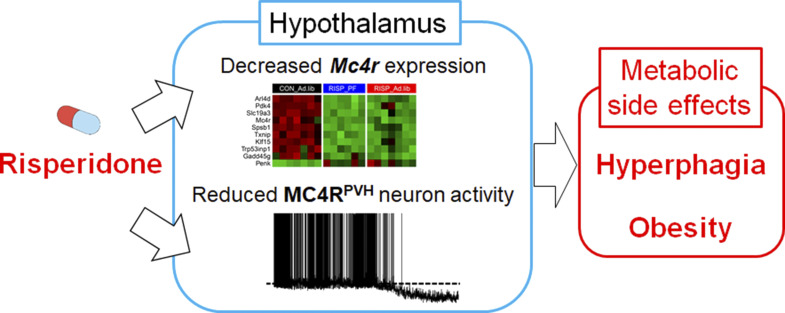
Li et al. develop a new mouse model that recapitulates risperidone-induced metabolic syndrome. Using a combination of transcriptomic, genetic, and electrophysiological approaches, they show that risperidone targets hypothalamic MC4Rs to cause weight gain and that setmelanotide, an MC4R agonist, mitigates its metabolic side effects.
Abstract
Atypical antipsychotics such as risperidone cause drug-induced metabolic syndrome. However, the underlying mechanisms remain largely unknown. Here, we report a new mouse model that reliably reproduces risperidone-induced weight gain, adiposity, and glucose intolerance. We found that risperidone treatment acutely altered energy balance in C57BL/6 mice and that hyperphagia accounted for most of the weight gain. Transcriptomic analyses in the hypothalamus of risperidone-fed mice revealed that risperidone treatment reduced the expression of Mc4r. Furthermore, Mc4r in Sim1 neurons was necessary for risperidone-induced hyperphagia and weight gain. Moreover, we found that the same pathway underlies the obesogenic effect of olanzapine—another commonly prescribed antipsychotic drug. Remarkably, whole-cell patch-clamp recording demonstrated that risperidone acutely inhibited the activity of hypothalamic Mc4r neurons via the opening of a postsynaptic potassium conductance. Finally, we showed that treatment with setmelanotide, an MC4R-specific agonist, mitigated hyperphagia and obesity in both risperidone- and olanzapine-fed mice.
Graphical Abstract
Introduction
Antipsychotics are essential medications for millions of patients combating a wide variety of neuropsychiatric conditions such as schizophrenia, bipolar disorder, and autism spectrum disorders (Meltzer, 2013). Despite their broad efficacy, many antipsychotic drugs, especially atypical antipsychotics (AAPs), have been associated with drug-induced metabolic syndrome, which is characterized by excessive weight gain, dyslipidemia, and insulin resistance (Gohlke et al., 2012). Although morbid obesity and type 2 diabetes typically take years to manifest in the general population, these conditions develop shortly after AAP treatment (Rojo et al., 2015). On the other hand, while considerable resources and efforts have been spent studying obesity and diabetes in the general population, relatively little progress has been made toward understanding or mitigating AAP-induced metabolic disturbances.
AAPs bind receptors for multiple neurotransmitters in the brain (Kaar et al., 2020). While the psychotropic effects are largely attributed to the blockade of dopamine D2 receptor (Drd2) and serotonin 2a receptor (Meltzer and Huang, 2008), the neural mechanisms that underlie their untoward metabolic effects remain poorly understood. Genome-wide association and candidate gene studies in human patients have identified a large number of risk alleles/genes that include the serotonin 2c receptor (Htr2c; Ellingrod et al., 2005), histamine H1 receptor (Kim et al., 2007; Kroeze et al., 2003), and α-2 adrenergic receptor (Park et al., 2006), just to name a few. However, few have been validated using genetic models in vivo (Lett et al., 2012). This is due, in part, to the difficulty of recapitulating human metabolic syndrome in animal models after chronic AAP treatment. For example, the half-life of AAPs such as olanzapine is much shorter in rodents (∼3 h) than that in humans (∼30 h; Mattiuz et al., 1997). Furthermore, their rapid degradation in liquid solutions hinders chronic drug delivery in animals (van der Zwaal et al., 2008). Finally, genetic backgrounds appear to be an important modifier for AAP-induced metabolic disturbances in mice (Zapata and Osborn, 2020).
Risperidone (Risperdal) is one of the most prescribed AAPs. It can cause significant weight gain in both youths and adults (Pozzi et al., 2020). In the current study, we develop a new mouse model that reliably reproduces risperidone-induced weight gain, adiposity, and glucose intolerance. We then use a combination of mouse genetic, transcriptomic, and electrophysiological approaches to investigate the molecular underpinnings of these metabolic perturbations.
Results and discussion
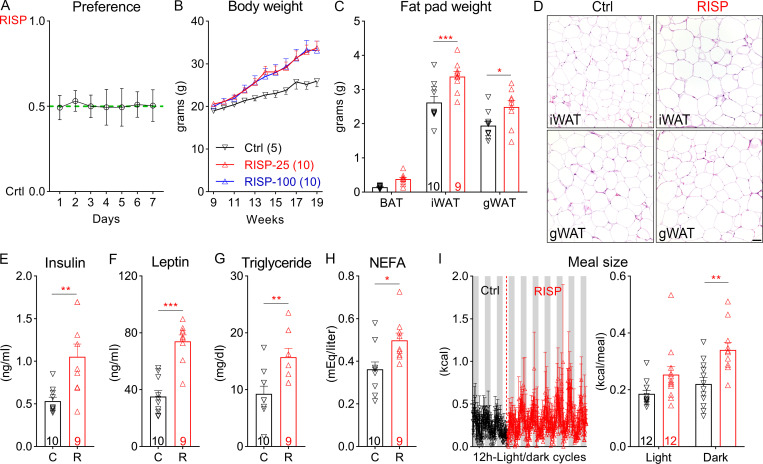
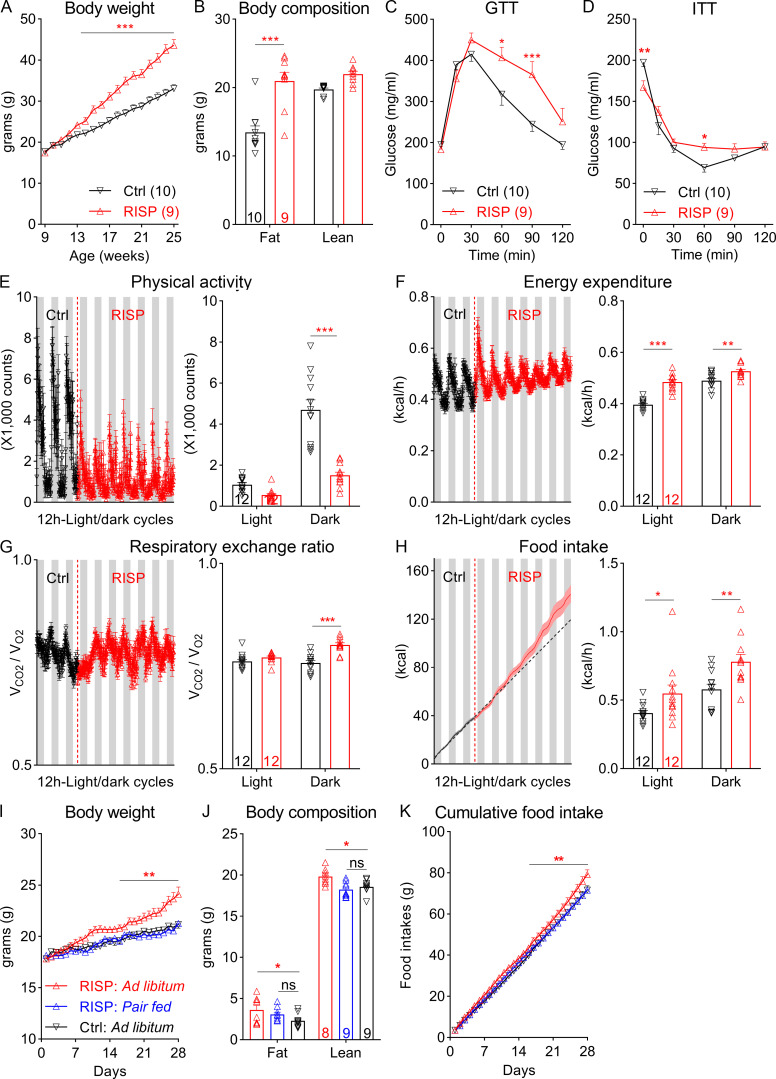
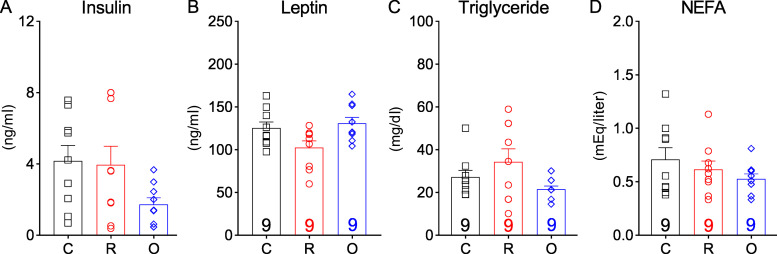
We developed a risperidone diet by compounding risperidone into a control diet (D09092903; Research Diets) so that the two diets have the same macronutrient composition and energy density (see Materials and methods). Moreover, when both diets were available in the same cage, mice did not show a preference between the two (Fig. S1 A), suggesting that the added risperidone did not significantly alter the valence of the control diet. We found that C57BL/6 mice fed the risperidone diet (25 mg/kg diet; D18041008; Research Diets) gained significantly more weight (26.18 ± 1.20 g vs. 15.41 ± 0.91 g) than those on the control diet over a 16-wk period (Fig. 1 A). Of note, a higher dose (100 mg/kg diet; D18041007; Research Diets) did not further exacerbate the weight gain (Fig. S1 B). Nuclear magnetic resonance analyses revealed that the excessive weight gain was largely due to an increase in fat mass (Fig. 1 B), which was evident in both the inguinal white adipose and gonadal white adipose tissues but not in the brown adipose tissue (BAT; Fig. S1 C). Consistent with this finding, H&E staining showed increased lipid accumulation in inguinal white adipose tissue and gonadal white adipose tissue of risperidone-fed mice (Fig. S1 D). Moreover, plasma levels of insulin, leptin, triglyceride, and nonesterified fatty acids (NEFAs) were significantly higher in these mice (Fig. S1, E–H).
