Abstract
Objectives:
Acute kidney injury (AKI) is a sudden episode of kidney failure or damage and the risk of AKI is determined by the complex interactions of patient factors. In this study, we aimed to find out which risk factors in hospitalized patients are more likely to indicate severe AKI.
Methods:
We constructed a retrospective cohort of adult patients from all inpatient units of a tertiary care academic hospital between November 2007 and December 2016. AKI predictors included demographic information, admission and discharge dates, medications, laboratory values, past medical diagnoses and admission diagnosis. We developed a machine learning-based knowledge mining model and a screening framework to analyze which risk predictors are more likely to imply severe AKI in hospitalized populations.
Results:
Among the final analysis cohort of 76,957 hospital admissions, AKI occurred in 7,259 (9.43 %) with 6,396 (8.31 %) at stage 1, 678 (0.88 %) at stage 2, and 185 (0.24 %) at stage 3. We compared the non-AKI (without AKI) vs any AKI (stages 1–3), and mild AKI (stage 1) vs severe AKI (stages 2 and 3), where the best cross-validated area under the receiver operator characteristic curve (AUC) were 0.81 (95 % CI, 0.79–0.82) and 0.66 (95 % CI, 0.62–0.71), respectively. Using the developed knowledge mining model and screening framework, we identified 33 risk predictors indicating that severe AKI may occur.
Conclusions:
This study screened out 33 risk predictors that are more likely to indicate severe AKI in hospitalized patients, which would help strengthen the early care and prevention of patients.
Keywords: Acute kidney injury (AKI), Risk predictors, Knowledge mining model, Severe AKI, Electronic medical records
1. Introduction
Acute Kidney Injury (AKI) is considered one of the most common complications of acute illness, affecting 11–12 % of all hospitalized patients worldwide with a mortality rate of ~10 % [1]. AKI is associated with significant short- and long-term morbidity and mortality [2]. Early prediction or prevention of AKI has profound clinical implications but remains a major challenge [3] because AKI is not a disease per se, but rather a loose collection of syndromes [4]. However, the timing of AKI and its clinical manifestations are not random, and they relate to both the type and severity of injury. Data-driven knowledge mining approaches that incorporate “big” electronic medical records (EMR) have presented a unique analytic opportunity for AKI [5].
AKI is related to multiple risk factors including intrinsic situations, exposure to nephrotoxins (e.g. non-steroidal anti-inflammatory drugs [6]), acute illnesses (e.g. sepsis [7]), and major surgeries (such as cardiopulmonary bypass and coronary angiography [8–10]). Intrinsic risk situations include susceptibilities of each individual patient (e.g. age [11]) and those associated with reduced kidney reserve or failure of other organs with known cross-talk with the kidneys (such as heart, liver and respiratory system) [12]. In recent years, there have been several reports regarding novel and previously unknown risk factors for AKI, such as hyperuricemia [13], obstructive sleep apnea [14], hypochloremia and hyperchloremia [15].
Existing studies mainly focused on forecasting tools for the early identification of patients at risk. Many researchers [11,16,17] used a small number of highly correlated risk factors based on existing knowledge to build prediction models, which may miss potential unknown risk factors. Especially in case of medications, which are modifiable risk factors for AKI, most of the past researches [18,19] only collected data on known nephrotoxic drugs. In particular, the recent AKI prediction work by Google published in Nature [20] utilized the whole EMR data from the U.S. Department of Veterans Affairs, but one of the problems with these deep learning models [21,22] may be the lack of interpretability. To the best of our knowledge, the differentiation study of AKI stages focuses only on the comparison of prediction performance, for example, some researchers found that the forecasting models displayed a stepwise increase in the area under the receiver operating characteristic (AUC) across all AKI stages, performing significantly stronger for stages 2/3 than “mild” stage 1 [5,18,23]. However, there is few studies on knowledge mining between AKI stages.
In view of the differences in the etiology and pathophysiology of AKI patients [24], this study used a knowledge mining model to analyze the clinical risk factor differences between AKI stages from the perspective of EMR, to improve prevention, early detection and clinical management.
2. Materials and methods
2.1. Study population
All adult patients (age at visit≥18) admitted to the University of Kansas Health System (a tertiary academic hospital) for two days or more from November 2007 to December 2016 were included in this retrospective observational cohort study, which included adult patients from all ICU, surgical, and general wards. Given that a patient may have multiple eligible hospital admissions or encounters and develop AKI during one but not another, this study was conducted at the encounter level with a total of 179,370 encounters. From those encounters, we excluded the ones (a) missing necessary data elements for outcome determination, i.e. less than two serum creatinine measurements and (b) had evidence of moderate or severe kidney dysfunction at admission based on lab measurements, i.e. estimated Glomerular Filtration Rate (eGFR) less than 60 mL/min/1.73 m2 or serum creatinine (SCr) level of >1.3 mg/dl within 24 h of hospital admission. eGFR was calculated with the Modification of Diet in Renal Disease (MDRD) equation and adjusted for race. The final analysis cohort contained 76,957 encounters.
The retrospective cohort was built using the University of Kansas Medical Center’s de-identified clinical data repository called HERON (Health Enterprise Repository for Ontological Narration) [25]. No approval by the Institutional Review Board was required for the study because all identifiers were removed and event date were shifted, meeting the de-identification criteria specified in the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. The de-identified data request for this study was approved by the HERON Data Request Oversight Committee.
2.2. AKI definition
We staged AKI for severity according to the SCr-based criteria described in the KDIGO (Kidney Disease Improving Global Outcomes) clinical practice guidelines [26] (see Table 1). Baseline SCr level was defined as the most recent SCr value within two-day window prior to admission if available; otherwise the first SCr value after admission was used as the baseline. Then all pairs of SCr levels measured between admission and discharge were evaluated on a rolling basis to determine the occurrence of AKI.
Table 1.
The KDIGO staging system for AKI.
| AKI Stage | Serum Creatinine (SCr) Criteria |
|---|---|
| Stage 1 | Increase >26.4 μmol/L (0.3 mg/dL) with 48 h or 1.5–1.9 times baseline within 7 days |
| Stage 2 | Increase 2.0–2.9 times baseline |
| Stage 3 | Increase creatinine >354 μmol/L (4.0 mg/dL) or 3 times baseline |
2.3. Clinical variables
For each hospital encounter in the final analysis cohort, we extracted time stamped clinical data on demographics, vital signs, medications, laboratory values, past medical diagnoses, and admission diagnoses. Details of the 1,888 clinical variables considered are available in Table 2. It is important to note that SCr and eGFR were not included as predictive variables because they were used to determine the outcome variable, and we aimed to focus on the contribution of other factors. Laboratory values were categorized as unknown, less than reference normal range, within normal range, or greater than the reference normal range. As show in Table B.1, the vital signs of patients were discretized into groups based on medical knowledge.
Table 2.
Clinical variables extracted for the study cohort.
| Feature Category | #of Variables | Details |
|---|---|---|
| Demographics (Demo) | 3 | Age, race, gender; |
| Vitals (Vitals) | 5 | BMI, diastolic BP, systolic BP, pulse, temperature; |
| Lab tests (Lab) | 14 | Albumin, ALT, AST, Ammonia, Calcium, BUN, Bilirubin, CK-MB, CK, Glucose, Lipase, Platelets, Troponin, WBC; |
| Admission diagnoses (DRG) | 315 | University Health System Consortium (UHC) APR-DRG; (e.g. liver transplant, heart &/or lung transplant, etc.) |
| Medications (MED) | 1271 | All medications are mapped to RxNorm ingredient; (e.g. lithium carbonate, pentostatin, ospemifene, oxybutynin, etc.) |
| Medical History (CCS) | 280 | ICD9 codes mapped to CCS major diagnoses. (e.g. Nervous system congenital anomalies, other congenital anomalies, etc.) |
Medication exposure included inpatient (i.e. dispensed during stay) and outpatient medications (i.e. medication reconciliation and prior outpatient prescriptions). All medication names were normalized by mapping to RxNorm ingredient. Admission diagnosis, i.e. all patient refined diagnosis related group (APR-DRG), were collected from the University Health System Consortium (UHC; http://www.vizientinc.com) data source in HERON. Patient past medical history was captured as major diagnoses (ICD-9 codes grouped according to the Clinical Classifications Software (CCS) diagnosis categories by the Agency for Healthcare Research and Quality.
