Abstract
The DNA viruses, Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), are members of the gammaherpesvirus subfamily, a group of viruses whose infection is associated with multiple malignancies, including cancer. The primary host for these viruses is humans and, like all herpesviruses, infection with these pathogens is lifelong. Due to the persistence of gammaherpesvirus infection and the potential for cancer formation in infected individuals, there is a driving need to understand not only the biology of these viruses and how they remain undetected in host cells but also the mechanism(s) by which tumorigenesis occurs. One of the methods that has provided much insight into these processes is proteomics. Proteomics is the study of all the proteins that are encoded by a genome and allows for i) identification of existing and novel proteins derived from a given genome, ii) interrogation of protein-protein interactions within a system, and iii) discovery of druggable targets for the treatment of malignancies. In this chapter, we explore how proteomics has contributed to our current understanding of gammaherpesvirus biology and their oncogenic processes, as well as the clinical applications of proteomics for the detection and treatment of gammaherpesvirus-associated cancers.
Keywords: KSHV, EBV, herpesvirus, proteomics, oncogenic, virus-host interactions
Introduction
Herpesviruses are DNA viruses that replicate their genomes in the nucleus of host cells. These viruses establish lifelong infections and have two phases of their lifecycle, including a lytic phase and a latent phase. The quiescent latent cycle expresses few viral proteins, while the lytic cycle includes the expression of most of the viral genes and results in viral replication. At least eight species of human herpesviruses have been characterized to date, and they are classified into three distinct subfamilies: α, β, and γ. The α-herpesviridae subfamily includes herpes simplex virus type 1 (HSV-1), herpes simplex virus type 2 (HSV-2), and varicella-zoster virus (VZV). The β-herpesviridae subfamily includes human cytomegalovirus (HCMV), by which congenital infection causes the most considerable burden of disease, as well as human herpesvirus-6 and human herpesvirus-7 (HHV-6 and HHV-7). The γ-herpesviridae subfamily includes Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), both of which are viruses linked to tumorigenesis (Jha et al., 2016). EBV is the etiological agent of Burkitt’s lymphoma (BL), nasopharyngeal carcinoma (NPC), post-transplant lymphoma (PTL), and Hodgkin’s lymphoma. KSHV infection is associated with its namesake Kaposi’s sarcoma (KS) as well as primary effusion lymphoma (PEL) and the plasmablastic variant of multicentric Castleman’s disease (MCD).
Currently, clinically available inhibitors for herpesviruses target the viral DNA polymerase, a vital protein necessary for the completion of the herpesvirus lifecycle (Pagano et al., 2018). The most common class of these inhibitors are nucleoside analogs, which resemble nucleosides produced by cells, but their incorporation results in termination of DNA replication. However, acyclic nucleoside phosphonates (ANPs) such as cidofovir can also be used to treat herpesvirus infections (De Clercq and Holy, 2005). Collectively, these anti-herpesvirus drugs target the lytic lifecycle of the virus, as they become active only once the viral thymidine kinase (a lytic protein) has phosphorylated the compound. Presently, for treatment of KS and EBV-associated cancers in HIV-positive individuals, highly active antiretroviral therapy (HAART) is used. Although these drugs do not directly target KSHV or EBV, they help to improve immune function by targeting HIV (Cheung et al., 2005; Manners et al., 2018). Likewise, PTL can be treated by stopping the use of immunosuppressive agents to help restore the patient’s immune system and therefore limit tumor progression; however, this approach may result in graft rejection. Thus, the generation of novel therapeutics for gammaherpesvirus infections and their associated cancers is of paramount importance.
Proteomics can offer insights that genomics or traditional laboratory techniques fail to elucidate, such as protein-protein interactions and the presence or modulation of post-translational modifications (PTMs). Through an understanding of i) the proteins contained within gammaherpesvirus particles, ii) which proteins these viruses interact with during infection, and iii) how these viruses modulate the protein content within the host, new therapeutic strategies against KSHV and EBV infections and their associated cancers can be developed. In this chapter, we review proteomic methodologies have been utilized in understanding gammaherpesvirus biology and their roles in tumorigenesis, and highlight proteomic studies that identified key viral proteins and their interaction networks throughout the viral lifecycle. Additionally, we explore in-depth the cancers associated with gammaherpesvirus infection and how proteomics is beneficial to the diagnosis and treatment of these virally-associated cancers. Finally, we conclude by discussing the future potential of proteomics for cancer research and viral oncology.
Proteomics approaches discussed in this chapter
Proteomics is the characterization of all of the proteins encoded by the genome of a given organism, tissue, or cell type (Larance and Lamond, 2015). Since proteins are responsible for cellular phenotypes, it is important to characterize individual protein binding partners as well as their functional roles in disease progression. Proteomics uses several approaches to determine protein-protein interactions (PPIs), whether they are direct or indirect. In this section, we discuss proteomic approaches that have been applied towards further characterizing gammaherpesvirus PPIs, either virus-virus or virus-host, as well as some of the drawbacks to each of these approaches.
Mass spectrometry
The most used and accepted method of proteomics is mass spectrometry (MS). MS measures the mass-to-charge ratio of a given sample. This measured ratio provides peptide predictions of what the sample identity could be by comparing actual sample spectra against central databases such as SwissProt, NCBI, NIST, and predicted peptide sequences. Major search engines include Mascot-Matrix Science, Sequest-John Yeats, and X-Tandem-GPM. These search engines compare peptides against decoy libraries to calculate false discovery rates, and software programs such as MAXQUANT/Perseus are used for quantitative proteomics analysis. Accordingly, each search engine uses different methods for scoring and removing false positives.
LC-MS and MALDI-MS
LC-MS and MALDI-MS are the two basic types of MS. In liquid chromatography-tandem mass spectrometry (LC-MS/MS), a protein sample is digested into peptides and then separated by reverse phase chromatography. Following chromatographic separation, an energy source ionizes the molecules. These ionized molecules strike a detector and further separate as they pass through a set of magnets based on their mass-to-charge ratio (Aebersold and Mann, 2016). In contrast to LC-MS/MS, the matrix-assisted laser desorption ionization (MALDI)-MS approach uses single positively-charged ions generated by a laser. Samples are spotted on a substrate with a matrix that assists ionization where a laser vaporizes and ionizes the sample. Ionized molecules are injected into the mass spectrometer, where determinations of molecular charge and mass are then made (Aebersold and Mann, 2003). Readouts from this assay are given in the composition of the sample determined from the spectra. As MALDI-MS relies on single positively-charged ions, the loss of a charge can interfere with the identification, rendering the prediction less accurate. For a novice to the practice of MS, MALDI-MS may represent a better approach to attempt to answer questions regarding protein interactions. Even though MALDI-MS is more time consuming, the samples are salvageable after processing on the machine, unlike when running LC-MS. MALDI-MS is also ideal when working with a complex mixture containing peptides that are low in abundance. However, a noteworthy caveat of both of these techniques is that accuracy relies on the purity of the protein isolated.
Proteomic approaches to detect protein complexes
A commonly used proteomic method to confirm the high-throughput data generated from MS is antibody-based immunoaffinity purification (IP) of protein complexes, also known as pull-down assays, followed by immunoblot analysis that detects the antibody used in the pull-down (Larance and Lamond, 2015) (Fig. 1). These IP/pull-down assays utilize the “bait and prey” system. The “bait proteins” are prepared with tags to fish for and detect the “prey proteins.” Here, we will highlight three antibody affinity assays commonly used to detect PPIs.
Figure 1.
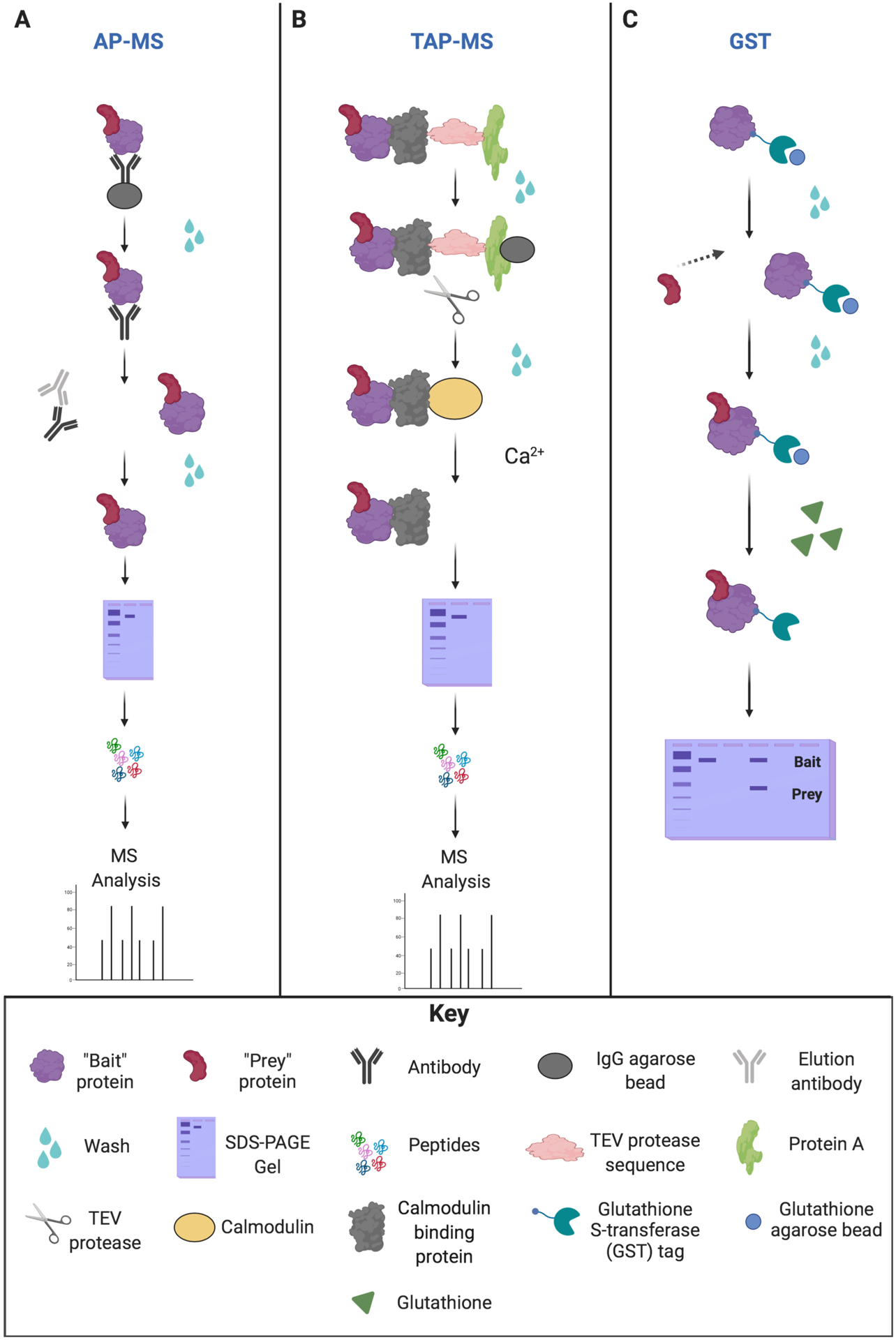
Overview of the different affinity pull-down approaches. (A) Affinity purification mass spectrometry (AP-MS) approach. AP-MS involves immobilization of “bait” protein on beads/resins, and then unbound proteins are washed away. The protein complexes are then eluted from the beads via antibody incubation. Protein complexes are purified and then separated by SDS-PAGE. Separated proteins are digested into small peptides, and the bait-prey interaction is analyzed by LC-MS/MS. (B) Tandem affinity purification (TAP-MS) approach. TAP-MS employs two different affinity purification steps for a protein complex containing the TAP-tagged protein. The first step immobilizes “bait” protein on an IgG bead/resin for protein A. Any unbound proteins are washed away. The protein complex is then immobilized on calmodulin-containing resins for the second affinity purification for calmodulin-binding protein (CBP). The addition of the tobacco etch virus (TEV) protease induces cleavage at the TEV recognition site. The addition of calcium (Ca2+) elutes the bound protein complex from the CBP. Protein complexes are purified and then separated by SDS-PAGE. Separated proteins are subsequently digested into small peptides that can be identified by LC-MS/MS. (C) Glutathione S-transferase (GST) pull-down assay. The recombinant fusion protein containing both GST and the “bait” is fixed to beads, and unbound proteins are washed away. The “prey” protein is added to the GST-tagged “bait” protein on beads to bind, and unbound “prey” proteins are washed away. Next, the bound protein complex is incubated with free glutathione, allowing for protein elution from the beads. Protein complexes are denatured by heating under reducing conditions. SDS-PAGE is then performed to separate the proteins, and the bait-prey interaction is analyzed by western blot.
The first assay is affinity purification mass spectrometry (AP-MS), by which PPIs are detected for a protein of interest that was purified from cell lysates using antibodies to precipitate the specific protein and its interacting partners (Fig. 1a). These precipitated samples are separated by the molecular weights of proteins in the immunoprecipitate using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein band(s) are prepared for MS analysis to determine if an interaction occurs and, if so, at which amino acid location. In-gel and in-solution digestion of proteins are commonly used preparation strategies prior to LC-MS/MS analysis (Dunham et al., 2012; Gingras et al., 2007). However, limitations of AP-MS include that it does not indicate whether the PPI is direct or indirect, nor the abundance or stoichiometry of the protein.
Tandem affinity purification coupled with MS (TAP-MS) can be performed to circumvent the issue of determining whether an interaction is direct or indirect. TAP-MS is similar in concept to epitope tagging except that it uses sequential tags. The first tag is the IgG Sepharose tag that is eventually cleaved off by the tobacco etch virus (TEV) protease (Fig. 1b). The second tag is the calmodulin-binding protein (CBP) that binds to calmodulin. Calcium addition elutes calmodulin off the column, leaving only the CBP, the bait protein, and any other protein(s) that interact with the bait protein in this pull-down assay (Gingras et al., 2007).
Glutathione S-transferase (GST) affinity chromatography is a commonly used research tool over the last several decades that enables the purification of proteins and, ultimately, the determination of the biological function of proteins, by use of recombinant GST-fusion proteins. To produce these recombinant proteins, the functional GST protein of 26 kDa can be tagged at either the N- or C-terminus of the recombinant protein and integrated into an expression vector. Purification of the GST-tagged fusion protein is achieved through the binding of GST to its substrate, glutathione (Dunham et al., 2012; Harper and Speicher, 2011) (Fig. 1c). GST is similar to AP-MS and TAP-MS; however, GST requires an additional step to permit the binding of the protein of interest with the substrate after the extraction of the GST-fusion protein.
Proteomic approaches to detect direct PPIs
Another commonly used proteomic method to determine PPIs is yeast two-hybrid (Y2H). Unlike AP-MS, Y2H can detect direct PPIs. Y2H involves transfecting yeast cells with two different plasmids, each containing a different reporter protein (such as fluorescent proteins or a LacZ reporter cassette) and the suspected binding protein. The first plasmid is the bait plasmid that contains a DNA binding domain (DBD) as well as coding sequences for protein X. The second plasmid is the prey plasmid that contains a transcription activation domain (AD) as well as coding sequences for protein Y. Upon interaction between proteins X and Y, the DBD and AD domains reconstruct, resulting in the transcription of the reporter gene (Brückner et al., 2009).
Quantitative techniques
Isotope-coded affinity tag (ICAT) is a method that labels cysteine-rich proteins for quantification via LC-MS/MS. The cysteine residues are recognized by the ICAT’s cysteine-specific reactive group that is connected to an isotope-coded linker (heavy or light can be used for comparison) that also contains a biotin affinity tag on the end of the linker (Aslam et al., 2017). It is through the biotin affinity tag that these labeled proteins are isolated (Tao and Aebersold, 2003). ICAT is ideal for complex mixtures of proteins as it is highly automated, and peptides are directly sequenced using MS/MS (Gygi et al., 1999). However, ICAT data can be biased by its reliance on cysteine-rich proteins.
Unlike ICAT, stable isotopic labeling with amino acids in cell culture (SILAC) labels the whole cellular proteome, not just cysteine-rich proteins, by using non-radioactive isotopic labeling. Amino acid isotopes based on molecular weight are labeled as either “light” or “heavy,” added to cell culture media, and then inserted into new polypeptide chains within the cultured cells. This allows for MS-based detection of differences in protein abundance as well as regulation of gene expression, cell signaling, and PTMs. A limitation of using SILAC for proteomic experimentation is that it restricts the user to the examination of biological systems under a very controlled condition (Chahrour et al., 2015). Therefore, these labeling methods are not suitable for use with primary tissue, such as patient samples, but would be suitable for endogenous labeling of cell lines.
Protein arrays/chips
Protein microarrays, also known as protein chips, detect protein interactions from a small amount of sample and permit the investigation of multiple interactions on a single chip, saving both time and sample material (Aslam et al., 2017). Protein microarrays are similar in concept to ELISAs in that both utilize antibodies to probe against the desired protein of interest. There are three types of protein microarrays: functional microarrays, analytical microarrays, and reverse-phase microarrays. Functional protein microarrays utilize antibodies to detect isolated proteins. Analytical microarrays use a chip that is pre-coated with a capture antibody prior to the addition of the protein and fluorophore detection, similar to a sandwich ELISA. In reverse-phase microarrays, a whole-cell lysate can be added to the chip (Sutandy et al., 2013).
Three-dimensional structure-based proteomic approaches to detect PPIs
Nuclear magnetic resonance (NMR) is a technique which provides dynamics of structure, such as distances between atoms, the locations of atoms within a molecule, and images of protein structure. NMR gathers information on how atoms move by exploiting the magnetic properties of specific atomic nuclei and their responses to powerful magnetic fields. NMR is often paired with liquid chromatography (LC) or high-performance liquid chromatography (HPLC) (Aslam et al., 2017). One major limitation of NMR is that the limit of detection is 30 kDa, as the size of the magnet limits which proteins can be analyzed using this method.
