Abstract
The coronavirus disease 2019 (COVID-19) pandemic is caused by newly discovered severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2). One of the striking targets amongst all the proteins in coronavirus is the main protease (Mpro), as it plays vital biological roles in replication and maturation of the virus, and hence the potential target. The aim of this study is to repurpose the Food and Drug Administration (FDA) approved molecules via computer-aided drug designing against Mpro (PDB ID: 6Y2F) of SARS CoV-2 due to its high x-ray resolution of 1.95 Å as compared to other published Mprostructures. High Through Virtual Screening (HTVS) of 2456 FDA approved drugs using structure-based docking were analyzed. Molecular Dynamics simulations were performed to check the overall structural stability (RMSD), Cα fluctuations (RMSF) and protein-ligand interactions. Further, trajectory analysis was performed to assess the binding quality by exploiting the protein-residue motion cross correlation (DCCM) and binding free energy (MM/GBSA). Tenofovir, an antiretroviral for HIV-proteases and Terlipressin, a vasoconstrictor show stable RMSD, RMSF, better MM/GBSA with good cross correlation as compared to the Apo and O6K. Moreover, the results show concurrence with Nelfinavir, Lopinavir and Ritonavir which have shown significant inhibition in in vitro studies. Therefore, we conclude that Tenofovir and Terlipresssin might also show protease inhibition but are still open to clinical validation in case of SARS-CoV 2 treatment.
Communicated by Ramaswamy H. Sarma
Keywords: Main protease (Mpro), molecular mechanics/Generalized87 Born model and solvent accessibility (MM/GBSA), molecular dynamics (MD), dynamic cross-correlation matrix (DCCM), severe acute respiratory syndrome coronavirus 2 (SARS-CoV2)
1. Introduction
An unusual strain of CoV, SARS-CoV-2, caused an atypical pneumonia outbreak in late December 2019 in Wuhan, China, and was termed Coronavirus disease 2019 (COVID-19). The World Health Organization has stated COVID-19 a pandemic, and urgent treatments are required as the world is stepping into a phase that is beyond containment. All the efforts are being made to curb the spread of the disease, but it is difficult to develop new medications for a rapidly mutating virus like SARS CoV 2. Drug repurposing is a new promising approach in which already approved and available drugs are being used to fight against rare diseases (Strittmatter, 2014). The broad-spectrum antiviral drugs are ideally suited for such outbreaks; the main motive here is not to necessarily understand all mechanisms of action of a broad-spectrum antiviral drug but instead choosing a drug with minimum side effects. Earlier repurposing therapy was successful in diseases such as HIV/AIDS (Zidovudine), erectile dysfunction (Sildenafil), Parkinson’s disease (Atomoxetine), Breast cancer (Raloxifene), Epilepsy (Topiramate). Advantages in using a previously available drug is that the chance of failure is reduced, the time and cost required to develop a new drug is also reduced (Xue et al., 2018). Various computational and experiment-based studies are conducted on drug repurposing, for example, gene association, pathway mapping, molecular docking, clinical analysis, novel data sources, in vitro and in vivo screening, etc (Pushpakom et al., 2019). According to a study Ombitasvir, a Hepatitis C polymerase inhibitor, was ‘repurposed’ to target and prevent beta-tubulin driven breast cancers (Karuppasamy et al., 2017). Further, these days computer-aided studies are being conducted which are quite beneficial and reduce the manpower and related resources.
In silico studies are useful in target identification and protein structure determination; with the help of computer-aided drug designing, the structure of the target is identified (Prajapat, Sarma, Shekhar, Avti, et al., 2020). Recently, scientists have started exploiting computational resources to explore the mechanisms of action, or the efficacy of a drug towards a target. Experimental studies capture some properties, but in order to analyze the interactions between the inhibitor and protein, and to access the conformational changes at the molecular levels, computational simlations are encouraged (Boopathi et al., 2020). Recently, the efficiency of an antiviral drug, Darunavir was analyzed against the binding site i.e. the major flap region of the protease (HIV-1). Thus, computer aided drug designing and analysis can provide an insight into various unknown mechanisms (Karuppasamy et al., 2017; Purohit & Sethumadhavan, 2009). Docking and molecular dynamic studies are proved to be excellent tools in designing new drugs (Verma et al., 2017). In the case of COVID-19, many studies have provided a blueprint about the structure of the virus. Coronaviruses (CoVs) are enveloped viruses containing positive-strand RNA and have spikes of glycoproteins that project from their viral envelopes and give them a corona or halo-like appearance. Strains of CoV were the cause of severe acute respiratory syndrome (SARS) epidemic in 2002 and the Middle East respiratory syndrome (MERS) epidemic in 2012. The viral genome is the largest known RNA genome, it ranges from 26 to 32 kb, and the viral particle has a diameter of 125 nm (González-Andrade et al., 2016). The CoV proteins help in replication, and some of them also contribute to the interactions between the host and the virus. This genome act as mRNA on infecting a cell and leads to synthesis of 2 long polypeptides that are responsible for further infection. It produces structural and non-structural proteins. Structural protein includes membrane protein (M), envelope protein (E), spike protein (S), nucleocaspid protein (N) and proteases. These proteins are present in viral membrane and comprise different functions, like the RNA genome assembly is carried by N protein and viral assembly by M and E protein. The NTD of the nucleocaspid protein aids in the formation of ribonucleoprotein complex that is necessary for viral replication. According to an in silico study, theophylline and pyrimidone derivatives can be considered as the probable inhibitors of RNA binding to the NTD of N protein (Prajapat, Sarma, Shekhar, Prakash, et al., 2020; Sarma, Kaur, et al., 2020; Sarma, Prajapat, et al., 2020). The trimeric glycoprotein S guides the entry of the virus into the host cells (Prajapat, Sarma, Shekhar, Avti, et al., 2020; Zhavoronkov et al., 2020). The CoVs share structural similarities, but they undergo genetic evolutions that provide new targets for antiviral vaccines and therapies. Various targets have been explored; many researchers are targeting these proteins which play a key role in viral replication because of their different mechanisms of action. Different studies are performed on the basis of structural aspects and variant functions and one of the potential and highly conserved targets is protease. Targeting protease can obstruct with its post-translational modifications and thus prevent the viral invasion into the human body. A characteristic CoV contains at least six open reading frames (ORFs) in its genome (Mousavizadeh & Ghasemi, 2020) which are responsible for encoding the non-structural proteins (NSPs). The two large non-structural viral polyproteins, PP1a and PP1ab are produced due to the translation of ORF1a and ORF1ab. The viral-encoded proteases are used for processing of NSPs to their functional forms. The two major proteases that have gained importance these days and that are potential targets for COVID-19 are the papain-like protease (PLpro) and a 3-chymotrypsin-like protease (3CLpro), which are also known as the main protease (Mpro) (Báez-Santos et al., 2015). For the replication and transcription of virus Mproand PLpro play essential roles, cysteine and histidine are responsible for cleavage of the NSPs. The main protease 3CLpro that is detected in the COVID-19 resembles 96% to the SARS COV; it has two identical subunits that form a dyad, which together forms the active sites responsible for its cleaving property. Mpro is a ∼ 306 aa long protein, and PLpro has a catalytic core domain of 316 amino acids responsible for cleavage of replicase substrates (Holmes, 2003). Mpro cleaves the polyprotein’s C-terminal and slice at eleven sites, whereas PLpro works at the N-terminal, cleaving three specific sites in the polyprotein (Liu et al., 2020). As both 3CLpro and PLpro are cysteine proteases, covalent inhibitors with high potencies can be potentially developed for precise targeting (Liu et al., 2020). Similarly, sixteen NSPs are cleaved and processed that have an essential role in the formation of the replicase complex, which is further exploited by the virus for replication and transcription. PLpro can remove ubiquitin from the ubiquitinated proteinase from the interferons and interferon regulatory factor 3 (IRF3) of host cell, so that the interferons are not produced and thus the innate immune system is compromised. Deubiquitination can inactivate the pathway for nuclear factor κ-light-chain enhancer of activated B cells (NFκB), this causes suppression of immune system of the host. Therefore, proteases can be a potential target to cure patients of COVID-19 and these are highly conserved sequences. Various studies are performed to identify therapeutics against SARS COV and MERS COV due to similarity in their structures. Proteases are targeted earlier in various viral diseases, for example in case of HIV and Hepatitis C, and with sudden outbreak of COVID-19 there is an urgent need for the treatment.
The process of drug repurposing can be beneficial in this situation since development is a lengthy procedure, and it might take a long time to develop drugs that mainly target COVID-19. Therefore, previously available medicines, already approved by the FDA, are widely being tested for various targets in COVID-19. Medications that are recently considered for antiviral treatment include hydroxychloroquine (HCQ), Lopinavir, Ritonavir, Ledipasvir, Velpatasvir, and Nelfinavir. Although treatment with hydroxychloroquine has recorded a decline in the number of cases, the safety profile is also necessary, and therefore few studies have discussed this point. The benefit of the HCQ + Azithrol treatment is research in progress, as a lot of uncertainty prevails (Sarma, Kaur, et al., 2020). Through in silico research (Liu & Wang, 2020) showed that the binding active site for Lopinavir/Ritonavir is conserved between SARS-COV-Mpro and 2019-n COV Mpro. Lopinavir and Ritonavir are used to target protease in HIV, and they were successful in inhibiting the main protease; these drugs can form H-bonds with amino acid Thr24-Asn28, Asn119 and thus can interfere with the COVID-19 binding.
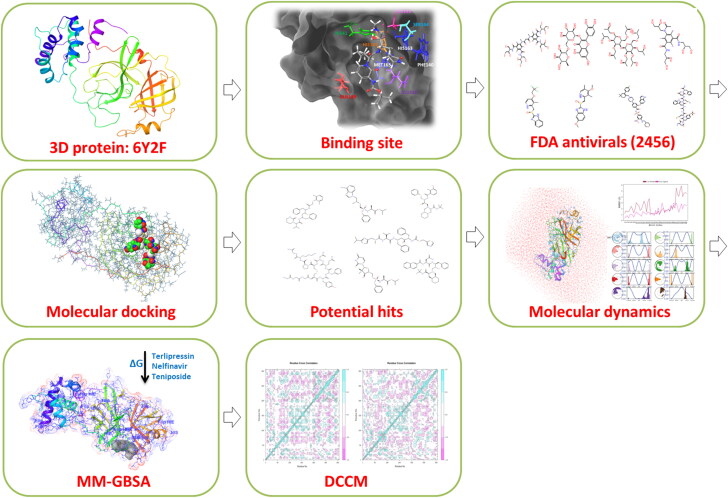
A 3-D model of 2019-n COV was presented by Chen et al. (2020), which depicted the conserved cleavage sites of 3CLpro as in SARS-COV, and they also used Ledipasvir or Velpatasvir as a repurposing therapy. Further (Xu et al., 2020), using homology modeling and docking studies proposed that Nelfinavir might be a potential inhibitor against n2019-n COV Mpro. Liponavir and Saquinavir have been proved to block CoV protease, although Lopinavir is a more potent inhibitor than Saquinavir. Most drugs involved in repurposing to target COVID-19 are known to target critical components in the viral lifecycle. For example, medications like Umifenovir and chloroquine hinder the entry of viral particles into the host cells. Lopinavir/Ritonavir, ASC09, or Darunavir block viral replication by inhibiting 3CL protease. Remdesivir, Favipiravir, Ribavirin is proved to inhibit viral RNA synthesis (Harrison, 2020). The novelty of the study is in the approach applied for the high throughput virtual screening of potent ligands. The study focusses on the utilization of computational methods to screen potent lead within FDA approved molecules exhibiting efficient binding profile with SARS-CoV-2 M pro protein. This was assessed using the combinatorial scheme of analysis of binding free energy trend in MD simulations, protein-ligand binding profile and conformational dynamics-based approach of correlated protein-residues interaction. The MD simulation output of protein-ligand frames helped to understand the conformational alterations during the entire 100 ns simulation duration. The DCCM analysis was performed to understand the diverse correlated motion of the aminoacid residues in the Mpro protein. This study shows noticeable conformational shifts in Mpro which are induced by FDA candidate molecules in comparison with the unbound (apo) and co-crystallized ligand (O6K) binding dynamics. A brief illustration of the methodology is depicted in Figure 1.
Figure 1.
Schematic representation of the study design illustration.
