Abstract
Volumetric estimates of subcortical and cortical structures, extracted from T1‐weighted MRIs, are widely used in many clinical and research applications. Here, we investigate the impact of the presence of white matter hyperintensities (WMHs) on FreeSurfer gray matter (GM) structure volumes and its possible bias on functional relationships. T1‐weighted images from 1,077 participants (4,321 timepoints) from the Alzheimer's Disease Neuroimaging Initiative were processed with FreeSurfer version 6.0.0. WMHs were segmented using a previously validated algorithm on either T2‐weighted or Fluid‐attenuated inversion recovery images. Mixed‐effects models were used to assess the relationships between overlapping WMHs and GM structure volumes and overall WMH burden, as well as to investigate whether such overlaps impact associations with age, diagnosis, and cognitive performance. Participants with higher WMH volumes had higher overlaps with GM volumes of bilateral caudate, cerebral cortex, putamen, thalamus, pallidum, and accumbens areas (p < .0001). When not corrected for WMHs, caudate volumes increased with age (p < .0001) and were not different between cognitively healthy individuals and age‐matched probable Alzheimer's disease patients. After correcting for WMHs, caudate volumes decreased with age (p < .0001), and Alzheimer's disease patients had lower caudate volumes than cognitively healthy individuals (p < .01). Uncorrected caudate volume was not associated with ADAS13 scores, whereas corrected lower caudate volumes were significantly associated with poorer cognitive performance (p < .0001). Presence of WMHs leads to systematic inaccuracies in GM segmentations, particularly for the caudate, which can also change clinical associations. While specifically measured for the Freesurfer toolkit, this problem likely affects other algorithms.
Keywords: Alzheimer's disease, FreeSurfer, gray matter segmentation, white matter hyperintensities
Presence of WMHs leads to systematic inaccuracies in GM segmentations, which can also change clinical associations. While specifically measured for the Freesurfer toolkit, this problem likely affects other algorithms.
1. INTRODUCTION
White matter hyperintensities (WMHs) are defined as areas of increased signal on T2‐weighted (T2w) and Fluid‐attenuated inversion recovery (FLAIR) magnetic resonance images (MRIs) (Raman, Kantarci, Murray, Jack, & Vemuri, 2016). WMHs are associated with a variety of underlying pathologies, such as amyloid angiopathy, arteriosclerosis, axonal loss, blood–brain barrier leakage, degeneration, demyelination, gliosis, hypoperfusion, hypoxia, and inflammation (Abraham et al., 2016). WMHs are commonly present in the otherwise asymptomatic aging population but are at higher prevalence in many diseases such as Alzheimer's disease (AD), diabetes, frontotemporal dementia, HIV, lewy body dementia, mild cognitive impairment (MCI), obesity, Parkinson's disease, and vascular dementia (Appelman et al., 2009; Barber et al., 1999; Caroppo et al., 2014; Dadar et al., 2018; Debette & Markus, 2010; Gouw et al., 2008; Kandiah et al., 2013; Sudre et al., 2017).
On T1‐weighted (T1w) MRI sequences, WMHs appear hypointense with respect to the normal‐appearing white matter, with intensities that can be very similar to cortical and subcortical gray matter (GM) (Dadar et al., 2017). The T1w intensity of WMHs is also associated with severity of damage to the tissue, with areas of higher damage appearing more hypointense (Dadar, Maranzano, Ducharme, & Collins, 2019).
As T1w images are the most commonly used structural MRI sequences in clinical and neuroscience applications, especially for purposes of segmentation and estimation of volumes for all or specific structures of interest (Mateos‐Pérez et al., 2018), the similarity in T1w intensity profiles of WMHs and GM gives rise to an important methodological question: can T1w MRI‐based GM structure segmentation differentiate between WMHs and GM? If not, how much of WMHs will be tagged as GM in their segmentation estimates, and if so, is this error systematic enough to bias results?
In the context of Multiple Sclerosis (MS) patients, the impact of hypointense MS lesions on tissue classification and GM structure segmentation is well‐established (Chard, Jackson, Miller, & Wheeler‐Kingshott, 2010; Gelineau‐Morel et al., 2012; González‐Villà et al., 2016; González‐Villà, Oliver, Huo, Lladó, & Landman, 2019). Simulating lesions with varying volumes and intensity ranges, Chard et al showed that both GM and WM volumes estimated by SPM were affected by presence of WM lesions (Chard et al., 2010). Similarly, using both real and simulated data, Gelineau‐Morel showed that presence of WM lesions can influence volume and shape estimations of GM measures in segmentations from FSL FAST (Gelineau‐Morel et al., 2012). In the context of MS, lesion filling approaches are generally employed before tissue classification and GM segmentation steps, to avoid the segmentations errors caused by presence of MS lesions (Battaglini, Jenkinson, & Stefano, 2012; Chard et al., 2010; Gelineau‐Morel et al., 2012; González‐Villà et al., 2019; Prados et al., 2016; Valverde et al., 2015; Valverde, Oliver, & Lladó, 2014).
To answer this question, we propose our study of WMHs segmentation bias in subcortical and cortical GM structures. More specifically, we investigated (a) whether there was any systematic overlap between WMHs and GM segmentations, and therefore volumetric biases; and (b) whether this overlap affected clinical findings (i.e., associations with cognitive scores). To these ends we used two segmentation tools, the first being FreeSurfer, one of the most commonly used publicly available brain segmentation tools (Fischl, 2012), and a previously validated tool for WMHs segmentation on multi‐contrast MRIs (Dadar et al., 2017), both applied on longitudinal data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database.
2. METHODS
2.1. Participants
We used longitudinal data from 1,077 participants (4,321 individual images from various timepoints) from the ADNI‐1, ADNI‐2, and ADNI‐GO database (adni.loni.usc.edu) that had T1w and either T2w/PDw or FLAIR MRIs available. The ADNI was launched in 2003 as a public‐private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. The study was approved by the institutional review board of all participating sites and written informed consent was obtained from all participants before inclusion in the study.
2.2. MRI acquisition and preprocessing
Table 1 summarizes MR imaging parameters for the data used in this study (for more details, see http://adni.loni.usc.edu/methods/documents/mri‐protocols/).
TABLE 1.