In contrast, X-ray crystallography does not have the same size limitation as NMR. In X-ray crystallography, proteins of any size can be analyzed, and this technique can measure up to sub-atomic (<1.0Å) levels. X-ray crystallography is considered the “gold standard” technique for the determination of three-dimensional protein structure. Diffraction patterns from the X-rays produce the repeating unit that comprises the crystal, size, and packing symmetry. X-ray crystallography can be useful for a wide range of applications, including studying protein-nucleic acid complexes and immune complexes in viral systems. The three-dimensional protein structure produced by X-ray crystallography can provide detailed information about protein-ligand interactions and enzyme mechanics, which are critical considerations for drug design (Khakurel et al., 2019). However, one major caveat to this technique is that the proteins must be highly purified and crystallized before their exposure to the X-rays.
Data repositories and databases
Several resources and databases are available from both predictive proteomic data and experimental proteomics, and encompass both known and novel protein pathways. Uniprot and Genecard are databases for everyday use. There are also databases for specific PTMs; for example, PhosphositePlus and UbiSite are both widely used for phosphorylation and ubiquitination, respectively. Databases such as the Biological General Repository for Interaction Datasets (BioGRID), the Molecular INTeraction database, and IntAct contain information regarding viral protein interactions in complexes, and are publicly available (Huttlin et al., 2015). Based on the list of provided genes and the available interactions using the IntAct database, users can visualize virus-protein interaction networks to gain a greater understanding of the biological system under investigation. Centralized data repositories allow for the data and results to be accessible to proteomics researchers and biologists alike, thus facilitating the dissemination of proteomics data.
Contributions of proteomics towards the discovery of KSHV and EBV virion composition
General structure of gammaherpesvirus particles
As previously mentioned, herpesviruses including KSHV and EBV are DNA viruses that replicate in the nucleus of host cells. The large double-stranded DNA genome of herpesviruses is located within a central core of the virus particle. The core containing the genomic information is encased by an icosahedral capsid, and the capsid is then surrounded by an amorphous region called the tegument (Fig. 2). The tegument contains virally-encoded proteins which are critical for early-stage infection as well as virus assembly and release from host cells (Guo et al., 2010; Lange and Damania, 2020). Finally, herpesvirus virions are encapsulated by a lipid envelope derived from host cell membranes, which contain embedded glycoproteins (Manservigi and Cassai, 1991).
Figure 2.
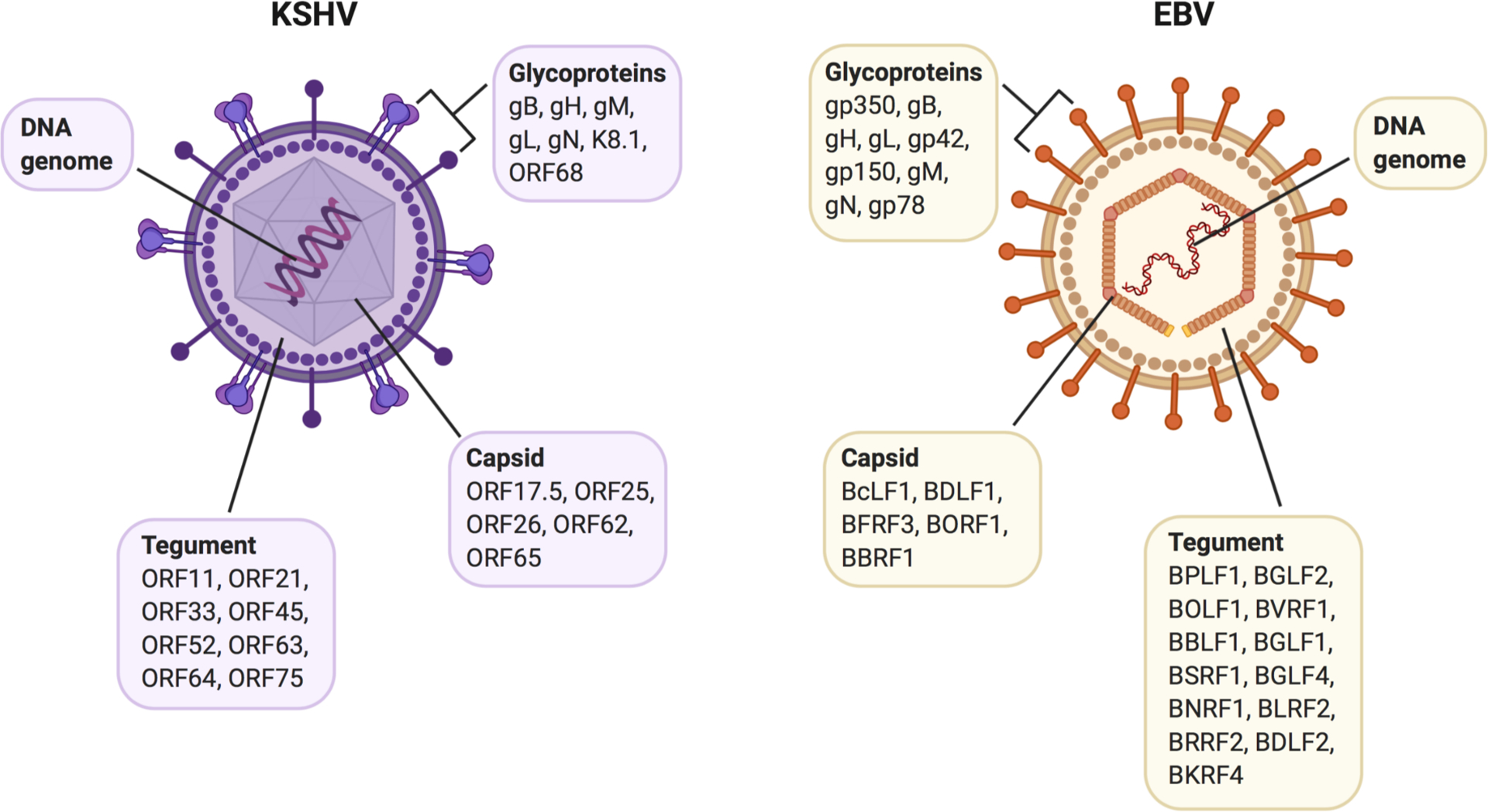
Virion-associated proteins of KSHV and EBV. Viral proteins include capsid, tegument, and envelope glycoproteins. Cellular (host) proteins associated with the virus particles are omitted for clarity.
Although DNA is the genetic material of gammaherpesviruses, the proteins located within the virus particle are just as important for virus propagation and survival. As stated above, many of these virion proteins are encoded by the viral genome and are located either inside (tegument and capsid proteins) or on the outside (membrane-bound glycoproteins) of the virus particle. Vital for virus attachment, entry, replication, assembly, and egress, these proteins have fascinated virologists for decades. This section will describe the contributions proteomic approaches have made in discovering and characterizing the proteins in KSHV and EBV virions.
Proteomics and KSHV virion composition
KSHV ORF45 is a viral protein encoded by an immediate early gene and was shown by Zhu and Yuan in 2003 to be a tegument protein associated with virus particles. KSHV virions were purified, lysed, resolved using a NuPAGE gel, and silver-stained to visualize protein bands. A band corresponding to the molecular weight of ORF45 was observed, and this band was then extracted and analyzed via MS to determine protein identity. The results confirmed that the ORF45 protein is contained within KSHV virus particles (Zhu and Yuan, 2003). This proteomics approach was complemented by several other methods, including immunoblotting, which demonstrated ORF45 presence alongside known KSHV structural proteins, sucrose gradient purification of virus particles, which isolated ORF45 protein within the same fraction as the known virion protein K8.1, and electron microscopy, which showed ORF45 staining of KSHV virions. Further experimentation suggested that ORF45 was a tegument protein; trypsin treatment of virus particles did not result in the degradation of ORF45 until the viral envelope had been compromised, and ORF45 was found in the tegument pellet when surface glycoproteins were liberated via detergent treatment. Thus, ORF45 was identified as a KSHV tegument protein and provides an example of how proteomics can be used to complement more targeted methods for protein identification.
A few years later, Zhu and colleagues undertook a global proteomics approach to identify a larger number of KSHV virion proteins. KSHV virus particles were purified, the proteins were separated out by SDS-PAGE, and the resulting bands were subjected to HPLC-MS analysis. Excitingly, analysis of the protein data revealed 24 proteins associated with KSHV virus particles, as well as a few cellular proteins. Identified virion proteins in this study included capsid proteins (ORF17.5, ORF25, ORF26, ORF62, ORF65), envelope glycoproteins (gB, gH, gM, gL, gN, K8.1, ORF68), tegument proteins (ORF11, ORF21, ORF33, ORF45, ORF52, ORF63, ORF64, ORF75), and ORF6, ORF7, and ORF27 (Zhu et al., 2005). Interestingly, the cellular proteins β-actin and myosin were found in the tegument of KSHV virus particles, although whether packaging of these proteins was selective or non-selective was not investigated in this study. Taken together, the data obtained from these proteomic methodologies greatly advanced our understanding of the proteins encompassing the KSHV virion and set the stage for subsequent functional studies.
Around the same time, Bechtel et al. also undertook a global study of the proteins incorporated into KSHV virions. KSHV virus particles were purified and their associated proteins were separated on an SDS-PAGE gel. Subsequent bands were then analyzed using MALDI-MS. Results yielded 18 proteins of both viral and cellular origin associated with the KSHV virions. Identified proteins included capsid proteins (ORF25, ORF26, ORF62), membrane-embedded glycoproteins (gB, gH), tegument proteins (ORF21, ORF33, ORF63, ORF75), and ORF24 (Bechtel et al., 2005b). As most of these data overlap with the findings of Zhu et al., these two studies together present a comprehensive description of the proteins that are found within KSHV virions (Fig. 2). Interestingly, Bechtel et al. also discovered cellular proteins such as actin and tubulin inside the KSHV virus particles, although again, whether packaging of these proteins was intentional or random due to high abundance during virion assembly was not determined.
Similar proteomic approaches were employed to determine the virion-associated proteins of murine gammaherpesvirus 68 (MHV68), which is a closely-related virus to KSHV and EBV and is often representative of gammaherpesvirus infection and pathogenesis (Virgin et al., 1997). LC-MS/MS analysis on proteins extracted from purified MHV68 virions revealed capsid, tegument, and envelope proteins, many of which were homologues of other gammaherpesvirus virion proteins (Bortz et al., 2003). Cellular β-actin was also found inside MHV68 virions, suggesting a commonality across gammaherpesvirus assembly mechanisms. A following study also used LC-MS/MS to address the same question for MHV68, and these results greatly improved the existing dataset, more than doubling the number of proteins shown to be associated with MHV68 virions from 14 to 31 (Vidick et al., 2013).
Proteomics and EBV virion composition
A systematic study to describe the proteins contained within mature EBV virus particles was undertaken by Johanssen and colleagues using a MS-based approach. EBV virions were purified, the proteins were separated out on a gel, and the resulting bands were analyzed via LC-MS/MS. The data analysis uncovered capsid, tegument, and envelope proteins of EBV, as well as host proteins within the virion. The EBV capsid proteins identified were BcLF1, BDLF1, BFRF3, BORF1, and BBRF1; tegument proteins included BPLF1, BGLF2, BOLF1, BVRF1, BBLF1, BGLF1, BSRF1, BGLF4, BNRF1, BLRF2, BRRF2, BDLF2, and BKRF4; and glycoproteins detected in the EBV envelope were gp350, gB, gH, gL, gp42, gp150, gM, gN, and gp78 (Johannsen et al., 2004). Some of these findings confirmed previous speculation or observations made by others, while many of these proteins were novel additions to the knowledge surrounding EBV virion protein composition. Additionally, cellular proteins such as tubulin and actin were found to be associated with EBV capsids, mirroring the cellular protein composition found in KSHV and MHV68 virions. Overall, these data substantially contributed to our understanding of the protein composition of EBV virus particles (Fig. 2).
Interestingly, more recent data suggest that cytoskeletal proteins are not the only cellular components incorporated into gammaherpesvirus virions. EBV was shown to utilize the macroautophagy pathway in cells to obtain the outer membrane during virion maturation (Nowag et al., 2014). The autophagic protein LC3-II was detected in concentrated EBV virus preparations, and immunoelectron microscopy analysis revealed autophagic membranes present in virus envelopes via LC3 staining. Thus, these data indicate that EBV hijacks the autophagy pathway for lytic viral production and incorporates proteins involved in autophagy, such as LC3-II, into the virus particle during membrane acquisition.
Global proteomic approaches can be used to identify not only proteins present within virus particles, but also proteins expressed during the lytic cycle of gammaherpesviruses. Traylen et al. used proteomics to globally assess the proteins that were expressed during lytic EBV replication in cell culture. BL cells undergoing lytic viral replication were isolated, and the cellular and viral proteins present were then identified and quantified using MS. This screen resulted in 44 lytic EBV proteins, as none of these proteins had previously been linked to latency (Traylen et al., 2015). Many of these identified lytic proteins were capsid, tegument, or envelope proteins as established by others. However, the MS analysis uncovered three proteins not previously demonstrated to be detectable by this method: BBRF2 (involved in viral egress), BFLF1 (involved in viral maturation), and BSLF1 (involved in viral replication). Thus, proteomic approaches can be used to detect i) viral proteins that compose virus particles, ii) cellular proteins that are incorporated into virus particles, and iii) viral proteins associated with either the latent or the lytic phase of herpesvirus lifecycles.
Proteomics and immunity
One of the hallmarks of oncogenic gammaherpesviruses is the ability to evade host immune recognition and downregulate the host immune response. Accordingly, these viruses encode multiple immunomodulatory proteins to achieve immune escape. K5, one of the immune evasion proteins encoded by KSHV, was first discovered through a genomic screen searching for viral proteins that downregulated MHC-I expression on host cells (Coscoy and Ganem, 2000). However, a more thorough understanding of proteins that were affected by K5 expression was lacking. To address this knowledge gap, comparative proteomics was used to assess differences in protein expression between cells expressing K5 versus cells lacking K5 (Bartee et al., 2006). This methodology employed SILAC, which incorporated “light” amino acids into the cells lacking K5 and “heavy” amino acids into the cells expressing K5, thus allowing identification and differentiation of sample proteins in combined MS analyses. Results of this approach identified MHC-I, activated leukocyte cell adhesion molecule (ALCAM), bone marrow stromal antigen 2 (BST-2), and syntaxin-4 as genes that were downregulated in cells expressing K5 compared to the control cells. This observed downregulation of ALCAM, BST-2, and syntaxin-4 in the presence of K5 was then independently verified using immunoblotting, immunofluorescence, and flow cytometry techniques. In sum, this study identified novel targets for the KSHV immunomodulatory protein K5 and demonstrated for the first time the utility of quantitative proteomics for this purpose.
Proteomics can also be used to evaluate immune system recognition and antibody responses to gammaherpesvirus infection. Zheng et al. designed protein microarrays containing 92 KSHV and 82 EBV proteins embedded on a chip. These microarrays were then used to screen plasma isolated from healthy donors, EBV-negative B cell lymphoma patients, and KS/HIV-positive patients. Recognition of the viral proteins by IgG and IgA antibodies was assessed. Results showed that ORF73 and ORF38 were the most-recognized antigens by IgG in the KS/HIV-positive samples only, confirming the utility of ORF73 for KSHV serology testing and identifying ORF38 as a novel serology marker for KSHV infection (Zheng et al., 2011). In contrast to KSHV, which is not a ubiquitous virus, EBV-IgG reactivity was detected in all but one individual. EBNA1, EBNA3B/3C, and the capsid protein BFRF3 were among the EBV antigens most recognized by IgG antibodies. Additionally, IgA reactivity against EBV BCRF1, EBNA1, and the early protein BRRF1 was detected in 100% of the samples. Altogether, this study demonstrates how proteomics can be used to design a specific and sensitive assay to measure humoral immunity against gammaherpesvirus infection and to determine the viral antigens predominantly recognized by the immune system.
Contributions of proteomics towards elucidating virus-host PPIs
There are several proteomic approaches used to explore virus-host interactions. Identifying virus-host interactions at sequential steps of the viral lifecycle may reveal virus-dependent signaling pathways that could be targeted by novel antiviral compounds to prevent or treat virus-induced diseases. In the following sections, we highlight examples of proteomic approaches used to uncover some of the gammaherpesvirus-host interactions.
Protein interactions associated with latency in gammaherpesviruses
Both EBV and KSHV attach to host cells via clathrin-mediated endocytosis. KSHV binds to cell surface heparin sulfate-like moieties (Wang et al., 2001), and enters cells via envelope glycoproteins, cellular integrins, the cystine-glutamate transporter (xCT), and CD98. Similarly, EBV binds to B cells via gp350/220, and then fuses with the host cell plasma membrane via gH, gL, and gB (Hutt-Fletcher, 2007). Using co-IP assays, Wang et al. identified neuropilin 1 (NRP1) as a novel EBV cellular entry factor that interacts with EBV gB. Interestingly, knockdown of NRP1 in NPC cells resulted in a significant decrease in EBV entry, suggesting that NRP1 is involved in EBV infection of epithelial cells in the nasopharynx (Wang et al., 2015). Subsequently, the same group identified Ephrin receptor A2 (EphA2) as an entry receptor for EBV on epithelial cells. By using co-IP, GST pull-down, and immunofluorescence assays, the authors showed that EphA2 interacted with the EBV fusion machinery (gH/gL/gB), similar to NRP1 (Zhang et al., 2018).
Once KSHV makes its way to the nucleus, ORF73, also known as the latency-associated nuclear antigen (LANA), attaches the viral episome to the host cell chromosome, where the virus remains in the latent cycle until reactivated (Aneja and Yuan, 2017). Each host cell harbors multiple copies of this KSHV episome, which is replicated alongside cellular DNA during cell division, allowing for viral persistence (Cesarman et al., 2019). During latent infection, infectious virions are not produced; however, a few genes are transcribed during KSHV latency (Dittmer et al., 1998). These latency-associated transcripts, or latent genes, include ORF73/LANA, ORF72/v-Cyclin, ORF71/v-FLIP, Kaposins, and microRNAs (miRNAs), which all function to help maintain the growth and survival of the infected cell (Mesri et al., 2010; Purushothaman et al., 2015). In EBV, the homolog to LANA is Epstein-Barr nuclear antigen-1 (EBNA-1).
