buffer, pH 7.4): λmax 278 nm (ε = (5.0 ± 0.1) × 104), λmax 646 nm (ε = (2.7 ± 0.1) × 105). The molecular weight of the conjugate was determined by MALDI-TOF analysis, and differences in MW were used to calculate the number of Hcy-TMX bound per albumin. MS (MALDI TOF) m/z: calculated for HSA-Cy5-Hcy-TMX, 67.889 kDa, found 68.476 kDa; 587 Da the error between calculated and found MS (MW for 2 residues of Hcy-TMX). Note: the calculated value is obtained as 67.303 kDa, (found MW of HSA-Cy5) + 586 Da (calculated MW of Hcy-TMX).
Preparation of spin Labeled HSA conjugate
The solution (0.2 mL, 0.5 mM, 0.1 μmol) of the HSA-Cy5-Hcy-TMX conjugate in PBS buffer (pH 7.4) was mixed with 5-fold excess of (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanesulfonothioate) (MTSL) in DMSO (10 μL, 50 mM, 0.5 μmol). The reaction mixture was incubated at 37 °C for 17 h. Ellman's test showed that no free sulfhydryl groups were left.94 Subsequently, low molecular weight materials were removed from solutions of protein conjugates by ultrafiltration at 9000 rpm twenty four times using a Millipore ultrafiltration tube (Amicon Centriprep YM30, Millipore, Bedford, MA). Then, the protein conjugate was purified by SEC utilizing a Sephadex-G 150 superfine column, freeze-dried and stored at 4 °C.
The yield of HSA-Cy5-Hcy-TMX-MTSL ∼60%. The amount of nitroxyl label per protein molecule for the protein using EPR method calculated to be: 3.3. UV-vis HSA-Cy5-Hcy-TMX-MTSL (PBS, pH 7.4, ε M−1 cm−1) λmax 278 nm (ε = (5.0 ± 0.1) × 104), λmax 646 nm (ε = (2.7 ± 0.1) × 105). MS (MALDI TOF) m/z. Calculated for HSA-Cy5-Hcy-TMX-MTSL, 68.661 kDa found 69.031 kDa; 370 Da the error between calculated and found MS (MW for 3 residues of MTSL). Note: the calculated value is obtained as 68.476 kDa, (found MW of HSA-Cy5-Hcy-TMX) + 185 Da (calculated MW of MTSL residue).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This study was financially supported by the Russian Science Foundation (grant No 19-74-20123), the Ministry of Science and Education of the Russian Federation (grant 14.W03.31.0034), and partially by Russian Foundation for Basic Research (no. 18-04-00393). We thank the Joint Center for genomic, proteomic and metabolomics studies (ICBFM SB RAS) for obtaining mass-spectra.
Electronic supplementary information (ESI) available: Spectroscopic materials and methods, additional spectra (UV-vis absorption, fluorescence, NMR), preparation of HSA modified by sulfo-Cy5 dye (HSA-Cy5), size exclusion chromatography (SEC), and in vitro cytotoxicity assay. See DOI: 10.1039/c9md00516a
References
- Chaudhury C. Mehnaz S. Robinson J. M. Hayton W. L. Pearl D. K. Roopenian D. C. Anderson C. L. J. Exp. Med. 2003;197:315–322. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. T. Cameron J. Plumridge A. Evans L. Sleep D. Sandlie I. J. Biol. Chem. 2013;288:24277–24285. doi: 10.1074/jbc.M113.463000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. T. Dalhus B. Cameron J. Daba M. B. Plumridge A. Evans L. Brennan S. O. Gunnarsen K. S. Bjørås M. Sleep D. Sandlie I. Nat. Commun. 2012;3:610. doi: 10.1038/ncomms1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V. Damschroder M. M. Cook K. E. Li Q. Gao C. Wu H. Dall'Acqua W. F. J. Biol. Chem. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Sun T. Jiang C. Acta Pharm. Sin. B. 2018;8:34–50. doi: 10.1016/j.apsb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Chen X. Chem. Soc. Rev. 2016;45:1432–1456. doi: 10.1039/c5cs00158g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F.-F. Zhang X.-H. Theranostics. 2017;7:3667–3689. doi: 10.7150/thno.19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzi A. Linciano S. Angelini A. MedChemComm. 2019;10:1068–1081. doi: 10.1039/c9md00018f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. A. Ther. Delivery. 2015;6:265–268. doi: 10.4155/tde.14.124. [DOI] [PubMed] [Google Scholar]
- Elsadek B. Kratz F. J. Controlled Release. 2012;157:4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- Sleep D. Cameron J. Evans L. R. Biochim. Biophys. Acta. 2013;1830:5526–5534. doi: 10.1016/j.bbagen.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Liu F. Mu J. Xing B. Curr. Pharm. Des. 2015;21:1866–1888. doi: 10.2174/1381612821666150302115411. [DOI] [PubMed] [Google Scholar]
- Larsen M. T. Kuhlmann M. Hvam M. L. Howard K. A. Mol. Cell. Ther. 2016;4:1–12. doi: 10.1186/s40591-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitskiy V. A. Khan H. Popova T. V. Chubarov A. S. Zakharova O. D. Akulov A. E. Shevelev O. B. Zavjalov E. L. Koptyug I. V. Moshkin M. P. Silnikov V. N. Ahmad S. Godovikova T. S. Bioorg. Med. Chem. Lett. 2017;27:3925–3930. doi: 10.1016/j.bmcl.2017.05.084. [DOI] [PubMed] [Google Scholar]
- Rhaese S. Von Briesen H. Rübsamen-Waigmann H. Kreuter J. Langer K. J. Controlled Release. 2003;92:199–208. doi: 10.1016/s0168-3659(03)00302-x. [DOI] [PubMed] [Google Scholar]
- Khramtsov V. V. Antioxid. Redox Signaling. 2018;28:1365–1377. doi: 10.1089/ars.2017.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J. N. Engl. J. Med. 2003;349:2290–2292. doi: 10.1056/NEJMp038193. [DOI] [PubMed] [Google Scholar]
- Kishimoto S. Matsumto K. Saito K. Enomoto A. Matsumoto S. Mitchell J. B. Nallathmby D. Krishna M. C. Antioxid. Redox Signaling. 2017:1–39. [Google Scholar]
- Swartz H. M. Williams B. B. Zaki B. I. Hartford A. C. Jarvis L. A. Chen E. Y. Comi R. J. Ernstoff M. S. Hou H. Khan N. Swarts S. G. Flood A. B. Kuppusamy P. Acad. Radiol. 2014;21:197–206. doi: 10.1016/j.acra.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovsky G. Rao N. U. M. Lukszo J. Brasch R. C. Z. Naturforsch., B: Anorg. Chem., Org. Chem. 1986;41:1170–1177. [Google Scholar]
- Popova T. V. Khan H. Chubarov A. S. Lisitskiy V. A. Antonova N. M. Akulov A. E. Shevelev O. B. Zavjalov E. L. Silnikov V. N. Ahmad S. Godovikova T. S. Bioorg. Med. Chem. Lett. 2018;28:260–264. doi: 10.1016/j.bmcl.2017.12.061. [DOI] [PubMed] [Google Scholar]
- Pavićević A. Luo J. Popović-Bijelić A. Mojović M. Eur. Biophys. J. 2017;46:773–787. doi: 10.1007/s00249-017-1257-z. [DOI] [PubMed] [Google Scholar]
- Gruian C. M. Rickert C. Nicklisch S. C. T. Vanea E. Steinhoff H. J. Simon S. ChemPhysChem. 2017;18:634–642. doi: 10.1002/cphc.201600886. [DOI] [PubMed] [Google Scholar]
- Park D. S. Petersen C. E. Ha C. E. Harohalli K. Feix J. B. Bhagavan N. V. IUBMB Life. 1999;48:169–174. doi: 10.1080/713803501. [DOI] [PubMed] [Google Scholar]
