Abstract
Non-alcoholic fatty liver disease (NAFLD) is a widespread threat to human health. However, the present screening methods for NAFLD are time-consuming or invasive. The present study aimed to assess the potential of microRNAs (miRNAs/miRs) in serum extracellular vesicles (EVs) as a biomarker of NAFLD. C57BL/6J mice were fed either a 12-week high-fat diet (HFD) or standard chow to establish NAFLD and control groups, respectively. Serum samples were obtained from the mouse model of NAFLD, as well as 50 patients with NAFLD and 50 healthy individuals, and EVs were extracted and verified. Using reverse transcription-quantitative PCR, the mRNA expression level of selected miRNAs in the serum and EVs was analyzed. In order to determine the diagnostic value, receiver operating characteristic (ROC) curves were used. The mice treated with HFD showed notable hepatic steatosis and higher concentrations of serum alanine aminotransferase (ALT). There was also a significant decrease in the expression levels of miR-135a-3p, miR-129b-5p and miR-504-3p, and an increase in miR-122-5p expression levels in circulating EVs in mice treated with HFD and patients with NAFLD. There were also similar miR-135a-3p and miR-122-5p expression patterns in the serum. ROC analysis demonstrated that miR-135a-3p in circulating EVs was highly accurate in diagnosing NAFLD, with the area under the curve value being 0.849 (95% CI, 0.777–0.921; P<0.0001). Bioinformatics analysis indicated that dysregulated miR-135a-3p was associated with ‘platelet-derived growth factor receptor signaling pathway’ and ‘AMP-activated protein kinase signaling pathway’. In summary, circulating miR-135a-3p in EVs may serve as a potential non-invasive biomarker to diagnose NAFLD. This miRNA was a more sensitive and specific biological marker for NAFLD compared with ALT.
Keywords: miR-135, extracellular vesicles, non-alcoholic fatty liver disease, biomarkers
Introduction
Non-alcoholic fatty liver disease (NAFLD) is caused by metabolic stress and genetic susceptibility. NAFLD includes simple steatosis and NA steatohepatitis (NASH) and may include hepatocellular carcinoma (HCC) (1,2). The global prevalence of NAFLD has been estimated to be 25% in recent years (3). Therefore, NAFLD has emerged as an important global public health issue, particularly in the Chinese population (4,5). Increasing evidence has indicated that NAFLD also contributes to a number of cardiovascular complications, in addition to liver-associated complications (6,7). The prevalence of coronary, cerebrovascular and peripheral vascular disease is remarkably higher among patients with NAFLD than among those in the general population (8). Currently, liver biopsy is the standard method to diagnose NAFLD. Computed tomography scans and ultrasound have also been used for the detection of NAFLD; however, these methods have a number of disadvantages, including financial cost, invasiveness and sampling variability (9). Therefore, a simpler, non-invasive strategy for the evaluation of NAFLD is required.
Recently, microRNAs (miRNAs/miRs), a type of small (19–25 nucleotides in length) non-coding RNA that regulates post-transcription gene expression levels, have been considered as a promising molecular marker for clinical application (10). In the liver, miRNAs are abundantly expressed and regulate various functions, including cell death, cell proliferation and developmental timing (11). Several studies have identified miRNA profiles that are altered in NAFLD. Dysregulated circulating miRNAs, including miR-27b, miR-122 and miR-21, were found to differentiate patients with NAFLD from healthy controls (12–14). However, miRNAs expressed by circulating dysfunctional cells can affect circulating miRNAs (15).
Extracellular vesicles (EVs), membrane-derived vesicles 100–1,000 nm in size, are secreted by numerous types of cell, including tumor and hepatic cells (16). EVs contain nucleic acids and proteins and are abundant in various types of bodily fluid, such as blood, ejaculates, urine, cerebrospinal fluid, saliva and breast milk (17). Accumulating evidence (18) has demonstrated that miRNAs in EVs in serum are more stable than serum circulating miRNAs, and are protected from endogenous RNases degradation (19). This indicates that miRNAs derived from serum EVs are preferable as diagnostic biological markers, compared with serum circulating miRNAs. Numerous studies have suggested the importance of EV-derived miRNAs in the diagnosis of cancer (20–22); however, further investigation is still required.
Our previous study identified dysregulated miRNA expression levels in EVs secreted by hepatic cells in mice fed high-fat diet (HFD) compared with mice fed chow diet (CD) using microarray analysis (23). It is still not known whether liver EV-derived miRNAs can be used to discriminate between patients with NAFLD and healthy controls. In the present study, the expression levels of four selected miRNAs were investigated based on our previous study. The aim of the present study was to identify the diagnostic potential of EV-derived miRNAs as a non-invasive method for the diagnosis of NAFLD.
Materials and methods
Study subjects
Serum samples were collected from 50 patients with NAFLD and 50 healthy individuals at The First Affiliated Hospital of Nanjing Medical University (Nanjing, China) between January and August 2018. The presence of FLD was assessed using ultrasonography. Patients were included in the study if they had confirmed fatty liver disease with no history of significant alcohol consumption, viral hepatitis, autoimmune hepatitis, metabolic diseases or hepatotoxic medication (e.g. amiodarone). Clinical information, including sex, age, body mass index and biochemical measurements were obtained from each patient. The Ethics Committees of Nanjing Medical University approved the present study and all participants provided written informed consent.
Animal experiments
The Model Animal Research Center of Nanjing University provided male C57BL/6J mice, (age, 4–5 weeks; weight, 18–22 g; n=40) which were kept in a pathogen-free barrier facility at 22°C, with 50% relative humidity and under a 12-h light/dark cycle. The animals were randomly assigned either into the normal control (chow diet; CD) or NAFLD group (HFD; 5.56 kcal/g; fat, 58 kcal%; hydrogenated coconut oil, 54%; carbohydrate, 25.5 kcal%) for 12 weeks, as described previously (24). The mice were weighed and euthanized by cervical dislocation at the end of week 12. The intact livers were rapidly excised from the abdominal cavity and washed with PBS for primary hepatic cell culture, RNA extraction or histopathology. The Animal Experiment Ethics Committee of Nanjing Medical University approved the study (approval no. IACUC-1601176).
Histopathology
Fresh liver tissue samples were washed with PBS and placed in 10% buffered formalin at 4°C for 24 h. After gradient dehydration in ethanol and infiltration in xylene for 30 min, liver samples were sectioned into 5-µm slices and stained using hematoxylin and eosin (hematoxylin for 10 min; and eosin for 15 sec at room temperature) to assess organization and architecture. In addition, Masson's trichrome and Sirius red staining was performed for extracellular matrix and fibrosis evaluation using commercially available kits (Beijing Solarbio Science & Technology Co., Ltd.), according to the manufacturer's protocols. Cell apoptosis was determined using the TUNEL method, as previously described (25). Neovascularization within the liver parenchyma and nodules was also determined using CD31 staining (Abcam; cat. no. ab182981; 1:2,000; 4°C overnight). All stained slides were observed using a Zeiss Axioskop 40® upright research light microscope with an original magnification, ×40 (Zeiss AG). Histological evaluation was performed in a blinded manner by two independent pathologists. In order to define the stage of steatosis, fibrosis, ballooning and parenchymal inflammation, NASH Activity Score (NAS) (26) was calculated.
