Abstract
The main-protease (Mpro) catalyzes a crucial step for the SARS-CoV-2 life cycle. The recent SARS-CoV-2 presents the main protease (MCoV2pro) with 12 mutations compared to SARS-CoV (MCoV1pro). Recent studies point out that these subtle differences lead to mobility variances at the active site loops with functional implications. We use metadynamics simulations and a sort of computational analysis to probe the dynamic, pharmacophoric and catalytic environment differences between the monomers of both enzymes. So, we verify how much intrinsic distinctions are preserved in the functional dimer of MCoV2pro, as well as its implications for ligand accessibility and optimized drug screening. We find a significantly higher accessibility to open binding conformers in the MCoV2pro monomer compared to MCoV1pro. A higher hydration propensity for the MCoV2pro S2 loop with the A46S substitution seems to exercise a key role. Quantum calculations suggest that the wider conformations for MCoV2pro are less catalytically active in the monomer. However, the statistics for contacts involving the N-finger suggest higher maintenance of this activity at the dimer. Docking analyses suggest that the ability to vary the active site width can be important to improve the access of the ligand to the active site in different ways. So, we carry out a multiconformational virtual screening with different ligand bases. The results point to the importance of taking into account the protein conformational multiplicity for new promissors anti MCoV2pro ligands. We hope these results will be useful in prospecting, repurposing and/or designing new anti SARS-CoV-2 drugs.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV, COVID 19, metadynamics simulation, multiconformational drug targeting, quantum calculations
Introduction
In December 2019, a cluster of pneumonia cases caused by a novel virus emerged from a local seafood and animal market of Wuhan city, the capital of Hubei province in China. The first genome sequencing analysis indicated that the genes of the novel virus share 80% nucleotide sequence identity to severe acute syndrome coronavirus (SARS-CoV). Further, RNA-based sequencing analysis showed that the virus genome was highly similar to BatCoVRaTG13, a bat coronavirus (CoV), with a sequence identity of 96.2% (Zhou et al., 2020). Therefore, this novel virus has been named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses, and pneumonia caused by it is called coronavirus disease 2019 (COVID-19). Currently, the COVID-19 has evolved to be transmitted by human-to-human and has spread rapidly around the world. People were submitted to social distancing and many countries have adopted quarantine regimes to stop the disease dissemination. Although an increasing number of different vaccines are being approved by regulatory agencies and administered worldwide, national health authorities continue to recommend the use of medical countermeasures and efforts to develop more effective drugs, given the limitations and uncertainties inherent in vaccines, especially in terms of new SARS-CoV-2 variants (Coronavirus disease (COVID-19): Vaccines). The growing set of these variants in different lineages has raised concerns about the susceptibility of SARS-CoV-2 to escape neutralizing antibodies, decreasing the effectivity of vaccines and increasing the risk of reinfection (Zhou et al., 2021). In this sense, it is urgent to screen antivirals as complementary strategies in the treatment or prophylaxis of COVID-19.
One of the best-characterized drug targets among CoVs is the viral 3 C–like protease (3CLpro or main protease Mpro) encoded by NSP5 (Figure 1). The polyprotein pp1ab and pp1a (encoded by ORF1ab and ORF1a) are cleaved by papain–like protease (PLpro) and the 3CLpro, to produce NSPs such as RNA-dependent RNA polymerase and helicase. Thus, such enzymes play an essential role in viral replication and its inhibition interrupts the viral life cycle (Zumla et al., 2016). Further, Mpro has no human homologous with a similar cleavage specificity, which contributes to the idea that inhibitors for this enzyme are unlikely to be toxic (Xiong et al., 2021).
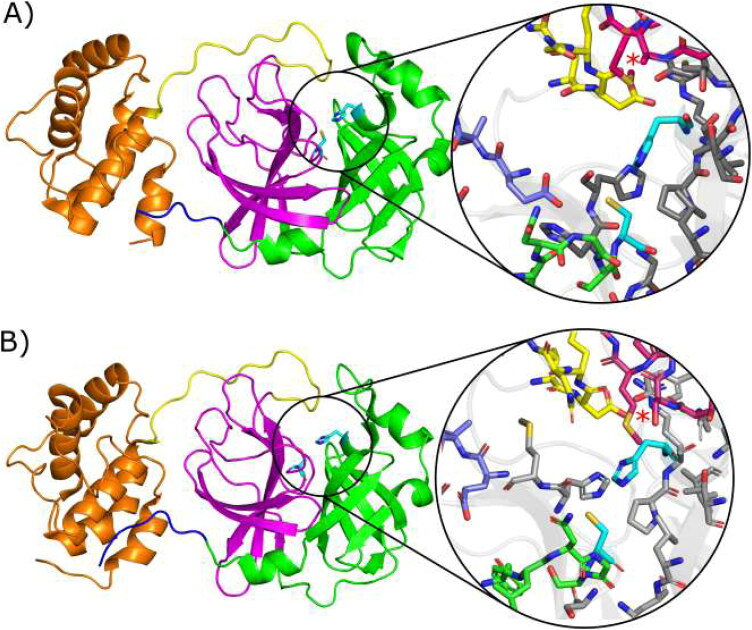
Figure 1.
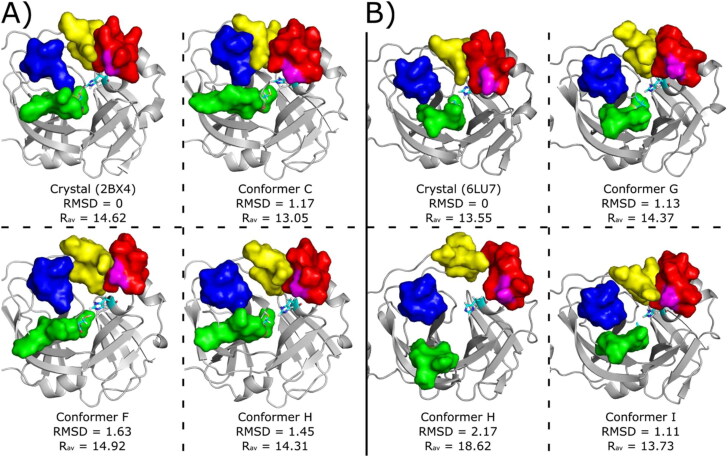
Crystallographic structures showing the mains proteases: (A) MCov1Pro (PDB code 2BX4) from SARS-CoV and (B) MCov2Pro (PDB code 6LU7) from SARS-CoV-2. Domains I, II, III, Linker and N-Finger are shown in cartoons coloured in green, magenta, orange, yellow and blue, respectively. The active pocket is highlighted for each structure and S11, S12, S2 and S4 loops are shown in blue, green, pink and yellow sticks. Cyan sticks represent the catalytic dyad (His41 and Cys145) in both cases and the region of the mutation Ala46/Ser46 is depicted by red stars. Gray sticks represent remaining residues around 5 Å from the catalytic dyad.
Mpro is highly conserved among CoVs, sharing over 90% sequence identity, differing only 12 residues between the SARS-CoV Mpro(MCoV1pro) and SARS-CoV-2 Mpro(MCoV2pro) (Anand et al., 2002; Wang et al., 2016; Xue et al., 2008; Yang et al., 2003). This enzyme is composed of three domains (Figure 1): (i) domain I, residues 8–101; (ii) domain II, residues 102–184; and (iii) domain III, residues 201–303. Domains I and II have an antiparallel β-barrel structure, and domain III contains five α-helices grouped into an antiparallel globular cluster and linked to domain II by a loop region (residues 185–200). The substrate binding-site is in the deep cleft between domains I and II and formed by the catalytic dyad His41 and Cys145 (Jin et al., 2020).
Recent conventional molecular dynamics (MD) studies have provided new perspectives about the flexibility of the MCoV2pro and MCoV1pro binding sites, such as the assistance of the different loops on this flexibility, and its correlation with dimerization and ligand attachment (Bzówka et al., 2020; Suárez & Díaz, 2020). The flexible loop C44-P52 seems to regulate the entrance of potential inhibitors into the binding site (Bzówka et al., 2020). Significant differences were found in this same study concerning both the flexibility of this loop and the accessibility to different chemical groups to the MCoV1pro and MCoV2pro respective active sites. In the same survey, a bioinformatic analysis showed that such a loop has a potential mutability that does not affect folding. Still, it may increase the inability of putative inhibitors to access the binding pocket, which can be a challenge for drug development. Another recent virtual screening/MM-PBSA study points to the loops around the MCoV2pro active site as the major pharmacophoric hot spots (Mittal et al., 2020). In other MD study, initial insights have been obtained about simultaneous changes in the dynamics of the loops around the active site and the interdomain mobility, both at the dimerization and at substrate association, for MCoV2pro (Bzówka et al., 2020). These findings have been associated with the experimental evidence that suggests reciprocal influences of the dimerization at the substrate binding and catalysis (Hilgenfeld, 2014; Shi et al., 2004), as well as of the substrate binding at the dimerization affinity for MCoV1pro in turn (Cheng et al., 2010).
The importance to confront the coronaviruses Mpro’s characteristic mobility and proper conformational space to optimize drug screening procedures has encouraged a crescent number of MD studies with a higher simulation time (the longer, up to now, a 100 µs simulation (Shaw)). Recently, a 15.14 µs unsupervised adaptive sampling molecular dynamics (asMD) for the dimeric MCoV2pro (in which these 15.14 µs have been sampled on iterations of subsequent sets of 10 ns simulations and using a bias algorithm to select the seeding for each iteration from the previously less visited configurations) has explored a substantially higher conformational space than this same longer and single one 100 µs simulation (Jaffrelot Inizan et al., 2021). This illustrates two principles: the power of adequately chosen unconventional high sampling simulation methods compared to equilibrium techniques to probe the conformational space of proteins; the fact that the proper choice of an enhanced sampling method compatible to the collective variable of interest provides a more meaningful sampling (and in a computationally cheaper way) than huge equilibrium simulations. In fact, there are significant energetic barriers between the representative conformational states, difficult to cross by equilibrium molecular dynamics (even at the micro or millisecond scales), a notoriously simpler task for ‘smartly’ chosen enhanced sampling approaches at substantially lower simulation time scales (Bernardi et al., 2015; Lazim et al., 2020; Rodriguez-Bussey et al., 2016).
The loop flexibility and the ‘breathing’ movements at the active site have been related as determinant to the substrate and antiviral accessibility for key viral proteases (Cheng et al., 2010; Tóth & Borics, 2006). In this way, the differential mobility at such loops is a crucial issue for understanding the differences in the biology, infectivity and pharmacological attributes from one viral strain to the other. For the coronaviruses’ main proteases, for which the major pharmacophoric subsites are located around such loops, these issues are still more determinant. The adequate comprehension of the intrinsic mobility of such loops, its correlation with the active site ligand accessibility and the way in which such features are modulated by single amino acid substitutions at different regions, will be crucial on drug screening procedures considering potential new Mpro variants. Conventional MD studies have pointed glimpses about the dynamics of such loops and their correlation with substrate accessibility and inter-domain motion in SARS-CoV main proteases (Bzówka et al., 2020; Cheng et al., 2010; Mittal et al., 2020). However, this mobility is represented mainly by high amplitude movements, not wholly accessible by conventional equilibrium simulations. In this sense, perturbative simulation techniques have already proved to be efficient at the characterization of the free energy landscape (FEL) related to the movements of these loops and their correlations with the whole protein dynamics (Lima et al., 2020; Mahanti et al., 2016; Tóth & Borics, 2006).
In this study, we carry out computational comparison between the intrinsic conformational/dynamics space of such loops in MCoV1pro and MCoV2pro monomers, followed by the exploration of the sampled MCoV2pro’s active site space on a extensive flexible virtual screening approach. For this end, we perform both well-tempered as nontempered metadynamics simulations (MetaDy) for each protein using as collective variables a pair of angles involving the four major active site loops and that have shown the higher mobility and pharmacophoric influences on crystallographic structures with different ligands (Barducci et al., 2008). In sequence, we analyse and compare the conformational, energetics, mutational and physicochemical attributes accessed by the MetaDy and by bioinformatics tools for the two proteins.
Between working with monomeric or dimeric form for the MetaDy studies, we strategically chose the first one. In this regard, we found a precedent in the use of Mpro monomers as drug targets, but with different approaches (Bolcato et al., 2020). The fact is that there is strong evidence that the active form of Mpros is dimeric, although a monomer-dimer equilibrium coexists (Xiong et al., 2021). Moreover, substrate binding to monomers can favor the dimeric state (Cheng et al., 2010). Even though only dimers are enzymatically active, some authors propose Mpro catalytic cycle models in which up (activated) and down (inactivated) states can be found at equilibrium in monomers (see figure 8 in Cheng et al., 2010 and figure 29 in Wan et al., 2020). But, only up monomers are capable of dimerization, especially those with bound substrates. Another recent study raised a ‘structure-based recognition’ hypothesis where the structural heterogeneity (more than the induced-fit theory) may be the main determinant of the greater diversity of Mpro's proteolytic activity, known to be able to recognize both N- and C-termini cleavage sites (Behnam, 2021). All in all, it seems to us that monomers have the potential to be alternative drug targets besides dimers, as they allow selection of ligands that can bind at a wider number of relevant conformational states, enabling both the virtual screening of competitive inhibitors (monomers near up state) and allosteric inhibitors (monomers near down state).