- Roshchupkina G. I. Bobko A. A. Bratasz A. Reznikov V. A. Kuppusamy P. Khramtsov V. V. Free Radical Biol. Med. 2008;45:312–320. doi: 10.1016/j.freeradbiomed.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustarini D. Colombo G. Garavaglia M. L. Astori E. Portinaro N. M. Reggiani F. Badalamenti S. Aloisi A. M. Santucci A. Rossi R. Milzani A. Dalle-Donne I. Free Radical Biol. Med. 2017;112:360–375. doi: 10.1016/j.freeradbiomed.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. Hyodo F. Matsumoto A. Koretsky A. P. Sowers A. I. Mitchell J. B. Krishna M. C. Clin. Cancer Res. 2006;12:2455–2462. doi: 10.1158/1078-0432.CCR-05-2747. [DOI] [PubMed] [Google Scholar]
- Estrela J. M. Ortega A. Obrador F. Crit. Rev. Clin. Lab. Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- Swartz H. M. Williams B. B. Zaki B. I. Hartford A. C. Jarvis L. A. Chen E. Y. Comi R. J. Ernstoff M. S. Hou H. Khan N. Swarts S. G. Flood A. B. Kuppusamy P. Acad. Radiol. 2014;21:197–206. doi: 10.1016/j.acra.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehtala J. G. Kulczar C. Lavan M. Knipp G. Wei A. Bioconjugate Chem. 2015;26:941–949. doi: 10.1021/acs.bioconjchem.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubarov A. S. Zakharova O. D. Koval O. A. Romaschenko A. V. Akulov A. E. Zavjalov E. L. Razumov I. A. Koptyug I. V. Knorre D. G. Godovikova T. S. Bioorg. Med. Chem. 2015;23:6943–6954. doi: 10.1016/j.bmc.2015.09.043. [DOI] [PubMed] [Google Scholar]
- Caspersen M. B. Kuhlmann M. Nicholls K. Saxton M. J. Andersen B. Bunting K. Cameron J. Howard K. A. Ther. Delivery. 2017;8:511–519. doi: 10.4155/tde-2017-0038. [DOI] [PubMed] [Google Scholar]
- Nakamura H. Kikuchi S. Kawai K. Ishii S. Sato S. Pure Appl. Chem. 2018;90:745–753. [Google Scholar]
- Era H., Terada S., Minami T., Takahashi T. and Arikawa T., in Heterogenity of Commercially Available Human Serum Albumin Products: Thiol Oxidation and protein Carbonylation, Birmingham, UK, in 37th Congress of IUPS, 2013 [Google Scholar]
- Miyamura S. Imafuku T. Anraku M. Taguchi K. Yamasaki K. Tominaga Y. Maeda H. Ishima Y. Watanabe H. Otagiri M. Maruyama T. J. Pharm. Sci. 2016;105:1043–1049. doi: 10.1016/j.xphs.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Watanabe H. Imafuku T. Otagiri M. Maruyama T. J. Pharm. Sci. 2017;106:2195–2203. doi: 10.1016/j.xphs.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Oettl K. Marsche G. Methods Enzymol. 2010;474:181–195. doi: 10.1016/S0076-6879(10)74011-8. [DOI] [PubMed] [Google Scholar]
- Kolb H. C. Finn M. G. Sharpless K. B. Angew. Chem., Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Tatiparti K. Sau S. Gawde K. A. Lyer A. K. Int. J. Mol. Sci. 2018;19:1–21. doi: 10.3390/ijms19030838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. Sachin K. Lee H. J. Kim D. W. Lee H. S. Bioconjugate Chem. 2012;23:2256–2261. doi: 10.1021/bc300364z. [DOI] [PubMed] [Google Scholar]
- Kolb H. C. Sharpless K. B. Drug Discovery Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- Moses J. E. Moorhouse A. D. Chem. Soc. Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- Agard N. J. Prescher J. A. Bertozzi C. R. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- van Dijk M. Rijkers D. T. S. Liskamp R. M. J. van Nostrum C. F. Hennink W. E. Bioconjugate Chem. 2009;20:2001–2016. doi: 10.1021/bc900087a. [DOI] [PubMed] [Google Scholar]
- Hoyle C. E. Lowe A. B. Bowman C. N. Chem. Soc. Rev. 2010;39:1355–1387. doi: 10.1039/b901979k. [DOI] [PubMed] [Google Scholar]