2. Material and methods
2.1. Computing system
The in silico experiments and analysis were proceeded on Linux Ubuntu OS 18.04.02 LTS (Acer Predator Helios 300 laptop), the Maestro, Schrödinger version 2019-3 was used. The GPU unit utilized for molecular modeling and dynamic simulations was Nvidia GeForce GTX 1660 Ti (6GB).
2.2. Protein structure and docking enrichment
The X-RAY crystal structure of the target of our interest was obtained from RCSB PDB (PDB ID: 6Y2F) by Zhang et al. (2020). The 3 D-structure obtained possessed a resolution of 1.95 Å and 306 amino acids. The validation of the structure was performed through SAVES v5.0 which predicts that according to the Ramachandran plot 88.8% (231a.a.) is the most favored region, 10% (26 a. a.) is the additional allowed region, 0.4% (1 a. a.) is the generously allowed region and 0.8% (2 a.a.) lie in the areas disallowed (Figure S1). Therefore, the server’s comprehensive report states that protease is a good structure for conducting further studies. For the protein preparation, Maestro version 10.2 was used, the OPLS3 force field was set to a minimum, and water molecules ≥ 5.0 Ao were excluded from ruling out any disturbance in the binding areas.
A cross-docking analysis between the native and modeled structure prepared for docking and simulations provides an RMSD (Root Mean Square Deviation) difference of 0.113 Å; the lower resolution makes it a compatible structure (Figure S2). Decoys and actives-based enrichment analysis for appropriate interaction cut-off were performed using provided decoy and active sets of HIV-1 protease (PDB: 1XL2) from DUDE decoys (Mysinger et al., 2012). This HIV-1 protease decoy list was selected because of a similarity index of 19.23% in the protease of HIV and SARS-CoV 2, and this has been verified through Culstal omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) by allingning the sequences (Figure S3). The decoy-actives screening was performed using the Glide XP algorithm outputs subjected to Molecular mechanics/Generalized87 Born model and solvent accessibility (MM/GBSA) calculations using prime for the selected protease protein model (PDB: 6Y2F). The ranked hits obtained from MM/GBSA values were sorted from high stable to low stable compounds. Enrichment calculations were performed from the Maestro tool Enrichment calculator.
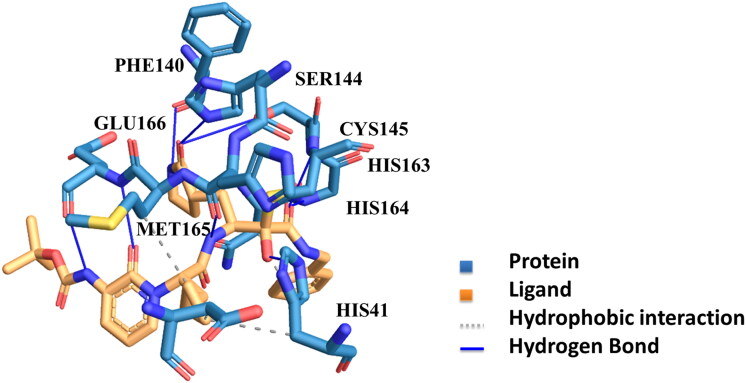
Figure 2.
The Substrate Binding Cleft of Protease: The figure depicts the hydrophobic interactions and hydrogen bonding developed between the ligand and the protein’s binding site.
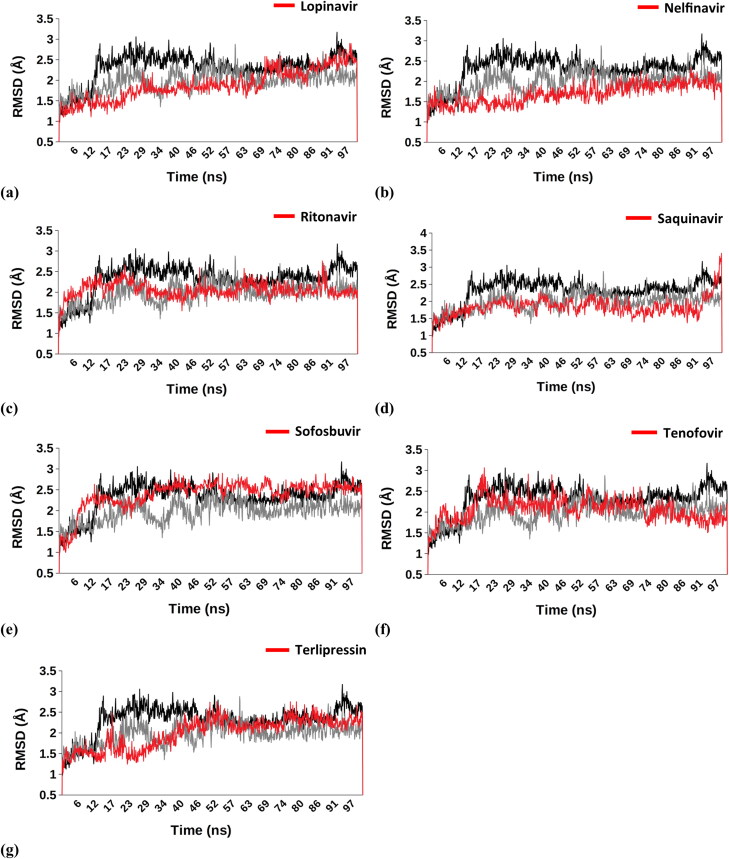
Figure 3.
Comparision-Cα RMSD – (a) Lopinavir (1.882 Å ± 0.375), (b) Nelfinavir (1.687 Å ± 0.251), (c) Ritonavir (2.048 Å ± 0.197), (d) Saquinavir (1.838 Å ± 0.269), (e) Sofosbuvir (2.395 Å ± 0.344), (f) Tenofovir (2.059 Å ± 0.277), (g) Terlipressin (1.998 Å ± 0.367); Apo (2.319 Å ± 0.363); O6K (1.976 Å ± 0.267).
2.3. Ligand preparation, primary screening and MM/GBSA (binding free energy)
FDA-approved drug dataset was taken from the Drug bank database that is a freely accessible platform. A total of 2456 drugs were selected for virtual screening against protease available until 27-03-2020 (DrugBank, n.d.). Further, the ligand was modified by ligprep in which OPLS 2003 force field was adjusted to obtain a ligand with minimum energy to make the ligand suitable for docking.