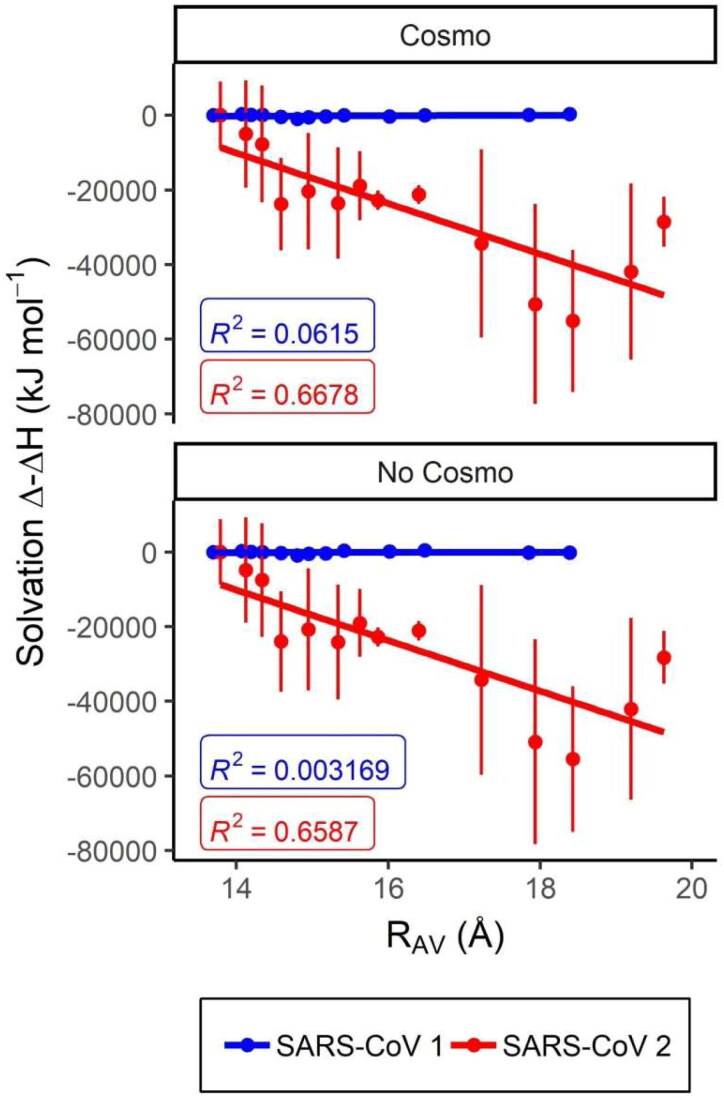
Figure 8.
Variation in the hydration enthalpy with the adoption of broader conformations for MCoV2pro and MCoV1pro. Semi-empirical QM estimated hydration enthalpy variation with the RAv value in angstroms for the metadynamics representative conformers. The plots were carried considering the cluster centroids from Figure 3 ordered according to the crescent RAv value and performed as window plots on a three-by-three mode. The respective values, correlations and deviation bars estimated with and without the COSMO implicit solvation are depicted for the MCoV1pro (blue) and MCoV2pro (red).
That is why we implement the two types of MetaDy aforementioned: well-tempered and nontempered. The former, its time-smoothed bias allows (at relatively short simulation times) an exploration of conformational states not so far from the starting point (the crystallographic conformation of the monomer in the dimer), keeping it closer to the up state. The latter, its free bias enables (at the same simulation time) exploration of conformational states more unrestrictedly, giving the monomer freedom to move further away from the crystallographic conformation, consequently, more chances of reaching down states. For both kinds of simulations and for both systems we have inspected the evolution of the potential of mean force (PMF) output files in order to properly calculate the free energy landscapes (FEL) only when (and immediately so) the FEL convergence was obtained (see the Theoretical Methods).
However, in order to analyse the extensibility and/or the adaptability of our findings with monomers to functional dimer, we also perform and compare the conformational statistics relevant to the binding site accessibility and activity with those obtained in freely available multiscale MCov2Pro dimer simulations (Jaffrelot Inizan et al., 2021; Shaw).
We complement such MetaDy simulations with quantum mechanics (QM) methods to analyze and compare the enthalpies of solvation and electronic density descriptors of the active site of each MPro's main conformers, in an effort to raise hypothesis with more detailed chemical explanations about the greater flexibility of the active site found for MCoV2pro compared to MCoV2pro, as well as about the low propensity of catalytic activity for the monomers.
Then, we evaluate the implications for the differences at the MCov1Pro and MCov2Pro conformational space (mainly around at active site) on the druggability of a compound for which the differential affinity between these proteins is already reported. Finally, from the representative conformers of the three less energetic clusters accessed by the MCov2Pro MetaDy, as well as the MD equilibrated crystallographic conformation, we carry out a multiconformer virtual screening strategy using three robust ligand databases: one for druggable compounds (DrugBank) (Wishart et al., 2018), one for commercially available compounds (ZINC15) (Irwin et al., 2012). and the other for natural compounds (SistematX) (Scotti et al., 2018). The major results point to the higher conformational promiscuity for the MPro from the current viruses compared to the 2009 one, the hydration changes due mainly the A46S substitution between both proteins as a crucial participant on such effect, as well as possibility of gain in recall when this robust conformational coverage could be explored on flexible virtual screening approaches. We hope these results can be useful for the rational design of new and more effective anti-SARS-CoV-2 drugs.
Theoretical methods
Pharmacophoric subsite analyses
Mpro PDB structures from SARS-CoV and SARS-CoV-2 available on 4 May 2020 were aligned and analyzed with the software Pymol. A qualitative analysis of all ligands bound to the active site of these structures was performed, focusing on protein–ligand interactions and on the diversity and chemical properties of ligands that bind to each subsite. Protein–ligand interactions in available PDBs were also analyzed with nAPOLI (Fassio et al., 2020).
Equilibrium molecular dynamics procedure
To explore the free energy landscape of the Mpro protomers from SARS-CoV and SARS-CoV-2 it was necessary to previously carry out an equilibrium MD stage. For this, the X-ray structures from the MCoV1pro (PDB code 2BX4) and MCoV2pro P(PDB code 6LU7) were used as start geometries (for this last one, previously removing the crystallographic ligand). MD simulations were performed in NAMD software version 2.12 using FF14SB AMBER force field (Maier et al., 2015; Phillips et al., 2005). Further, both monomers were inserted in a cubic water box of 20 Å containing TIP3P waters and ions to the neutrality and then, such simulations were performed according to the following parameters: (i) periodic boundary conditions, restriction of vibration in covalent bonds involving hydrogen atoms, HOH angle and OH bond distance of TIP3P water molecules (SHAKE algorithm); (iii) time steps equal to 2 fs; (iv) electrostatic interaction cutoff of 9.0 A for all steps of the simulation. Before equilibrium simulation in the NPT ensemble, the geometries were optimized by sequential stages of minimization, heating and pressurization. Then, the geometries were subjected to the equilibrium stage for 10 ns with a constant pressure and temperature of 1.0 atm and 310 K, respectively.
Metadynamics procedures and analyses
The MetaDy simulations were performed in NAMD 2.12 software using ff99SB AMBER force field at 300 K and 1 atm (Lindorff-Larsen et al., 2010; Phillips et al., 2005). The height of the Gaussians for the MetaDy was set to 0.42 kJ ·mol−1, added every 2 ps with a width of 1.77 grids. All the systems used the same set of collective variables (CVs). The first CV was the angle between the center of mass of the loops (CVang1): 166–171 (S11)/17–22, 25–32 (β-sheet)/45–53 (S2), varying between −20° and 160° with an increment of 7°. The second CV was the angle between the center of mass of the loops (CVang2): 140–145 (S12)/17–22, 25–32 (β-sheet)/186–191 (S4, S5), varying between −20° and 160° with an increment (grid) of 7°. Harmonic restraints were used on the β-sheet 17–22, 25–32. For the well-tempered MetaDy, the bias temperature was 1490 K. The convergence of each MetaDy simulation (both well-tempered as nontempered) and for each system was properly checked according to the protocol described by Incerti et al. (2017), i.e. calculating the progress of the free energy landscapes (FEL) along the time and verifying about the convergence of the major minima (Supporting Information Figure S11). The equations for MetaDy projections were used similarly to Brandt et al. (2016). Similarly to recent works of our group, we used the colony method adapted by Martínez et al. to estimate the differences of conformational entropy between MCoV1pro and MCoV2pro over the CVs space (Martínez et al., 2009; Rocha & Lima, 2019). Animated gifs of both MetaDy for both Mpro can be seen in Supporting Information.
Potential of Mean Force Maps and Projections of the MetaDy at a Single Collective Variable
The resulting trajectories were visualized and analyzed with VMD software and the potential maps were made using ‘home-made’ R scripts. The MetaDy projections were made following the methodology of the similar work carried out by Brandt et al. (2016). The projections of the MetaDy simulations trajectories onto collective variables (CVs) was calculated as follows:
| (1) |
Where the projection ω for CVang1 is a function of a given value of CVang1 (x) and the minimum in CVang2 at this specific CVang1(x) value. Similarly, for CVang2, the projection β is a function of a given value of CVang2 (y) and the minimum in CVang1 at this specific CVang2(y) value.
Clustering procedures and obtaining of representative conformers
The well-tempered and nontempered trajectories were merged, aligned and first divided into energetic states between 4 to 84 and 4 to 42 kJ mol−1 for MCov1Pro and MCov2Pro, respectively, and then, subdivided into minor energy terms of 10.5 kJ mol−1. A clustering hierarchical algorithm was applied to extract representative geometries from each one of the minima energy ranges. Additionally, adjustments in the epsilon term of the algorithm were carried out to improve the number of clusters. Then, the centroids of each cluster were sampled and manually filtered to reach all the conformational sets of the active site. In this procedure, when two or more centroids belonging to different energetic states were significantly closer at the two-dimensional FEL map, only the less energetic centroid was considered. At last, the filtered geometries were used as input for the next steps of this study.
CV space entropy estimation
The colony method adapted by Martínez et al. was applied to estimate the differences of conformational entropy between MCov1Pro and MCov2Pro at the CVang1 vs. CVang2 space (Martínez et al., 2009). Briefly, this method assumes that the conformational entropy can be recovered over the molecular dynamics simulations by clustering the microstates in the 6D phase space, which consists of coordinates in X, Y and Z vs. velocities in X, Y and Z. This 6D phase space can be approximated to a 2D phase space simply by representing the coordinates by representative coordinates (in this case, the MetaDy collective variables were used). Then, this approximated space is recursively divided into grids with decreasing steps, in which each bin can be assumed as a microstate. For each step of grid division, the number of occupied microstates is recovered and applied to a Boltzmann like approximation as:
| (2) |
where kB is the Boltzmann constant, ωCvsCov1 and ωCvsCov2 are the respective number of occupied bins for the MCov1Pro and MCov2Pro active sites at the CVang1 vs. CVang2 space and ΔSCVsCov1-2 is the configurational entropy at this same space from MCov1Pro to MCov2Pro. Such ΔSCVsCov1-2 as a function of the ratio of occupied bins between each system for different grid subdivisions reaches a maximum when the FEL subdivision is significant, but not excessive. At the adapted colony method, this maximum is taken as the more accurate estimation for the entropy difference between both systems. For a deeper background, we suggest the study carried by Martínez et al. (2009). Here, this adapted colony method entropy variation was recovered using an in-house script similar to previous works of the group (Rocha & Lima, 2019).
Semi-empirical quantum calculations
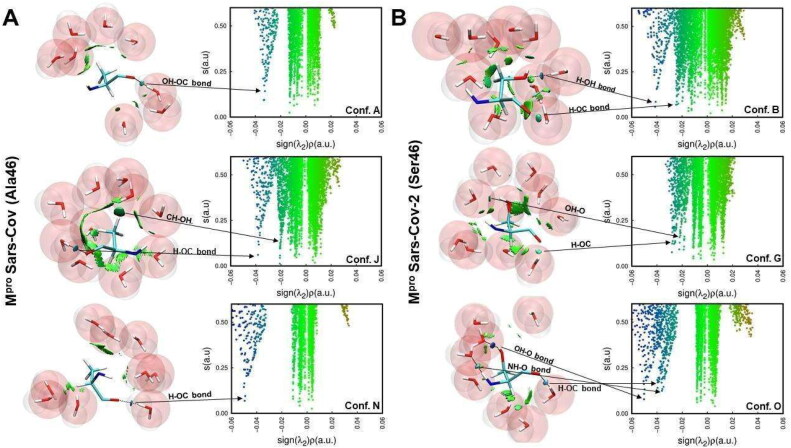
The enthalpies of solvation were estimated by using the PM7 semi-empirical method available on MOPAC 2016 software by having the centroid geometries of each cluster as input (see Metadynamics Procedures and Analyses section) (Stewart, 2013; Stewart Computational Chemistry). The MOZYME method and COSMO implicit solvent model were used under a dielectric constant of 78.4 (Klamt & Schüürmann, 1993; Stewart, 1996). We performed NCI calculations to explore the intermolecular interactions of water molecules around the Ala46 residue in SARS-CoV and Ser46 in SARS-CoV-2. The different conformations of both molecular structures of Mpro were used, taking into account their opening movement: closed structure (A, B), partially open (J, G) and fully open (N, O) for SARS-CoV and SARS–CoV-2, respectively.
The calculations were carried out considering a solvation shell with 2.500 water molecules around the protein structure against protein alone at the vacuum. So, the enthalpy of solvation was calculated according to
| (3) |
Electronic density quantum descriptors for the catalytic residues
Reactivity descriptors (RDs) are quantities that can be produced through mathematical manipulations of the calculated electronic structure of a molecular structure (Geerlings et al., 2003). RDs, such as Electrophilic Attack Susceptibility (EAS), Nucleophilic (NAS) and local hardness, can be used to profile the enzymatic catalysis trends (Grillo, Urquiza-Carvalho, Bachega, et al., 2020; Grillo, Urquiza-Carvalho, Chaves, et al., 2020). The same RDs were used in the present study, calculated by the PRIMoRDiA software (Grillo, Urquiza-Carvalho, Rocha, 2020), that contains these new implementations and efficiently parses the large outputs from Quantum Chemistry Packages. Thus, in this work we characterize the propensity of the proton transfer from Cys145 to His41 using the local hardness, and the nucleophilicity of the Cys145, towards to a peptide substrate that would bind with the protein, using the EAS RD.