Figure S1.
Risperidone treatment alters energy metabolism. (A) Ratio of daily intake of the RISP diet (25 mg/kg) over that of the Ctrl diet when both diets were available in the cage for 7 d; n = 9. (B) Body weight of wild-type C57BL/6 mice fed the Ctrl, RISP-25 (25 mg/kg), or RISP-100 (100 mg/kg) diet; n = 5–10; two-way ANOVA; F(20, 220) = 2.71; P < 0.001. (C) Fat pad weight of mice after 16-wk treatment of the Ctrl or RISP diet. n = 9 or 10; F(2, 34) = 409.2; two-way ANOVA; P < 0.001. (D) H&E staining in mice after 16-wk treatment of the Ctrl or RISP diet. Scale bar is 100 µm. (E–H) Plasma levels of insulin (E), leptin (F), triglyceride (G), and NEFA (H) in mice after 16-wk treatment of the Ctrl or RISP diet. n = 9 or 10. (I) Meal size. Left: traces of continuous measurement in metabolic cages; right: summarized daily average (binned into 12-h light and dark phases) before (black) and after (red) the dietary switch; n = 12; two-way ANOVA; F(1, 22) = 7.452; P = 0.012. *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Two-way ANOVA with Sidak’s post hoc tests in A–C and I. Unpaired t test in E–H. All data were verified in at least two independent experiments. C, control; gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue; R, risperidone. Data are presented as mean ± SEM.
Figure 1.
Chronic exposure to risperidone causes obesity and glucose intolerance in C57BL/6 mice. (A) Body weight of wild-type C57BL/6 mice fed either the Ctrl or the risperidone (RISP) diet; n = 9 or 10; two-way ANOVA; F(16, 272) = 25.53; P < 0.001. (B) Body composition of mice in A after 16-wk treatment with the Ctrl or RISP diet; n = 9 or 10; two-way ANOVA; F(1, 17) = 13.9; P = 0.002. (C) GTT after 16-wk treatment with the Ctrl or RISP diet; n = 9 or 10; two-way ANOVA; F(5, 85) = 8.224; P < 0.001. (D) ITT after 16-wk treatment with the Ctrl or RISP diet; n = 9 or 10; two-way ANOVA; F(5, 85) = 6.334; P < 0.001. (E–H) Left: traces of continuous measurement in metabolic cages; right: summarized daily average (binned into 12-h light and dark phases) before (black) and after (red) the dietary switch. (E) Physical activity, F(1, 22) = 41.49; P < 0.001. (F) EE, F(1, 22) = 26.64; P < 0.001. (G) Respiratory exchange ratio, F(1, 22) = 8.01; P = 0.01). (H) Food intake, F(1, 22) = 5.982; P < 0.05; n = 12. (I–K) A 28-d PF experiment. Body weight (I; n = 8 or 9; two-way ANOVA; F[56, 643] = 9.425; P < 0.001) and food intake (K; two-way ANOVA; F[56, 588] = 3.597; P < 0.001) were measured daily, and body composition (J; n = 8 or 9; two-way ANOVA; F[2, 46] = 7.505; P = 0.002) was measured at the end of the experiment. *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Two-way ANOVA with Sidak’s post hoc tests in A–K. All data were verified in at least two independent experiments. RER, respiratory exchange ratio. Data are presented as mean ± SEM.
Concomitant with obesity were perturbations in glucose homeostasis. For example, risperidone-fed mice exhibited a deficit in glucose clearance in a glucose tolerance test (GTT; Fig. 1 C). Despite augmented insulin levels, these mice had higher levels of plasma glucose under the fed state and were resistant to the glucose-lowering effect of insulin in an insulin tolerance test (ITT; Fig. 1 D). Collectively, these findings show that dietary supplementation of risperidone reproduces key symptoms of risperidone-induced metabolic syndrome in humans.
To determine risperidone’s effects on energy intake and expenditure, we monitored individual mice before and after risperidone treatment using a fully automated metabolic chamber system (PhenoMaster; TSE Systems). Mice were fed the control diet during acclimation and the first 3 d of the recording before being switched to the risperidone diet for the next 6 d. The lethargic effect of AAPs is deemed a potential contributor to weight gain in humans (Wichniak et al., 2011). As expected, upon risperidone exposure, physical activity plummeted during the dark phase of a day when mice were otherwise most active (Fig. 1 E). Meanwhile, indirect calorimetry analyses showed that energy expenditure (EE; Fig. 1 F), as well as the respiratory exchange ratio (Fig. 1 G), increased after mice were fed the risperidone diet. Remarkably, mice quickly developed hyperphagia following the dietary switch, as they consumed more food during both light and dark phases of a day (Fig. 1 H). Furthermore, meal pattern analyses showed that meal sizes increased in risperidone-fed mice (Fig. S1 I), whereas meal frequency or duration did not change (data not shown). Together, our data demonstrate that risperidone rapidly alters multiple metabolic parameters in mice.
Using a pair-feeding paradigm, we investigated whether hyperphagia was the driving force behind weight gain. When fed ab libitum, individually housed mice on the risperidone diet gained significantly more weight and adiposity than those fed the control diet over a 28-d period (Fig. 1, I–K). In the pair-fed (PF) group, however, overeating was avoided by restricting risperidone-fed mice to the same amount of food consumed by those fed the control diet. We found that pair feeding completely prevented the risperidone-induced increases in body weight and adiposity (Fig. 1, I and J), suggesting that hyperphagia is the primary cause of obesity in risperidone-fed mice.
To uncover the molecular mechanisms behind risperidone-induced hyperphagia, we employed whole-transcriptome RNA sequencing (RNA-seq) to investigate how risperidone globally alters gene transcription in the hypothalamus—a critical regulator of food intake (Xu et al., 2011). To this end, we dissected hypothalami from mice fed either the risperidone (RISP_Ad.lib; n = 7) or the control (Ctrl_Ad.lib; n = 7) diet for 28 d, along with those from a third group of “PF” mice (RISP_PF; n = 6) that were given the risperidone diet but limited to the same amount that Ctrl_Ad.lib mice had consumed. At the time of tissue collection, weight gain was significantly higher in RISP_Ad.lib mice (6.15 ± 0.79 g; one-way ANOVA; P < 0.001) but was comparable between Ctrl_Ad.lib (3.71 ± 0.33 g) and RISP_PF mice (3.06 ± 0.37 g).
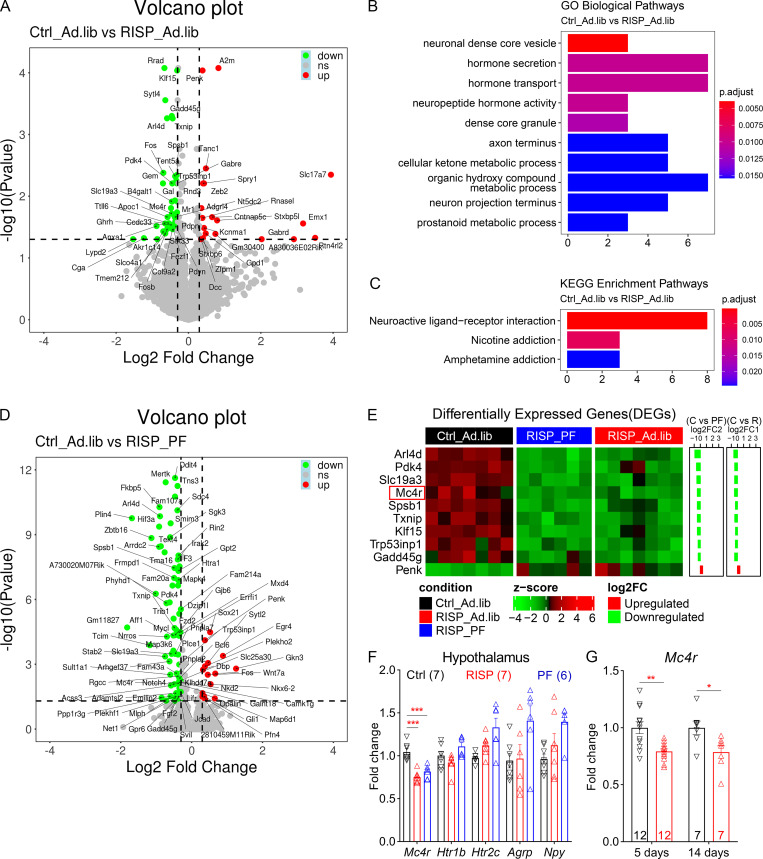
Comparative transcriptomic analyses identified 65 genes that were differentially expressed (DEGs) in RISP_Ad.lib mice (compared with Ctrl_Ad.lib mice; Fig. 2 A). Among them, Gene Ontology analyses demonstrated enrichment of genes involved in hormone secretion, transport, and release (Fig. 2 B). Furthermore, Kyoto Encyclopedia of Genes and Genomes pathway mapping showed that the identified DEGs are characteristic of neuroactive ligand–receptor interaction and overlap with those implicated in nicotine or amphetamine addiction (Fig. 2 C). Meanwhile, similar analyses found 97 DEGs in RISP_PF mice (compared with Ctrl_Ad.lib mice; Fig. 2 D) that were also exposed to risperidone but did not gain extra weight. Cross-referencing the two DEG sets identified 10 common genes that were regulated in the same way (both up-regulated and down-regulated) in RISP_Ad.lib and RISP_PF mice (Fig. 2 E), suggesting that their expression responded directly to chronic risperidone treatment. Interestingly, the melanocortin 4 receptor (Mc4r)—a critical regulator for appetite in mice and humans (Huszar et al., 1997; Yeo et al., 1998)—was among the genes that were down-regulated in the two risperidone-fed groups (Fig. 2 E). Consistent with the RNA-seq results, quantitative PCR (qPCR) analyses confirmed the decrease of Mc4r mRNAs in both RISP_Ad.lib and RISP_PF mice (Fig. 2 F). The reduction of Mc4r can be detected as early as 5 d following risperidone treatment (Fig. 2 G). In contrast, the hypothalamic expression of other appetite regulators such as agouti-related peptide, neuropeptide Y, Htr2c, orserotonin 1b receptor was not affected after chronic risperidone treatment (Fig. 2 F).
