Abstract
Rho guanine nucleotide exchange factors (RhoGEFs) integrate cell signaling inputs into morphological and functional responses. However, little is known about the endothelial repertoire of RhoGEFs and their regulation. Thus, we assessed the expression of 81 RhoGEFs (70 homologous to Dbl and 11 of the DOCK family) in endothelial cells. Further, in the case of DH-RhoGEFs, we also determined their responses to VEGF exposure in vitro and in the context of tumors. A phylogenetic analysis revealed the existence of four groups of DH-RhoGEFs and two of the DOCK family. Among them, we found that the most abundant endothelial RhoGEFs were: Tuba, FGD5, Farp1, ARHGEF17, TRIO, P-Rex1, ARHGEF15, ARHGEF11, ABR, Farp2, ARHGEF40, ALS, DOCK1, DOCK7 and DOCK6. Expression of RASGRF2 and PREX2 increased significantly in response to VEGF, but most other RhoGEFs were unaffected. Interestingly murine endothelial cells isolated from tumors showed that all four phylogenetic subgroups of DH-RhoGEFs were altered when compared to non-tumor endothelial cells. In summary, our results provide a detailed assessment of RhoGEFs expression profiles in the endothelium and set the basis to systematically address their regulation in vascular signaling.
Keywords: RhoGEF, Rho GTPases, Rac, Cdc42, VEGF, Endothelial cell cytoskeleton, Tumor angiogenesis
Graphical Abstract
1. Introduction
Angiogenesis requires dynamic adjustments in cell morphology that rely on the tight control of cytoskeletal proteins [1]. The integration of angiogenic signals determines the specific sites where membrane projections must be generated in orchestration with concurrent retractions at the opposite pole of the cell. This process is further associated with spatially controlled adhesion and release from extracellular substrates. Altogether, these changes determine how endothelial cells acquire a sprouting phenotype and later develop a tubular morphology in response to external angiogenic cues [2-5]. In this manner, continuous cytoskeletal adjustments occur throughout the angiogenic process, including sprouting, cell migration, tubulogenesis and stabilization [6,7].
The plasticity of the cytoskeleton is partially granted by a group of molecular switches that provide rapid responses in precise subcompartments. In particular, Rho GTPases are included in this category. These molecules are essential regulators of cytoskeletal dynamics, and therefore, their participation throughout the angiogenic response is most likely essential [8,9]. Filopodia emerge from tip cells in response to agonists that promote the activation of Cdc42, a Rho GTPase particularly linked to cell polarity that has been implicated in lumen morphogenesis [10,11]. Whereas lamellipodia are continuously assembled at sites where Rac is activated and stress fibers are formed in response to RhoA. These cytoskeletal dynamics controlled by Rho GTPases are further refined by the existence of twenty different Rho GTPases, whose activation is catalyzed by guanine nucleotide exchange factors (GEFs) [12-14].
RhoGEFs, constitute a group of several tens of complex multi-domain proteins that interact with inactive GDP-bound Rho GTPases promoting the release of GDP, which is exchanged for GTP, turning Rho GTPases into an active conformation with high affinity for their cognate cytoskeletal-remodeling effectors. Changes in expression of some of these molecules have been linked to cancer progression [13]. Thus, RhoGEFs are currently being explored as therapeutic targets, particularly through the use of small molecules that inhibit their catalytic domain [15-19]. As part of an angiogenic response, RhoGEFs are likely to integrate different inputs to translate these molecular cues into a responsive cellular morphology [20]. The most diverse family of RhoGEFs includes proteins with homology to Dbl. This group according to the SMART database [21] includes sixty three members encoded in the human genome. In addition there are seven members that are homologous to Dbl [13,14,22]. The second group of Cdc42 and Rac activators includes eleven proteins of the DOCK family, a subgroup that works in a functional complex with ELMO scaffolds [23]. Therefore, to systematically assess the role of Rho GTPases at every step of the angiogenic process, it is critical to initially determine their expression pattern in the endothelium and its responses to angiogenic signals.
The overall importance of RhoGEFs in the vascular system has been recently highlighted by reports analyzing their expression in arteries and vascular smooth muscle cells in the context of hypertension [24] and in endothelial cells by mining public microarray data [25]. Specifically, Cario-Toumaniantz, and colleagues analyzed the expression of twenty eight RhoGEFs specific for RhoA. The study reported that sixteen of these were decreased in hypertensive rats and a subgroup of nine were downregulated in vascular smooth muscle cells treated with Angiotensin II via a pathway that required Rho kinase [24]. A second study used multiple microarrays from public databases to compare the expression of RhoGEFs and RhoGAPs in human endothelial cells and other cell types [25]. This in silico analysis highlighted that a group of RhoGEFs was particularly enriched in the endothelium. They discussed their findings in the context of endothelial adhesion and revealed that many potentially important regulators of this process have not been studied in endothelial cells [25]. Accordingly, here we undertook a systematic approach focused directly on the expression of RhoGEFs to define the repertoire of potential direct activators of Rho GTPases endogenously expressed in various endothelial cell types. We also explored whether changes were noted in response to VEGF and in context of the tumor microenvironment.
2. Material and methods
2.1. Bioinformatic tools
The list of human and mouse RhoGEFs with a predicted Dbl-homology domain was obtained from the SMART genomic database (http://smart. embl-heidelberg.de) [21]. Seven additional DH-RhoGEFs not included in the SMART database were also incorporated in our analysis based on a previously reported comprehensive list [22]. The corresponding sequences of cDNAs and open reading frames of DH-RhoGEFs, as well as members of the DOCK family and ELMO scaffolds were obtained from the NCBI/nucleotide database and used to design primers with the Primer 3 web-based interface (http://bioinfo.ut.ee/primer3-0.4.0/primer3/). The specificity of each pair of selected primers was confirmed by BLAST (http://www.blast.ncbi.nlm.nih.gov/). The phylogenetic relations among the multi-domain family of DH-RhoGEFs and DOCKs were revealed by comparison of the protein sequence of their DH domains (also available at SMART) or their DHR2, respectively, aligned with ClustalW (http:// www.ebi.ac.uk/Tools/msa/clustalw2/) and visualized as a phylogenetic tree with the FigTree v1.4.0 platform (http://tree.bio.ed.ac.uk/software/ figtree/). The structures of representative complexes of DH-PH modules crystallized with their cognate paradigmatic Rho GTPases (RhoA, Rac1 and Cdc42) and the DHR2 domains for the DOCK GEFs in complex with Rac or Cdc42 were analyzed with the CN3D program, available from the NCBI web site (www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). This program was used to identify the amino acids at the ITSN1/Cdc42-, ARHGEF11/RhoA-, Vav1/Rac1-, DOCK2/Rac1- and DOCK9/Cdc42-contact interfaces within 3 Å using the following structures available at the NCBI/structure web page (http://www.ncbi.nlm.nih.gov/structure/): ITSN1/Cdc42 (MMDB ID: 97,009) [26], ARHGEF11/RhoA (MMDB ID: 31,086) [27], Vav1/Rac1 (MMDB ID: 65,476) [28], DOCK2/Rac1 (MMDB ID: 90,722) [29] and DOCK9/Cdc42 (MMDB ID: 76,807) [30].
2.2. Cell culture
Human microvascular endothelial cells (HMEC-1) were maintained in MCDB-131 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 25 mM L-glutamine (Invitrogen), 10 ng/ml epidermal growth factor (GIBCO cat. 13,247–051), 2.5 ng/ml bFGF basic fibroblast growth factor (R&D Systems, cat. 234-FSE/CF) and 1 μg/ml hydrocortisone (Sigma-Aldrich, St. Louis, MO). EGFP-tagged Lewis lung carcinoma cells (LLC-GFP) were cultured in Dulbecco's modified Eagle's Media (cat. D7777-10XL Sigma- Aldrich, St. Louis, MO) supplemented with 10% FBS. Human umbilical vein endothelial cells (HUVECs) were a kind gift from Dr. Alejandro Zentella (Biomedical Research Institute, National Autonomous University of Mexico). Endothelial cells from Immortomice (IMEC) were isolated from SV40-TAg-transgenic mice purchased from Charles River, as previously described [31] and grown in Dulbecco's modified Eagle's Media (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS fetal bovine serum (Invitrogen). HUVECs were used at passages 3 or 4, all other cultured endothelial cells were used within passages 4 to 6.