Bioinformatics ΔΔG Estimation for MCov1Pro and MCov2Pro Single Residue Permutations
In order to evaluate the impact of amino acid permutations from MCov1Pro to MCov2Pro and the reverse, we predicted the effect of single point mutations on the stability of their MD simulation representative conformers. Usually, this effect is measured by changes in the alterations of folding energies (ΔΔG) between the wild-type and a mutant. In this article, we performed the ΔΔG estimation for MCov1Pro and MCov2Pro amino acid permutations using CUPSAT (Parthiban et al., 2006). We chose this software among many others because it accepts as input a generated PDB file (not deposited on Protein Data Bank) and it is more sensible to mutations, that is, it presents higher differences in the predicted ΔΔG values along the MD conformers than other methods.
We submitted representative conformers of MCov1Pro and MCov2Pro MD simulations to CUPSAT for predicting reverse substitutions from their enzymes at the 12 amino acid substitutions from the first to the second: 35(T/V), 46 (A/S), 65 (S/N), 86 (L/V), 88 (R/K), 94 (S/A), 134 (H/F), 180 (K/N), 202(L/V), 267 (A/S), 285 (T/A), 286 (I/L). We also performed reverse substitutions from MCov2Pro and MCov1Pro: 35 (V/T), 46 (S/A), 65 (N/S), 86 (V/L), 88 (K/R), 94 (A/S), 134 (F/H), 180 (N/K), 202 (S/A), 285 (A/T), 286 (L/I). All the results regarding the effect of single mutations are presented in Supporting Information Figure S1.
Protein: solvent noncovalent interaction estimations
Noncovalent interactions (NCIs) play a pivotal role in biological environments (Johnson et al., 2010; Williams et al., 2004). These interactions depend on the electronic effects of electron density and can be estimated from quantum chemistry calculations (Christensen et al., 2016; Rozengurt and Heppel, 1975). We carried out these calculations concerning the solvent interactions at position 46 for both enzymes using NCIPLOT that produces isosurfaces of topological quantities of electron density (Boto et al., 2020). This points out the type of intermolecular interactions this region of the system does, be it an atom, part of the molecule or residue.
Ligand virtual screening and docking methodologies
In order to find a selective hit for the Mpro active site, a hierarchical virtual screening cascade was performed in two sequential steps: (i) structure-based pharmacophore screening and (ii) ensemble docking. The following ligand databases were used in the virtual screening: (i) Drugbank, (ii) ZINC15 database and (iii) SistematX (Irwin et al., 2012; Scotti et al., 2018; Wishart et al., 2018).
Virtual Screening library preparation
The virtual screening libraries were selected considering three main perspectives: (i) fast clinical application; which includes FDA drug approved and investigational drugs; (ii) synthesis facilitated; here, we adopt the perspective of natural products; and (iii) long-term clinical application; includes virtual screening compounds from ZINC database, which includes compounds without biological activity described. For the ZINC database, compounds were retrieved using the following parameters: (i) Representation: 3D; (ii) Reactivity: Standard; (iii) Purchasability: In stock; (iv) pH: Reference Models; (v) Charge: Neutral. Further, the remaining compounds were subjected to a multiconformational search by using OMEGA software (Hawkins et al., 2010), which was set to find 50 low energy conformers for each compound.
Structure-based pharmacophore screening
In an attempt to increase the chance to find a selective hit into the ZINC database, which in turn has more than 1 million compounds, the binding pose of N3 inhibitor (PDB ID 6LU7) was used as a pharmacophore model. For this, we used the ROCS software (OpenEye Scientific) (Hawkins et al., 2007) to build a shape query. In the ROCS search, the conformers of the VS library were aligned with the shape query and a similarity score (Tanimoto Combo [TC]) was estimated. A high score means that the compound is similar to the shape query, thus, once the N3 is a potent inhibitor of Mpro from SARS-CoV-2, it is expected that similar compounds to its query also have a similar biological activity. The remaining compounds of the pharmacophore screening as well as the Drugbank and Sistematx compounds were subjected to molecular docking procedure.
Preparation of the receptors and ligands
The X-ray structure of the Mpro (PDB ID 6LU7) as well as the MetaDy conformers G, H and I were used as receptors for the molecular docking step. The geometries of these receptors were prepared according to the FF14SB force field by using the tLeap software, included in the Ambertools package; ligands were prepared considering the AM1-BCC atomic charges by using the Antechamber software (Maier et al., 2015).
Molecular docking procedures
In order to validate the docking protocol, the GOLD and Molegro Virtual Docker (MVD) softwares were used. In summary, we use the 6XHM and 6XHL crystallographic structures and the MetaDy conformers (Jones et al., 1997; Thomsen & Christensen, 2006). The structures were aligned according to the backbone and the active site was defined considering the same area of the virtual screening stage. The receptors and ligands were prepared according to the protocol presented in the previous section. For the GOLD, the standard configuration was used, with the exception, only, of the total executions of the genetic algorithm, which was modified to 30. In this validation protocol, in order to make the different docking score metrics from GOLD and MVD comparable we have created a ‘normalized relative score’ function. First, once the best scores at the MVD metric are negative, while for GOLD they are positive, we have taken the negative of the GOLD scores. Second, once our major interest in this validation was to verify the docking score variation at the MetaDy decoys concerning the redocking at the respective crystallographic structures, we have computed the variation of each score related to the proper crystallographic pose (6XHM for MCov2Pro decoys and 6XHL for MCov1Pro). Finally, as the significant score amplitudes for both softwares are also distinct, we have divided the score values recovered at each respective software by our data amplitude using that same program (i.e. dividing by the higher score variation considering together both MCov1Pro as MCov2Pro). While this normalized relative score function is shown and discussed along the validation section at the principal text, the ‘raw’ values for the respective GOLD and MVD docking outputs are also depicted at the Supporting Information Figure S13.
We also have used Molegro Virtual Docker, v. 6.0.1 (MVD) (Molexus IVS Rørth Ellevej 3, Odder, Denmark) (Thomsen & Christensen, 2006), two scoring functions Moldock and rerank were selected (Molexus IVS Rørth Ellevej 3, Odder, Denmark) and GPU screening search algorithm was performed. The following parameter settings of software package were used (Score function: MolDock and rerank scores; Ligand evaluation: Internal ES, Internal HBond, Sp2–Sp2 Torsions, all checked; Number of runs: 10 runs; Algorithm: GPU screening; Maximum Interactions: 1500; Simultaneous evaluation: 256; Max. number of poses returned: 10). The docking procedure was performed using a center grid: x = 78.5; Y = 65; z = 58, with 16 A in radius and 0.38 in resolution to cover the ligand-binding site of the Mpro structure.
Rescore
The Table 1 shows the first 10 hits of each of the three bases (Drugbank, ZINC15 SistematX) when the screening/docking is performed only on the crystallographic pose (Rigid) and when it is performed for the crystallographic and the three lower energy poses of MetaDy (Flexible).
Table 1.
Molegro’s docking scores.
| DB | Hit | Rigid |
Flexible |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Score | ID | Min | ID | Max | ID | Average | ID | Boltzmann | ||
| Drugbank | 1 | DB12206 | –160.29 | DB14792 | –199.41 | DB04461 | –140.98 | DB14792 | –157.98 ± 37.65 | DB14792 | –123.06 |
| 2 | DB15435 | –147.03 | DB04461 | –187.63 | DB13648 | –139.29 | DB04461 | –157.57 ± 26.08 | DB04461 | –115.81 | |
| 3 | DB12234 | –142.19 | DB12760 | –161.48 | DB07458 | –137.00 | DB13648 | –144.44 ± 6.90 | DB12760 | –99.71 | |
| 4 | DB03638 | –141.22 | DB14850 | –160.13 | DB03777 | –129.61 | DB07458 | –142.20 ± 5.79 | DB14850 | –98.88 | |
| 5 | DB03777 | –139.90 | DB06809 | –158.79 | DB14850 | –129.25 | DB14850 | –141.37 ± 16.47 | DB06809 | –98.05 | |
| 6 | DB06976 | –137.83 | DB07796 | –158.24 | DB07456 | –128.45 | DB11829 | –140.97 ± 16.43 | DB07796 | –97.71 | |
| 7 | DB06228 | –137.57 | DB11913 | –153.19 | DB15403 | –128.13 | DB03777 | –139.29 ± 8.79 | DB11913 | –94.60 | |
| 8 | DB12983 | –136.97 | DB15011 | –152.47 | DB11769 | –127.96 | DB07456 | –136.55 ± 9.12 | DB15011 | –94.16 | |
| 9 | DB07146 | –136.83 | DB11829 | –152.45 | DB14792 | –125.86 | DB11913 | –136.46 ± 18.49 | DB11829 | –94.16 | |
| 10 | DB12054 | –135.30 | DB13648 | –152.28 | DB15343 | –125.02 | DB11769 | –134.27 ± 6.39 | DB13648 | –94.05 | |
| ZINC15 | 1 | 20617839 | –170.76 | 252495534 | –156.96 | 17026643 | –140.71 | 17026643 | –141.66 ± 1.24 | 252495534 | –96.93 |
| 2 | 72273058 | –143.96 | 33937156 | –156.19 | 14246524 | –128.13 | 20617839 | –139.77 ± 13.52 | 33937156 | –96.45 | |
| 3 | 17026319 | –143.68 | 15881778 | –155.36 | 69656209 | –127.14 | 40080363 | –135.90 ± 14.67 | 15881778 | –95.94 | |
| 4 | 408616086 | –141.33 | 20617839 | –153.74 | 20617839 | –126.74 | 69656209 | –132.57 ± 8.59 | 20617839 | –94.94 | |
| 5 | 65099268 | –141.14 | 71747809 | –152.73 | 96099223 | –125.72 | 96099223 | –131.61 ± 7.20 | 71747809 | –94.32 | |
| 6 | 11972252 | –138.95 | 949898 | –152.62 | 9217657 | –123.82 | 85746622 | –129.94 ± 14.59 | 949898 | –94.26 | |
| 7 | 20490081 | –138.93 | 62166167 | –151.31 | 12419204 | –121.40 | 3632671 | –129.30 ± 22.61 | 62166167 | –93.45 | |
| 8 | 17026643 | –137.53 | 21155835 | –150.27 | 65502553 | –121.30 | 575443705 | –128.94 ± 13.65 | 21155835 | –92.81 | |
| 9 | 24992790 | –137.39 | 9265260 | –150.15 | 41127019 | –121.14 | 14246524 | –128.79 ± 0.72 | 9265260 | –92.74 | |
| 10 | 8705677 | –136.96 | 54318925 | –150.09 | 14991101 | –120.56 | 46915378 | –128.21 ± 15.35 | 252495534 | –96.93 | |
| SistematX | 1 | 15629 | –155.37 | 14420 | –175.94 | 15553 | –150.70 | 14178 | –153.38 ± 11.62 | 14420 | –108.61 |
| 3 | 21181 | –151.88 | 21418 | –166.16 | 18176 | –142.34 | 14543 | –151.29 ± 9.56 | 21418 | –102.59 | |
| 4 | 18925 | –142.53 | 18339 | –163.55 | 14543 | –141.04 | 18176 | –150.13 ± 11.46 | 18339 | –100.98 | |
| 5 | 13895 | –140.96 | 20761 | –163.50 | 14094 | –135.82 | 21181 | –149.95 ± 19.49 | 20761 | –100.95 | |
| 6 | 17386 | –140.61 | 18176 | –163.29 | 14544 | –135.72 | 14544 | –144.33 ± 10.23 | 18176 | –100.83 | |
| 7 | 15117 | –137.73 | 21181 | –162.27 | 14545 | –133.78 | 21047 | –143.91 ± 18.07 | 21181 | –100.26 | |
| 8 | 18823 | –136.14 | 13947 | –161.60 | 19827 | –131.19 | 14420 | –142.06 ± 29.57 | 13947 | –99.79 | |
| 9 | 14543 | –135.97 | 14083 | –161.60 | 15629 | –130.35 | 14083 | –141.16 ± 18.06 | 14083 | –99.78 | |
| 10 | 18338 | –135.69 | 15534 | –160.42 | 18925 | –128.50 | 15629 | –140.66 ± 9.60 | 15534 | –99.05 | |
Rigid vs. flexible (MetaDy) virtual screening results for MCoV2pro (see section Rescore for more details).
From these results, four metrics were generated:
the score according to the LOWEST score among the three MetaDy selected conformations (Flexible-Min.);
the score according to the ‘LOWEST HIGH’ score among the conformations (that is, the lowest maximum of energy - Flexible-Max.);
the score according to ARITHMETIC AVERAGE of the scores among the three conformations (Flexible-Average);
the score according to the BOLTZMANN AVERAGE among the three conformations (Flexible-Boltzmann).
In this selection, if a ligand appears as a hit in the five selection criteria (Rigid, Flexible-Min, Flexible-Max, Flexible-Average, Flexible-Boltzman) it is colored red in the matrix; if it appears in four it is colored in magenta; if it appears in three it is colored in yellow and if it appears in two it is colored in green. If it only appears in the rigid conformation, it is not colored.
Results and discussion
Pharmacophoric variance of crystallographic structures
To examine the differences between Mpro from SARS–CoV and SARS–CoV-2 from a drug discovery perspective, we first analyzed available PDB structures of ligands in complex with these enzymes (see PDB list in Supporting Information). We have also carried out this analysis to define the collective variables (CV) to be used at our MetaDy experiments.
