Summary
Ustilago maydis is the causal agent of maize smut disease. During the colonization process, the fungus secretes effector proteins that suppress immune responses and redirect the host metabolism in favor of the pathogen. As effectors play a critical role during plant colonization, their identification and functional characterization are essential to understanding biotrophy and disease.
Using biochemical, molecular, and transcriptomic techniques, we performed a functional characterization of the U. maydis effector Jasmonate/Ethylene signaling inducer 1 (Jsi1).
Jsi1 interacts with several members of the plant corepressor family Topless/Topless related (TPL/TPR). Jsi1 expression in Zea mays and Arabidopsis thaliana leads to transcriptional induction of the ethylene response factor (ERF) branch of the jasmonate/ethylene (JA/ET) signaling pathway. In A. thaliana, activation of the ERF branch leads to biotrophic susceptibility. Jsi1 likely activates the ERF branch via an EAR (ET‐responsive element binding‐factor‐associated amphiphilic repression) motif, which resembles EAR motifs from plant ERF transcription factors, that interacts with TPL/TPR proteins.
EAR‐motif‐containing effector candidates were identified from different fungal species, including Magnaporthe oryzae, Sporisorium scitamineum, and Sporisorium reilianum. Interaction between plant TPL proteins and these effector candidates from biotrophic and hemibiotrophic fungi indicates the convergent evolution of effectors modulating the TPL/TPR corepressor hub.
Keywords: EAR motif, ethylene response factor, jasmonate/ethylene (JA/ET) signaling, Jsi1, Topless, Ustilago maydis
Introduction
The biotrophic fungus Ustilago maydis causes smut disease on maize (Zea mays). During colonization, the fungus secretes manipulative molecules, termed effectors, that interfere with the host’s cellular machinery to suppress plant defense responses, redirect development, and enhance nutrient access (Win et al., 2012). As effectors play a critical role during plant colonization, their identification and functional characterization are essential to understanding the process of plant–pathogen interaction. In U. maydis, 467 genes were classified as putative secreted proteins (Lanver et al., 2017). To date, only a few have been characterized as effector proteins, and they have diverse functions during the biotrophic phase (Djamei et al., 2011; Redkar et al., 2015; Ma et al., 2018).
Plants coordinate pathogen‐specific immune responses through an elaborate crosstalk between hormone signaling pathways. Activation of salicylic acid (SA) signaling usually leads to activation of immune responses against biotrophic and hemibiotrophic pathogens. By contrast, jasmonate (JA) signaling leads to activation of immune responses to necrotrophic pathogens. Whereas ethylene (ET) signaling can be synergistic with JA signaling, SA and JA signaling are generally antagonistic to one another (Pieterse et al., 2012). In Arabidopsis thaliana, two major branches of the JA signaling pathway have been described. The MYC branch, controlled by MYC‐type transcription factors (TFs), is associated with wound response and defense against herbivorous insects. The ET response factor (ERF) branch is associated with resistance to necrotrophic pathogens. This branch is regulated by members of the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) family of TFs, like ERF1 and OCTADECANOID‐RESPONSIVE ARABIDOPSIS59 (ORA59), and leads to the transcriptional upregulation of PLANT DEFENSIN1.2 (PDF1.2), a well‐known marker of JA/ET signaling. The ERF branch is co‐regulated by JA and ET signaling (Lorenzo et al., 2003; McGrath et al., 2005; Dombrecht et al., 2007; Pré et al., 2008). Some evidence of SA–JA antagonism was shown in monocots, where overexpression of the key SA regulator NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES1 (OsNPR1) is followed by strong induction of SA‐responsive genes and suppression of JA‐responsive genes (Yuan et al., 2007). On the other hand, a dichotomy in resistance against biotrophic and necrotrophic pathogens is not quite so simple, as previous studies demonstrated that ET signaling could suppress Cochliobolus miyabeanus infection in A. thaliana but promote it in Oryza sativa (Völz et al., 2020).
Pathogens evolved strategies to manipulate defense hormone signaling to render plants more susceptible to infection. Effector proteins from Pseudomonas syringae interfere with activity of repressors of the JA signaling, leading to transcriptional activation of JA responses and, thus, promoting bacterial proliferation (Gimenez‐Ibanez et al., 2014; Yang et al., 2017). How fungal pathogens manipulate JA signaling is only poorly understood. The hemibiotrophic fungal pathogens Fusarium oxysporum f.sp. conglutinans and F. oxysporum f.sp. matthioli produce different JA conjugates and exhibit reduced virulence in the coronatine insensitive1 (coi1) mutant, indicating that JA signaling is involved in promoting Fusarium infection (Cole et al., 2014). Furthermore, previous studies identified JA signaling as a target for both a mutualistic fungus (Plett et al., 2014) and a pathogenic fungus (Patkar et al., 2015).
The A. thaliana TPL/TPR corepressor family is involved in several plant processes, including JA and auxin signaling (Szemenyei et al., 2008; Pauwels et al., 2010) and defense responses (Zhu et al., 2010). TPL/TPR proteins contain several conserved domains. The N‐terminal portion contains LIS1 homology (LisH), C‐terminal to LisH (CTLH), and CT11‐RanBPM (CRA) domains. The C‐terminal portion contains two WD40 domains (Martin‐Arevalillo et al., 2017). TPL/TPRs can interact with transcriptional regulators via short repression domains (Causier et al., 2012b) which include the ethylene‐responsive element binding‐factor‐associated amphiphilic repression (EAR) motif, defined by a consensus sequence of either LxLxL or DLNxxP (Kagale et al., 2010). Proteins with an LxLxL motif have been found to interact with the N‐terminal portion of TPL/TPR proteins (Szemenyei et al., 2008; Pauwels et al., 2010). By contrast, proteins with a DLNxxP motif interact with the C‐terminal portion of TPL/TPR proteins (Liu et al., 2019), but it is not known which of the two WD40 domains is responsible for the interaction with the DLNxxP motif, and the contribution of the C‐terminal portion to the transcriptional repression activity of the TPL/TPR proteins is also unclear. XopD, an effector possessing two LxLxL EAR motifs, was identified in the plant pathogen Xanthomonas euvesicatoria (Kim et al., 2013). XopD binds SlERF4, and its EAR motif is required for suppression of the plant immune response. Additionally, an effector from Ralstonia solanacearum, PopP2, possesses an LxLxL EAR motif that is required for avirulence and protein stability (Segonzac et al., 2017). No EAR‐motif‐containing effectors have been reported in any Ustilago pathosystem so far.
Here, we demonstrate that the U. maydis effector Jsi1 possesses a DLNxxP motif that interacts with the second WD40 domain of TPL/TPRs. Upon expression in A. thaliana, Jsi1 leads to induction of genes related to the ERF branch of JA/ET signaling, suggesting that binding to the second WD40 domain of TPL/TPRs may trigger this branch of the JA/ET signaling pathway. In addition, A. thaliana plants expressing Jsi1 are more susceptible to P. syringae infection, which would correlate with the induction of the ERF branch. In maize, Jsi1‐dependent interaction with TPL/TPRs leads to induction of ERF genes that could be associated with ERF‐branch activation in maize. Jsi1 could activate the ERF branch by interfering with the activity of endogenous DLNxxP‐motif‐containing ERF TFs. The identification of unrelated effector proteins from different fungal species with a DLNxxP motif and validation of the interaction between Magnaporthe oryzae, Sporisorium scitamineum, and Sporisorium reilianum effectors with TPL/TPRs indicate the convergent evolution of a strategy to manipulate this signaling hub in plants.
Material and Methods
Plant material, growth conditions, and plasmids
Zea mays cv Early Golden Bantam (EGB; Olds Seeds, Madison, WI, USA) was used for infection with U. maydis. Maize were grown in a glasshouse (16 h : 8 h, light : dark cycle, 28°C : 20°C). Nicotiana benthamiana plants were grown in a growth chamber (16 h : 8 h, light : dark cycle, 22°C, 60% humidity). Arabidopsis thaliana β‐estradiol inducible lines XVE‐jsi1‐mCherry and control XVE‐mCherry lines were created by transfer DNA insertion in Col‐0 background. Arabidopsis thaliana plants were grown in a growth chamber (12 h : 12 h, light : dark cycle, 21°C, 60% humidity). All plasmids used in this work are provided in Supporting Information Table S1. Detailed cloning, gene accession numbers, virulence assay and phytohormone measurements are provided in Methods S1.
Secretion experiments in axenic culture and in planta
Ustilago maydis strain AB33Potef jsi1‐3xHA was generated through insertion of plasmid pUG‐Potef‐Jsi1‐3xHA into the ip locus of AB33 according to Aichinger et al. (2003). We performed the secretion assay according to Brachmann et al. (2001). Mouse monoclonal anti‐hemagglutinin (HA; Sigma Aldrich) and anti‐actin (Invitrogen) antibodies were used for Western blot. The experiment was repeated with three independent transformant strains with similar results.
To visualize protein secretion in planta, we generated the SG200Δjsi1Pcmu1 Jsi1mCherry strain by integrating Jsi1‐mCherry under control of the cmu1 promoter in the ip locus. In addition, we built a nonsecreted version of the Jsi1‐mCherry strain (SG200Pcmu1 Jsi127641mCherry). We independently infected both strains in 7‐d‐old maize seedlings. mCherry fluorescence signal was detected using confocal microscopy at 3 d postinfection (dpi).