2.4. Data preprocessing
Only the most recently recorded vitals and labs before the AKI prediction point (i.e. 24 h prior to AKI event or last normal SCr for non-AKI cases) were used for each encounter. Meanwhile, instead of any imputation of missing numerical values, missing values were captured as a separate category because information may be contained in the choice to not perform a particular test [16]. To enhance interpretability, the above multivalued variables and ethnicity (as a dummy variable) are converted into multiple binary variables. Medical history was defined as true if it happened before the AKI prediction point. Hence, Medical history and admission diagnosis were all binary variables (i.e. presence or absence). Medication variables were defined as the number of days taken within 7-days before the AKI point. To enhance interpretability, 15 unknown medical ingredients were removed (e.g., 1,2,6-hexanetriol and 3,7-dimethyloctane-1,7-diol). The final number of features become 1945.
2.5. Knowledge mining model
In the knowledge mining model, we mainly applied two methods: eXtreme Gradient Boosting (XGBoost) [27] and SHapley Additive exPlanations (SHAP) [28,29] (see Methods A.1 and A.2). XGBoost is a machine learning technique that assembles weak prediction models (typically decision trees) to build a prediction model, in which the parameters were trained and obtained by 10-fold cross-validation scheme. SHAP method was used to explain the XGBoost prediction results, namely it would transform the original nonlinear prediction model to the summing effects of all variable attributions while approximating the output risk for each patient, thus we can easily interpret the impact of the predictors on each patient from SHAP explanation.
As shown in Fig. 1(A), we applied cross-validation strategy to obtain the more stable scores (namely, the weighted average SHAP values), where the weight was the area under the receiver operating characteristic curve (AUC) [30] derived from XGBoost model in each fold and the original score was derived from SHAP interpretation for entire data set. A positive weight (i.e., weighted SHAP value) means that the presence of the variable increases the likelihood of a positive outcome for this sample. A negative weight suggests that the presence of the variable decreases the likelihood of the positive outcome in this patient. If a weight is close to 0, this suggests that the model does not consider the variable useful for estimating the likelihood.
Fig. 1. Overall framework flowchart.
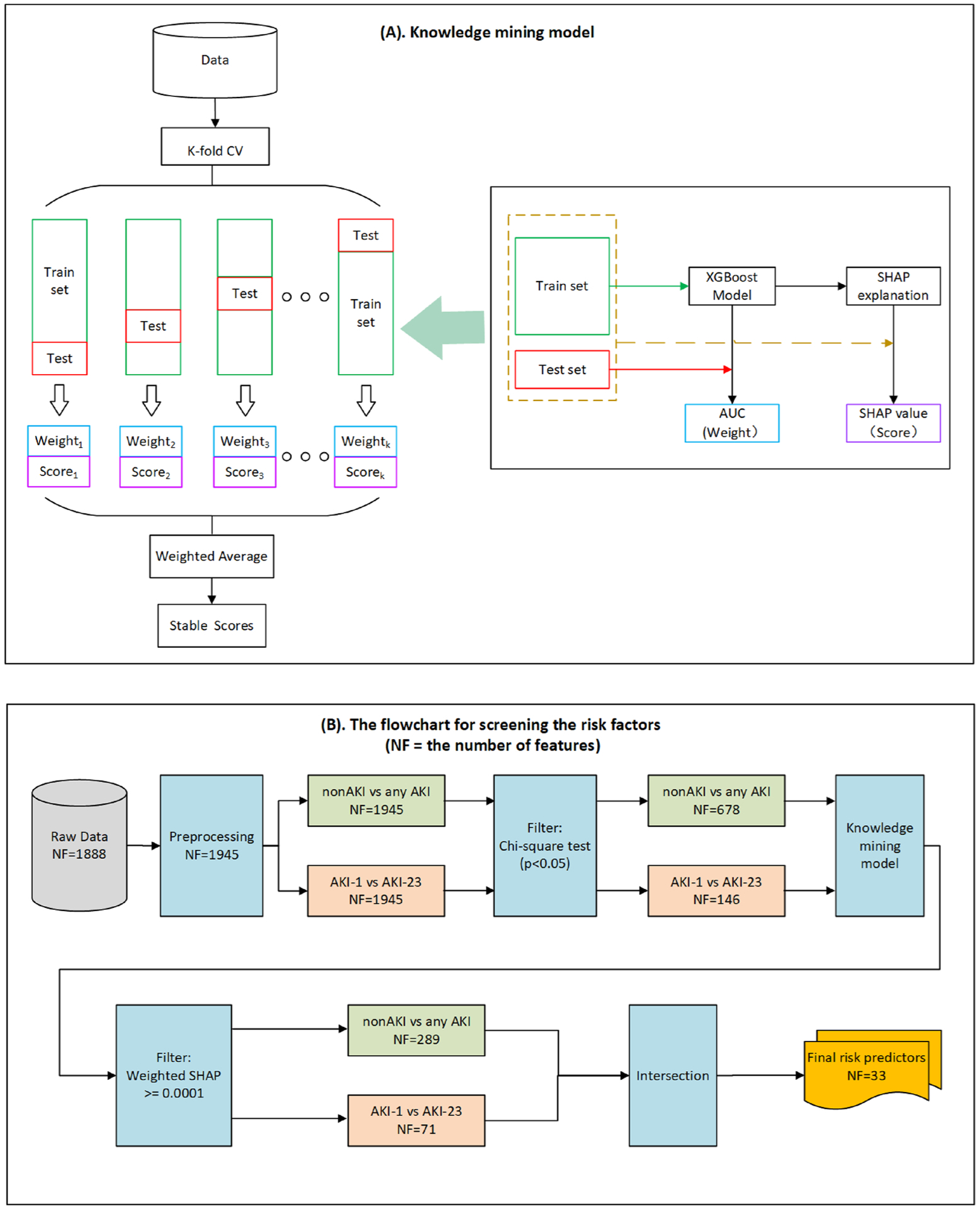
(A) The knowledge mining model; and (B) The flowchart for screening the risk factors.
2.6. Risk predictor screening process and statistical analysis
As shown in Fig. 1(B), we analyzed two classification tasks, namely the non-AKI (without AKI) vs any AKI, and AKI stage 1 (AKI-1) vs AKI stages 2 and 3(AKI-23). For better knowledge mining, a univariate filtering method (i.e., chi-square test, p value <0.05) for categorical variables is first performed to reduce the EMR variable dimension. Next, based on the above knowledge mining model, we obtained the weighed SHAP value of each feature for each patient, and calculated the average value of the weighted SHAP values when the features are present. Then, a second filtering mechanism was established to filter out factors that have a very small classification prediction effect and risk-free positive effects, such as normal BMI (body mass index) and the standard WBC (white blood cell count) lab test. Finally, the intersection of risk factors obtained from the two classification tasks was used to screen out the final risk predictors, which were more likely to indicate severe AKI in the inpatient population.