For the purpose of virtual screening, the glide docking module of Schrodinger was utilized to obtain accurate results. Docking was conducted in three different progressions; High-Throughput Virtual Screening (HTVS, Standard-Precision), Standard Precision (SP), and extra-precision (XP) using the glide program to screen selected 2456 drugs. The percentage of output for HTVS was set at 50%, for SP at 25%, and to improve the quality of selection, an enriched sampling mode and flexible docking were selected in XP with a percentage output of 25%. After docking, all the compounds were verified through MM/GBSA free energy score (Sarma, Shekhar, et al., 2020).
For calculating the free energy between the target and the ligand prime, MM/GBSA was used. To organize the results output, docking score was considered, and for molecular docking binding, free energy between ligand and receptor was measured through MM/GBSA in VSGB solvation at the OPLS3 force field. Molecular docking was employed through Schrodinger docking suits (Schrodinger Maestro, New York, NY, USA) (Bowers et al., 2006).
2.4. Molecular dynamics simulation
MD simulation was performed by utilizing Desmond (D E Shaw Research (DESRES)), through this protein/ligand complexes were analyzed (Mysinger et al., 2012; Zhang et al., 2020). The 3-site model TIP3P was used to achieve high computational efficiency considering the actual geometry of water molecules. A periodic cuboidal box of 10x10x20 0A was filled with protein-drug/ligand complexes, counter ions like 29 Na+ and 29 Cl- were added to neutralize the system. The number of water molecules added to the simulation were 10,505 molecules. The simulation was planned for 100 ns with Isothermal–isobaric (NPT) ensemble class possessing Nose-Hoover thermostat 310 K and Martyna-Tobias kleinbarostat at 1.0132 Pa. Desmond was used for analyzing the trajectory data.
2.4.1. MM/GBSA and DCCM
MM/GBSA was calculated using trajectory analysis. The experiment was set up at an interval of 1 ns for each protein-ligand complex using script thermal_mmgbsa.py available from schrodinger.com.
Dynamic cross-correlation matrix (DCCM) depicts the magnitude of the correlation coefficient, which depends on the degree to which the fluctuations of the system are associated. The cross-corelation was computed through the MD trajectory of the protease and ligand complex. Major characteristics of DCCMs comprise strong relativity along the diagonal, spreading out from diagonal and off-diagonal cross-relationships.The correlated motions of residues were analyzed to define the quality of protein-ligand complexes. The residue cross covariance matrix was calculated for atomic fluctuations procured through MD trajectories. Bio3D package in R studio was used to obtain insights from the MD trajectories of protein-ligand complexes (Grant et al., 2006).
3. Results
3.1. Protein binding sites
After analyzing the stereochemical quality of the protein structure through procheck, 6Y2F was selected and explored. The number of residues included in the structure was 303 a.a., with a resolution of 1.950A. The structure of the protein comprises three domains one, two, and three. The active site is expected somewhere in between six-stranded β barrels in domain one and domain two; domain three is usually concerned with the dimerization of the complex via interacting with amino acid GLU290 and ARG4 of two protomers (Shi & Song, 2006). A typical substrate sequence that is necessary to bind to the protein for Mpro is Leu-Gln (Ser, Ala, Gly) (Ziebuhr et al., 2000). The molecular dynamics simulation study of the available structure at RCSB bound with an inhibitor O6K (α- ketoamide) shows that the inhibitor binds to the protein through hydrogen bonding that depicts a major role in ligand binding, the interaction is also observed through water bridges, the inhibitor’s carbonyl oxygen bonds through a hydrogen bond to the main chain amide of the amino acid Glu166. The amino acids involved in the interaction include GLU166 (highest), GLN189 (second highest), HIS41, HIS164, HIS163, SER144, PHE140, MET165, and CYS145 Figure 2. The highest contact was with the residue number GLU166; the distance between the hydrogen atom and acceptor atom was 1.98, and an angle acquired was 168.61 degrees (Salentin et al., 2015). In order to narrow the site of interaction, a receptor grid was generated with dimensions 10 × 10 x 100A.
3.2. Enrichment analysis
Out of the total ligands (active (47) + decoy (200)) input of 247 molecules, the number of ranked actives was 30 in the top 100 molecules. The screened percent actives on the base of MM/GBSA energy gave ROC of 0.60 and BEDROC (α = 160.9) = 0.514, with 2 ranked actives in top 10 entries, the 1st ranked compound is active with MM/GBSA −69.30 kcal/mol (top 1%) and the second active that is listed at 5th position has −60.845 kcal/mol (top 5%) MM/GBSA. So, in the upcoming sections, the hits will be classified based on their scores equal or above −60.845 MM/GBSA score to reduce the percentage of false positives in our screening.
3.3. Virtual screening and molecular dynamics
In order to discover the probable binders to the target, an FDA approved library of the drugs was selected for screening against SARS-COV-2 protease. The molecular docking based virtual screening of 2456 ligands depicted good docking energy and secure interactions with the residues. The binding free energy attained via molecular docking of the drug molecules was analyzed and compared to the MM/GBSA (-69.30 kcal/mol and −60.845 kcal/mol) obtained through enrichment analysis crystallized inhibitors structures to protease molecule as actives. The details of the docking score and MM/GBSA ΔGbind score are compiled in Table 1. The lower values of binding free energies scores depicted the stable conformation attained by these protein complexes. The stability of the docked complexes was examined through molecular dynamics simulations, which was executed to ascertain the equilibrium of the docked compounds. Further, eight compounds were filtered, which included Terlipressin, Nelfinavir, Teniposide, Tenofovir, Saquinavir, Sofosbuvir, Lopinavir, and Ritonavir.
Table 1.