First, we have superposed 48 MCoV1pro and 109 MCoV2pro structures complexed with ligands and profiled the intermolecular interactions at the subsites S1, S2, S4, S5 and S1’ using nAPOLI (Figure 2(A)) (Fassio et al., 2020). The only amino acid distinction in the binding site region between MCoV1pro and MCoV2pro is in position 46 at the loop 44–53, where alanine is found in SARS–CoV and serine in SARS–CoV-2. This substitution is relatively far from the binding pocket. The nAPOLI analyses revealed hydrophobic contacts among only four SARS-CoV ligands (PDB codes 3V3M, 4TWW, 4TWY, 5C5O) and Ala46. However, previous studies have suggested a substantial indirect influence of this substitution at the active site entrance dynamics (Brandt et al., 2016; Domingo et al., 2016). Indeed, analysis of the crystallographic B-factors at different MCoV2pro complexes points to the loop 44–53 at domain I and its neighbor, the loop 184–193 at domain II, as mobility hotspots at the enzyme active site (Figure 2(B)).
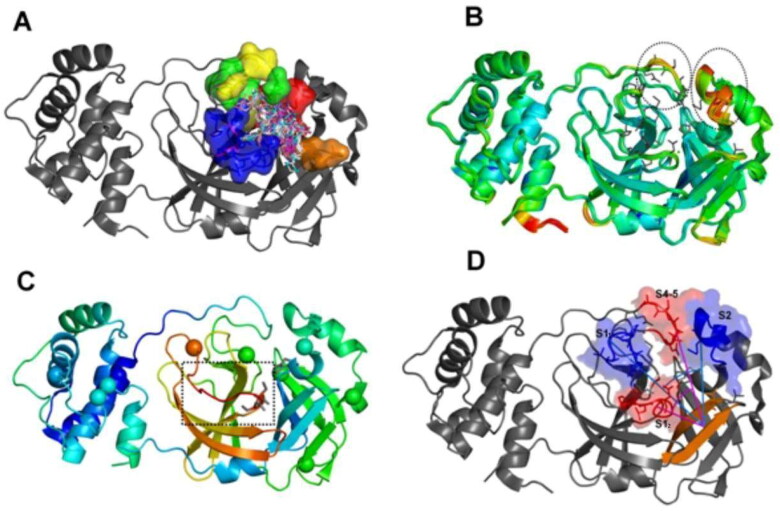
Figure 2.
The setting of the metadynamics collective variables based on intrinsic mobility of the pharmacophore subsites. (A) Superposition of crystallographic poses of different MPro ligands (depicted as sticks) at the enzyme active site. The five major MPro pharmacophoric subsites are depicted on surface representations and with different colors (blue for the S1 subsite; red for S2; green for S4; yellow for S5 and orange for the S11); (B) Superposition of 52 PDB structures of the SARS–CoV-2 (MCoV2pro) colored by the B-factors (blue:green:red scale, where red represents higher values) and highlighting the local hot spots at the subsites S2, S4 and S5 (dashed circles). Pharmacophore side chains are depicted in sticks; (C) Structure of the SARS-CoV1 Mpro (PDB: ID 2BX4) colored by the same B-factor scale and highlighting the significant local hot spots at the subsite S1 (dashed square). The Cα atoms of the twelve positions substituted between SARS-CoV1 and 2 are depicted as vdw spheres; (D) Angular collective variables used at our MetaDy experiments: CVang1 (cyan) between respective geometric centers at one half of S1 (S1’ in blue), the S1 prime bound β-hairpin (orange) and S2 (blue); CVang2 (magenta) between geometric centers at the other half of S1 (S1’ in red), the same β-hairpin (orange) and the S4-5 region (red).
The residues with most varying behavior were Met49, Met165 and Gln189, which impact in the shape of the S2 pocket, and Asn142, located in the S1 pocket. Consequently, high plasticity is observed especially for the S2 pocket. Moreover, such residues are located on a set of four distinct loops, which in turn belongs to different pharmacophoric subsites, delimitate the same S2-1 pocket walls and encompass mobility hot spots according to the experimental B-factors (Figure 2(B,C)).
Hence, our integrated analysis of structure superposition, pharmacophoric and B-factor profiling points to two approximately orthogonal angles, involving these four highly flexible loops as adequate angular collective variables (CV angle’s) for the MetaDy experiments (Figure 2(D)). These two CV angle’s encompass an angle involving loops 44–53 (S2) and 166–173 (S11) (i.e. CVang1) and other involving loops 140–147 (S12) and 184–193 (S4–5) (i.e. CVang2), described at the Methods section. They entail the four loops involved at crucial ligand contacts and whose intrinsic dynamics seem to be more naturally involved in the deformation of the substrate pocket and ligand adequation.
Metadynamics simulations suggest significantly higher intrinsic flexibility for the MProCov2 monomer active site compared to MProCov1
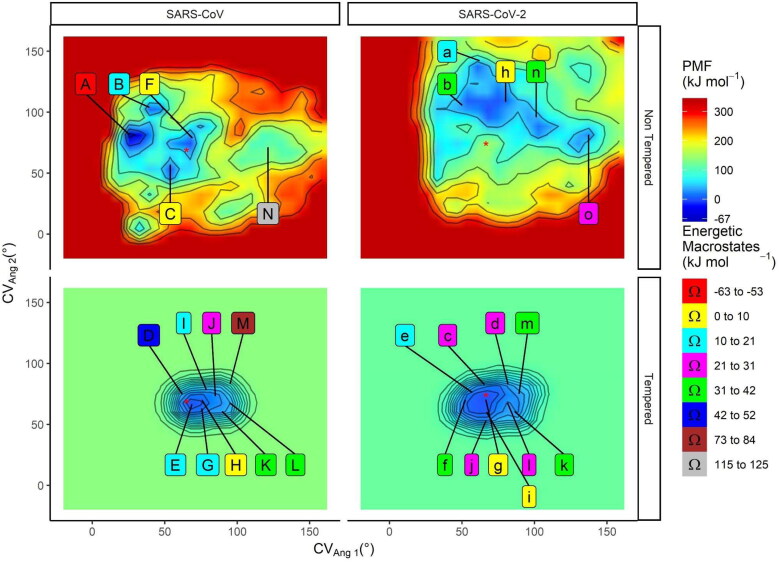
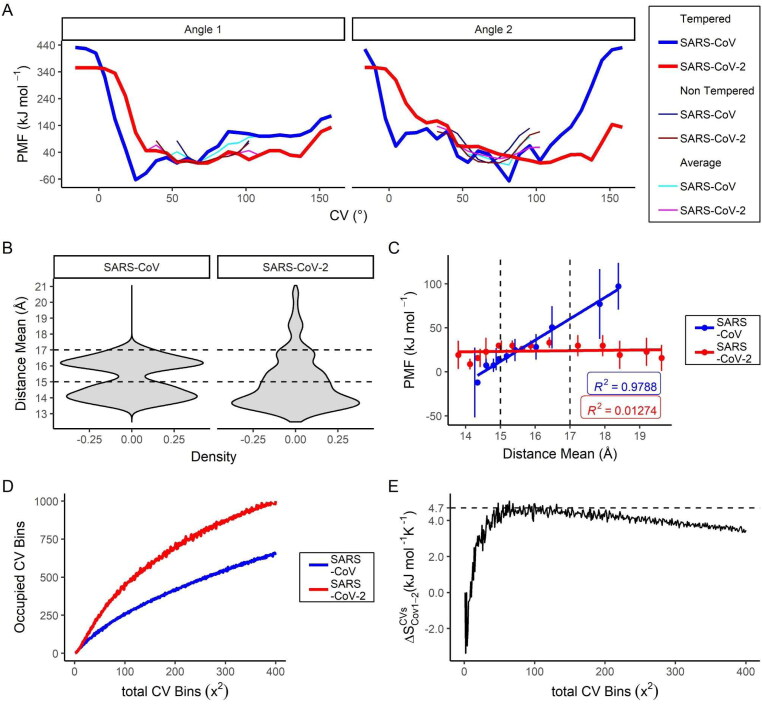
Using MetaDy protocols with different attenuation degrees (i.e. well-tempered and nontempered MetaDy - wtMetaDy and ntMetaDy) we have been able to detail conformations both immediately close to the crystallographic one as highly distant from this starting configuration for each protease. For all the MetaDy procedures, It was observed the desirable increase in conformational abrangency related to the equilibrium MD with the same simulation interval (Supporting Information Figure S12). However, the two highly attenuated wtMetaDy simulations trend to sample more precisely the local minimum around the crystallographic structure (closer to the original dimmeric and up context), while the two ntMetaDy simulations sample more freely the distant minima Figure 3 and Supporting Information Figure S12), some of them, according to will be discussed below, probably monomeric typical and at the down state. Taking together the resulting free energy landscapes for each procedure, as well as their respective linearization, a more restricted conformational space was observed for MCoV1pro compared to MCoV2pro (Figures 3 and 4(A)). In fact, MCoV1pro is considerably more confined at a deeper minima close to the crystallographic structure. MCoV2pro, conversely, can easily access a wide range of minima spreading majorly at the CVang1 dimension, although the coverage around the CVang2 is also higher than for MCoV1pro, mostly at low CVang1 values (Figures 3 and 4(A)).
Figure 3.
Free energy landscape and representative cluster centroids from the SARS-CoV and SARS-CoV-2 Mpro MetaDy. Potential of mean force (PMF) maps for the SARS-CoV and SARS-CoV-2 Mpro for nontempered (above) and tempered (below) MetaDy. The zeropoint of the energetic scale is arbitrarily chosen, setting a 0 kJ · mol−1 value at the local minimum closer to the crystallographic structure (structure coordinates depicted as a red star). Capital and lowercase letters are used to point the location of each representative conformers sampled for MCoV1pro and MCoV2pro, respectively. The energetic window from each macrostate is described on a ‘valueL ≤ window range < valueR’ scheme, being valueL the left term and valueR the right one.
Figure 4.
Accessibility to wider active site configurations and higher conformational promiscuity for the SARS-CoV-2 Mpro compared to the SARS-CoV one. (A) MetaDy projections at the CVang1 and CVang2 collective variables; (B) Density for the average width of the active site (RAv) considering the energetic macrostates until 10 kcal · mol−1 for the respective MCoV1pro and MCoV2pro enzymes. Dashed lines delimit the two major modes observed; (C) Variation of the PMF free energy as a function of the mean of the same RAv value on a three-by-three scale window plot considering the representative MetaDy conformers clusterized according to Figure 3. Correlation coefficients (R2) are shown. Dashed lines delimit the same regions as in C. (D) Occupied microstates on the respective PMF maps for the MCoV1pro and MCoV2pro as estimated by the adapted colony method; (E) Conformational entropy variation at the CVang1 vs. CVang2 phase space (ΔSCVsCoV1-2) between the MCoV1pro and MCoV2pro as estimated by the adapted colony method from the plots in (D).
To estimate the consequences for ligand accessibility, we have analyzed the average active site width as the mean distance between the Cα atoms of the active site loops (RAv) (Figure 4(B,C)). This measure has already proved to be well correlated with ligand accessibility/resistance in previous studies (Konar et al., 2019; Lima et al., 2020). First, we have measured the Rav statistics for conformations with the potential of mean force (PMF) at the 20% more favorable values from our MetaDy analyses (i.e. between −63 and +53 kJ · mol−1). The data show a bimodal distribution for the MCoV1pro low energy conformers, one mode centered at a closer and the other one at a partially opened active site (Figure 4(B), left). MCoV2pro, conversely, presents just one well-defined mode at the closer conformations, but with additional open conformations, in which all of them share almost the same energetic state (Figure 4(B)). In contrast, opened states seem to be less favorable for MCoV1pro (Figure 4(C)), clearly resulting in entropic disadvantages due to conformational inaccessibility according to our adapted colony method estimation (Figure 4(D,E)) (Martínez et al., 2009). Altogether, these results point to the higher flexibility of MCoV2pro compared to MCoV1pro. A recent classical molecular dynamics study suggests that such increased plasticity can be an important mechanism for stabilization of the enzyme-substrate complex by Mpro (Suárez & Díaz, 2020). This results may also be in line with the structural heterogeneity of the Mpro catalytic sites found by (Behnam, 2021).
The region which comprises the β-sheet and S1, S2, S4 and S5 pockets seems to form a key flexible neighborhood important for the activity of the Mpro. Recent computational studies have shown S2 loop as a regulator to access the active site (Bzówka et al., 2020), which agrees with the high flexibility suggested by our crystals analysis and observed in our MetaDy simulations. Clearly, the exchange of A46S at the S2 loop (from MCoV1pro to MCoV2pro) can someway decrease entropic disadvantages and allows MCoV2pro to access more open conformations, facilitating the access to the binding site. To better explore this, we systematically sampled representative frames of each macrostate for both proteases, trying to encompass the whole range of the Mpro’s conformational multiplicity. These representative conformations were labeled alphabetically (Figure 3) and submitted to deeper flexibility comparison and quantum chemical analyses. We will return to this point later.
Finally, it is worth highlighting that MCov2Pro shows increased movement correlations over the active site opening. To better understand how the internal motions of both MPros are affected by the mutations, we analyzed the movement correlations of the loops around the active-site (Supporting Information Tables S1 and S2) and the protein domains. Briefly, numbers between 0 and 0.5 suggest no motion dependency between two regions, whereas numbers bigger than 0.5 up to 1 imply movement correlation. In that way, an increase in the internal movements is observed in MCov2Pro mainly in motions that correlate the domain III and S2 loop with the rest of the enzyme. So, the opening movement in MCov2Pro is more interconnected between the active-site loops and the domain III in which S2 loop can be the regent of this choreography. Recent MD studies suggested that rotation events in domain III caused by S2 loop are important to the active site opening (Suárez & Díaz, 2020). This movement can be associated with the activity of the enzyme in a remarkable way. Experimental and computational studies suggested homodimerization as an important step for the enzymatic activation of MPro (Lin et al., 2008; Suárez & Díaz, 2020). In this way, a deeper understanding of the stabilization mechanisms by which MCov2Pro access opened conformations without the substrate binding can be useful for rational drug design.