- Nair D. P. Podgórski M. Chatani S. Gong T. Xi W. Fenoli C. R. Bowman C. N. Chem. Mater. 2014;26:724–744. [Google Scholar]
- Rostovtsev V. V. Green L. G. Fokin V. V. Sharpless K. B. Angew. Chem., Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Meldal M. Tomøe C. W. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- Eriksson U. J. Borg L. A. Diabetologia. 1991;34:325–331. doi: 10.1007/BF00405004. [DOI] [PubMed] [Google Scholar]
- Pisanti F. A. Frascatore S. Vuttariello E. Grillo A. Biochem. Med. Metab. Biol. 1987;37:265–267. doi: 10.1016/0885-4505(87)90036-3. [DOI] [PubMed] [Google Scholar]
- Papaccio G. Pisanti F. A. Frascatore S. Diabetes. 1986;35:470–474. doi: 10.2337/diab.35.4.470. [DOI] [PubMed] [Google Scholar]
- Espeel P. Prez F. Adv. Polym. Sci. 2015;269:105–132. [Google Scholar]
- Espeel P. Du Prez F. E. Eur. Polym. J. 2015;62:247–272. [Google Scholar]
- Jakubowski H., Homocysteine in Protein Structure/Function and Human Disease: Chemical Biology of Homocysteine-containing Proteins, Springer-Verlag Wien, 2013, pp. 1–166 [Google Scholar]
- Mehta V. D. Kulkarni P. V. Mason R. P. Constantinescu A. Antich P. P. Bioconjugate Chem. 1994;5:257–261. doi: 10.1021/bc00027a011. [DOI] [PubMed] [Google Scholar]
- Shagufta W. Ahmad I. Eur. J. Med. Chem. 2018;143:515–531. doi: 10.1016/j.ejmech.2017.11.056. [DOI] [PubMed] [Google Scholar]
- Hui A. M. Zhang W. Chen W. Xi D. Purow B. Friedman G. C. Fine H. A. Cancer Res. 2004;64:9115–9123. doi: 10.1158/0008-5472.CAN-04-2740. [DOI] [PubMed] [Google Scholar]
- Hu R. Hilakivi-Clarke L. Clarke R. Oncol. Lett. 2015;9:1495–1501. doi: 10.3892/ol.2015.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S. Holz M. K. Signal Transduction Insights. 2016;10:1–7. doi: 10.4137/STI.S29901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Chin J. Im C. Y. Yoo E. K. Woo S. Hwang H. J. Cho J. H. Seo K. A. Song J. Hwang H. Kim K. H. Kim N. D. Yoon S. K. Jeon J. H. Yoon S. Y. Jeon Y. H. Choi H. S. Lee I. K. Kim S. H. Cho S. J. Eur. J. Med. Chem. 2016;120:338–352. doi: 10.1016/j.ejmech.2016.04.076. [DOI] [PubMed] [Google Scholar]
- Parney I. F. Chang S. M. Cancer J. 2003;9:149–156. doi: 10.1097/00130404-200305000-00003. [DOI] [PubMed] [Google Scholar]
- He W. Liu R. Yang S. H. Yuan F. Anti-Cancer Drugs. 2015;26:293–300. doi: 10.1097/CAD.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham C. D. Kaza N. Klocke B. J. Yancey Gillespie G. Shevde L. A. Carroll S. L. Roth K. A. J. Neuropathol. Exp. Neurol. 2016;75:946–954. doi: 10.1093/jnen/nlw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence A. M. Peterson R. A. Scharnhorst J. D. Silbergeld D. L. Rostomily R. C. J. Neurooncol. 2004;70:91–95. doi: 10.1023/b:neon.0000040837.68411.97. [DOI] [PubMed] [Google Scholar]
- Patel S. Dibiase S. Meisenberg B. Flannery T. Patel A. Dhople A. Cheston S. Amin P. Int. J. Radiat. Oncol., Biol., Phys. 2012;82:739–742. doi: 10.1016/j.ijrobp.2010.12.053. [DOI] [PubMed] [Google Scholar]
- Balça-Silva J. Matias D. Do Carmo A. Dubois L. G. Gonçalves A. C. Girao H. Silva Canedo N. H. Correia A. H. De Souza J. M. Sarmento-Ribeiro A. B. Lopes M. C. Moura-Neto V. Oncol. Rep. 2017;38:1341–1352. doi: 10.3892/or.2017.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmaksoud B. El-Azony A. J. Cancer Prev. Curr. Res. 2016;6:1–6. [Google Scholar]