Yeast two‐hybrid assay
We performed yeast two‐hybrid (Y2H) assays with the Matchmaker™ GAL4 Two hybrid system (Clontech®, Mountain View, CA, USA) following the manufacturer's protocol. We fused the GAL4 activation domain of the prey vector pGG446 (modified version of pGADT7) to the genes Jsi127641, Jsi1m27641 , ZmERF4, and yellow fluorescent protein (YFP). We fused the GAL4 binding domain from the bait vector pGG187 (modified version of pGBKT7) to the genes ZmTPL1, ZmTPL2, ZmTPL3, TPL (AT1G15750), TPR1 (AT1G80490), TPR2 (AT3G16830), TPR4 (AT3G15880), YFP, and N and C‐terminal portions of the different topless orthologues. We transformed the combinations of pGG446 and pGG187 vectors carrying the different genes in the yeast strains Y187 (MAT α) and AH109 (MAT a), respectively. We selected diploid yeast after mating for growth on (SD)−Leu/−Trp and (SD)−Leu/−Trp/−His plates at 28°C for 4 d. We repeated the experiments twice from independent mating events.
Co‐immunoprecipitation assay in N. benthamiana and Z. mays
We infiltrated 4‐wk‐old N. benthamiana leaves with Agrobacterium tumefaciens carrying different genes cloned into an expression vector as described (Ma et al., 2012). Cultures carrying the different gene combinations were infiltrated in six leaves (three plants, two leaves from each plant). A 450 mg sample of tissue powder was suspended in 2 ml cold extraction buffer for protein extraction (50 mM Hepes pH 7.5, 100 mM sodium chloride, 10% v/v glycerol, 1 mM EDTA, 0.1% v/v Triton X‐100, 2% polyvinylpolypyrrolidone, 1 mM dithiothreitol, 1 mM phenylmethanesulfonylfluoride, and EDTA‐Free Protease Inhibitor cocktail; Roche). Protein pull‐down was performed using the μMACS™ MicroBeads system from Miltenyi Biotech (Bergisch Gladbach, Germany) following the manufacturer's instructions.
We quantified protein signals of ZmTPL1, ZmERF4, and the different versions of Jsi1 in the input. The protein signals of ZmTPL1, ZmERF4, and the different versions of Jsi1 were normalized to the respective Rubisco (Ponceau) protein signal. Fold change (FC) for each protein was shown relative to the normalized protein value observed in ZmTPL1 co‐expressed with YFP or Jsi1 and ZmERF4. Quantification of pulled‐down proteins signal of ZmERF4 and the different versions of Jsi1 were normalized to their respective pulled‐down ZmTPL1 protein signal. FC for each protein was shown relative to the value in ZmTPL1 co‐expressed with Jsi1 and ZmERF4. FC ± SD values are means of three biological replicates for all the experiments.
In the case of maize, U. maydis strains SG200Pcmu1 jsi1‐3xHA, SG200Pcmu1 jsi1m‐3xHA, and SG200Pcmu1‐SPcmu1(1‐22)‐mCherry‐3xHA were generated by integration of the different constructs into the ip locus of SG200. We infected 7‐d‐old seedlings with each strain (30 plants per strains). Infected tissue was collected 7 dpi. The co‐immunoprecipitation (Co‐IP) protocol was the same as for N. benthamiana.
We detected the immunoprecipitated proteins with anti‐MYC (Sigma Aldrich), anti‐HA, anti‐mCherry (Abcam), or anti‐green fluorescent protein (GFP; Miltenyi Biotech) antibodies depending on the experiment. The TPL‐specific antibody was raised using a small peptide, CNEQLSKYGDTKSAR, selected from a conserved region of the TPL/TPR proteins. The polyclonal antibody was produced in rabbit by Eurogentec (Seraing, Belgium). We repeated each experiment three times.
Arabidopsis thaliana RNA‐sequencing sample collection
Arabidopsis thaliana seeds from XVE‐jsi1‐mCherry‐1/2 and XVE‐mCherry lines were grown vertically on square plates containing Murashige & Skoog medium for 7 d. Arabidopsis thaliana seedlings were transferred to square plates with the same media containing 5 µM β‐estradiol and incubated for 6 h. Three independent replicates for each genotype were collected. Mock treatment was only performed for the control line to confirm that the concentration of β‐estradiol used for the experiment did not itself alter gene‐expression.
RNA‐sequencing analysis
We removed adapter sequences and performed quality trimming using Trimmomatic (Bolger et al., 2014). Reads were mapped to the reference genome using Star, v.2.7.0e (Dobin et al., 2013) with the parameter outFilterMismatchNoverLmax 0.05. We input the bam files to R v.3.5.1 using the package R/samtools. We obtained the genome annotation from Araport11 and gene models and read counts per gene were obtained with the packages Genomic Features and Genomic Alignments, respectively. We removed low‐expressed genes and analyzed 28 843 for differential expression using DESeq2 after performing regularized log transformation (Love et al., 2014). We compared all the replicates from β‐estradiol‐induced XVE‐jsi1‐mCherry lines with the replicates from control XVE‐mCherry lines with and without β‐estradiol induction and kept genes with log FC ˃ 1.5 and adjusted P < 0.05. We performed Gene Ontology (GO)‐term analysis for biological processes using the ThaleMine tool (Krishnakumar et al., 2014). The data sets were deposited in National Center for Biotechnology Information’s (NCBI's) Gene Expression Omnibus and are accessible through GEO Series accession no. GSE142128 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142128).
To assess the significance of enrichment for TF binding sites, we first determined the direct target genes of ERF3, 4, 7, 8, 10, 11 and ZAT10 using available DNA affinity purification sequencing (DAP‐seq) data (O’Malley et al., 2016). We overlapped each list of putative direct target genes with genes upregulated upon jsi1 induction (FC ˃ 1.5, P < 0.05). We determined the significance of the overlapping genes with Fisher's exact test using the R package geneoverlap function newGOM (https://github.com/shenlab‐sinai/GeneOverlap).
Reverse transcription PCR for RNA‐sequencing validation
Total RNA was extracted from three independent replicates from each A. thaliana line (XVE‐jsi1‐mCherry‐1 and 2 and XVE‐mCherry) using the same protocol for RNA‐sequencing (RNA‐seq) samples. Complemetary DNA (cDNA) was generated from total RNA using the iScript cDNA synthesis kit (Bio‐Rad). We performed quantitative reverse transcription (qRT)‐PCR using FastStart Universal SYBR Green Master mix (Roche) according to the manufacturer's instructions. The relative amount of amplicons in the samples were calculated with the method (Livak & Schmittgen, 2001) with actin2 (AT3G18780) as the reference gene (Czechowski et al., 2005). We calculated FC in the expression level of each gene in the XVE‐jsi1‐mCherry lines compared with the XVE‐mCherry line, and data are represented for each jsi1‐mCherry line as the mean of three replicates. We calculated statistically significant differences in gene expression between each jsi1‐mCherry line and mCherry line using ANOVA followed by Dunnett’s multiple comparison test with P < 0.05.
For evaluation of the induction of the ERF branch in maize, 10 EGB seedlings infected with SG200 were collected at 4 and 6 dpi. Mock seedlings were infected with water, and tissue was collected for each time point. Three independent replicates were performed for infected and mock tissue at each time point. RNA extraction, cDNA synthesis, and qRT‐PCR and data analysis were performed as previously described but using a cyclin‐dependent kinase (CDK; GRMZM2G149286) as the reference gene (Lin et al., 2014). Primers used for RT‐PCR are described in Table S2.
Biolistic transformation of maize for localization and gene induction analysis
We bombarded 6‐d‐old maize leaves with 1.6 µm gold particles coated with 5 µg of each plasmid as described by Djamei et al., (2011). Fluorescence emission was observed 1 d after transformation by confocal microscopy. For gene induction analysis, we bombarded 7 µg of the corresponding plasmids (35S‐Jsi1‐mcherry or 35S‐Jsi1m‐mcherry) into 12‐d‐old maize leaves. Samples were harvested 10 h after bombardment for RNA extraction and qRT‐PCR.
Identification of putative secreted effector proteins with a DLNxxP motif
We downloaded predicted protein sequences of the different plant pathogens from EnsemblFungi (https://fungi.ensembl.org/index.html) or NCBI (https://www.ncbi.nlm.nih.gov/). To identify putative secreted effector proteins with a DLNxxP motif, we searched for the DLNxxP motif in all predicted proteins from the different fungal species using CLC Main Workbench 7.7.2 (Qiagen). Among all the DLNxxP‐motif‐containing proteins, we searched for those with a predicted secretion signal (SignalIP‐5.0), lacking transmembrane domains (Tmhmm v.2.0 from http://www.cbs.dtu.dk/services/), and no predicted enzymatic domains (InterPro, https://www.ebi.ac.uk/interpro/beta/).
Results
Jsi1 interacts with the C‐terminal portion of Topless
As EAR‐motif‐containing effectors can be important for the establishment of plant‐pathogen interactions, we screened putative U. maydis effector proteins for the presence of the DLNxxP EAR motif and identified the gene jsi1 (UMAG_01236), located in the effector cluster 2A (Table 1; Fig. S1a; Kämper et al., 2006). Jsi1 is transcriptionally induced during biotrophy and its expression peaks 4 dpi (Fig. S1b). To test if Jsi1 without signal peptide (Jsi127641) can interact with TPL, we cloned three Z. mays TPL orthologues: ZmTpl1, ZmTpl2 and ZmTpl3 (Fig. S1c). Jsi127641 interacts with all three ZmTPLs in Y2H assays (Figs 1a, S1d). To identify which TPL domain interacts with Jsi1, we split ZmTPL1 into its N‐terminal portion comprising the LisH, CTLH, and CRA domains (ZmTPL1Nt) and C‐terminal portion containing the WD40 domains (ZmTPLCt). We found that Jsi127641 interacts with ZmTPLCt in Y2H assays (Fig. 1a). To identify which of the two WD40 repeats is responsible for this interaction, we further divided ZmTPLCt into two fragments, each containing a single WD40 repeat (WD40‐1 and WD40‐2) and tested them for interaction by Y2H assays. Jsi127641 specifically interacted with WD40‐2 (Fig. 1b). To determine if Jsi1 is able to interact with ZmTPL/TPR proteins in maize, we created a U. maydis strain expressing Jsi1‐3xHA under the control of the strong biotrophy‐induced cmu1 promoter to increase the protein expression level of Jsi1 during infection. In addition, we raised an anti‐TPL antibody that was tested for specificity with TPL proteins from different plant species (Fig. S1e). We immunoprecipitated Jsi1‐3xHA from infected maize seedlings and were able to detect co‐immunoprecipitated TPL/TPR proteins by Western blot (Figs 1c, S2). To test the specificity of the Jsi1/TPL interaction, we mutated the DLNxxP EAR motif in Jsi1 to AHNxxP (Jsi1m). We found that Jsi1m did not interact with ZmTPL1 in either Y2H or in planta Co‐IP assays (Fig. 1a,c), indicating a critical role for the DLNxxP EAR motif in the interaction between Jsi1 and TPL/TPR proteins.