Scanner information and MRI acquisition parameters for ADNI1, and ADNI2/GO datasets
| Sequence | T1w | T2w/PDw or FLAIR | |||||
|---|---|---|---|---|---|---|---|
| ADNI1 | Manufacturer | GE | Siemens | Philips | GE | Siemens | Philips |
| Number of subjects | 331 | 281 | 55 | 331 | 281 | 55 | |
| Number of Timepoints | 1,342 | 1,062 | 155 | 1,342 | 1,062 | 155 | |
| Field strength | 1.5 T | 1.5 T | 1.5 T | 1.5 T | 1.5 T | 1.5 T | |
| Slice thickness | 1.2 mm | 1.2 mm | 1.2 mm | 3 mm | 3 mm | 3 mm | |
| No. of slices | 160 | 160 | 170 | 56 | 48 | 48 | |
| Field of view | 260 mm | 240 mm | 240 mm | 260 mm | 240 mm | 240 mm | |
| Scan matrix | 192 × 192 cm2 | 192 × 192 cm2 | 192 × 192 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | |
| Repetition time (TR) | 3,000 ms | 2,400 ms | Shortest | 3,000 ms | 3,000 ms | 3,000 ms | |
| Echo time (TE) | Min full | 3.5 ms | User defined (3 ms) | Min full/100 ms | 12/97 ms | User defined | |
| Flip angle | 8 | 8 | 8 | ‐ | 150 | 90 | |
| ADNI2/GO | Manufacturer | GE | Siemens | Philips | GE | Siemens | Philips |
| Number of subjects | 12 | 349 | 134 | 12 | 349 | 134 | |
| Number of Timepoints | 24 | 1,196 | 455 | 24 | 1,196 | 455 | |
| Field strength | 3 T | 3 T | 3 T | 3 T | 3 T | 3 T | |
| Slice thickness | 1.2 mm | 1.2 mm | 1.2 mm | 5 mm | 5 mm | 5 mm | |
| No. of slices | 200 | 176 | 170 | 42 | 35 | 35 | |
| Field of view | 260 mm | 256 mm | 256 mm | 220 mm | 220 mm | 220 mm | |
| Scan matrix | 256 × 256 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | 256 × 256 cm2 | |
| Repetition time (TR) | 7.2 ms | 2,300 ms | Shortest | 11,000 ms | 9,000 ms | User defined | |
| Echo time (TE) | Min full | 2.98 ms | Shortest | 147 ms | 90 ms | User defined | |
| Flip angle | 11 | 9 | 9 | ‐ | 150 | 150 | |
Note: T2w/PDw or FLAIR column presents the parameters for T2w/PDw and FLAIR acquisitions in ADNI1, and ADNI2/GO datasets, respectively.
2.3. GM segmentations
All T1w images were identically processed using FreeSurfer version 6.0.0 (recon‐all ‐all). FreeSurfer is an open source software (https://surfer.nmr.mgh.harvard.edu/) that provides a full processing stream for structural T1w data (Fischl, 2012). The final segmentation output (aseg.mgz) was then used to obtain structure masks and volumes based on the look up table available at https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/AnatomicalROI/FreeSurferColorLUT.
2.4. WMH segmentations
T1w, T2w/PDw, and FLAIR scans were pre‐processed as follows: (a) image denoising (Manjón, Coupé, Martí‐Bonmatí, Collins, & Robles, 2010); (b) intensity inhomogeneity correction; and (c) intensity normalization to a 0–100 range. For each subject, the T2w, PDw, or FLAIR scans were then co‐registered to the structural T1w scan of the same timepoint using a 6‐parameter rigid registration and a mutual information objective function (Dadar, Fonov, Collins, & Initiative, 2018). We used a previously validated WMH segmentation method that employs a set of location and intensity features in combination with a random forests classifier to detect WMHs using either T1w + FLAIR or T1w + T2w/PDw images. The training library consisted in manual, expert segmentations of WMHs from 100 subjects from ADNI (not included in the current sample) (Dadar, Pascoal, et al., 2017; Dadar, Maranzano, Misquitta, et al., 2017; Dadar, Maranzano, et al., 2018). WMHs were automatically segmented at all timepoints and then co‐registered to the T1w images using the obtained rigid registrations, in order to assess overlaps between WMHs and GM segmentations.
2.5. Cognitive evaluations
All subjects received a comprehensive battery of clinical assessments and cognitive testing based on a standardized protocol (adni.loni.usc.edu) (Petersen et al., 2010). At each visit, participants underwent a series of assessments including the Alzheimer's Disease Assessment Scale‐13 (ADAS13) (Mohs & Cohen, 1987), which was used to assess cognitive performance.
2.6. Statistical analyses
Overlaps between GM and WMH segmentations were calculated (number of overlapping voxels in mm3) for each subcortical and cortical GM region. The following mixed effects models were used to assess whether the WMH‐GM overlaps were associated with overall WMH burden, controlling for age and sex.
| (1) |
Since both WMHs and GM volumes are highly correlated with age, and WMHs might have a different prevalence between men and women, we controlled for age and sex, to account for the likelihood that (a portion of) the relationships might be driven by intercorrelations between these factors.
Mixed effects models were also used to assess the relationships between GM volumes and age and diagnostic cohort, and GM volumes and cognition, once using the GM volume estimates obtained directly from the FreeSurfer segmentation, and once after removing the regions overlapping with the WMH segmentations.
| (2) |
| (3) |
All volumes were normalized by the individual's intracranial volume. Total WMH loads and WMH_GM overlaps were log‐transformed to obtain normal distributions. All mixed effects models included Subject as categorical random variables, and Scanner Manufacturer and Field Strength as categorical fixed effects to account for any potential variabilities caused by contrast differences in the images from different scanners. Scanner Manufacturer denotes a categorical variable contrasting scans from Siemens Healthcare (Erlangen, GER), Philips Medical Systems (Best, Netherlands), and GE Healthcare (Milwaukee, WI) manufacturers. The results were corrected for multiple comparisons using the false discovery rate (FDR) controlling method with a significance threshold of p = .05 (Benjamini & Hochberg, 1995).
2.7. Data and code availability statement
Data used in this article is available at http://adni.loni.usc.edu/. FreeSurfer and the WMH segmentation pipeline used are also publicly available at https://surfer.nmr.mgh.harvard.edu/and http://nist.mni.mcgill.ca/?p=221, respectively.
3. RESULTS
3.1. Study participants
Preprocessed and registered images were visually assessed for quality control (presence of imaging artifacts, failure in registrations). WMH segmentations were also visually assessed for missing hyperintensities or over‐segmentation. Either failures resulted in the participant being removed from the analyses. All MRI processing, segmentation and quality control steps were blinded to clinical outcomes. All cases passed co‐registration QC. Figure 1 summarizes the QC information for the subjects that were excluded. The final sample included 1,077 participants (4,321 timpoints) with WMH and FreeSurfer segmentations.
FIGURE 1.