As LANA regulates latency in immune cells, further elucidation of LANA-specific associations via proteomics may uncover interactions critical for the development of cancer. To determine if LANA is arginine-methylated, Campbell et al. screened a library consisting of eight of the nine mammalian protein arginine methyltransferase (PRMT) recombinant viral proteins to determine if they could function as methylation transferases of LANA in vitro by a methylation (3H autoradiograph) assay. Using GST fusion protein mutants, the authors uncovered a single PRMT1 methylation site on LANA at Arg-20 and confirmed this finding by mutating LANA at this site (R20K) and comparing methylation levels to those of wild-type (WT) LANA. Results of this study showed that LANA R20K was considerably less methylated than WT LANA and, additionally, that PRMT1 associated with LANA in the cells, further suggesting that PRMT1 methylates LANA at Arg-20 (Campbell et al., 2012).
Like LANA, the EBV homolog EBNA-1 is present in both the latent and lytic lifecycles of the virus. To understand some critical interacting regulators of EBNA-1, Holowaty and colleagues utilized TAP-tagged EBNA-1. This TAP-tagged EBNA-1, and subsequent confirmation with co-IP, identified USP7/HAUSP, a cellular deubiquitinating protease, as a stable interacting protein with EBNA-1 (Holowaty et al., 2003). Other top interacting proteins identified were casein kinase II subunit α (CSNK2A2), HAUSP/USP7, NAP1, template-activating factor-Iβ/SET, CK2, and PRMT5. This study demonstrates the successful use of an alternative proteomic tagging technique for the identification of cellular proteins bound by EBNA-1. Adding to these findings, EBNA-1 was shown to also interact with the cellular ribosomal proteins RPL4 and nucleolin, facilitating stability of the EBV genome during latency (Shen et al., 2016). Finally, Wang et al. recently demonstrated that EBNA-1 interacts with and inhibits the cellular protein STUB1, which helps to suppress EBV lytic gene expression and maintain viral genomic stability in latently-infected host cells (Wang et al., 2020).
Upon EBV infection of B cells, EBNA-2 and EBNA-LP help to establish the latency program as well as maintain the growth and survival of the host cells (Kempkes et al., 1995; Szymula et al., 2018). Using two-dimensional PAGE and MALDI-MS techniques, Schlee et al. compiled a list of cellular proteins that were modulated by EBNA-2 in B cells. Results included the upregulation of protein synthesis and degradation factors as well as cell growth factors, while proteins involved in apoptosis and cytoskeleton arrangement were downregulated (Schlee et al., 2004). Additionally, co-IP and MS-based assays revealed that truncated EBNA-LP interacts with multiple subunits of the protein phosphatase PP2A in an inhibitory manner (Chelouah et al., 2018; Garibal et al., 2007). Thus, by inhibiting PP2A, EBV exerts both anti-apoptotic and pro-oncogenic effects in infected cells.
Latent membrane protein 1 (LMP-1) is an EBV transmembrane protein important for both successful EBV infection as well as primary B cell activation (Kieser and Sterz, 2015; Pratt et al., 2012). LMP-1 is a functional homolog of cellular CD40 (Graham et al., 2010), and is required for EBV-mediated B cell transformation (Soni et al., 2007). To identify novel PPIs between LMP-1 and cellular proteins, Song et al. utilized a Y2H strategy and identified interferon regulatory factor 7 (IRF7) as a top interacting protein with LMP-1 (Song et al., 2008). Co-IP confirmed IRF7 to interact with LMP-1, and the authors hypothesized that this interaction might function to evade host immune detection by silencing transcriptional activation of virus-inducible cellular genes. Using pull-down assays, Bentz et al. demonstrated that LMP-1 interacts with Ubc9, a cellular factor involved in protein modification. Furthermore, LMP-1 was shown to increase the sumoylation (a PTM) of host proteins through Ubc9, which led to an increase in cellular migration and oncogenic potential (Bentz et al., 2011). Additionally, using a global proteomics approach, DeKroon et al. examined the cellular proteins that were modulated by EBV LMP-1 and LMP-2A expression. The authors grouped the resulting data into pathways, and the cellular areas most affected by LMP-1 and LMP-2A included ubiquitination, vesicle formation/trafficking (including extracellular vesicles, which will be discussed in a later section), and cellular migration (DeKroon et al., 2018). Finally, Tworkoski and Raab-Traub performed a kinase screen to identify cellular kinases that were activated by LMP-1. Results of the screen demonstrated that LMP-1 phosphorylates and activates insulin-like growth factor 1 receptor (IGF1R), allowing for increased cell proliferation and loss of contact inhibition, both of which are characteristic of transformed cells (Tworkoski and Raab-Traub, 2015). Collectively, these examples demonstrate how proteomic approaches can be applied to uncover details in gammaherpesvirus lifecycles that might otherwise go unnoticed using an alternative approach.
Comparable to LMP-1 of EBV, KSHV encodes the transmembrane protein K1, which is necessary for transforming B cells and is similar to a B cell receptor (BCR) in its immunoreceptor tyrosine-based activation motif (ITAM) (Wen and Damania, 2010b). Proteomics has enabled insights into critical interactions which allow for K1 to regulate KSHV persistence in host cells. Lee et al. used KSHV-infected cell lysates labeled with [35S] methionine-cysteine and the GST pull-down method to identify K1 binding proteins. Results of these assays revealed Lyn, Syk, Grb2, Vav, and PLCy2 as binding partners of tyrosine-phosphorylated K1 (Lee et al., 2005). Using TAP-MALDI-TOF, we found that K1 resists cellular apoptosis by interacting with heat shock protein 90 (Hsp90), and we confirmed this result through reverse co-IPs, confocal microscopy, and pharmacological inhibition of Hsp90 in both 293 cells and B cells (Wen and Damania, 2010a). Additionally, in a separate study, we performed TAP-MS to identify K1 binding partners. We identified 5’ adenosine monophosphate-activated protein kinase (AMPK) as a K1 interacting protein and found that this K1-AMPK interaction enhanced cell survival during periods of stress (Anders et al., 2016). Using pull-downs of GST fusion proteins containing K1 followed by LC-MS, Choi et al. determined that K1 interacted with cellular PYCR and showed that this K1-PYCR interaction increased cell growth in vitro and increased tumor size in mice (Choi et al., 2020). Altogether, these studies helped to reveal intricate ways K1 interacts with cellular proteins to enable resistance to apoptosis, promote cellular growth and survival, increase angiogenesis, and facilitate tumor development.
KSHV expresses four proteins similar to the cellular interferon regulatory factors (IRFs) called viral IRFs (vIRFs), which help to antagonize the host cell antiviral response (Giffin and Damania, 2014). vIRF3, also called LANA-2, is encoded by ORF K10.5 and is the only vIRF expressed during latency (Rivas et al., 2001). Using pull-down assays, Lubyova et al. found that vIRF3 interacts with cellular IRF3 and IRF7. Further experimentation suggested that vIRF3 enhances IRF3 and IRF7 innate immune signaling, but the effects that these interactions might have on viral latency, as well as tumor formation, were not addressed (Lubyova et al., 2004). The same group subsequently performed Y2H and GST pull-down assays in PEL cells and found that vIRF3 associated with the c-Myc suppressor protein MM-1alpha as well as the CDK4 promoter region (Lubyova et al., 2007).
In vitro pull-down of GST fusions of vIRF1 revealed both IRF1 and p300 as vIRF1 interacting partners, and identified a location on vIRF1 involved in overcoming the cellular interferon response and cell death (Burysek et al., 1999a). Nakamura and colleagues found vIRF1 to interact with p53 by using co-IP deletion mutants, and this interaction hindered growth arrest (Nakamura et al., 2001). Similarly, Seo et al. identified this vIRF1–p53 interaction by using GST pull-down assays, in vitro co-IP, and domain mapping. A luciferase reporter assay with vIRF1 revealed that the vIRF1–p53 interaction regulated p53 and p21 gene transcription (Seo et al., 2001). Finally, we used MS to identify vIRF1 interacting proteins and found that HERC5, an ISG15 ligase, interacted with vIRF1, and that this interaction resulted in an increase in KSHV reactivation and the number of infectious viral particles (Jacobs et al., 2015).
vIRF2 is another KSHV homolog of cellular IRFs. Burysek et al. made a GST fusion protein containing vIRF2 and performed an in vitro binding assay. Results of this assay determined that vIRF2 interacts with cellular IRF1, IRF2, ICSBP, the N-terminal domain of RelA (p65), and the C-terminal domain of p300 (Burysek et al., 1999b). A couple of years later, Burysek and Pitha used the GST pull-down method along with 35S-labeling to identify vIRF2 protein interactions and found that protein kinase R (PKR) interacted with vIRF2. Studies showed that vIRF2 inhibited PKR autophosphorylation and phosphorylation of downstream PKR substrates as measured by a kinase assay (Burýšek and Pitha, 2001).
K15 is a latent KSHV protein, and several groups have utilized proteomics to understand the molecular interactions of how K15 modulates host cells to aid in virus survival and persistence. Sharp et al. found that K15 binds the cellular protein HAX-1 by using Y2H, GST pull-down assays, and co-IPs of the C-terminal domain of K15 (Sharp et al., 2002). As the receptor function of K15 is constitutively active once phosphorylated, Abere et al. examined potential phosphorylated K15 (pK15) binding partners. IP-MS of pK15 identified phosphatidylinositol 4-phosphate 3-kinase (PI3K)-C2α as a pK15 interacting protein. The authors then confirmed this pK15 interaction with PI3K-C2α using reverse co-IP in 293 cells, GST pull-down assays, and confocal microscopy. Functionally, siRNA depletion of PI3K-C2α was found to reduce KSHV lytic replication (Abere et al., 2018).
Protein interactions associated with reactivation from latency in gammaherpesviruses
Similar to other herpesviruses, KSHV can reactivate from latency under conditions that are stressful to the host or the surrounding cellular environment, such as inflammation, hypoxia, infection, or immunosuppression (Purushothaman et al., 2015). In culture, KSHV-infected cells can be chemically induced to reactive from latency by using drugs such as TPA and sodium butyrate (Aneja and Yuan, 2017; Miller et al., 1997). KSHV encodes a replication and transcription activator known as RTA or ORF50. RTA is a switch that is both necessary and sufficient to initiate the lytic cycle of KSHV, and K8/Kb-ZIP is an immediate-early gene that suppresses RTA function (Izumiya et al., 2003). By applying proteomic methods such as affinity chromatography and Y2H, several research groups demonstrated that K8/Kb-ZIP interacts with HDAC1/2 (Martínez and Tang, 2012; Tomita et al., 2004). This interaction with HDAC1/2 likely keeps the chromosome condensed and inaccessible for RTA binding to occur.
Conserved among all herpesviruses is an mRNA transcript accumulation (MTA) protein, which is a viral regulatory protein that, like RTA, allows for lytic replication to occur (Majerciak and Zheng, 2015). In KSHV, this protein is ORF57, and the EBV homolog is also called MTA or EB2. By interacting with an assortment of cellular proteins, ORF57 promotes the expression of viral genes. However, it was unclear at which stage of the viral lifecycle ORF57 was functionally active: pre- or post-transcription. Boyne et al. addressed this gap in knowledge by determining if ORF57 associates with either the pre-transcriptional human transcription/export (hTREX) mRNA processing complex or the post-transcriptional exon junction complex (EJC). Immunoaffinity assays showed that only hTREX components (Aly, CBP80, TAP, and UAP56) interacted with MTA and not the EJC components, suggesting that the hTREX, not the EJC, is recruited to intronless viral transcripts (Boyne et al., 2008).
In addition to MTA, EBV also encodes RTA and ZTA, which are two viral transcription factors essential for lytic replication (Chevallier-Greco et al., 1986). Through co-IP and LC-MS assays, EBV ZTA was found to interact with several cellular heat shock proteins, RNA binding proteins, and single-stranded DNA binding proteins (Wiedmer et al., 2008). One of these proteins, the mitochondrial DNA binding protein mtSSB, was shown to enhance EBV replication. Interestingly, when a mutation known to inhibit EBV replication was introduced into ZTA, mtSSB was no longer able to associate with ZTA, thus demonstrating that this mutation reduced viral replication due to lack of mtSSB binding. Along the same lines, Zhou et al. employed IP- and MS-based approaches to characterize the ZTA protein interaction network in BL cells. Both viral and cellular proteins were shown to interact with ZTA, two of which were of the nuclear factor of activated T cells family (NFATC1 and NFATC2). By binding to these cellular NFAT proteins, EBV modulates NFAT signaling, potentially as a mechanism to control ZTA expression levels in infected cells (Zhou et al., 2020).
To gain a perspective of how EBV modulates host cells on a global scale, Calderwood and colleagues utilized the Y2H system to create an “ORFeome” interaction data set consisting of both EBV-EBV and EBV-host interactions (Calderwood et al., 2007). HOMER3 was one of the 13 human proteins that interacted with several EBV proteins. HOMER3 interacted with nine EBV baits, including BFRF1 (nuclear membrane homolog of the alphaherpesvirus UL34), BILF1 (gp64; a G-protein coupled receptor), BLLF1 (gp350/220), BKRF2 (gL), BBRF3 (gM), BDLF3 (gH; formerly gp85), the replication and transcription factor BLRF1, the capsid binding protein BGLF2, and the tegument protein BFLF2.
Using AP-MS, we identified host proteins that interacted with the KSHV immunomodulatory protein vIL6. Hypoxia upregulated protein 1 (HYOU1) was shown to be a novel vIL6-interacting protein, and HYOU1 expression enhanced cell survival, migration, and signaling (Giffin et al., 2014). Similarly, but on a global scale, Davis et al. used AP-MS to identify over 500 interactions between KSHV and host cell proteins. Of note, ORF24 was shown to interact with cellular RNA polymerase II (RNA pol II). This interaction was not limited to KSHV, as the ORF24 homologues in EBV, HCMV, and MHV68 all interacted with RNA pol II as well, suggesting a conserved function among these herpesviruses. Finally, the KSHV ORF24–host RNA pol II interaction was necessary for the transcription of late genes to occur, indicating that the completion of the KSHV life cycle depends on this viral interaction with the host cell (Davis et al., 2015).
Similar global proteomic techniques have been employed for studying EBV modulation of host cell proteins during infection. Using a global and comparative proteomics approach, Ersing et al. quantified thousands of cellular and EBV proteins that were modulated in BL cells during the lytic phase of the EBV lifecycle. Noteworthy findings included viral targeting of B cell receptors for degradation by an EBV early protein, and activation of innate immune signaling pathways in the B cells (Ersing et al., 2017). The authors then extended these findings for inclusion in comparative analyses with other related herpesviruses. Results of this comparison showed that eight cellular RNA binding proteins were downregulated by both EBV and HCMV, while neuroligins 1 and 4X were downregulated by three human herpesviruses (EBV, HCMV, and KSHV), suggesting that these cell adhesion proteins might play a global role in herpesvirus biology.
Contributions of proteomics towards elucidating virus-virus PPIs
To date, much has been learned about the protein interactions that occur between herpesviruses and host cells. In contrast, information regarding the interactions that occur between herpesvirus proteins, and in particular KSHV, is lacking, in part due to the challenges of robustly growing KSHV in culture (Uetz et al., 2004). To help elucidate the interactions among herpesvirus proteins, Uetz and colleagues generated protein interaction maps for KSHV and VZV through Y2H bait and prey arrays. For KSHV, the authors examined over 12,000 viral protein interactions utilizing both fragment and full-length proteins. Results of this screen identified 123 KSHV protein-protein interactions that were non-redundant, and ~30% of these interactions were novel (Uetz et al., 2006). Of note, ORF50/RTA, the latent-to-lytic switch, was found to interact with ORF75 by several methodologies.
In an effort to better understand the protein-protein interactions that involve KSHV tegument proteins, Rozen et al. investigated global protein-protein interactions within KSHV virus particles using Y2H and co-IP assays. Results of these studies showed that ORF45 interacted with multiple tegument proteins, including ORF11, ORF21, ORF27, ORF33, ORF63, ORF64, and ORF75 (Rozen et al., 2008). Next, the authors used co-IP assays to examine the interaction network of ORF64 and ORF52 with other KSHV tegument components. ORF64 interacted with the most tegument proteins (ORFs 11, 21, 27, 33, 45, 52, 63, 64N, 64M, and 75) compared to ORF53, which interacted with only ORF45 and ORF75. Thus, these data indicate potentially essential roles for ORF45 and ORF64 in the KSHV virion, as their interaction networks were the most extensive. Additionally, Rozen et al. examined potential ORF64 and ORF52 interactions with the glycoproteins gB, gH, gM, gL, and gN by using co-IP and immunoblotting for the FLAG-tagged glycoproteins. ORF52 interacted with glycoproteins gM and gN, while ORF64 interacted with glycoproteins gB, gH, and gM, further strengthening the observations that ORF64 is promiscuous when it comes to interactions and could, therefore, be of significant importance for viral infectivity.
Finally, Fossum and colleagues systematically tested all three herpesvirus subfamilies by Y2H to determine if interactions are conserved between viral protein homologs. They added the interactomes of HSV-1, murine CMV, and EBV to their existing dataset of KSHV and VZV. The results indicated that viral homolog functionality could be more conserved than sequence (Fossum et al., 2009). Overall, studies involving proteomics have given researchers a clearer picture of the viral signaling network, allowing for additional insights into viral infectivity and persistence.