Docking score and MM/GBSA, ΔGbind (kcal/mol).
| Sr. no. | Compound names | Glide docking score | MM/GBSA ΔGbind |
|---|---|---|---|
| 1. | Angiotensin II | –6.966 | –98.338 |
| 2. | Sofosbuvir | –7.001 | –85.6 |
| 3. | Paclitaxel | –10.1 | –83.022 |
| 4. | Nelfinavir | –6.197 | –79.902 |
| 5. | Octreotide | –7.795 | –78.405 |
| 6. | Ritonavir | –9.37 | –76.259 |
| 7. | Floctafenine | –8.078 | –72.19 |
| 8. | Terlipressin | –10.332 | –71.868 |
| 9. | Flavin adenine dinucleotide | –8.849 | –70.809 |
| 10. | Lomitapide | –7.323 | –70.01 |
| 11. | Carfilzomib | –7.606 | –69.032 |
| 12. | Riboflavin | –8.67 | –68.72 |
| 13. | Idarubicin | –9.097 | –66.23 |
| 14. | Rutin | –11.287 | –64.905 |
| 15. | Tenofoviralafenamide | –7.266 | –64.704 |
| 16. | Delavirdine | –7.237 | –63.6 |
| 17. | Imidurea | –7.316 | –62.075 |
| 18. | Pimozide | –7.807 | –61.9 |
| 19. | Lopinavir | –7.252 | –61.363 |
| 20. | Daunorubicin | –8.403 | –61.26 |
| 21. | Saquinavir | –7.872 | –60.87 |
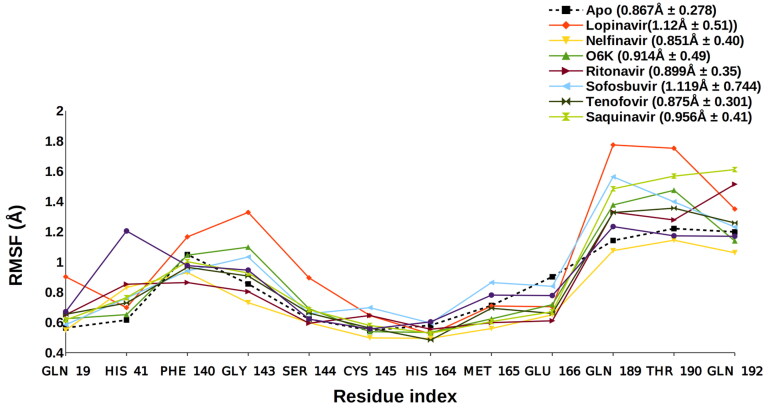
We conducted 100 nanoseconds molecular dynamic simulation (MDS) for each protein to understand the conformational changes that the protein undergoes after interacting with different ligands. The statistics obtained from the trajectory aided in investigating the Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuations (RMSF). The structure of proteins, conformational versatility, and stability were analyzed by comparing the RMSD of each ligand-bound complex with the unbound form (Apo) and reported inhibitor (O6K)-protease complex. During the MDS, the Cα-RMSD for respective drug molecule-Mpro complexes is aligned with the RMSD spectra of Apo form and O6K-bound form of the Mpro Figure 3. The average RMSD value for Apo form and O6K-bound Mpro was observed to be 2.319 and 1.976 Å, respectively. The Cα-RMSF fluctuations of binding site residues were analyzed to determine the outcome of the ligands on the protein’s active site residues dynamics. The combined RMSF spectra for the simulated complexes can be seen in Figure 4.
Figure 4.
Side Chain RMSF – Apo (0.867 Å ± 0.278), Lopinavir (1.12 Å ± 0.51), Nelfinavir (0.851 Å ± 0.40), O6K (0.914 Å ± 0.49), Ritonavir (0.899 Å ± 0.35), Sofosbuvir (1.119 Å ± 0.744), Tenofovir (0.875 Å ± 0.301), Saquinavir (0.956 Å ± 0.41), Terlipressin (0.96 Å ± 0.342).
3.4. MD trajectory analysis
3.4.1. MM/GBSA
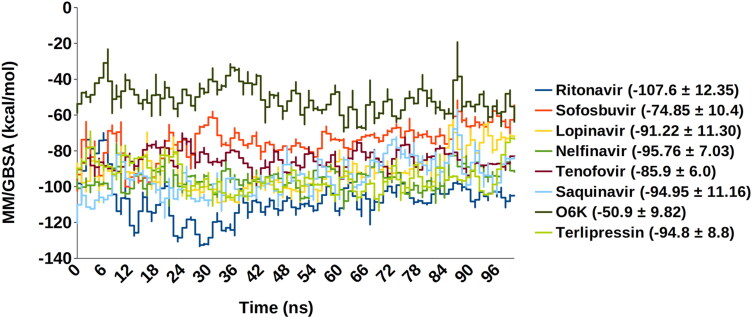
To evaluate the energetics of binding of protease to the different ligands, the energy parameters were calculated using protein-ligand complexes from the MD simulation trajectory. The respective average energies of the docked complexes in the simulated system ranged from −107.6 to −51 kcal/mol. The remaining compounds show stable and low binding free energy (ΔGbind) as compared to the native inhibitor (O6K). Figure 5 represents an exact comparison between the original inhibitor and drugs, which provides us with a clear iteration about the distinction in protein-ligand complexes’ energy states.
Figure 5.
MM/GBSA-Saquinavir (0–94.95 ± 11.16), Ritonavir (–107.6 ± 12.35), Tenofovir (–85.9 ± 6.0), Sofosbuvir (–74.85 ± 10.4), Lopinavir (–91.22 ± 11.30), Nelfinavir (–95.76 ± 7.03), O6K (–50.9 ± 9.82), Terlipressin (–94.8 ± 8.8).
3.4.2. DCCM
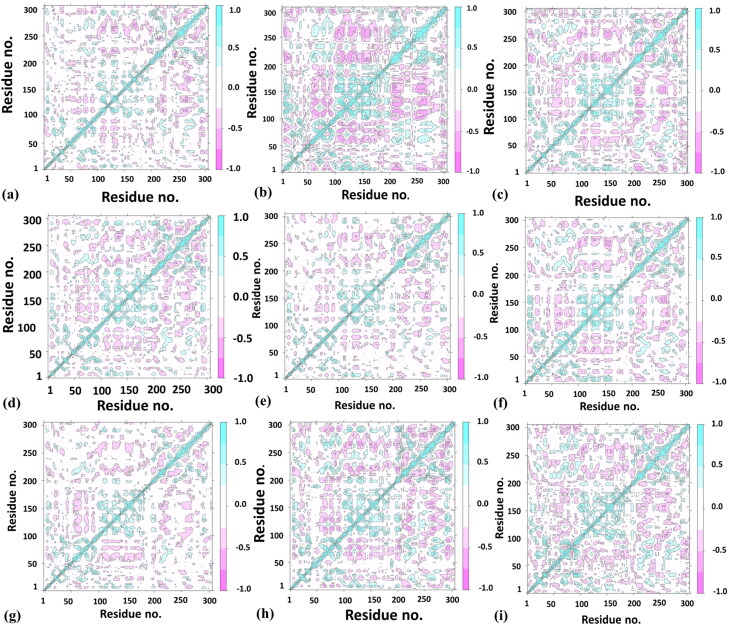
Dynamic cross-correlation matrix (DCCM) analysis represents a 2 D matrix representing correlation in the residue motions for MD simulations’ timeline. The densely colored regions show a respective positive and negative correlation between the protein residues, whereas the uncolored regions depict no correlation in residue movement. The highly correlated matrix represents strong interaction of the drug molecule with the protease binding site, bringing coordinated motions in the overall protein structure. Figure 6 depicts that Lopinavir shows the best and high correlation among protease residues compared with O6K, the original inhibitor, Nelfinavir, Saquinavir, Terlipressin, and Tenofovir were seen showing enhanced correlations. Whereas, reduced correlations were noticed for protease residues in complex with Sofosbuvir and Ritonavir, which describes the protease molecule’s reduced activity.