Figure 2.
A hypothalamic gene expression signature associated with chronic risperidone treatment. (A) Volcano plot showing 65 DEGs between Ctrl_Ad.lib and RISP_Ad.lib mice; green, down-regulated, red up-regulated. (B and C) Gene Ontology (GO; B) and Kyoto Encyclopedia of Genes and Genomes (KEGG; C) analyses of DEGs in A showing enriched biological pathways and physiological functions. (D) Volcano plot showing 97 DEGs between Ctrl_Ad.lib and RISP_PF mice; green, down-regulated, red up-regulated. (E) Heat-map of the expression of 10 common DEGs that were down-regulated or up-regulated in both RISP_PF and RISP_Ad.lib mice. Each row shows the relative expression of a DEG in individual mice. (F and G) qPCR analyses of hypothalamic gene expression. (G) Expression of Mc4r in risperidone-fed mice was 79% compared with that in mice fed the control diet. *, P < 0.05; **, P < 0.01; ***, P < 0.001. One-way ANOVA with Sidak’s post hoc tests in F. Unpaired t test in G. Data in F and G were verified in three independent experiments. Agrp, agouti-related peptide; C, control; FC, fold change; p.adjust, adjusted P value; Htr1b, serotonin 1b receptor; Npy, neuropeptide Y; R, risperidone. Data are presented as mean ± SEM in F and G.
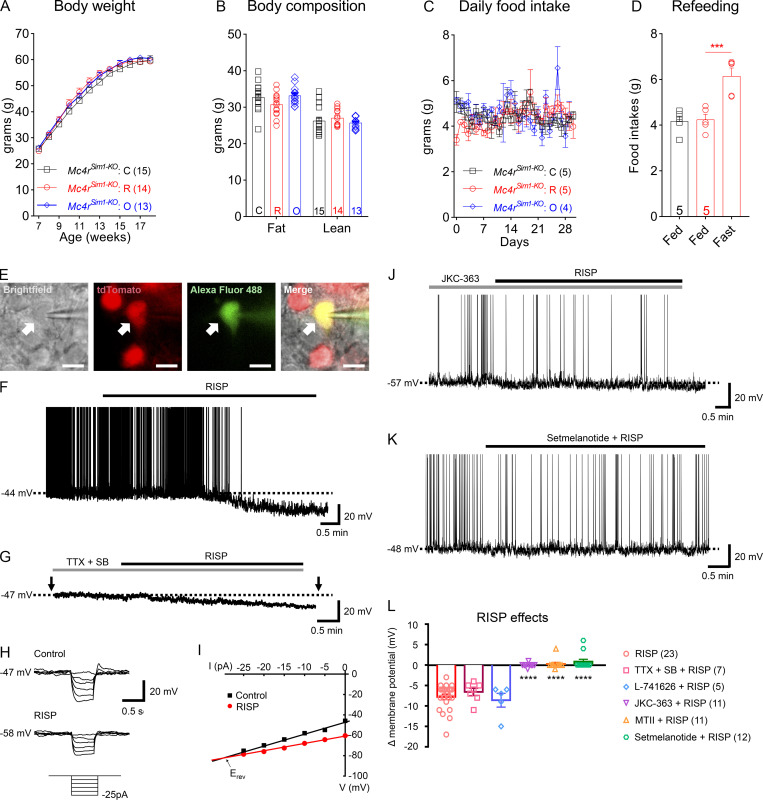
We investigated whether the down-regulation of Mc4r in risperidone-fed mice contributed to their hyperphagia and obesity. Inside the brain, Mc4r acts on hypothalamic single-minded homolog 1 (Sim1)–expressing neurons to regulate food intake (Balthasar et al., 2005). To this end, we generated Mc4rSim1-KO mice in which Mc4r was selectively deleted in Sim1-Cre–expressing neurons. We found that risperidone’s effect on weight gain was absent in these mice, as young Mc4rSim1-KO mice on the risperidone diet gained similar weights compared with those fed the control diet (Fig. 3 A). Consistent with this finding, adiposity (Fig. 3 B) as well as plasma levels of insulin, leptin, triglyceride, and NEFA were comparable between the two groups (Fig. S2, A–D). Importantly, different from wild-type mice, risperidone treatment did not increase food intake in Mc4rSim1-KO mice (Fig. 3 C). In comparison, an overnight fast significantly boosted food intake in these mice (Fig. 3 D), suggesting that they were still capable of consuming more food when fed the risperidone diet. Altogether, our findings suggest that risperidone-induced obesity and hyperphagia require MC4Rs in Sim1 neurons. Additionally, we investigated whether these receptors could underlie the obesogenic effect of other AAPs. To this end, we tested olanzapine—another commonly used AAP that causes the most weight gain in human patients (Meltzer, 2013). We showed previously that wild-type mice fed an olanzapine diet developed hyperphagic obesity (Lord et al., 2017). In contrast, the same diet did not change food intake, body weight, or adiposity in Mc4rSim1-KO mice (Fig. 3, A–C). Likewise, plasma levels of insulin, leptin, triglyceride, and NEFA remained constant in olanzapine-fed Mc4rSim1-KO mice (Fig. S2, A–D). Collectively, these findings raise the intriguing possibility that risperidone and olanzapine target a common hypothalamic pathway to induce hyperphagia and weight gain.
Figure 3.
Risperidone acts on hypothalamic MC4Rs/Mc4r-expressing neurons to cause weight gain. (A) Body weight of Mc4rSim1-KO mice fed the control (C), risperidone (R), or olanzapine (O) diet; n = 14 or 15; two-way ANOVA; F(22, 427) = 2.12; P = 0.46. (B) Body composition of mice in A after 10-wk treatment of the C, R, or O diet; n = 14 or 15; two-way ANOVA; F(1, 54) = 0.4843; P = 0.49. (C) Daily food intake in the first 28 d of treatment with the C, R, or O diet; n = 4 or 5; two-way ANOVA; F(30, 240) = 1.022; P = 0.44. (D) Fast-induced refeeding in risperidone-fed Mc4rSim1-KO mice; n = 5; one-way ANOVA; F(2, 12) = 0.3575; ***, P < 0.001. (E) Bright-field, fluorescent illumination (TRITC [tetramethylrhodamine isothiocyanate] for Mc4r-T2A-Cre::tdTomato; FITC for Alexa Fluor 488) and the merged image of a targeted MC4RPVH neuron for whole-cell patch-clamp experiments. Arrows indicate the targeted cell. Scale bars are 10 µm. (F) Acute effects of RISP (10 µM) on MC4RPVH neurons. (G) Acute effects of RISP in the presence of tetrodotoxin (TTX; 0.5 µM), kynurenic acid (1 mM), and picrotoxin (50 µM). SB, synaptic blocker. (H) Voltage responses to hyperpolarizing current steps (from −25 pA to 0 pA, applied as indicated by arrows in G. (I) Current-voltage relationship demonstrates RISP-induced decreases of input resistance. Erev = reversal potential. (J) Pretreatment with JKC-363 (20 nM, an MC4R antagonist) blocked the acute effects of RISP. (K) Co-treatment with setmelanotide (100 nM, an MC4R agonist) blocked the acute effects of RISP. (L) Bar graphs summarize the acute effects of RISP. The dashed line in F, G, J, and K indicates resting membrane potential. Data are presented as mean ± SEM. ****, P < 0.0001. One-way ANOVA with Sidak’s post hoc tests. All data were verified in at least two independent experiments.
Figure S2.
Chronic treatment of risperidone or olanzapine did not change plasma levels of insulin, leptin, triglyceride, or NEFA in Mc4rSim1-KO mice. (A–D) Plasma levels of insulin (A), leptin (B), triglyceride (C), and NEFA (D) in mice treated with the control (C), risperidone (R), or olanzapine (O) diet; n = 9. One-way ANOVA with Sidak’s post hoc tests. All data were verified in at least two independent experiments. Data are presented as mean ± SEM.