2.3. Isolation of tumor endothelial cells
Lewis lung tumors were generated by subcutaneous injection of nu/nu mice (6–8 weeks) with LLC-GFP cells (5 × 105 cells/100 μl). Tumors were allowed to develop for 16 days, at which time, mice were injected intravenously with Lycopersicon esculentum biotinylated-lectin (100 μl) (Vector laboratories). Tumors were surgically removed, fragmented and incubated in DMEM containing 0.5 mg/ml type I collagenase (Sigma, cat. C0130) at 37 °C for 15 min. Single cell suspensions were then washed with FBS-supplemented medium, followed by serum-free medium and incubated with Streptavidin-Dynabeads (100 μl) for 15 min on a rocker platform. Labeled endothelial cells were isolated with a magnetic column and washed three times prior to RNA extraction with the RNeasy kit (Qiagen). Experiments with mice were approved by the UCLA's Committee for Animal Research.
2.4. Transcript analysis of RhoGEFs
Expression of all DH-RhoGEFs listed in the human and mouse genomes, and human DOCKs and ELMOs, was assessed by RT-PCR using total RNA isolated from endothelial cells (TRIzol, Invitrogen) and retrotranscribed (Superscript lII kit, Invitrogen). In some cases, as indicated, HMEC-1 were stimulated with VEGF 165 (100 ng/ml; human, recombinant, Calbiochem cat. PF074). Importantly, prior to expression analysis of all human and mouse RhoGEFs, to confirm the quality and concentration of the cDNA used, each cDNA preparation was initially optimized. This optimization included the development of a curve of amplification of β-actin that was repeated with different dilutions of endothelial cDNA until samples provided equivalent levels of starting cDNA. In the case of samples prepared from tumor endothelial cells, controls to assess potential contamination from other cell types were performed. PCR assays were conducted using JumpStart RED Taq ReadyMix PCR Reaction Mix kit (Sigma-Aldrich). PCR products were evaluated by 2% agarose gels. Images were captured with a BioDoc-It imaging system (UVP Inc., Upland, CA).
2.5. Western blot analysis of selected RhoGEFs
Confluent cultures of HMEC-1 were incubated in the absence of serum for 12 h and stimulated or not with 100 ng/ml of VEGF for the indicated times. Total protein lysates were prepared with TBS-Triton (50 mM Tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1% Triton X-100) containing protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 mM β-glycerophosphate, 1 mM NaF and 1 mM sodium orthovanadate). Lysates were diluted with 4X Laemmli sample buffer containing β-mercaptoethanol and boiled for 5 min. Equal amounts of protein were resolved on SDS-PAGE gels, transferred to Immobilon membranes (Millipore), blocked with 5% non-fat dry milk in TBS-Tween and incubated overnight at 4 °C, on a rocking platform, with the following antibodies, at the indicated dilutions, in blocking buffer: LARG (H-70) (ARHGEF12, 1/1000), sc-25,638; p115/Lsc RhoGEF (H-165) (ARHGEF1, 1/1000), sc-20,804; Trio (H-120) (1/3000), sc-28,564, all from SANTA CRUZ BIOTECHNOLOGY; or in TBS-T: Anti-β-PIX (ARHGEF7, 1/5000) Chemicon; Lfc (K-17) (GEF-H1-ARHGEF2, 1/5000), sc-9334, SANTA CRUZ BIOTECHNOLOGY; PREX1 (1/5000) HPA001927, SIGMA; Intersectin1 (1/3000) ab118262, Abcam; Vav2 (C64H2) (1/5000) #2848, Cell Signaling Technology. Secondary antibodies were incubated in blocking buffer for 1 h and developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore).
2.6. Statistical analysis
Densitometric quantitation was performed with ImageJ software. Data are presented as the means and SEM of three to five independent experiments as indicated in figure legends. Statistical analysis was performed using Prism software V6.0. Statistical significance was considered for values of p<0.05 using the Student's t test for paired data or one-way ANOVA followed by Bonferroni's multiple comparison test.
3. Results
3.1. Endothelial expression of DH-RhoGEFs
To investigate endothelial expression of all RhoGEFs homologous to Dbl encoded in the human and mouse genomes, we initially considered all proteins with an identifiable consensus Dbl-homologous domain (DH-domain) as listed by the genomic SMART database (http://smart.embl-heidelberg.de) [21]. We also included 7 additional Dbl-homologous RhoGEFs not listed in the SMART database [22], 11 GEFs of the DOCK family and three ELMO scaffolds [23]. The complete coding sequence of each one was obtained from NCBI database and optimal primers were designed with Primer 3, a free online primer design tool (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (supplemental Table 1). Expression of the initial group of sixty three RhoGEFs was assessed in human microvascular endothelial cells (HMEC-1). As shown in Fig. 1, forty seven of these DH-RhoGEFs were detected by RT-PCRs after thirty cycles of amplification. Interestingly, the most abundant RhoGEFs have been only superficially studied in the endothelium and, in several cases, little information was available in the literature (Fig. 1, right panel). Primers that gave no amplification on endothelial RNA were further tested for their efficiency in other cell types to determine their ability to amplify endogenously expressed RhoGEFs (results not shown). Our secondary validation indicated that a subset of RhoGEFs was either not expressed by endothelial cells under non-stimulated conditions or was below the limits of detection of the PCR system used.
Fig. 1.
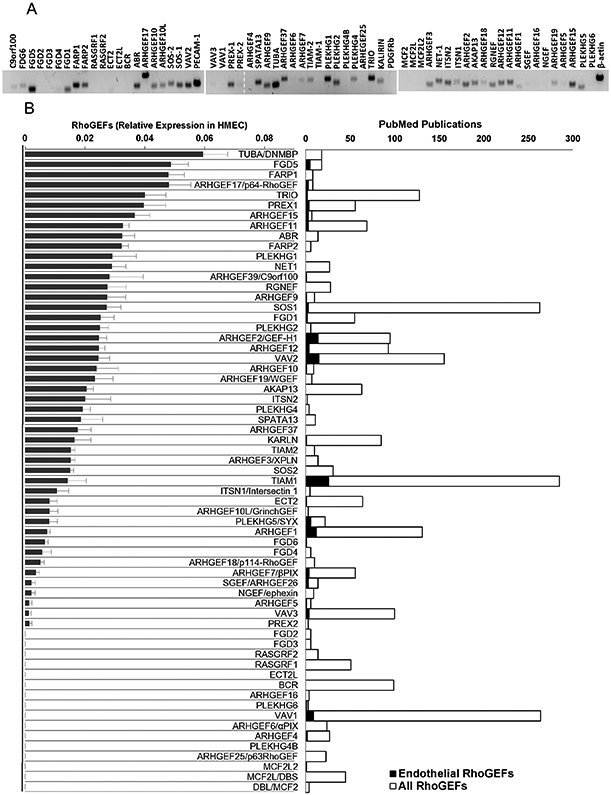
RhoGEFs (Rho guanine nucleotide exchange factors) expressed in endothelial HMEC-1. A, Representative gel showing the expression of 63 human RhoGEFs (containing a Dbl-homologous domain as listed by the genomic SMART database (http://smart.embl-heidelberg.de)) detected by RT-PCR (using 30 cycles of amplification) in serum starved endothelial HMEC-1. The primers used are listed in supplemental Table 1. Positive and negative controls included in the test were PECAM-1 (CD31, positive control), PDGFRb (negative control) and actin. B, The graph on the left shows the normalized expression of DH-RhoGEFs in endothelial HMEC-1, results from three independent experiments. The graph on the right shows the number of publications indexed in PubMed that mention, either in the title or abstract, the indicated RhoGEF, restricted or not to endothelial cells in the search. Statistical analysis for each of the phylogenetic subgroups of DH-RhoGEFs is presented in Figs. 3 to 8. For members of the DOCK family results, statistics are presented in Fig. 9.