To obtain an intuitive visualization for AKI cohorts, t-Distributed Stochastic Neighbor Embedding (t-SNE) [31] was applied to visualize the high-dimensional EMR data by giving each sample a location in a two-dimensional map, which is better than existing techniques at creating a single map that reveals structure at many different scales. The use of networks to integrate different datasets has been proposed as a viable path toward elucidating the origins of specific diseases [32]. Therefore, we utilized the EMR data to show the risk predictor correlation network between AKI-1 and AKI-23. In this study, we applied the Chi-square test to calculate the p-value to quantify the strength of correlation between features. Two-tailed p < 0.05 denoted statistical significance for all comparisons. Data extraction and all analyses were performed using Python 3.7 software.
3. Results
3.1. Characteristics of the study cohort
Among the final analysis cohort of 76,957 hospital admissions, AKI occurred in 7,259 (9.43 %) encounters, with 6,396 (8.31 %) at stage 1, 678 (0.88 %) at stage 2, and 185 (0.24 %) at stage 3. Distribution of patient demographic variables among AKI stages 1–3 and non-AKI encounters is listed in Table 3. Most demographic variables (expect Asian and other race) were statistically different between AKI and non-AKI encounters (p< = 0.05). And the incidence of AKI in male patients is slightly higher than that of females. The percentage of missing values in vital signs and lab tests is shown in Table B.2.
Table 3.
Clinical demographics of final cohort stratified by AKI status.
| Characteristic n (%) | AKI-1 (n = 6,396) | AKI-2 (n = 678) | AKI-3 (n = 185) | non-AKI (n = 69,698) | P value |
|---|---|---|---|---|---|
| Age, year | |||||
| 18–25 | 303(4.74) | 29(4.28) | 25(13.51) | 4596(6.59) | <0.001 |
| 26–35 | 514(8.04) | 44(6.49) | 23(12.43) | 7339(10.53) | <0.001 |
| 36–45 | 711(11.12) | 76(11.21) | 25(13.51) | 8601(12.34) | 0.004 |
| 46–55 | 1218(19.04) | 157(23.16) | 35(18.92) | 14374(20.62) | 0.016 |
| 56–64 | 1672(26.14) | 185(27.29) | 49(26.49) | 16192(23.23) | <0.001 |
| >64 | 1978(30.93) | 187(27.58) | 28(15.14) | 18596(26.68) | <0.001 |
| Race | |||||
| White | 4791(74.91) | 487(71.83) | 130(70.27) | 53177(76.30) | <0.001 |
| African American | 918(14.35) | 111(16.37) | 36(19.46) | 9336(13.39) | 0.003 |
| Asian | 45(0.70) | 7(1.03) | 2(1.08) | 600(0.86) | 0.302 |
| Other | 642(10.04) | 73(10.77) | 17(9.19) | 6585(9.45) | 0.079 |
| Gender | |||||
| Male | 3822(59.76) | 378(55.75) | 109(58.92) | 37850(54.31) | <0.001 |
Note: P value for the comparison of any AKI and non-AKI group was obtained by using Chi-square test.
3.2. Knowledge mining evaluation
Fig. 2(A) shows an exponential decline trend in the importance of clinical characteristics, where the feature weights are the average weighed SHAP values of those samples with features. To further assess the discrimination of the top-ranked features for AKI prediction, we used the XGBoost prediction model (parameters shown in Table B.3) to conduct a series of prediction experiments by including different numbers of top-k features (i.e. k = 1–140). From Fig. 2(B), we can see that the best cross-validated area under the receiver operator characteristic curve (AUC) for non-AKI vs any AKI, and mild AKI (AKI-1) vs severe AKI (AKI-23) were 0.81 (95 % CI, 0.79–0.82) and 0.66 (95 % CI, 0.62–0.71), respectively.
Fig. 2.
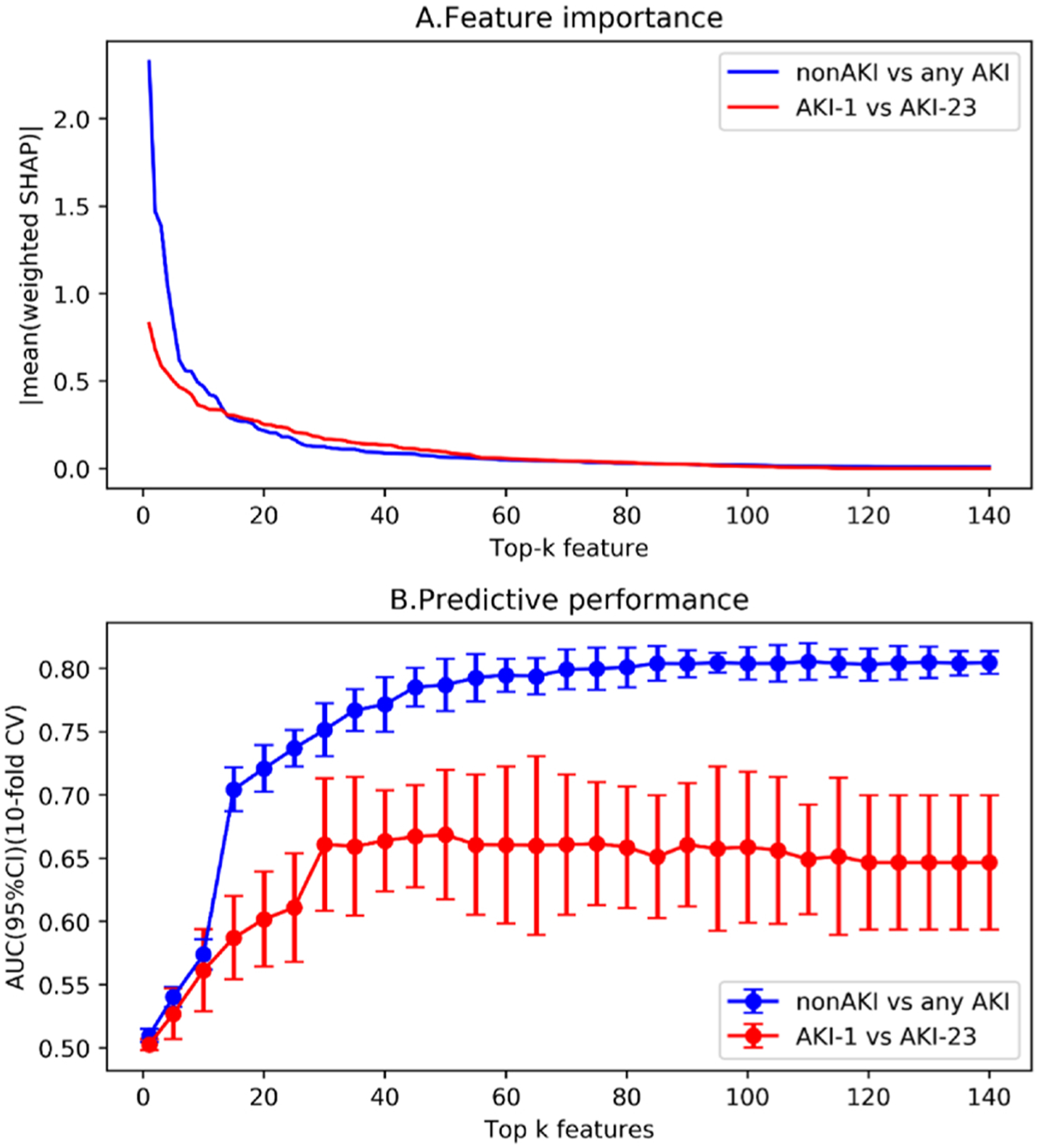
The feature importance trend and prediction performance of top-ranking features for nonAKI vs any AKI and AKI-1 vs AKI-23, respectively.