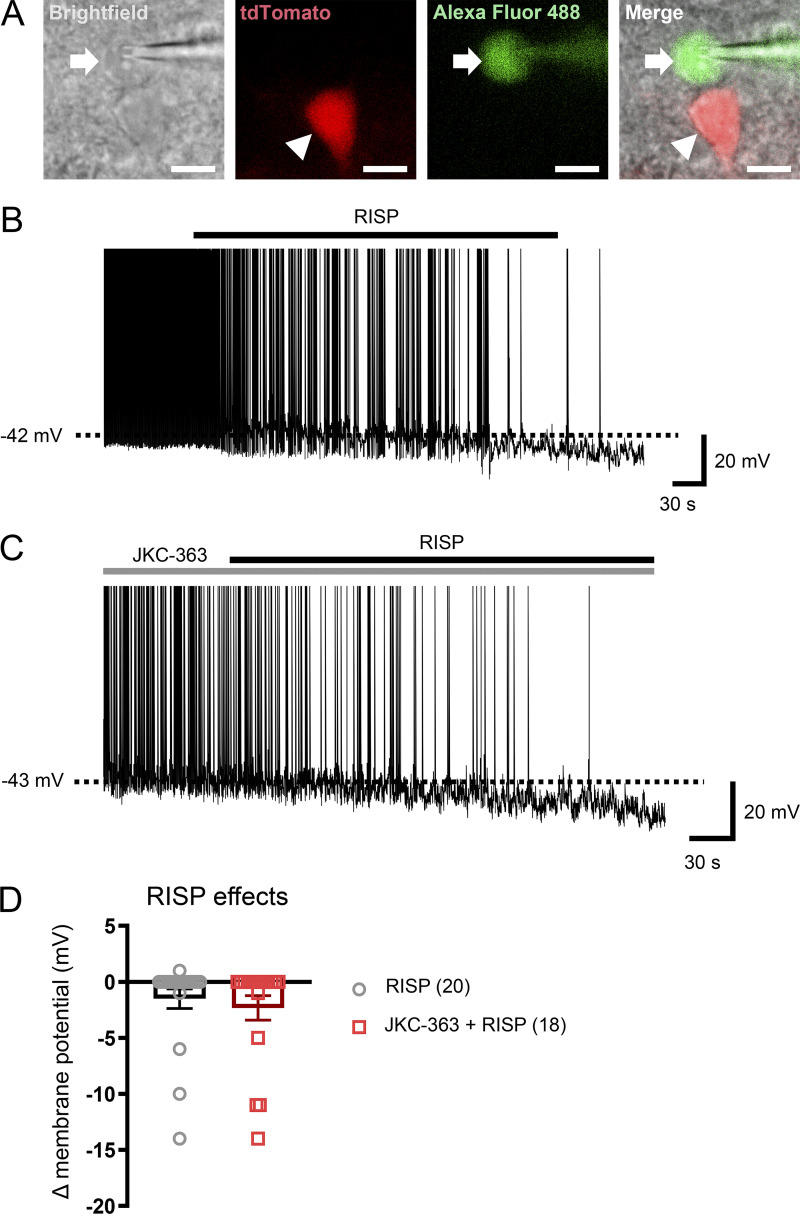
In addition to MC4Rs, activities of MC4R-expressing neurons in the paraventricular nucleus of the hypothalamus (PVH) directly regulate food intake (Garfield et al., 2015; Li et al., 2019). As a result, we tested the effects of risperidone on the electrical properties of PVH MC4R-expressing neurons (MC4RPVH neurons). We conducted whole-cell patch-clamp recordings of MC4RPVH neurons on acutely prepared hypothalamic slices from Mc4r-T2A-Cre::tdTomato mice (Fig. 3 E). We found that bath applications of risperidone (10 µM) hyperpolarized membrane potentials of MC4RPVH neurons by −7.8 ± 0.7 mV (n = 23 of 49 cells, 46.9%, from −48.7 ± 1.4 mV to −56.5 ± 1.4 mV; Fig. 3, F and L). Risperidone-induced inhibition of MC4RPVH neurons occurred in a concentration-dependent manner, where the half-maximal effective concentration was ∼4.5 µM. Additionally, risperidone inhibited MC4RPVH neurons in the presence of tetrodotoxin (0.5 µM) and a cocktail of synaptic blockers (1 mM kynurenic acid and 50 µM picrotoxin; Fig. 3, G and L), which was consistent with a postsynaptic effect. Moreover, risperidone-induced hyperpolarization was accompanied by 20.3 ± 1.7% decrease of input resistance (n = 24; from 1,420.9 ± 74.4 MΩ to 1,132.4 ± 65.4 MΩ) with a reversal potential of −93.8 ± 2.5 mV (n = 24; Fig. 3, H, I, and L). These findings suggest that risperidone inhibited MC4RPVH neurons via the opening of a putative potassium conductance. Notably, the acute inhibitory effects of risperidone on MC4RPVH neurons were not affected by the pretreatment of hypothalamic slices with 1 µM L-741626, a Drd2 antagonist, but were prevented by 20 nM JKC-363, an MC4R antagonist (Fig. 3, J and L). Moreover, many AAPs, including risperidone, exhibit inverse agonist properties at other G protein–coupled receptors (Rauser et al., 2001). As a result, we tested whether the acute effects of risperidone could be blunted by competitive binding with an MC4R agonist. Indeed, we found that coapplications of either MTII (100 nM) or setmelanotide (100 nM) completely blocked the risperidone-induced hyperpolarization of MC4RPVH neurons (Fig. 3, K and L; note: one neuron was depolarized after cotreatment of MTII and risperidone, and two neurons were depolarized after cotreatment of setmelanotide and risperidone in Fig. 3 L). Risperidone also inhibited a smaller percentage of PVH non-MC4R neurons (Fig. S3 A) by −10.0 ± 2.3 mV (n = 3 of 20 cells; 15.0%, from −46.3 ± 5.4 mV to −56.3 ± 3.4 mV; Fig. S3, B and D). However, different from MC4RPVH neurons, pretreatment with JKC-363 failed to block risperidone-induced hyperpolarization in PVH non-MC4R neurons (Fig. S3, C and D). Collectively, these findings suggest that risperidone acutely inhibits the activity of MC4RPVH neurons via its interaction with MC4Rs.
Figure S3.
Risperidone inhibits PVH non-MC4R neurons independently of MC4R. (A) Bright-field, fluorescent illumination (TRITC [tetramethylrhodamine isothiocyanate] for Mc4r-T2A-Cre::tdTomato; FITC for Alexa Fluor 488) and the merged image for whole-cell patch-clamp experiments. Arrows indicate the targeted cell. Arrowheads indicate MC4R-positive cells. Scale bars are 10 µm. (B) Acute effects of RISP (10 µM) on PVH non-MC4R neurons. (C) Acute effects of RISP in the presence of JKC-363 (20 nM). (D) Bar graphs summarize the acute effects of RISP. The dashed line in B and C indicates resting membrane potential. Data are presented as mean ± SEM. Unpaired t test. All data were verified in at least two independent experiments.
Inhibition of MC4RPVH neuron activity led to hyperphagia and obesity in mice (Garfield et al., 2015; Li et al., 2019). Given our observation that MC4R agonists blocked risperidone’s inhibitory effect on MC4RPVH neurons, we investigated whether treatment with an MC4R agonist would ameliorate metabolic syndrome in risperidone-fed mice. Notably, setmelanotide is a selective MC4R agonist that binds and activates the human MC4R with nanomolar potency (Collet et al., 2017). It reduces food intake and body weight in animal models of diet-induced obesity and humans with leptin receptor deficiency (Chen et al., 2015; Clément et al., 2018; Kievit et al., 2013). Importantly, unlike other MC4R agonists (e.g., MTII), it does not cause an acute or long-term increase in blood pressure or heart rate in obese primates or humans (Chen et al., 2015; Kievit et al., 2013). As a result, it was recently approved for the treatment of several rare genetic disorders of obesity.
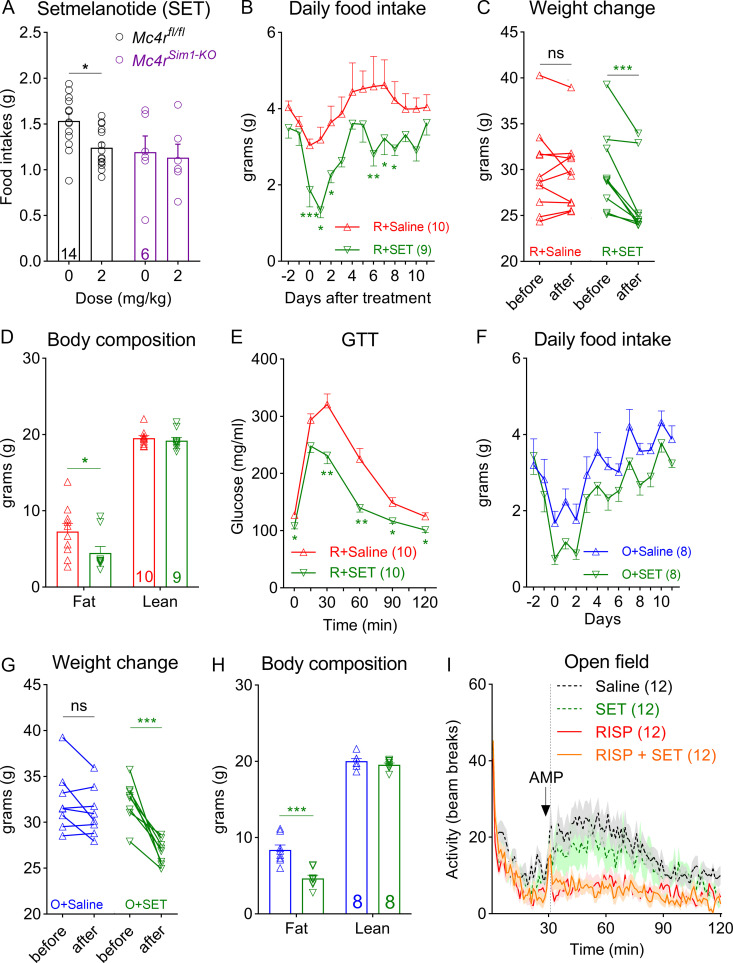
Prior to the experiment, we first established that the anorexigenic effect of setmelanotide (2 mg/kg) depended on the endogenous MC4Rs. For example, a single i.p. dose of setmelanotide suppressed fast-induced refeeding in control mice (Mc4rfl/fl). However, such an effect was absent in Mc4rSim1-KO mice (Fig. 4 A). We fed C57BL/6 mice the risperidone diet for 8 wk to induce hyperphagia and obesity. Using implanted osmotic mini-pumps (model 1002; Alzet), we then treated them with either setmelanotide (2 mg/kg/d) or vehicle (saline) while they were still being fed the risperidone diet. We found that infusion of setmelanotide attenuated hyperphagia in risperidone-fed mice (Fig. 4 B). As a result, body weight significantly decreased after the 14-d treatment (Fig. 4 C; −3.42 ± 1.14 g or −11.16% ± 3.72%). Consistent with this finding, nuclear magnetic resonance analyses revealed a selective decrease of fat mass in these mice (Fig. 4 D). In comparison, body weight or adiposity did not change in mice treated with saline (Fig. 4, C and D). In addition to improvements in body weight, setmelanotide-treated mice exhibited accelerated glucose clearance in a GTT (Fig. 4 E). Moreover, similar experiments showed that setmelanotide treatment also alleviated hyperphagia and obesity in olanzapine-fed mice (Fig. 4, F–H). Collectively, these findings show that setmelanotide treatment was effective in reversing AAP-induced metabolic perturbations. Finally, we investigated whether cotreatment with setmelanotide interfered with risperidone’s psychotropic effect using a rodent model of schizophrenia (Van Swearingen et al., 2013). It is well known that amphetamine triggers schizophrenia-like psychotic episodes in healthy human subjects (Janowsky and Risch, 1979), and this effect is amplified in schizophrenic patients (Angrist et al., 1980). Similarly, amphetamine (2.5 mg/kg, i.p.) induced hyperactivity in C57BL/6 mice, an effect that was greatly attenuated by pretreatment with risperidone (Fig. 4 I). Importantly, we found that cotreatment with setmelanotide did not diminish the ability of risperidone to suppress amphetamine-induced hyperlocomotion in an open field test (Fig. 4 I).