3.2. Structural aspects and phylogenetic distribution of endothelial DH-RhoGEFs
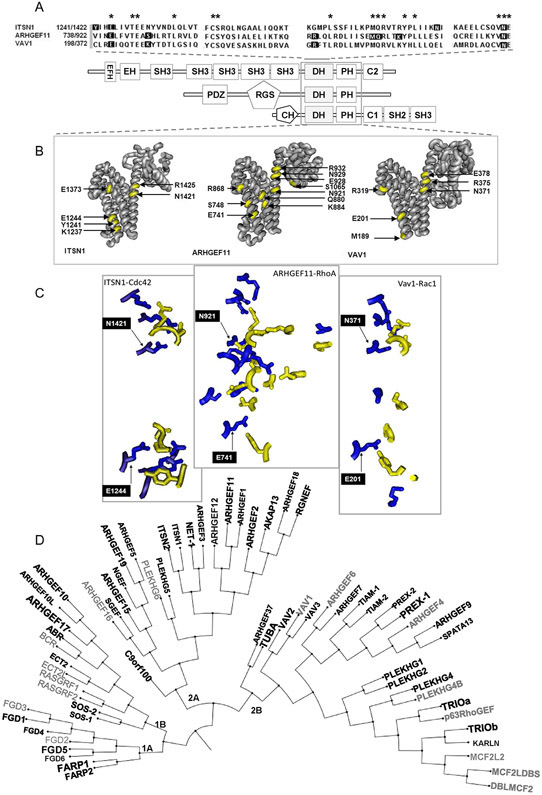
To gain additional insight into the physiological relevance of this high diversity of endothelial RhoGEFs, we first explored the potential interactions between well-characterized examples of Cdc42-, Rac- and Rho-GEFs, crystallized in complexes with their cognate GTPases. The overall structure DH-PH-module/GTPase complex of intersectin-1/ Cdc42, PDZ-RhoGEF (ARHGEF11)/RhoA and VAV1/Rac1 complexes, analyzed with CN3D, revealed that the three DH domains contain various conserved amino acids as expected from their consensus DH-sequence (Fig. 2A) and displayed a similar alpha helical structure (Fig. 2B and C). However, only a couple of conserved amino acids were present at their interfaces of interaction with their cognate GTPases, suggesting a wide potential variability on their interactions. As shown in Fig.2A, these two conserved amino acids (ITSN1-E1244 and -N1421; ARHGEF11-E741 and -N921 and VAV1-E201 and -N371), flanking the consensus DH domain, are more distant at the interacting interface between ITSN1/Cdc42 and Vav1/Rac1 with respect to ARHGEF11/RhoA (Fig. 2B and C). Thus, considering this diversity, we performed multiple alignments of the human DH domains and obtained the RhoGEF phylogenetic tree shown in Fig. 2D, in which those RhoGEFs detected in HMEC cells are indicated with black bold letters, whereas a bigger font indicates higher levels of expression of these RhoGEFs as shown in Fig. 1. This analysis revealed that the phylogenetic tree of RhoGEFs contains two main groups, each one subdivided into two branches with further subdivisions in each. All the four RhoGEF main branches 1A, 1B, 2A and 2B contributed to the diversity of RhoGEFs expressed by endothelial cells. We used this phylogenetic organization to systematically study endothelial RhoGEFs. Specifically, we assessed their expression patterns in human endothelial umbilical vein cells (HUVECs), endothelial cells isolated from Immortomice (IMEC) and endothelial cells isolated from Lewis lung carcinoma (LLC) tumors.
Fig. 2.
Phylogenetic organization of RhoGEFs based on the structure of their DH domains. A, Upper panel, alignment of the DH domains (catalytic domains) of ITSN1, ARHGEF11 and VAV1. Their predicted structures are shown under the aligned sequences. These RhoGEFs are known activators of Cdc42, RhoA and Rac1, respectively. Residues in the DH sequences shown on a black background correspond to those present at the interacting interface within 3 Å of distance between the DH domain and the GTPase of the complexes shown in B and C, in which only the interacting amino acids are shown, highlighting those conserved residues (E and N) that flank the DH domain. D, Phylogenetic tree of human RhoGEFs based on alignment of their DH domains. The RhoGEFs indicated in black fonts correspond to those whose expression was detected in HMEC-1 (shown in Fig. 1). Also the size of the font correspond to their relative level of expression (bigger size, higher expression). Listed in gray are those not detected in the endothelium.
3.3. Expression of DH-RhoGEFs in normal and tumor endothelial cells
The comparative expression analysis of the initial group of 63 DH-RhoGEFs from the SMART database is shown in Figs. 3-6. Importantly, the evaluation was performed in both normal and tumor endothelial cells. These figures also showed their predicted structural characteristics, highlighting the presence of the Dbl-homology (DH) domain.
Fig. 3.
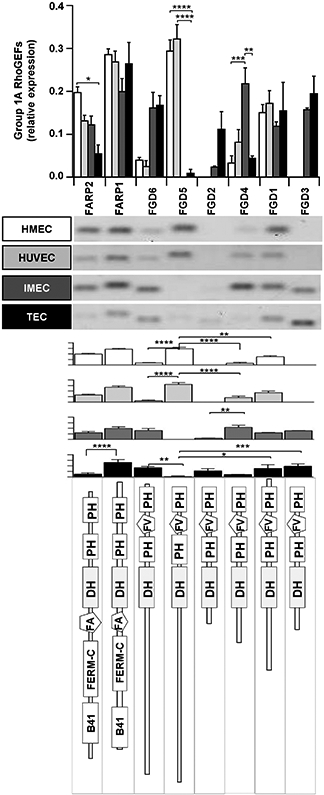
Comparative analysis of expression for group 1A of RhoGEFs (according to the phylogenetic tree shown in Fig. 2D). Shown are human (HMEC, HUVEC) and mouse endothelial cells (IMEC), and tumor-derived endothelial cells (TEC) isolated from LLC tumors. The results are shown in distinct graphs to facilitate comparison of individual RhoGEFs among different endothelial cells (upper graph) or the group of RhoGEFs in each endothelial cell type (graphs shown below representative RT-PCR results). The bottom panel shows the predicted structure of each RhoGEF. Normalized relative expression of this group of RhoGEFs is shown ±SEM (error bars) for n = 5 (HMEC-1), n = 3 (HUVEC), n = 3 (IMEC) and n = 4 (TEC) independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Fig. 6.