Table 1.
Fungal effector proteins possessing an DLNxxP motif.
| Protein ID | Length (aa) | SignalP a | DLNxxP | Species | Lifestyle | ||
|---|---|---|---|---|---|---|---|
| Score signal | Cleavage site (aa) | Location (aa) | Sequence motif | ||||
| UMAG_01236 | 641 | 0.8706 | 26 | 39–44 | DLNELP | Ustilago maydis | Biotrophic |
| UMAG_01237 | 633 | 0.9817 | 21 | 36–41 | DLNKLP | ||
| UMAG_05303 | 193 | 0.991 | 21 | 53–58 | DLNFHP | ||
| UMAG_02826 | 399 | 0.9677 | 22 | 251–256 | DLNIAP | ||
| Sr10432 | 120 | 0.9884 | 23 | 104–109 | DLNKHP | Sporisorium reilianum | Biotrophic |
| Sr10312 | 631 | 0.9564 | 23 | 36–41 | DLNEIP | ||
| Sr13382 | 289 | 0.9961 | 21 | 48–53 | DLNQPP | ||
| SPSC_03537 | 653 | 0.9437 | 20 | 27–32 | DLNKIP | Sporisorium scitamineum | Biotrophic |
| PTTG_28402 | 394 | 0.9663 | 31 | 57–62 | DLNSIP | Puccinia triticina | Biotrophic |
| PTTG_07660 | 442 | 0.9345 | 24 | 135–140 | DLNGTP | ||
| PTTG_27442 | 229 | 0.9321 | 23 | 38–43 | DLNEFP | ||
| PTTG_27452 | 529 | 0.9315 | 23 | 38–43 | DLNEFP | ||
| PTTG_27005 | 407 | 0.921 | 25 | 30–35 | DLNLPP | ||
| PTTG_26956 | 418 | 0.728 | 25 | 312–317 | DLNDRP | ||
| PTTG_05870 | 213 | 0.8818 | 21 | 77–82 | DLNNVP | ||
| PTTG_26367 | 313 | 0.6855 | 19 | 40–45 | DLNEYP | ||
| PTTG_25346 | 473 | 0.7534 | 23 | 37–42 | DLNAFP | ||
| CSEP0438 | 227 | 0.9855 | 26 | 145–150 | DLNYYP | Blumeria graminis | Biotrophic |
| FOXG_20822 | 81 | 0.993 | 17 | 23–28 | DLNRDP | Fusarium oxysporum | Hemibiotrophic |
| MGG_15391 | 222 | 0.8834 | 23 | 94–99 | DLNKAP | Magnaporthe oryzae | Hemibiotrophic |
| MGG_05887 | 247 | 0.9944 | 16 | 131–136 | DLNKVP | ||
Program used to bioinformatically predict the secretion signal in proteins (http://www.cbs.dtu.dk/services/signalP/). aa, amino acids.
Fig. 1.
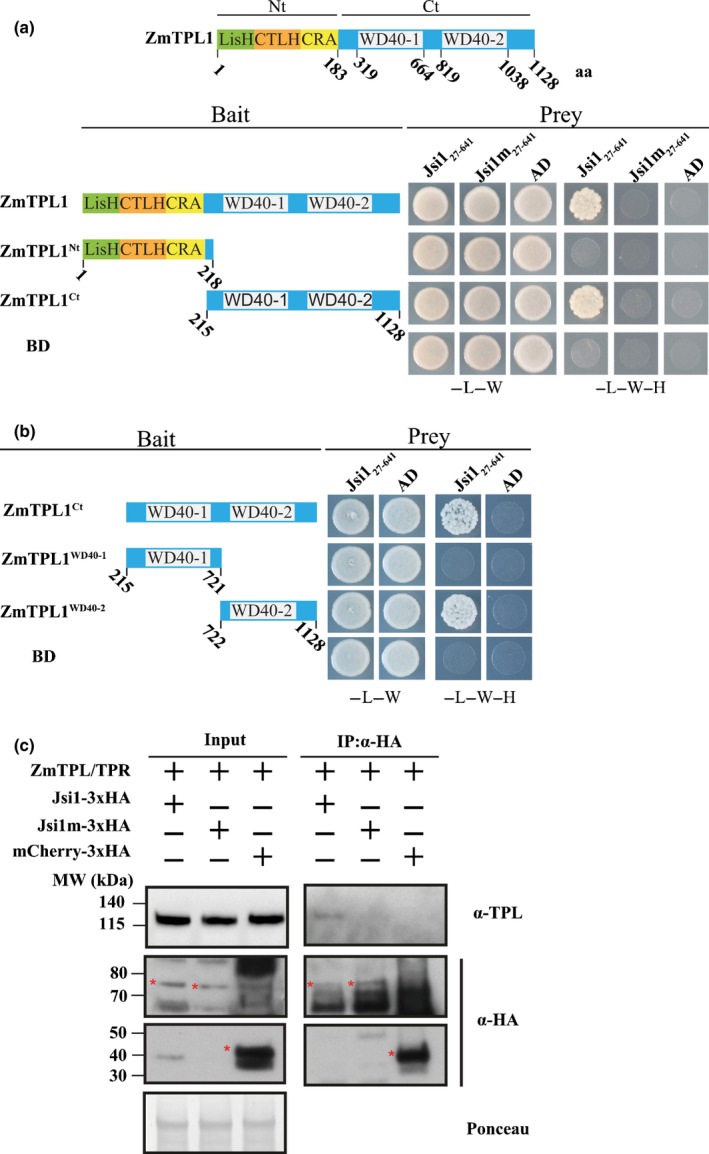
Jasmonate/Ethylene signaling inducer 1 (Jsi1) interacts with the second WD40 domain of ZmTPL1 through its DLNxxP motif. (a) Yeast two‐hybrid (Y2H) assay with Jsi127641 or the mutant version Jsi1m27641 as prey and full‐length ZmTPL1 or its N and C‐terminal regions (ZmTPL1Nt and ZmTPL1Ct) as bait. (b) ZmTPL1WD401 and ZmTPL1WD402 each containing one of the WD40 repeats were used as baits to test which WD40 domain interacts with Jsi1 in Y2H assay. As a negative control, we used enhanced yellow fluorescent protein fused to the GAL4‐binding domain (BD) and GAL4 activation domain (AD). −L, −W and −H indicate medium lacking leucine, tryptophan, and histidine, respectively. (c) Co‐immunoprecipitation (Co‐IP) assay showing that Jsi1 interacts with ZmTPL/TPRs in Zea mays. We infected maize seedlings with Ustilago maydis strains expressing Jsi1‐3xHA, Jsi1m‐3xHA and mCherry‐3xHA and performed a Co‐IP using anti‐hemagglutinin (HA) antibody. Topless (TPL)‐specific antibody shows that endogenous maize Topless/Topless related (TPL/TPR) proteins are co‐purified with Jsi1‐3xHA but not with Jsi1m‐3xHA or mCherry‐3xHA. Red asterisks indicate the full‐length proteins of Jsi1‐3xHA, Jsi1m‐3xHA and mCherry‐3HA. Ponceau staining was used to ensure equal loading. To detect mCherry, membranes were exposed between 15 and 30 min, whereas for Jsi1 and Jsi1m the membranes were exposed longer, between 3 and 4 h.
Jsi1 is a secreted effector located in the nucleus of maize cell leaves
To test whether Jsi1 is secreted, we integrated a version of Jsi1‐3xHA into U. maydis strain AB33, which is commonly used to study effector secretion (Tollot et al., 2016). As expected for a secreted protein, we detect Jsi1‐3xHA in the culture supernatant by Western blot. Actin, which served as a lysis control, was only present in whole‐cell extracts (Fig. 2a). To confirm that Jsi1 is secreted in planta, we expressed a Jsi1‐mCherry fusion protein in strain SG200Δjsi1, where the endogenous jsi1 locus was deleted. To increase protein levels for visualization, jsi1‐mCherry was expressed by the cmu1 promoter. Jsi127641‐mCherry, without signal peptide, was used as a negative control. We observed Jsi1‐mCherry fluorescence around and outside the fungal hyphae whereas Jsi127641‐mCherry was localized inside the fungal hyphae, indicating that Jsi1 is secreted by U. maydis in planta (Fig. 2b). To determine the subcellular localization of Jsi1 in maize cells, we transiently co‐transformed a Jsi127641‐mCherry construct with a GFP‐nuclear localization signal construct as a nuclear marker into maize leaves. Using confocal microscopy, we found Jsi127641‐mCherry signal inside the plant nucleus 1 d after biolistic transformation (Fig. 2c). To test whether ZmTPL1 colocalizes with Jsi1 in maize cells, we co‐transformed a ZmTPL1‐GFP construct with Jsi127641‐mCherry. ZmTPL1‐GFP signal emission overlapped with the Jsi127641‐mCherry signal in the nucleus, indicating co‐localization of both proteins (Fig. 2d).