Primary hepatic cell isolation
Hepatic cells were isolated using collagenase (type IV; Sigma-Aldrich; Merck KGaA) perfusion, as described previously (27). In brief, after the portal vein was cannulated, the liver was perfused for 5 min with calcium-free HEPES buffer (0.33 mM; pH, 7.6), followed by perfusion for 8 min using collagenase (0.025%) and calcium chloride (0.075%) at 37°C. Following enzymatic digestion, the solution was centrifuged (50 × g for 2 min at 4°C) to obtain hepatic cells. Freshly isolated hepatic cells were maintained in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% exosome-depleted FBS and 1% penicillin/streptomycin. The cells were incubated at 37°C in a humidified chamber with 5% CO2 for 24 h.
Biochemical assays
Blood was collected from animals via cardiac puncture following sacrifice. Serum from animals and human subjects was obtained via centrifugation of whole blood (2,000 × g for 15 min at room temperature) and stored at −80°C. Serum biochemical markers [alanine aminotransferase (ALT), alkaline phosphatase (ALP), aspartate aminotransferase (AST), albumin (ALB), globulin, γ-glutamyl transpeptidase, lactate dehydrogenase (LDH), creatine kinase (CK), hydroxybutyrate dehydrogenase, total bilirubin, cholesterol (CHOL), triglyceride (TG), high-density lipoprotein (HDL), low-density lipoprotein (LDL), lipoprotein a (Lpa), fasting blood-glucose (GLU0), creatinine (Cr) and uric acid] characterizing liver disease metabolic features were measured using a biochemical analyzer (Roche Diagnostics).
Bioinformatics analysis
Putative target genes of the selected miRNAs (miR-135a-3p and miR-504-3p) were predicted using two databases, with a pure algorithm (miRDB: mirdb.org and TargetScan: www.targetscan.org/mamm_31) and overlapping target genes were selected for further functional analysis. In order to investigate the biological function of the predicted genes, Gene Ontology (GO) classification and Kyoto Encyclopedia Genes and Genomes (KEGG) enrichment were performed according to gene annotation using Database for Annotation, Visualization and Integrated Discovery (www.kegg.jp). Fisher's exact test, modified by the false discovery rate, was used to evaluate the statistical significance of enrichment.
EVs isolation
EVs from in vitro hepatocellular culture medium were separated using differential centrifugation and filtration. Briefly, cell culture medium was centrifuged at 10,000 × g for 30 min at 4°C to deplete the cells and debris. Subsequently, the supernatant was filtered using a 0.22-µm pore membrane and centrifuged for 70 min at 100,000 × g at 4°C to pellet the EVs. The pellets were then suspended in 100 µl PBS. Using ExoQuick reagent (System Biosciences, LLC), EVs from mouse or human serum were extracted according to the manufacturer's protocol. In brief, the EVs were isolated by adding EV extraction reagent and centrifuging for 20 min at 10,000 × g at 4°C. The resulting EV pellets were stored at −80°C for subsequent experiments.
Western blot analysis
Using RIPA buffer (Beyotime Institute of Biotechnology), whole mouse primary hepatic cell extracts and EV proteins were prepared and measured using a BCA protein assay kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. The extracted proteins (20 µg per lane) were separated using 10% SDS-PAGE, transferred onto PVDF membranes and blocked with 5% non-fat milk in TBS-0.05% Tween 20 for 2 h at room temperature then incubated with primary antibodies against tumor susceptibility 101 (TSG101; 1:1,000; cat. no. ab125011; Abcam), CD9 (1:1,000; cat. no. ab92726; Abcam), heat shock protein 70 (HSP70; 1:1,000; cat. no. 33-3800; Invitrogen; Thermo Fisher Scientific, Inc.), calreticulin (1:1,000; cat. no. ab92516; Abcam) and GAPDH (1:3,000; cat. no. ab8245; Abcam) overnight at 4°C. Subsequently, the membrane was incubated with secondary antibody [goat anti-rabbit IgG (HRP), cat. no. sc-2004; goat anti-mouse IgG (HRP), cat. no. sc-2005; 1:10,000; Santa Cruz Biotechnology, Inc.] for 1 h, then a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific, Inc.) was used to visualize the proteins according to the manufacturer's instructions. BandScan5.0 software (Glyko, Inc.) was used to analyze protein expression level.
Transmission electron microscopy (TEM)
In total, 5 µl EV preparation was fixed with 5 µl 4% paraformaldehyde at 4°C for 30 min and added onto formvar-coated copper grids to settle for 30 min. Subsequently, the samples were fixed with 1% glutaraldehyde for 5 min at 25°C, then stained with 2% uranyl oxalate for 5 min at 25°C and 1.8% methyl cellulose uranyl acetate for 10 min on ice in the dark, embedded in epoxy resin and polymerized at 35°C for 12 h, 45°C for 12 h and 60°C for 24 h. Residual liquid was removed from the grid with filter paper and the samples were visualized at 120 kV using the FEI Tecnai G2 Spirit Bio TWIN TEM (FEI; Thermo Fisher Scientific, Inc.). Images were acquired using an AMT 2k CCD camera (Advanced Microscopy Techniques, Corp.).
Nanoparticle tracking analysis (NTA)
In order to identify the diameter and concentration of EVs, Nanosight NS300 system (Malvern Instruments, Ltd.) fitted with a fast video capture was used. The images of sample movement were captured for 60 sec at room temperature. In order to analyze the videos and measure particle concentrations and size distribution, the NanoSight LM10 system (NanoSight Ltd.) was used. Each sample was evaluated ≥3 times.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from EVs and cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions and quantitated using the NanoDrop 2000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). TRIzol® LS (Invitrogen; Thermo Fisher Scientific, Inc.) was also used to extract serum RNA. A total of 1 pM cel-miR-39 RNA oligonucleotide was added to each sample as a spike-in control when extracting RNA from EVs or serum. A Taqman® miRNA RT kit and miRNA-specific stem-loop primers (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used to detect the expression levels of miRs, according to the manufacturer's instructions. In order to detect the expression levels of miRNA, the miR-X miRNA RT-qPCR SYBR kit (Clontech Laboratories, Inc.) was used for RT-qPCR with the Step One Plus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following program: Initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 sec and annealing and extension at each annealing temperature at 60°C for 30 sec. Relative expression level of each miRNA were normalized to cel-miR-39 and via the 2−ΔΔCq method (28). The following primer sequences were used for RT-qPCR: cel-miR-39 forward (F), 5′-ACACTCCAGCTGGGGTCACCGGGTGTAAATC-3′ and reverse (R), 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAGCTGA−3′; miR-122-5p F, 5′-ACACTCCAGCTGGGTGGAGTGTGACAATGG-3′ and R, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACACC−3′; miR-129-5p F, 5′-ACACTCCAGCTGGGCTTTTTGCGGTCTGG-3′ and R, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCAAGCCC-3′; miR-135a-3p F, 5′-ACACTCCAGCTGGGTATAGGGATTGGAGCC-3′ and R, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCCACGG-3′; and miR-504-3p F, 5′-ACACTCCAGCTGGGGGGAGTGCAGGGCAG-3′ and R, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAAACCCT-3′. Universal reverse primer: 5′-TGGTGTCGTGGAGTCG-3′.