Fig. 3 shows the visualization of the patient-patient scatter plot obtained by t-SNE. In the two subgraphs on top (Fig. 3a and b), blue dots stand for non-AKI patients, red stars indicate any AKI patients. In the two subgraphs on the bottom (Fig. 3c and d), blue dots represent mild AKI-1 population, red stars denote severe AKI-23 population. Furthermore, in the two subgraphs on the left (Fig. 3a and c), we used raw feature data to visualize patient-patient scatterplot with unsupervised learning; in the two subgraphs on the right (Fig. 3b and d), we applied the weighted SHAP value obtained by the knowledge mining model that incorporates the importance of feature prediction, that is, illustrating the distribution between samples in a supervised learning manner. In short, compared with unsupervised learning, supervised learning methods are more suitable for AKI research. More importantly, it illustrates the validity of the characteristic risk information expressed by the weighted SHAP value.
Fig. 3.
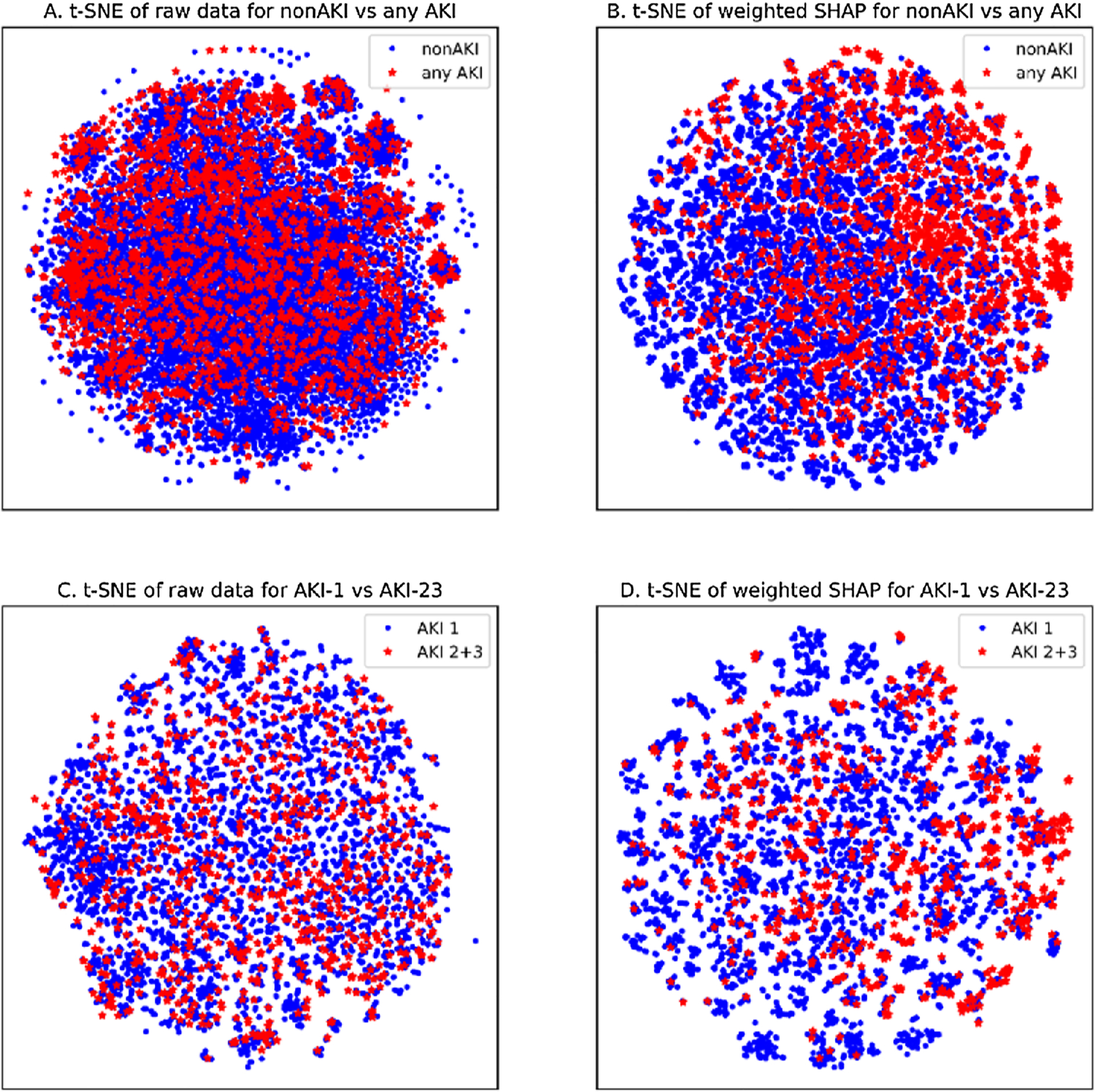
Visualization by t-SNE method.
3.3. Screening for risk factors causing severe AKI
As shown in Fig. 1(B), we took the intersection of two kinds of risk feature sets (i.e., 289 and 71 for non-AKI vs any AKI and AKI-1 vs AKI-23, respectively), then obtained 33 risk predictors that not only distinguish severe AKI, but also are the predictors in discriminating non-AKI from any AKI. For those risk factors that appeared only in non-AKI vs any AKI but not in AKI-1 vs AKI-23 (see Fig. B.1, which shows the feature importance of the top 100 risk factors), they may also be risk factors for AKI, but their ability to distinguish between mild AKI and severe AKI is not obvious. Table 4 and Table B.4 list the characteristics of the 33 risk factors finally screened out from the AKI-1 vs AKI-23 and non-AKI vs any AKI classification tasks, respectively.
Table 4.
The characteristics of the 33 risk factors in AKI-1 vs AKI-23.