Fig. 2.
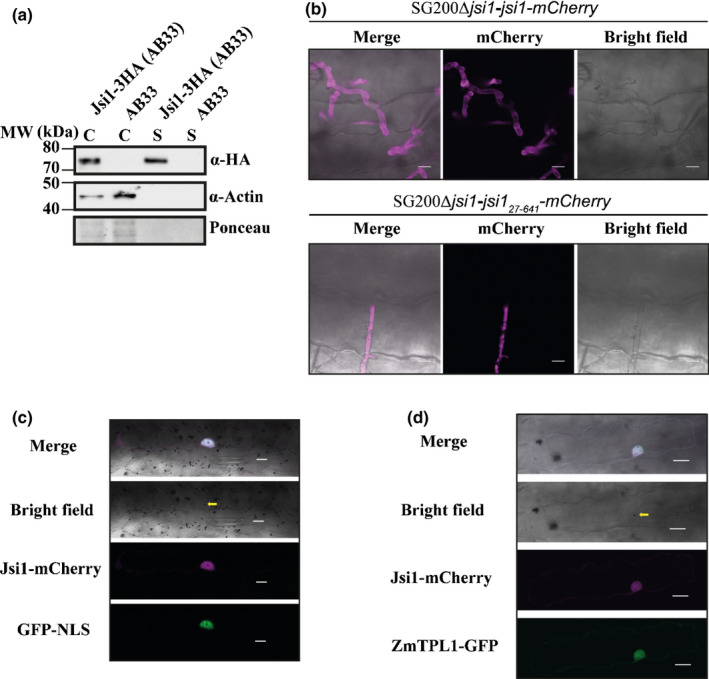
Jasmonate/Ethylene signaling inducer 1 (Jsi1) is a secreted effector that is targeted to the plant cell nucleus. (a) Jsi1 is secreted in axenic culture. We expressed Jsi1‐3xHA in the Ustilago maydis strain AB33. Proteins from filamentous cells and culture supernatants were subjected to Western blot analysis using anti‐hemagglutinin (HA) and anti‐actin antibodies. We used actin as a control of cell lysis, and it was only detected in whole‐cell extracts (C). Jsi1‐3xHA was detected in whole cell extracts (C) and culture supernatants (S). (b) Jsi1‐mCherry is secreted by U. maydis in maize. Confocal images of infected maize leaves 3 d postinfection with SG200Δjsi1‐Jsi1‐mCherry and SG200Δjsi1‐Jsi127641‐mCherry (a nonsecreted version of Jsi1). Bars, 10 µm. (c) Jsi1 localizes to the nucleus of maize cells. Maize cells expressing Jsi127641‐mCherry and green fluorescent protein–nuclear localization signal (GFP‐NLS) as a nuclear marker after biolistic transformation of leaves. (d) Jsi1 and ZmTPL1 co‐localize in the nucleus of maize leaf cells. Maize cells expressing Jsi127641‐mCherry and ZmTPL1‐GFP. The yellow arrow indicates the transformed cell with the gold particle inside the nucleus. Bars, 20 µm.
Jsi1 belongs to the U. maydis cluster 2a, which was previously shown to cause a mild hypervirulence phenotype in maize when deleted (Kämper et al., 2006). To test whether Jsi1 contributes to virulence, we infected maize seedlings with three independent strains mutated in the jsi1 locus (SG200Δjsi1 1 to 3). SG200Δjsi1 mutant strains showed no impaired ability to filament on charcoal, a prerequisite for infection (Fig. S1f). Plants infected with the mutant strains did not show any significant changes in symptom development 12 dpi when compared with plants infected with SG200 (Fig. S1g), which may indicate redundancy with other effectors possessing an EAR motif (Table 1).
Jsi1 activates jasmonate/ethylene and salicylic acid signaling in A. thaliana
Since Jsi127641 also binds to TPL/TPR proteins from A. thaliana (Fig. S3a), we were able to study which pathways are manipulated by Jsi1 in planta. We generated two independent A. thaliana lines expressing Jsi127‐641‐mCherry under the control of the estradiol‐inducible XVE system (XVE‐jsi1‐mCh 1 and 2) as well as an XVE‐mCherry (XVE‐mCh) control line. We confirmed expression of the transgenes 6 h after β‐estradiol induction by Western blotting (Fig. S3b) and then subjected induced samples to RNA‐seq. Principal component analysis of the resulting transcriptomes showed that replicates from the two Jsi1 lines group together and are separate from the replicates of the control line, which also clustered (Fig. S3c). Using cutoffs of FC > 1.5 and P < 0.05, we identified 1090 differentially expressed genes (DEGs) in Jsi1 lines relative to the control, 915 of which were upregulated and 175 were downregulated. The more than five times higher number of upregulated than downregulated genes is consistent with the model that Jsi1 interferes with the repressor‐function of TPL/TPR proteins. GO‐term analysis for biological processes show several categories related to ‘responses to different stimulus’. Within these, ‘responses to stress’, ‘defense responses’, ‘responses to external stimulus’, and ‘response to biotic stimulus’ were the major categories with 26% to 12% of the total DEGs. This indicates that Jsi1 induces plant immune responses in A. thaliana (Fig. S4a). In addition, we identified two GO categories related to hormone responses: ‘response to salicylic acid’ and ‘ethylene response genes’.
In the ET response gene category, 14 DEGs belong to the AP2/ERF family of TFs. Seven of these belong to the B3 group of the ERF subfamily, which are characterized as being positive regulators of transcription. Of these, ERF2, ERF5, ERF6 and ERF107 have been associated with defense responses against necrotrophic infections and are positive regulators of the defense response gene PDF1.2 (McGrath et al., 2005; Moffat et al., 2012; Ju et al., 2017) (Fig. 3a; Table S3). Two other TFs of the B3 group, ERF1 and ORA59, were found to be transcriptionally controlled by JA and ET and induce PDF1.2 expression (Lorenzo et al., 2003; Pré et al., 2008). Even though we did not find ERF1 and ORA59 to be upregulated upon jsi1 induction (Fig. S3e), a comparison of genes upregulated by Jsi1 and those found to be induced by ERF1 and ORA59 in previous studies show a 20% and 30% overlap, respectively (Fig. 3b). Among the upregulated genes shared by Jsi1 with ERF1 and/or ORA59, we note the defense‐related genes OSM34, PR5 and PDF1.2 (Table S3). In addition, Jsi1 also induces three 1‐aminocyclopropane‐1‐carboxylate synthases (ACSs), ACS2, ACS6, and ACS11, and MAP KINASE KINASE 9 (MKK9), which are involved in ET biosynthesis (Xu et al., 2008; Tsuchisaka et al., 2009) (Table S3). We validated the expression of some ERFs, ACSs, MKK9, and defense‐related genes by qRT‐PCR upon jsi1 expression (Fig. S3 e). Taken together, our results show that Jsi1 induces the expression of several ERFs, genes related with ET synthesis and defense genes, including PDF1.2, indicating that Jsi1 induces the ERF branch of the JA/ET signaling pathways.
Fig. 3.
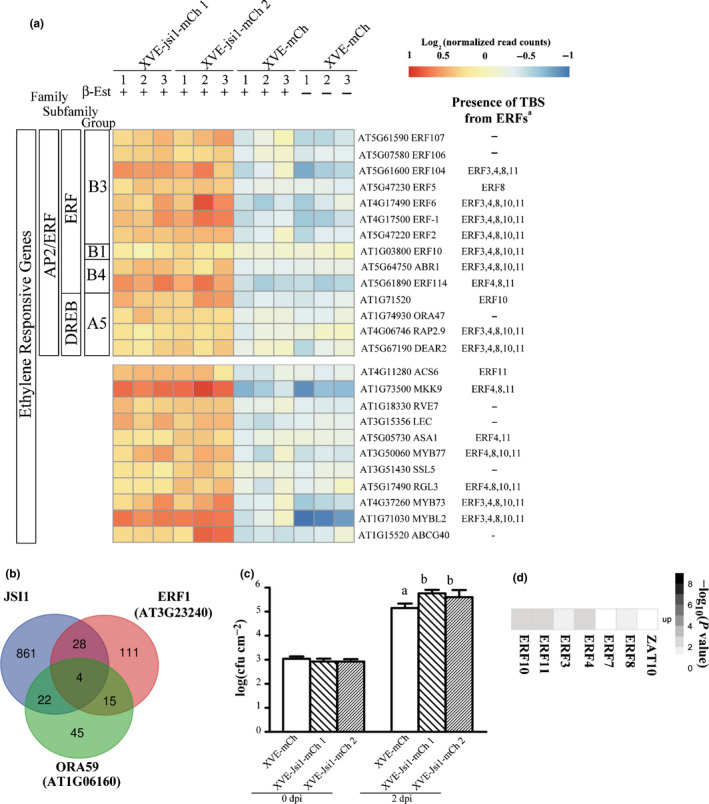
Jasmonate/Ethylene signaling inducer 1 (Jsi1) activates jasmonate/ethylene signaling leading to biotrophic susceptibility. (a) Heat map from RNA sequencing showing ethylene‐responsive genes. Numbers under the lines represent the replicate number. †Genes enriched in transcription binding sites (TBSs) from ethylene response factors (ERFs) with repression activity. (b) Venn diagram showing transcriptionally induced genes shared by Arabidopsis thaliana plants expressing either Jsi1, ERF1 or ORA59. (c) Pseudomonas syringae pv tomato (Pst) DC3000 proliferate better in A. thaliana plants expressing Jsi1. Infected leaves were collected at 0 d postinfection (dpi) and 2 dpi to quantify bacterial proliferation. The graph shows one representative replicate of three repeated experiments. Different letters indicate statistically significant differences among the different genotypes, which were calculated by Tukey's honestly significant difference post‐hoc test (P < 0.05). log(CFU cm−2) ± SD: log scale of colony forming units per square centimeter. (d) Genes upregulated upon jsi1 induction are enriched in TBSs of ERFs with a DLNxxP motif. Matrix summarizing the overlap enrichment between putative direct target genes of ERFs and ZAT10 from previously available DNA affinity purification sequencing data and genes upregulated upon jsi1 induction. Significance of enrichment of TBSs for each TF was determined by Fisher’s exact test (P < 0.05).