Statistical analysis
The data are presented as the mean ± SEM (n≥3). The differences between two groups were analyzed using unpaired Student's t-test. Receiver operating characteristic (ROC) curves were generated based on the expression level of each miRNA to determine the utility of each miRNA for diagnosing NAFLD. GraphPad Prism 7 (GraphPad Software, Inc.) and SPSS v22.0 software (IBM Corp) were used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Liver steatosis is induced by HFD
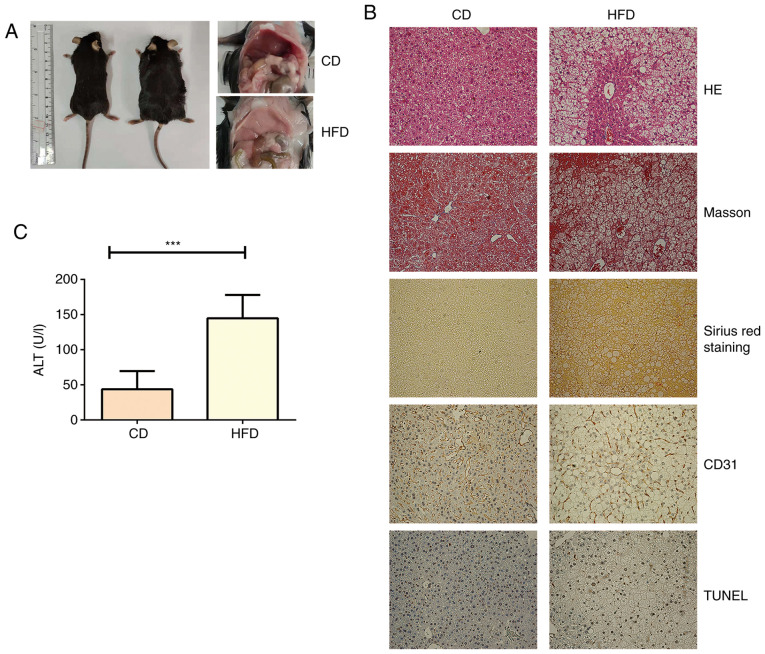
After feeding for 12 weeks, the liver from mice in the control group showed normal morphology with red, shiny and smooth appearance, whereas the liver from mice fed HFD exhibited a slightly yellow color, smooth tight membranes and soft texture (Fig. 1A). The biochemical analyses showed that mice fed HFD exhibited significantly higher AST, albumin, ALP, CHOL, HDL and LDL levels compared with in mice fed CD (Table I). In addition, the mice fed HFD exhibited notable hepatic steatosis, liver injury, hepatocellular ballooning, fibrosis, angiogenesis and cell death (Fig. 1B and Table SI). Serum ALT levels were also significantly increased in mice fed HFD compared with in mice fed CD, suggesting that a HFD induced hepatocyte damage (Fig. 1C).
Figure 1.
Characterization of HFD-induced NAFLD model. (A) Representative images of livers from mice fed HFD or CD. (B) Liver paraffin sections from C57BL/6 mice (n=5) fed HFD or CD for 12 weeks were used to assess liver damage (HE), fibrosis by detecting collagen deposition (Masson and Sirius red staining), cell death (TUNEL staining) and pathological angiogenesis (CD31 staining). (C) Serum ALT levels were measured in mice fed HFD or CD (n=5). ***P<0.001. HFD, high-fat diet; CD, chow diet; HE, hematoxylin-eosin; NAFLD, non-alcoholic fatty acid liver disease; NAS, non-alcoholic steatohepatitis activity score; ALT, alanine aminotransferase.
Table I.
Biochemical characteristics of HFD mice and controls.
| Parameter | CD (n=5) | HFD (n=5) | P-value |
|---|---|---|---|
| AST, U/l | 144.80±10.06 | 205.00±11.93 | 0.0048 |
| ALB, g/l | 28.22±0.37 | 26.22±0.42 | 0.0070 |
| ALP, U/l | 77.00±4.65 | 52.60±2.66 | 0.0019 |
| CHOL, mg/dl | 2.56±0.08 | 4.31±0.18 | <0.0001 |
| TG, mmol/l | 0.95±0.06 | 0.76±0.08 | 0.1011 |
| HD, mmol/l | 1.50±0.06 | 1.96±0.06 | 0.0005 |
| LDL, mmol/l | 0.32±0.02 | 0.68±0.05 | 0.0001 |
Data are presented as the mean ± SD. AST, aspartate aminotransferase; ALB, albumin; ALP, alkaline phosphatase; CHOL, cholesterol; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; HFD, high-fat diet; CD, chow diet.
Isolation and characterization of hepatocyte-released EVs
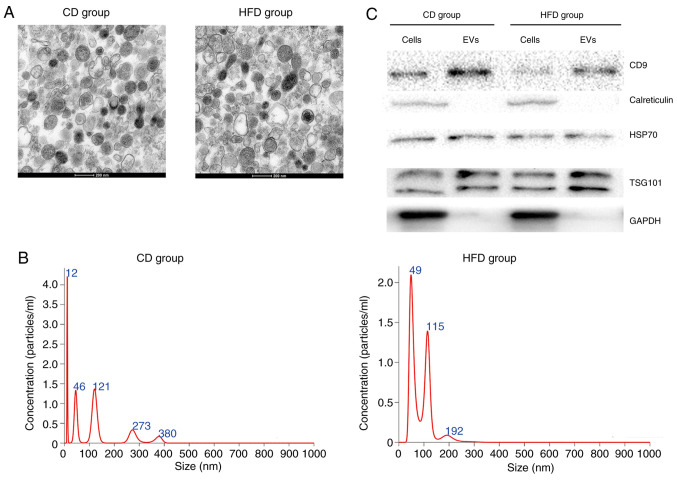
TEM images showed that isolated EVs demonstrated typical morphology with an exosomal characteristic bilayer membrane (Fig. 2A). The size distribution of EVs was measured using NTA (Fig. 2B), which was within the normal range of EV sizes. Equal amounts of protein from EVs and primary hepatocytes were separated; expression levels of exosomal markers CD9, HSP70 and TSG101 were higher in EVs compared with in primary hepatocytes (Fig. 2C). In addition, calreticulin and GAPDH were expressed in primary hepatocytes, but not expressed in EVs (Fig. 2C).
Figure 2.
Characterization of hepatocyte-released EVs. (A) Representative transmission electron microscopy images of morphology of EVs isolated from mice fed HFD or CD. (B) Size distribution of EVs was analyzed using nanoparticle tracking analysis. (C) Markers of EVs (CD9, calreticulin, HSP70, TSG101 and GAPDH) were measured in cells and EVs using western blot analysis. EVs, extracellular vesicles; HFD, high-fat diet; CD, chow diet; HSP70, heat shock protein 70; TSG101, tumor susceptibility 101.
miR-129-5p, miR-135a-3p and miR-504-3p expression levels decrease in serum EVs in mice fed HFD
According to our previous study (23), C57BL/6J mice were fed HFD for 12 weeks for induction of an NAFLD model. EVs were collected from the culture medium of hepatocytes from mice fed HFD or CD. There was a marked difference in the expression levels of several miRNAs in the hepatocyte-derived EVs from mice fed HFD compared with mice fed CD (Table II). From these miRNAs, three human and mouse homologous miRNAs were selected, with fold change >2 and P<0.05 (miR-129-5p, miR-135a-3p and miR-504-3p) for further investigation. miR-122-5p, which served as a good indicator of NAFLD in previous studies (12,15), was also included for verification.