Figure 6.
DCCM analysis – (a) Apo, (b) Lopinavir, (c) Nelfinavir, (d) O6K, (e) Ritonavir, (f) Saquinavir, (g) Sofosbuvir, (h) Tenofovir, (i)Terlipressin.
4. Discussion
The virtual screening in this study facilitated a brief understanding of the biochemically crucial residues defining the binding site of the main protease (Mpro) enzyme. This binding cavity’s coordinates were obtained from the bound ligand (O6K) in the very crystal structure utilized for the present study, i.e. PDB ID: 6Y2F. The results were enriched with the enrichment analysis help using the reported actives binding the Mpro binding cleft, out of which a standard value was set (-60.845 kcal/mol); this lower limit value aided in the selection of the top seven compounds with potent interaction scores. The compounds were also analyzed by comparing them to the Apo state of Mpro and formerly bound inhibitor O6K. The substrates obtained belong to anti-cancerous and antiviral drug class, as summarized in Table 1. The topmost seven hit compounds selected against Mpro are as follows:
Nelfinavir is an antiretroviral drug that is a potent protease inhibitor and is used in the treatment of HIV; Lopinavir belongs to the class of viral protease inhibitors and is usually incorporated as a combination therapy with Ritonavir, which is a peptidomimetic agent used in inhibition of HIV-1 and HIV-2 proteases. Tenofovir is also an antiviral drug used against HIV and chronic hepatitis B. Terlipressin is an anti-tumor and hypotensive drug used in the treatment of carcinoid tumors and lowering the blood pressure; Saquinavir is an antiretroviral drug, it usually works in combination therapy with Lopinavir/Ritonavir to enhance its effects and has a terminal half-life of 17 hrs, it displays a high binding affinity to MPro (-9.0 kcal/mol) (Joshi et al., 2020). Sofosbuvir is an antiviral drug used in treating Hepatitis C. The mentioned inhibitors and their interactions with the binding residues of MPro are summarized in Table 2.
Table 2.
Comparison of protein-ligand interaction point of contacts.
| Sr. no. | Compound | Point of contact |
|---|---|---|
| 1. | O6K (α-ketoamide inhibitor) |
GLU166, HIS41, HIS164, HIS163, SER144, PHE140, MET165, and CYS145 (hydrogen bonds and water bridges), GLN189 |
| 2. | Terlipressin | GLU47, GLN189, GLU166 (Hydrogen bonds and water bridges) |
| 3. | Nelfinavir | GLN192, HIS41, HIS164, ASN142, GLU166 (Water bridges and H-bonding) |
| 4. | Tenofovir | GLN192, GLU166, GLY143, HIS41, THR190, CYS145: (little hydrophobic but majorly Water bridges and H-bonding) |
| 5. | Saquinavir | HIS164, GLN189, GLU166 (Water bridges and H-bonding) |
| 6. | Sofosbuvir | GLN189, GLU166 (Water bridges and H-bonding) |
| 7. | Lopinavir | MET49, HIS41, HIS164, GLU166,GLN189 (little hydrophobic but majorly H-bonding and Water bridges ) |
| 8. | Ritonavir | GLU166, GLY143 (H-bonding and Water bridges) |
An in silico study noticed that Nelfinavir and Entecavir could aid in targeting SARS-CoV 2. The improved binding and better interaction with the help of an anchor site is described by Huynh et al. (2020); the amino acid residues contacting with Nelfinavir and Entecavir include THR26, GLY143, HIS41, GLN189, and GLU166 (Huynh et al., 2020). Nelfinavir is one of the top drugs in this study, which also shows similar protein-ligand interactions (HIS41, GLU166). It possesses the most stable RMSD (2 Å) and RMSF (0.851 Å) even when compared to the O6K and Apo forms, the RMSF of O6K and Apo was 0.941 Å and 0.867 Å, respectively. The MM/GBSA score of its MD trajectory is comparatively good −95.76 kcal/mol compared to O6K −50.9 kcal/mol. The DCCM analysis also shows an enhanced correlation in residue motions. Moreover, Nelfinavir is also reported recently in various in vitro studies as a probable treatment for SARS CoV 2. Yamamoto et al. (2020) suggest that the effective concentrations of Nelfinavir to inhibit SARS Cov 2 would be 1.13 μM and 1.76 μM, respectively, at 50 percent and 90 percent inhibition. Similarly, a systemic screening of HIV inhibitors concludes that MesilateViracept (Nelfinavir) strongly inhibits S‐n‐and S‐o‐mediated cell fusion with full inhibition at a concentration of 10 μM (Musarrat et al., 2020).
A recent study about the repurposing of the antiviral drugs targeting Mpro reveals that their top three drugs include a combination therapy: Ritonavir-Lopinavir, Raltegravir, and Tipranavir, the residues found interacting with the protease were GLU166, HIS41, HIS164, GLN192, CYS142, ARG188. These repurposed drugs combination can be useful in targeting SARS-CoV 2 (Kumar et al., 2020). Lopinavir is an antiretroviral protease inhibitor that is already used for treating HIV since the 2000s. Ritonavir, along with other drugs, is a highly active antiretroviral therapy prescribed for treating HIV; therefore, it is preferred in combination with other protease inhibitors. In our study, Lopinavir and Ritonavir are amongst the top compounds which show good binding with the protease. Ritonavir and Lopinavir show a stable, less fluctuating RMSD than the unbound and native inhibitor and depicts a stable Cα-RMSF. Ritonavir possesses the best MM/GBSA score −107.6 kcal/mol, and Lopinavir also has a high score of −91.22 kcal/mol. Further, looking at the DCCM analysis results, we can predict that Lopinavir is highly correlated with protease residues whereas, Ritonavir shows reduced relation with the target.