Determination of the protein content of exosomes secreted from gammaherpesvirus-infected cells
Exosomes are approximately 40–150 nm in diameter and deliver cargo such as proteins and miRNAs to nearby cells. Exosomes are classified as a form of extracellular vesicles (EVs) and originate from internal membrane budding of late endosomes within the cell. These exosomes can then either fuse with the lysosome for content degradation, or with the multivesicular body (MVB) for content release at the plasma membrane (Fig. 3). Additionally, exosomes carry trafficking proteins, such as tetraspanins, that can be utilized for exosome identification and experimental isolation. In the context of viral infection, it has been shown that EBV-infected cells can produce exosomes containing viral miRNAs (Pegtel et al., 2010). These viral miRNAs can then be delivered to neighboring cells, where modulation of cell signaling and oncogenic pathways can occur (Fig. 3).
Figure 3.
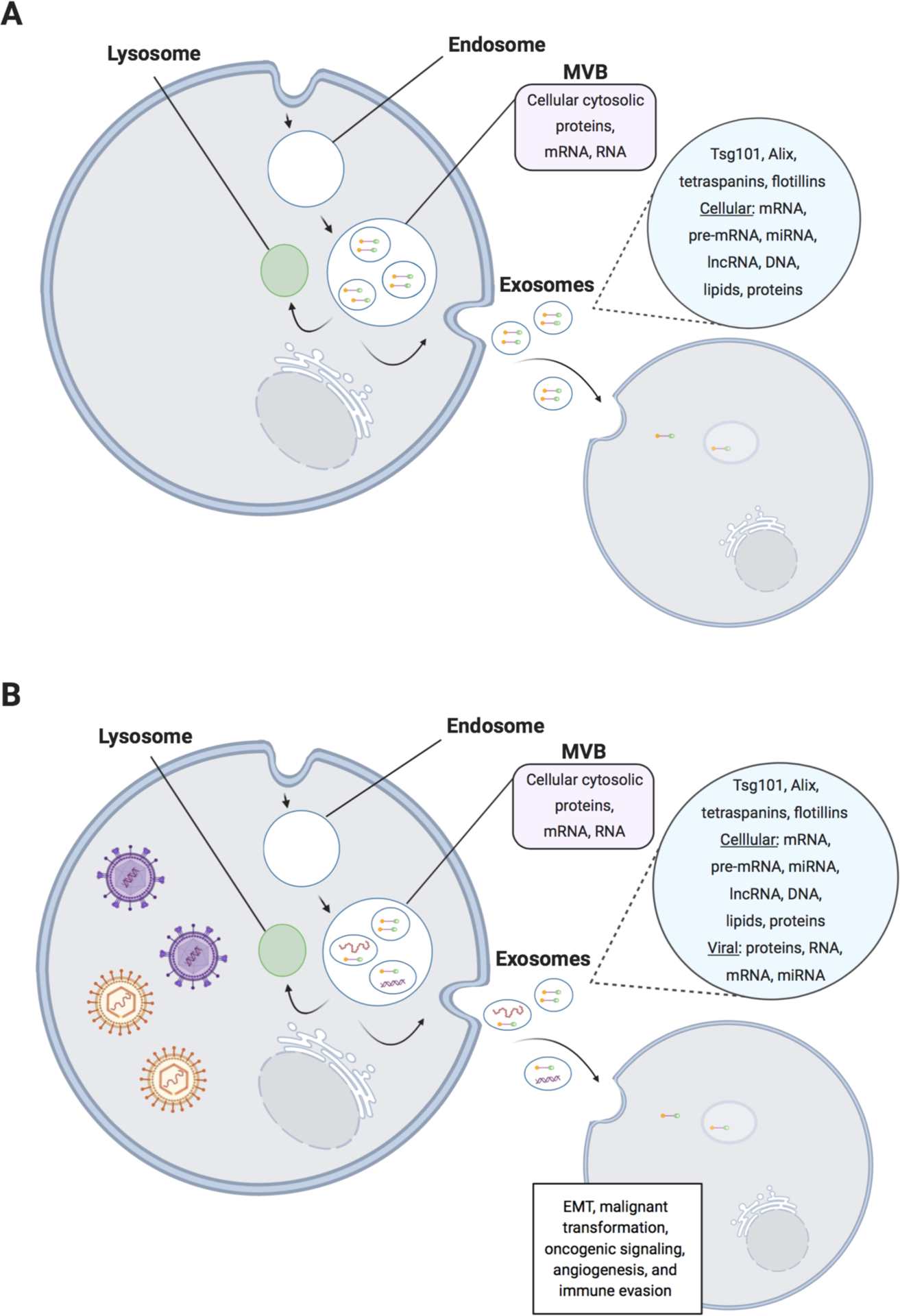
Differences in exosome content secretion from uninfected and gammaherpesvirus-infected cells. (A) Exosome biogenesis and secretion from uninfected cells. Multivesicular bodies (MVBs) are formed by fusion with early endosomes. Next, the MVBs fuse either with the lysosome for degradation, or with the plasma membrane, which releases the exosomal content into the extracellular space. The exosomal content contains proteins encoded by tumor susceptibility gene 101 (Tsg101), apoptosis-linked gene-2 interacting protein X (Alix), tetraspanins, and flotillins, which can all be used for exosome identification. Additionally, multiple RNA species such as cellular mRNA, microRNA (miRNA), and long non-coding RNA (lncRNA) can be found within exosomes. Finally, exosomes can also contain DNA and lipids. (B) Exosome biogenesis and secretion from gammaherpesvirus-infected cells. In addition to the above content found in exosomes secreted from uninfected cells, viral proteins and viral nucleic acids can also be found within exosomes from virus-infected cells. The content of these exosomes often influences the surrounding cellular environment to result in epithelial-to-mesenchymal transition (EMT), malignant transformation, oncogenic signaling, angiogenesis, and immune evasion.
Latent membrane protein 1 (LMP-1) of EBV has been detected in EVs from infected cells (Houali et al., 2007; Meckes et al., 2010). Similarly, LMP-1 was identified in EVs harvested from mice bearing NPC (Flanagan et al., 2003). Additionally, it was found that LMP-1 was packaged into EVs via a mechanism involving the cellular protein TRAF2, and that these LMP-1/TRAF2 complexes were involved in oncogenic NF-κB signaling (Verweij et al., 2015). As mentioned above, numerous studies have detected cellular and viral miRNAs in EVs, and it is thought these miRNAs are intentional to sidestep immune detection and to alter cellular functions in favor of cellular survival and, subsequently, the development of cancer (Chugh et al., 2013; Rainy et al., 2016; Schwab et al., 2015). Overall, gammaherpesviruses utilize multiple strategies to commandeer exosomes to aid in their continued propagation (Sadeghipour and Mathias, 2017).
To interrogate if exosome content was altered by infection with EBV or KSHV, Meckes et al. purified exosomes taken from i) an uninfected B cell line, ii) cells infected with EBV, iii) cells infected with KSHV, and iv) cells dually infected with EBV and KSHV, and compared exosomal components by MS data processing (Meckes et al., 2013). Additionally, the authors confirmed the LC-MS/MS data with immunoblotting and unique clustering pattern analysis. The resulting data suggested that EBV and KSHV infection influences exosome content, as over 360 proteins were identified in the virally-infected exosomes that were absent in the exosomes generated from uninfected cells.
Implications of gammaherpesvirus modulation of exosome content
Gammaherpesviruses escape the antiviral activities of the host immune response in part by interfering with cellular signaling pathways designed to alert the cell to invading pathogens (Liu et al., 2019). A recent study suggested that gammaherpesviruses might also escape immune detection by using exosomes. McNamara et al. analyzed the EV delivery system in the context of gammaherpesvirus infection by isolating EVs from both a KSHV-positive PEL cell line (BCBL1) and KSHV-positive PEL patient fluid and comparing them to the EVs present in normal plasma. KSHV-infected EVs circumvented the innate immune response and did not induce activation of antiviral cellular responses, including INF-β, TLR, RIG-I, or the cGAS/STING pathway (McNamara et al., 2019). Additionally, KSHV-infected EVs were shown to activate MEK/ERK signaling, which resulted in endothelial cell activation, proliferation, and migration. Thus, there are several functional advantages for KSHV to utilize EVs, including immune evasion and the modulation of the surrounding cellular environment in favor of viral spread.
Additionally, Meckes et al. analyzed KHSV- and EBV-modified exosome components with Ingenuity Pathway Analysis (IPA) software in order to determine if the exosome alterations exhibited oncogenic properties. While the IPA data in this study exposed differences in exosome cargo based on viral influence, the data also revealed similarities in exosomal content-mediated alteration of signaling cascades involved in cellular survival and the synthesis of proteins (Meckes et al., 2013). These data from Meckes and colleagues further support previous studies, which altogether demonstrate that modification of exosome cargo by KSHV or EBV can dysregulate cellular functions and lead to tumor development (Fig. 3).
Clinical implementation of proteomics for the diagnosis and treatment of KSHV- and EBV-associated cancers
Cancers associated with oncogenic human herpesviruses
KSHV
As mentioned previously, KSHV infection is associated with several different cancers, including Kaposi’s sarcoma (KS), multicentric Castleman’s disease (MCD), and primary effusion lymphoma (PEL) (see Table 1). KS is a highly vascularized endothelial cell tumor, which results in characteristic reddish-brown lesions on the skin. KS predominantly manifests on the extremities (arms/hands and legs/feet) or mucosal surfaces of infected individuals, but can sometimes be found internally on organs such as the spleen and liver (Siegel et al., 1969; Valls et al., 1991). KS is more prevalent in males and is the most common KSHV-associated cancer (Goncalves et al., 2017b). Interestingly, it has been shown that KSHV prevalence is not ubiquitous across populations, in contrast to many human herpesviruses (Gao et al., 1996). Instead, KSHV exhibits the highest prevalence in sub-Saharan Africa (Goncalves et al., 2017a), and KS is the most frequently occurring cancer in some of these countries (Dollard et al., 2010; Parkin, 2006).
Table 1.
Cancers associated with oncogenic human herpesviruses and their treatments
| Cancer | Origin/description | Treatment |
|---|---|---|
| KSHV | ||
| Kaposi’s sarcoma | Highly vascularized endothelial cell tumor, usually found on the extremities, often associated with HIV infection | HAART; systemic therapy in HAART-resistant cases (pegylated liposomal doxorubicin, pomalidomide) |
| Multicentric Castleman’s disease | Plasmablastic tumor in lymph nodes, excessive inflammatory profile | Rituximab (anti-CD20 antibody); rituximab plus doxorubicin; valganciclovir plus zidovudine |
| Primary effusion lymphoma | B cell lymphoma, body cavities fill with malignant B cell effusions | Chemotherapeutic cocktails (CHOP, dose-adjusted EPOCH); HAART |
| EBV | ||
| Post-transplant lymphoma | B cell lymphoma, occurs in immunocompromised individuals after organ transplant | Tapering or removal of immunosuppressive drugs |
| Burkitt’s lymphoma | Germinal center B cell tumor, highly proliferative | Chemotherapy |
| Hodgkin’s lymphoma | Only some forms associated with EBV infection, characterized by presence of Hodgkin/Reed-Sternberg cells (B cells) | Chemotherapy and radiotherapy |
| Nasopharyngeal carcinoma | Epithelial cell tumor of the nasopharynx, specific geographic distribution | Surgery to remove the tumor, followed by radiotherapy |
While there are several different forms of MCD, the plasmablastic variant of MCD is the form associated with KSHV infection (KSHV-MCD) (Soulier et al., 1995). This lymphoproliferative cancer originates in lymph node plasmablasts and is often characterized by excessive inflammation, including robust production of the pro-inflammatory cytokine IL-6 (Nishimoto et al., 2005; Oksenhendler et al., 2000; Yoshizaki et al., 1989). Interestingly, many KSHV-MCD cases co-exist with KS in the same tissue (Naresh et al., 2008). In contrast to KS, where KSHV mainly exists in its latent (non-replicative) form in infected cells, KSHV-MCD exhibits a more pronounced lytic phenotype (Asahi-Ozaki et al., 2006). These data suggest that more virus replication is occurring in KSHV-MCD compared to KS, which might contribute to the aggressiveness of this cancer.
PEL is a highly aggressive but rare form of non-Hodgkin lymphoma (NHL) characterized by KSHV-infected B cells undergoing rapid proliferation. PEL is a body cavity malignancy where anatomical sites surrounding the heart (pericardial), lungs (pleural), and abdomen (peritoneal) become filled with these B cell effusions (Nador et al., 1996). PEL normally presents as an effusion tumor with no solid mass, but there have been reports of solid PEL tumors in the literature (Carbone et al., 2005; Chadburn et al., 2004). Each PEL cell contains 50–100 copies of the KSHV genome (Renne et al., 1996), which are passed along in the form of episomes to dividing daughter cells. Most patients with PEL are co-infected with EBV (Boulanger et al., 2005; Nador et al., 1996), but the implications of this co-infection for PEL disease progression and outcome are largely unknown.
It is worthwhile to mention that, although KSHV can infect anyone, symptomatic KSHV infection and the development of the above-mentioned cancers usually occur in the context of immunosuppression (Thakker and Verma, 2016). Examples of immunosuppression include HIV co-infection or transplant recipients receiving immunosuppressive drugs to help acceptance of the graft. One treatment avenue for these KSHV-associated cancers is the reinstatement of immunocompetence, and this will be discussed further in the next section.
EBV
Like KSHV, EBV is an oncogenic virus, and is associated with four cancers that will be discussed herein: post-transplant lymphoma (PTL), Burkitt’s lymphoma (BL), Hodgkin’s lymphoma (HL), and nasopharyngeal carcinoma (NPC) (Raab-Traub, 2007) (see Table 1). As its name suggests, PTL occurs in patients who have undergone bone marrow or solid organ transplant and have suppressed immune function, usually due to irradiation therapy and/or immunosuppressive drugs. PTL is a B cell lymphoma, arising in immunosuppressed individuals largely due to the lack of a virus-clearing cytotoxic T cell response (Green and Michaels, 2013; Meij et al., 2003; Smets et al., 2002). The likelihood of PTL developing after transplant depends on many factors, including the age of the organ recipient, whether the recipient was EBV-positive and/or CMV-positive at the time of transplant, and the type of organ received (Allen and Preiksaitis, 2009; Cockfield, 2001). Finally, children exhibit a higher occurrence of PTL compared to adults; children are more likely to be EBV-negative at the time of transplant, and primary EBV infection has been shown to be an important driver in the manifestation of PTL (Ho et al., 1988; Ho et al., 1985).
Burkitt’s lymphoma (BL) was originally characterized by Denis Burkitt, who observed unusual facial tumors in the jaws of children in Uganda (Burkitt, 1958). BL is a highly proliferative B cell tumor of germinal center origin. The epidemiology of this disease suggests an association with malaria, as BL endemic regions closely overlap those of malaria (Burkitt, 1971). Unlike the causative agent of malaria (P. falciparum), EBV cannot be spread by mosquitoes. However, recent data suggest that infection with P. falciparum malaria is a risk factor for BL, due to dysregulation of activation-induced cytidine deaminase (AID) function in B cells and subsequent oncogenesis (Robbiani et al., 2015; Thorley-Lawson et al., 2016; Torgbor et al., 2014). Although first described in children, BL can develop in both children and adults. Accordingly, BL accounts for 1–5% of all non-Hodgkin lymphomas in adults and is observed more frequently in males (Boerma et al., 2004; Linch, 2012). BL can be either EBV-positive or EBV-negative, and EBV-positive BL will be further discussed in this section.
HL is characterized by the presence of distinct cells called Hodgkin/Reed-Sternberg (HRS) cells, which are B cells originating from various stages of development (Kuppers et al., 1994). There are many different forms of HL, only some of which are associated with EBV infection. Studies have shown that EBV infection of HRS cells and their precursors can contribute to the development of HL by providing anti-apoptotic signals, thus allowing the B cells to proliferate and escape programmed cell death (Bechtel et al., 2005a; Chaganti et al., 2005; Mancao et al., 2005).
NPC is an epithelial tumor of the nasopharynx that, similar to BL, displays a distinct geographic distribution (Raab-Traub, 2007). NPC is most prevalent in southern China, southeast Asia, parts of Africa, and Alaska (Mahdavifar et al., 2016). Aside from EBV infection, other risk factors for NPC include alcohol use, tobacco use, and HPV infection (Lubin et al., 2009; Mork et al., 2001). Establishing cell lines and in vivo models for NPC has proven difficult over the years, hindering the ability to understand this cancer and to develop novel treatments (Cheung et al., 1999; Tsao et al., 2017). Recently, however, several new tumor xenografts and cell lines were added to the NPC research pipeline, providing hope for further progress on elucidating NPC biology and methods to target this malignancy (Lin et al., 2018; Yip et al., 2019).
Current treatment methodologies
Antiretroviral therapy and immune suppression reduction
Many, but not all, of the cancers associated with KSHV and EBV infection arise in HIV-positive or otherwise immunocompromised individuals. Therefore, one of the main treatment options for some of these cancers, such as KS and PEL, revolves around improving immune function. For HIV-positive patients, this is achieved through the use of highly active antiretroviral therapy (HAART). By lowering HIV viral load and improving T cell count and function, the HIV-infected individual’s immune system is more prepared to fight the cancer. In most KS cases, treatment with HAART is sufficient, and surgical resection of the lesions as well as radiotherapy are rarely used (Yarchoan and Uldrick, 2018). HAART treatment alone has been shown to improve KS disease burden and lengthen survival time in patients (Bower et al., 2009; Mosam et al., 2012). Additionally, it has been suggested that HAART treatment can lead to better control of KSHV replication and a more robust antibody response towards KSHV, which could in turn improve KS disease progression (Wilkinson et al., 2002). Finally, HAART has been shown to reduce the risk of death by over 80% in HIV-positive men diagnosed with either KS or NHL (Tam et al., 2002). Unfortunately, not all HIV-positive KS patients will respond to HAART. In instances of HAART-resistant KS, systemic therapies such as pegylated liposomal doxorubicin (Northfelt et al., 1998) and pomalidomide (Polizzotto et al., 2016) can be used (Yarchoan and Uldrick, 2018).
The mainstay treatment for PTL is the lessening or removal of drugs that suppress the immune system post-transplant. This approach is more effective in pediatric patients compared to adults, and sometimes fails due to tumor heterogeneity/unresponsiveness, advanced disease stage, advanced patient age, or complications with rejection (Allen and Preiksaitis, 2009; Reshef et al., 2011). Interestingly, this immunosuppression reduction approach is most successful when the tumor is still under EBV control (Green and Michaels, 2013).