Literature simulations for the MCov2Pro dimer recover a significant amount of the monomeric active site flexibility for the protomers as sampling increases
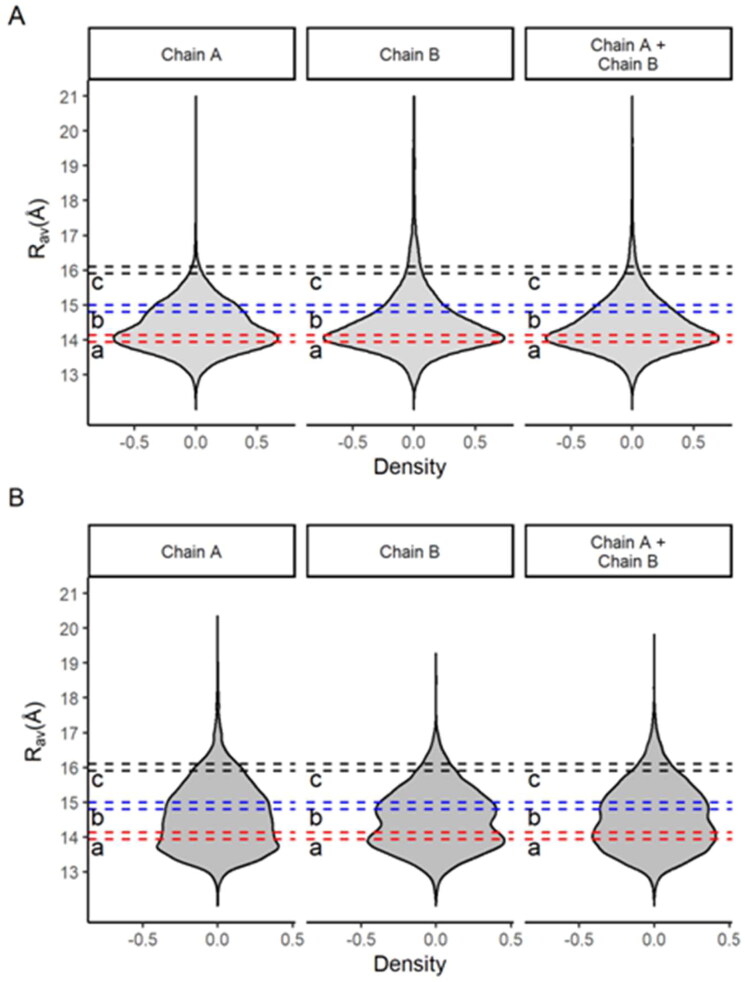
We analyze the Rav density distribution of two long multi-microsecond simulations recently published for MCoV2pro dimers (Jaffrelot Inizan et al., 2021; Shaw). The Rav metric (average active site width) is a measure of the active site openness, and it was taken for each chain and both together in the dimer (Figure 5). It is noticed that for both simulations the Rav density distributions are asymmetric with positive skewness, indicating a broader flexibility of the active sites. But, the unsupervised adaptive sampling simulation reveals an even more remarkable positive spread of these distributions, which reinforces its greater efficiency for the purpose of conformational explorations.
Figure 5.
Density for the average active site width (RAV) for the respective protomers A and B (chain A and chain B), as well for both on lower and higher sampling multi-microsecond simulations for the MCov2Pro dimer. (A) Conventional multi-microsecond simulation from the DESRES group (Shaw). (B) Tinker-HP unsupervised adaptive sampling multi-microsecond simulations (Jaffrelot Inizan et al., 2021). The dashed sections in different colors depict different conformational spaces representatively sampled based on the joint analysis from our monomeric MetaDy and the literature dimeric MDs.
The important thing here is the comparison of these MCoV2pro dimer simulations (Figure 5) with those of our MetaDy’s monomer (Figure 4(B)). We perceive equivalent profiles in the distributions for both dimers and monomers, indicating that our MetaDy simulations were able to perform similar samples from the conformational space of the active sites. In spite of this general equivalence, it is also quite evident that in the monomer the distribution expands more at high Rav values, indicating active sites even more open than in the dimers.
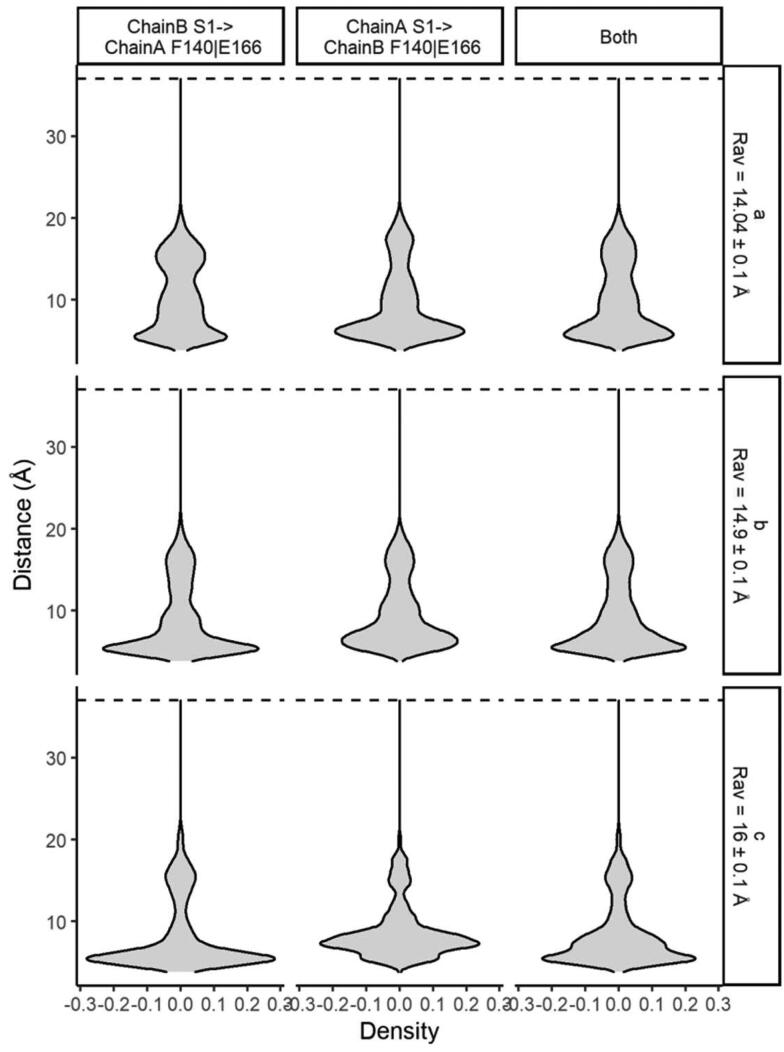
This is consistent with other results evidencing that, in the dimeric state, the stabilization of the N-finger close to the active site makes it more cohesive and prone to subtract-processing (Behnam, 2021; Cheng et al., 2010; Jaffrelot Inizan et al., 2021). Moreover, we show that even if the active sites of the dimers expand, the proximity of the N-finger to the active site remains almost constant or rather increases, preserving its catalytic capacity (Figure 6). Conversely, at monomeric states, the constrictions by the N-finger from the neighbor protomer (as well the dimeric environment as a whole) is no longer present, allowing wider and more diversal conformations for new virtual screening opportunities.
Figure 6.
Relative density distributions for the interchain distance between the S1 residue from the N-finger of a monomer to the F140|E166 residues at the catalytic pocket from the other at different average active site width values (RAV) on a multi-microsecond unsupervised adaptive sampling MD for the MCov2Pro dimer. Analysis carried at the freely available data from the Tinker-HP simulation described by Jaffrelot Inizan et al. (2021). From top to the bottom, the different sections with increasing average RAv values from Figure 5 are depicted.
Open conformers present nonideal catalytic environment in MCoV2pro monomer according quantum calculations
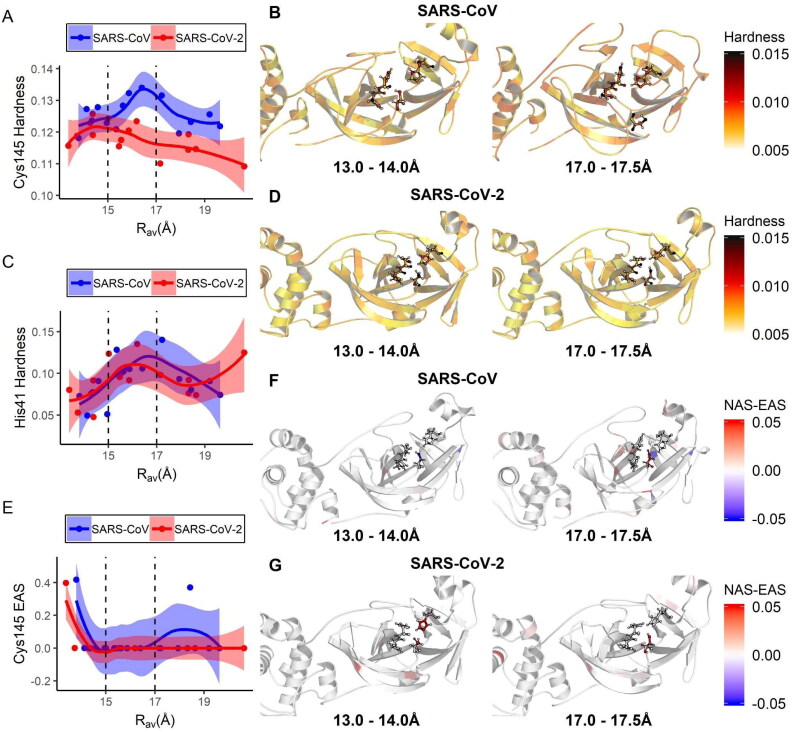
The expected reaction mechanism for the catalytic dyad of Mpros is the typical for cysteine proteases: His41 receives a proton from Cys145 which, therefore, can cleave the substrate by nucleophilic attack (Blanchard et al., 2004). Thus, a residual chemical environment favorable for catalysis is expected even before substrate binding (Grillo, Urquiza-Carvalho, Chaves, et al., 2020). Hence, we estimated electronic density descriptors related to local hardness (HA), such as nucleophilic and electrophilic attack susceptibility (respectively, NAS and EAS) for the catalytic site in the representative conformers sampled by MetaDy. These same descriptors have already shown good correlations with the catalytic path in our previous study (Grillo, Urquiza-Carvalho, Bachega, et al., 2020).
The average His41 local hardness for both Mpro presents a peak at intermediary values of RAv (between 15 and 17 Å), presenting also moderate intensity at closer conformations (being subtly higher for MCoV1pro at this last region) (Figure 7(C)). Uniquely for MCoV2pro, an increasing behavior at more open conformations (above 19 Å) is still observed for this descriptor. It is not accompanied by an equal increase on Cy145 local hardness or EAS (see below). This suggests that such increasing behavior for this unique His41 descriptor for MCoV2pro at broader conformations is not catalytically relevant. Interestingly, we observed a clear difference in the local hardness behavior for Cys145 between the two proteases (Figure 7(A,E)). In that case, the hardness of Cys145 seems to increase as the binding site of MCoV1pro opens, forming a peak at approximately the same RAv values for this same descriptor for His41. The Cys145 local hardness for MCoV2pro, conversely, presents a moderate plateau at closer conformations and decreases gradually and progressively according to the increase in RAv. The Cys145 EAS behavior for both Mpros increases approximately at the same extension according to the RAv diminishes below 15 Å, but presents a second peak around broader conformations (about 18 Å) just for MCoV1pro (Figure 7(E)). This second peak, that is absent in MCoV2pro, presents its initial phase in superposition to the respective Cys145 and His41 peaks at MCoV1pro.
Figure 7.
Variation of electronic quantum chemical descriptors related to catalysis with the average active site opening for the SARS-CoV and SARS-CoV-2 main proteases. In (A), (C) and (E), are depicted the respective plots for the average values of local hardness of His41 and Cys145, as well the Cys145 electrophilic attack susceptibility (EAS) according to the increase in RAv. Dashed lines highlight the superior deviation limit for the two ensemble modes depicted in Figure 4(B). In (B) and (D), respective images of the MCoV1pro and MCoV2pro active sites colored according to atomic local hardness are shown for representative conformers contained on each RAv ensemble. In (F) and (G), the same is illustrated coloring according to the nucleophilic attack susceptibility (NAS) to EAS behavior. In every case, the residues His41, Cys145 and neighbors are depicted on ball and stick representations.
Taken together, these results suggest that the monomeric MCoV1pro can combine with a significant amount the descriptors important for proton exchange and nucleophilic attack (the two initial steps at the catalysis) in both close or mildly open conformations (Figure 7(B,D,F)). In contrast, MCoV2pro presents a reasonably superposition in high values both for His41 and Cys145 local hardness, as for Cy145 EAS just in closer active site modes. For this protease, the binding site opening seems to disrupt the optimum catalytic environment. In that sense, although MCoV2pro can easier access more opened modes (Figures 3, 4, 5 and 10), these new conformers seem to be catalytically inactive (Figure 7). As already discussed on the last topic, this effect is probably compensated at the dimer by the increase of the proximity to the N-finger from the neighbor protomer according to the active site opening at this environment (Figure 6). We will turn later to the probable importance of the accessibility to these wider conformations (even than less catalytically active at the monomer) for protein processivity, as well their potential for new approaches of drug screening and design for MCov2Pro.
Figure 10.