Table II.
Differential miR signature of extracellular vesicles derived from hepatocytes of high-fat diet mice compared with chow diet mice.
| miR | P-value | Fold change |
|---|---|---|
| mmu-miR-129b-5p | 0.041 | 0.133 |
| mmu-miR-135a-3p | 0.050 | 0.431 |
| mmu-miR-1927 | 0.033 | 0.144 |
| mmu-miR-2137 | 0.021 | 0.498 |
| mmu-miR-3070-2-3p | 0.045 | 0.114 |
| mmu-miR-3102-3p.2-3p | 0.019 | 2.197 |
| mmu-miR-504-3p | 0.048 | 0.184 |
| mmu-miR-5131 | 0.038 | 0.584 |
| mmu-miR-5622-3p | 0.026 | 0.433 |
| mmu-miR-6904-5p | 0.044 | 0.170 |
| mmu-miR-7018-5p | 0.030 | 0.569 |
| mmu-miR-7218-5p | 0.040 | 0.196 |
| mmu-miR-7219-5p | 0.034 | 0.422 |
| mmu-miR-7221-5p | 0.037 | 0.634 |
| mmu-miR-7654-3p | 0.032 | 0.469 |
| mmu-miR-7654-5p | 0.010 | 0.481 |
miR, microRNA.
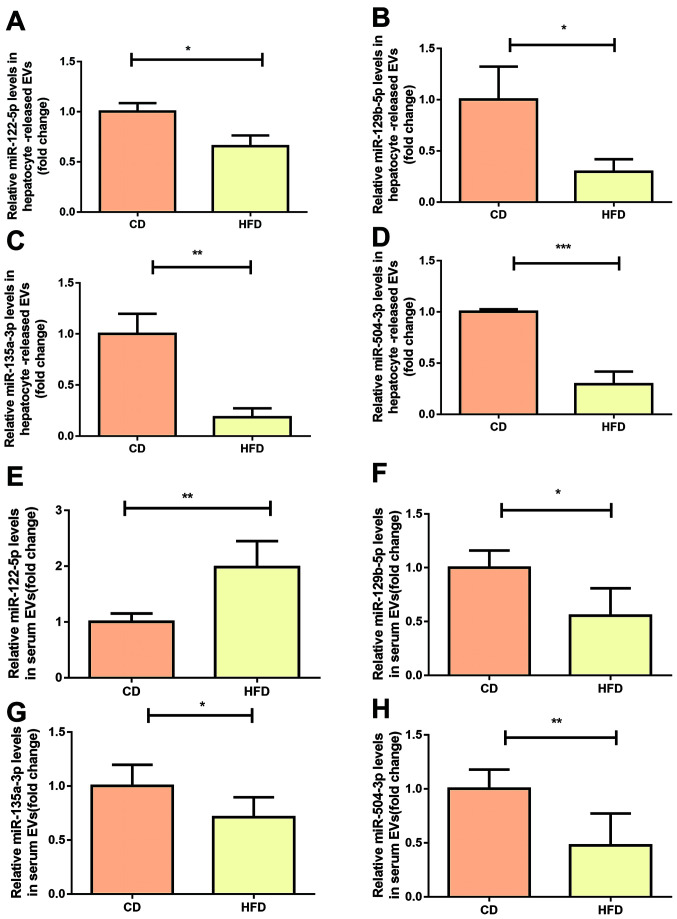
In order to verify our previous microarray results (23), the expression levels of miR-129-5p, miR-135a-3p and miR-504-3p was measured in hepatocellular EVs using RT-qPCR. The results showed that all three miRNAs exhibited lower expression levels in hepatocellular EVs from mice fed HFD compared with in mice fed CD, showing consistency with our previous microarray results (Fig. 3B-D). The expression levels of miR-122-5p were slightly decreased in hepatocellular EVs derived from mice fed HFD (Fig. 3A). In order to investigate whether these miRNAs were present in the liver, the expression levels of candidate miRNAs in hepatocytes was investigated. miR-122-5p and miR-504-3p were decreased in primary hepatocytes from mice fed HFD, while differences in miR-129-5p and miR-135a-3p were not significant (Fig. S1). Then, it was investigated whether the selected miRNAs were differentially expressed in serum or hepatocyte-derived EVs. Expression levels of miR-504-3p, miR-135a-3p and miR-129-5p were significantly decreased in serum from mice fed HFD compared with in mice fed CD (Fig. 3F-H). However, there was increased miR-122-5p expression in serum EVs from HFD mice compared with CD mice (Fig. 3E).
Figure 3.
miR signatures of hepatocyte-released and serum-derived EVs in mice fed HFD or CD. (A) miR-122-5p, (B) miR-129-5p, (C) miR-135a-1-3p and (D) miR-504-3p were verified using RT-qPCR in hepatocyte-released EVs. Expression levels of (E) miR-122-5p, (F) miR-129-5p, (G) miR-135a-1-3p and (H) miR-504-3p were detected in serum-derived EVs isolated from mice fed HFD or CD mice using RT-qPCR (n=5/group). Data are presented as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001. RT-q, reverse transcription-quantitative; miR, microRNA; HFD, high-fat diet; CD, chow diet; EVs, extracellular vesicles.
miR-135a-3p and miR-504-3p expression levels decrease in serum EVs from patients with NAFLD
In order to investigate the expression levels of selected miRNAs, 50 healthy individuals with no signs of liver disease and 50 patients with NAFLD were recruited. The clinical features of subjects are shown in Table III. The patients were sex- and age-matched. The patients with NAFLD and healthy controls did not exhibit significantly different levels of ALP, CK, hydroxybutyrate dehydrogenase, total bilirubin, total CHOL, LDL, Lpa, total protein, ALB, globulin, ALB/globulin, GLU0 and Cr. However, patients with NAFLD had notably higher AST, γ-glutamyl transpeptidase, LDH, TG and urea levels and lower HDL levels compared with healthy controls.
Table III.