- Agudelo D. Sanyakamdhorn S. Nafisi S. Tajmir-Riahi H. A. PLoS One. 2013;8:e60250. doi: 10.1371/journal.pone.0060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyakamdhorn S. Agudelo D. Bekale L. Tajmir-Riahi H. A. Colloids Surf., B. 2016;145:55–63. doi: 10.1016/j.colsurfb.2016.04.035. [DOI] [PubMed] [Google Scholar]
- Chubarov A. S. Shakirov M. M. Koptyug I. V. Sagdeev R. Z. Knorre D. G. Godovikova T. S. Bioorg. Med. Chem. Lett. 2011;21:4050–4053. doi: 10.1016/j.bmcl.2011.04.119. [DOI] [PubMed] [Google Scholar]
- Linkova M. G. Kuleshova N. D. Knunyants I. L. Russ. Chem. Rev. 1964;33:493–507. [Google Scholar]
- du Vigneaud V. Patterson W. I. Hunt M. J. Biol. Chem. 1938;126:217–231. [Google Scholar]
- Benesch R. Benesch R. E. J. Am. Chem. Soc. 1956;78:1597–1599. [Google Scholar]
- Leanza W. J. Chupak L. S. Tolman R. L. Marburg S. Bioconjugate Chem. 1992;3:514–518. doi: 10.1021/bc00018a009. [DOI] [PubMed] [Google Scholar]
- Kunzmann M. H. Staub I. Böttcher T. Sieber S. A. Biochemistry. 2011;50:910–916. doi: 10.1021/bi101858g. [DOI] [PubMed] [Google Scholar]
- Kratz F. Müller I. A. Ryppa C. Warnecke A. ChemMedChem. 2008;3:20–53. doi: 10.1002/cmdc.200700159. [DOI] [PubMed] [Google Scholar]
- Likhtenshtein G. I., Yamauchi J., Nakatsuji S., Smirnov A. I. and Tamura R., Nitroxides: Applications in Chemistry, Biomedicine, and Materials Science, Wiley-VCH, 2008, p. 419 [Google Scholar]
- Cornell C. N. Chang R. Kaplan L. J. Arch. Biochem. Biophys. 1981;209:1–6. doi: 10.1016/0003-9861(81)90250-2. [DOI] [PubMed] [Google Scholar]
- Brewer C. F. Riehm J. P. Anal. Biochem. 1967;18:248–255. [Google Scholar]
- Berliner L. J. Grunwald J. Hankovszky H. O. Hideg K. Anal. Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- Berliner L. J. Ann. N. Y. Acad. Sci. 1983;414:153–161. doi: 10.1111/j.1749-6632.1983.tb31681.x. [DOI] [PubMed] [Google Scholar]
- Peters T., All About Albumin, Academic Press, San Diego, 1996, p. 432 [Google Scholar]
- Wang Y. Wang S. Huang M. Curr. Pharm. Des. 2015;21:1831–1836. doi: 10.2174/1381612821666150302113906. [DOI] [PubMed] [Google Scholar]
- Schwartz M. Clin. Chim. Acta. 1982;124:213–223. doi: 10.1016/0009-8981(82)90389-8. [DOI] [PubMed] [Google Scholar]
- Jones R. G. A. Liu Y. Halls C. Thorpe S. J. Longstaff C. Matejtschuk P. Sesardic D. J. Pharm. Biomed. Anal. 2011;54:74–80. doi: 10.1016/j.jpba.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Moradi N. Ashrafi-Kooshk M. R. Chamani J. Shackebaei D. Norouzi F. J. Mol. Liq. 2018;249:1083–1096. [Google Scholar]
- Díaz N. Suárez D. Sordo T. L. Merz K. M. J. Med. Chem. 2001;44:250–260. doi: 10.1021/jm000340v. [DOI] [PubMed] [Google Scholar]
- Rehman M. Khan A. Curr. Pharm. Des. 2015;21:1785–1799. doi: 10.2174/1381612821666150304161201. [DOI] [PubMed] [Google Scholar]
- Kluger R. Tsui W. C. Can. J. Biochem. 1980;58:629–632. doi: 10.1139/o80-086. [DOI] [PubMed] [Google Scholar]
- Benson J. R. Baum M. Br. J. Cancer. 1996;74:993–994. doi: 10.1038/bjc.1996.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Stoll S. Schweiger A. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Stoll S. Schweiger A. Biol. Magn. Reson. 2007;27:299–321. [Google Scholar]
- Janatova J. Fuller J. K. Hunter M. J. J. Biol. Chem. 1968;243:3612–3622. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.