To test whether U. maydis is able to induce genes connected with the ERF branch in maize, we searched for genes orthologous to those induced by Jsi1 in A. thaliana and tested their expression by qRT‐PCR. We selected ZmERF1, ZmERF1a, ZmERF2 and ZmERF105 (which belong to the B3 group of the AP2/ERF family), ZmERF12 (which belongs to the B1 group of the AP2/ERF family characterized as repressors of transcription) (Du et al., 2014), and ZmACS6, ZmPR5 and ZmOSM34. Gene induction was evaluated during U. maydis infection at 4 and 6 dpi where Jsi1 expression is relatively high (Figs 4a, S1b). Most genes tested were found to be significantly induced at 4 and 6 dpi, with the exceptions of ZmERF2 and ZmOSM34 (which were not found to be induced) and ZmERF1 (which was significantly induced only at 4 dpi) (Fig. 4a). These data indicate that U. maydis infection induces genes associated with the ERF branch in maize. To test whether Jsi1 is able to induce these genes in maize, we used detached maize leaves to express Jsi1 and Jsi1m under the 35S promoter via biolistic bombardment and evaluated gene expression 10 h after bombardment. ZmERF1, ZmERF1a and ZmPR5 were induced by expression of Jsi1‐mCherry compared with Jsi1m‐mCherry in three replicates, whereas ZmERF105 was induced in two replicates, and ZmERF12 and ZmACS6 were only induced in a single replicate (Fig. 4b). Our results demonstrate that Jsi1 induces genes connected with the ERF branch in maize, and the DLNxxP EAR motif, which is required for interaction with TPL/TPRs, plays an important role in this induction.
Fig. 4.
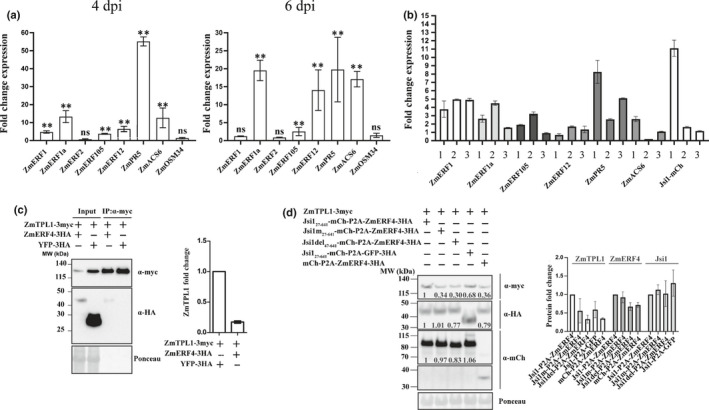
Induction of ethylene response factor (ERF)‐branch genes upon Ustilago maydis infection and Jasmonate/Ethylene signaling inducer 1 (Jsi1) overexpression. (a) Quantitative reverse transcription (qRT)‐PCR evaluation of maize orthologues of ERF branch upon U. maydis infection at 4 d postinfection (dpi) and 6 dpi. Fold change (FC) ± SD is relative to the expression in maize plants without U. maydis infection and normalized to the cyclin‐dependent kinase (CDK) RNA expression values. Values shown are the means of three replicates. Statistically significant differences between genes expressed in U. maydis‐infected maize tissue and mock were calculated using Mann–Whitney test (**, P < 0.01; ns, not significant). (b) qRT‐PCR evaluation of maize orthologues to the ERF branch upon Jsi1‐mcherry overexpression via biolistic bombardment. Each bar represents an independent biological replicate, three replicates per gene. FC ± SD is relative to the expression observed upon Jsi1m‐mcherry expression and normalized to the CDK RNA expression values. Values shown are the means of two technical replicates. (c) ZmERF4 interacts and destabilizes ZmTPL1 in Nicotiana benthamiana. We performed co‐immunoprecipitation (Co‐IP) using anti‐myc antibody. Co‐IP shows interaction between ZmERF4 and ZmTPL1. Quantification of the ZmTPL1 protein signal in the input in presence of yellow fluorescent protein (YFP) or ZmERF4 was represented as FC ± SD. (d) Jsi1 interferes with the destabilization of ZmTPL1 mediated by ZmERF4 in N. benthamiana. Protein signal quantification of ZmTPL1, ZmERF4, and the different versions of Jsi1 in the input were normalized to the respective Rubisco signal (Ponceau staining). FC for each protein was expressed relative to the normalized protein value observed in ZmTPL1 co‐expressed with YFP (c) or Jsi1 and ZmERF4 (d). FC ± SD values represented in the bar graph are means of three biological replicates.
Regarding the GO category of SA‐response genes, we identified 39 upregulated genes, including GRX480, ALD1 and WRKY70, which have been described as SA‐regulated genes (Li et al., 2004; Herrera‐Vásquez et al., 2014; Cecchini et al., 2015) (Fig. S6). In addition, we observed a cell‐death phenotype 3 d after expression of Jsi1 in A. thaliana plants (Fig. S5c). The cell‐death phenotype was also observed for prolonged expression of Jsi1 in N. benthamiana leaves 5 dpi (Fig. S5b). We further found that cell‐death induction correlates with the presence of the TPL/TPRs interaction EAR motifs, as shown in maize plants locally overexpressing Jsi1 after 10 dpi with the Foxtail mosaic virus‐based overexpression system and in N. benthamiana leaves (Fig. S5a,b) (Bouton et al., 2018). Activation of SA signaling could be due to recognition of the Jsi1–TPL interaction by the plant immune system, as previously reported (Gawehns, 2014), or the Jsi1–TPL interaction could interfere with the repressive activity of TPL‐interacting transcriptional regulators involved in suppressing the SA signaling pathway.
Jsi1 promotes biotrophic susceptibility in A. thaliana
In order to assess whether the transcriptional changes observed in the JA/ET and SA signaling pathways correlated with changes in SA and JA hormone levels, we measured the levels of SA, JA and JA‐Ile, the bioactive form of JA, in A. thaliana shoots. The two A. thaliana XVE‐jsi1‐mCh lines expressing jsi1 showed a significant increase in SA compared with the XVE‐mCh line, whereas we could not detect JA‐Ile or JA in either the XVE‐jsi1‐mCh lines or in the control (Fig. S3d). The lack of JA and JA‐Ile indicates activation of the ERF branch by Jsi1 is independent of the hormone itself. Activation of SA signaling should lead to repression of JA/ET signaling, as extensive crosstalk between these two signaling pathways has been reported (Caarls et al., 2015), and would increase resistance to biotrophic infection. To test how Jsi1 expression in A. thaliana impacts biotrophic susceptibility, we tested the XVE‐jsi1‐mCh and XVE‐mCh control lines after estradiol treatment for their susceptibility towards the hemibiotrophic pathogen Pst DC3000. We sprayed jsi1‐expressing and control lines with 150 nM estradiol to avoid the cell‐death phenotype associated with prolonged expression of jsi1 during Pst DC3000 infection (Fig. S5c). Jsi1‐expressing lines were more susceptible to Pseudomonas syringae pv tomato (Pst) DC3000 infection than the control (Fig. 3c), indicating that activation of SA signaling in this context does not interfere with biotrophic susceptibility.
Jsi1 may alter the repressing activity of ethylene response factors
The activation of JA/ET and SA signaling by Jsi1 could be a consequence of its interaction with TPL/TPR proteins, leading to interference with the repressive activity of endogenous DLNxxP‐containing transcriptional regulators. In A. thaliana, 67 transcriptional regulators were identified with a predicted DLNxxP motif from the B1 group of AP2/ERF and C2H2 families (Kagale et al., 2010). TFs that interact with TPL/TPRs are mainly negative regulators of transcription (Causier et al., 2012b). We therefore focused on genes that are upregulated in the Jsi1‐expressing lines that could be targets for DLNxxP‐containing TFs. Using previously available DAP‐seq data, we first determined genome‐wide putative direct target genes of six ERFs from the B1 subfamily, ERF3, 4, 7, 8, 10 and 11, and ZAT10 from the C2H2 family (O'Malley et al., 2016). Next, we compared the list of putative target genes for each TF with those genes upregulated in the jsi1‐expressing lines. Significant enrichment for each TF was evaluated by Fisher’s exact test (P < 0.05). Except for ERF7 and ZAT10, transcriptional targets of the other ERFs were strongly enriched for genes de‐repressed by Jsi1. In total, 269 of the 915 genes upregulated by jsi1 expression possess at least one transcription binding site (TBS) for an ERF (Fig. 3d; Table S4). GO‐term analysis of these upregulated genes showed 26 categories, 24 of which were previously identified in the GO‐term analysis of the Jsi1 RNA‐seq (Fig. S4b). In fact, from the 25 upregulated genes identified in the RNA‐seq that respond to ET, 72% possess TBSs for ERFs and only 33% of the 39 upregulated SA‐responsive genes possess TBSs for ERFs (Figs 3a, S6). This indicates that Jsi1 may regulate the expression of several ET‐responsive genes by altering the repressive activity of ERFs via interference with AtTPL/TPR proteins. Regarding the SA‐responsive genes, some of them might be regulated by ERFs. However, other unknown TFs whose interaction with TPL is altered by Jsi1 cannot be excluded.