Clinical and biochemical characteristics.
| Parameter | Healthy controls, n=50 | Patients with NAFLD, n=50 | P-value |
|---|---|---|---|
| Sex | |||
| Male | 36 (72%) | 39 (78%) | |
| Female | 14 (28%) | 11 (22%) | |
| Age, years | 42.60±1.67 | 42.24±1.50 | 0.8727 |
| AST, U/l | 22.95±0.84 | 26.63±1.23 | 0.0153 |
| ALP, U/l | 79.55±2.56 | 78.59±2.66 | 0.7958 |
| GGT, U/l | 25.60±1.94 | 50.38±6.10 | 0.0002 |
| LDH, U/l | 164.50±2.98 | 175.00±3.47 | 0.0231 |
| CK, U/l | 115.70±13.72 | 118.20±14.64 | 0.9060 |
| HBDH, U/l | 106.30±3.79 | 103.00±2.37 | 0.5102 |
| TBIL, µmol/l | 13.44±0.60 | 12.64±0.62 | 0.3536 |
| TC, µmol/l | 4.89±0.12 | 5.09±0.15 | 0.2961 |
| TG, µmol/l | 1.40±0.11 | 2.47±0.37 | 0.0061 |
| HDL, µmol/l | 1.27±0.03 | 1.15±0.04 | 0.0272 |
| LDL, µmol/l | 3.19±0.10 | 3.27±0.10 | 0.5662 |
| Lpa, mg/l | 210.10±41.60 | 126.00±23.25 | 0.1154 |
| TP, g/l | 73.13±0.75 | 71.71±0.52 | 0.1211 |
| ALB, g/l | 45.73±0.42 | 45.01±0.34 | 0.1871 |
| GLB g/l | 27.90±0.45 | 26.70±0.45 | 0.0638 |
| A/G | 1.66±0.03 | 1.71±0.04 | 0.2861 |
| GLU0, mmol/l | 5.55±0.19 | 5.69±0.32 | 0.7048 |
| Urea, mmol/l | 5.36±0.16 | 5.16±0.17 | 0.4004 |
| Cr, µmol/l | 69.32±1.54 | 68.17±1.63 | 0.6099 |
| UA, µmol/l | 328.60±9.25 | 373.30±13.46 | 0.0074 |
Data are presented as the mean ± SD. AST, aspartate aminotransferase; ALP, alkaline phosphatase; GGT, γ-glutamyl transpeptidase; LDH, lactate dehydrogenase; CK, creatine kinase; HBDH, hydroxybutyrate dehydrogenase; TBIL, total bilirubin; TC, total cholesterol; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lpa, lipoprotein a; TP, total protein; ALB, albumin; GLB, globulin; A/G, albumin/globulin; GLU0, fasting blood-glucose; Cr, creatinine; UA, uric acid; NAFLD, non-alcoholic fatty liver disease.
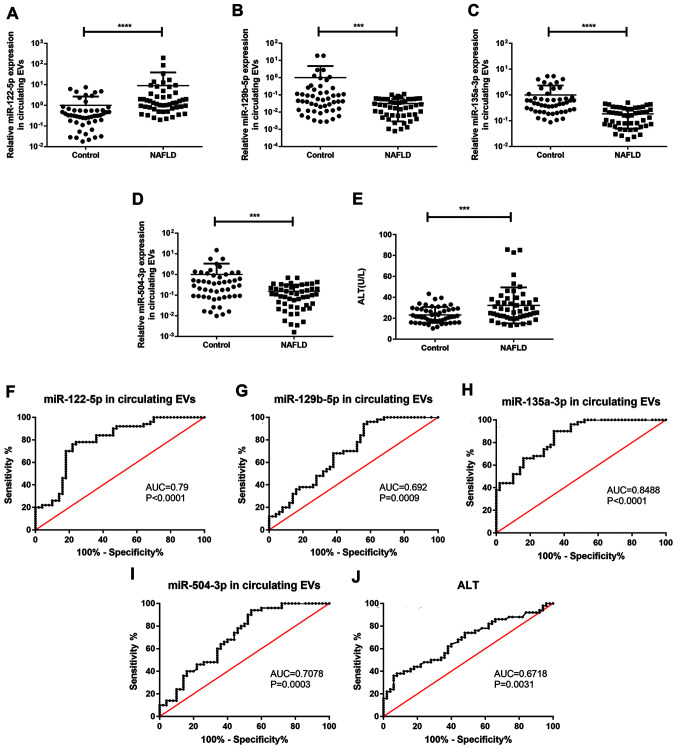
As EVs protect circulating miRNA from degradation by RNase, it was hypothesized that miRNA expression levels in EVs from serum would detect NAFLD progression more effectively compared with those in total serum. Using RT-qPCR and cel-miR-39 as an exogenous normalizer, it was found that miR-135a-3p, miR-129b-5p and miR-504-3p expression levels in EVs isolated from the serum of patients with NAFLD were markedly decreased compared with in the healthy controls, confirming this hypothesis. In addition, miR-122-5p in circulating EVs and serum ALT expression levels were significantly increased in patients with NAFLD (Fig. 4A-E). In order to evaluate the diagnostic potential of candidate miRNAs, ROC curve analysis was performed. miR-135a-3p exhibited the highest area under the curve (AUC; 0.849; 95% CI, 0.777–0.921), while miR-122-5p and miR-504-3p had AUC values of 0.790 (95% CI, 0.700–880) and 0.708 (95% CI, 0.606–0.810), respectively. The AUC of the traditional diagnostic indicator ALT was 0.672 (95% CI, 0.567–0.778; Fig. 4F-J). These data indicated that miR-135a-3p in EVs may be a promising diagnostic indicator and were more sensitive than ALT.
Figure 4.
Expression levels and ROC curves of screened miRs in serum EVs. Reverse transcription-quantitative PCR was used to determine mRNA expression levels of (A) miR-122-5p, (B) miR-129-5p, (C) miR-135a-3p and (D) miR-504-3p in serum EVs and (E) serum ALT levels in patients with NAFLD (n=50) and control subjects (n=50). ROC curves and AUC for serum EV-derived (F) miR-122-5p, (G) miR-135a-3p, (H) miR-129-5p, (I) miR-504-3p and (J) ALT levels in patients with NAFLD. Data are presented as the mean ± SD. ***P<0.001, ****P<0.0001. ROC, receiver operator characteristic; miR, microRNA, EVs, extracellular vesicles; AUC, area under the curve; ALT, alanine aminotransferase; NAFLD, non-alcoholic fatty liver disease.
Subsequently, the presence of these miRNAs in serum from patients with NAFLD was investigated. miR-135a-3p expression levels were markedly lower, whereas those of miR-122-5p were notably increased in patients with NAFLD compared with healthy controls. However, there was no notable difference in expression levels of miR-129-5p and miR-504-3p in serum between patients with NAFLD and healthy controls (Fig. S2A-D). Thus, in NAFLD, some miRNAs were specifically enriched in EVs, and detection of serum exosomes was a more favorable diagnostic method than direct detection of serum miRNA for NAFLD.