Figure 4.
Setmelanotide mitigates hyperphagia and obesity in risperidone- and olanzapine-fed mice. (A) Refeeding after an i.p. dose of setmelanotide (SET; 2 mg/kg) or saline in Mc4rfl/fl and Mc4rSim1-KO mice after an overnight fast; n = 6–14; two-way ANOVA; F(1, 36) = 4.651; P > 0.999. (B) Daily food intake in risperidone-fed wild-type mice treated with SET or saline (mini-pumps); n = 9 or 10; two-way ANOVA; F(1, 15) = 14.46; P = 0.002. (C) Changes in body weight before and after the 14-d treatment with SET or saline; n = 9; two-way ANOVA; F(2, 25) = 9.995; P < 0.001. (D) Body composition in risperidone-fed mice after the 14-d treatment with SET or saline; n = 9 or 10; two-way ANOVA; F(2, 50) = 4.215; P = 0.034. (E) GTT in risperidone-fed mice after the 14-d treatment with SET or saline; n = 10; F (18, 90) = 5.378; two-way ANOVA; P < 0.001. (F) Daily food intake in olanzapine-fed wild-type mice treated with SET or saline (mini-pumps); n = 8; two-way ANOVA; F(1, 14 = 7.734; P = 0.015. (G) Changes in body weight before and after the 14-d treatment with SET or saline; n = 8; two-way ANOVA; F(1, 14) = 14.17; P = 0.002. (H) Body composition in olanzapine-fed mice after the 14-d treatment with SET or saline; n = 8; two-way ANOVA; F(1, 14) = 16.70; P = 0.0011. (I) Open field test measuring amphetamine-induced hyperlocomotion in mice pretreated with saline, SET, RISP, or RISP + SET; n = 12; two-way ANOVA; F(357, 5236) = 1.683; P < 0.001. Saline versus RISP, P = 0.002; RISP versus RISP + SET, P > 0.999. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Two-way ANOVA with Sidak’s post hoc tests. All data were verified in at least two independent experiments.
General discussion and conclusions
Antipsychotic drug-induced metabolic syndrome is a pressing clinical problem affecting millions of patients. However, difficulty in modeling their metabolic effects in laboratory animals has significantly hindered relevant mechanistic studies. For instance, previous efforts to chronically administer risperidone to rodents via either i.p. or s.c. injections or using peanut-butter pills only produced a modest impact on body weight and adiposity (Kaur and Kulkarni, 2002; Li et al., 2013; Ota et al., 2002). Furthermore, due to its potent sedative effect, an acute dose of risperidone often renders mice immobile for hours, which confounds subsequent metabolic analyses. In the current study, we developed a risperidone diet that reliably caused excessive weight gain, adiposity, and glucose intolerance in C57BL/6 mice. Notably, the amount of risperidone administered daily through the diet is comparable to that via other routes (Kaur and Kulkarni, 2002; Li et al., 2013). However, dietary supplementation produced more pronounced metabolic disturbances. The mechanism behind the increased efficacy is less clear. It is possible that risperidone is more stable when mixed with the diet (van der Zwaal et al., 2008). Moreover, oral delivery may allow for interactions between risperidone and the gut microbiota, which could play a potential role in weight gain (Morgan et al., 2014). Finally, intake of risperidone via multiple meals throughout a day may produce a “slow-release” effect so that mice can better cope with its sedative effect.
The success in modeling risperidone-induced metabolic syndrome provided us with a unique opportunity to precisely characterize the pathophysiology underlying risperidone-induced metabolic perturbations. We found that risperidone treatment profoundly altered energy intake and expenditure in C57BL/6 mice. For example, hyperphagia developed shortly after risperidone exposure. Most notably, food intake significantly increased during the light phase of a day when mice were usually inactive. It remains to be determined whether risperidone treatment perturbs the circadian regulation of metabolism, as eating at the “wrong” time could also contribute to weight gain and metabolic dysfunction (Pickel and Sung, 2020). Both the increase in food intake and decrease in physical activity contributed to a positive energy balance. On the other hand, risperidone acutely increased EE, which protected against weight gain. The increase in EE could be due to risperidone’s effect on thermogenic gene expression. Indeed, it has been shown that risperidone treatment induces uncoupling protein 1 (UCP1) expression in the BAT (Li et al., 2013; Ota et al., 2002). Additionally, risperidone may promote thermogenesis by enhancing activities of the sympathetic nerves that innervate the BAT (Kalkman and Loetscher, 2003). Nevertheless, the modest rise in EE was not sufficient to counterbalance the extra energy intake. Indeed, results from the PF experiments revealed that hyperphagia alone accounted for most of the weight gain in risperidone-fed mice and therefore should be the focus for prospective therapeutics treating risperidone-induced obesity.
To uncover candidate genes and pathways behind risperidone-induced hyperphagia, we studied how risperidone treatment altered the transcriptomic landscape in the hypothalamus—an essential regulator of food intake. Using comparative transcriptomic analyses, we identified a hypothalamic gene expression signature associated with chronic risperidone exposure. It consisted of 10 genes whose expression responded directly to risperidone treatment. Among them, Mc4r plays a critical role in appetite control in both humans and rodents. Of note, human genetic variants near the Mc4r locus have been associated with AAP-induced weight gain (Malhotra et al., 2012); however, whether or not Mc4r plays a role in the underlying pathophysiology has not been investigated. We found that risperidone’s obesogenic effect depended on Mc4r in Sim1 neurons. Unlike in wild-type mice, risperidone treatment did not increase food intake or body weight in Mc4rSim1-KO mice. Of note, a limitation of the Mc4rSim1-KO model is that these mice develop hyperphagic obesity. As a result, a ceiling effect could exist that prevented these mice from eating more food and gaining extra weight when fed the risperidone diet. To mitigate this confounder, we began our chronic feeding study in young (7-wk-old) Mc4rSim1-KO mice, when they had not yet become severely obese (∼25 g). During the first 8 wk of the treatment, risperidone-fed wild-type mice gained 20.71% more weight than those fed the control diet, whereas risperidone-fed Mc4rSim1-KO mice gained only 2.62% more weight during the same period. Similarly, we measured daily food intake in young Mc4rSim1-KO mice when their intake (∼4 g/d) was equivalent to that of weight-matched wild-type mice. We found that risperidone did not increase food intake in young Mc4rSim1-KO mice. Meanwhile, hyperphagia could still be induced in these mice by an overnight fast. Collectively, these findings support the notion that risperidone-induced hyperphagia and weight gain were blunted in Mc4rSim1-KO mice. Interestingly, we showed that the same pathway was necessary for similar metabolic effects of olanzapine, another AAP that is widely used for many mood disorders. Our findings therefore raise the intriguing possibility that despite their distinct receptor-binding profiles in the brain, different AAPs may target a common hypothalamic pathway to cause weight gain. However, it is important to note that our data do not support the notion that risperidone acts exclusively on MC4Rs to cause weight gain. Rather, given its promiscuous binding profiles, we suspect risperidone’s metabolic effects are likely due to its interactions with multiple central and peripheral receptor pathways (Lett et al., 2012).
Our study provides the first experimental evidence linking deficits in hypothalamic MC4R signaling to AAP-induced metabolic syndrome. We uncovered two mechanisms whereby risperidone impairs energy balance through MC4Rs. First, haploinsufficiency of Mc4r leads to hyperphagia and obesity (Vaisse et al., 1998; Yeo et al., 1998). We found that risperidone treatment decreased Mc4r mRNAs in the hypothalamus. The reduction could be detected as early as 5 d after risperidone exposure and occurred before the onset of obesity. An earlier study suggested that Mc4r was a transcriptional target of the transcription factor nescient helix-loop-helix 2 (Nhlh2; Wankhade and Good, 2011). However, Nhlh2 expression did not change in response to risperidone treatment. Therefore, future work is warranted to investigate the transcriptional or the post-transcriptional mechanisms whereby risperidone regulates Mc4r expression. Second, chemogenetic silencing of MC4RPVH neurons resulted in hyperphagia and obesity in mice (Garfield et al., 2015; Li et al., 2019). Remarkably, whole-cell patch-clamp recording demonstrated for the first time that risperidone acutely inhibited the activity of genetically labeled MC4RPVH neurons. The hyperpolarizing effect was observed in nearly half (46.9%) of the neurons recorded and can be prevented by pretreatment with an MC4R blocker, JKC-363. Of note, while risperidone still inhibited a smaller percentage of recorded PVH non-MC4R neurons (15%), risperidone-induced hyperpolarization in these neurons cannot be blocked by JKC-363. Moreover, the Drd2 is a common target of antipsychotic drugs and is responsible for their antipsychotic effects (Lett et al., 2012). Interestingly, Drd2 is expressed by some MC4R neurons (Oude Ophuis et al., 2014). Therefore, it raises the possibility that risperidone may regulate activities of MC4RPVH neurons through Drd2. However, L-741626, a dopamine D2 receptor antagonist, did not prevent risperidone-induced inhibition of MC4RPVH neurons, suggesting that risperidone’s effect on MC4RPVH neuron activity is independent of Drd2.