Terlipressin is a vasoconstrictor which binds to vascular smooth muscle cell receptors V1 (Saner et al., 2007). It helps to alleviate systolic pulmonary artery pressure in patients of liver cirrhosis (Altintas et al., 2004). It shows a stable RMSD that equilibrates after 63 ns and almost overlaps with Apo form. The average RMSF value of Terlipressin is 0.96 Å, which is better than the Apo, O6K, and Lopinavir (1.12 Å), a reported SARS CoV 2 inhibitor. The DCCM analysis of the Terlipressin-bound Mpro residues reveals the strongest correlation among the residue motions, which attributes for the most robust interaction.
Tenofovir and Sofosbuvir, are described in a recent molecular docking study inhibiting the RNA dependant RNA polymerase (RdRp) of SARS CoV 2 (Elfiky, 2020). Tenofovir, an anti-retroviral drug, shows tight bonding with the Mpro and exhibits steady RMSD at and beyond 75 ns around 1.9 Å (2.06 Å ± 0.27). This lowering of protein RMSD is in synergy with the lowest observed deviation in the 100 ns MD trajectory’s free energy change. This is attributed to its efficient occupancy of the binding site with the least conformational fluctuation. Tenofovir possesses relatively low RMSF of around 0.899 Å, 0.956 Å, and 0.875 Å, respectively, with the residues in contact Figure 4. This local RMSF contact profile extends to explain good correlation in residue motions and intense interaction; see DCCM of Tenofovir in Figure 6.
Saquinavir and Sofosbuvir show stable RMSD and Cα-RMSF values compared to the initially reported inhibitor (O6K) and the unbound form (Apo). Moreover, all these possess a better MM/GBSA score of the MD trajectories when compared to Apo and O6K. According to the DCCM results, the least correlated amongst them was Sofosbuvir, whereas Saquinavir was correlated.
The above discussion is based on the approximations of MD analysis, and this predicts the possibility of the discussed drugs with the active site of SARS CoV-2 Mpro (PDB ID: 6Y2F). However, the screened drugs should be tested, and their in vitro potential should be revealed for further validation of the results.
5. Conclusion and future aspects
Considering the spread of infectious respiratory viral disease and rapidly mutating nature of the virus, there is a need to develop medication that can stop the rapid viral spread during the pandemics. An in silico drug repurposing research is conducted here to detect a potential inhibitor to target the key protein protease of SARS-CoV 2. A group of compounds representing a higher binding affinity towards protease active site were selected after high throughput virtual screening the entire FDA approved drug library. The top compounds obtained in our study are already in in vitro studies, such as Ritonavir (involved in highly active antiretroviral therapy), Lopinavir (involved in combination therapy with Ritonavir), and Nelfinavir (involved in combination therapy with Ritonavir). Terlipressin shows the closest similarity to these drugs, identical to the outcome of their trajectories, and provides a better result according to the Apo form and O6K. Extensive analysis of Terlipressin reveals marginally superior ligand activity as deduced from DCCM and interaction profile compared to the unbound, O6K, and reported antivirals. Moreover, Tenofovir was likewise observed to be a potent ligand for the Mpro inhibition site, given that it displays decent interaction and correlation among Mpro residues upon contact. The results obtained in this study using computational tools efficiency can lead to drug discovery against new diseases and might constrain the viral protease. Bearing in mind the time required to advanced therapeutics during the pandemics, drug repurposing seems to be the most alluring and upfront method in targeting SARS CoV 2. Such in silico trails can be useful in designing and testing drugs against SARS CoV 2 and enhance the drug discovery pipeline, predicting the therapeutic failures and diminishing undesired side effects. Further, a compound library targeting protease protein can also be developed for rapid screening against other deadly viruses, in which proteases serve as a crucial protein for the viral transmission.
Acknowledgment
We are obliged to Dr. Prajwal Nandekar, Mr. Vinod Devaraji, and Mr. Nripendra Bhatta of Schrodinger Corporation for their appreciated support, technical guidance, and logistic help.
Disclosure statement
The authors have no known opposing commercial concerns or private associations that might affect the work reported in this paper.
References
- Altintas, E., Akkus, N., Gen, R., Helvaci, M. R., Sezgin, O., & Oguz, D. (2004). Effects of Terlipressin on systolic pulmonary artery pressure of patients with liver cirrhosis: An echocardiographic assessment. World Journal of Gastroenterology, 10(15), 2278. 10.3748/wjg.v10.i15.2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos, Y. M., John, S. E. S., & Mesecar, A. D. (2015). The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research, 115, 21–38. 10.1016/j.antiviral.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., & Gregersen, B. A. (Eds.). (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters [Paper presentation]. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06 (pp. 43–43), Tampa, FL, USA. 10.1109/SC.2006.54 [DOI] [Google Scholar]
- Chen, Y. W., Yiu, C.-P., & Wong, K.-Y. (2020). Prediction of the 2019-nCoV 3C-like protease (3CLpro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. 10.12688/f1000research.22457.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DrugBank . (n.d.). https://www.drugbank.ca/
- Elfiky, A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592. . 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Andrade, M., Rodríguez-Sotres, R., Madariaga-Mazón, A., Rivera-Chávez, J., Mata, R., Sosa-Peinado, A., Del Pozo-Yauner, L., & Arias-Olguín, I. I. (2016). Insights into molecular interactions between CaM and its inhibitors from molecular dynamics simulations and experimental data. Journal of Biomolecular Structure & Dynamics, 34(1), 78–91. 10.1080/07391102.2015.1022225 [DOI] [PubMed] [Google Scholar]
- Grant, B. J., Rodrigues, A. P., ElSawy, K. M., McCammon, J. A., & Caves, L. S. (2006). Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics, 22(21), 2695–2696. 10.1093/bioinformatics/btl461 [DOI] [PubMed] [Google Scholar]
- Harrison, C. (2020). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology, 38(4), 379–381. 10.1038/d41587-020-00003-1 [DOI] [PubMed] [Google Scholar]