To test whether Jsi1 can interfere with the interaction between ERFs and TPL/TPR proteins, we cloned ZmERF4, which was found to be induced in maize during U. maydis infection (Lanver et al., 2018) and possesses a DLNxxP and an LxLxL motif. We found that ZmERF4 interacted with ZmTPL1 both by Co‐IP in N. benthamiana and in a Y2H assay, where the second WD40 domain of ZmTPL1 was required for interaction (Figs 4c, S7a). Co‐expression of ZmERF4 with ZmTPL1 leads to destabilization of ZmTPL1, as protein amounts in the input were c. 80% lower than ZmTPL1 expressed with YFP (Fig. 4c). To test whether Jsi127641 can interfere with ZmERF4‐mediated destabilization of ZmTPL1, we co‐expressed ZmTPL1 and ZmERF4 cloned in frame with Jsi127641, Jsi1m27641, a version with a deletion at the N‐terminus including the EAR motif (Jsi1del47641), and an mCherry control. ZmERF4 was separated from the different Jsi1 versions by the porcine teschovirus‐1 2A co‐translational skipping motif to produce equimolar amounts of both proteins upon polycistronic expression (Kim et al., 2011). Jsi127641 interferes with the destabilization of ZmTPL1 by ZmERF4, as ZmTPL1 protein levels in presence of Jsi127641 are higher than ZmTPL1 protein levels in the presence of ZmERF4 with Jsi1m27641, Jsi1del47641, or the mCherry control (Fig. 4d). To test whether the interference of Jsi1 on ZmERF4 activity is due to a competition for its binding to ZmTPL1, we performed a Co‐IP experiment between ZmERF4 and ZmTPL1 in the presence of either Jsi1, Jsi1m27641 or Jsi1del47641. However, we did not find that Jsi1 competes with ZmERF4 for its binding to ZmTPL1, as the amount of ZmERF4 co‐precipitated with ZmTPL1 does not change when co‐expressed with the different versions of Jsi1 (Fig. S7b). These results suggest that Jsi1 might interfere with ZmERF4 activity via an unknown mechanism that involves TPL/TPR interaction and is associated with the upregulation of genes mainly connected with JA/ET signaling.
Conserved EAR‐motif from different fungi effectors is responsible for interaction with corresponding TPL/TPR
TPL/TPR proteins are highly conserved between different plant species (Causier et al., 2012a), so we asked whether DLNxxP‐motif‐containing effectors from various fungal pathogens with different hosts also use a similar strategy to manipulate TPL/TPR signaling. We performed a motif search analysis across published proteomes of plant pathogenic fungi to identify putative secreted effectors with a DLNxxP motif. We searched for effectors with a DLNxxP motif in the smut proteomes of Ustilago hordei, Ustilago bromivora, S. scitamineum and S. reilianum and identified additional effector candidates (Table 1). We performed the same search in plant pathogenic fungi with different lifestyles and from different fungal divisions. Based on sequence availability, we selected the obligate biotroph Puccinia triticina from the Basidiomycota division and from the Ascomycota division the biotrophic Blumeria graminis, the hemibiotrophic F. oxysporum and M. oryzae, and the necrotrophic pathogens Botrytis cinerea, Sclerotinia sclerotiorum and Bipolaris maydis. In all of the pathogens examined, with the exception of the necrotrophic pathogens, we found at least one predicted secreted protein possessing a DLNxxP motif after the secretion signal (Table 1), indicating that effectors possessing a DLNxxP motif are mainly found in pathogens with biotrophic and hemibiotrophic lifestyles. To test whether these effectors can interact with TPL, we selected Sr10312 from S. reilianum, SPSC_03537 from S. scitamineum, and MGG_15391 from M. oryzae. As the M. oryzae effector belongs to a strain that infects rice (Oryza sativa) and S. scitamineum infects sugarcane (Saccharum hybrid), we cloned a rice TPL gene OsTPL1 and a sugarcane TPL gene Sh_TPR3 (Fig. S1c). We fused the effectors MGG_1539124222, Sr1031223631 and SPSC_0353721653 to mCherry and OsTPL1, ZmTPL1, and Sh_TPR3 to GFP. In addition, we mutated the DLNxxP motif of these effectors to AHNxxP (MGG_15391m24222, Sr10312m23631, and SPSC_03537m21653) to test the relevance of the EAR motif in the interaction with TPL. We performed Co‐IP assays by co‐expressing each effector and its mutated version with their respective host TPL protein in N. benthamiana. MGG_1539124222, Sr1031223631 and SPSC_0353721653 co‐immunoprecipitated with their respective TPL protein. Sr10312m23631 and SPSC_03537m21653 mutant versions did not pull down their respective TPL proteins as efficiently as their wild‐type versions, indicating that the EAR motif is responsible for their interaction with TPL (Fig. 5a). Thus, our data indicate that the DLNxxP motif from different fungal pathogens is responsible for interaction with the corresponding host TPL, suggesting that these pathogens have convergently evolved a strategy to manipulate the host‐signaling pathway by mimicking an endogenous host motif.
Fig. 5.
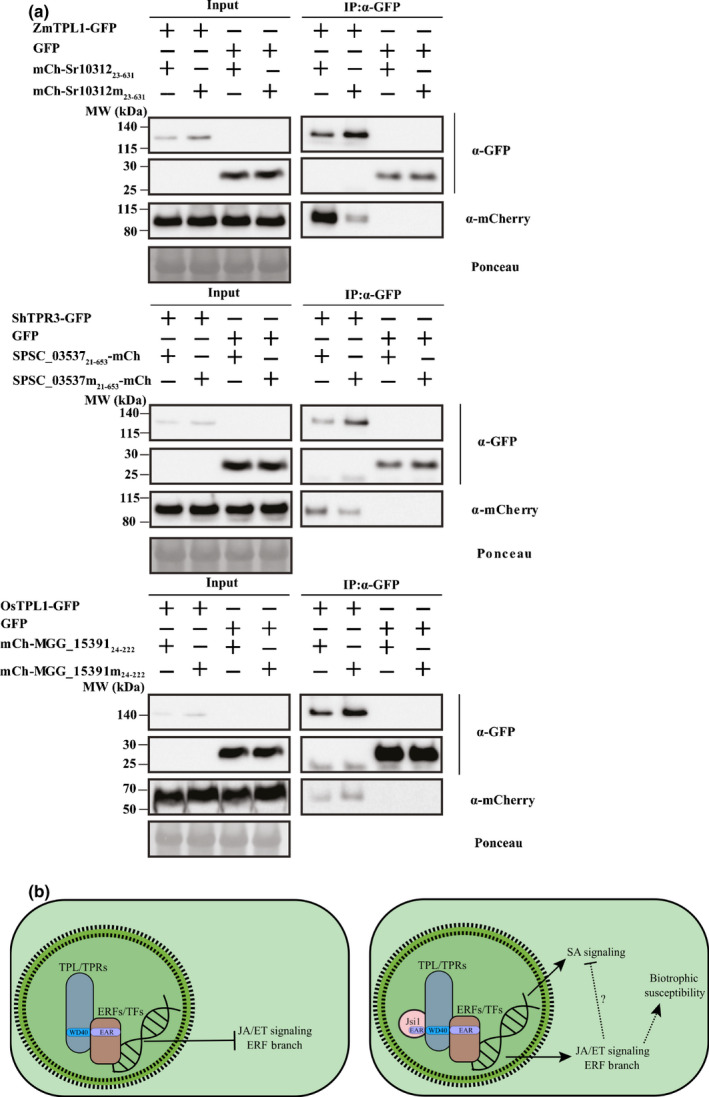
DLNxxP‐motif containing effectors of different fungal pathogens interacts with Topless (TPL). (a) We co‐infiltrated Sr10312, SPSC_03537, MGG_15391 and their versions mutated in the ethylene‐responsive element binding factor‐associated amphiphilic repression (EAR) motif (Sr10312m, SPSC_03537m and MGG_15391m) with their respective TPL proteins in Nicotiana benthamiana leaves. Co‐immunoprecipitated proteins were detected with anti‐GFP and anti‐mCherry antibodies. GFP, green fluorescent protein. (b) Jasmonate/Ethylene signaling inducer 1 (Jsi1) hijacks jasmonate/ethylene (JA/ET) signaling by interaction with TPL/Topless related (TPR) corepressors. Left panel: in the absence of the fungal Jsi1 effector, interaction between plant ethylene response factors (ERFs) and unknown transcription factors (TFs) possessing a DLNxxP with the second WD40 domain of TPL/TPR corepressor proteins may lead to repression of the ERF branch of the JA/ET signaling. Right panel: Jsi1 may interfere with the activity of ERFs and other unknown TFs, leading to activation of the ERF branch of the JA/ET signaling. Activation of salicylic acid (SA) signaling by Jsi1 could be due to activation of the plant immune system by recognition of the Jsi1‐TPL/TPRs interaction. On the other hand, SA signaling activation could also be due to Jsi1 interfering with the activity of ERFs. However, SA signaling cannot repress the ERF branch of the JA signaling as repressive ERF activity is blocked by the interaction between Jsi1 and TPL/TPRs. Therefore, a not fully activated SA defense pathway cannot lead to inactivation of the ERF branch, which may lead to biotrophic susceptibility in planta.