In addition to Drd2, risperidone directly binds other G protein–coupled receptors and functions as an inverse agonist (Lett et al., 2012). Our observation that risperidone-induced hyperpolarization was blocked by two MC4R agonists (MTII and setmelanotide) suggests that risperidone could also act as an inverse agonist for MC4R and compete with its endogenous ligand α–melanocyte-stimulating hormone for direct binding. Nevertheless, evidence supporting such a direct interaction is lacking, which is a limitation of the current study. Similarly, the intracellular signaling cascade that mediates risperidone’s effect within MC4RPVH neurons remains to be determined. We found that risperidone-induced hyperpolarization involved the opening of a postsynaptic potassium conductance. Interestingly, Cone and colleagues recently reported that regulation of firing activity of MC4RPVH neurons was independent of Gs α-subunit signaling but was mediated by ligand-induced coupling of the inwardly rectifying potassium channel 7.1 (Ghamari-Langroudi et al., 2015). Therefore, it remains to be determined whether risperidone hijacks the same pathway to inhibit MC4RPVH neurons. It is worth mentioning that, in addition to MC4RPVH neurons, MC4Rs in other hypothalamic and extra-hypothalamic neurons have been implicated in the regulation of food intake and body weight (Cui and Lutter, 2013; Rossi et al., 2011). Risperidone treatment could have similar effects on Mc4r expression and/or activity in these neurons. Therefore, risperidone-induced metabolic deficits could be the result of MC4Rs/MC4R neuron deficiency at multiple brain nodes.
Finally, mechanism-based pharmacotherapies against risperidone- or other AAP-induced metabolic syndromes have been lacking. For many youths and adults taking risperidone, lifestyle changes, nutritional consulting, and commonly used anti-obesity medications only provide limited relief. Our findings that hypothalamic MC4Rs mediate the obesogenic effect of two commonly prescribed AAPs suggest that MC4R is a novel therapeutic target for AAP-induced weight gain. Indeed, we demonstrated that setmelanotide, an MC4R-specific agonist, was effective in reversing hyperphagia and obesity in both risperidone- and olanzapine-fed mice. Mechanistically, we showed that setmelanotide blocked risperidone-induced hyperpolarization of MC4RPVH neurons. We believe that disinhibition of these neurons by setmelanotide treatment contributes to the relief of metabolic syndrome in risperidone-fed mice. Moreover, since risperidone inhibited MC4RPVH neurons in a concentration-dependent manner, we suspect that the therapeutic effect of setmelanotide may also be dose dependent. Notably, setmelanotide has just been approved for treating rare genetic obesity disorders in humans (Clément et al., 2018; Kühnen et al., 2016). Results from recent clinical trials have shown that it is well tolerated for long-term use with no detrimental effects on blood pressure or heart rate (Haws et al., 2020). However, for setmelanotide to become a potential therapy for risperidone-induced weight gain, it must not compromise risperidone’s psychotropic efficacy (Liu et al., 2014). To this end, we showed that cotreatment of setmelanotide did not diminish risperidone’s ability to suppress schizophrenia-like hyperlocomotion in mice. These findings raise hope that risperidone’s psychotropic and metabolic effects are mediated by distinct neural pathways and therefore may be targeted separately. Furthermore, our findings suggest that setmelanotide has the potential to become a novel mechanism-based therapy for millions of affected patients.
Materials and methods
All mice were housed in a temperature-controlled room with a 12-h light/12-h dark cycle (lights on at 6 a.m., lights off at 6 p.m.) in the animal facility of University of Texas (UT) Southwestern Medical Center. Mice were provided standard chow (No. 2016; Harlan Teklad) as well as water ad libitum unless otherwise noted. C57BL/6 mice were purchased from the UT Southwestern Rodent Breeding Core Facility. All mouse experiments were performed in female mice, as AAP treatment produced stronger metabolic phenotypes in female mice than in males (Zapata and Osborn, 2020). Sim1-Cre+/−; Mc4rfl/fl and Mc4rfl/fl littermates were maintained on a C57BL/6 background. The chronic drug infusion study was conducted under isoflurane anesthesia followed by carprofen analgesia. Setmelanotide (2 mg/kg/d) was administered using s.c. osmotic mini-pumps (Alzet). All mouse experiments were approved by the Institutional Animal Care and Use Committee of the UT Southwestern Medical Center.
Diets
All diets were prepared by Research Diets and shared the same macronutrients (21% kcal protein, 34% kcal carbohydrate, and 45% kcal fat), ingredient composition (Casein 800 kcal, DL-Methionine 12 kcal, cornstarch 200 kcal, maltodextrin 400 kcal, sucrose 695 kcal, corn oil 450 kcal, lard 1,305 kcal, and vitamin mix 40 kcal), and energy density (4.76 kcal/g). Risperidone (Cat# AC459390010; Acros Organics) was mixed into a control diet D09092903 to make the risperidone diets D18041008 (25 mg/kg) and D18041007 (100 mg/kg). Similarly, olanzapine (Cat# 356941G; ChemPacific) was mixed into the same control diet D09092903 to make the olanzapine diet as we described previously (Lord et al., 2017). All diets are available for purchase at the vendor.
Metabolic phenotype analysis
We used magnetic-resonance whole-body composition analyzer (EchoMRI) to analyze a mouse’s body composition (fat mass, lean mass, and water content). For chronic studies, body weight and food intake were measured weekly. The acute effects of risperidone on energy intake and expenditure were measured using an indirect calorimetric system (PhenoMaster; TSE Systems) in the Metabolic Phenotyping Core of UT Southwestern Medical Center.
Histology
Mouse adipose tissue was fixed in 4% paraformaldehyde overnight and then dehydrated through a series of ethanol baths with ascending concentrations (up to 100%). Paraffin embedding, sectioning, and H&E staining were performed by the Histo Pathology Core of the UT Southwestern Medical Center.
GTT and ITT
For the GTT, mice were fasted for 7 h with water provided ad libitum from 8 a.m. on the experimental day. During GTT, blood glucose levels were monitored at 0, 15, 30, 60, 90, and 120 min after an i.p. dose of glucose (dextrose; 1.0 g/kg body weight). Blood glucose was taken from the tail vein and analyzed using a Glucometer (Johnson & Johnson). For the ITT, blood glucose levels were monitored at 0, 15, 30, 60, 90, and 120 min after an i.p. dose of insulin (1 U/kg body weight).
Hormone measurements
Serum leptin and insulin levels were measured by ELISA kits (Cat# 22-LEPMS-E01 and Cat# 80-INSMSU-E01, respectively) from Alpco Diagnostics. Serum levels of NEFAs were measured using an in vitro enzymatic colorimetric method with an HR series NEFA-HR(2) kit (Cat# 999–34691, 995–34791, 991–34891, 993–35191, 276–76491, 997–76491; WAKO Diagnostics). Triglyceride was measured by Glycerol-3-phosphate oxidase and N-(3-sulfopropyl)-3-methoxy-5-methylaniline method using the L-Type Triglyceride M kit (Cat# 994–02891, 990–02991, 992–02892, 998–02992, 464–01601; WAKO Diagnostics). Blank and quality controls were added in every measurement. All experimental procedures were conducted according to manufacturers’ manuals.
qPCR
Total RNA was isolated by the Direct-zol RNA Kit (Zymo) according to the manufacturer’s recommendations. 3 μg of total RNA was used as the template for cDNA synthesis (Invitrogen). Real-time PCR was performed using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), and qPCR reactions were performed in triplicate on a 384-well PCR microplate for CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Taqman probes used include (agouti-related peptide, Mm00475829_g1; serotonin 1b receptor, Mm00439377_s1; Htr2c, Mm00434127_m1; Mc4r, Mm00457483_s1; neuropeptide Y, Mm01410146_m1; and Tbp, Mm00446973_m1; Thermo Fisher Scientific). The relative expression levels of each gene were normalized to the housekeeping gene TATA-box binding protein (Tbp). The specificity of amplified genes was verified by dissociation curves. RNA expression data were analyzed using the comparative cycle threshold method.
Feeding studies
For the refeeding experiment, Mc4rSim1-KO mice and their littermate controls were habituated with daily handling and i.p. injections of saline for 3 d before the fasting-refed test. Mice were fasted for 18 h (from 4 p.m. to 10 a.m.) before receiving an i.p. dose of saline or setmelanotide (Cat# HY-19870; MedChem Express; 2 mg/kg dissolved in saline). 30 min after the injection, a chow pellet was given to singly housed mice. Food consumption was monitored at 30, 60, 120, and 240 min.
For the PF experiment, 30 mice were individually housed and fed with the control diet for a week before being randomly divided into three groups with similar numbers. Mice in the control group continued to be fed with the control diet. Mice in the RISP_Ad.lib group were switched to the risperidone diet. Mice in the RISP_PF group were given the risperidone diet but were restricted to the same amount that the control mice had consumed.
Amphetamine-induced hyperlocomotion
The amphetamine-induced hyperlocomotion test was performed by the Rodent Behavior Core in the Peter O’Donnell Jr. Brain Institute at UT Southwestern Medical Center. Briefly, C57BL/6 mice were habituated for the open field test with daily single i.p. injections of saline for 3 d. On the third day of habituation, all mice were given D-amphetamine (2.5 mg/kg) 30 min after the saline injection. Following the habituation, all mice were randomly divided into four groups: (1) mice in the saline group received an i.p. dose of saline; (2) mice in the setmelanotide group received an i.p. dose of setmelanotide (2 mg/kg); (3) mice in the RISP group received an i.p. dose of risperidone (0.2 mg/kg); and (4) mice in the RISP + setmelanotide group received both risperidone (0.2 mg/kg) and setmelanotide (2 mg/kg). Locomotor activity (beam breaks) was immediately recorded after the first injection. An i.p. dose of D-amphetamine (2.5 mg/kg) was administered 30 min after the first injection.
RNA-seq and bioinformatics
Total RNA from the hypothalamus was extracted using the Direct-zol RNA Kits (Zymo) according to the manufacturer’s recommendations. RNA quality was validated using the Agilent 2100 Bioanalyzer. 150-bp paired-end reads were sequenced on the DNBseq platform in BGI Americas Corporation. Full sequencing data are available online at National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE158751). We obtained an average of 2.5 million mapped reads per sample with 90% unique mapped efficiency. The paired-end clean reads were aligned to the mouse reference genome GRCm38_mm10 (release99) using STAR (Dobin et al., 2013), and each gene’s reads numbers were counted by FeatureCounts (Liao et al., 2014). DEG analysis was performed by DEseq2 package (Love et al., 2014), in which gene length and dispersion bias were corrected. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P value <0.05 and logFold >0.3 by DESeq2 were defined as DEGs.