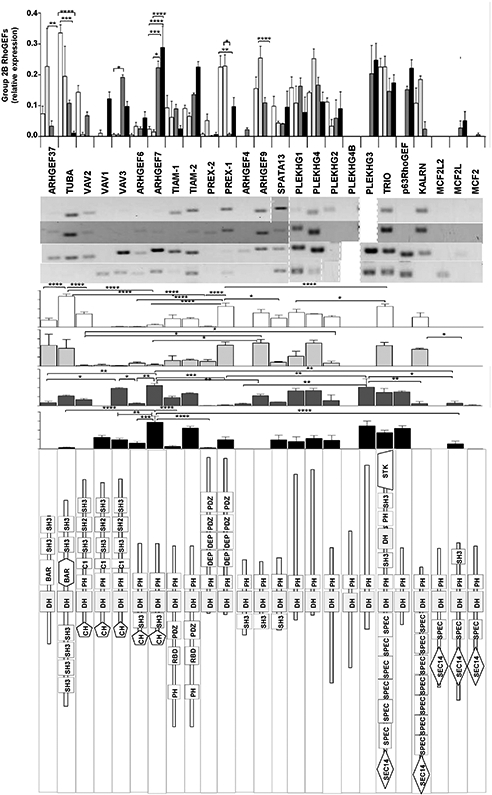
Expression profile for group 2B of RhoGEFs. The upper graph compares individual RhoGEFs among different endothelial cells. The images show representative RT-PCRs (middle panel) for the indicated endothelial cell types. The structure of this group of RhoGEFs is shown in the lower panel. Normalized relative expression is shown ±SEM (error bars) for n = 5 (HMEC-1), n = 3 (HUVEC), n = 3 (IMEC) and n = 4 (TEC) independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Group 1A includes eight RhoGEFs, structurally distinguishable as two types (Fig. 3): FARPs (1 and 2), both detectable in normal endothelial cells, and FGDs (1 to 6) which showed a diverse pattern of expression between human and mouse endothelial cells. FGD1 was similarly expressed, whereas FGD5 was highly expressed exclusively by human endothelial cells while FGD3 was present in mouse endothelium. The only RhoGEF in this group that was differentially expressed by tumor-derived endothelial cells was FGD2. In tumor endothelial cells, FARP2 was significantly reduced, whereas FARP1 and FGD3 were the most abundant.
Group 1B includes eleven RhoGEFs, nine of these were detected by at least one type of endothelial cell (Fig. 4). Structurally, four types of similar RhoGEFs are distinguishable in this branch: the first group contains RasGEF domains (Sos1/2 and RasGRF1/2), the second group contains a RhoGAP domain (BCR and ABR), the third group shows one GEF with two BRCT domains in tandem (ECT2) and the fourth group includes the remaining RhoGEFs, which putatively lack a PH domain. RasGRF1 and ECT2L were absent in all endothelial cells tested, whereas BCR was most abundant in murine endothelial cells and ARHGEF10 was differentially decreased in tumor endothelial cells (Fig. 4).
Fig. 4.
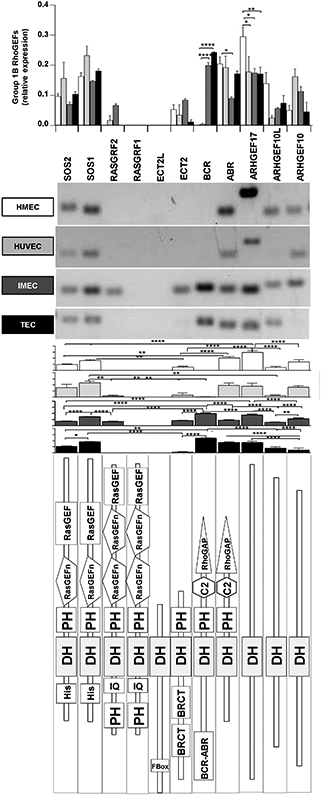
Endothelial expression for group 1B of RhoGEFs. The comparison of individual RhoGEFs among different endothelial cells is shown in the upper graph, whereas results from each endothelial cell type are presented below representative RT-PCRs. Subtypes of endothelial cells are indicated. The predicted structure of each RhoGEF is shown in the bottom panel. Normalized relative expression is shown ±SEM (error bars) for n = 5 (HMEC-1), n = 3 (HUVEC), n = 3 (IMEC) and n = 4 (TEC) independent experiments; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Group 2A comprises 20 RhoGEFs, 19 of them expressed by at least one type of endothelial cell (the only exception was ARHGEF16, which was not detectable at all) (Fig. 5). They can be classified into five structurally distinguishable groups: one formed by the very complex intersectins (ITSN1 and ITSN2) characterized by the presence of five SH3 domains at the amino terminal region, a second group constituted by the G12/13-protein regulated family of RGS-RhoGEFs (ARHGEF1 (p115RhoGEF), ARHGEF11 (PDZRhoGEF) and ARHGEF12 (LARG)), characterized by the presence of an RGS domain towards the amino-terminal extension of the DH-PH module, a third group characterized by the presence of an SH3 domain towards the carboxyl extension of the DH-PH module (ARHGEF16, SGEF, NGEF, ARHGEF19, and ARHGEF5), a fourth group constituted by three RhoGEFs with a C1 domain towards the amino of the DH-PH module (ARHGEF2 (GEFH1), AKAP13 and RGNEF), a fifth group having just the prototypical DH-PH module (c9orf100, PLEKHG6, PLEKHG5, NET1, ARHGEF3, and ARHGEF18) and a sixth group that only includes ARHGEF15 containing the defining DH domain but putatively lacking a PH domain. In tumor endothelial cells, the most highly expressed RhoGEFs were ARHGEF11 (PDZRhoGEF), intersectin2 (ITSN2) and AKAP13, whereas the expression of ARHGEF15, ARHGEF19, PLEKHG5 and ARHGEF2 (GEFH1) was strongly reduced.
Fig. 5.
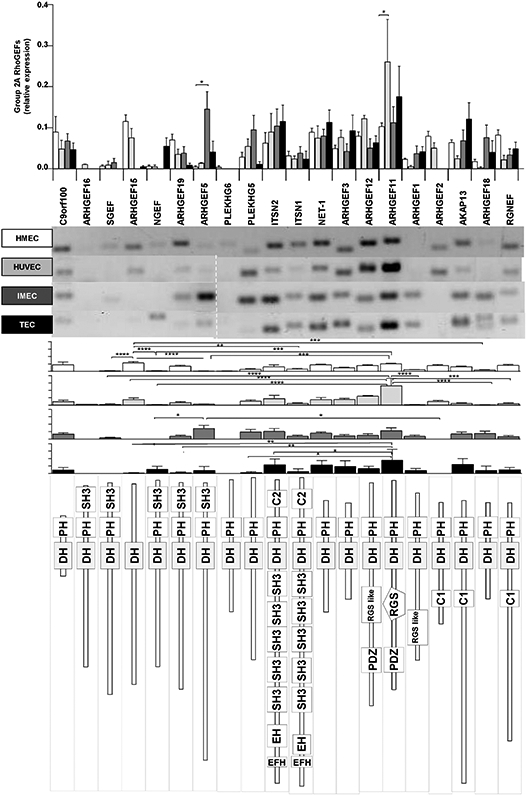
Comparative expression for group 2A of RhoGEFs. Expression of individual RhoGEFs compared among different endothelial cells is shown in the upper graph. Representative RT-PCR results are shown in the middle panel followed by histogram representation for each endothelial cell types (indicated by shades of grey). The predicted structure of each RhoGEF is displayed in the lower panel. Normalized relative expression is shown ± SEM (error bars) for n = 5 (HMEC-1), n = 3 (HUVEC), n = 3 (IMEC) and n = 4 (TEC) independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Group 2B includes 24 RhoGEFs, 21 of these were detected in at least one type of endothelial cell, whereas the three members of the group of MCF2 were absent (MCF2L2, MCF2L and MCF2) (Fig. 6). They can be classified into eight structurally distinguishable groups: the first includes the two RhoGEFs with a BAR domain (ARHGEF37 and Tuba), which is located in the region where commonly a PH domain is found in most DH-RhoGEFs. The second group includes the VAV family (VAV1 VAV2 and VAV3), characterized by the presence of a CH domain towards the amino-terminal extension of the DH-PH module and a carboxyl-terminal region containing C1, SH2 and SH3 domains. The third group also contains a CH domain, in this case preceding an SH3 domain that flanks the amino-terminal region of the DH-PH module (ARHGEF6 and ARHGEF7). The fourth group includes TIAM1 and TIAM2, characterized by an amino-terminal region containing a PH domain followed by RBD and PDZ domains preceding the DH-PH module. The fifth group is formed by P-Rex1 and P-Rex2, which are characterized by a complex amino-terminal region which, after the DH-PH module contains two DEP domains followed by two PDZ domains. The sixth group contains three RhoGEFs with an amino-terminal SH3 domain preceding the DH-PH module (ARHGEF4, ARHGEF9 and SPATA13). The seventh group includes 5 RhoGEFs with an amino SEC14 domain followed by one or various SPEC domains preceding the DH-PH module (TRIO, KALRN, MCFs) and the eighth group with four RhoGEFs that only contain the defining DH-PH module. In tumor endothelial cells, VAV1 and VAV2 were differentially expressed compared to other endothelial cells, whereas tumor endothelial cells were the only ones expressing VAV1. In contrast, VAV2 was expressed by all endothelial cells except from those isolated from tumors. In these tumor endothelial cells, the highly expressed RhoGEFs of branch 2B were ARHGEF7 and TIAM2, whereas the expression of Tuba, VAV2 and ARHGEF9 was strongly reduced in these cells.