Biological function analysis
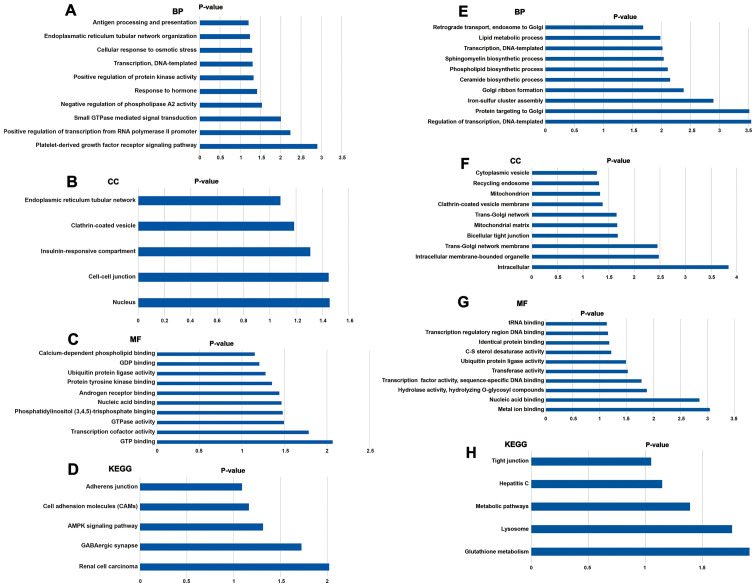
KEGG and GO analysis were used to investigate the key biological associations, signaling pathways and molecular functions of target genes of miR-135a-3p and miR-504-3p. For miR-135a-3p, the most significantly enriched GO terms were associated with ‘platelet-derived growth factor receptor signaling pathway’, ‘cell-cell junction’ and ‘GTP binding’ (Fig. 5A-C). The significantly enriched KEGG pathways had functions associated with ‘AMP-activated protein kinase (AMPK) signaling pathway’ (Fig. 5D). The target genes of miR-504-3p were significantly enriched in the GO terms ‘nucleotide-binding, oligomerization domain-like receptor signaling pathway’, ‘glutamatergic synapse’, ‘inflammatory responses’ and ‘ion transmembrane transport’ (Fig. 5E-G). According to the KEGG enrichment analysis, the most significantly enriched pathway for the target genes of miR-504-3p was ‘glutathione metabolism’ (Fig. 5H). Furthermore, ‘lysosome’ and ‘metabolic pathways’ were also highly enriched.
Figure 5.
Enriched GO terms and KEGG pathways of target genes. (A-C) Functional analysis of the potential targets of miR-135 using GO. (D) KEGG pathways enriched in miR-135 target genes. (E-G) GO function enrichment of miR-504 target genes. (H) KEGG pathway enrichment analysis of target genes of miR-504. BP, biological process; CC, cellular component; MF, molecular function; GO, Gene Ontology; KEGG, Kyoto Encyclopedia Genes and Genomes; miR, microRNA.
Discussion
NAFLD is a chronic liver disease and widely occurs in adult and children. During the past 20 years, the prevalence of NAFLD in China has doubled (3). The overall prevalence rate of NAFLD for adults in China has been ~15%, although there are regional variations (3). Moreover, the prevalence of NAFLD is 2.1% in children and 68.2% in obese children (3,5). Measurement of the serum AST/ALT ratio and AFP have been suggested as indicators of NAFLD; however, normal ALT or AFP levels can be detected in patients with steatosis and early-stage HCC (29–31). There are limitations to the existing diagnostic approaches (such as unfeasible serum markers and imaging techniques); therefore, tissue biopsy continues to be the primary NASH diagnostic standard (32–35), although biopsy is invasive and sampling errors can occur (36). In addition, biopsy cannot be used to predict disease progression (37). Thus, potential stable and non-invasive markers for liver disease progression are increasingly investigated.
During lipotoxicity, hepatocytes release a high number of EVs that affect various target cells (including macrophages, Kuffer cells and hepatic stellate cells) and improve key processes associated with NAFLD pathogenesis (such as fibrosis, angiogenesis and immune regulation) (38,39). EVs generated and released during NAFLD progression exhibit a special antigenic composition (such as C-X-C motif chemokine ligand 10, C16:0 ceramide and inositol requiring enzyme 1α), which reflects characteristic pathological changes in its progression, the number of miRNAs and proteins released from the liver increases (40). In addition, the level of EVs is dynamic and associated with variations in liver histopathology. Thus, in the present study well-defined and stable miRNAs were investigated to demonstrate whether Evs-derived miRNAs may be effective non-invasive biomarkers to determine NAFLD progression.
Previous studies have revealed that serum miR-16, miR-34a, miR-21 and miR-122 expression levels increase in patients with NAFLD (15,41). Furthermore, circulating EVs exhibit increased expression levels of asialoglycoprotein receptor 1, carboxylesterase 1 and liver miRNAs (miR-192 and miR-122) in patients with NAFLD, while miR-192 and miR-122 expression levels decrease in the liver (42–44). EVs protect miRNAs from RNase degradation (18); therefore, both miRNA biological marker sensitivity and specificity may be enhanced by enriching EVs.
In the present study, EV-derived miR-504-3p and miR-135a-3p expression levels were notably lower in patients with NAFLD compared with healthy controls. ROC analysis indicated that EV-derived miR-135a-3p and miR-504-3p may be sensitive biological markers for diagnosis of NAFLD, particularly miR-135a-3p, which exhibited the highest AUC and was more sensitive than ALT. However, there was no significant difference in serum circulating miR-504-3p expression levels between patients with NAFLD and healthy individuals. Total miRNAs from circulating serum may contain endogenous cellular miRNAs originating from debris from various cells; cell lysis and artifacts may influence expression levels of serum circulating miRNA (15). Recent studies have shown that exosomal miRNA is stable in serum because exosomes serve a protective function against degradation from enzymes, such as RNAse (15,18). Therefore, most studies use EVs to enhance the sensitivity and specificity of miRNA biological markers (45,46). Regarding the difference between miRNAs from hepatocytes and hepatocyte-derived EVs, a more notable change was observed between patients with NAFLD and healthy individuals in hepatocyte-derived EVs, suggesting that EVs derived from hepatocytes may selectively increase the expression of certain miRNAs to reflect the condition of the body (47). Hence, miRNAs from hepatocyte-derived EVs may be a more sensitive biological marker than miRNAs from hepatocytes. When comparing the expression level patterns of miRNAs from serum EVs and hepatocyte-derived EVs, consistency was found between these two indicators. As serum EVs are more accessible, miRNAs derived from serum EVs may be used to determine NAFLD progression. Consistent with previous studies (48,49), miR-122-5p was increased in serum EVs and decreased in the liver of mice fed HFD. Hepatocytes may serve as a key source of circulating EVs and miR-122 is predominantly encapsulated in EVs in animals with NAFLD (42). Following stress or damage, hepatocytes release these miRNAs in EVs during NAFLD progression, which may explain the decrease in the expression levels of miR-122 in liver from patients with NAFLD (49). In renal fibrosis, the expression levels of exosomal miR-29c are negatively associated with tubulointerstitial fibrosis score (50). Severity of liver injury also dictates EV-derived miRNA compartmentalization (51). Thus, it is important to investigate the use of EVs to reflect the activity and severity of NAFLD in future.
Hepatic cells may simultaneously serve as an EV-mediated intercellular source of communication and as a target (52). The effect of EV-derived miR-135a-3p and miR-504-3p is important during the development of NAFLD. Dysregulated expression of miR-504 is found in numerous types of tumor (such as HCC). Overexpression of miR-504 attenuates the invasion and proliferation of HCC cells by targeting frizzled class receptor 7, thus inhibiting the Wnt/β-catenin signaling pathway, which is key to the metabolism and pathology of the liver (53,54). A tumor suppressing effect of miR-504 in HCC, mediated by negatively regulating the expression levels of CDK16, has also been reported by Li et al (55). In addition to miR-504, miR-135a also suppresses the development of tumors by suppressing C-C motif chemokine receptor 2 expression levels in ovarian cancer (56). Furthermore, miR-135a can predict and prognose patients with urothelial carcinoma of the bladder (57). The results of the present study confirmed the diagnostic potential of miR-135a-3p and miR-504-3p in NAFLD. In addition, KEGG pathway analysis predicted the potential roles of miR-135a-3p and miR-504-3p in regulating the AMPK and metabolic pathways, which are involved in NAFLD (58).