Electrophysiology
Whole-cell patch-clamp recordings from MC4RPVH neurons were maintained in hypothalamic slice preparations from Mc4r-T2A-iCre::tdTomato mice (Garfield et al., 2015), and data analysis was performed as previously described (Sohn et al., 2016). Mc4r-T2A-iCre::tdTomato mice were housed in a temperature-controlled room with a 12-h light/12-h dark cycle (lights on at 7 a.m., lights off at 7 p.m.) in the animal facility of the Korea Advanced Institute of Science and Technology. Mouse protocols for electrophysiology experiments were approved by the Institutional Animal Care and Use Committee of Korea Advanced Institute of Science and Technology. Briefly, 3–5-wk-old male and female mice were deeply anesthetized with isoflurane inhalations and transcardially perfused with a modified ice-cold artificial CSF (ACSF; described below), in which an equimolar amount of sucrose was substituted for NaCl. The mice were then decapitated, and the entire brain was removed and immediately submerged in ice-cold, carbogen-saturated (95% O2 and 5% CO2) ACSF (126 mM NaCl, 2.8 mM KCl, 1.2 mM MgSO4, 2.5 mM CaCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, and 5 mM glucose). A brain block containing the hypothalamus was made. Coronal sections (250 µm) were cut with a Leica VT1200S Vibratome and then incubated in oxygenated ACSF at room temperature for at least 1 h for recovery. Slices were transferred to the recording chamber and allowed to equilibrate for 10–20 min before recording. The slices were bathed in oxygenated ACSF (32–34°C) at a flow rate of ∼4 ml/min. The pipette solution for whole-cell recording was modified to include an intracellular dye (Alexa Fluor 488) for whole-cell recording: 120 mM K-gluconate, 10 mM KCl, 10 mM Hepes, 5 mM EGTA, 1 mM CaCl2, 1 mM MgCl2, 2 mM MgATP, and 0.03 mM Alexa Fluor 488 hydrazide dye, pH 7.3. Epifluorescence was briefly used to target fluorescent cells, at which time the light source was switched to infrared differential interference contrast imaging to obtain the whole-cell recording (Nikon Eclipse FN1 equipped with a fixed stage and an optiMOS scientific CMOS camera). Electrophysiological signals were recorded using an Axopatch 700B amplifier (Molecular Devices), low-pass filtered at 2–5 kHz, and analyzed offline on a PC with pCLAMP programs (Molecular Devices). Recording electrodes had resistances of 4–6 MΩ when filled with the K-gluconate internal solution. Input resistance was assessed by measuring voltage deflection at the end of the response to hyperpolarizing rectangular current pulse steps (500 ms of −25 to 0 pA). Membrane potential values were not compensated to account for junction potential (−8 mV). Solutions containing risperidone were typically perfused for ∼5 min. A drug effect was required to be associated temporally with peptide application, and the response had to be stable within a few minutes. A neuron was considered depolarized or hyperpolarized if a change in membrane potential was at least 2 mV in amplitude. MC4RPVH neurons from male and female mice had comparable basic electrical properties (resting membrane potential, cell capacitance, and input resistance) and responses to risperidone, and thus we pooled data from either sex for further analyses.
Quantification and statistical analyses
Replicate information is indicated in the figure legends. All results are presented as mean ± SEM and analyzed using statistical tools implemented in Prism (GraphPad version 8). Statistical analyses were performed using the Student’s t test and regular one-way or two-way ANOVA. Differences with P < 0.05 were considered to be significant: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Data availability
The transcriptomic data are now deposited in the Gene Expression Omnibus database under accession number GSE158751.
Online supplemental material
Fig. S1 demonstrates additional metabolic characterization of risperidone-fed mice. Fig. S2 depicts plasma levels of insulin, leptin, triglyceride, and NEFA in Mc4rSim1-KO mice treated with either the control, risperidone, or olanzapine diet. Fig. S3 demonstrates whole-cell patch-clamp recordings of PVH non-MC4R neurons and their responses to acute risperidone treatment.
Acknowledgments
The project was inspired by a conversation with Dr. Ted Abel, director of the Iowa Neuroscience Institute, during his visit to the UT Southwestern Medical Center. We thank members of the UT Southwestern Metabolic Phenotyping Core. We thank Priscilla Saenz at the Rodent Behavior Core in the Peter O’Donnell Jr. Brain Institute at UT Southwestern Medical Center for performing the behavioral experiments. We thank Claire Liu for helping with the literature search and proofreading the manuscript.
The authors were supported by U.S. National Institutes of Health grants R01 DK114036 to C. Liu; F32DK116427 to S.C. Wyler, and K01AA024809 to L. Jia and National Research Foundation of Korea grant 2019R1A2C2005161 to J.-W. Sohn. C. Liu was also supported by an American Heart Association Scientist Development Grant 16SDG27260001, a UT Southwestern Medical Center Pilot and Feasibility Award, and a Grossman Endowment Award for Excellence in Diabetes Research.
Author contributions: L. Li, E.-S. Yoo, J.-W. Sohn, and C. Liu designed the experiments. X. Li conducted the first body weight study using the risperidone diet in mice. L. Li, E.-S. Yoo, X. Li, X. Chen, R. Wan, and A.G. Arnold collected data. L. Li, E.-S. Yoo, S.C. Wyler, S.G. Birnbaum, L. Jia, J.-W. Sohn, and C. Liu analyzed the data. L. Li, E.-S. Yoo, J.-W. Sohn, and C. Liu wrote the manuscript.
References
- Angrist, B., Rotrosen J., and Gershon S.. 1980. Responses to apomorphine, amphetamine, and neuroleptics in schizophrenic subjects. Psychopharmacology (Berl.). 67:31–38. 10.1007/BF00427592 [DOI] [PubMed] [Google Scholar]
- Balthasar, N., Dalgaard L.T., Lee C.E., Yu J., Funahashi H., Williams T., Ferreira M., Tang V., McGovern R.A., Kenny C.D., et al. 2005. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 123:493–505. 10.1016/j.cell.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Chen, K.Y., Muniyappa R., Abel B.S., Mullins K.P., Staker P., Brychta R.J., Zhao X., Ring M., Psota T.L., Cone R.D., et al. 2015. RM-493, a melanocortin-4 receptor (MC4R) agonist, increases resting energy expenditure in obese individuals. J. Clin. Endocrinol. Metab. 100:1639–1645. 10.1210/jc.2014-4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément, K., Biebermann H., Farooqi I.S., Van der Ploeg L., Wolters B., Poitou C., Puder L., Fiedorek F., Gottesdiener K., Kleinau G., et al. 2018. MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med. 24:551–555. 10.1038/s41591-018-0015-9 [DOI] [PubMed] [Google Scholar]
- Collet, T.H., Dubern B., Mokrosinski J., Connors H., Keogh J.M., Mendes de Oliveira E., Henning E., Poitou-Bernert C., Oppert J.M., Tounian P., et al. 2017. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab. 6:1321–1329. 10.1016/j.molmet.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H., and Lutter M.. 2013. The expression of MC4Rs in D1R neurons regulates food intake and locomotor sensitization to cocaine. Genes Brain Behav. 12:658–665. 10.1111/gbb.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingrod, V.L., Perry P.J., Ringold J.C., Lund B.C., Bever-Stille K., Fleming F., Holman T.L., and Miller D.. 2005. Weight gain associated with the -759C/T polymorphism of the 5HT2C receptor and olanzapine. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 134B:76–78. 10.1002/ajmg.b.20169 [DOI] [PubMed] [Google Scholar]
- Garfield, A.S., Li C., Madara J.C., Shah B.P., Webber E., Steger J.S., Campbell J.N., Gavrilova O., Lee C.E., Olson D.P., et al. 2015. A neural basis for melanocortin-4 receptor-regulated appetite. Nat. Neurosci. 18:863–871. 10.1038/nn.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamari-Langroudi, M., Digby G.J., Sebag J.A., Millhauser G.L., Palomino R., Matthews R., Gillyard T., Panaro B.L., Tough I.R., Cox H.M., et al. 2015. G-protein-independent coupling of MC4R to Kir7.1 in hypothalamic neurons. Nature. 520:94–98. 10.1038/nature14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke, J.M., Dhurandhar E.J., Correll C.U., Morrato E.H., Newcomer J.W., Remington G., Nasrallah H.A., Crystal S., Nicol G., and Allison D.B.. Adipogenic and Metabolic Effects of APDs Conference Speakers . 2012. Recent advances in understanding and mitigating adipogenic and metabolic effects of antipsychotic drugs. Front. Psychiatry. 3:62. 10.3389/fpsyt.2012.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haws, R., Brady S., Davis E., Fletty K., Yuan G., Gordon G., Stewart M., and Yanovski J.. 2020. Effect of setmelanotide, a melanocortin-4 receptor agonist, on obesity in Bardet-Biedl syndrome. Diabetes Obes. Metab. 22:2133–2140. 10.1111/dom.14133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar, D., Lynch C.A., Fairchild-Huntress V., Dunmore J.H., Fang Q., Berkemeier L.R., Gu W., Kesterson R.A., Boston B.A., Cone R.D., et al. 1997. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 88:131–141. 10.1016/S0092-8674(00)81865-6 [DOI] [PubMed] [Google Scholar]
- Janowsky, D.S., and Risch C.. 1979. Amphetamine psychosis and psychotic symptoms. Psychopharmacology (Berl.). 65:73–77. 10.1007/BF00491982 [DOI] [PubMed] [Google Scholar]