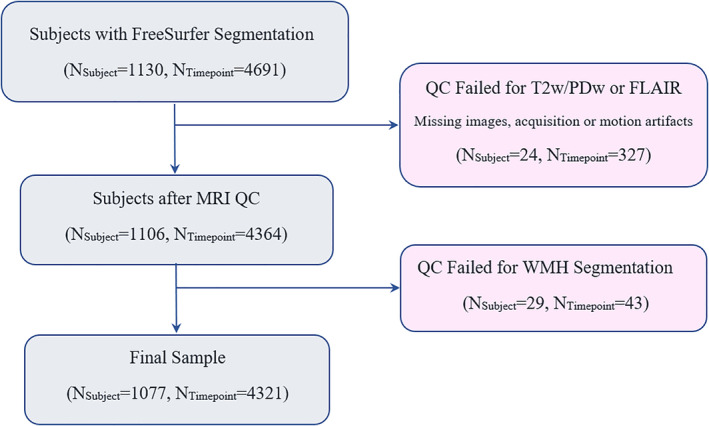
Flowchart of subjects in the study
3.2. Segmentations
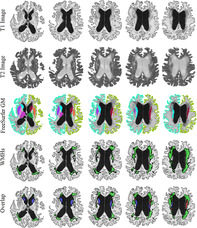
Figure 2 shows an example of a subject with a high degree of overlap between FreeSurfer caudate segmentations and WMH segmentations. The first two rows show axials slices of T1w and T2w images, respectively. The third and fourth rows show the FreeSurfer GM and WMH segmentations overlaid on the T1w images, respectively. The last row shows overlapping areas between caudate segmentations and WMH segmentations in red, the areas only segmented as caudate in blue, and the areas only segmented as WMHs in green. Note the intensity of the T1w hypointensities (i.e., T2w WMHs), ranging from slightly hypointense compared with the normal appearing WM, to iso‐intense to cortical and subcortical GM, and finally approaching CSF intensity ranges. Based solely on T1w images, it would be very challenging to differentiate WMHs from GM, particularly in areas where the two tissue types share a boundary. This is exactly the area where over‐segmentation occurs (i.e., the red regions are always at the border of caudate and WMH regions), with FreeSurfer segmenting the caudate more generously, including some of the neighboring WM.
FIGURE 2.
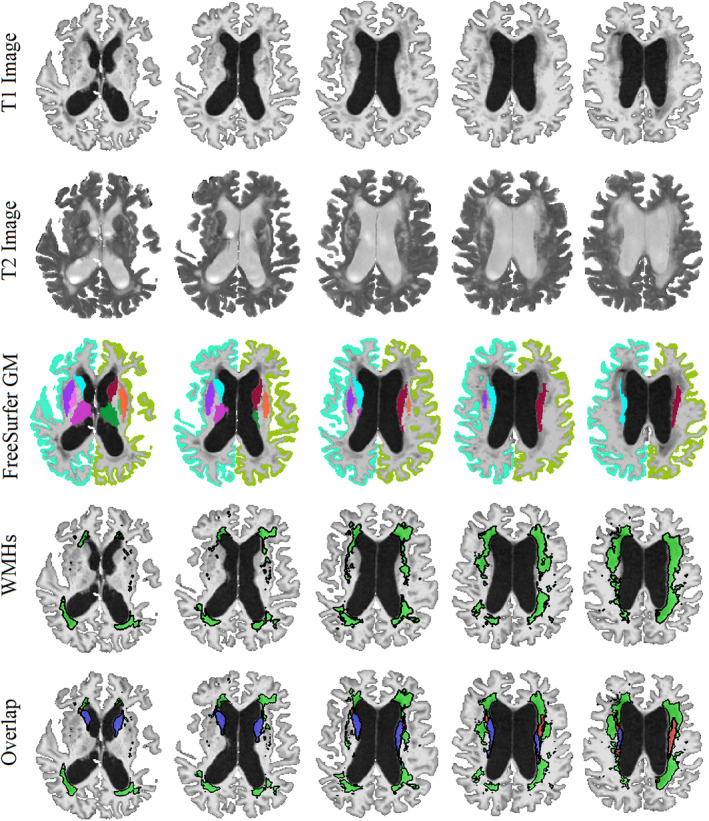
Example of a case with high WMH and caudate segmentation overlap. First and second rows: axial slices showing the T1 and T2 images, respectively (note the hyperintense WMH areas on T2 images and the corresponding hypointensities in T1 images). Third row: FreeSurfer GM segmentations overlaid on T1 images. Fourth row: WMH segmentations overlaid on T1 images. Last row: the overlap between caudate and WMH segmentations. GM = Gray Matter. WMH = White Matter Hyperintensity. Blue = Caudate. Green = WMHs. Red = The overlapping voxels between caudate and WMH segmentations
3.3. Segmentation overlap
Table 2 shows the average amount of overlap between WMHs and FreeSurfer segmentations for each GM structure, as well as their association with the overall WMH burden, controlling for age and sex (Equation (1)). Caudate segmentations had by far the highest percentage of overlapping WMHs (6% of the mean caudate volume). The overlapping volumes were significantly related to the overall WMH burden for bilateral caudate, cerebral cortex, putamen, thalamus, pallidum, accumbens area, and the right hippocampus (p < .0002) (Figure 3). Figure 2 shows the associations for the top six regions. In addition, we observed an overall trend of smaller GM and WMH overlaps for measures driven from 3 T scanners, with left caudate, right cerebral cortex, and putamen reaching significance (Table S1). GM volumes derived from Siemens and Philips scanners also had smaller overlaps with WMH segmentations in comparison with GE in general, with bilateral cerebral cortex, putamen, and pallidum passing the significance threshold after correction for multiple comparisons in both, and caudate only for Philips versus GE comparisons. GM volumes derived from Philips scanners also had smaller overlaps with WMH segmentations in comparison with Siemens, with left putamen, bilateral pallidum, and right amygdala and ventral diencephalon passing the threshold for significance.
TABLE 2.