Discussion
Jsi1 interacts with the C‐terminus of TPL and hijacks jasmonate/ethylene signaling
Pathogens have developed diverse strategies to activate JA defense signaling, including producing bioactive forms or mimics of JA or effector proteins that activate JA signaling. Activation of JA signaling antagonizes SA signaling, promoting biotrophic susceptibility (Howe et al., 2018). In rice, the negative SA–JA signal crosstalk seems to be conserved (Yuan et al., 2007), but each hormone provides resistance to pathogens with different lifestyles (De Vleesschauwer et al., 2014). The SA–JA crosstalk also seems to be present in maize, but it has not been fully elucidated (Ziemann et al., 2018). Doehlemann et al. (2008) revealed that U. maydis infection of maize induces JA signaling and downregulates SA signaling. The induction of several members of the AP2/ERF family upon establishment of biotrophy by U. maydis suggests that induction of the ERF branch of the JA/ET defense signaling is beneficial for U. maydis infection. Nevertheless, its role in promoting biotrophic susceptibility in maize remains unknown. Here, we identified the U. maydis effector Jsi1 that activates JA/ET signaling. Jsi1 activates the ERF branch of the JA/ET defense signaling pathway by interacting with TPL/TPR proteins. In A. thaliana, Jsi1 induces the ERFs ERF2, ERF5, ERF6 and ERF107, which are associated with resistance to necrotrophic pathogens and activation of the JA defense signaling pathway. Jsi1 also induces PDF1.2, further supporting the idea that Jsi1 activates the ERF branch of the JA signaling pathway. Finally, A. thaliana plants expressing Jsi1 are more susceptible to biotrophic infection, which also correlates with the activation of the ERF branch of the JA/ET signaling. In maize, overexpression of Jsi1 induces the expression of ZmERF1, ZmERF1, and ZmPR5, which were also induced during U. maydis infection. Taken together, these results demonstrate that Jsi1 contributes to the activation of the ERF‐branch of the JA/ET signaling pathway, which may promote fungal infection.
We have shown that Jsi1 interacts with TPL/TPRs via the second WD40 domain near their C‐terminus and that this interaction is dependent on the DLNxxP motif. This interaction induces the ERF branch of the JA/ET defense signaling pathway. ERF TFs with a DLNxxP motif were previously described to interact with AtTPL/TPRs (Causier et al., 2012b). For AtERF3 and AtERF4, the DLNxxP motif is essential for their repressive activity (Ohta et al., 2001). In addition, AtERF4 and AtERF9 can suppress the expression of PDF1.2 and are negative regulators of resistance to necrotrophic pathogens, indicating that they act as negative regulators of the JA defense signaling (McGrath et al., 2005; Maruyama et al., 2013). The significant enrichment of TBSs of several ERFs with repressor activity in 269 out of the 915 genes upregulated upon Jsi1 induction and the ability of Jsi1 to interfere with the destabilization of ZmTPL1 by ZmERF4 show that ERFs with a DLNxxP motif are likely involved in repression of the ERF branch of the JA/ET signaling pathways.
Overexpression of ZmERF4 correlates with ZmTPL1 destabilization, suggesting that the repressive activity of ERFs is pernicious for the plant. Stabilization of ZmTPL1 in the presence of Jsi1 could indicate a protective effect, as we could not observe competition between Jsi1 and ZmERF4 for binding to ZmTPL1. It has been shown that TPL indirectly interacts with Histone Deacetylase 19 and both proteins are involved in transcriptional repression (Long et al., 2006). Therefore, Jsi1 may bind the C‐terminus of TPL and inhibit the interaction of other unknown repressor components required for ZmERF4’s repressive activity.
Activation of salicylic acid signaling by Jsi1 expression does not lead to repression of jasmonate/ethylene signaling in A. thaliana
Jsi1 expression in A. thaliana leads to activation of the SA signaling pathway, as evidenced by the upregulation of several SA responsive genes and an increase in total SA levels. In addition, prolonged expression of Jsi1 leads to a cell‐death phenotype across the different plant species we tested, which may be connected to activation of SA signaling. Studying an effector function in planta, separated from the context of the rest of the effectome, may reveal complex responses derived from initial effector action and subsequent recognition responses by the plant immune system which would usually be counteracted by other effectors in the natural context (Thordal‐Christensen, 2020). It was reported that interaction between Six8, a F. oxysporum effector, and TPL leads to activation of SA defense signaling in A. thaliana (Gawehns, 2014). Therefore, the activity of Jsi1 on TPL may also trigger the plant immune system.
Another explanation is that the interaction between Jsi1 and TPL/TPR proteins could interfere with the repressive activity of TFs with a DLNxxP motif, leading to activation of the SA signaling pathway. SA‐signaling‐based inhibition of JA/ET signaling has been previously demonstrated (Caarls et al., 2015) and would lead to increased resistance towards biotrophic interactions. However, A. thaliana plants expressing Jsi1 are more susceptible to Pst DC3000 infection. Furthermore, PDF1.2 and several ERFs related to activation of JA/ET signaling are upregulated by Jsi1, indicating that JA/ET signaling cannot be repressed by SA signaling. One explanation could be a potential role of EAR‐motif‐containing ERFs in mediating the SA repression of JA/ET signaling. It was previously reported that the promoter regions of genes induced by methyl jasmonate are enriched in a GCC‐box motif, the DNA binding motif of ERFs. In addition, it was shown that the GCC‐box is sufficient for transcriptional suppression by SA and that SA leads to degradation of ORA59, a positive regulator of the ERF branch (Van der Does et al., 2013). In summary, Jsi1 activates both JA/ET and SA‐responsive genes but SA antagonism on JA/ET signaling, which may be dependent on the ERFs–TPL/TPRs interaction cannot be exerted as a consequence of the interaction of Jsi1 with TPL/TPRs (Fig. 5b).
Effectors of diverse biotrophic and hemibiotrophic fungi convergently evolved
Plant host proteins targeted by effectors are under selective pressure to evade manipulation by the pathogen. On the other hand, if central regulators like TPL/TPRs interact with many endogenous host proteins via a specific motif, like the DLNxxP motif, it becomes nearly impossible to mutate the binding sites without tremendous fitness costs to the plant. This is likely why effectors from diverse biotrophic and hemibiotrophic fungi, including Sr10312 and SPSC_03537 from S. reilianum and S. scitamineum, respectively, may have convergently evolved the DLNxxP motif to interfere with the transcriptional control of the co‐repressors from the Topless family.
Author contributions
AD conceived the original research plan. AD and MD designed and coordinated the experimental work. DA, IF, JB, JM, K‐SC, KZ, MB, MD, LMSJ, IS, RB, SU and YP‐H contributed to the experimental work. AD, K‐SC and MD wrote the manuscript. MD and K‐SC contributed equally to this work.
Supporting information
Fig. S1 jsi1 is part of effector cluster 2A and its deletion has no detectable contribution to the virulence of U. maydis.
Fig. S2 Co‐IP assay showing that Jsi1 interacts with ZmTPL/TPRs in Z. mays.
Fig. S3 Jsi1 induces the ERF‐branch in A. thaliana.
Fig. S4 GO‐term analysis for biological process of genes differentially expressed in A. thaliana lines expressing Jsi1.
Fig. S5 Prolonged expression of Jsi1 leads to a cell‐death phenotype in Z. mays, N. benthamiana and A. thaliana.
Fig. S6 Heat map from RNA‐seq showing the GO category for SA responsive genes.
Fig. S7 ZmERF4 binds to the C‐terminal of TPL but Jsi1 does not interfere with the ZmERF4 binding to ZmTPL1.
Methods S1 Gene accession numbers, plasmid cloning, virulence assay in maize, phytohormone measurements, Pseudomonas syringae pv. tomato (Pst) DC3000 infection assay in A. thaliana, biolistic transformation of maize for virus‐mediated effector overexpression experiments.
Table S1 Constructs used in this study.
Table S2. List of primers used for RT‐PCR.
Table S3 Genes upregulated in A . thaliana XVE‐Jsi1 lines upon Jsi1 expression.
Table S4 Gene upregulated in A . thaliana XVE‐Jsi1mCh lines upon Jsi1 expression nriched for ERFs transcription‐binding sites.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We would like to acknowledge Fernando Navarrete for useful comments during the development of this project; Dr Ari Pekka Mähönen and Mikko Herpola for providing us the XVE inducible cassette; Rothamsted Research Limited and Dr Kostya Kanyuka for kindly providing us the vector PV101 for virus‐based protein expression in maize; Dr J. Matthew Watson for editing; GMI/IMBA/IMP service facilities, especially Dr Robert Heinen and Ms Zuzana Dzupinkova from the Molecular Biology services, Mr Pawel Pasierbek from the BioOptics facility, and Mr Mathias Madalinski from the Protein Chemistry Core facility for their excellent technical support, and the Plant Sciences Facility at the Vienna BioCenter Core Facilities GmbH; we thank also Dr Diaa El‐Din Daghma and Dr Jochen Kumlehn for technical support. This work was supported by the European Research Council under the European Union’s Seventh Framework Program (FP7/2007‐2013)/ERC grant agreement no. GA335691 (‘Effectomics’), the Austrian Science Fund (FWF; I 3033‐B22, P27818‐B22) and the Austrian Academy of Sciences. IF and KZ were supported by the Deutsche Forschungsgemeinschaft (INST 186/822‐1).