Conformers used for ligand virtual screening and docking studies. The conformers were taken from the cluster centroids at the range between 0 and 10 kJ/mol in Figure 3. (A) MCoV1pro Crystallographic pose (PDB code 2BX4) and conformers C, F and H retrieved from metaDy simulations. (B) MCoV2pro crystallographic pose (PDB code 6LU7) and conformers G, H and I retrieved from metaDy simulations. For MCoV1pro only the conforme H is from well-tempered MetaDy, while for MCoV2pro only the conformer with this same label is from nontempered MetaDy. For all structures: RMSD (in Å) relative to crystal structure; Rav - average active site width (in Å); S11, S12, S2 and S4 loops are represented by surfaces coloured in blue, green, red and yellow, respectively. Catalytic residues His41 e Cys145 are represented in cyan sticks and residue Ala46 (MCoV1pro) or Ser46 (MCoV2pro) is represented in magenta surface. Animated gifs of both MetaDy for both Mpro can be seen in Supporting Information.
The impact of the mutations on the stability of open conformations in both MPros
We tried to get a glimpse of the impact on the protein structure stabilization at closer Vs wider active site conformations for each one of the 12 aa substitutions between both studied MPros. For this, we used bioinformatic tools to predict the protein stability changes for each point mutation that transforms MCov1Pro in MCov2Pro (MCov1 → MCov2) and also the opposite way (MCov2 → MCov1) (Parthiban et al., 2006). We did these calculations for each representative conformer sampled by MetaDy and plotted the results as a function of Rav (Supporting Information Figure S1).
For closer or mildly open active site conformations, it can be noticed equal incidence of stabilization both for MCov1 → MCov2 as for MCov2 → MCov1 substitutions. This is consonant with previous bioinformatics studies using just crystallographic like conformations (Bzówka et al., 2020), that shows the 12 substitutions at the MPro from SARS-CoV to SARS-CoV-2 do not stabilize or destabilize the native structure as a whole. According the Rav increases, however, stabilization effects for MCov1 > MCov2 and/or destabilization for MCov2 > MCov1 permutations prevail, especially considering five MCov1 > MCov2 substitutions: T35V, A46S, S65N, H134F and I286L (Supporting Information Figure S1). Between them, only I286L is located at the dimerization domain (III), being the other four located at the catalytic region (domains I and II).
K180N is the only substitution that seems to stabilize wider Rav values with the MCov2 → MCov1 transformation. The position 180 is located at the center of the hinge between the domains I and II. Once our analyses show higher correlations in MCov1Pro than in MCov2Pro between the Rav value and the tumbling of the domain I related to domain II (Supporting Information Tables S1 and S2), the influence of this region at this same tumbling is consonant with the role of this position on such stabilization exclusively for MCov1Pro.
The Role of A46S Mutation on the Attenuation of the Energetic Disadvantages for Binding Site Opening in MCoV2pro
Suárez and Díaz already observed greater exposure of the hydrophobic gorge of MCoV2pro active site because of the interdomain opening movements (Bzówka et al., 2020). Moreover, the mutation that occurs around the binding site (A46S) is located exactly in the S2 loop that presents increased mobility in MCoV2pro. This loop also shows increased movement correlations over the active site opening, suggesting a key role for binding site exposure motions (Supporting Information Tables S1 and S2). Additionally, A46S changing (a hydrophobic to hydrophilic substitution) seems to significantly impact on the open conformations stability in both Mpros (Supporting Information Figure S1(B)).
Thus, hydration can have a key effect to explain the differences in the conformational set of both Mpros. To check this hypothesis, we used semi-empirical quantum chemistry methods based on Neglect of Diatomic Differential Overlap (NDDO) formalism, which are a good option for predicting heat of formation for biological systems (Stewart, 2004). We estimated the variation of hydration enthalpy for each representative conformer sampled by MetaDy simulations (Figure 8) and we observed no enthalpic variation over the binding site opening for MCoV1pro, but significant decrease on this same enthalpy with this opening in MCoV2pro. The Ala/Ser exchange at S2 loop possibly improves the chances of hydrophilic contacts with the solvent and contributes to decreased hydration penalties. Noncovalent interaction (NCI) calculations that can verify the influence of the different binding site modes on the hydration of the A46S position seem to corroborate this interpretation (Figure 9). Basically, the polar S46 in MCov2Pro allows favorable solvent interactions at the wider conformations, while the hydrophobic A46 in MCov1Pro seems to promote unfavorable clathrates at the same condition (see higher discussions on the SI text) (Homans, 2007; Makhatadze & Privalov, 1996; Maurer & Oostenbrink, 2019).
Figure 9.
Hydration effects on the residue 46 of MCoV1pro and MCoV2pro. For each plot, on the left, is presented the 3D representation of noncovalent interactions, while on the right is shown the promolecular density. (A) Noncovalent interactions between A46 and water molecules of three conformers (A, J and N) from MCoV1pro; (B) Noncovalent interactions between S46 and water molecules of three conformers (B, G and O) from MCoV2pro.
Our results agree with recent MD studies which pointed to the importance of Ser46 (Bzówka et al., 2020; Suárez & Díaz, 2020). Therefore, Ser46 seems to work like a weak lock that releases the S2 loop by breaking H–bonds with the enzyme and interacting with environmental water molecules, at the same time that it elegantly decreases the solvation penalties when opening the binding site. Then, we were curious about the druggability and enzymatic activity impact of these opened conformations in both Mpros, but, especially in monomers of MCoV2pro (see docking analyses below).
Relative advantages for the MCoV2pro active site conformational promiscuity concerning ligand accessibility and adaptability
The evolutionary reasons by which the enzyme monomer can adopt a wide range of stable open conformations for the MCoV2pro, compared to the older MCoV1pro is not clear yet, although this may have to do with the greater heterogeneity of the MPro proteolytic recognition process (Behnam, 2021). In order to better understand how the greater conformational variety of the active site of these proteases can orchestrate their differences on ligand accessibility and/or catalytic activities, we redocked the PF-00835231 ligand into the crystallographic structures of MCoV1pro (PDB code 6XHL) and MCoV2pro (PDB code 6XHM), as well as into their respective most representative and energetically favorable MetaDyn's conformers (G,H,I) and (C,F,H) (Figures 10 and 11).
Figure 11.
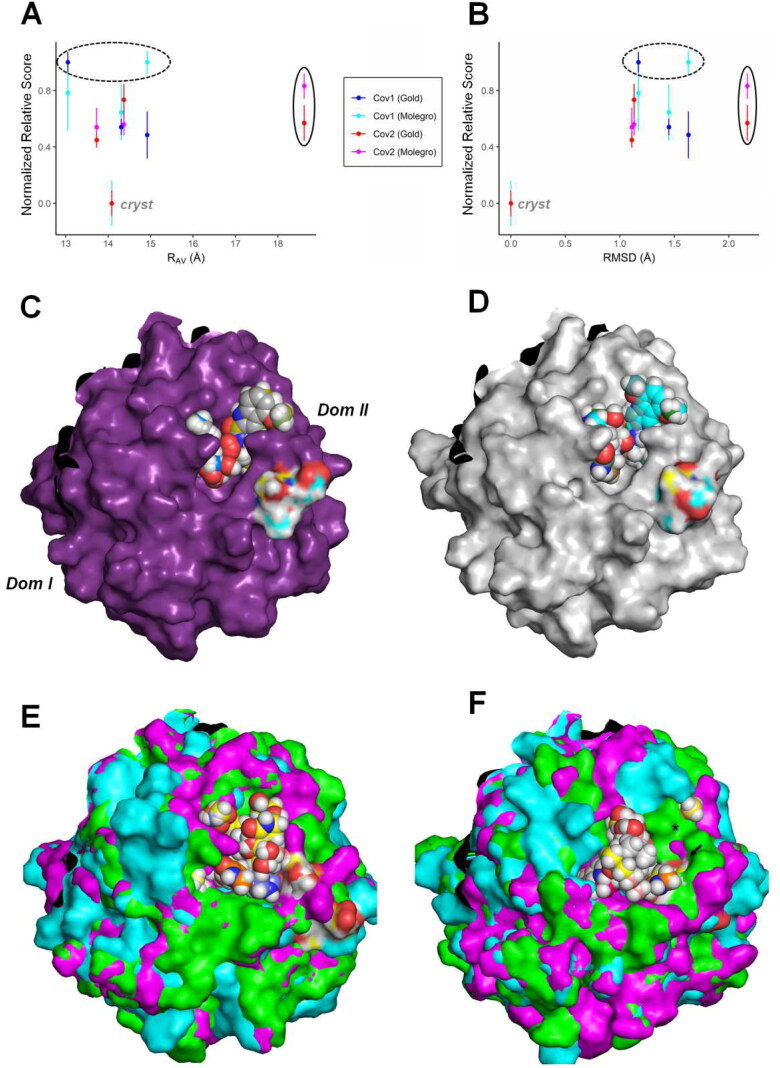
Analysis of the active site accessibility for the covalent inhibitor PF-00835231 on MCov2Pro and MCov1Pro in different conformations by virtual docking procedures. At the top, plots relating redocking score statistics (best three poses) with the active site dimensions are depicted for the ligand PF-00835231 (Boras et al., 2020; Hoffman et al., 2020), considering both crystallographic structures as the MetaDy decoys for MCov1Pro and MCov2Pro. The different score metrics from the GOLD and Molegro virtual docker (MVD) softwares, are normalized for comparison according to the docking methodology. (A) Variation of the docking normalized score according to the increase in the active site Rav value; (B) Variation according to the active site RMSD considering the crystal conformation. The respective values for the crystallographic redocking (Crystal), the less favorable scores (dashed circle) and the scores for wider conformations (solid circle) are highlighted. (C, D), redocking poses for the crystallographic conformations for MCoV2pro and MCoV1pro, respectively. The atomic colors on the surface highlight the region of Ser46 and Ala46 on MCoV2pro and MCoV1pro, respectively. At the bottom, 3D plot depicts the redocking of PF-00835231 into superimposed conformer structures of MCoV2pro (E) and MCoV1pro (F). The black asterisk and the little black arrow indicate a region poorly accessed in MCoV1pro compared to MCoV2pro and Ala46, respectively. (See Figure 10).
PF-00835231 is a potent ketone-based covalent inhibitor for both MCoV1pro and MCoV2pro with IC50 about 0.004 and 0.00027 µM, respectively (Boras et al., 2020; Hoffman et al., 2020). It is interesting (at the same time a conundrum) so high variation in affinity (i.e. one magnitude order) of the proteases for this same ligand since the only difference bordering all the active site regions is the A46S substitution at the loop C44-P52, a region that does not contact directly the ligand. In fact, no notorious differential contact with the ligand can be glimpsed at the respective 6XHL and 6XHM PDB structures. In this way, we have inferred that the differences in dynamics and conformational subspaces for the active site of both enzymes (differences for which our analyses point to a great participation of the A46S substitution) can play a pivotal role in this phenomena and carried out the differential docking analysis described below.
At the top of Figure 11, we see plots of a comparative redocking score metric for both Mpros conformers considering two different softwares and algorithms: GOLD and the Molegro Virtual Docker (MVD) (Jones et al., 1997; Thomsen & Christensen, 2006). It can be noticed, for both proteins, than the crystallographic conformation (with active site Rav measure around 14 Å) presents the more favorable scores, as well the lower RMSD considering the ligand heavy atoms at the respective crystal structures (close to 0 Å) for both softwares (Figure 11(A–D) and Supporting Information Figure S13). Although the variant MetaDy major decoys have, all of them, less favorable docking scores related to the crystal structures and considering both programs, it can be noticed two apparent advantageous features for MCoV2pro compared to MCoV1pro. First the ‘worst of all’ scores are always found between the mildly open or most closed MCoV1pro conformers (Figure 11(A,B), dashed circles). Second, it can be noted that with the wider MCoV2pro conformations (here represented by the MCoV2pro MetaDy decoy H), this protein gain a new option of ligand access with similar affinity than the average for the remained conformations (Figure 11(A,B), solid circles). In other words, the apparent higher conformational entropy gained for the MCoV2pro active site compared to MCoV1pro and depicted at Figure 4(D,E) seems to be reflected also on a gain in entropy related to paths of ligand access. This can be better glimpsed if we compare the structural visualization of all the more favorable docking poses (here, considering just the GOLD poses to avoid visual pollution) for all the conformations in both proteins (Figure 11(E,F)). It can be noted that the higher diversity of wider poses in MCoV2pro allows a higher exploration of the active site in all its extension for the ligand access, while for the closer MCoV1pro conformations, some regions are less accessed (asterisk in Figure 11(F)). In fact, a previous QM/MM study has already shown for other similar covalent inhibitor (the aldehyde derivative GC373) than the primary and noncovalent access to the MCoV2pro active site occurs at a wider and noncatalytic conformation (Ramos-Guzmán et al., 2021). Following this initial binding, the active site better fit to the ligand and the first catalytic steps happen. In this way, this higher facility to adopt wider and ligand accessible conformations could confer to the MCoV2pro enzyme advantages related both to substrate affinity as to catalysis mechanism compared to MCoV1pro. If the ligand were a peptide substrate, for instance, we could conceive that it would have easier access by a higher amount of paths to this first noncovalent event in MCoV2pro than in MCoV1pro, increasing the chances of proteolysis. It is expected that this apparently entropic facilitation of access to the MCoV2pro active site could reflect both at a diminishing of the catalytic free energy (and hence on a higher catalytic efficiency and the consequent facilitation to the first Cys145 nucleotide attack that could justify the higher affinity to the PF-00835231 covalent inhibitor), as (and maybe fortunately) on a higher multiplicity of accessible conformational microstates to be explored in drug screening. In this sense, we look for an exploration of such accessibility on a multiconformational ligand screening strategy on the last part of this study.