| ID [Name] | AKI-1 n (%) | AKI-23 n (%) | Weighted SHAP Median [IQR] | P value |
|---|---|---|---|---|
| race_2[’African American’] | 918(14.35) | 147(17.03) | 0.0466[0.0364–0.0561] | 0.0415 |
| BMI_4[’>30.0 obese’] | 2655(41.51) | 440(50.99) | 0.0994[0.0731–0.1281] | <0.001 |
| Temperature_4 [’99.5–104.0 Fever’] | 195(3.05) | 57(6.61) | 0.4134[0.3486–0.528] | <0.001 |
| DBP_1[’<80’] | 4812(75.24) | 616(71.38) | 0.0019[0.0005–0.0028] | 0.0161 |
| DBP_3[’90–99 stage 1 hypertension’] | 383(5.99) | 85(9.85) | 0.1313[0.103–0.1624] | <0.001 |
| SBP_3[’140–159 stage 1 hypertension’] | 1208(18.89) | 198(22.94) | 0.0147[0.0094–0.0224] | 0.0054 |
| SBP_4[’>160 stage 2 hypertension’] | 224(3.50) | 47(5.45) | 0.1677[0.1282–0.2377] | 0.0063 |
| Lab13_3[’WBC, more than the standard value’] | 2252(35.21) | 371(42.99) | 0.0507[0.0354–0.0685] | <0.001 |
| Lab2_3[’AST, more than the standard value’] | 1389(21.72) | 214(24.80) | 0.0405[0.028–0.0559] | 0.0451 |
| Lab9_3[’Glucose, more than the standard value’] | 1095(17.12) | 177(20.51) | 0.0685[0.056–0.0887] | 0.0159 |
| CCS131[’Conduction disorders’] | 373(5.83) | 33(3.82) | 0.0032[−0.0034–0.0077] | 0.0198 |
| CCS182[’Septicemia (except in labor)’] | 710(11.10) | 123(14.25) | 0.0509[0.0388–0.069] | 0.0076 |
| CCS202[’Fracture of upper limb’] | 88(1.38) | 20(2.32) | 0.1513[0.1108–0.2049] | 0.0460 |
| CCS219[’Cancer of liver and intrahepatic bile duct’] | 107(1.67) | 28(3.24) | 0.2555[0.1902–0.3351] | 0.0021 |
| CCS271[’Biliary tract disease’] | 236(3.69) | 46(5.33) | 0.1640[0.1218–0.2432] | 0.0246 |
| CCS71[’Skin and subcutaneous tissue infections’] | 482(7.54) | 85(9.85) | 0.0439[0.0322–0.0607] | 0.0209 |
| DRG0[’liver transplant’] | 126(1.97) | 31(3.59) | 0.2544[0.193–0.359] | 0.0032 |
| DRG261[’infect & parasitic disease’] | 110(1.72) | 37(4.29) | 0.3050[0.2228–0.4054] | <0.001 |
| DRG262[’post-op/trauma infect proc’] | 62(0.97) | 23(2.67) | 0.1127[0.0801–0.1479] | <0.001 |
| DRG263[’septicemia & dissem infect’] | 359(5.61) | 66(7.65) | 0.0476[0.0375–0.066] | 0.0207 |
| MED1008[’vasopressin’] | 126(1.97) | 26(3.01) | 0.1191[0.0834–0.1827] | 0.0394 |
| MED1009[’diphenhydramine’] | 3591(56.14) | 503(58.29) | 0.0035[−0.0058–0.0138] | 0.0421 |
| MED1086[’tazobactam’] | 1899(29.69) | 377(43.69) | 0.1613[0.1062–0.23] | <0.001 |
| MED1115[’erythropoietin’] | 69(1.08) | 16(1.85) | 0.1446[0.076–0.2252] | 0.006 |
| MED1142[’acetazolamide’] | 58(0.91) | 5(0.58) | 0.0029[0.0008–0.0077] | 0.0052 |
| MED1152[’albumin’] | 1328(20.76) | 213(24.68) | 0.0415[0.0332–0.057] | 0.0097 |
| MED302[’morphine’] | 2650(41.43) | 411(47.63) | 0.0084[0.0108–0.0217] | 0.0199 |
| MED321[’vancomycin’] | 2005(31.35) | 398(46.12) | 0.1758[0.1231–0.2372] | <0.001 |
| MED516[’glucose’] | 4119(64.40) | 654(75.78) | 0.0644[0.044–0.0799] | <0.001 |
| MED593[’homatropine’] | 34(0.53) | 8(0.93) | 0.0347[0.0187–0.0456] | 0.0097 |
| MED852[’budipine hcl’] | 2176(34.02) | 342(39.630) | 0.0050[−0.0076–0.0163] | 0.0003 |
| MED864[’hydroxyethyl starch 130–0.4’] | 97(1.52) | 30(3.48) | 0.2433[0.195–0.3249] | 0.0004 |
| MED943[’insulin, isophane’] | 235(3.67) | 36(4.17) | 0.0204[0.0112–0.0373] | 0.0130 |
Abbreviations:AKI = acute kidney injury, IQR = interquartile ranges, AKI-1 = AKI stage 1 or mild AKI, AKI-23 = AKI stages 2 and 3 or severe AKI, SHAP = SHapley Additive exPlanations.
Fig. 4 shows the weighted SHAP summary plot of the 33 risk factors in the mild AKI vs severe AKI classification, where the higher the SHAP value of a feature, the higher risk of AKI due to this feature, each dot is a person on each feature’s line, dots are colored by the feature value for that person and pile up vertically to show density. Particularly, for binary variables (i.e., {0,1}), red dot represents a value of 1, and blue represents a value of 0; for medications, red dot indicates use of this medicine in the past week, and blue dot implies the drug has not been used in the past week. From Fig. 4, we can see that most of the selected risk factors have a higher risk effect in predicting the occurrence of severe AKI.
Fig. 4. The weighted SHAP summary plot for the 33 risk predictors.
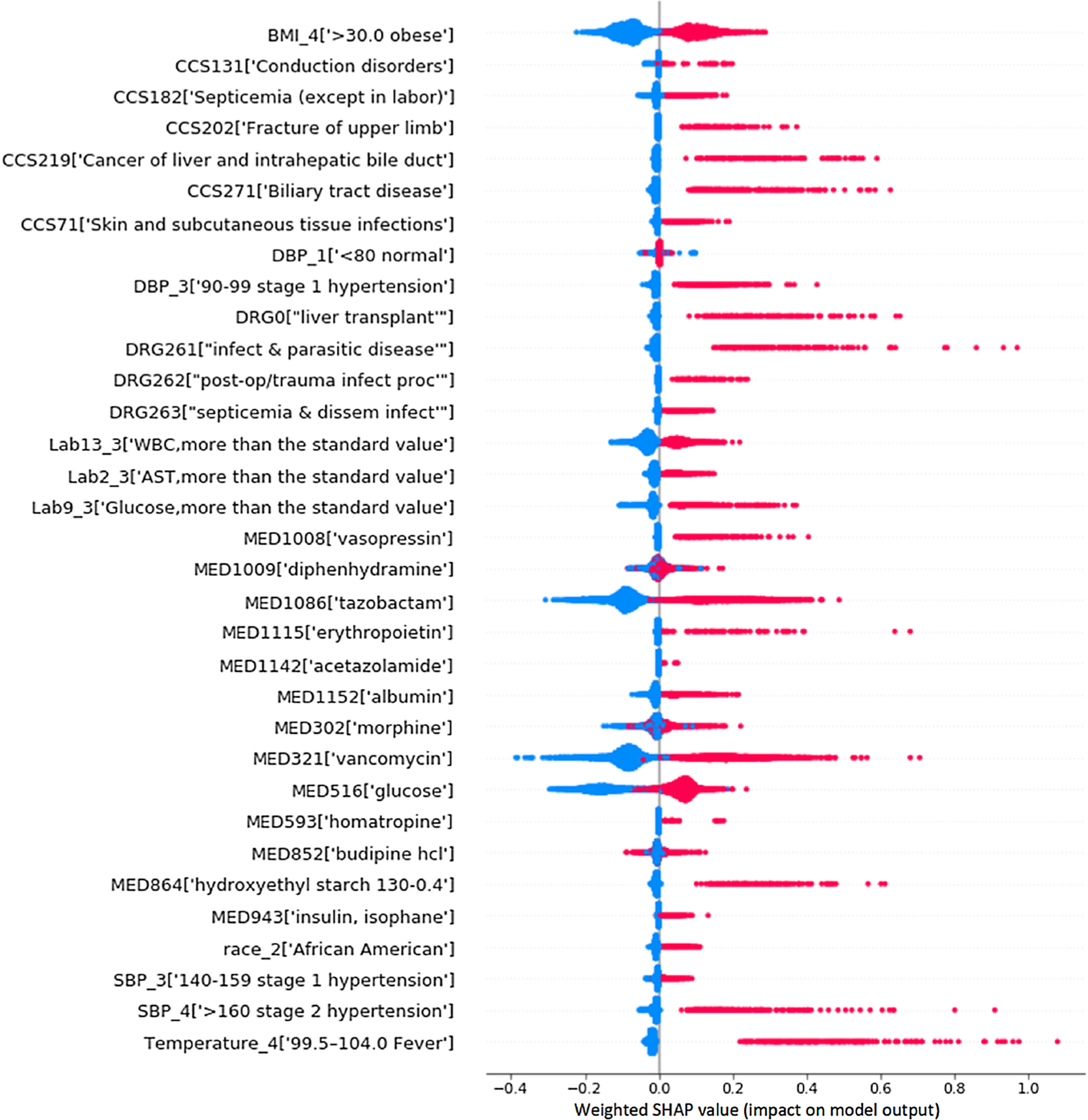
(The higher the weighted SHAP value of a feature, the higher its impact on the risk of AKI. Each dot represents a person. Dots are colored by the feature value for that person and piled up vertically to show density. For binary (0/1) variables, most of the points with the value of 1 indicating that the variable exists are on the right; on the contrary, 0 means that there is no variable and most of the points are on the left. For the drug variable, the points that have taken the drug in the past week are concentrated on the right, and the points that have not taken the drug are concentrated on the left.