Overlaps between WMHs and FreeSurfer GM segmentations, and their associations with overal WMH burden. The regions are sorted based on effect size. Significant results after FDR correction are indicated in bold font
| Structure name | FreeSurfer label | Volume (mm3) | Overlap with WMH (mm3) | Percentage of overlap (%) | Association with WMH | |
|---|---|---|---|---|---|---|
| T stat | p value | |||||
| Caudate—right | 50 | 3,532.9 ± 596.2 | 217.2 ± 271.0 | 6.153 | 46.57 | <.0001 |
| Caudate—left | 11 | 3,395.9 ± 556.4 | 222.9 ± 283.9 | 6.565 | 46.02 | <.0001 |
| Cerebral cortex—left | 3 | 202,864 ± 25,017 | 73.8 ± 240.5 | 0.036 | 31.81 | <.0001 |
| Cerebral cortex—right | 42 | 203,763 ± 25,182 | 79.8 ± 218.8 | 0.039 | 30.03 | <.0001 |
| Putamen—right | 51 | 4,235.6 ± 644.1 | 21.9 ± 74.4 | 0.518 | 23.02 | <.0001 |
| Putamen—left | 12 | 4,194.8 ± 648.3 | 17.4 ± 56.1 | 0.416 | 21.20 | <.0001 |
| Thalamus—left | 10 | 6,598.1 ± 729.3 | 1.01 ± 5.72 | 0.015 | 12.25 | <.0001 |
| Thalamus—right | 49 | 6,499.9 ± 711.3 | 0.55 ± 4.07 | 0.008 | 8.76 | <.0001 |
| Pallidum—right | 52 | 1829.0 ± 256.6 | 0.85 ± 3.50 | 0.047 | 8.63 | <.0001 |
| Accumbens area—left | 26 | 405.1 ± 86.6 | 0.23 ± 1.96 | 0.057 | 7.80 | <.0001 |
| Pallidum—left | 13 | 1863.1 ± 255.1 | 0.45 ± 2.50 | 0.024 | 5.99 | <.0001 |
| Accumbens area—right | 58 | 452.7 ± 90.7 | 0.17 ± 0.88 | 0.038 | 4.80 | <.0001 |
| Hippocampus—right | 53 | 3,572.6 ± 600.3 | 0.22 ± 2.24 | 0.006 | 3.70 | .0002 |
| Ventral diencephalon—right | 60 | 3,754.9 ± 459.5 | 0.10 ± 1.78 | 0.003 | 1.98 | .05 |
| Amygdala—right | 54 | 1,469.7 ± 319.3 | 0.04 ± 0.88 | 0.003 | 1.59 | .11 |
| Amygdala—left | 18 | 1,275.3 ± 299.1 | 0.01 ± 0.23 | 0.0001 | 0.77 | .44 |
| Ventral diencephalon—left | 28 | 3,782.2 ± 473.9 | 0.06 ± 2.03 | 0.002 | 0.74 | .45 |
| Hippocampus—left | 17 | 3,476.3 ± 564.9 | 0.17 ± 4.77 | 0.005 | 0.35 | .72 |
FIGURE 3.
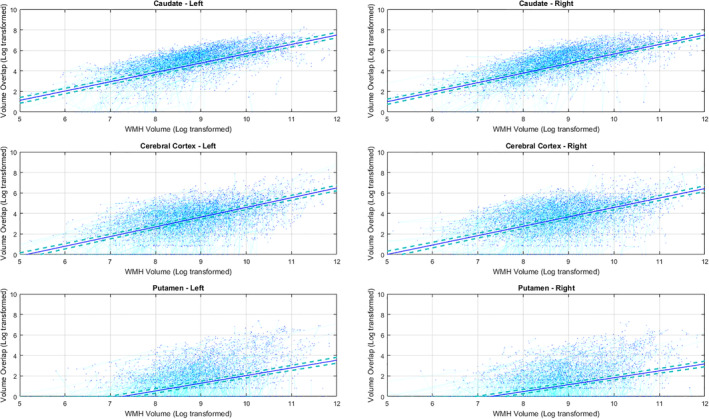
The association between overlapping GM and WMH volumes and overall WMH burden (Table 2). Subjects with higher WMH loads also have greater amounts of WMH overlap with FreeSurfer GM segmentations in bilateral caudate, cerebral cortex, and putamen. GM = Gray Matter. WMH = White Matter Hyperintensity
To better demonstrate how much the misclassification of WMHs as caudate might increase caudate volume estimates, we have also plotted volume overlaps for caudate without log‐transformation (i.e., volumes in mm3) in Figure 4. In extreme cases, for individuals with very high WMH burden (log transformed value of 11, equivalent to 50,000 mm3), caudate volume can be over‐estimated by more than 1,000 mm3, equivalent to 30% of the average caudate volume.
FIGURE 4.
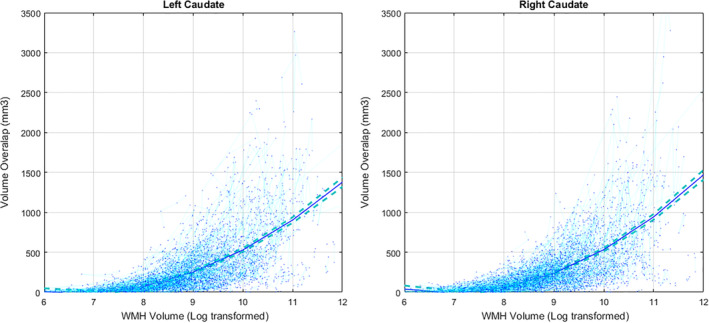
The association between overlapping Caudate and WMH volumes (in mm3) and overall WMH burden. Subjects with higher WMH loads also have greater amounts of WMH overlap with FreeSurfer caudate segmentations. WMH, White Matter Hyperintensity
3.4. Associations
Uncorrected caudate volumes increased with age (p < .0001, Table S3), and MCI and AD groups had slightly higher volumes than the normal aging (NA) cohort (although not significant), whereas the corrected caudate volumes decreased with age (p < .0001), and MCI and AD groups had slightly lower volumes than the NA group (only the AD vs. NA difference was significant with p = .009). The uncorrected and corrected results were similar in terms of effect size and direction of associations for other regions, with the corrected volumes having slightly larger effects sizes for putamen. Table S2 shows the associations between GM volumes and age for all regions (Equation (2)), before and after removing the voxels overlapping with WMHs (Table 3).
TABLE 3.
Associations between uncorrected and corrected caudate volumes, age, and diagnostic cohort. Significant results after FDR correction are indicated in bold font
| Structure name | Age | MCI vs NA | AD vs NA | ||||
|---|---|---|---|---|---|---|---|
| T stat | p value | T stat | p value | T stat | p value | ||
| Caudate—right | Uncorrected | 4.05 | <.0001 | 0.53 | .60 | 0.65 | .52 |
| Corrected | −5.04 | <.0001 | −1.23 | .21 | −2.38 | .01 | |
| Caudate—left | Uncorrected | 5.85 | <.0001 | 0.81 | .42 | 0.31 | .76 |
| Corrected | −7.90 | <.0001 | −1.43 | .15 | −2.59 | .009 | |
Uncorrected caudate volumes were not significantly associated with ADAS13 scores (Table 4), whereas lower corrected caudate volumes were significantly associated with higher ADAS13 scores (i.e., poorer cognitive performance, p < .0001). The uncorrected and corrected results were similar in terms of effect size and direction of associations for other regions, with the corrected volumes having slightly larger effects sizes for putamen. Table S3 shows the associations between ADAS13 and GM volumes for all regions, before and after removing the voxels overlapping with WMHs. Figure 5 shows the associations between uncorrected and corrected caudate volumes and ADAS13 scores.