Exploration of the Significant Active Site Flexibility for MCov2Pro on a MultiConformational Strategy for anti-SARS–CoV-2 Drug Development
Whatever the exact reasons for these Mpro flexibilities we are evidencing here, the realisation of more open conformations sheds light on new possibilities for drug design targeting MCoV2pro, especially with monomers. Leastwise two possible ways can be considered for the development of ligands targeting the protease monomers: (i) the planning of allosteric or mixed inhibitors that are able to avoid entropic penalties by favorable binding modes at any open state of the enzyme; (ii) the design of enthalpically directed orthosteric inhibitors that are able to overcome entropic penalties by strong contacts with the crystallographic conformation. Besides that, we investigate a third way for drug development targeting MCoV2pro monomers, which regards the overstabilization of its open conformations. The presence of an allosteric or mixed inhibitor able to displace the conformational equilibrium to these open conformations should decrease the proteolytic activity of the enzyme. Beyond the active site, the dimerization pocket seems to be a potential binding spot for this kind of ligand, as already suggested by Suárez and Díaz and by our analyses (Suárez & Díaz, 2020). In our analyses, this can be inferred by the high correlation between the dimerization domain tumbling and the active site loops movements, specially for MCov2Pro (Supporting Information Table S2), as well the influence of substitutions at the position 286 at the domain III on the stabilization of wider conformations on the active site (Supporting Information Figure S1(I)). For the case of a ligand that would prevent the enzyme from adopting the crystallographic mode by binding on the active site, this molecule should have higher affinity for opening states.
In order to test this hypothesis, as well the potentiality of the MCov2Pro higher abrangency on the conformational space for drug discover, we perform a multiconformational target ligand virtual screening and docking with the aim of targeting MCoV2pro monomers from three bases: DrugBank, Zinc15 and SistematX (Irwin et al., 2012; Scotti et al., 2018; Wishart et al., 2018). The first two are canonical bases of ligands. The latter is a base of natural products and secondary metabolites hosted at the Federal University of Paraiba, Brazil. We use Molegro Virtual Docker (MVD) for the preparation of complexes, cavity prediction and docking (see Table 1) (Thomsen & Christensen, 2006).
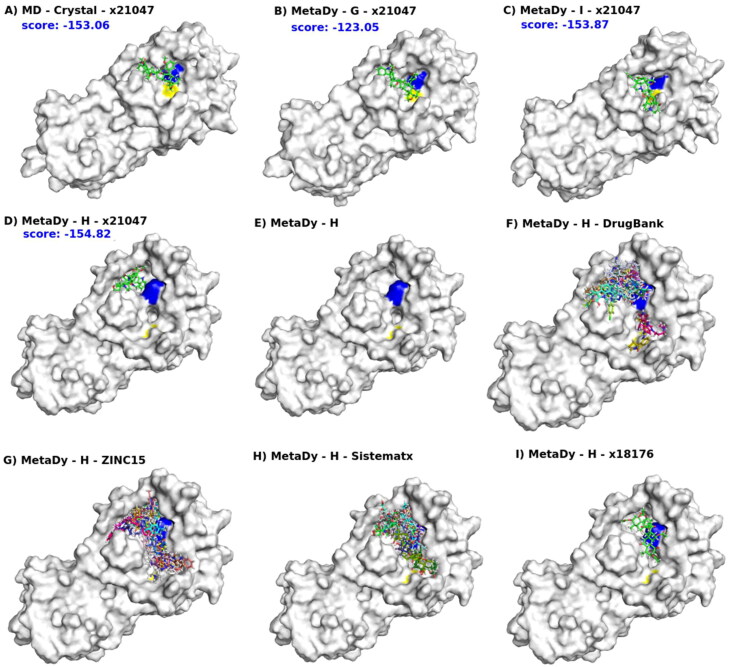
In Figure 12(A–D) we see the docking of ligand x21181 (one of our best consensus scores) into four conformers: MD crystal (i.e. the last snapshot from the equilibrium MD simulation from the crystallographic structure), wtMetaDy G and I and ntMetaDy H (see also Figure 10). The blue and yellow spots on the molecular surface indicate the presence of the catalytic dyad (His41 and Cys145, respectively). These conformers (MD crystal, G, I, H) are in ascending order of RMSD, having the crystallographic structure as a reference. Hence, they represent targets in increasing order of deviation from the protomers of the dimer (the crystal form). It is interesting to see how this ligand was able to assume different poses at all four targets. In the first three, it is positioned more directly on the catalytic site core, as a candidate for competitive inhibitor. But, the last one (H conformer) occupied a newly formed pocket (or subpocket) just above His41 (Figure 12(D)), and now it also becomes a candidate for allosteric inhibitor, given that it may stabilize an inactive state and even interfere with dimerization. In fact, the conformer H differs from all other conformers and crystal structures, with a big conformational change that affects mostly the regions of the S2, S4 and S5 subsites, drastically changing the shape of this region, and possibly allowing larger moieties to occupy these pockets. The docking score value for this ligand in H conformer is equivalent (or rather higher) than in other conformers. It is worth noting that such H conformer becomes bicavitary, with His41 in the middle of the two cavities (Figure 12(E)).
Figure 12.
Multi-conformer flexible docking’s hits explore the MCov2Pro active site space in different ways. For all, the blue and yellow patches on the molecular surface indicate the presence of the catalytic dyad (His41 and Cys145, respectively). In (A), a representative conformer of MD at equilibrium from the crystallographic structure (MD - crystal). In (B, C), the representative conformers G and I from a well-tempered MetaDy, respectively. In (D), the representative conformer H from a nontempered MetaDy. Conformers G, I and H are in increasing order of RMSD in relation to the crystallographic structure (see Figure 10). Also, from (A) to (D) we show the docking of the same ligand x21181 into MD crystal, G, I and H conformes, with respective scores in blue. In (E), the H conformer highlighting its bicavitary site. Note that the dyad residues are separated and this opens a cavity in between. From (F) to (H), superimposed docking of several ligands selected from DrugBank, ZINC15 and Sistemamatx, respectively. In (I), an example of ligand (x18176) that binds both cavities of the H conformer. (See Figure 10).
In this sense, it is significant to pay more attention to the docking into H like conformers. It is possible to verify that the H conformer was the most open of all between the low energy conformations, which implied even in the separation of the two residues that integrate the catalytic dyad (colored spot on surface), making the monomer certainly inactive. But, this separation induces the formation of an unexpected cavity between them. Thus, a ligand that could bind strongly in this state (even enzymatically inactive), could shift the equilibrium from this nonfunctional state, acting as a kind of allosteric inhibitor. Curiously, the overlap of the various ligands with better docking scores from Drugbank, ZINC and SistematX shows that there are preferences in the exploration for one of these two cavities around His45 (being more evident in the results from Drugbank) (Figure 12(F–H)). However, it is possible to find ligands that sought to cover both cavities, like x18176 (Figure 12(I)). Even if this conformation is not relevant for direct contacts with eventual ligands, the easier access to open conformations by MCoV2pro should be considered due to entropic penalties associated with stabilization of closed conformations (Figure 4(D,E)).
In this work, we gather evidence that the facility for MCoV2pro to access more open conformations, even if not initially catalytic, could confer processivity advantages to this enzyme. In fact, the openness of active-site is well correlated with the S2 loop movements, which is quite distant from the dimerization interface. So, it is reasonable to suggest that this set of open conformations could persist some way, even if residual, after dimerization. If this is true, an energetically facilitated transition between closed and open conformations can particularly favor the cycle involving substrate entrance, adaptation to the catalytic pose, catalysis, product liberation and diffusional searching for a new substrate, as well the inactive monomer/active dimer cycle. Indeed, similar accessibility to the ‘open and close’ movements at the binding site loops has already been shown functionally advantageous in previous studies in our group (Costa et al., 2019; Lima et al., 2020). In particular for viral Mpro, whose facility to bind on different points at the polyprotein sequence in a timely manner is crucial for the viral replication, such enhanced processivity could be determinant. If true, why not explore such ‘breathing’ movements in the search of innovative inhibitors for these viral proteases? Animated gifs illustrating such movements of both MetaDy for both Mpro can be seen in Supporting Information.
It is essential to point out that the docking tests performed here were only a proof of concept in terms of indicating the potentiality of the intrinsic conformational variety of MCoV2pro monomers as alternative druggable targets, rather than an in-depth study of drug discovery. Our present work (already quite extensive) was not designed to go deep into such study. But, we intend to do it in a second article (in progress) that will also include in vitro validations.
Conclusion
Through a combination of perturbative molecular dynamics, pharmacophoric analyses and quantum mechanics calculations, we brought light to motion, druggability and activity differences between Mpro monomers from both SARS-CoV and SARS-CoV-2. A new set of highly open conformations emerges from MCoV2pro which seems to be facilitated by solvation effects. Also, NCI calculations suggested that the hydrophobic to hydrophilic A46S substitution from MCoV1pro to MCoV2pro at the more flexible loop S2 participates significantly on such solvation improvement. Comparison with dimer statistics, suggests some extensibility of this same active site flexibility at the dimer. Our quantum calculations revealed, also, a disturbed chemical environment of the active site for highly open states of MCoV2pro monomers, indicating loss of proteolytic capacity at this state.
The reasons why these inactive and flexible conformers would be more remarkable in the MCoV2pro monomer than MCoV1pro are still under debate, and this work is part of it. Whatever, the rising of these open conformations affect directly on the pharmacophoric exposition of the MCoV2pro active site, reflecting on possible new drug development strategies. Our preliminary docking studies in different conformers were able to show promising possibilities in the virtual screening of new ligands, whether competitive inhibitors for monomers closer to the up state (near to the protomer in the dimer) or allosteric inhibitors for those closer to the down state, with more open but inactive catalytic sites. Even such nonfunctional monomers should still be considered as drug targets, since a ligand that binds strongly in these down states could stabilize it and even displace the monomer-dimer balance and shut off viral replication.
We believe that our comparative simulations on Mpro monomers of SARS-CoV and SARS-CoV-2 have generated intriguing results and raised hypotheses that should be considered by the scientific community in the search for new antiviral drugs against COVID19.
Supplementary Material
Acknowledgements
The authors thank Dr. Peter Göttig for rich suggestions. The authors also acknowledge the physical structure and computational support provided by Universidade Federal da Paraíba (UFPB); Universidade Federal de Itajubá (Unifei); Laboratório Nacional de Computação Científica – LNCC, Petrópolis/RJ, Brazil (Project “Prospecção e testes in vitro de inibidores de proteínas associadas ao vírus SARS-Cov 2 por meio do uso conjunto de ferramentas de bioinformática, simulação molecular, química quântica e aprendizado de máquina - qcbiocovid19” Supercomputer SDumont); Centro Nacional de Processamento de Alto Desempenho em São Paulo (CENAPAD-SP); Núcleo de Processamento de Alto Desempenho of Universidade Federal do Rio Grande do Norte (NPAD/UFRN).
Disclosure statement
No potential conflict of interest was reported by the authors.
Author’s contributions
L.H.F.L. and G.B.R. designed research; R.E.O.R., E.J.F.C., P.H.C., L.S.C.C., I.B.G., L.E.G.C., F.C.G., C. H.S., M.T.S., A.D.C., K.S.M., A.V.W., R.S.F., G.B.R. and L.H.F.L. performed research; R.E.O.R., E.J.F.C., I.B.G., L.E.G.C., R.S.F., G.B.R., C.H.S. and L.H.F.L. analyzed data; and R.E.O.R., E.J.F.C., G.B.R., C.H.S. and L.H.F.L. wrote the paper.
References
- Anand, K., Palm, G. J., Mesters, J. R., Siddell, S. G., Ziebuhr, J., & Hilgenfeld, R. (2002). Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. The EMBO Journal, 21(13), 3213–3224. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barducci, A., Bussi, G., & Parrinello, M. (2008). Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Physical Review Letters, 100(2), 020603. 10.1103/PhysRevLett.100.020603 [DOI] [PubMed] [Google Scholar]
- Behnam, M. A. M. (2021). Protein structural heterogeneity: A hypothesis for the basis of proteolytic recognition by the main protease of SARS-CoV and SARS-CoV-2. Biochimie, 182, 177–184. 10.1016/j.biochi.2021.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, R. C., Melo, M. C. R., & Schulten, K. (2015). Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochimica et Biophysica Acta, 1850(5), 872–877. 10.1016/j.bbagen.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, J. E., Elowe, N. H., Huitema, C., Fortin, P. D., Cechetto, J. D., Eltis, L. D., & Brown, E. D. (2004). High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chemistry & Biology, 11(10), 1445–1453. 10.1016/j.chembiol.2004.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcato, G., Bissaro, M., Pavan, M., Sturlese, M., & Moro, S. (2020). Targeting the coronavirus SARS-CoV-2: Computational insights into the mechanism of action of the protease inhibitors lopinavir, ritonavir and nelfinavir. Scientific Reports, 10, 20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras, B., Jones, R. M., Anson, B. J., Arenson, D., Aschenbrenner, L., Bakowski, M. A., Beutler, N., Binder, J., Chen, E., Eng, H., Hammond, H., Hammond, J., Haupt, R. E., Hoffman, R., Kadar, E. P., Kania, R., Kimoto, E., Kirkpatrick, M. G., Lanyon, L., … Allerton, C. (2020). Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. bioRxiv. The Preprint Server for Biology, 1–31. 10.1101/2020.09.12.293498 [DOI] [Google Scholar]
- Boto, R. A., Peccati, F., Laplaza, R., Quan, C., Carbone, A., Piquemal, J.-P., Maday, Y., & Contreras-García, J. (2020). NCIPLOT4: Fast, Robust, and Quantitative Analysis of Noncovalent Interactions. Journal of Chemical Theory and Computation, 16(7), 4150–4158. 10.1021/acs.jctc.0c00063 [DOI] [PubMed] [Google Scholar]
- Brandt, A. M. L., Batista, P. R., Souza-Silva, F., Alves, C. R., & Caffarena, E. R. (2016). Exploring the unbinding of Leishmania (L.) amazonensis CPB derived-epitopes from H2 MHC class I proteins. Proteins: Structure, Function, and Bioinformatics, 84(4), 473–487. 10.1002/prot.24994 [DOI] [PubMed] [Google Scholar]
- Bzówka, M., Mitusińska, K., Raczyńska, A., Samol, A., Tuszyński, J. A., & Góra, A. (2020). Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. International Journal of Molecular Sciences, 21, 3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.-C., Chang, G.-G., & Chou, C.-Y. (2010). Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophysical Journal, 98(7), 1327–1336. 10.1016/j.bpj.2009.12.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A. S., Kubař, T., Cui, Q., & Elstner, M. (2016). Semiempirical quantum mechanical methods for noncovalent interactions for chemical and biochemical applications. Chemical Reviews, 116(9), 5301–5337. 10.1021/acs.chemrev.5b00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus disease (COVID-19): Vaccines . 2020. https://www.who.int/news-room/q-a-detail/coronavirus-disease-(covid-19)-vaccines.