Fig. 5 presents the correlation network for the 33 features and AKI label in mild AKI vs severe AKI classification, where features (nodes) are connected to one another by edges if they exhibit a certain degree of clinical correlation. For clear visualization, we made a tradeoff between the number of associations included in the network and the clarity with which these associations can be appreciated; for example, a cutoff value 0.05 was specified so that only links satisfying p< = 0.05 were kept [33]. In addition, link color indicates correlation strength (i.e., 0.05, 0.01 and 1e-5); node color identifies the feature category; node size is proportional to feature prevalence. The network diagram not only presents a strong correlation between the 33 risk factors and AKI label, but also shows that there is a strong correlation between the 33 risk factors.
Fig. 5.
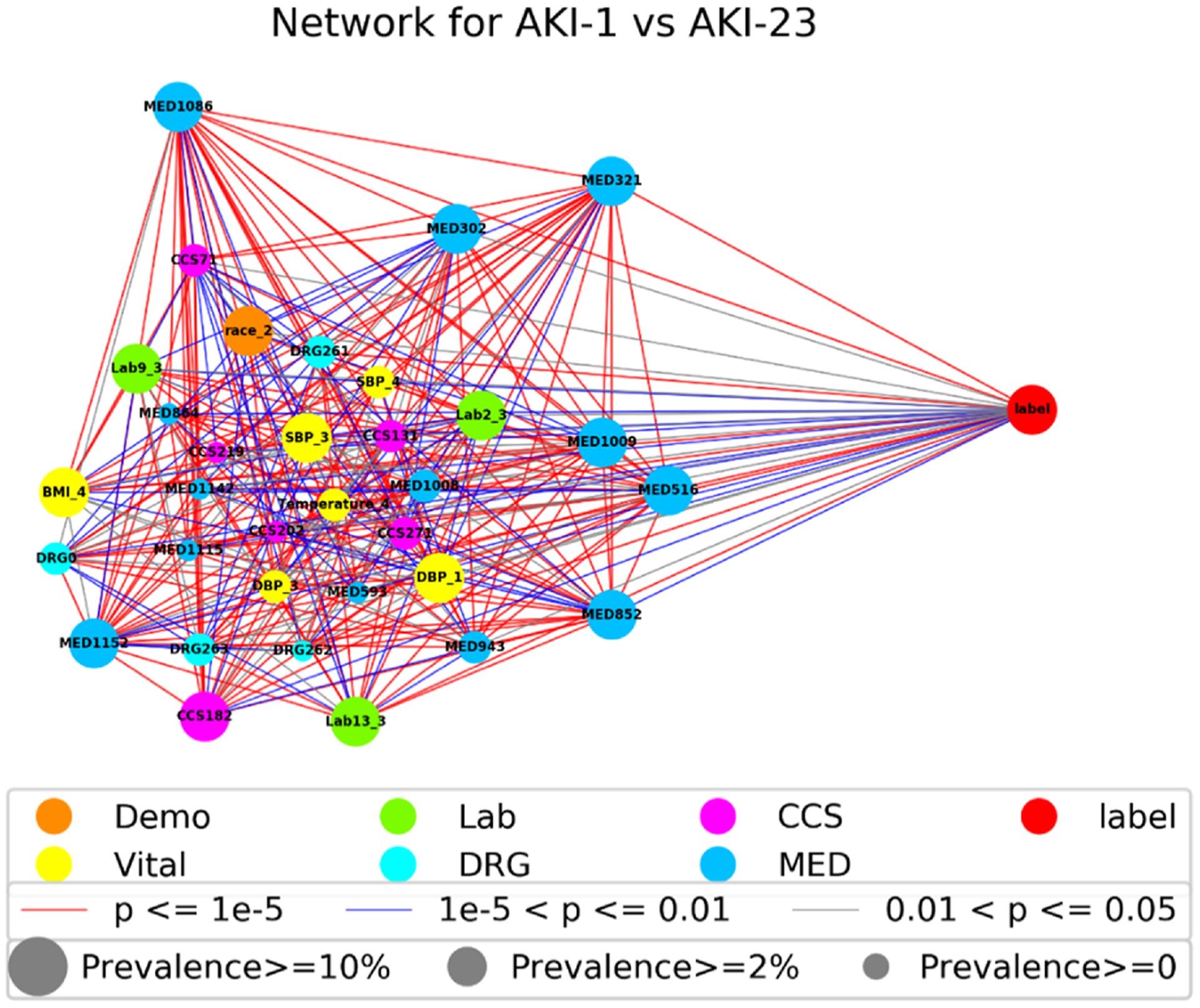
The correlation network of the 33 risk factors for AKI-1 vs AKI-23.
4. Discussion
Current international guidelines recommend risk assessment for AKI for the purpose of preventing kidney injury progression and severity [34]. In order to prevent the occurrence of severe AKI or reduce the duration and severity of AKI as early as possible, in this study, we established a knowledge mining method to screen those risk predictors that may cause severe AKI in hospitalized patients from the perspective of high-dimensional EMR characteristics.
Although some researchers focused solely on improving prediction performance at the expense of model interpretability, model transparency is essential for many clinical applications, which will accelerate the widespread adoption of such methods in clinical practice. In our model, the SHAP values were applied to explain the individualized feature attributes and demonstrated better agreement with human knowledge. For example, as shown in Fig. 4 and Table 4, obesity, fever, and hypertensions were screened as the risk predictors for severe AKI.
Our model identified diastolic blood pressure (DBP) below 80 to be associated with AKI risk. From Fig. 4, we can see that although most of the normal BP samples are in the middle of the axis, that is, the risk is close to 0, there are still a small number of samples showing a certain risk. We speculate that these samples belong to the hypotension group, and the blood pressure variables may have a U-shape relationship with AKI, but in our study the hypotensive group of patients with DBP < 80 was not subdivided further. For lab tests, high WBC, aspartate amino-transferase (AST) and glucose values above the standard reference range show high risk for severe AKI. For medical history (CCS), conduction disorders, septicemia, fracture of upper limb, cancer of liver and intrahepatic bile duct, skin and subcutaneous tissue infections, and biliary tract disease, show strong risk discrimination in AKI-1 vs AKI-23. The screened admission diagnoses (DRG) risk factors include liver transplant, infect & parasitic disease, post-op/trauma infect proc, and septicemia & disseminated infection.