TABLE 4.
Associations between uncorrected and corrected caudate volumes and ADAS13. Significant results after FDR correction are indicated in bold font
| Structure name | Uncorrected | Corrected | ||
|---|---|---|---|---|
| T stat | p value | T stat | p value | |
| Caudate—right | 0.27 | .78 | −6.47 | <.0001 |
| Caudate—left | 0.66 | .50 | −9.27 | <.0001 |
FIGURE 5.
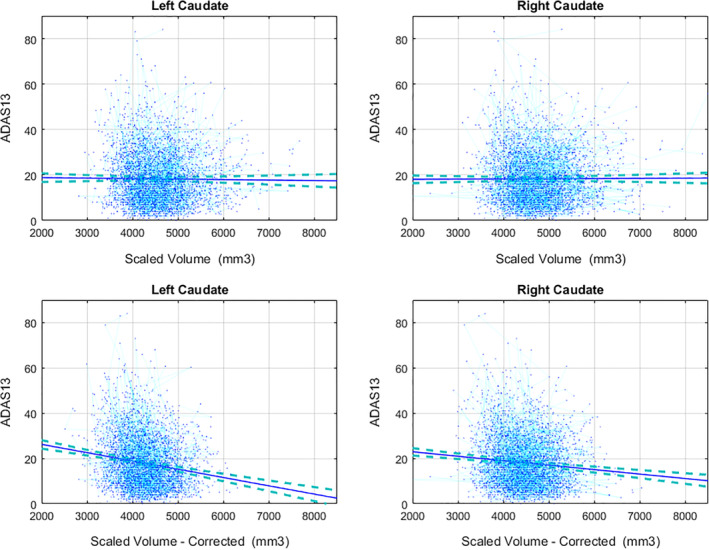
The association between uncorrected and corrected Caudate volumes ADAS13 scores. Uncorrected volumes were not associated with ADAS13 scores, whereas lower corrected caudate volumes were significantly associated with higher ADAS13 scores
4. DISCUSSION
In this study, we investigated the impact of WMHs on GM segmentations, specifically for the Freesurfer segmentation tool, in order to determine whether WMHs might lead to systematic segmentation errors in certain GM regions, and whether such errors might impact associations between GM volumes and clinical outcomes. We found such errors in a number of regions, and in the caudate in particular it propagated to the association with clinical variables.
We found that overlapping voxel volumes between WMH and GM segmentations were significantly associated with overal WMH burden (Table 2 and Figure 3), indicating higher error rates for subjects with high WMH loads. This affected both cortical and subcortical structures, and in particular the caudates bilaterally. Uncorrected, caudate volumes showed a significant increase with age, which is highly unlikely to be a real effect given that all regions, except the caudate, have been shown to decline in late‐life cognitively healthy individuals (Potvin, Mouiha, Dieumegarde, Duchesne, & Initiative, 2016a, 2016b). This was further improbable given that the population of this study consists of not only aging individuals but also patients with MCI and AD, which are known to have increasing levels of atrophy across cortical areas. In contrast, the corrected caudate volumes significantly decreased with age, as could be expected. Given that WMHs are highly prevalent in the periventricular regions (i.e., white matter areas surrounding the caudate), the significant increase estimate is likely due to the fact that WMH burden increases with age and AD and MCI patients tend to have higher WMH loads and faster WMH progression.
Along the same line, corrected caudate volumes showed significant differences between AD and cognitively healthy cohorts, as well as significant associations with cognitive performance in the expected directions, whereas the uncorrected volumes were not significantly different and in the opposite direction (Table 3). This again highlights the fact that if uncorrected for overlapping WMHs, estimates of caudate volumes can lead to incorrect or unreliable findings, particularly in populations with high prevalence of WMHs.
While other GM regions also had overlaps with WMHs that were significantly associated with the overal WMH burden (e.g., cerebral cortex, putamen), the amount and percentage of these overlaps were not nearly as high as caudate (17, 22, 80, and 74 mm3 vs. 217 and 223 mm3 or 0.4%, 0.5%, 0.04%, and 0.04% vs. 6.1% and 6.5%), and therefore, they did not affect their overall estimates and associations with age, diagnosis, and ADAS13. However, this might not be the case in other populations, where WMHs are more prevalent, or have different distributions. Therefore, WMHs should be accurately segmented and used to correct GM segmentations wherever such errors occur, and not only for caudate.
The current analysis has the underlying assumption that any overlapping voxels between WMH segmentations (based on T2w/PD and FLAIR images) and FreeSurfer segmentations (based only on T1w images) were WMHs that were misclassified as GM by FreeSurfer. This is a reasonable assumption, since subcortical structures appear hypointense on T2w and FLAIR images, whereas WMHs appear hyperintense. The fact that subcortical structures and WMHs have contrasting intensity profiles in T2w and FLAIR images (i.e., hypointense vs. hyperintense, respectively) and similar intensity profiles in T1w images (hypointense for both) makes it unlikely that they would be incorrectly classified as WMHs, as the WMH segmentations are mainly driven by the intensity profile in T2w and FLAIR sequences. Additionally, the WMH segmentation method used here has been developed and extensively validated for use in multi‐site and multi‐scanner studies, and has been previously used in several multi‐site datasets, including ADNI (Anor, Dadar, Collins, & Tartaglia, 2020; Dadar et al., 2019; Dadar et al., 2020; Dadar, Camicioli, Duchesne, Collins, & Initiative, 2020; Dadar, Gee, Shuaib, Duchesne, & Camicioli, 2020; Misquitta et al., 2018; Sanford et al., 2019). The training library used consists of manually segmented labels from the same dataset (ADNI), to ensure optimal classifier performance (Dadar, Maranzano, Misquitta, et al., 2017). Further, all automatic WMH segmentations were visually assessed by an expert, and cases that failed this QC step were removed from this analysis.
FreeSurfer is one of the most widely used publicly available brain segmentation tools. Large databases such as the UK Biobank provide GM structure volumes derived from FreeSurfer to researchers. In line with our findings, other researchers have reported a positive association between WMH load and FreeSurfer caudate volumes in the UK biobank participants (Morys, Dadar, & Dagher, 2020) and in another large sample of cognitively healthy individuals (Potvin, Dieumegarde, Duchesne, & Initiative, 2017). Studies investigating a larger age range report a U‐shape curve for caudate volumes, decreasing from early adulthood to the 60s, and then increasing afterwards (Fjell et al., 2009, 2013; Goodro, Sameti, Patenaude, & Fein, 2012; Pfefferbaum et al., 2013; Potvin et al., 2016b; Walhovd et al., 2011). Given that WMHs generally occur in this same age range (i.e., after 60s), these results are also likely due to the segmentation errors caused by presence of WMHs in older participants.