However, there are certain limitations to the present study. NAFLD is not an isolated disease but can occur as a complication of insulin resistance and other metabolic disorders (59). Thus, a variety of tissues (such as pancreas, muscle and adipose tissue) are impacted by the associated oxidative stress and fatty acid flux (59), thereby promoting immune cell activation and migration to these sites. Therefore, extrahepatic release of EVs may hide NAFLD pathogenesis. In addition, there is a lack of specific liver molecular markers; further studies are required to investigate the association between circulating EVs and its source from the liver. Furthermore, there was a small number of subjects enrolled into the present study; a prospective cohort study is required to verify the diagnostic value of miR-135a-3p and miR-504-3p. In addition, the target genes of the selected miRNAs and their roles in the development of NAFLD were not investigated. The functions of miR-135a-3p and miR-504-3p should be determined in future studies.
In summary, the present study suggested that downregulated expression levels of circulating EV-derived miR-135a-3p was a potential novel and non-invasive marker to diagnose NAFLD; however further investigation is required.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Gongqian Jiao (The First Affiliated Hospital of Nanjing Medical University) for providing serum samples from patients with NAFLD.
Glossary
Abbreviations
- RT-q
reverse transcription-quantitative
- TEM
transmission electron microscopy
- NTA
nanoparticle tracking analysis
- miR
microRNA
- CD
chow diet
- HFD
high fat diet
- EVs
extracellular vesicles
- AMPK
AMP-activated protein kinase
- TSG101
tumor susceptibility 101
- HSP70
heat shock protein 70
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant nos. 81830023 and 81600640).
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81830023 and 81600640).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TY and QF worked on the conception and design of the study, HMJ wrote the manuscript. HMJ, YQ, ZYS, YQH and SC performed the experiments. HMJ, YWL, RG and MS collected and analyzed data. HMJ, SC and MS confirmed the authenticity of all the raw data. All authors contributed to the preparation of the manuscript, and read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committees of Nanjing Medical University approved the present study and all participants provided written informed consent. All animal experiments in this study were approved by The Animal Experiment Ethics Committee of Nanjing Medical University (approval no. IACUC-1601176).
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing interests.
References
- 1.Albhaisi S, Sanyal A. Recent advances in understanding and managing non-alcoholic fatty liver disease. F1000Res. 2018;7 doi: 10.12688/f1000research.14421.1. F1000 Faculty Rev-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucas C, Lucas G, Lucas N, Krzowska-Firych J, Tomasiewicz K. A systematic review of the present and future of non-alcoholic fatty liver disease. Clin Exp Hepatol. 2018;4:165–174. doi: 10.5114/ceh.2018.78120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi Z. Non-alcoholic fatty liver disease-A global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Fan JG, Farrell GC. Epidemiology of non-alcoholic fatty liver disease in China. J Hepatol. 2009;50:204–210. doi: 10.1016/j.jhep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Fan JG. Epidemiology of alcoholic and nonalcoholic fatty liver disease in China. J Gastroenterol Hepatol. 2013;28(Suppl 1):S11–S17. doi: 10.1111/jgh.12036. [DOI] [PubMed] [Google Scholar]
- 6.Adams LA, Harmsen S, St Sauver JL, Charatcharoenwitthaya P, Enders FB, Therneau T, Angulo P. Nonalcoholic fatty liver disease increases risk of death among patients with diabetes: A community-based cohort study. Am J Gastroenterol. 2010;105:1567–1573. doi: 10.1038/ajg.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsiki N, Athyros VG, Karagiannis A, Mikhailidis DP. Managing the combination of non-alcoholic fatty liver disease and metabolic syndrome. Expert Opin Pharmacother. 2012;13:287–290. doi: 10.1517/14656566.2012.644449. [DOI] [PubMed] [Google Scholar]
- 8.Byrne CD, Targher G. NAFLD: A multisystem disease. J Hepatol. 2015;62(1 Suppl):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Mazhar K. The future of nonalcoholic fatty liver disease treatment. Med Clin North Am. 2019;103:57–69. doi: 10.1016/j.mcna.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 10.He Z, Yang JJ, Zhang R, Li HT, Wu L, Jiang F, Jia WP, Hu C. Circulating miR-29b positively correlates with non-alcoholic fatty liver disease in a Chinese population. J Dig Dis. 2019;20:189–195. doi: 10.1111/1751-2980.12716. [DOI] [PubMed] [Google Scholar]
- 11.DiStefano JK, Gerhard GS. Circulating microRNAs in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2016;10:161–163. doi: 10.1586/17474124.2016.1125290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99–103. doi: 10.1016/j.cca.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Liu CH, Ampuero J, Gil-Gomez A, Montero-Vallejo R, Rojas Á, Muñoz-Hernández R, Gallego-Durán R, Romero-Gómez M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J Hepatol. 2018;69:1335–1348. doi: 10.1016/j.jhep.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonso MB, Rodrigues PM, Simao AL, Castro RE. Circulating microRNAs as potential biomarkers in non-alcoholic fatty liver disease and hepatocellular carcinoma. J Clin Med. 2016;5:30. doi: 10.3390/jcm5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Deng F, Yang S, Wang D, Chen X, Zhong S, Zhao J, Tang J. Exosomal microRNA: A novel biomarker for breast cancer. Biomark Med. 2018;12:177–188. doi: 10.2217/bmm-2017-0305. [DOI] [PubMed] [Google Scholar]
- 17.Huang G, Lin G, Zhu Y, Duan W, Jin D. Emerging technologies for profiling extracellular vesicle heterogeneity. Lab Chip. 2020;20:2423–2437. doi: 10.1039/D0LC00431F. [DOI] [PubMed] [Google Scholar]
- 18.Ibáñez F, Ureña-Peralta JR, Costa-Alba P, Torres JL, Laso FJ, Marcos M, Guerri C, Pascual M. Circulating MicroRNAs in extracellular vesicles as potential biomarkers of alcohol-induced neuroinflammation in adolescence: Gender differences. Int J Mol Sci. 2020;21:6730. doi: 10.3390/ijms21186730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48–56. doi: 10.1038/cgt.2016.77. [DOI] [PubMed] [Google Scholar]
- 20.Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155–161. doi: 10.1002/jhbp.524. [DOI] [PubMed] [Google Scholar]
- 21.Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018;15:9584–9592. doi: 10.3892/ol.2018.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Ni M, Su Y, Wang H, Zhu S, Zhao A, Li G. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur Urol Focus. 2018;4:412–419. doi: 10.1016/j.euf.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Fu Q, Li Y, Jiang H, Shen Z, Gao R, He Y, Liu Y, Xu K, Yang T. Hepatocytes derived extracellular vesicles from high-fat diet induced obese mice modulate genes expression and proliferation of islet β cells. Biochem Biophys Res Commun. 2019;516:1159–1166. doi: 10.1016/j.bbrc.2019.06.124. [DOI] [PubMed] [Google Scholar]