For medication, drugs with known nephrotoxicity such as tazobactam [35] and vancomycin [36] show high weighted SHAP values in AKI-1 vs AKI-23 classification. And diphenhydramine (MED1009) has been identified to show certain degree of predictability for severe AKI. Although people generally do not think that diphenhydramine is the main source of renal insufficiency, some studies [37] have found that diphenhydramine can cause AKI in some susceptible people (such as the elderly). In addition, existing evidence regarding administration of albumin-containing fluids have been conflicting [37,38], for example, Lee et al. found that administration of 20 % exogenous albumin immediately before surgery increases urine output during surgery and reduces the risk of AKI after off-pump coronary artery bypass surgery in patients with a preoperative serum albumin level of less than 4.0 g/dl [38], but in patients with shock, hyperoncotic albumin has been associated with a fivefold increased risk of AKI [39]. In this study, albumin (MED1152) shows strong risk predictability for severe AKI, which may be due to poor physical conditions pushing patients to a higher risk. Meanwhile, there have been numerous studies showing that hydroxyethyl starch solutions may cause AKI [40], and our study found that this risk factor may also cause severe AKI.
To explain the interaction of many factors from an individual perspective, we randomly selected three patients (Patient A, B and C) from non-AKI, mild AKI, and severe AKI samples, respectively. Table B.5 shows the relevant information of the top five positive and negative impact factors for the three patients. For non-AKI patient A, WBC, albumin and calcium lab tests are abnormal, but glucose, tazobactam and vancomycin drugs are not used. Although mild AKI patient B has multiple risk factors, he is neither feverish nor obese. For patient C with severe AKI, he has suffered from stage 2 hypertension, which is the driving risk factor (weighted SHAP = 0.2864) for severe AKI.
It is worth noting that knowledge mining method can identify factors with strong predictive ability, but these factors are not necessarily causal inducers. More specifically, some medicines by themselves do not increase risk for AKI, but the disease that is treated by the medicine increases the risk of AKI. For example, in Table 4 insulin was screened as an important risk predictor for severe AKI, and presumably this is just a marker for diabetes, i.e. patients with diabetes or diabetic nephropathy are at higher risk for severe AKI. Hence, our knowledge mining method is only a hypothesis generator, but whether a factor (e.g., drug) truly cause increase in the risk of AKI requires rigorous demonstration from clinical experiments, and further work is necessary to investigate the nature of the association.
Our analysis has a few limitations. First, risk factor profiles were learned based on a single-center data, and external validation in other institutions would reinforce validity [41]. Second, we limited the analysis to patients with a minimum eGFR (estimated glomerular filtration rate) of 60 mL/min/1.73m2 and normal serum creatinine on the day of admission at hospital admission. We acknowledge that patients with reduced eGFR have an increased risk of developing AKI; however, we made the decision to focus on hospital-acquired AKI. Third, we did not include service unit as a risk factor and only selected certain lab tests based on previous literature for AKI prediction. Finally, since our study was not limited to the ICU (intensive care unit), we did not include urine output criteria as a predictor nor using it to define AKI.
5. Conclusions
Early prediction or prevention of AKI has profound clinical implications, and data-driven approaches that incorporate “big” EMR has presented a unique analytic opportunity for AKI. We used 9 years of EMR data, including 76,957 eligible hospital encounters, and established a knowledge mining model to screen the risk predictors that are more likely to imply severe AKI. The results of this study provide potential and opportunities to enhance the prevention of severe AKI in clinical care.
Supplementary Material
Summary points.
What was already known on the topic
Acute kidney injury (AKI) is an adverse event associated with significant short- and long-term morbidity and mortality;
The treatment of AKI is usually poor; therefore, efforts have been made to detect and prevent AKI early;
There has been growing interest in harnessing electronic health records for the AKI prediction in hospitalized patients.
What this study added to our knowledge
We established a knowledge mining model, namely combined eXtreme Gradient Boosting (XGBoost) model and SHapley Additive exPlanations (SHAP) method;
This study used a knowledge mining model to screen out 33 risk factors that are more likely to indicate severe AKI;
The results of this study provide potential and opportunities to enhance the prevention of severe AKI in clinical care.
Acknowledgements
This research was partially supported by the Youth Science Fund of the National Natural Science Foundation of China (Grant No. 61802149), the Major Research Plan of the National Natural Science Foundation of China (Key Program, Grant No. 91746204), the Fundamental Research Funds for the Central Universities (Grant No. 21618315), the Science and Technology Development in Guangdong Province (Major Projects of Advanced and Key Techniques Innovation, Grant No. 2017B030308008), and Guangdong Engineering Technology Research Center for Big Data Precision Healthcare (Grant No. 603141789047). ML was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH) under award number R01DK116986. The clinical dataset used for analysis described in this study was obtained from the University of Kansas Medical Center (KUMC) HERON clinical data repository which is supported by institutional funding and by the KUMC Clinical Translational Science Award (CTSA) grant UL1TR002366 from NIH.
The authors are grateful to the reviewers for their valuable suggestions.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijmedinf.2020.104270.
References
- [1].Al-jaghbeer M, Dealmeida D, Bilderback A, Ambrosino R, Kellum JA, Clinical decision support for in-hospital AKI, J. Am. Soc. Nephrol 29 (2018) 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kate RJ, Perez RM, Mazumdar D, Pasupathy KS, Nilakantan V, Prediction and detection models for acute kidney injury in hospitalized older adults, BMC Med. Inform. Decis. Mak 16 (2016) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Flechet M, Güiza F, Schetz M, Wouters P, Vanhorebeek I, Derese I, Gunst J, Spriet I, Casaer M, Van den Berghe G, Meyfroidt G, AKIpredictor, an online prognostic calculator for acute kidney injury in adult critically ill patients: development, validation and comparison to serum neutrophil gelatinase-associated lipocalin, Intensive Care Med. 43 (2017) 764–773. [DOI] [PubMed] [Google Scholar]
- [4].Kellum JA, Prowle JR, Paradigms of acute kidney injury in the intensive care setting, Nat. Rev. Nephrol 14 (2018) 217–230. [DOI] [PubMed] [Google Scholar]
- [5].Sutherland SM, Chawla LS, Kane-Gill SL, Hsu RK, Kramer AA, Goldstein SL, Kellum JA, Ronco C, Bagshaw SM, Utilizing electronic health records to predict acute kidney injury risk and outcomes: workgroup statements from the 15 th ADQI consensus conference, Can. J. Kidney Heal. Dis 3 (2016) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cronin RM, VanHouten JP, Siew ED, Eden SK, Fihn SD, Nielson CD, Peterson JF, Baker CR, Ikizler TA, Speroff T, et al. , National Veterans Health Administration inpatient risk stratification models for hospital-acquired acute kidney injury, J. Am. Med. Inform. Assoc 22 (2015) 1054–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malhotra R, Kashani KB, Macedo E, Kim J, Bouchard J, Wynn S, Li G, Ohno-Machado L, Mehta R, A risk prediction score for acute kidney injury in the intensive care unit, Nephrol. Dial. Transplant 32 (2017) 814–822. [DOI] [PubMed] [Google Scholar]