3.4. 3.4 Effect of VEGF on endothelial DH-RhoGEFs expression
In order to assess the potential effect of VEGF-dependent signaling in the regulation of the initial group of 63 DH-RhoGEFs included in the SMART database, we stimulated HMEC-1 with VEGF-165 (100 ng/ml) for 1, 3 or 12 h or left unstimulated (Fig. 7A and B). First, we confirmed that total RNA and the corresponding cDNA used for each experimental condition was of adequate quality (Fig. 7C and D). We then verified that in our samples VEGF signaling promoted the expression of HLX-1, previously recognized to respond to VEGF treatment [32-34] (Fig. 7E). Our results revealed that RasGRF2 and P-Rex2 were strongly stimulated by VEGF, showing more than a two-fold increase in response to the growth factor (Fig. 7A). In addition, FGD5, ITSN1, AKAP13, VAV3, ARHGEF6, ARHGEF7, P-REX1 and PLEKHG1 showed lower, but significant and reproducible increases (Fig. 7A and B). No changes were detected in the other RhoGEFs expressed by endothelial cells when exposed to VEGF.
Fig. 7.
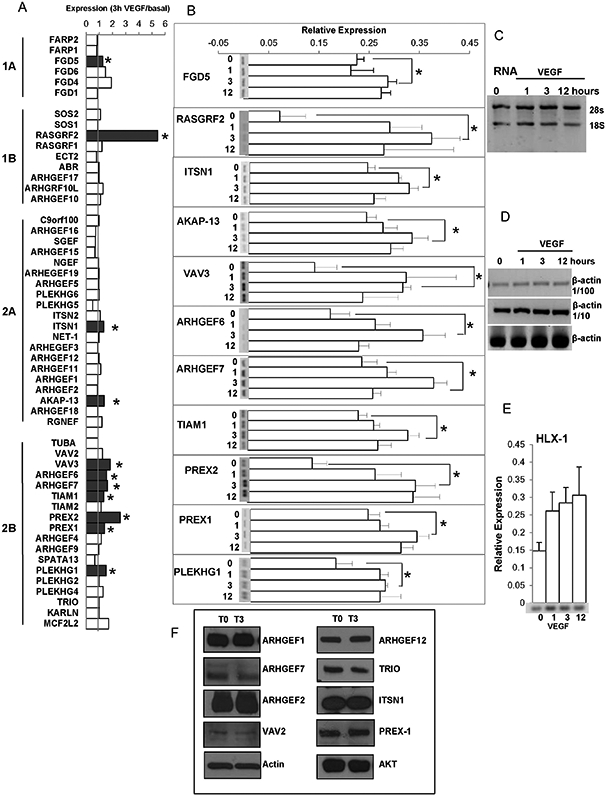
Effect of VEGF on the transcriptional levels of RhoGEFs in HMEC-1. A, Relative expression of the indicated human RhoGEFs in HMEC-1 cells stimulated with VEGF, 100 ng/ml for 3 h. B, Effect of VEGF on the expression of the indicated RhoGEF in HMEC-1 stimulated for 1, 3 or 12 h, or non-stimulated (0). A representative RT-PCR gel is shown for those RhoGEFs whose expression was significantly induced by VEGF. Bars represent the mean ± SEM (error bars) for n = 3 independent experiments; *p < 0.05. C, Representative gel showing the quality of RNA used for the analysis of expression of RhoGEFs in HMEC-1 endothelial cells. D, Representative gel showing the expression of actin (used as control to calibrate the cDNA used to assess expression of all the RhoGEFs indicated in A). E, Effect of VEGF on expression of HLX-1 (positive control, indicator of VEGF-dependent gene expression) in HMEC-1. F, Representative results of three independent experiments showing expression, at the protein level, of the indicated selected RhoGEFs in HMEC cells under non-stimulated conditions (T0) or stimulated with VEGF (100 ng/ml) for 3 h (T3); actin and AKT were used as loading controls.
Although functional regulation of RhoGEFs occurs at the protein level via multiple mechanisms including protein–protein interactions, posttranslational modifications and response to second messengers at different subcellular locations [35,36], transcriptional changes are an obvious mode of regulatory control. Our data indicate that VEGF stimulation promotes transcriptional changes of a small subgroup of RhoGEFs. We also analyzed protein levels of selected RhoGEFs that were either regulated (ITSN1, ARHGEF7/β-Pix, P-Rex1,) or not (ARHGEF1/p115RhoGEF, ARHGEF12/LARG, TRIO, ARHGEF2/GEFH1 and VAV2) by VEGF at the mRNA level. As shown in Fig. 7F the protein expression levels of these RhoGEFs were not modified after 3 h of treatment with VEGF, indicating that the temporality on the regulation of RhoGEFs transcripts does not directly correlate with their regulation at the protein level, likely due to different kinetics of synthesis and degradation not evident, in the case of proteins, at the time tested.
3.5. Expression of 7 “atypical” DH-RhoGEFs
The data shown so far was confined to the 63 human DH-RhoGEFs included in the SMART database, which bases its classification in a manually corrected Hidden Markov Model to predict the existence of a RhoGEF domain in a given protein sequence [21]. Since other DH-RhoGEFs not included in the SMART database have also been considered bona fide RhoGEFs [22], we assessed whether these additional 7 “atypical” DH-RhoGEFs were expressed in HMEC endothelial cells. As shown in Fig. 8, ARHGEF40 is abundantly expressed in these cells, followed by ALS2, PLEKHG3, ARHGEF38 and ARHGEF33; whereas obscurin and PLEKHG7 were undetectable by our RT-PCR assays. From this group, ARHGEF40 (also known as Solo) has been involved in mechanical sensing promoting endothelial cell reorientation in response to cyclic stretch [37]; whereas the endothelial functions of all the others are completely unknown.
Fig. 8.
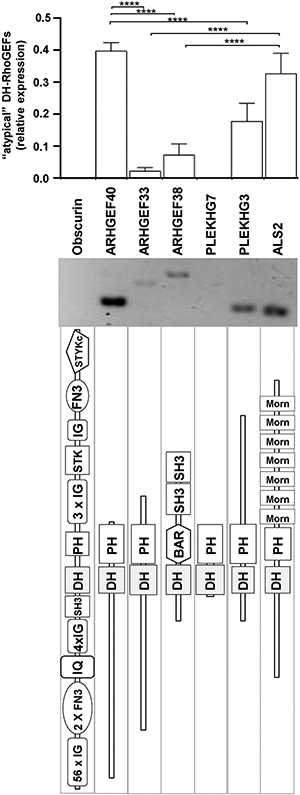
Transcriptional levels of “atypical” DH-RhoGEFs in HMEC. Results correspond to DH-RhoGEFs not considered in the SMART genomic database [21], but included in a comprehensive list of DH-RhoGEFs [22]. The graph shows normalized expression for this group of RhoGEFs. Bars represent the mean and SEM (error bars) of four independent experiments, ****p < 0.0001. The middle panel shows a representative result, the lower panel shows the predicted structure of each RhoGEF.