Specifically for this algorithm, our study emphasizes the need for correcting FreeSurfer GM volume estimates for WMHs, particularly for the caudate. However, it is likely that other algorithms exhibit the same behavior. Evidence can be found in the literature, for example in the works by Goodro et al. and Pfefferbaum et al. using FSL volumes (Goodro et al., 2012; Pfefferbaum et al., 2013). Developers and users alike should therefore be aware of the possibility of systematic bias from not taking into account WMHs in GM segmentation from T1w images.
In addition to the consequences the presence of WMHs might have on the GM structure borders, depending on the segmentation segmentation technique, they might also impact gray level distribution, and could thus lead to an overall bias and a global shift of the borders in partial volume areas. Previous work by Gelineau‐Morel et al. and also Chard et al. in Multiple Sclerosis patients has demonstrated this effect on FAST from FSL and SPM5 tissue segmentations, respectively (Ashburner et al., 2014; Chard et al., 2010; Gelineau‐Morel et al., 2012; Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012). In other words, misclassification of lesions as GM might lead to an overall increase in the mean GM intensities, causing GM boundaries to shift toward lighter (WM) regions, further increasing overall GM volume estimates (Chard et al., 2010; Gelineau‐Morel et al., 2012). In the context of MS, lesion filling approaches are generally employed before tissue classification and GM segmentation steps, to avoid the segmentations errors caused by presence of MS lesions (Battaglini et al., 2012; Chard et al., 2010; Gelineau‐Morel et al., 2012; González‐Villà et al., 2019; Prados et al., 2016; Valverde et al., 2014; Valverde et al., 2015).
The WMHs that are observed in the aging and neurodegenerative populations have a different pathogenesis from MS lesions. They generally co‐occur with (extensive) atrophy, and have more varying loads and less well‐shaped borders than MS lesions (Caligiuri et al., 2015). Another important factor in dealing with WMHs in the aging populations is the fact that the MRI contrast between GM and WM tissues decreases with age, making the distinction between the boundaries more challenging. Most lesion filling methods have assessed the impact of lesion filling in simulated lesions on young healthy brains. While these results might be generalizable to MS patients, the presence of extensive atrophy, differences in the intensity ranges and distributions of WMHs, and the lower GM/WM contrast in the aging populations, makes their results less likely to generalize to the aging cohorts. Therefore, further validation is necessary to demonstrate the impact of lesion filling methods in aging and neurodegenerative populations.
FreeSurfer GM segmentation pipeline performs T1w hypointensity segmentation simultaneously and tries to capture as many of the WMHs as possible to avoid misclassifying them as GM. Therefore, the segmentation errors that occur when using FreeSurfer might be much more subtle than FSL and SPM, where lesion filling methods have been applied and proven to be useful in the context of MS. In fact, a previous study by Guo et al, has assessed the impact of lesion filling in improving FreeSurfer, FSL‐SIENAX, SPM and SPM‐CAT segmentations in MS patients, showing that while lesion filling changed the output of FSL‐SIENAX and SPM, FreeSurfer output was not affected since it already takes WM hypointensities into account (Guo, Ferreira, Fink, Westman, & Granberg, 2019).
More recently, certain tissue classification and GM segmentation tools such as SPM12 (Ashburner et al., 2014, p. 12) and FAST from FSL (Jenkinson et al., 2012) have provided multi‐channel segmentation options, allowing users to perform segmentations with multiple MRI sequences (e.g., T2w and PDw). Such multi‐channel approaches might help reduce the segmentation errors caused by WMHs, and further studies investigating their performance in presence of WMHs are warranted.
In conclusion, the presence of WMHs can lead to systematic errors in GM segmentations in certain regions, particularly in the caudate, which, if not corrected, can impact findings in populations with high WMH prevalence.
Supporting information
Table S1 The effect of field strength and scanner manufacturer on WMH and FreeSurfer GM segmentation overlaps. Significant results after FDR correction are indicated in bold font.
Table S2 Associations between uncorrected and corrected GM volumes and ADAS13. Significant results after FDR correction are indicated in bold font.
Table S3 Associations between uncorrected and corrected GM volumes, age, and diagnostic cohort. Significant results after FDR correction are indicated in bold font.
ACKNOWLEDGMENTS
MD is supported by a scholarship from the Canadian Consortium on Neurodegeneration in Aging in which SD and RC are co‐investigators. The Consortium is supported by a grant from the Canadian Institutes of Health Research with funding from several partners including the Alzheimer Society of Canada, Sanofi, and Women's Brain Health Initiative. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Dadar M, Potvin O, Camicioli R, Duchesne S, for the Alzheimer's Disease Neuroimaging Initiative. Beware of white matter hyperintensities causing systematic errors in FreeSurfer gray matter segmentations!. Hum Brain Mapp. 2021;42:2734–2745. 10.1002/hbm.25398
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-ontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding information Canadian Consortium on Neurodegeneration in Aging; Women's Brain Health Initiative; Sanofi; Alzheimer Society of Canada; Canadian Institutes of Health Research
DATA AVAILABILITY STATEMENT
Data used in this article is available at http://adni.loni.usc.edu/. Links to the pipelines used for GM and WMH segmentations (which are both publicly available) are provided within the text of the manuscript.