Chemotherapy and radiotherapy
Chemotherapy has been a frontline treatment for cancer for decades. As more chemotherapeutic agents are developed and brought into the clinic, the combinations in which these drugs can be used against different malignancies constantly changes. Drugs such as prednisone or methotrexate can be added to chemotherapeutic regimens to lessen inflammation and reduce the side effects from the other drugs. Chemotherapy cocktails aim to be potent enough to act against the tumor at a concentration that is not toxic to the vital tissues of the patient. The combination therapies CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) and dose-adjusted EPOCH (cyclophosphamide, doxorubicin, vincristine, prednisone, and etoposide) are used as treatments for PEL, usually in combination with HAART (Goncalves et al., 2017b). Similar chemotherapeutic regimens are used in the treatment of BL; generally, EBV-positive and EBV-negative BL are treated alike, except in the context of immunotherapy or clinical trials (Dunleavy, 2018; Kanakry and Ambinder, 2013). For HIV-positive lymphoma patients, providers must carefully consider any potential drug interactions between HAART and the combination chemotherapy being used. The composition of the drug cocktails, as well as the timing of drug administration, can be modified as needed to help prevent adverse events from occurring (Yarchoan and Uldrick, 2018).
As a solid tumor, treatment for NPC includes surgery to remove the tumor mass if possible, followed by radiotherapy. Radiotherapy has been recognized as the frontline treatment for NPC, but has proven difficult to manage due to the proximity of the tumor to anatomical sites such as the brainstem, brain, salivary glands, and retinas (Holliday and Frank, 2016). Although efficacious, radiotherapy alone can be hindered by cytotoxic side effects, metastasis, or radio-resistance (Au et al., 2018; Yeo et al., 2018). For this reason, combining radiotherapy with chemotherapy or immunotherapy may improve patient outcomes.
Immunotherapy
Both KSHV and EBV are B cell tropic; thus, many of the cancers associated with these viruses are B cell lymphomas. As such, one drug that has shown marked results in the clinic is the monoclonal antibody rituximab. Rituximab targets the CD20 antigen on B cells, but this antibody can also have an effect on some CD20-negative B cell lymphomas, possibly by altering signaling pathways in the tumor microenvironment (Carbone et al., 2015). PEL cells usually do not express normal B cell markers, including CD20, and rituximab has been largely unusable against this lymphoma in the context of KSHV infection (Nador et al., 1996). In contrast, rituximab has shown extremely promising results against KSHV-MCD, with remission achieved in over 90% of patients in a phase II trial (Bower et al., 2007). The safety profile of this drug combined with its clinical success resulted in rituximab being recommended as the primary treatment for KSHV-MCD, and the five-year progression-free survival after rituximab treatment has been estimated to be 70–90% (Goncalves et al., 2017a; Pria et al., 2017). Additionally, rituximab can be combined with doxorubicin for treatment of KSHV-MCD (Yarchoan and Uldrick, 2018). Finally, rituximab recently showed promise for treatment of EBV-PTL. Two clinical studies demonstrated a complete response in 70% or more of PTL patients, and the drug was well-tolerated (Trappe et al., 2017; Zhu et al., 2019).
Immunotherapy is still in its infancy in the oncology world, and many exciting treatment avenues remain in the context of gammaherpesvirus-associated cancers. One of these avenues is anti-PD1 therapy, which has shown incredible promise in the treatment of other cancers such as melanoma and metastatic non-small cell lung cancer (Hamid et al., 2013; Sui et al., 2018). Current efforts are underway to evaluate the safety and efficacy of anti-PD1 therapy for KS as well as EBV-associated NPC and HL (Broussard and Damania, 2019; Cao et al., 2019; Carbone et al., 2018; Makowska et al., 2019).
Antiviral drugs
As both KSHV and EBV are herpesviruses, drugs targeting the herpesvirus lifecycle can be used in an attempt to control the underlying cause of the cancer. These drugs include ganciclovir and its pro-drug valganciclovir, as well as zidovudine. KSHV lytic genes ORF36 and ORF21 phosphorylate ganciclovir and zidovudine, respectively, to toxic tri-phosphorylated forms, thus disabling herpesvirus replication (Cannon et al., 1999; Gustafson et al., 2000). Ganciclovir and valganciclovir have been used for treatment of KSHV-MCD, with varying rates of success (Casper et al., 2004; Kantarci et al., 2016; Speicher et al., 2014; Uldrick et al., 2011). However, using valganciclovir in combination with zidovudine resulted in 86% of KSHV-MCD patients demonstrating a positive clinical response (Uldrick et al., 2011). Ganciclovir is also included in treatment for PTL, although no clinical studies have definitively demonstrated efficacy of ganciclovir in this context (Green and Michaels, 2013). It should be noted that these drugs only target the lytic phase of the virus lifecycle, and do not affect the latent virus reservoir. As such, treatment with these antiviral drugs can control but not eradicate the virus, and recurrence can occur.
Taken together, although different treatment options exist for KSHV- and EBV-associated cancers, for many patients these are not the solution. These cancers can develop resistance to therapy or become too aggressive for these treatments to have much effect. Despite our above arsenal, the prognosis for some of these cancers remains poor. Hence, new diagnostic and treatment options are increasingly sought after, and proteomic analyses offer a powerful tool for cancer therapeutic discovery.
Clinical uses of proteomics in cancer diagnosis and treatment
Proteomics in cancer diagnosis and detection
Proteomic approaches encompass a rapidly expanding and exciting field for cancer diagnosis and personalized treatment. Many cancers are detected later, often after secondary spread to distal tissues. Implementation of biomarker assays using blood or saliva samples that can detect cancers early while still in the formative stages would greatly improve clinical outcomes for cancer patients and significantly ease healthcare burdens. To date, no biomarker assays have been clinically established for KSHV- or EBV-associated cancers, but potential candidates are in development. Using LC-MS and MALDI-TOF approaches, Gloghini et al. found an array of proteins that were specifically secreted by PEL cell lines and were also identified in PEL patient samples and KSHV-associated solid tumors (Gloghini et al., 2014). Examples of these specifically secreted proteins included catalase, which promotes B cell growth, and cellular nucleic acid-binding protein (CNBP), which is involved in transcription regulation. Thus, PEL “secretomes” are enriched with PEL-specific proteins that could potentially be used for diagnostic purposes. Additionally, multiple NHL studies have been undertaken to examine the serum and/or plasma of patients prior to lymphoma diagnosis in the hopes of uncovering novel protein biomarkers for this class of cancer (Vendrame and Martínez-Maza, 2011). Increased levels of cytokines such as IL-6 and TNFα, cytokine receptors such as soluble CD27 and CD30, and immunoglobulins such as circulating free light chains have all been implicated as potential NHL biomarkers (Breen et al., 2011; Landgren et al., 2010; Rabkin et al., 2011). Finally, analyses of the NPC proteome, secretome, and patient serum proteome using various MS-based approaches have demonstrated that protein profiling can be used to distinguish NPC from non-cancer samples, and identified CCL5 as a novel NPC therapeutic candidate and biomarker (Cao et al., 2010; Feng et al., 2011; Ho et al., 2006; Lin et al., 2013).
Although MS-based proteomic approaches have the capacity to yield many novel cancer biomarkers in the lab, overall, their clinical translation has been disappointing. These clinical failures can be due to study design, discrepancies in sample collection/quality, the assays used, or control issues (You et al., 2018). While reliable biomarkers are still in development for KSHV- and EBV-associated cancers, proteomics has nevertheless been successfully used to identify diagnostic biomarkers for other cancers with clinical success. Prostate cancer (PCa) often progresses undetected until the later stages of malignancy, when treatment is often futile (Catalona, 2018). Therefore, it was of great interest to develop a biomarker assay that could detect PCa prior to the onset of symptoms. Currently, the measurement of prostate-specific antigen (PSA) levels in the blood is used for PCa screening. PSA is a protein secreted by epithelial cells in the prostate, and higher PSA levels in the serum are indicative of PCa (Catalona et al., 1991). Additionally, this screening assay can detect PCa prior to palpable disease (Stormont et al., 1993). Using radioimmunoassays, Killian et al. showed in 1986 that PSA was the most reliable biomarker for PCa progression, and the PSA assay has progressively developed since this discovery (Killian et al., 1986). Although somewhat controversial due to potential over-diagnosis and unnecessary treatment, PSA remains one of the most widely-used cancer biomarker assays and has led to a decline in PCa deaths (Catalona, 2018).
The glycoprotein alpha-fetoprotein (AFP) is the most commonly used and studied biomarker for hepatocellular carcinoma (HCC) (Carr et al., 2018). While not all HCC cases can be detected via AFP screening, it has been estimated that 25% of non-viral HCC can be identified earlier using AFP plus imaging compared to imaging techniques alone (Worland et al., 2018). However, AFP levels in the serum can be elevated for reasons other than cancer, such as benign liver disease, and for this reason the specificity of elevated AFP for HCC diagnosis remains a concern. To help improve this specificity issue, a post-translationally modified isoform of AFP called AFP-L3 can be used, which better differentiates HCC from benign liver disorders (Aoyagi et al., 2002). Recently, a high-throughput protein microarray that requires minimal sample volume was developed to detect both AFP and AFP-L3 (Wu et al., 2018). Although not widely implemented, examining AFP-L3 serum levels can potentially improve HCC diagnosis.
Like AFP, carcinoembryonic antigen (CEA) is a protein normally produced by fetal gastrointestinal cells but can be present at low levels in adults (Kim et al., 2018). Elevated CEA levels can be indicative of cancer, including colorectal cancer (CRC). CEA is not specific enough to be useful as a diagnostic marker for CRC, but it has shown promise alongside imaging as a marker for CRC recurrence post-resection (Nicholson et al., 2015). Additionally, it has been suggested that CEA levels measured at the time of surgery for CRC hepatic metastases are indicative not only of prognosis but also the choice of supplemental chemotherapeutic strategies (Chiang et al., 2019). Finally, conjugation of tagged antibodies to CEA proteins can allow for better imaging to help guide surgical procedures post-metastasis, and radiolabeled anti-CEA antibodies can be used for delivery of tumor-specific therapy (Meredith et al., 1996; Meyer et al., 2009; Tiernan et al., 2013).
Cancer antigen 125 (CA-125) is commonly used as a biomarker for ovarian cancer. Like prostate cancer, ovarian cancer often progresses undetected until later stages when achieving cure is unlikely, highlighting the need for earlier detection methods (Scholler and Urban, 2007). CA-125, a membrane-bound glycoprotein, was first established as an ovarian cancer biomarker through radioimmunoassays performed in the 1980’s (Canney et al., 1984). As with most biomarkers, there are issues with specificity, as conditions other than ovarian cancer can lead to elevated CA-125 levels in the blood. Despite this, CA-125 screening is commonly used as a biomarker i) alongside imaging for diagnosis of suspected ovarian cancer cases, ii) for monitoring response to treatment post-diagnosis, and iii) for monitoring disease recurrence post-surgery. There have been recent attempts to improve the sensitivity and specificity of CA-125 screening for ovarian cancer. One such attempt used quantitative proteomics methods to identify candidate autoantibodies highly expressed in ovarian cancer but not healthy controls (Karabudak et al., 2013). These autoantibody markers were shown to be specific across a range of ovarian cancer disease stages as well as CA-125 levels. Thus, combining CA-125 screening with alternative screening methods such as autoantibody markers could improve testing efficiency.
Although not an exhaustive list of cancer biomarkers currently used in the clinic, these data demonstrate how analysis of proteins that differ between healthy vs. cancer patients can be developed into assays that enable earlier detection of malignancies and improved survival outcomes for patients.
Proteomics in cancer treatment and drug discovery
There have been numerous studies to date that have used proteomic approaches to identify novel targets for treatment of KSHV- and EBV-associated cancers. We recently utilized multiplexed inhibitor bead-mass spectrometry (MIB/MS) to profile the kinome of PEL cells against healthy B cell controls. This method runs cell lysates through a column containing kinase inhibitors attached to beads. The beads bind functionally expressed kinases, which can then be eluted off, identified, and quantified using MS. This kinome screen resulted in identification of several kinases that were significantly upregulated in PEL cells but not in healthy B cells. We went on to demonstrate that inhibition of one these kinases, Tyro3, resulted in decreased PEL cell survival in vitro and decreased tumor burden in vivo (Wong et al., 2019). Thus, inhibition of Tyro3 represents a novel therapeutic strategy for PEL.
Morphoproteomics is a novel approach to large-scale protein analysis; it combines histology, molecular biology, and chemistry to pinpoint specific pathways which drive a specific cancer’s phenotype, thus enabling identification of druggable targets in a personalized manner (Tan, 2008). Using this morphoproteomic approach in a published case study, Salihi et al. identified cyclo-oxygenase 2 (COX2) and Enhancer of Zeste homologue 2 (EZH2) as drivers of the pathogenic phenotype exhibited by a patient with KS (Salihi et al., 2016). They were then able to recommend personalized therapy based on these findings, which included treatment with metformin and/or celecoxib, both of which can inhibit COX2/EZH2 expression and KSHV replication.
While proteomic analyses can identify novel therapeutic targets in cancer, these techniques can also be used to help discern novel treatment options in the context of an existing druggable target. For example, heat shock protein 90 (Hsp90) is known to play a role in tumorigenesis, including KSHV- and EBV-associated lymphomas (Calderwood et al., 2006). Given this knowledge, Nayar et al. inhibited Hsp90 in PEL cells and examined the resulting Hsp90 “interactome,” looking for proteins that interacted with Hsp90, which could then be targeted. Interacting proteins were identified using unbiased affinity capture, which utilized beads conjugated to the Hsp90 inhibitor, followed by MS analysis of the captured proteins. The screen revealed multiple anti-apoptotic proteins interacting with Hsp90, leading to the hypothesis that inhibition of apoptosis was driving the PEL phenotype. Cell viability assays showed that PEL cell death was induced by inhibiting members of the BCL family of anti-apoptotic proteins, and that combinatorial therapy against both Hsp90 and BCL proteins resulted in a synergistic effect on PEL cell death (Nayar et al., 2013). Thus, probing protein-protein interactions can uncover targetable oncogenic pathways or novel drug treatment combinations for virus-associated malignancies.
To date, there are a multitude of reports using proteomics to uncover novel treatment avenues for EBV-associated cancers, especially for NPC. Here, we will highlight some of these reports for NPC, as well as EBV-BL. Using two-dimensional gel electrophoresis (2DGE) and subsequent MS, Tang et al. identified amyloid-β precursor protein (APP) as a secreted protein from an NPC cell line. They then went on to show that antibody blocking of secretory APP resulted in decreased tumor growth and migration, suggesting that APP helps to drive NPC progression and thus warrants further investigation into APP as a novel therapeutic target (Tang et al., 2010). Other groups have performed comparative analyses using a nasopharyngeal (NP) epithelial cell line alongside an NPC cell line to identify differentially expressed proteins between the healthy and cancerous cells. 2DGE/MS analysis showed that the NPC cells upregulated glycolysis compared to the control NP cell line, as expression of the glycolytic enzymes PKM2 and TPI1 was increased in NPC cells as well as glucose uptake and the secretion of lactic acid (Huang et al., 2015). Thus, targeting the glycolytic pathway in NPC is a possible therapeutic target, since NPC cells rely more heavily on glycolysis compared to normal NP epithelial cells. A similar comparative approach was employed by Li and colleagues, who analyzed differential protein expression in the stroma of NPC vs. normal NP mucosa (NNM) isolated from patients. Laser capture microdissection was used to separate stroma cells from NPC or NNM patient tissue, and then the proteins were separated using 2DGE and identified using MALDI-TOF-MS. Periostin, a protein involved in cell communication and signaling, was significantly upregulated in the stroma of NPC compared to NNM (Li et al., 2012). Overexpression of periostin in an NPC cell line led to increased invasion, and analysis of NPC patient samples showed that increased expression of periostin was correlated with poorer survival outcomes. Taken together, these data suggest that increased periostin levels are involved in NPC metastasis and that expression of periostin can help predict NPC survival. These findings were later confirmed by an independent cohort study, highlighting the potential of periostin as a novel target for NPC (Wei et al., 2018).
Comparative proteomic analyses have also been used in EBV-BL to identify virally regulated proteins that could potentially be targeted. 2DGE/MS analysis in an EBV-transformed BL cell line showed downregulation of members of the COMM domain (COMMD) family of proteins, which negatively regulate NF-κB signaling pathway components (Gkiafi and Panayotou, 2011). The authors hypothesized that downregulation of COMMD proteins and the subsequent upregulation of NF-κB signaling was one mechanism by which EBV infection results in oncogenesis, although this hypothesis was not formally tested in this study. While intriguing, more work is needed to elucidate the role of COMMD proteins in EBV tumor formation and the potential of these proteins as therapeutic targets.
Phosphoproteomic analyses can reveal signaling pathways important in cancer and help identify key regulatory proteins in these pathways. Rituximab and obinutuzumab are both anti-CD20 antibodies; however, they recognize distinct epitopes. Obinutuzumab has been shown to reduce tumor burden in rituximab-resistant EBV-positive BL in vivo and result in enhanced survival compared to rituximab treatment in rituximab-susceptible mice (Awasthi et al., 2015). To help unravel the mechanism behind these different treatment outcomes, Awasthi et al. used a global phosphoproteomics approach. BL cell lines, including EBV-positive BL, were treated with either rituximab or obinutuzumab and subsequent phosphorylated peptides were enriched using metal oxide affinity chromatography and analyzed via MS. Results revealed many differentially phosphorylated proteins, including proteins involved in B cell receptor signaling and NK cell-mediated cytotoxicity. One of these B cell signaling proteins, PLGC-2, was upregulated after treatment with obinutuzumab vs. rituximab, and knockdown of PLGC-2 resulted in increased BL cell growth and cell viability after obinutuzumab treatment compared to rituximab treatment (Awasthi et al., 2017). These data suggest that the differences in treatment outcomes between these two anti-CD20 antibodies could be explained in part by differential phosphorylation signatures on B cell receptor signaling molecules and invite future investigations of these signaling molecules as therapeutic targets. For NPC, a comparative phosphoproteomic analysis showed that EphA2 phosphorylation was significantly increased in metastatic NPC compared to non-metastatic NPC, and a phosphokinase antibody array revealed that AKT/Stat3 signaling was responsible for phospho-EphA2-mediated NPC invasiveness (Li et al., 2019). Therefore, inhibition of phospho-EphA2 and potentially other molecules in the AKT/Stat3 pathway are possible targets for treatment of NPC.