- Costa, L. S. C., Mariano, D. C. B., Rocha, R. E. O., Kraml, J., Silveira, C. H. d., Liedl, K. R., de Melo-Minardi, R. C., & Lima, L. H. F. d. (2019). Molecular dynamics gives new insights into the glucose tolerance and inhibition mechanisms on β-glucosidases. Molecules, 24(18), 3215. 10.3390/molecules24183215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D. E. Shaw Research, “Molecular Dynamics Simulations Related to SARS-CoV-2,” D. E. Shaw Research Technical Data, 2020. http://www.deshawresearch.com/resources_sarscov2.html.
- Domingo, L. R., Ríos-Gutiérrez, M., & Pérez, P. (2016). Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules, 21(6), 748. 10.3390/molecules21060748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassio, A. V., Santos, L. H., Silveira, S. A., Ferreira, R. S., & de Melo-Minardi, R. C. (2020). nAPOLI: A graph-based strategy to detect and visualize conserved protein-ligand interactions in large-scale. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 17, 1317–1328. [DOI] [PubMed] [Google Scholar]
- Geerlings, P., De Proft, F., & Langenaeker, W. (2003). Conceptual density functional theory. Chemical Reviews, 103(5), 1793–1873. 10.1021/cr990029p [DOI] [PubMed] [Google Scholar]
- Grillo, I. B., Urquiza-Carvalho, G. A., Bachega, J. F. R., & Rocha, G. B. (2020). Elucidating enzymatic catalysis using fast quantum chemical descriptors. Journal of Chemical Information and Modeling, 60(2), 578–591. 10.1021/acs.jcim.9b00860 [DOI] [PubMed] [Google Scholar]
- Grillo, I. B., Urquiza-Carvalho, G. A., Chaves, E. J. F., & Rocha, G. B. (2020). Semiempirical methods do Fukui functions: Unlocking a modeling framework for biosystems. Journal of Computational Chemistry, 41(9), 862–873. 10.1002/jcc.26148 [DOI] [PubMed] [Google Scholar]
- Grillo, I. B., Urquiza-Carvalho, G. A., & Rocha, G. B. (2020). PRIMoRDiA: A software to explore reactivity and electronic structure in large biomolecules. Journal of Chemical Information and Modeling, 60(12), 5885–5890. 10.1021/acs.jcim.0c00655 [DOI] [PubMed] [Google Scholar]
- Hawkins, P. C. D., Skillman, A. G., & Nicholls, A. (2007). Comparison of shape-matching and docking as virtual screening tools. Journal of Medicinal Chemistry, 50(1), 74–82. 10.1021/jm0603365 [DOI] [PubMed] [Google Scholar]
- Hawkins, P. C. D., Skillman, A. G., Warren, G. L., Ellingson, B. A., & Stahl, M. T. (2010). Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. Journal of Chemical Information and Modeling, 50(4), 572–584. 10.1021/ci100031x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld, R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, R. L., Kania, R. S., Brothers, M. A., Davies, J. F., Ferre, R. A., Gajiwala, K. S., He, M., Hogan, R. J., Kozminski, K., Li, L. Y., Lockner, J. W., Lou, J., Marra, M. T., Mitchell, L. J., Murray, B. W., Nieman, J. A., Noell, S., Planken, S. P., Rowe, T., … Taggart, B. (2020). Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. Journal of Medicinal Chemistry, 63(21), 12725–12747. 10.1021/acs.jmedchem.0c01063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homans, S. W. (2007). Water, water everywhere–except where it matters? Drug Discovery Today, 12(13–14), 534–539. 10.1016/j.drudis.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Incerti, M., Russo, S., Callegari, D., Pala, D., Giorgio, C., Zanotti, I., Barocelli, E., Vicini, P., Vacondio, F., Rivara, S., Castelli, R., Tognolini, M., & Lodola, A. (2017). Metadynamics for perspective drug design: Computationally driven synthesis of new protein-protein interaction inhibitors targeting the EphA2 receptor. Journal of Medicinal Chemistry, 60(2), 787–796. 10.1021/acs.jmedchem.6b01642 [DOI] [PubMed] [Google Scholar]
- Irwin, J. J., Sterling, T., Mysinger, M. M., Bolstad, E. S., & Coleman, R. G. (2012). ZINC: A free tool to discover chemistry for biology. Journal of Chemical Information and Modeling, 52(7), 1757–1768. 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrelot Inizan, T., Célerse, F., Adjoua, O., El Ahdab, D., Jolly, L.-H., Liu, C., Ren, P., Montes, M., Lagarde, N., Lagardère, L., Monmarché, P., & Piquemal, J.-P. (2021). High-resolution mining of the SARS-CoV-2 main protease conformational space: Supercomputer-driven unsupervised adaptive sampling. Chemical Science, 12(13), 4889–4907. https://doi.org/10.1039/d1sc00145k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of M from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Johnson, E. R., Keinan, S., Mori-Sánchez, P., Contreras-García, J., Cohen, A. J., & Yang, W. (2010). Revealing noncovalent interactions. Journal of the American Chemical Society, 132(18), 6498–6506. 10.1021/ja100936w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G., Willett, P., Glen, R. C., Leach, A. R., & Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology, 267(3), 727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- Klamt, A., & Schüürmann, G. (1993). COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. Journal of the Chemical Society, Perkin Transactions, 2(5), 799–805. 10.1039/P29930000799 [DOI] [Google Scholar]
- Konar, S., Sinha, S. K., Datta, S., & Ghorai, P. K. (2019). Probing the effect of glucose on the activity and stability of β-glucosidase: An all-atom molecular dynamics simulation investigation. ACS Omega, 4(6), 11189–11196. 10.1021/acsomega.9b00509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazim, R., Suh, D., & Choi, S. (2020). Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. International Journal of Molecular Sciences, 21(17), 1–20. https://doi.org/10.3390/ijms21176339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, L. H. F. d., Fernandez-Quintéro , M. L., Rocha , R. E. O., Mariano , D. C. B., Melo-Minardi, R. C. de, & Liedl, K. R. (2020). Conformational flexibility correlates with glucose tolerance for point mutations in β-glucosidases - A computational study. Journal of Biomolecular Structure and Dynamics, 39(5) 1621–1634. [DOI] [PubMed] [Google Scholar]
- Lin, P.-Y., Chou, C.-Y., Chang, H.-C., Hsu, W.-C., & Chang, G.-G. (2008). Correlation between dissociation and catalysis of SARS-CoV main protease. Archives of Biochemistry and Biophysics, 472(1), 34–42. 10.1016/j.abb.2008.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O., & Shaw, D. E. (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics, 78(8), 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanti, M., Bhakat, S., Nilsson, U. J., & Söderhjelm, P. (2016). Flap dynamics in aspartic proteases: A computational perspective. Chemical Biology & Drug Design, 88(2), 159–177. 10.1111/cbdd.12745 [DOI] [PubMed] [Google Scholar]
- Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhatadze, G. I., & Privalov, P. L. (1996). On the entropy of protein folding. Protein Science, 5(3), 507–510. 10.1002/pro.5560050312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, L.Nascimento, A. S., Nunes, F. M., Phillips, K., Aparicio, R., Dias, S. M. G., Carolina, A., Figueira, M., Lin, J. H., Nguyen, P., Apriletti, J. W., Neves, F. A. R., Baxter, J. D., Webb, P., Skaf, M. S., & Polikarpov, I. (2009). Gaining ligand selectivity in thyroid hormone receptors via entropy. Proceedings of the National Academy of Sciences of the United States of America, 106, 20717–20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer, M., & Oostenbrink, C. (2019). Water in protein hydration and ligand recognition. Journal of Molecular Recognition, 32, e2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure and Dynamics, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthiban, V., Gromiha, M. M., & Schomburg, D. (2006). CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Research., 34, W239–W242. 10.1093/nar/gkl190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kalé, L., & Schulten, K. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26(16), 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Guzmán, C. A., Ruiz-Pernía, J. J., & Tuñón, I. (2021). Multiscale simulations of SARS-CoV-2 3CL protease inhibition with aldehyde derivatives. Role of protein and inhibitor conformational changes in the reaction mechanism. ACS Catalysis, 11(7), 4157–4168. 10.1021/acscatal.0c05522 [DOI] [PubMed] [Google Scholar]
- Rocha, R. E. O., & Lima, L. H. F. (2019). Cooperative hydrogen bonds and mobility of the non-aromatic ring as selectivity determinants for human acetylcholinesterase to similar anti-Alzheimer’s galantaminics: A computational study. Journal of Biomolecular Structure and Dynamics, 37(7), 1843–1856. 10.1080/07391102.2018.1470036 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bussey, I. G., Doshi, U., & Hamelberg, D. (2016). Enhanced molecular dynamics sampling of drug target conformations. Biopolymers, 105(1), 35–42. 10.1002/bip.22740 [DOI] [PubMed] [Google Scholar]
- Rozengurt, E., & Heppel, L. A. (1975). A Specific effect of external ATP on the permeability of transformed 3T3 cells. Biochemical and Biophysical Research Communications, 67(4), 1581–1588. 10.1016/0006-291X(75)90207-7 [DOI] [PubMed] [Google Scholar]
- Scotti, M., Herrera-Acevedo, C., Oliveira, T., Costa, R., Santos, S., Rodrigues, R., Scotti, L., & Da-Costa, F. (2018). SistematX, an online web-based cheminformatics tool for data management of secondary metabolites. Molecules, 23(1), 103. 10.3390/molecules23010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Wei, Z., & Song, J. (2004). Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: Defining the extra domain as a new target for design of highly specific protease inhibitors. Journal of Biological Chemistry, 279(23), 24765–24773. 10.1074/jbc.M311744200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart Computational Chemistry - MOPAC Home Page . http://OpenMOPAC.net.
- Stewart, J. J. P. (1996). Application of localized molecular orbitals to the solution of semiempirical self-consistent field equations. International Journal of Quantum Chemistry, 58(2), 133–146. [DOI] [Google Scholar]
- Stewart, J. J. P. (2004). Comparison of the accuracy of semiempirical and some DFT methods for predicting heats of formation. Journal of Molecular Modeling, 10(1), 6–12. 10.1007/s00894-003-0157-6 [DOI] [PubMed] [Google Scholar]
- Stewart, J. J. P. (2013). Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. Journal of Molecular Modeling, 19(1), 1–32. 10.1007/s00894-012-1667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez, D., & Díaz, N. (2020). SARS-CoV-2 Main Protease: A Molecular Dynamics Study. Journal of Chemical Information and Modeling, 60(12), 5815–5831. 10.1021/acs.jcim.0c00575 [DOI] [PubMed] [Google Scholar]
- Thomsen, R., & Christensen, M. H. (2006). MolDock: A new technique for high-accuracy molecular docking. Journal of Medicinal Chemistry, 49(11), 3315–3321. 10.1021/jm051197e [DOI] [PubMed] [Google Scholar]
- Tóth, G., & Borics, A. (2006). Closing of the flaps of HIV-1 protease induced by substrate binding: A model of a flap closing mechanism in retroviral aspartic proteases. Biochemistry, 45(21), 6606–6614. 10.1021/bi060188k [DOI] [PubMed] [Google Scholar]
- Wan, H., Aravamuthan, V., & Pearlstein, R. A. (2020). Probing the dynamic structure-function and structure-free energy relationships of the coronavirus main protease with biodynamics theory. ACS Pharmacology & Translational Science, 3(6), 1111–1143. 10.1021/acsptsci.0c00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Chen, C., Tan, W., Yang, K., & Yang, H. (2016). Structure of main protease from human coronavirus NL63: Insights for wide spectrum anti-coronavirus drug design. Scientific Reports, 6, 22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D. H., Stephens, E., O'Brien, D. P., & Zhou, M. (2004). Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angewandte Chemie International Edition, 43(48), 6596–6616. 10.1002/anie.200300644 [DOI] [PubMed] [Google Scholar]
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., … Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, M., Su, H., Zhao, W., Xie, H., Shao, Q., & Xu, Y. (2021). What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design. Medicinal Research Reviews. 10.1002/med.21783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., Gao, G. F., Anand, K., Bartlam, M., Hilgenfeld, R., & Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Dejnirattisai, W., Supasa, P., Liu, C., Mentzer, A. J., Ginn, H. M., Zhao, Y., Duyvesteyn, H. M. E., Tuekprakhon, A., Nutalai, R., Wang, B., Paesen, G. C., Lopez-Camacho, C., Slon-Campos, J., Hallis, B., Coombes, N., Bewley, K., Charlton, S., Walter, T. S., … Screaton, G. R. (2021). Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell, 184(9), 2348–2361.e6. 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A., Chan, J. F. W., Azhar, E. I., Hui, D. S. C., & Yuen, K.-Y. (2016). Coronaviruses - Drug discovery and therapeutic options. Nature Reviews Drug Discovery, 15(5), 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.