- [8].Jiang W, Teng J, Xu J, Shen B, Wang Y, Fang Y, Zou Z, Jin J, Zhuang Y, Liu L, Luo Z, Wang C, Ding X, Dynamic predictive scores for cardiac surgery-associated acute kidney injury, J. Am. Heart Assoc 5 (2016) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pablo Jorge-Monjas C, Bustamante-Munguira J, Lorenzo M, Heredia-Rodríguez M, Fierro I, Gómez-Sánchez E, Hernandez A, Álvarez FJ, Bermejo-Martin JF, Gómez-Pesquera E, Gómez-Herreras JI, Tamayo E, Predicting cardiac surgery-associated acute kidney injury: the CRATE score, J. Crit. Care 31 (2016) 130–138. [DOI] [PubMed] [Google Scholar]
- [10].Palomba H, De Castro I, Neto ALC, Lage S, Yu L, Acute kidney injury prediction following elective cardiac surgery: AKICS Score, Kidney Int. 72 (2007) 624–631. [DOI] [PubMed] [Google Scholar]
- [11].Kane-Gill SL, Sileanu FE, Murugan R, Trietley GS, Handler SM, Kellum JA, Risk factors for acute kidney injury in older adults with critical illness: a retrospective cohort study, Am. J. Kidney Dis 65 (2015) 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Leblanc M, Kellum JA, Gibney RTN, Lieberthal W, Tumlin J, Mehta R, Risk factors for acute renal failure: inherent and modifiable risks, Curr. Opin. Crit. Care 11 (2005) 533–536. [DOI] [PubMed] [Google Scholar]
- [13].Park S, Shin W, Lee EY, Gil H, Lee S, Lee S, Hong S, The impact of hyperuricemia on in-hospital mortality and incidence of acute kidney injury in patients undergoing percutaneous coronary intervention, Circ. J 75 (2011) 692–697. [DOI] [PubMed] [Google Scholar]
- [14].Dou L, Lan H, Reynolds DJ, Gunderson TM, Kashyap R, Gajic O, Caples S, Li G, Kashani KB, Association between obstructive sleep apnea and acute kidney injury in critically ill patients: a propensity-matched study, Nephron (2017) 137–146. [DOI] [PubMed] [Google Scholar]
- [15].Shao M, Li G, Sarvottam K, Wang S, Dyschloremia Is a Risk Factor for the Development of Acute Kidney Injury in Critically Ill Patients, 2016, pp. 1–13. [DOI] [PMC free article] [PubMed]
- [16].Matheny ME, Miller RA, Ikizler TA, Waitman LR, Denny JC, Schildcrout JS, Dittus RS, Peterson JF, Development of inpatient risk stratification models of acute kidney injury for use in electronic health records, Med. Decis. Making 30 (2010) 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kashani K, Acute kidney injury risk prediction. Annu. Updat. Intensive Care Emerg. Med. 2018, Springer, Cham, 2018, pp. 321–332. [Google Scholar]
- [18].Koyner JL, Carey KA, Edelson DP, Churpek MM, The development of a machine learning inpatient acute kidney injury prediction model, Crit. Care Med 46 (2018) 1070–1077. [DOI] [PubMed] [Google Scholar]
- [19].Koyner JL, Davison DL, Brasha-mitchell E, Chalikonda DM, Arthur JM, Shaw AD, Tumlin JA, Trevino SA, Bennett MR, Kimmel PL, Seneff MG, Chawla LS, Furosemide stress test and biomarkers for the prediction of AKI severity, J. Am. Soc. Nephrol 26 (2015) 2023–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tomašev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, Mottram A, Meyer C, Ravuri S, Protsyuk I, et al. , A clinically applicable approach to continuous prediction of future acute kidney injury, Nature 572 (2019) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Y, Yao L, Mao C, Srivastava A, Jiang X, Luo Y, Early prediction of acute kidney injury in critical care setting using clinical notes, 2018 IEEE Int. Conf. Bioinforma. Biomed. (2018) 683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pan Z, Du H, Ngiam KY, Wang F, Shum P, Feng M, A self-correcting deep learning approach to predict acute conditions in critical care, ArXiv Prepr. (2019). ArXiv1901.04364. [Google Scholar]
- [23].Koyner JL, Adhikari R, Edelson DP, Churpek MM, Development of a multicenter ward–Based AKI prediction model, Clin. J. Am. Soc. Nephrol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Levey AS, James MT, Acute kidney injury, Ann. Intern. Med 167 (2017) ITC66–ITC80. [DOI] [PubMed] [Google Scholar]
- [25].Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I, Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2), J. Am. Med. Inform. Assoc 17 (2010) 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fliser D, Laville M, Covic A, Fouque D, Vanholder R, Juillard L, Van Biesen W, A european renal best practice (ERBP) position statement on the Kidney Disease Improving Global Outcomes (KDIGO) clinical practice guidelines on acute kidney injury: part 1: definitions, conservative management and contrast-induced nephropathy, Nephrol. Dial. Transplant 27 (2012) 4263–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen T, Guestrin C, Xgboost: a scalable tree boosting system, Proc. 22nd Acm Sigkdd Int. Conf. Knowl. Discov. Data Min. (2016) 785–794. [Google Scholar]
- [28].Lundberg SM, Erion GG, Lee S, Consistent Individualized Feature Attribution for Tree Ensembles, (n.d.).
- [29].Lundberg SM, Lee S, A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst, 2017, pp. 4765–4774. [Google Scholar]
- [30].Bradley AP, The use of the area under the ROC curve in the evaluation of machine learning algorithms, Pattern Recognit. 30 (1997) 1145–1159. [Google Scholar]
- [31].Van Der Maaten LJP, Hinton GE, Visualizing high-dimensional data using t-sne, J. Mach. Learn. Res 9 (2008) 2579–2605. [Google Scholar]
- [32].Hidalgo CA, Blumm N, Barabási AL, Christakis NA, A dynamic network approach for the study of human phenotypes, PLoS Comput. Biol 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caldarelli G, Scale-free Networks: Complex Webs in Nature and Technology, Oxford University Press, 2007. [Google Scholar]
- [34].Khwaja A, KDIGO clinical practice guidelines for acute kidney injury, Nephron Clin. Pract 120 (2012) c179–c184. [DOI] [PubMed] [Google Scholar]
- [35].Navalkele B, Pogue JM, Karino S, Nishan B, Salim M, Solanki S, Pervaiz A, Tashtoush N, Shaikh H, Koppula S, et al. , Risk of acute kidney injury in patients on concomitant vancomycin and piperacillin–tazobactam compared to those on vancomycin and cefepime, Clin. Infect. Dis 64 (2016) 116–123. [DOI] [PubMed] [Google Scholar]
- [36].Minejima E, Choi J, Beringer P, Lou M, Tse E, Wong-Beringer A, Applying new diagnostic criteria for acute kidney injury to facilitate early identification of nephrotoxicity in vancomycin-treated patients, Antimicrob. Agents Chemother 55 (2011) 3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Frenette AJ, Bouchard J, Bernier P, Charbonneau A, Nguyen LT, Rioux J, Troyanov S, Williamson DR, Albumin Administration Is Associated With Acute Kidney Injury in Cardiac Surgery : A Propensity Score Analysis, 2014, pp. 1–11. [DOI] [PMC free article] [PubMed]
- [38].Lee E-H, Kim W-J, Kim J-Y, Chin J-H, Choi D-K, Sim J-Y, Choo S-J, Chung C-H, Lee J-W, Choi I-C, Effect of exogenous albumin on the incidence of postoperative acute kidney injury in patients undergoing off-pump coronary artery bypass surgery with a preoperative albumin level of less than 4.0 g/dl, Anesthesiol. J. Am. Soc. Anesthesiol 124 (2016) 1001–1011. [DOI] [PubMed] [Google Scholar]
- [39].Schortgen F, Girou E, Deye N, Brochard L, Group CS, et al. , The risk associated with hyperoncotic colloids in patients with shock, Intensive Care Med 34 (2008) 2157. [DOI] [PubMed] [Google Scholar]
- [40].Sigrist NE, Kälin N, Dreyfus A, Changes in serum creatinine concentration and acute kidney injury (AKI) grade in dogs treated with hydroxyethyl starch 130/0.4 from 2013 to 2015, J. Vet. Intern. Med 31 (2017) 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matheny ME, Ohno-Machado L, Resnic FS, Discrimination and calibration of mortality risk prediction models in interventional cardiology, J. Biomed. Inform 38 (2005) 367–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