Finally, proteomics can be used to uncover novel mechanisms of resistance in cancer cells and to identify targets that could reinstate susceptibility to radiotherapy or chemotherapy. An example of this approach is a 2018 study by Li et al., who identified mitogen-activated protein kinase 15 (MAPK15) as a novel mediator of radiotherapy resistance in NPC. In an effort to screen for possible modulators of radioresistance, peptides derived from radiosensitive and radioresistant NPC cell lysates were fractionated via reverse-phase HPLC and analyzed via MS. Differential protein expression between the susceptible and resistant cell lines was then compiled, and MAPK15 was the most abundant protein kinase in the radioresistant cells (Li et al., 2018). Subsequent knockdown of MAPK15 in the resistant cell line rendered the cells more susceptible to therapy, and overexpression of MAPK15 in the sensitive cell line enhanced resistance to therapy. Lastly, the mechanism driving this radioresistance was examined, and the data suggested that MAPK15 controls radioresistance by limiting the accumulation of reactive oxygen species and improving repair of damaged DNA. In sum, these elegant experiments indicate that inhibition of MAPK15 sensitizes treatment-resistant NPC to radiotherapy, providing an exciting therapeutic addition, which hopefully translates to clinical application.
Although beyond the scope of this chapter, MS-based proteomic approaches that screen not only the overall proteome but also the phosphoproteome, glycoproteome, kinome, interactome, and secretome of cancer are widely employed to identify novel therapeutic targets and drugs for many other cancer types aside from those associated with human gammaherpesvirus infection. We refer the reader to Dias et al. for a comprehensive review on this topic (Dias et al., 2016). Overall, this is an exciting time for cancer research. As we move forward towards personalized cancer medicine, studying the proteome of cancer on an individual basis will help lead to more targeted therapies with fewer off-target effects and improved patient outcomes.
Conclusions and future perspectives
Proteomic techniques, many of which are based in mass spectrometry or microarray technology, have substantially advanced our understanding of gammaherpesvirus biology and their associated malignancies. Proteomics has significantly aided in the identification of the proteins contained within KSHV and EBV virus particles and the exosomes secreted from infected cells, as well as the interaction network that exists between gammaherpesvirus proteins and host cells. Proteomic approaches have also led to a better understanding of how gammaherpesvirus proteins contribute to immune evasion, thus allowing successful viral persistence within infected cells, and how targeting both viral and host proteins can be beneficial from a therapeutic perspective.
As such, proteomic approaches represent a powerful methodology for pushing the fields of cancer diagnostics and treatment forward. Immunotherapy and personalized medicine are the future of oncology. Immune responses are influenced by different factors, including genetics and the surrounding environment. Hence, every individual’s immune system is different, and every individual’s cancer is different. A blanket approach to cancer treatment, therefore, has many shortcomings. Precision medicine treats each individual’s cancer as a stand-alone malignancy and enables targeted therapy with a lower chance of resistance. Ideally, genomics and proteomics will be combined to inform personalized cancer treatment. Using genomics, one can screen individual cancers and determine i) the genetic landscape of the cancer and how it might have formed, ii) if genetic mutations are present that would indicate or contraindicate certain treatment choices, and iii) the extent of genetic variation and the impact this variation could have on protein structure and function. Proteomic screening would complement this genetic information, allowing for i) identification of upregulated or dysregulated pathways in the cancer, which can inform treatment choices, ii) identification of proteins and/or post-translational modifications that are driving the cancer, leading to novel therapeutic targets, and iii) identification of resistant signatures, which could indicate selection of an alternative therapy. Comparative proteomic analyses can also be used to identify potential biomarkers for different cancers. The earlier cancer is detected, the better the prognosis, and since many cancers do not elicit symptoms until later stages of malignancy, having a sensitive and specific screening assay for each cancer would save many lives. By comparing cancerous tissue to normal tissue, proteomics can reveal differentially expressed proteins that might be present in blood or saliva prior to symptoms, which could indicate the beginnings of cancer formation. Although most biomarkers identified in the lab fail to translate to clinical application, the increasing improvement of the selectivity and sensitivity of proteomic approaches will enable identification of more robust candidates with better clinical success.
In conclusion, proteomics has greatly advanced our understanding of oncogenic human gammaherpesviruses and their associated cancers, leading to new discoveries in their biology, virus-host interactions, and treatment methodologies. We look forward to the future and the innovations that these powerful techniques will bring to the field of oncogenic viruses.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants CA019014, DE028211, CA096500, CA239583, and CA163217 to BD. DLC is supported by the Caroline H. and Thomas S. Royster Fellowship from the Graduate School at the University of North Carolina at Chapel Hill and a NCI diversity supplement. MCW is supported by NIH grant 5T32CA009156-44. BD is a Leukemia and Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Infectious Disease. We apologize to all the authors whose work we could not discuss due to length restrictions. All figures were made using BioRender.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Abere B, Samarina N, Gramolelli S, Ruckert J, Gerold G, Pich A and Schulz TF (2018). Kaposi’s Sarcoma-Associated Herpesvirus Nonstructural Membrane Protein pK15 Recruits the Class II Phosphatidylinositol 3-Kinase PI3K-C2alpha To Activate Productive Viral Replication. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R and Mann M (2003). Mass spectrometry-based proteomics. Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- Aebersold R and Mann M (2016). Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–55. [DOI] [PubMed] [Google Scholar]
- Allen U and Preiksaitis J (2009). Epstein-barr virus and posttransplant lymphoproliferative disorder in solid organ transplant recipients. Am J Transplant 9 Suppl 4, S87–96. [DOI] [PubMed] [Google Scholar]
- Anders PM, Zhang Z, Bhende PM, Giffin L and Damania B (2016). The KSHV K1 Protein Modulates AMPK Function to Enhance Cell Survival. PLoS Pathog 12, e1005985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja KK and Yuan Y (2017). Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front Microbiol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi Y, Mita Y, Suda T, Kawai K, Kuroiwa T, Igarashi M, Kobayashi M, Waguri N and Asakura H (2002). The fucosylation index of serum alpha-fetoprotein as useful prognostic factor in patients with hepatocellular carcinoma in special reference to chronological changes. Hepatol Res 23, 287. [DOI] [PubMed] [Google Scholar]
- Asahi-Ozaki Y, Sato Y, Kanno T, Sata T and Katano H (2006). Quantitative analysis of Kaposi sarcoma-associated herpesvirus (KSHV) in KSHV-associated diseases. J Infect Dis 193, 773–82. [DOI] [PubMed] [Google Scholar]
- Aslam B, Basit M, Nisar MA, Khurshid M and Rasool MH (2017). Proteomics: Technologies and Their Applications. J Chromatogr Sci 55, 182–196. [DOI] [PubMed] [Google Scholar]
- Au KH, Ngan RKC, Ng AWY, Poon DMC, Ng WT, Yuen KT, Lee VHF, Tung SY, Chan ATC, Sze HCK et al. (2018). Treatment outcomes of nasopharyngeal carcinoma in modern era after intensity modulated radiotherapy (IMRT) in Hong Kong: A report of 3328 patients (HKNPCSG 1301 study). Oral Oncol 77, 16–21. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Ayello J, Van de Ven C, Elmacken M, Sabulski A, Barth MJ, Czuczman MS, Islam H, Klein C and Cairo MS (2015). Obinutuzumab (GA101) compared to rituximab significantly enhances cell death and antibody-dependent cytotoxicity and improves overall survival against CD20(+) rituximab-sensitive/-resistant Burkitt lymphoma (BL) and precursor B-acute lymphoblastic leukaemia (pre-B-ALL): potential targeted therapy in patients with poor risk CD20(+) BL and pre-B-ALL. Br J Haematol 171, 763–75. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Rolland DCM, Ayello J, van de Ven C, Basrur V, Conlon K, Fermin D, Barth MJ, Klein C, Elenitoba-Johnson KSJ et al. (2017). A comparative global phosphoproteomics analysis of obinutuzumab (GA101) versus rituximab (RTX) against RTX sensitive and resistant Burkitt lymphoma (BL) demonstrates differential phosphorylation of signaling pathway proteins after treatment. Oncotarget 8, 113895–113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E, McCormack A and Fruh K (2006). Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog 2, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel D, Kurth J, Unkel C and Kuppers R (2005a). Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood 106, 4345–50. [DOI] [PubMed] [Google Scholar]
- Bechtel JT, Winant RC and Ganem D (2005b). Host and viral proteins in the virion of Kaposi’s sarcoma-associated herpesvirus. J Virol 79, 4952–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz GL, Whitehurst CB and Pagano JS (2011). Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J Virol 85, 10144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerma EG, van Imhoff GW, Appel IM, Veeger NJ, Kluin PM and Kluin-Nelemans JC (2004). Gender and age-related differences in Burkitt lymphoma--epidemiological and clinical data from The Netherlands. Eur J Cancer 40, 2781–7. [DOI] [PubMed] [Google Scholar]
- Bortz E, Whitelegge JP, Jia Q, Zhou ZH, Stewart JP, Wu TT and Sun R (2003). Identification of proteins associated with murine gammaherpesvirus 68 virions. J Virol 77, 13425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger E, Gerard L, Gabarre J, Molina JM, Rapp C, Abino JF, Cadranel J, Chevret S and Oksenhendler E (2005). Prognostic factors and outcome of human herpesvirus 8-associated primary effusion lymphoma in patients with AIDS. J Clin Oncol 23, 4372–80. [DOI] [PubMed] [Google Scholar]
- Bower M, Powles T, Williams S, Davis TN, Atkins M, Montoto S, Orkin C, Webb A, Fisher M, Nelson M et al. (2007). Brief communication: rituximab in HIV-associated multicentric Castleman disease. Ann Intern Med 147, 836–9. [DOI] [PubMed] [Google Scholar]
- Bower M, Weir J, Francis N, Newsom-Davis T, Powles S, Crook T, Boffito M, Gazzard B and Nelson M (2009). The effect of HAART in 254 consecutive patients with AIDS-related Kaposi’s sarcoma. Aids 23, 1701–6. [DOI] [PubMed] [Google Scholar]
- Boyne JR, Colgan KJ and Whitehouse A (2008). Recruitment of the Complete hTREX Complex Is Required for Kaposi’s Sarcoma–Associated Herpesvirus Intronless mRNA Nuclear Export and Virus Replication. PLoS Pathog 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen EC, Hussain SK, Magpantay L, Jacobson LP, Detels R, Rabkin CS, Kaslow RA, Variakojis D, Bream JH, Rinaldo CR et al. (2011). B-cell stimulatory cytokines and markers of immune activation are elevated several years prior to the diagnosis of systemic AIDS-associated non-Hodgkin B-cell lymphoma. Cancer Epidem Biomar 20, 1303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard G and Damania B (2019). KSHV: Immune Modulation and Immunotherapy. Front Immunol 10, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner A, Polge C, Lentze N, Auerbach D and Schlattner U (2009). Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci 10, 2763–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkitt D (1958). A sarcoma involving the jaws in African children. Br J Surg 46, 218–23. [DOI] [PubMed] [Google Scholar]
- Burkitt DP (1971). Epidemiology of Burkitt’s lymphoma. Proc R Soc Med 64, 909–10. [PMC free article] [PubMed] [Google Scholar]
- Burýšek L and Pitha PM (2001). Latently Expressed Human Herpesvirus 8-Encoded Interferon Regulatory Factor 2 Inhibits Double-Stranded RNA-Activated Protein Kinase. J Virol 75, 2345–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burysek L, Yeow WS, Lubyova B, Kellum M, Schafer SL, Huang YQ and Pitha PM (1999a). Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol 73, 7334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burysek L, Yeow WS and Pitha PM (1999b). Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2). J Hum Virol 2, 19–32. [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE et al. (2007). Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A 104, 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB and Ciocca DR (2006). Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci 31, 164–72. [DOI] [PubMed] [Google Scholar]
- Campbell M, Chang P-C, Huerta S, Izumiya C, Davis R, Tepper CG, Kim KY, Shevchenko B, Wang D-H, Jung JU et al. (2012). Protein Arginine Methyltransferase 1-directed Methylation of Kaposi Sarcoma-associated Herpesvirus Latency-associated Nuclear Antigen. J Biol Chem 287, 5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canney PA, Moore M, Wilkinson PM and James RD (1984). Ovarian cancer antigen CA125: a prospective clinical assessment of its role as a tumour marker. Br J Cancer 50, 765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JS, Hamzeh F, Moore S, Nicholas J and Ambinder RF (1999). Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J Virol 73, 4786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Wei Q, Tang X, Jia Y, Sun X, Li W, Hu Q and Chen X (2019). PD-1 and PD-L1 in locoregionally advanced nasopharyngeal carcinoma: Substudy of a randomized phase III trial. Head Neck 41, 1427–1433. [DOI] [PubMed] [Google Scholar]
- Cao SM, Yu JK, Chen QY, Li NW, Xiang YQ, Qian CN, Hu X, Zhang CQ, Xie D and Guo X (2010). Detection of nasopharyngeal carcinoma using surface-enhanced laser desorption and ionization mass spectrometry profiles of the serum proteome. Chin J Cancer 29, 721–8. [DOI] [PubMed] [Google Scholar]
- Carbone A, De Paoli P, Gloghini A and Vaccher E (2015). KSHV-associated multicentric Castleman disease: A tangle of different entities requiring multitarget treatment strategies. Int J Cancer 137, 251–61. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A and Carlo-Stella C (2018). Are EBV-related and EBV-unrelated Hodgkin lymphomas different with regard to susceptibility to checkpoint blockade? Blood 132, 17–22. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Vaccher E, Cerri M, Gaidano G, Dalla-Favera R and Tirelli U (2005). Kaposi’s sarcoma-associated herpesvirus/human herpesvirus type 8-positive solid lymphomas: a tissue-based variant of primary effusion lymphoma. J Mol Diagn 7, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr BI, Akkiz H, Uskudar O, Yalcin K, Guerra V, Kuran S, Karaogullarindan U, Altintas E, Ozakyol A, Tokmak S et al. (2018). HCC with low- and normal-serum alpha-fetoprotein levels. Clin Pract (Lond) 15, 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper C, Nichols WG, Huang ML, Corey L and Wald A (2004). Remission of HHV-8 and HIV-associated multicentric Castleman disease with ganciclovir treatment. Blood 103, 1632–4. [DOI] [PubMed] [Google Scholar]
- Catalona WJ (2018). Prostate Cancer Screening. Med Clin North Am 102, 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalona WJ, Smith DS, Ratliff TL, Dodds KM, Coplen DE, Yuan JJ, Petros JA and Andriole GL (1991). Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med 324, 1156–61. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Damania B, Krown SE, Martin J, Bower M and Whitby D (2019). Kaposi sarcoma. Nat Rev Dis Primers 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadburn A, Hyjek E, Mathew S, Cesarman E, Said J and Knowles DM (2004). KSHV-positive solid lymphomas represent an extra-cavitary variant of primary effusion lymphoma. Am J Surg Pathol 28, 1401–16. [DOI] [PubMed] [Google Scholar]
- Chaganti S, Bell AI, Pastor NB, Milner AE, Drayson M, Gordon J and Rickinson AB (2005). Epstein-Barr virus infection in vitro can rescue germinal center B cells with inactivated immunoglobulin genes. Blood 106, 4249–52. [DOI] [PubMed] [Google Scholar]
- Chahrour O, Cobice D and Malone J (2015). Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J Pharm Biomed Anal 113, 2–20. [DOI] [PubMed] [Google Scholar]
- Chelouah S, Cochet E, Couvé S, Balkaran S, Robert A, May E, Ogryzko V and Wiels J (2018). New Interactors of the Truncated EBNA-LP Protein Identified by Mass Spectrometry in P3HR1 Burkitt’s Lymphoma Cells. Cancers (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MC, Pantanowitz L and Dezube BJ (2005). AIDS-related malignancies: emerging challenges in the era of highly active antiretroviral therapy. The Oncologist 10, 412–426. [DOI] [PubMed] [Google Scholar]
- Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM and Lee JC (1999). Nasopharyngeal carcinoma cell line (C666–1) consistently harbouring Epstein-Barr virus. Int J Cancer 83, 121–6. [DOI] [PubMed] [Google Scholar]
- Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J and Sergeant A (1986). Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J 5, 3243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JM, Hung HY, You JF, Chiang SF, Lee CF, Chou HS, Lee WC and Chan KM (2019). Applicability of postoperative carcinoembryonic antigen levels in determining post-liver-resection adjuvant chemotherapy regimens for colorectal cancer hepatic metastasis. Medicine (Baltimore) 98, e17696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi UY, Lee JJ, Park A, Zhu W, Lee HR, Choi YJ, Yoo JS, Yu C, Feng P, Gao SJ et al. (2020). Oncogenic human herpesvirus hijacks proline metabolism for tumorigenesis. Proc Natl Acad Sci U S A 117, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh PE, Sin SH, Ozgur S, Henry DH, Menezes P, Griffith J, Eron JJ, Damania B and Dittmer DP (2013). Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog 9, e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockfield SM (2001). Identifying the patient at risk for post-transplant lymphoproliferative disorder. Transpl Infect Dis 3, 70–8. [DOI] [PubMed] [Google Scholar]