REFERENCES
- Abraham, H. M. A. , Wolfson, L. , Moscufo, N. , Guttmann, C. R. , Kaplan, R. F. , & White, W. B. (2016). Cardiovascular risk factors and small vessel disease of the brain: Blood pressure, white matter lesions, and functional decline in older persons. Journal of Cerebral Blood Flow and Metabolism, 36, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anor, C. J. , Dadar, M. , Collins, D. L. , & Tartaglia, M. C. (2020). The longitudinal assessment of neuropsychiatric symptoms in mild cognitive impairment and Alzheimer's disease and their association with White matter Hyperintensities in the National Alzheimer's coordinating Center's uniform data set. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 6(1), 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelman, A. P. , Exalto, L. G. , Van Der Graaf, Y. , Biessels, G. J. , Mali, W. P. , & Geerlings, M. I. (2009). White matter lesions and brain atrophy: More than shared risk factors? A systematic review. Cerebrovascular Diseases, 28, 227–242. [DOI] [PubMed] [Google Scholar]
- Ashburner, J. , Barnes, G. , Chen, C. , Daunizeau, J. , Flandin, G. , Friston, K. , … Moran, R. (2014). SPM12 manual. UK: Wellcome Trust Cent. Neuroimaging Lond. [Google Scholar]
- Barber, R. , Scheltens, P. , Gholkar, A. , Ballard, C. , McKeith, I. , Ince, P. , … O'Brien, J. (1999). White matter lesions on magnetic resonance imaging in dementia with Lewy bodies, Alzheimer's disease, vascular dementia, and normal aging. Journal of Neurology, Neurosurgery, and Psychiatry, 67, 66–72. 10.1136/jnnp.67.1.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini, M. , Jenkinson, M. , & Stefano, N. D. (2012). Evaluating and reducing the impact of white matter lesions on brain volume measurements. Human Brain Mapping, 33, 2062–2071. 10.1002/hbm.21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B: Methodological, 57, 289–300. [Google Scholar]
- Caligiuri, M. E. , Perrotta, P. , Augimeri, A. , Rocca, F. , Quattrone, A. , & Cherubini, A. (2015). Automatic detection of White matter Hyperintensities in healthy aging and pathology using magnetic resonance imaging: A review. Neuroinformatics, 13, 261–276. 10.1007/s12021-015-9260-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroppo, P. , Ber, I. L. , Camuzat, A. , Clot, F. , Naccache, L. , Lamari, F. , … Brice, A. (2014). Extensive White matter involvement in patients with frontotemporal lobar degeneration: Think Progranulin. JAMA Neurology, 71, 1562–1566. 10.1001/jamaneurol.2014.1316 [DOI] [PubMed] [Google Scholar]
- Chard, D. T. , Jackson, J. S. , Miller, D. H. , & Wheeler‐Kingshott, C. A. M. (2010). Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. Journal of Magnetic Resonance Imaging, 32, 223–228. 10.1002/jmri.22214 [DOI] [PubMed] [Google Scholar]
- Dadar, M. , Camicioli, R. , Duchesne, S. , Collins, D. L. , & Initiative, A. D. N. (2020). The temporal relationships between white matter hyperintensities, neurodegeneration, amyloid beta, and cognition. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, 12, e12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Fereshtehnejad, S.‐M. , Zeighami, Y. , Dagher, A. , Postuma, R. B. , & Collins, D. L. (2020). White matter hyperintensities mediate impact of dysautonomia on cognition in Parkinson's disease. Pract: Mov. Disord. Clin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Fonov, V. S. , Collins, D. L. , & Initiative, A. D. N. (2018). A comparison of publicly available linear MRI stereotaxic registration techniques. NeuroImage, 174, 191–200. [DOI] [PubMed] [Google Scholar]
- Dadar, M. , Gee, M. , Shuaib, A. , Duchesne, S. , & Camicioli, R. (2020). Cognitive and motor correlates of grey and white matter pathology in Parkinson's disease. NeuroImage: Clinical, 27, 102353. 10.1016/j.nicl.2020.102353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Maranzano, J. , Ducharme, S. , Carmichael, O. T. , Decarli, C. , & Collins, D. L. (2017). Validation of T1w‐based segmentations of white matter hyperintensity volumes in large‐scale datasets of aging. Human Brain Mapping, 39(3), 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Maranzano, J. , Ducharme, S. , Carmichael, O. T. , Decarli, C. , Collins, D. L. , & Initiative, A. D. N. (2018). Validation of T 1w‐based segmentations of white matter hyperintensity volumes in large‐scale datasets of aging. Human Brain Mapping, 39, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Maranzano, J. , Ducharme, S. , & Collins, D. L. (2019). White matter in different regions evolves differently during progression to dementia. Neurobiology of Aging, 76, 71–79. 10.1016/j.neurobiolaging.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Dadar, M. , Maranzano, J. , Misquitta, K. , Anor, C. J. , Fonov, V. S. , Tartaglia, M. C. , … Alzheimer's Disease Neuroimaging Initiative . (2017). Performance comparison of 10 different classification techniques in segmenting white matter hyperintensities in aging. NeuroImage, 157, 233–249. 10.1016/j.neuroimage.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadar, M. , Pascoal, T. , Manitsirikul, S. , Misquitta, K. , Tartaglia, C. , Brietner, J. , … Collins, D. L. (2017). Validation of a regression technique for segmentation of White matter Hyperintensities in Alzheimer's disease. IEEE Transactions on Medical Imaging, 36, 1758–1768. [DOI] [PubMed] [Google Scholar]
- Debette, S. , & Markus, H. S. (2010). The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: Systematic review and meta‐analysis. BMJ, 341, c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell, A. M. , Westlye, L. T. , Amlien, I. , Espeseth, T. , Reinvang, I. , Raz, N. , … Walhovd, K. B. (2009). Minute effects of sex on the aging brain: A multisample magnetic resonance imaging study of healthy aging and Alzheimer's disease. The Journal of Neuroscience, 29, 8774–8783. 10.1523/JNEUROSCI.0115-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell, A. M. , Westlye, L. T. , Grydeland, H. , Amlien, I. , Espeseth, T. , Reinvang, I. , … Walhovd, K. B. (2013). Critical ages in the life course of the adult brain: Nonlinear subcortical aging. Neurobiology of Aging, 34, 2239–2247. 10.1016/j.neurobiolaging.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelineau‐Morel, R. , Tomassini, V. , Jenkinson, M. , Johansen‐Berg, H. , Matthews, P. M. , & Palace, J. (2012). The effect of hypointense white matter lesions on automated gray matter segmentation in multiple sclerosis. Human Brain Mapping, 33, 2802–2814. 10.1002/hbm.21402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Villà, S. , Oliver, A. , Huo, Y. , Lladó, X. , & Landman, B. A. (2019). Brain structure segmentation in the presence of multiple sclerosis lesions. NeuroImage Clinical, 22, 101709. 10.1016/j.nicl.2019.101709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Villà, S. , Oliver, A. , Valverde, S. , Wang, L. , Zwiggelaar, R. , & Lladó, X. (2016). A review on brain structures segmentation in magnetic resonance imaging. Artificial Intelligence in Medicine, 73, 45–69. 