- Kaar, S.J., Natesan S., McCutcheon R., and Howes O.D.. 2020. Antipsychotics: Mechanisms underlying clinical response and side-effects and novel treatment approaches based on pathophysiology. Neuropharmacology. 172:107704. 10.1016/j.neuropharm.2019.107704 [DOI] [PubMed] [Google Scholar]
- Kalkman, H.O., and Loetscher E.. 2003. alpha2C-Adrenoceptor blockade by clozapine and other antipsychotic drugs. Eur. J. Pharmacol. 462:33–40. 10.1016/S0014-2999(03)01308-6 [DOI] [PubMed] [Google Scholar]
- Kaur, G., and Kulkarni S.K.. 2002. Studies on modulation of feeding behavior by atypical antipsychotics in female mice. Prog. Neuropsychopharmacol. Biol. Psychiatry. 26:277–285. 10.1016/S0278-5846(01)00266-4 [DOI] [PubMed] [Google Scholar]
- Kievit, P., Halem H., Marks D.L., Dong J.Z., Glavas M.M., Sinnayah P., Pranger L., Cowley M.A., Grove K.L., and Culler M.D.. 2013. Chronic treatment with a melanocortin-4 receptor agonist causes weight loss, reduces insulin resistance, and improves cardiovascular function in diet-induced obese rhesus macaques. Diabetes. 62:490–497. 10.2337/db12-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.F., Huang A.S., Snowman A.M., Teuscher C., and Snyder S.H.. 2007. From the Cover: Antipsychotic drug-induced weight gain mediated by histamine H1 receptor-linked activation of hypothalamic AMP-kinase. Proc. Natl. Acad. Sci. USA. 104:3456–3459. 10.1073/pnas.0611417104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze, W.K., Hufeisen S.J., Popadak B.A., Renock S.M., Steinberg S., Ernsberger P., Jayathilake K., Meltzer H.Y., and Roth B.L.. 2003. H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology. 28:519–526. 10.1038/sj.npp.1300027 [DOI] [PubMed] [Google Scholar]
- Kühnen, P., Clément K., Wiegand S., Blankenstein O., Gottesdiener K., Martini L.L., Mai K., Blume-Peytavi U., Grüters A., and Krude H.. 2016. Proopiomelanocortin Deficiency Treated with a Melanocortin-4 Receptor Agonist. N. Engl. J. Med. 375:240–246. 10.1056/NEJMoa1512693 [DOI] [PubMed] [Google Scholar]
- Lett, T.A., Wallace T.J., Chowdhury N.I., Tiwari A.K., Kennedy J.L., and Müller D.J.. 2012. Pharmacogenetics of antipsychotic-induced weight gain: review and clinical implications. Mol. Psychiatry. 17:242–266. 10.1038/mp.2011.109 [DOI] [PubMed] [Google Scholar]
- Li, X., Johnson M.S., Smith D.L. Jr., Li Y., Kesterson R.A., Allison D.B., and Nagy T.R.. 2013. Effects of risperidone on energy balance in female C57BL/6J mice. Obesity (Silver Spring). 21:1850–1857. 10.1002/oby.20350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.M., Madara J.C., Steger J.S., Krashes M.J., Balthasar N., Campbell J.N., Resch J.M., Conley N.J., Garfield A.S., and Lowell B.B.. 2019. The Paraventricular Hypothalamus Regulates Satiety and Prevents Obesity via Two Genetically Distinct Circuits. Neuron. 102:653–667.e6. 10.1016/j.neuron.2019.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu, C., Lee S., and Elmquist J.K.. 2014. Circuits controlling energy balance and mood: inherently intertwined or just complicated intersections? Cell Metab. 19:902–909. 10.1016/j.cmet.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.C., Wyler S.C., Wan R., Castorena C.M., Ahmed N., Mathew D., Lee S., Liu C., and Elmquist J.K.. 2017. The atypical antipsychotic olanzapine causes weight gain by targeting serotonin receptor 2C. J. Clin. Invest. 127:3402–3406. 10.1172/JCI93362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra, A.K., Correll C.U., Chowdhury N.I., Müller D.J., Gregersen P.K., Lee A.T., Tiwari A.K., Kane J.M., Fleischhacker W.W., Kahn R.S., et al. 2012. Association between common variants near the melanocortin 4 receptor gene and severe antipsychotic drug-induced weight gain. Arch. Gen. Psychiatry. 69:904–912. 10.1001/archgenpsychiatry.2012.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuz, E., Franklin R., Gillespie T., Murphy A., Bernstein J., Chiu A., Hotten T., and Kassahun K.. 1997. Disposition and metabolism of olanzapine in mice, dogs, and rhesus monkeys. Drug Metab. Dispos. 25:573–583. [PubMed] [Google Scholar]
- Meltzer, H.Y. 2013. Update on typical and atypical antipsychotic drugs. Annu. Rev. Med. 64:393–406. 10.1146/annurev-med-050911-161504 [DOI] [PubMed] [Google Scholar]
- Meltzer, H.Y., and Huang M.. 2008. In vivo actions of atypical antipsychotic drug on serotonergic and dopaminergic systems. Prog. Brain Res. 172:177–197. 10.1016/S0079-6123(08)00909-6 [DOI] [PubMed] [Google Scholar]
- Morgan, A.P., Crowley J.J., Nonneman R.J., Quackenbush C.R., Miller C.N., Ryan A.K., Bogue M.A., Paredes S.H., Yourstone S., Carroll I.M., et al. 2014. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One. 9:e115225. 10.1371/journal.pone.0115225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, M., Mori K., Nakashima A., Kaneko Y.S., Fujiwara K., Itoh M., Nagasaka A., and Ota A.. 2002. Peripheral injection of risperidone, an atypical antipsychotic, alters the bodyweight gain of rats. Clin. Exp. Pharmacol. Physiol. 29:980–989. 10.1046/j.1440-1681.2002.t01-1-03755.x [DOI] [PubMed] [Google Scholar]
- Oude Ophuis, R.J., Boender A.J., van Rozen A.J., and Adan R.A.. 2014. Cannabinoid, melanocortin and opioid receptor expression on DRD1 and DRD2 subpopulations in rat striatum. Front. Neuroanat. 8:14. 10.3389/fnana.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.M., Chung Y.C., Lee S.H., Lee K.J., Kim H., Byun Y.C., Lim S.W., Paik J.W., and Lee H.J.. 2006. Weight gain associated with the alpha2a-adrenergic receptor -1,291 C/G polymorphism and olanzapine treatment. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 141B:394–397. 10.1002/ajmg.b.30311 [DOI] [PubMed] [Google Scholar]
- Pickel, L., and Sung H.K.. 2020. Feeding Rhythms and the Circadian Regulation of Metabolism. Front. Nutr. 7:39. 10.3389/fnut.2020.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi, M., Ferrentino R.I., Scrinzi G., Scavone C., Capuano A., Radice S., Nobile M., Formisano P., Clementi E., Bravaccio C., et al. 2020. Weight and body mass index increase in children and adolescents exposed to antipsychotic drugs in non-interventional settings: a meta-analysis and meta-regression. Eur. Child Adolesc. Psychiatry. 10.1007/s00787-020-01582-9 [DOI] [PubMed] [Google Scholar]
- Rauser, L., Savage J.E., Meltzer H.Y., and Roth B.L.. 2001. Inverse agonist actions of typical and atypical antipsychotic drugs at the human 5-hydroxytryptamine(2C) receptor. J. Pharmacol. Exp. Ther. 299:83–89. [PubMed] [Google Scholar]
- Rojo, L.E., Gaspar P.A., Silva H., Risco L., Arena P., Cubillos-Robles K., and Jara B.. 2015. Metabolic syndrome and obesity among users of second generation antipsychotics: A global challenge for modern psychopharmacology. Pharmacol. Res. 101:74–85. 10.1016/j.phrs.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Rossi, J., Balthasar N., Olson D., Scott M., Berglund E., Lee C.E., Choi M.J., Lauzon D., Lowell B.B., and Elmquist J.K.. 2011. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13:195–204. 10.1016/j.cmet.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, J.-W., Oh Y., Kim K.W., Lee S., Williams K.W., and Elmquist J.K.. 2016. Leptin and insulin engage specific PI3K subunits in hypothalamic SF1 neurons. Mol. Metab. 5:669–679. 10.1016/j.molmet.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse, C., Clement K., Guy-Grand B., and Froguel P.. 1998. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 20:113–114. 10.1038/2407 [DOI] [PubMed] [Google Scholar]
- van der Zwaal, E.M., Luijendijk M.C., Adan R.A., and la Fleur S.E.. 2008. Olanzapine-induced weight gain: chronic infusion using osmotic minipumps does not result in stable plasma levels due to degradation of olanzapine in solution. Eur. J. Pharmacol. 585:130–136. 10.1016/j.ejphar.2007.11.078 [DOI] [PubMed] [Google Scholar]
- Van Swearingen, A.E., Walker Q.D., and Kuhn C.M.. 2013. Sex differences in novelty- and psychostimulant-induced behaviors of C57BL/6 mice. Psychopharmacology (Berl.). 225:707–718. 10.1007/s00213-012-2860-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wankhade, U.D., and Good D.J.. 2011. Melanocortin 4 receptor is a transcriptional target of nescient helix-loop-helix-2. Mol. Cell. Endocrinol. 341:39–47. 10.1016/j.mce.2011.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichniak, A., Skowerska A., Chojnacka-Wójtowicz J., Tafliński T., Wierzbicka A., Jernajczyk W., and Jarema M.. 2011. Actigraphic monitoring of activity and rest in schizophrenic patients treated with olanzapine or risperidone. J. Psychiatr. Res. 45:1381–1386. 10.1016/j.jpsychires.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Xu, Y., Elmquist J.K., and Fukuda M.. 2011. Central nervous control of energy and glucose balance: focus on the central melanocortin system. Ann. N. Y. Acad. Sci. 1243:1–14. 10.1111/j.1749-6632.2011.06248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G.S., Farooqi I.S., Aminian S., Halsall D.J., Stanhope R.G., and O’Rahilly S.. 1998. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 20:111–112. 10.1038/2404 [DOI] [PubMed] [Google Scholar]
- Zapata, R.C., and Osborn O.. 2020. Susceptibility of male wild type mouse strains to antipsychotic-induced weight gain. Physiol. Behav. 220:112859. 10.1016/j.physbeh.2020.112859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The transcriptomic data are now deposited in the Gene Expression Omnibus database under accession number GSE158751.