- Coscoy L and Ganem D (2000). Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A 97, 8051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W et al. (2015). Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57, 349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E and Holy A (2005). Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat Rev Drug Discov 4, 928–40. [DOI] [PubMed] [Google Scholar]
- DeKroon RM, Gunawardena HP, Edwards R and Raab-Traub N (2018). Global Proteomic Changes Induced by the Epstein-Barr Virus Oncoproteins Latent Membrane Protein 1 and 2A. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias MH, Kitano ES, Zelanis A and Iwai LK (2016). Proteomics and drug discovery in cancer. Drug Discov Today 21, 264–77. [DOI] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A and Ganem D (1998). A Cluster of Latently Expressed Genes in Kaposi’s Sarcoma-Associated Herpesvirus. J Virol 72, 8309–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard SC, Butler LM, Jones AM, Mermin JH, Chidzonga M, Chipato T, Shiboski CH, Brander C, Mosam A, Kiepiela P et al. (2010). Substantial regional differences in human herpesvirus 8 seroprevalence in sub-Saharan Africa: insights on the origin of the “Kaposi’s sarcoma belt”. Int J Cancer 127, 2395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham WH, Mullin M and Gingras AC (2012). Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics 12, 1576–90. [DOI] [PubMed] [Google Scholar]
- Dunleavy K (2018). Approach to the Diagnosis and Treatment of Adult Burkitt’s Lymphoma. J Oncol Pract 14, 665–671. [DOI] [PubMed] [Google Scholar]
- Ersing I, Nobre L, Wang LW, Soday L, Ma Y, Paulo JA, Narita Y, Ashbaugh CW, Jiang C, Grayson NE et al. (2017). A Temporal Proteomic Map of Epstein-Barr Virus Lytic Replication in B Cells. Cell Rep 19, 1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhang J, Chen WN and Ching CB (2011). Proteome profiling of Epstein-Barr virus infected nasopharyngeal carcinoma cell line: identification of potential biomarkers by comparative iTRAQ-coupled 2D LC/MS-MS analysis. J Proteomics 74, 567–76. [DOI] [PubMed] [Google Scholar]
- Flanagan J, Middeldorp J and Sculley T (2003). Localization of the Epstein-Barr virus protein LMP 1 to exosomes. J Gen Virol 84, 1871–1879. [DOI] [PubMed] [Google Scholar]
- Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S et al. (2009). Evolutionarily Conserved Herpesviral Protein Interaction Networks. PLoS Pathog 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J et al. (1996). KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med 2, 925–8. [DOI] [PubMed] [Google Scholar]
- Garibal J, Hollville E, Bell AI, Kelly GL, Renouf B, Kawaguchi Y, Rickinson AB and Wiels J (2007). Truncated form of the Epstein-Barr virus protein EBNA-LP protects against caspase-dependent apoptosis by inhibiting protein phosphatase 2A. J Virol 81, 7598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin L and Damania B (2014). KSHV: pathways to tumorigenesis and persistent infection. Adv Virus Res 88, 111–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin L, Yan F, Ben Major M and Damania B (2014). Modulation of Kaposi’s sarcoma-associated herpesvirus interleukin-6 function by hypoxia-upregulated protein 1. J Virol 88, 9429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B and Aebersold R (2007). Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol 8, 645–54. [DOI] [PubMed] [Google Scholar]
- Gkiafi Z and Panayotou G (2011). Comparative proteomic analysis implicates COMMD proteins as Epstein-Barr virus targets in the BL41 Burkitt’s lymphoma cell line. J Proteome Res 10, 2959–68. [DOI] [PubMed] [Google Scholar]
- Gloghini A, Volpi CC, Caccia D, Gualeni AV, Cilia AM, Carbone A and Bongarzone I (2014). Primary effusion lymphoma: secretome analysis reveals novel candidate biomarkers with potential pathogenetic significance. Am J Pathol 184, 618–30. [DOI] [PubMed] [Google Scholar]
- Goncalves PH, Uldrick TS and Yarchoan R (2017a). HIV-associated Kaposi sarcoma and related diseases. AIDS 31, 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves PH, Ziegelbauer J, Uldrick TS and Yarchoan R (2017b). Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr Opin HIV AIDS 12, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JP, Arcipowski KM and Bishop GA (2010). Differential B-lymphocyte regulation by CD40 and its viral mimic, latent membrane protein 1. Immunol Rev 237, 226–48. [DOI] [PubMed] [Google Scholar]
- Green M and Michaels MG (2013). Epstein-Barr virus infection and posttransplant lymphoproliferative disorder. Am J Transplant 13 Suppl 3, 41–54; quiz 54. [DOI] [PubMed] [Google Scholar]
- Guo H, Shen S, Wang L and Deng H (2010). Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1, 987–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Schinazi RF and Fingeroth JD (2000). Human herpesvirus 8 open reading frame 21 is a thymidine and thymidylate kinase of narrow substrate specificity that efficiently phosphorylates zidovudine but not ganciclovir. J Virol 74, 684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH and Aebersold R (1999). Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17, 994–9. [DOI] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS et al. (2013). Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 369, 134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S and Speicher DW (2011). Purification of proteins fused to glutathione S-transferase. Methods Mol Biol 681, 259–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DW, Yang ZF, Wong BY, Kwong DL, Sham JS, Wei WI and Yuen AP (2006). Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry serum protein profiling to identify nasopharyngeal carcinoma. Cancer 107, 99–107. [DOI] [PubMed] [Google Scholar]
- Ho M, Jaffe R, Miller G, Breinig MK, Dummer JS, Makowka L, Atchison RW, Karrer F, Nalesnik MA and Starzl TE (1988). The frequency of Epstein-Barr virus infection and associated lymphoproliferative syndrome after transplantation and its manifestations in children. Transplantation 45, 719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Miller G, Atchison RW, Breinig MK, Dummer JS, Andiman W, Starzl TE, Eastman R, Griffith BP, Hardesty RL et al. (1985). Epstein-Barr virus infections and DNA hybridization studies in posttransplantation lymphoma and lymphoproliferative lesions: the role of primary infection. J Infect Dis 152, 876–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday EB and Frank SJ (2016). Proton therapy for nasopharyngeal carcinoma. Chin Clin Oncol 5, 25. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J and Frappier L (2003). Protein Profiling with Epstein-Barr Nuclear Antigen-1 Reveals an nteraction with the Herpesvirus-associated Ubiquitin-specific Protease HAUSP/USP7. J Biol Chem 278, 29987–29994. [DOI] [PubMed] [Google Scholar]
- Houali K, Wang X, Shimizu Y, Djennaoui D, Nicholls J, Fiorini S, Bouguermouh A and Ooka T (2007). A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin Cancer Res 13, 4993–5000. [DOI] [PubMed] [Google Scholar]
- Huang PY, Zeng TT, Li MQ, Ban X, Zhu YH, Zhang BZ, Mai HQ, Zhang L, Guan XY and Li Y (2015). Proteomic analysis of a nasopharyngeal carcinoma cell line and a nasopharyngeal epithelial cell line. Tumori 101, 676–83. [DOI] [PubMed] [Google Scholar]
- Hutt-Fletcher LM (2007). Epstein-Barr virus entry. J Virol 81, 7825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K et al. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Lin SF, Ellison T, Chen LY, Izumiya C, Luciw P and Kung HJ (2003). Kaposi’s sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J Virol 77, 1441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Stopford CM, West JA, Bennett CL, Giffin L and Damania B (2015). Kaposi’s Sarcoma-Associated Herpesvirus Viral Interferon Regulatory Factor 1 Interacts with a Member of the Interferon-Stimulated Gene 15 Pathway. J Virol 89, 11572–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha HC, Banerjee S and Robertson ES (2016). The Role of Gammaherpesviruses in Cancer Pathogenesis. Pathogens (Basel, Switzerland) 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D and Kieff E (2004). Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101, 16286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakry JA and Ambinder RF (2013). EBV-related lymphomas: new approaches to treatment. Curr Treat Options Oncol 14, 224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci FE, Eren R, Gundogan C, Huq GE, Dogu MH and Suyani E (2016). A HHV-8 positive, HIV negative multicentric Castleman disease treated with R-CEOP chemotherapy and valganciclovir combination. J Infect Chemother 22, 483–5. [DOI] [PubMed] [Google Scholar]
- Karabudak AA, Hafner J, Shetty V, Chen S, Secord AA, Morse MA and Philip R (2013). Autoantibody biomarkers identified by proteomics methods distinguish ovarian cancer from non-ovarian cancer with various CA-125 levels. J Cancer Res Clin Oncol 139, 1757–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempkes B, Spitkovsky D, Jansen-Dürr P, Ellwart JW, Kremmer E, Delecluse HJ, Rottenberger C, Bornkamm GW and Hammerschmidt W (1995). B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J 14, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakurel KP, Angelov B and Andreasson J (2019). Macromolecular Nanocrystal Structural Analysis with Electron and X-Rays: A Comparative Review. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser A and Sterz KR (2015). The Latent Membrane Protein 1 (LMP1). Curr Top Microbiol Immunol 391, 119–49. [DOI] [PubMed] [Google Scholar]
- Killian CS, Emrich LJ, Vargas FP, Yang N, Wang MC, Priore RL, Murphy GP and Chu TM (1986). Relative reliability of five serially measured markers for prognosis of progression in prostate cancer. J Natl Cancer Inst 76, 179–85. [PubMed] [Google Scholar]
- Kim SS, Donahue TR and Girgis MD (2018). Carcinoembryonic Antigen for Diagnosis of Colorectal Cancer Recurrence. JAMA 320, 298–299. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R and Hansmann ML (1994). Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A 91, 10962–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren O, Goedert JJ, Rabkin CS, Wilson WH, Dunleavy K, Kyle RA, Katzmann JA, Rajkumar SV and Engels EA (2010). Circulating serum free light chains as predictive markers of AIDS-related lymphoma. J Clin Oncol 28, 773–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange P and Damania B (2020). Kaposi Sarcoma-Associated Herpesvirus (KSHV). Trends Microbiol 28, 236–237. [DOI] [PubMed] [Google Scholar]
- Larance M and Lamond AI (2015). Multidimensional proteomics for cell biology. Nat Rev Mol Cell Biol 16, 269–80. [DOI] [PubMed] [Google Scholar]
- Lee BS, Lee SH, Feng P, Chang H, Cho NH and Jung JU (2005). Characterization of the Kaposi’s sarcoma-associated herpesvirus K1 signalosome. J Virol 79, 12173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Xiao T, Yi HM, Yi H, Feng J, Zhu JF, Huang W, Lu SS, Zhou YH, Li XH et al. (2019). S897 phosphorylation of EphA2 is indispensable for EphA2-dependent nasopharyngeal carcinoma cell invasion, metastasis and stem properties. Cancer Lett 444, 162–174. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Li D, Xie Y, Shi J, Li G, Guan Y, Li M, Zhang P, Peng F et al. (2012). Periostin, a stroma-associated protein, correlates with tumor invasiveness and progression in nasopharyngeal carcinoma. Clin Exp Metastasis 29, 865–77. [DOI] [PubMed] [Google Scholar]
- Li Z, Li N, Shen L and Fu J (2018). Quantitative Proteomic Analysis Identifies MAPK15 as a Potential Regulator of Radioresistance in Nasopharyngeal Carcinoma Cells. Front Oncol 8, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Chang KP, Hsu CW, Chi LM, Chien KY, Liang Y, Tsai MH, Lin YT and Yu JS (2013). Low-molecular-mass secretome profiling identifies C-C motif chemokine 5 as a potential plasma biomarker and therapeutic target for nasopharyngeal carcinoma. J Proteomics 94, 186–201. [DOI] [PubMed] [Google Scholar]
- Lin W, Yip YL, Jia L, Deng W, Zheng H, Dai W, Ko JMY, Lo KW, Chung GTY, Yip KY et al. (2018). Establishment and characterization of new tumor xenografts and cancer cell lines from EBV-positive nasopharyngeal carcinoma. Nat Commun 9, 4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linch DC (2012). Burkitt lymphoma in adults. Br J Haematol 156, 693–703. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rao Y, Tian M, Zhang S and Feng P (2019). Modulation of Innate Immune Signaling Pathways by Herpesviruses. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JH, Purdue M, Kelsey K, Zhang ZF, Winn D, Wei Q, Talamini R, Szeszenia-Dabrowska N, Sturgis EM, Smith E et al. (2009). Total exposure and exposure rate effects for alcohol and smoking and risk of head and neck cancer: a pooled analysis of case-control studies. Am J Epidemiol 170, 937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubyova B, Kellum MJ, Frisancho AJ and Pitha PM (2004). Kaposi’s Sarcoma-associated Herpesvirus-encoded vIRF-3 Stimulates the Transcriptional Activity of Cellular IRF-3 and IRF-7. J Biol Chem 279, 7643–7654. [DOI] [PubMed] [Google Scholar]
- Lubyova B, Kellum MJ, Frisancho JA and Pitha PM (2007). Stimulation of c-Myc transcriptional activity by vIRF-3 of Kaposi sarcoma-associated herpesvirus. J Biol Chem 282, 31944–53. [DOI] [PubMed] [Google Scholar]
- Mahdavifar N, Towhidi F, Makhsosi BR, Pakzad R, Moini A, Ahmadi A, Lotfi S and Salehiniya H (2016). Incidence and Mortality of Nasopharynx Cancer and Its Relationship With Human Development Index in the World in 2012. World J Oncol 7, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V and Zheng ZM (2015). KSHV ORF57, a protein of many faces. Viruses 7, 604–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowska A, Braunschweig T, Denecke B, Shen L, Baloche V, Busson P and Kontny U (2019). Interferon beta and Anti-PD-1/PD-L1 Checkpoint Blockade Cooperate in NK Cell-Mediated Killing of Nasopharyngeal Carcinoma Cells. Transl Oncol 12, 1237–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancao C, Altmann M, Jungnickel B and Hammerschmidt W (2005). Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood 106, 4339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners O, Murphy JC, Coleman A, Hughes DJ and Whitehouse A (2018). Contribution of the KSHV and EBV lytic cycles to tumourigenesis. Curr Opin Virol 32, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manservigi R and Cassai E (1991). The glycoproteins of the human herpesviruses. Comp Immunol Microbiol Infect Dis 14, 81–95. [DOI] [PubMed] [Google Scholar]
- Martínez FP and Tang Q (2012). Leucine Zipper Domain Is Required for Kaposi Sarcoma-associated Herpesvirus (KSHV) K-bZIP Protein to Interact with Histone Deacetylase and Is Important for KSHV Replication. J Biol Chem 287, 15622–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R, Cheves A, Eason AB, Landis JT, Host KM et al. (2019). Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr., Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B and Raab-Traub N (2013). Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110, E2925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr., Shair KH, Marquitz AR, Kung CP, Edwards RH and Raab-Traub N (2010). Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107, 20370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meij P, van Esser JW, Niesters HG, van Baarle D, Miedema F, Blake N, Rickinson AB, Leiner I, Pamer E, Lowenberg B et al. (2003). Impaired recovery of Epstein-Barr virus (EBV)--specific CD8+ T lymphocytes after partially T-depleted allogeneic stem cell transplantation may identify patients at very high risk for progressive EBV reactivation and lymphoproliferative disease. Blood 101, 4290–7. [DOI] [PubMed] [Google Scholar]
- Meredith RF, Khazaeli MB, Plott WE, Grizzle WE, Liu T, Schlom J, Russell CD, Wheeler RH and LoBuglio AF (1996). Phase II study of dual 131I-labeled monoclonal antibody therapy with interferon in patients with metastatic colorectal cancer. Clin Cancer Res 2, 1811–8. [PubMed] [Google Scholar]
- Mesri EA, Cesarman E and Boshoff C (2010). Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer 10, 707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Gaya AM, Dancey G, Stratford MR, Othman S, Sharma SK, Wellsted D, Taylor NJ, Stirling JJ, Poupard L et al. (2009). A phase I trial of radioimmunotherapy with 131I-A5B7 anti-CEA antibody in combination with combretastatin-A4-phosphate in advanced gastrointestinal carcinomas. Clin Cancer Res 15, 4484–92. [DOI] [PubMed] [Google Scholar]
- Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S and Chang Y (1997). Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol 71, 314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork J, Lie AK, Glattre E, Hallmans G, Jellum E, Koskela P, Moller B, Pukkala E, Schiller JT, Youngman L et al. (2001). Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med 344, 1125–31. [DOI] [PubMed] [Google Scholar]
- Mosam A, Shaik F, Uldrick TS, Esterhuizen T, Friedland GH, Scadden DT, Aboobaker J and Coovadia HM (2012). A randomized controlled trial of highly active antiretroviral therapy versus highly active antiretroviral therapy and chemotherapy in therapy-naive patients with HIV-associated Kaposi sarcoma in South Africa. J Acquir Immune Defic Syndr 60, 150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Sald J and Knowles DM (1996). Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 88, 645–56. [PubMed] [Google Scholar]
- Nakamura H, Li M, Zarycki J and Jung JU (2001). Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 75, 7572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh KN, Rice AJ and Bower M (2008). Lymph nodes involved by multicentric Castleman disease among HIV-positive individuals are often involved by Kaposi sarcoma. Am J Surg Pathol 32, 1006–12. [DOI] [PubMed] [Google Scholar]
- Nayar U, Lu P, Goldstein RL, Vider J, Ballon G, Rodina A, Taldone T, Erdjument-Bromage H, Chomet M, Blasberg R et al. (2013). Targeting the Hsp90-associated viral oncoproteome in gammaherpesvirus-associated malignancies. Blood 122, 2837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BD, Shinkins B, Pathiraja I, Roberts NW, James TJ, Mallett S, Perera R, Primrose JN and Mant D (2015). Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev, Cd011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto N, Kanakura Y, Aozasa K, Johkoh T, Nakamura M, Nakano S, Nakano N, Ikeda Y, Sasaki T, Nishioka K et al. (2005). Humanized anti-interleukin-6 receptor antibody treatment of multicentric Castleman disease. Blood 106, 2627–32. [DOI] [PubMed] [Google Scholar]