10.1016/j.artmed.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Goodro, M. , Sameti, M. , Patenaude, B. , & Fein, G. (2012). Age effect on subcortical structures in healthy adults. Psychiatry Research: Neuroimaging, 203, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw, A. A. , van der Flier, W. M. , Fazekas, F. , van Straaten, E. C. , Pantoni, L. , Poggesi, A. , … Waldemar, G. (2008). Progression of white matter hyperintensities and incidence of new lacunes over a 3‐year period: The Leukoaraiosis and disability study. Stroke, 39, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Guo, C. , Ferreira, D. , Fink, K. , Westman, E. , & Granberg, T. (2019). Repeatability and reproducibility of FreeSurfer, FSL‐SIENAX and SPM brain volumetric measurements and the effect of lesion filling in multiple sclerosis. European Radiology, 29, 1355–1364. 10.1007/s00330-018-5710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). Fsl. NeuroImage, 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Kandiah, N. , Mak, E. , Ng, A. , Huang, S. , Au, W. L. , Sitoh, Y. Y. , & Tan, L. C. S. (2013). Cerebral white matter hyperintensity in Parkinson's disease: A major risk factor for mild cognitive impairment. Parkinsonism & Related Disorders, 19, 680–683. 10.1016/j.parkreldis.2013.03.008 [DOI] [PubMed] [Google Scholar]
- Manjón, J. V. , Coupé, P. , Martí‐Bonmatí, L. , Collins, D. L. , & Robles, M. (2010). Adaptive non‐local means denoising of MR images with spatially varying noise levels. Journal of Magnetic Resonance Imaging, 31, 192–203. [DOI] [PubMed] [Google Scholar]
- Mateos‐Pérez, J. M. , Dadar, M. , Lacalle‐Aurioles, M. , Iturria‐Medina, Y. , Zeighami, Y. , & Evans, A. C. (2018). Structural neuroimaging as clinical predictor: A review of machine learning applications. NeuroImage Clinical, 20, 506–522. 10.1016/j.nicl.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta, K. , Dadar, M. , Tarazi, A. , Hussain, M. W. , Alatwi, M. K. , Ebraheem, A. , … Tartaglia, M. C. (2018). The relationship between brain atrophy and cognitive‐behavioural symptoms in retired Canadian football players with multiple concussions. NeuroImage Clinical, 19, 551–558. 10.1016/j.nicl.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohs, R. C. , & Cohen, L. (1987). Alzheimer's disease assessment scale (ADAS). Psychopharmacology Bulletin, 24, 627–628. [PubMed] [Google Scholar]
- Morys, F. , Dadar, M., & Dagher, A. (2020). Obesity impairs cognitive function via metabolic syndrome and cerebrovascular disease: An SEM analysis in 15,000 adults from the UK Biobank. The Journal of Clinical Endocrinology and Metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, R. C. , Aisen, P. S. , Beckett, L. A. , Donohue, M. C. , Gamst, A. C. , Harvey, D. J. , … others . (2010). Alzheimer's disease neuroimaging Initiative (ADNI) clinical characterization. Neurology, 74, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum, A. , Rohlfing, T. , Rosenbloom, M. J. , Chu, W. , Colrain, I. M. , & Sullivan, E. V. (2013). Variation in longitudinal trajectories of regional brain volumes of healthy men and women (ages 10 to 85 years) measured with atlas‐based parcellation of MRI. NeuroImage, 65, 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin, O. , Dieumegarde, L. , Duchesne, S. , & Initiative, A. D. N. (2017). Normative morphometric data for cerebral cortical areas over the lifetime of the adult human brain. NeuroImage, 156, 315–339. [DOI] [PubMed] [Google Scholar]
- Potvin, O. , Mouiha, A. , Dieumegarde, L. , Duchesne, S. , & Initiative, A. s. D. N. (2016a). FreeSurfer subcortical normative data. Data in Brief, 9, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin, O. , Mouiha, A. , Dieumegarde, L. , Duchesne, S. , & Initiative, A. D. N. (2016b). Normative data for subcortical regional volumes over the lifetime of the adult human brain. NeuroImage, 137, 9–20. [DOI] [PubMed] [Google Scholar]
- Prados, F. , Cardoso, M. J. , Kanber, B. , Ciccarelli, O. , Kapoor, R. , Wheeler‐Kingshott, C. A. G. , & Ourselin, S. (2016). A multi‐time‐point modality‐agnostic patch‐based method for lesion filling in multiple sclerosis. NeuroImage, 139, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, M. R. , Kantarci, K. , Murray, M. E. , Jack, C. R. , & Vemuri, P. (2016). Imaging markers of cerebrovascular pathologies: Pathophysiology, clinical presentation, and risk factors. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, 5, 5–14. 10.1016/j.dadm.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, R. , Strain, J. , Dadar, M. , Maranzano, J. , Bonnet, A. , Mayo, N. E. , … Collins, D. L. (2019). HIV infection and cerebral small vessel disease are independently associated with brain atrophy and cognitive impairment. Aids, 33, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre, C. H. , Bocchetta, M. , Cash, D. , Thomas, D. L. , Woollacott, I. , Dick, K. M. , … Warren, J. D. (2017). White matter hyperintensities are seen only in GRN mutation carriers in the GENFI cohort. NeuroImage Clinical, 15, 171–180. 10.1016/j.nicl.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, S. , Oliver, A. , & Lladó, X. (2014). A white matter lesion‐filling approach to improve brain tissue volume measurements. NeuroImage Clinical, 6, 86–92. 10.1016/j.nicl.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, S. , Oliver, A. , Roura, E. , Pareto, D. , Vilanova, J. C. , Ramió‐Torrentà, L. , … Lladó, X. (2015). Quantifying brain tissue volume in multiple sclerosis with automated lesion segmentation and filling. NeuroImage Clinical, 9, 640–647. 10.1016/j.nicl.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd, K. B. , Westlye, L. T. , Amlien, I. , Espeseth, T. , Reinvang, I. , Raz, N. , … Fjell, A. M. (2011). Consistent neuroanatomical age‐related volume differences across multiple samples. Neurobiology of Aging, 32, 916–932. 10.1016/j.neurobiolaging.2009.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The effect of field strength and scanner manufacturer on WMH and FreeSurfer GM segmentation overlaps. Significant results after FDR correction are indicated in bold font.
Table S2 Associations between uncorrected and corrected GM volumes and ADAS13. Significant results after FDR correction are indicated in bold font.
Table S3 Associations between uncorrected and corrected GM volumes, age, and diagnostic cohort. Significant results after FDR correction are indicated in bold font.
Data Availability Statement
Data used in this article is available at http://adni.loni.usc.edu/. FreeSurfer and the WMH segmentation pipeline used are also publicly available at https://surfer.nmr.mgh.harvard.edu/and http://nist.mni.mcgill.ca/?p=221, respectively.
Data used in this article is available at http://adni.loni.usc.edu/. Links to the pipelines used for GM and WMH segmentations (which are both publicly available) are provided within the text of the manuscript.