References
- Aichinger C, Hansson K, Eichhorn H, Lessing F, Mannhaupt G, Mewes W, Kahmann R. 2003. Identification of plant‐regulated genes in Ustilago maydis by enhancer‐trapping mutagenesis. Molecular Genetics and Genomics 270: 303–314. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton C, King RC, Chen H, Azhakanandam K, Bieri S, Hammond‐Kosack KE, Kanyuka K. 2018. Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiology 177: 1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Weinzierl G, Kämper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis . Molecular Microbiology 42: 1047–1063. [DOI] [PubMed] [Google Scholar]
- Caarls L, Pieterse CM, Van Wees S. 2015. How salicylic acid takes transcriptional control over jasmonic acid signaling. Frontiers in Plant Science 6: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012b. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiology 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B. 2012a. TOPLESS co‐repressor interactions and their evolutionary conservation in plants. Plant Signaling and Behavior 7: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini NM, Jung HW, Engle NL, Tschaplinski TJ, Greenberg JT. 2015. ALD1 regulates basal immune components and early inducible defense responses in Arabidopsis . Molecular Plant–Microbe Interactions 28: 455–466. [DOI] [PubMed] [Google Scholar]
- Cole SJ, Yoon AJ, Faull KF, Diener AC. 2014. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine and leucine‐conjugated jasmonates. Molecular Plant Pathology 15: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Xu J, Höfte M. 2014. Making sense of hormone‐mediated defense networking: from rice to Arabidopsis . Frontiers in Plant Science 5: e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I et al. 2011. Metabolic priming by a secreted fungal effector. Nature 478: 395–398. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. Star: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, Stitt M, Pons‐Kühnemann J, Sonnewald U, Kahmann R. 2008. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis . The Plant Journal 56: 181–195. [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM et al. 2007. MYC2 differentially modulates diverse jasmonate‐dependent functions in Arabidopsis . Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Huang M, Zhang Z, Cheng S. 2014. Genome‐wide analysis of the AP2/ERF gene family in maize waterlogging stress response. Euphytica 198: 115–126. [Google Scholar]
- Gawehns FKK. 2014. The Fusarium oxysporum effector Six8 interacts with TOPLESS and induces an SNC1 mediated stunting phenotype in A. thaliana.. In Function and targets of Fusarium oxysporum effectors. PhD thesis, Universiteit van Amsterdam, Amsterdam, the Netherlands. [Google Scholar]
- Gimenez‐Ibanez S, Boter M, Fernández‐Barbero G, Chini A, Rathjen JP, Solano R. 2014. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis . PLoS Biology 12: e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera‐Vásquez A, Carvallo L, Blanco F, Tobar M, Villarroel‐Candia E, Vicente‐Carbajosa J, Salinas P, Holuigue L. 2014. Transcriptional control of glutaredoxin GRXC9 expression by a salicylic acid‐dependent and NPR1‐independent pathway in Arabidopsis . Plant Molecular Biology Reporter 33: 624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ. 2018. Modularity in jasmonate signaling for multistress resilience. Annual Review of Plant Biology 69: 387–415. [DOI] [PubMed] [Google Scholar]
- Ju S, Go YS, Choi HJ, Park JM, Suh MC. 2017. DEWAX transcription factor is involved in resistance to Botrytis cinerea in Arabidopsis thaliana and Camelina sativa . Frontiers in Plant Science 8: e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. 2010. Genome‐wide analysis of ethylene‐responsive element binding factor‐associated amphiphilic repression motif‐containing transcriptional regulators in Arabidopsis. Plant Physiology 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J, Kahmann R, Bölker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus‐1 in human cell lines, zebrafish and mice. PLoS ONE 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J‐G, Stork W, Mudgett MB. 2013. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host & Microbe 13: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng CY, Moreira W, Mock SA et al. 2014. Araport: the Arabidopsis information portal. Nucleic Acids Research 43: D1003–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D, Müller AN, Happel P, Schweizer G, Haas FB, Franitza M, Pellegrin C, Reissmann S, Altmüller J, Rensing SA et al. 2018. The biotrophic development of Ustilago maydis studied by RNA‐seq analysis. Plant Cell 30: 300–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D, Tollot M, Schweizer G, Presti LL, Reissmann S, Ma LS, Schuster M, Tanaka S, Liang L, Ludwig N et al. 2017. Ustilago maydis effectors and their impact on virulence. Nature Reviews Microbiology 15: 409–421. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell 16: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Jiang L, Liu Y, Lv Y, Dai H, Zhao H. 2014. Genome‐wide identification of housekeeping genes in maize. Plant Molecular Biology 86: 543–554. [DOI] [PubMed] [Google Scholar]
- Liu X, Galli M, Camehl I, Gallavotti A. 2019. RAMOSA1 ENHANCER LOCUS2‐mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiology 179: 348–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis . Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez‐Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lukasik E, Gawehns F, Takken FL. 2012. The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. In: Bolton MD, Thomma BPHJ, eds. Plant fungal pathogens. New York, NY, USA: Humana Press, 61–74. [DOI] [PubMed] [Google Scholar]
- Ma LS, Wang L, Trippel C, Mendoza‐Mendoza A, Ullmann S, Moretti M, Carsten A, Kahnt J, Reissmann S, Zechmann B et al. 2018. The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose‐binding maize proteins. Nature Communications 9: e1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Arevalillo R, Nanao MH, Larrieu A, Vinos‐Poyo T, Mast D, Galvan‐Ampudia C, Brunoud G, Vernoux T, Dumas R, Parcy F. 2017. Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression. Proceedings of the National Academy of Sciences, USA 114: 8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K‐i, Sato T, Yamaguchi J. 2013. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Science 213: 79–87. [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. 2005. Repressor and activator‐type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome‐wide screen of Arabidopsis transcription factor gene expression. Plant Physiology 139: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR. 2012. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et‐mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SSC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme‐Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar RN, Benke PI, Qu Z, Constance Chen YY, Yang F, Swarup S, Naqvi NI. 2015. A fungal monooxygenase‐derived jasmonate attenuates host innate immunity. Nature Chemical Biology 11: 733–740. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E et al. 2010. NINJA connects the co‐repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon‐Reyes A, Van Wees SC. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Plett JM, Daguerre Y, Wittulsky S, Vayssières A, Deveau A, Melton SJ, Kohler A, Morrell‐Falvey JL, Brun A, Veneault‐Fourrey C et al. 2014. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proceedings of the National Academy of Sciences, USA 111: 8299–8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, Hoser R, Schilling L, Zechmann B, Krzymowska M, Walbot V, Doehlemann G. 2015. A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell 27: 1332–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Newman TE, Choi S, Jayaraman J, Choi DS, Jung GY, Cho H, Lee YK, Sohn KH. 2017. A conserved EAR motif is required for avirulence and stability of the Ralstonia solanacearum effector PopP2 in planta . Frontiers in Plant Science 8: e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin‐dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen H. 2020. A holistic view on plant effector‐triggered immunity presented as an iceberg model. Cellular and Molecular Life Sciences 77: 3963–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollot M, Assmann D, Becker C, Altmüller J, Dutheil JY, Wegner C‐E, Kahmann R. 2016. The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis . PLoS Pathogens 12: e1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A. 2009. A combinatorial interplay among the 1‐aminocyclopropane‐1‐carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana . Genetics 183: 979–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon‐Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T et al. 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völz R, Park J‐Y, Kim S, Park S‐Y, Harris W, Chung H, Lee Y‐H. 2020. The rice/maize pathogen Cochliobolus spp. infect and reproduce on Arabidopsis revealing differences in defensive phytohormone function between monocots and dicots. The Plant Journal 103: 412–429. [DOI] [PubMed] [Google Scholar]
- Win J, Chaparro‐Garcia A, Belhaj K, Saunders D, Yoshida K, Dong S, Schornack S, Zipfel C, Robatzek S, Hogenhout S et al. 2012. Effector biology of plant‐associated organisms: concepts and perspectives. Cold Spring Harbor Symposia on Quantitative Biology 77: 235–247. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D. 2008. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. Journal of Biological Chemistry 283: 26996–27006. [DOI] [PubMed] [Google Scholar]
- Yang L, Teixeira PJPL, Biswas S, Finkel OM, He Y, Salas‐Gonzalez I, English ME, Epple P, Mieczkowski P, Dangl JL. 2017. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host & Microbe 21: 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D et al. 2007. Functional analysis of rice NPR1‐like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnology Journal 5: 313–324. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. 2010. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proceedings of the National Academy of Sciences, USA 107: 13960–13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann S, van der Linde K, Lahrmann U, Acar B, Kaschani F, Colby T, Kaiser M, Ding Y, Schmelz E, Huffaker A et al. 2018. An apoplastic peptide activates salicylic acid signalling in maize. Nature Plants 4: 172–180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 jsi1 is part of effector cluster 2A and its deletion has no detectable contribution to the virulence of U. maydis.
Fig. S2 Co‐IP assay showing that Jsi1 interacts with ZmTPL/TPRs in Z. mays.
Fig. S3 Jsi1 induces the ERF‐branch in A. thaliana.
Fig. S4 GO‐term analysis for biological process of genes differentially expressed in A. thaliana lines expressing Jsi1.
Fig. S5 Prolonged expression of Jsi1 leads to a cell‐death phenotype in Z. mays, N. benthamiana and A. thaliana.
Fig. S6 Heat map from RNA‐seq showing the GO category for SA responsive genes.
Fig. S7 ZmERF4 binds to the C‐terminal of TPL but Jsi1 does not interfere with the ZmERF4 binding to ZmTPL1.
Methods S1 Gene accession numbers, plasmid cloning, virulence assay in maize, phytohormone measurements, Pseudomonas syringae pv. tomato (Pst) DC3000 infection assay in A. thaliana, biolistic transformation of maize for virus‐mediated effector overexpression experiments.
Table S1 Constructs used in this study.
Table S2. List of primers used for RT‐PCR.
Table S3 Genes upregulated in A . thaliana XVE‐Jsi1 lines upon Jsi1 expression.
Table S4 Gene upregulated in A . thaliana XVE‐Jsi1mCh lines upon Jsi1 expression nriched for ERFs transcription‐binding sites.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