3.6. Expression of the DOCK family of Rac- and Cdc42-GEFs and their binding partners (ELMOs)
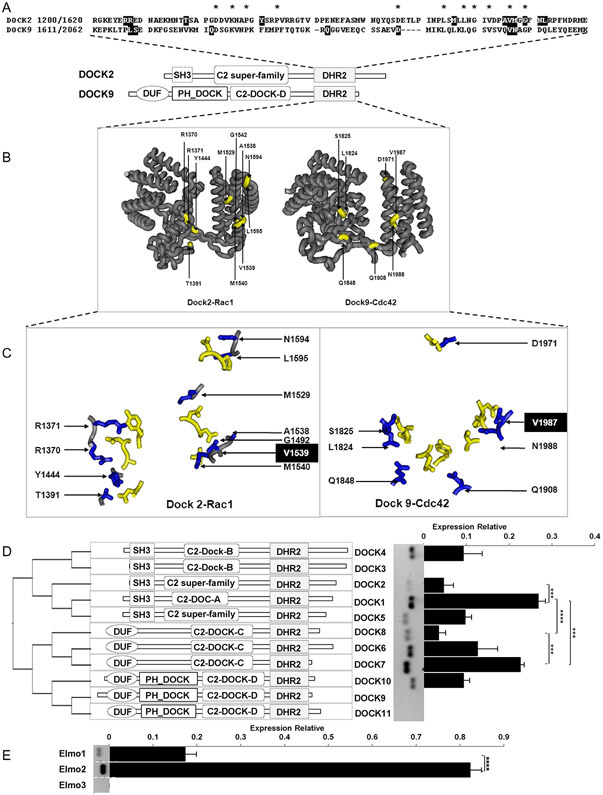
The DOCK family of GEFs constitutes a non-conventional group of Rac and Cdc42 activators that partners with ELMO scaffolds to form functional complexes [23]. We analyzed the phylogenetic relations of these GEFs based on the homologies of their DHR2 domain, this group of GEFs is distributed in two phylogenetic groups further subdivided into two subgroups each (Fig. 9). Their structural characteristics, in complex with Rac or Cdc42, revealed an overall similar shape but having just one amino acid (V1539 in DOCK2 and V1987 in DOCK9) conserved at the interface with the GTPase (Fig. 9). We then assessed their expression at the mRNA level in HMEC endothelial cells. DOCK1 and DOCK7 are conspicuously expressed, followed by DOCK6, DOCK4 and DOCK10 and, at lower levels, we detected DOCK2 and DOCK8. In contrast, DOCK3, DOCK9 and DOCK11 were undetectable. Among the ELMO family, ELMO2, was 4 times higher than ELMO1, whereas ELMO3 was undetected.
Fig. 9.
Comparative expression of Rac- and Cdc42-guanine exchange factors for the DOCK family and ELMOs (binding partners) in HMEC. A, Alignment of the DHR2 catalytic domain of DOCK2 and DOCK9. Residues of the DHR2 sequences shown with a black background correspond to those at the interacting interface within 3 Å of distance between the DHR2 domain and the GTPase of the complexes shown in B and C, in which the interacting amino acids are highlighted. D. Phylogenetic organization of the DOCK family of Rac- and Cdc42-GEFs based on the structure of their DHR2 domains (left panel). The central panel shows the predicted structure of the DOCK family, organized according to their phylogenetic relationships. Comparative expression of all members of the DOCK family is shown in the right panel. E. Comparative expression of ELMOs. The graphs in D and E show normalized relative expression of DOCKs and ELMOs respectively. Bars represent mean ± SEM of four independent experiments; ***p < 0.001, ****p < 0.0001.
4. Discussion
The systematic identification of endothelial RhoGEFs constitutes the first step towards clarifying their respective roles as regulators of endothelial cytoskeletal dynamics. These multidomain proteins putatively integrate multiple angiogenic signals that result in spatially constrained adjustments of cell shape. They also participate in the maintenance of homeostasis by regulating permeability and intracellular calcium levels [38,39]. In blood vessels responding to angiogenic factors, Rho GTPases contribute to create actin nucleation sites, assembling dynamic cytoskeletal structures critical to initiate the formation of vascular sprouts. Here, we analyzed the expression of all RhoGEFs (direct activators of Rho GTPases, identified by their DH-domain present in all RhoGEF homologous to Dbl) in human and mouse endothelial cells and in tumor-derived endothelial cells. Intriguingly, we found that 52, out of 70 DH-RhoGEFs, and 8 out of 11 GEFs of the DOCK family and 2 out of 3 ELMOs were expressed by endothelial cells. These results set the basis for future systematic studies of subgroups of endothelial RhoGEFs, most of which have been poorly characterized in the context of angiogenesis.
Phylogenetic analysis of the DH domains revealed the existence of two main groups of RhoGEFs, each subdivided into two branches with multiple members each. Although the overall structure of different DH domains is similar, the interface of interaction with Rho GTPases varies widely: only two GTPase-interacting amino acids are conserved in the catalytic DH of intersectin-1, ARHGEF11 and VAV1. All the phylogenetic groups of RhoGEFs were expressed by endothelial cells. Interestingly, some of these RhoGEFs showed changes in expression in tumor-derived endothelial cells. FGD2 and VAV1 were singularly increased by endothelial cells isolated from tumors. In contrast, FARP2, ARHGEF10, ARHGEF15, ARHGEF19, PLEKHG5, ARHGEF2, TUBA, VAV2 and ARHGEF9 showed reduced expression in tumor endothelial cells. These results indicate that endothelial cells adjust their repertoire of RhoGEFs as part of their angiogenic response and, potentially, the action of some of these RhoGEFs might be linked to the integration of angiogenic signals that initiate endothelial sprouting. PLEKHG5 (also known as Syx), one of the GEFs whose expression was reduced in tumor-derived endothelial cells, has been found to maintain endothelial cell junctions via a mechanism that is sensitive to VEGF [40]. Thus, it might be speculated that its reduction in tumors is potentially linked to a decrease on vascular stability. RhoGEFs that were found decreased in tumor-derived endothelium include: FGD5, TUBA, ARHGEF15, VAV2, ARHGEF10 and PLEKHG4. Interestingly, FGD5 and ARHGEF15 have both been implicated in Cdc42 activation in response to VEGF [41] and are considered positive regulators in the maintenance of endothelial adhesions [42,43]. In addition, FGD5 controls developmental angiogenesis [44]. The endothelial functions of TUBA have not been studied but in epithelial cells it plays a role in lumen formation [45,46]. In tumor endothelial cells, we found that FGD6 was increased with respect to FGD5, while in HMEC and HUVEC the expression of FGD5 was higher. Since both RhoGEFs have similar structure, perhaps their activities might be redundant. While the endothelial effects of FGD6 are presently unknown, it is likely that it coordinates polarity via Cdc42 and membrane recycling as per its reported function in osteoclasts [47].
We tested the effect of VEGF on the expression of DH-RhoGEFs in human microvascular endothelial cells. First, we confirmed that VEGF signaling stimulated gene expression by demonstrating the increased levels of HLX-1, a transcription factor whose expression is known to be stimulated by VEGF signaling [32-34]. Interestingly, VEGF increased the expression of RasGRF2 and P-Rex2 by at least two fold. Lower, but significant upregulation in response to VEGF was also noted in FGD5, ITSN1, VAV3, ARHGEF6, ARHGEF7, TIAM1, P-REX1, and PLEKHG1 transcripts. Other RhoGEFs did not show changes in expression in response to VEGF, providing a certain degree of specificity to the response by the growth factor.