- 24.Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Mastroiaco V, Vetuschi A, Sferra R, Barnabei R, et al. MicroRNA expression analysis in high fat diet-induced NAFLD-NASH-HCC progression: Study on C57BL/6J mice. BMC Cancer. 2016;16:3. doi: 10.1186/s12885-015-2007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasci I, Mas MR, Vural SA, Deveci S, Comert B, Alcigir G, Mas N, Akay C, Bozdayi M, Yurdaydin C, et al. Pegylated interferon-alpha plus taurine in treatment of rat liver fibrosis. World J Gastroenterol. 2007;13:3237–3244. doi: 10.3748/wjg.v13.i23.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA, NASH Clinical Research Network (CRN) Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: Distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loréal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Angulo P. GI epidemiology: Nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2007;25:883–889. doi: 10.1111/j.1365-2036.2007.03246.x. [DOI] [PubMed] [Google Scholar]
- 30.Mofrad P, Contos MJ, Haque M, Sargeant C, Fisher RA, Luketic VA, Sterling RK, Shiffman ML, Stravitz RT, Sanyal AJ. Clinical and histologic spectrum of nonalcoholic fatty liver disease associated with normal ALT values. Hepatology. 2003;37:1286–1292. doi: 10.1053/jhep.2003.50229. [DOI] [PubMed] [Google Scholar]
- 31.Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, et al. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30:871–873. doi: 10.1093/annonc/mdy510. [DOI] [PubMed] [Google Scholar]
- 32.Deffieux T, Gennisson JL, Bousquet L, Corouge M, Cosconea S, Amroun D, Tripon S, Terris B, Mallet V, Sogni P, et al. Investigating liver stiffness and viscosity for fibrosis, steatosis and activity staging using shear wave elastography. J Hepatol. 2015;62:317–324. doi: 10.1016/j.jhep.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Khov N, Sharma A, Riley TR. Bedside ultrasound in the diagnosis of nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20:6821–6825. doi: 10.3748/wjg.v20.i22.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers RP, Pomier-Layrargues G, Kirsch R, Pollett A, Beaton M, Levstik M, Duarte-Rojo A, Wong D, Crotty P, Elkashab M. Discordance in fibrosis staging between liver biopsy and transient elastography using the FibroScan XL probe. J Hepatol. 2012;56:564–570. doi: 10.1016/j.jhep.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Myers RP, Pomier-Layrargues G, Kirsch R, Pollett A, Duarte-Rojo A, Wong D, Beaton M, Levstik M, Crotty P, Elkashab M. Feasibility and diagnostic performance of the FibroScan XL probe for liver stiffness measurement in overweight and obese patients. Hepatology. 2012;55:199–208. doi: 10.1002/hep.24624. [DOI] [PubMed] [Google Scholar]
- 36.Angulo P. Long-term mortality in nonalcoholic fatty liver disease: Is liver histology of any prognostic significance? Hepatology. 2010;51:373–375. doi: 10.1002/hep.23677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dongiovanni P, Meroni M, Longo M, Fargion S, Fracanzani AL. miRNA Signature in NAFLD: A turning point for a non-invasive diagnosis. Int J Mol Sci. 2018;19:3966. doi: 10.3390/ijms19123966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorairaj V, Sulaiman S, Abu N, Abdul Murad N. Extracellular vesicles in the development of the non-alcoholic fatty liver disease: An update. Biomolecules. 2020;10:1494. doi: 10.3390/biom10111494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banales JM, Feldstein AE, Sanger H, Lukacs-Kornek V, Szabo G, Kornek M. Extracellular vesicles in liver diseases: Meeting report from the international liver congress 2018. Hepatol Commun. 2019;3:305–315. doi: 10.1002/hep4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Martinez I, Alen R, Rada P, Valverde AM. Insights into extracellular vesicles as biomarker of NAFLD pathogenesis. Front Med (Lausanne) 2020;7:395. doi: 10.3389/fmed.2020.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuka M, Kishikawa T, Yoshikawa T, Yamagami M, Ohno M, Takata A, Shibata C, Ishibashi R, Koike K. MicroRNAs and liver disease. J Hum Genet. 2017;62:75–80. doi: 10.1038/jhg.2016.53. [DOI] [PubMed] [Google Scholar]
- 42.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Povero D, Eguchi A, Niesman IR, Andronikou N, de Mollerat du Jeu X, Mulya A, Berk M, Lazic M, Thapaliya S, Parola M, et al. Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Sci Signal. 2013;6:ra88. doi: 10.1126/scisignal.2004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35:532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18:5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 46.Mori MA, Ludwig RG, Garcia-Martin R, Brando BB, Kahn CR. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang F, Chen Q, Wang W, Ling Y, Yan Y, Xia P. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J Hepatol. 2020;72:156–166. doi: 10.1016/j.jhep.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lv LL, Cao YH, Ni HF, Xu M, Liu D, Liu H, Chen PS, Liu BC. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am J Physiol Renal Physiol. 2013;305:F1220–F1227. doi: 10.1152/ajprenal.00148.2013. [DOI] [PubMed] [Google Scholar]
- 51.Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devhare PB, Ray RB. Extracellular vesicles: Novel mediator for cell to cell communications in liver pathogenesis. Mol Aspects Med. 2018;60:115–122. doi: 10.1016/j.mam.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quan H, Li B, Yang J. MicroRNA-504 functions as a tumor suppressor in hepatocellular carcinoma through inhibiting Frizzled-7-mediated-Wnt/β-catenin signaling. Biomed Pharmacother. 2018;107:754–762. doi: 10.1016/j.biopha.2018.07.150. [DOI] [PubMed] [Google Scholar]
- 54.Tian Y, Mok MT, Yang P, Cheng AS. Epigenetic activation of Wnt/β-catenin signaling in NAFLD-associated hepatocarcinogenesis. Cancers (Basel) 2016;8:76. doi: 10.3390/cancers8080076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol. 2018;52:1603–1612. doi: 10.3892/ijo.2018.4313. [DOI] [PubMed] [Google Scholar]
- 56.Duan S, Dong X, Hai J, Jiang J, Wang W, Yang J, Zhang W, Chen C. MicroRNA-135a-3p is downregulated and serves as a tumour suppressor in ovarian cancer by targeting CCR2. Biomed Pharmacother. 2018;107:712–720. doi: 10.1016/j.biopha.2018.08.044. [DOI] [PubMed] [Google Scholar]
- 57.Inamoto T, Uehara H, Akao Y, Ibuki N, Komura K, Takahara K, Takai T, Uchimoto T, Saito K, Tanda N, et al. A Panel of MicroRNA signature as a tool for predicting survival of patients with urothelial carcinoma of the bladder. Dis Markers. 2018;2018:5468672. doi: 10.1155/2018/5468672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esquejo RM, Salatto CT, Delmore J, Albuquerque B, Reyes A, Shi Y, Moccia R, Cokorinos E, Peloquin M, Monetti M, et al. Activation of Liver AMPK with PF-06409577 Corrects NAFLD and lowers cholesterol in rodent and primate preclinical models. EBioMedicine. 2018;31:122–132. doi: 10.1016/j.ebiom.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watt MJ, Miotto PM, De Nardo W, Montgomery MK. The liver as an endocrine organ-linking NAFLD and insulin resistance. Endoc Rev. 2019;40:1367–1393. doi: 10.1210/er.2019-00034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.