- Northfelt DW, Dezube BJ, Thommes JA, Miller BJ, Fischl MA, Friedman-Kien A, Kaplan LD, Du Mond C, Mamelok RD and Henry DH (1998). Pegylated-liposomal doxorubicin versus doxorubicin, bleomycin, and vincristine in the treatment of AIDS-related Kaposi’s sarcoma: results of a randomized phase III clinical trial. J Clin Oncol 16, 2445–51. [DOI] [PubMed] [Google Scholar]
- Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J and Munz C (2014). Macroautophagy Proteins Assist Epstein Barr Virus Production and Get Incorporated Into the Virus Particles. EBioMedicine 1, 116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel JP and Agbalika F (2000). High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric castleman disease in HIV-infected patients. Blood 96, 2069–73. [PubMed] [Google Scholar]
- Pagano JS, Whitehurst CB and Andrei G (2018). Antiviral Drugs for EBV. Cancers 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM (2006). The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118, 3030–44. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T and Middeldorp JM (2010). Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107, 6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzotto MN, Uldrick TS, Wyvill KM, Aleman K, Peer CJ, Bevans M, Sereti I, Maldarelli F, Whitby D, Marshall V et al. (2016). Pomalidomide for Symptomatic Kaposi’s Sarcoma in People With and Without HIV Infection: A Phase I/II Study. J Clin Oncol 34, 4125–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt ZL, Zhang J and Sugden B (2012). The latent membrane protein 1 (LMP1) oncogene of Epstein-Barr virus can simultaneously induce and inhibit apoptosis in B cells. J Virol 86, 4380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pria AD, Pinato D, Roe J, Naresh K, Nelson M and Bower M (2017). Relapse of HHV8-positive multicentric Castleman disease following rituximab-based therapy in HIV-positive patients. Blood 129, 2143–2147. [DOI] [PubMed] [Google Scholar]
- Purushothaman P, Uppal T and Verma SC (2015). Molecular biology of KSHV lytic reactivation. Viruses 7, 116–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N (2007). EBV-induced oncogenesis. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, (ed. C.-F. G Arvin A, Mocarski E, et al. ). Cambridge: Cambridge University Press. [PubMed] [Google Scholar]
- Rabkin CS, Engels EA, Landgren O, Schuurman R, Camargo MC, Ruth P and Goedert JJ (2011). Circulating cytokine levels, Epstein-Barr viremia, and risk of acquired immunodeficiency syndrome-related non-Hodgkin lymphoma. Am J Hematol 86, 875–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainy N, Zayoud M, Kloog Y, Rechavi O and Goldstein I (2016). Viral oncomiR spreading between B and T cells is employed by Kaposi’s sarcoma herpesvirus to induce non-cell-autonomous target gene regulation. Oncotarget 7, 41870–41884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Lagunoff M, Zhong W and Ganem D (1996). The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol 70, 8151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef R, Vardhanabhuti S, Luskin MR, Heitjan DF, Hadjiliadis D, Goral S, Krok KL, Goldberg LR, Porter DL, Stadtmauer EA et al. (2011). Reduction of immunosuppression as initial therapy for posttransplantation lymphoproliferative disorder (bigstar). Am J Transplant 11, 336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Thlick AE, Parravicini C, Moore PS and Chang Y (2001). Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol 75, 429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Deroubaix S, Feldhahn N, Oliveira TY, Callen E, Wang Q, Jankovic M, Silva IT, Rommel PC, Bosque D et al. (2015). Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell 162, 727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen R, Sathish N, Li Y and Yuan Y (2008). Virion-Wide Protein Interactions of Kaposi’s Sarcoma-Associated Herpesvirus. J Virol 82, 4742–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghipour S and Mathias RA (2017). Herpesviruses hijack host exosomes for viral pathogenesis. Sem Cell Dev Biol 67, 91–100. [DOI] [PubMed] [Google Scholar]
- Salihi SA, Buryanek J, Rios AA and Brown RE (2016). Morphoproteomics Identifies Etiopathogenetic Correlates of HHV-8-Associated Kaposi’s Sarcoma and Provides Pathways with Therapeutic Options: A Case Study. Ann Clin Lab Sci 46, 537–43. [PubMed] [Google Scholar]
- Schlee M, Krug T, Gires O, Zeidler R, Hammerschmidt W, Mailhammer R, Laux G, Sauer G, Lovric J and Bornkamm GW (2004). Identification of Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) target proteins by proteome analysis: activation of EBNA2 in conditionally immortalized B cells reflects early events after infection of primary B cells by EBV. J Virol 78, 3941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler N and Urban N (2007). CA125 in ovarian cancer. Biomark Med 1, 513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab A, Meyering SS, Lepene B, Iordanskiy S, van Hoek ML, Hakami RM and Kashanchi F (2015). Extracellular vesicles from infected cells: potential for direct pathogenesis. Front Microbiol 6, 1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T, Park J, Lee D, Hwang SG and Choe J (2001). Viral interferon regulatory factor 1 of Kaposi’s sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. J Virol 75, 6193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Wang HW, Koumi A, Hollyman D, Endo Y, Ye H, Du MQ and Boshoff C (2002). K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol 76, 802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CL, Liu CD, You RI, Ching YH, Liang J, Ke L, Chen YL, Chen HC, Hsu HJ, Liou JW et al. (2016). Ribosome Protein L4 is essential for Epstein-Barr Virus Nuclear Antigen 1 function. Proc Natl Acad Sci U S A 113, 2229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JH, Janis R, Alper JC, Schutte H, Robbins L and Blaufox MD (1969). Disseminated visceral Kaposi’s sarcoma. Appearance after human renal homograft operation. JAMA 207, 1493–6. [PubMed] [Google Scholar]
- Smets F, Latinne D, Bazin H, Reding R, Otte JB, Buts JP and Sokal EM (2002). Ratio between Epstein-Barr viral load and anti-Epstein-Barr virus specific T-cell response as a predictive marker of posttransplant lymphoproliferative disease. Transplantation 73, 1603–10. [DOI] [PubMed] [Google Scholar]
- Song Y-J, Izumi KM, Shinners NP, Gewurz BE and Kieff E (2008). IRF7 activation by Epstein–Barr virus latent membrane protein 1 requires localization at activation sites and TRAF6, but not TRAF2 or TRAF3. Proc Natl Acad Sci U S A 105, 18448–18453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni V, Cahir-McFarland E and Kieff E (2007). LMP1 TRAFficking activates growth and survival pathways. Adv Exp Med Biol 597, 173–87. [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L et al. (1995). Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86, 1276–80. [PubMed] [Google Scholar]
- Speicher DJ, Sehu MM, Mollee P, Shen L, Johnson NW and Faoagali JL (2014). Successful treatment of iatrogenic multicentric Castleman’s disease arising due to recrudescence of HHV-8 in a liver transplant patient. Am J Transplant 14, 1207–13. [DOI] [PubMed] [Google Scholar]
- Stormont TJ, Farrow GM, Myers RP, Blute ML, Zincke H, Wilson TM and Oesterling JE (1993). Clinical Stage B0 or T1c prostate cancer: nonpalpable disease identified by elevated serum prostate-specific antigen concentration. Urology 41, 3–8. [DOI] [PubMed] [Google Scholar]
- Sui H, Ma N, Wang Y, Li H, Liu X, Su Y and Yang J (2018). Anti-PD-1/PD-L1 Therapy for Non-Small-Cell Lung Cancer: Toward Personalized Medicine and Combination Strategies. J Immunol Res 2018, 6984948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutandy FX, Qian J, Chen CS and Zhu H (2013). Overview of protein microarrays. Curr Protoc Protein Sci Chapter 27, Unit 27 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymula A, Palermo RD, Bayoumy A, Groves IJ, Ba Abdullah M, Holder B and White RE (2018). Epstein-Barr virus nuclear antigen EBNA-LP is essential for transforming naïve B cells, and facilitates recruitment of transcription factors to the viral genome. PLoS Pathog 14, e1006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam HK, Zhang ZF, Jacobson LP, Margolick JB, Chmiel JS, Rinaldo C and Detels R (2002). Effect of highly active antiretroviral therapy on survival among HIV-infected men with Kaposi sarcoma or non-Hodgkin lymphoma. Int J Cancer 98, 916–22. [DOI] [PubMed] [Google Scholar]
- Tan D (2008). Morphoproteomics: a novel approach to identify potential therapeutic targets in cancer patients. Int J Clin Exp Pathol 1, 331–2. [PMC free article] [PubMed] [Google Scholar]
- Tang CE, Guan YJ, Yi B, Li XH, Liang K, Zou HY, Yi H, Li MY, Zhang PF, Li C et al. (2010). Identification of the amyloid beta-protein precursor and cystatin C as novel epidermal growth factor receptor regulated secretory proteins in nasopharyngeal carcinoma by proteomics. J Proteome Res 9, 6101–11. [DOI] [PubMed] [Google Scholar]
- Tao WA and Aebersold R (2003). Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol 14, 110–8. [DOI] [PubMed] [Google Scholar]
- Thakker S and Verma SC (2016). Co-infections and Pathogenesis of KSHV-Associated Malignancies. Front Microbiol 7, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson D, Deitsch KW, Duca KA and Torgbor C (2016). The Link between Plasmodium falciparum Malaria and Endemic Burkitt’s Lymphoma-New Insight into a 50-Year-Old Enigma. PLoS Pathog 12, e1005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiernan JP, Perry SL, Verghese ET, West NP, Yeluri S, Jayne DG and Hughes TA (2013). Carcinoembryonic antigen is the preferred biomarker for in vivo colorectal cancer targeting. Br J Cancer 108, 662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M, Choe J, Tsukazaki T and Mori N (2004). The Kaposi’s sarcoma-associated herpesvirus K-bZIP protein represses transforming growth factor β signaling through interaction with CREB-binding protein. Oncogene 23, 8272–8281. [DOI] [PubMed] [Google Scholar]
- Torgbor C, Awuah P, Deitsch K, Kalantari P, Duca KA and Thorley-Lawson DA (2014). A multifactorial role for P. falciparum malaria in endemic Burkitt’s lymphoma pathogenesis. PLoS Pathog 10, e1004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappe RU, Dierickx D, Zimmermann H, Morschhauser F, Mollee P, Zaucha JM, Dreyling MH, Duhrsen U, Reinke P, Verhoef G et al. (2017). Response to Rituximab Induction Is a Predictive Marker in B-Cell Post-Transplant Lymphoproliferative Disorder and Allows Successful Stratification Into Rituximab or R-CHOP Consolidation in an International, Prospective, Multicenter Phase II Trial. J Clin Oncol 35, 536–543. [DOI] [PubMed] [Google Scholar]
- Traylen C, Ramasubramanyan S, Zuo J, Rowe M, Almohammad R, Heesom K, Sweet SM, Matthews DA and Sinclair AJ (2015). Identification of Epstein-Barr Virus Replication Proteins in Burkitt’s Lymphoma Cells. Pathogens 4, 739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao SW, Tsang CM and Lo KW (2017). Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkoski K and Raab-Traub N (2015). LMP1 promotes expression of insulin-like growth factor 1 (IGF1) to selectively activate IGF1 receptor and drive cell proliferation. J Virol 89, 2590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Dong Y-A, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E et al. (2006). Herpesviral Protein Networks and Their Interaction with the Human Proteome. Science 311, 239–242. [DOI] [PubMed] [Google Scholar]
- Uetz P, Rajagopala SV, Dong YA and Haas J (2004). From ORFeomes to protein interaction maps in viruses. Genome Res 14, 2029–33. [DOI] [PubMed] [Google Scholar]
- Uldrick TS, Polizzotto MN, Aleman K, O’Mahony D, Wyvill KM, Wang V, Marshall V, Pittaluga S, Steinberg SM, Tosato G et al. (2011). High-dose zidovudine plus valganciclovir for Kaposi sarcoma herpesvirus-associated multicentric Castleman disease: a pilot study of virus-activated cytotoxic therapy. Blood 117, 6977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls C, Canas C, Turell LG and Pruna X (1991). Hepatosplenic AIDS-related Kaposi’s sarcoma. Gastrointest Radiol 16, 342–4. [DOI] [PubMed] [Google Scholar]
- Vendrame E and Martínez-Maza O (2011). Assessment of pre-diagnosis biomarkers of immune activation and inflammation: insights on the etiology of lymphoma. J Proteome Res 10, 113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij FJ, de Heus C, Kroeze S, Cai H, Kieff E, Piersma SR, Jimenez CR, Middeldorp JM and Pegtel DM (2015). Exosomal sorting of the viral oncoprotein LMP1 is restrained by TRAF2 association at signalling endosomes. J Extracell Vesicles 4, 26334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidick S, Leroy B, Palmeira L, Machiels B, Mast J, Francois S, Wattiez R, Vanderplasschen A and Gillet L (2013). Proteomic characterization of murid herpesvirus 4 extracellular virions. PLoS One 8, e83842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H. W. t., Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ and Speck SH (1997). Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71, 5894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Akula SM, Pramod NP, Zeng L and Chandran B (2001). Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J Virol 75, 7517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Zhang H, Zhang JP, Li Y, Zhao B, Feng GK, Du Y, Xiong D, Zhong Q, Liu WL et al. (2015). Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun 6, 6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Du S, Zhu C, Wang C, Yu N, Lin Z, Gan J, Guo Y, Huang X, He Y et al. (2020). STUB1 is targeted by the SUMO-interacting motif of EBNA1 to maintain Epstein-Barr Virus latency. PLoS Pathog 16, e1008447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YC, Yang SF, Chang SL, Chen TJ, Lee SW, Chen HS, Lin LC and Li CF (2018). Periostin overexpression is associated with worse prognosis in nasopharyngeal carcinoma from endemic area: a cohort study. Onco Targets Ther 11, 3205–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KW and Damania B (2010a). Hsp90 and Hsp40/Erdj3 are required for the expression and anti-apoptotic function of KSHV K1. Oncogene 29, 3532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KW and Damania B (2010b). Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett 289, 140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmer A, Wang P, Zhou J, Rennekamp AJ, Tiranti V, Zeviani M and Lieberman PM (2008). Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J Virol 82, 4647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J, Cope A, Gill J, Bourboulia D, Hayes P, Imami N, Kubo T, Marcelin A, Calvez V, Weiss R et al. (2002). Identification of Kaposi’s sarcoma-associated herpesvirus (KSHV)-specific cytotoxic T-lymphocyte epitopes and evaluation of reconstitution of KSHV-specific responses in human immunodeficiency virus type 1-Infected patients receiving highly active antiretroviral therapy. J Virol 76, 2634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JP, Stuhlmiller TJ, Giffin LC, Lin C, Bigi R, Zhao J, Zhang W, Bravo Cruz AG, Park SI, Earp HS et al. (2019). Kinome profiling of non-Hodgkin lymphoma identifies Tyro3 as a therapeutic target in primary effusion lymphoma. Proc Natl Acad Sci U S A 116, 16541–16550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland T, Harrison B, Delmenico L and Dowling D (2018). Hepatocellular Carcinoma Screening Utilising Serum Alpha-Fetoprotein Measurement and Abdominal Ultrasound Is More Effective than Ultrasound Alone in Patients with Non-viral Cirrhosis. J Gastrointest Cancer 49, 476–480. [DOI] [PubMed] [Google Scholar]
- Wu M, Liu H, Liu Z, Liu C, Zhang A and Li N (2018). Analysis of serum alpha-fetoprotein (AFP) and AFP-L3 levels by protein microarray. J Int Med Res 46, 4297–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchoan R and Uldrick TS (2018). HIV-Associated Cancers and Related Diseases. N Engl J Med 378, 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo ELL, Li YQ, Soo KC, Wee JTS and Chua MLK (2018). Combinatorial strategies of radiotherapy and immunotherapy in nasopharyngeal carcinoma. Chin Clin Oncol 7, 15. [DOI] [PubMed] [Google Scholar]
- Yip YL, Lin WT, Deng W, Tsang CM and Tsao SW (2019). Chapter 5 - Establishment of Nasopharyngeal Carcinoma Cell Lines, Patient-Derived Xenografts, and Immortalized Nasopharyngeal Epithelial Cell Lines for Nasopharyngeal Carcinoma and Epstein–Barr Virus Infection Studies. In Nasopharyngeal Carcinoma, (eds Lee AWM Lung ML and Ng WT), pp. 85–107: Academic Press. [Google Scholar]
- Yoshizaki K, Matsuda T, Nishimoto N, Kuritani T, Taeho L, Aozasa K, Nakahata T, Kawai H, Tagoh H, Komori T et al. (1989). Pathogenic significance of interleukin-6 (IL-6/BSF-2) in Castleman’s disease. Blood 74, 1360–7. [PubMed] [Google Scholar]
- You J, Kao A, Dillon R, Croner LJ, Benz R, Blume JE and Wilcox B (2018). A large-scale and robust dynamic MRM study of colorectal cancer biomarkers. J Proteomics 187, 80–92. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, Dong XD, Li SB, Du Y, Xiong D et al. (2018). Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3, 1–8. [DOI] [PubMed] [Google Scholar]
- Zheng D, Wan J, Cho YG, Wang L, Chiou CJ, Pai S, Woodard C, Zhu J, Liao G, Martinez-Maza O et al. (2011). Comparison of humoral immune responses to Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus using a viral proteome microarray. J Infect Dis 204, 1683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Heesom K, Osborn K, AlMohammed R, Sweet SM and Sinclair AJ (2020). Identifying the Cellular Interactome of Epstein-Barr Virus Lytic Regulator Zta Reveals Cellular Targets Contributing to Viral Replication. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CY, Zhao SS, Wang XK, Wang L, Wang FY, Fang S, Liu ZX, Guan LX, Liu YC, Ding Y et al. (2019). Outcome of Rituximab-Based Treatment for Post-Transplant Lymphoproliferative Disorder After Allogeneic Hematopoietic Stem Cell Transplantation: A Single-Center Experience. Ann Transplant 24, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Chong JM, Wu L and Yuan Y (2005). Virion proteins of Kaposi’s sarcoma-associated herpesvirus. J Virol 79, 800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX and Yuan Y (2003). The ORF45 protein of Kaposi’s sarcoma-associated herpesvirus is associated with purified virions. J Virol 77, 4221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]