Some RhoGEFs reported as enriched in endothelium by the in silico analysis performed by van Buul and colleagues [25] were not detected by this strategy. The reason(s) for this discrepancy most likely rely on differences in the methods and probes used to detect the transcripts. In addition, it is important to highlight that van Buul and colleagues organized their list of endothelial RhoGEFs based on the relative expression level of the transcript when compared to other cell types. Using this approach, a RhoGEF highly expressed by endothelial cells as well as by other cell types would be low in the list. In contrast, our results highlight expression of endothelial RhoGEFs based on their relative abundance and independently of their potential expression in other cell types. Also we found differences in species that can also contribute to some of the discrepancies. For example, BCR, was reported to be highly expressed in endothelial cells and we found it undetectable in human endothelial cells, but present in murine endothelial cells. Importantly, in cases when expression was undetected, we confirmed the effectiveness of the primers using other cell types. Although our primers were designed to detect RhoGEFs regardless of possible splicing variants, considering the contrast of our result on BCR compared to the report of van Buul and colleagues, it is also possible that differences in the sensitivity of the techniques used or the splice variants, might contribute to the differences.
Our results indicate that most GEFs of the DOCK family are expressed by HMEC endothelial cells. The endothelial functions of this group of GEFs with specificity for Rac and Cdc42 have remained elusive. Very recently, Mavria and colleagues revealed that members of this group of GEFs are involved in capillary lumen morphogenesis in response to VEGF. Based on an RNAi screen of endothelial cells, they found that their ability to recapitulate the different steps of lumen morphogenesis in a 3D co-culture assay is strongly compromised when the expression of DOCK4 is reduced. Further studies revealed that DOCK4 initiates the process by directing the formation of lateral filopodia via a cascade of GTPases initiated by the activation of Rac, which leads the activation of Cdc42 downstream of SGEF, a DH-RhoGEF that activates RhoG [48]. Regarding the expression of ELMOs, the binding partners of the DOCK family, we found that ELMO2 was highly abundant. These adaptors of the DOCK family contribute to assemble functional complexes that enable connection with cell-surface receptors and other signaling mediators. ELMO2 has not been studied in endothelial cells, whereas ELMO1 is considered part of a complex that protects from apoptosis [49] and controls vasculogenesis in zebrafish [50].
According to their expression and structural characteristics, various RhoGEFs might have a redundant function in endothelial cells. Likely candidates include FARP1 and FARP2; FGD1 and FGD6; SOS1 and SOS2; ITSN1 and ITSN2; and RGS-RhoGEFs (in particular, ARHGEF11 (PDZ-RhoGEF) and ARHGEF12 (LARG)), which are pairs of RhoGEFs with similar structure and found to be expressed in all endothelial cells. Others with similar structure, such as Vav1/2/3, TIAM1/2 and P-Rex1/2 are differentially expressed by endothelial cells, thus their function is likely not redundant. In the case of the RGS-RhoGEFs, known to be activated through direct interactions between GTP-bound Galpha12/13 and their RGS domains, it has been recently reported that while inactivation of PDZ-RhoGEF is viable, combination of PDZ-RhoGEF and LARG double knockouts are embryonic lethal. Interestingly, these double knockouts exhibit developmental vascular defects [51].
Most investigations on the endothelial functions of RhoGEFs have mainly focused on examples of individual RhoGEFs studied in the context of in vitro and in vivo angiogenesis elicited by specific stimuli. For instance, in response to sphingosine-1-phosphate, S1P2R activates RhoC partially through a G12/G13 dependent mechanism mediated by LARG (ARHGEF12) and this leads to RhoC activation. Through this pathway, S1P2 negatively regulates angiogenic responses [52]. Interestingly, sphingosine-1-phosphate also activates Gi-coupled S1PR which activates Rac via TIAM, exerting a positive control on endothelial adhesions, thus controlling endothelial permeability [53]. This rather complex pattern of expression and functions is only possible through the activity of multiple RhoGEFs. For example, we found that in endothelial HMEC-1, both stromal cell-derived factor/CXCL12 and VEGF are able to activate Rac, cell migration and in in vitro angiogenesis, acting on CXR4 and VEGFR receptors, respectively. In the first case, the GPCR-dependent pathway involves a critical participation of the RacGEF P-Rex1, which is not involved in the cascade activated by VEGF [54]. In the case of VEGF, it is believed that it activates Rac in endothelial cells via Vav2 [55].
The results presented here open a plethora of possibilities regarding the potential involvement of multiple RhoGEFs in angiogenic signaling cascades. They also provide guidance to future studies aiming at exploring the intricacies of signal integration critical to assemble cytoskeletal structures that lead the formation of new blood vessels.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.vph.2015.10.003.
Supplementary Material
Acknowledgments
We acknowledge J. Silvio Gutkind, who kindly provided us several anti-RhoGEF antibodies and Estanislao Escobar-Islas, Margarita Valadez-Sánchez, David Pérez-Rangel and Jaime Estrada-Trejo for their technical assistance. This work was supported by Consejo Nacional de Ciencia y Tecnologia (CONACyT) grants 152434 (to J.V.P.) and 79429 (to G.R.C.) and by a University of California Institute for Mexico and the United States, UC-Mexus-Conacyt collaborative grant to JVP and MLIA. RHG is a graduate student supported by a CONACyT fellowship.
References
- 1.Senger DR, Davis GE, Angiogenesis, Cold Spring Harb. Perspect Biol 3 (8) (2011) a005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iruela-Arispe ML, Davis GE, Cellular and molecular mechanisms of vascular lumen formation, Dev. Cell 16 (2) (2009) 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams RH, Alitalo K, Molecular regulation of angiogenesis and lymphangiogenesis. Nature reviews, Mol. Cell. Biol 8 (6) (2007) 464–478. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Jain RK, Molecular mechanisms and clinical applications of angiogenesis, Nature 473 (7347) (2011) 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potente M, Gerhardt H, Carmeliet P, Basic and therapeutic aspects of angiogenesis, Cell 146 (6) (2011) 873–887. [DOI] [PubMed] [Google Scholar]
- 6.Abraham S, et al. , VE-cadherin-mediated cell–cell interaction suppresses sprouting via signaling to MLC2 phosphorylation, Curr. Biol 19 (8) (2009) 668–674. [DOI] [PubMed] [Google Scholar]
- 7.Giannotta M, Trani M, Dejana E, VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity, Dev. Cell 26 (5) (2013) 441–454. [DOI] [PubMed] [Google Scholar]
- 8.Cascone I, et al. , Temporal and spatial modulation of rho GTPases during in vitro formation of capillary vascular network. Adherens junctions and myosin light chain as targets of Rac1 and RhoA, J. Biol. Chem 278 (50) (2003) 50702–50713. [DOI] [PubMed] [Google Scholar]
- 9.Hoang MV, Whelan MC, Senger DR, Rho activity critically and selectively regulates endothelial cell organization during angiogenesis, Proc. Natl. Acad. Sci. U. S. A 101 (7) (2004) 1874–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacharidou A, et al. , Endothelial lumen signaling complexes control 3D matrix- specific tubulogenesis through interdependent Cdc42- and MT1-MMP-mediated events, Blood 115 (25) (2010) 5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh W, Mahan RD, Davis GE, Cdc42- and Rac1-mediated endothelial lumen formation requires Pak2, Pak4 and Par3, and PKC-dependent signaling, J. Cell Sci 121 (Pt 7) (2008) 989–1001. [DOI] [PubMed] [Google Scholar]
- 12.Hall A, Rho family GTPases, Biochem. Soc. Trans 40 (6) (2012) 1378–1382. [DOI] [PubMed] [Google Scholar]