- Holmes, K. V. (2003). SARS coronavirus: A new challenge for prevention and therapy. The Journal of Clinical Investigation, 111(11), 1605–1609. 10.1172/JCI18819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, T., Wang, H., & Luan, B. (2020). In silico exploration of molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. Journal of Physical Chemistry Letters. 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- Joshi, R., Giri, A. P., Kulkarni, M. J., Verma, S., Chaudhary, D., & Deshmukh, N. (2020). Rationale based selection and prioritization of antiviral drugs for COVID-19 management. ChemRxiv.Preprint. 10.26434/chemrxiv.12429629.v1 [DOI] [Google Scholar]
- Karuppasamy, R., Verma, K., Sequeira, V. M., Basavanna, L. N., & Veerappapillai, S. (2017). An integrative drug repurposing pipeline: Switching viral drugs to breast cancer. Journal of Cellular Biochemistry, 118(6), 1412–1422. 10.1002/jcb.25799 [DOI] [PubMed] [Google Scholar]
- Kumar, Y., Singh, H., & Patel, C. N. (2020). In silico prediction of potential inhibitors for the Main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. Journal of Infection and Public Health, 13(9), 1210–1223. 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Morse, J. S., Lalonde, T., & Xu, S. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem: A European Journal of Chemical Biology, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., & Wang, X.-J. (2020). Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. Journal of Genetics and Genomics, 47(2), 119–121. 10.1016/j.jgg.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh, L., & Ghasemi, S. (2020). Genotype and phenotype of COVID-19: Their roles in pathogenesis. Journal of Microbiology, Immunology and Infection, 54(2), 159–163. 10.1016/j.jmii.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarrat, F., Chouljenko, V., Dahal, A., Nabi, R., Chouljenko, T., Jois, S. D., & Kousoulas, K. G. (2020). The anti‐HIV Drug NelfinavirMesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARS‐CoV‐2 Spike (S) glycoprotein warranting further evaluation as an antiviral against COVID‐19 infections. Journal of Medical Virology, 92(10), 2087–2095. 10.1002/jmv.25985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger, M. M., Carchia, M., Irwin, J. J., & Shoichet, B. K. (2012). Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. Journal of Medicinal Chemistry, 55(14), 6582–6594. 10.1021/jm300687e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., Kumar, S., Bhattacharyya, A., Kumar, H., Bansal, S., & Medhi, B. (2020). Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology, 52(1), 56–65. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat, M., Sarma, P., Shekhar, N., Prakash, A., Avti, P., Bhattacharyya, A., Kaur, H., Kumar, S., Bansal, S., Sharma, A. R., & Medhi, B. (2020). Update on the target structures of SARS-nCoV-2: A systematic review. Indian Journal of Pharmacology, 52(2), 142. 10.4103/ijp.IJP_338_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, R., & Sethumadhavan, R. (2009). Structural basis for the resilience of Darunavir (TMC114) resistance major flap mutations of HIV-1 protease. Interdisciplinary Sciences, Computational Life Sciences, 1(4), 320–328. 10.1007/s12539-009-0043-8 [DOI] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: Progress, challenges and recommendations. Nature Reviews. Drug Discovery, 18(1), 41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F., & Schroeder, M. (2015). PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saner, F. H., Canbay, A., Gerken, G., & Broelsch, C. E. (2007). Pharmacology, clinical efficacy and safety of Terlipressin in esophagealvarices bleeding, septic shock and hepatorenal syndrome. Expert Review of Gastroenterology & Hepatology, 1(2), 207–217. 10.1586/17474124.1.2.207 [DOI] [PubMed] [Google Scholar]
- Sarma, P., Kaur, H., Kumar, H., Mahendru, D., Avti, P., Bhattacharyya, A., Prajapat, M., Shekhar, N., Kumar, S., Singh, R., Singh, A., Dhibar, D. P., Prakash, A., & Medhi, B. (2020). Virological and clinical cure in COVID‐19 patients treated with hydroxychloroquine: A systematic review and meta‐analysis. Journal of Medical Virology, 92(7), 776–785. 10.1002/jmv.25898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Prajapat, M., Avti, P., Kaur, H., Kumar, S., & Medhi, B. (2020). Therapeutic options for the treatment of 2019-novel coronavirus: An evidence-based approach. Indian Journal of Pharmacology, 52(1), 1–5. 10.4103/ijp.IJP_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., & Kumar, S. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure & Dynamics, 39(8), 2724–2732. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., & Song, J. (2006). The catalysis of the SARS 3C‐like protease is under extensive regulation by its extra domain. The FEBS Journal, 273(5), 1035–1045. 10.1111/j.1742-4658.2006.05130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter, S. M. (2014). Overcoming drug development bottlenecks with repurposing: Old drugs learn new tricks. Nature Medicine, 20(6), 590–591. 10.1038/nm.3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, K., Kannan, K., Shanthi, V., Sethumadhavan, R., Karthick, V., & Ramanathan, K. (2017). Exploring β‐tubulin inhibitors from plant origin using computational approach. Phytochemical Analysis: PCA, 28(3), 230–241. 10.1002/pca.2665 [DOI] [PubMed] [Google Scholar]
- Xu, Z., Peng, C., Shi, Y., Zhu, Z., Mu, K., & Wang, X. (2020). Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 10.1101/2020.01.27.921627 [DOI] [Google Scholar]
- Xue, H., Li, J., Xie, H., & Wang, Y. (2018). Review of drug repositioning approaches and resources. International Journal of Biological Sciences, 14(10), 1232–1244. 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, N., Matsuyama, S., Hoshino, T., & Yamamoto, N. (2020). Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. BioRxiv. https://doi.org10.1101/2020.04.06.026476 [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov, A., Zagribelnyy, B., Zhebrak, A., Aladinskiy, V., Terentiev, V., & Vanhaelen, Q. (2020). Potential non-covalent SARS-CoV-2 3C-like protease inhibitors designed using generative deep learning approaches and reviewed by human medicinal chemist in virtual reality. ChemRxiv. Preprint. 10.13140/RG.2.2.13846.98881 [DOI] [Google Scholar]
- Ziebuhr, J., Snijder, E. J., & Gorbalenya, A. E. (2000). Virus-encoded proteinases and proteolytic processing in the Nidovirales. The Journal of General Virology, 81(Pt 4), 853–879. 10.1099/0022-1317-81-4-853 [DOI] [PubMed] [Google Scholar]