- 13.Cook DR, Rossman KL, Der CJ, Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease, Oncogene (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goicoechea SM, Awadia S, Garcia-Mata R, I'm coming to GEF you: regulation of RhoGEFs during cell migration, Cell Adhes. Migr (2014) 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesland A, et al. , Small molecule targeting Cdc42–intersectin interaction disrupts Golgi organization and suppresses cell motility, Proc. Natl. Acad. Sci. U. S. A 110 (4) (2013) 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang X, et al. , Small-molecule inhibitors targeting G-protein-coupled rho guanine nucleotide exchange factors, Proc. Natl. Acad. Sci. U. S. A 110 (8) (2013) 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evelyn CR, et al. , High-throughput screening for small-molecule inhibitors of LARG-stimulated RhoA nucleotide binding via a novel fluorescence polarization assay, J. Biomol. Screen 14 (2) (2009) 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vives V, et al. , Pharmacological inhibition of Dock5 prevents osteolysis by affecting osteoclast podosome organization while preserving bone formation, Nat. Commun 6 (2015) 6218. [DOI] [PubMed] [Google Scholar]
- 19.Blangy A, Fort P, Targeting the Dbl and dock-family RhoGEFs: a yeast-based assay to identify cell-active inhibitors of Rho-controlled pathways, The Enzymes, 33 Pt A2013. 169–191. [DOI] [PubMed] [Google Scholar]
- 20.Kather JN, Kroll J, Rho guanine exchange factors in blood vessels: fine-tuners of angiogenesis and vascular function, Exp. Cell Res 319 (9) (2013) 1289–1297. [DOI] [PubMed] [Google Scholar]
- 21.Letunic I, Doerks T, Bork P, SMART: recent updates, new developments and status in 2015, Nucleic Acids Res. 43 (Database issue) (2015) D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal M, Dvorsky R, Ahmadian MR, Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins, J. Biol. Chem 288 (6) (2013) 4486–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurin M, Cote JF, Insights into the biological functions of dock family guanine nucleotide exchange factors, Genes Dev. 28 (6) (2014) 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cario-Toumaniantz C, et al. , RhoA guanine exchange factor expression profile in arteries: evidence for a Rho kinase-dependent negative feedback in angiotensin II-dependent hypertension. American journal of physiology, Cell. Physiol 302 (9) (2012) C1394–C1404. [DOI] [PubMed] [Google Scholar]
- 25.van Buul JD, Geerts D, Huveneers S, Rho GAPs and GEFs: controlling switches in endothelial cell adhesion, Cell Adhes. Migr 8 (2) (2014) 108–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapp GT, et al. , Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair, Proc. Natl. Acad. Sci. U. S. A 109 (14) (2012) 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derewenda U, et al. , The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle, Structure 12 (11) (2004) 1955–1965. [DOI] [PubMed] [Google Scholar]
- 28.Chrencik JE, et al. , Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1, J. Mol. Biol 380 (5) (2008) 828–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni K, et al. , Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors, J. Biol. Chem 286 (28) (2011) 25341–25351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. , Activation of rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor, Science 325 (5946) (2009) 1398–1402. [DOI] [PubMed] [Google Scholar]
- 31.He H, et al. , Endothelial cells provide an instructive niche for the differentiation and functional polarization of M2-like macrophages, Blood 120 (15) (2012) 3152–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbert SP, Cheung JY, Stainier DY, Determination of endothelial stalk versus tip cell potential during angiogenesis by H2.0-like homeobox-1, Curr. Biol 22 (19) (2012) 1789–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Testori J, et al. , The VEGF-regulated transcription factor HLX controls the expression of guidance cues and negatively regulates sprouting of endothelial cells, Blood 117 (9) (2011) 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweighofer B, et al. , The VEGF-induced transcriptional response comprises gene clusters at the crossroad of angiogenesis and inflammation, Thromb. Haemost 102 (3) (2009) 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossman KL, Der CJ, Sondek J, GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nature reviews, Mol. Cell. Biol 6 (2) (2005) 167–180. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt A, Hall A, Guanine nucleotide exchange factors for Rho GTPases: turning on the switch, Genes Dev. 16 (13) (2002) 1587–1609. [DOI] [PubMed] [Google Scholar]
- 37.Abiko H, et al. , Rho guanine nucleotide exchange factors involved in cyclic- stretch-induced reorientation of vascular endothelial cells, J. Cell Sci 128 (9) (2015) 1683–1695. [DOI] [PubMed] [Google Scholar]
- 38.Beckers CM, et al. , ROCK2 primes the endothelium for vascular hyperpermeability responses by raising baseline junctional tension, Vasc. Pharmacol 70 (2015) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egea-Guerrero JJ, et al. , Role of L-type Ca(2+) channels, sarcoplasmic reticulum and Rho kinase in rat basilar artery contractile properties in a new model of subarachnoid hemorrhage, Vasc. Pharmacol 72 (2015) 64–72. [DOI] [PubMed] [Google Scholar]
- 40.Ngok SP, et al. , VEGF and angiopoietin-1 exert opposing effects on cell junctions by regulating the rho GEF Syx, J. Cell Biol 199 (7) (2012) 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurogane Y, et al. , FGD5 mediates proangiogenic action of vascular endothelial growth factor in human vascular endothelial cells, Arterioscler. Thromb. Vasc. Biol 32 (4) (2012) 988–996. [DOI] [PubMed] [Google Scholar]
- 42.Ando K, et al. , Rap1 potentiates endothelial cell junctions by spatially controlling myosin II activity and actin organization, J. Cell Biol 202 (6) (2013) 901–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kusuhara S, et al. , Arhgef15 promotes retinal angiogenesis by mediating VEGF-induced Cdc42 activation and potentiating RhoJ inactivation in endothelial cells, PLoS One 7 (9) (2012), e45858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng C, et al. , Endothelial cell-specific FGD5 involvement in vascular pruning defines neovessel fate in mice, Circulation 125 (25) (2012) 3142–3158. [DOI] [PubMed] [Google Scholar]
- 45.Bryant DM, et al. , A molecular network for de novo generation of the apical surface and lumen, Nat. Cell Biol 12 (11) (2010) 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin Y, et al. , Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation, J. Cell Biol 189 (4) (2010) 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steenblock C, et al. , The Cdc42 guanine nucleotide exchange factor FGD6 coordinates cell polarity and endosomal membrane recycling in osteoclasts, J. Biol. Chem 289 (26) (2014) 18347–18359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abraham S, et al. , A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis, Nat. Commun 6 (2015) 7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaker K, et al. , The bipartite rac1 guanine nucleotide exchange factor engulfment and cell motility 1/dedicator of cytokinesis 180 (elmo1/dock180) protects endothelial cells from apoptosis in blood vessel development, J. Biol. Chem 290 (10) (2015) 6408–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epting D, et al. , The Rac1 regulator ELMO1 controls vascular morphogenesis in zebrafish, Circ. Res 107 (1) (2010) 45–55. [DOI] [PubMed] [Google Scholar]
- 51.Mikelis CM, et al. , PDZ-RhoGEF and LARG are essential for embryonic development and provide a link between thrombin and LPA receptors and rho activation, J. Biol. Chem 288 (17) (2013) 12232–12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Del Galdo S, et al. , The activation of RhoC in vascular endothelial cells is required for the S1P receptor type 2-induced inhibition of angiogenesis, Cell. Signal 25 (12) (2013) 2478–2484. [DOI] [PubMed] [Google Scholar]
- 53.Singleton PA, et al. , Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P(1) receptor, PI3 kinase, Tiam1/Rac1, and alpha-actinin, FASEB J. 19 (12) (2005) 1646–1656. [DOI] [PubMed] [Google Scholar]
- 54.Carretero-Ortega J, et al. , Phosphatidylinositol 3,4,5-triphosphate-dependent Rac exchanger 1 (P-Rex-1), a guanine nucleotide exchange factor for Rac, mediates angiogenic responses to stromal cell-derived factor-1/chemokine stromal cell derived factor-1 (SDF-1/CXCL-12) linked to Rac activation, endothelial cell migration, and in vitro angiogenesis, Mol. Pharmacol 77 (3) (2010) 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrett TA, Van Buul JD, Burridge K, VEGF-induced Rac1 activation in endothelial cells is regulated by the guanine nucleotide exchange factor Vav2, Exp. Cell Res 313 (15) (2007) 3285–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.