Abstract
The human family of Potassium (K+) Channel Tetramerization Domain (KCTD) proteins counts 25 members, and a significant number of them are still only partially characterized. While some of the KCTDs have been linked to neurological disorders or obesity, a growing tally of KCTDs are being associated with cancer hallmarks or involved in the modulation of specific oncogenic pathways. Indeed, the potential relevance of the variegate KCTD family in cancer warrants an updated picture of the current knowledge and highlights the need for further research on KCTD members as either putative therapeutic targets, or diagnostic/prognostic markers. Homology between family members, capability to participate in ubiquitination and degradation of different protein targets, ability to heterodimerize between members, role played in the main signalling pathways involved in development and cancer, are all factors that need to be considered in the search for new key players in tumorigenesis. In this review we summarize the recent published evidence on KCTD members’ involvement in cancer. Furthermore, by integrating this information with data extrapolated from public databases that suggest new potential associations with cancers, we hypothesize that the number of KCTD family members involved in tumorigenesis (either as positive or negative modulator) may be bigger than so far demonstrated. 
Video abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-021-00737-8.
Keywords: KCTD family, BTB domain, Cancer, Oncogene, Tumor suppressor, KCASH family, KCTD15, Cul3, Ubiquitination
Background
The human family of Potassium (K+) Channel Tetramerization Domain (KCTD) proteins counts 25 members which are only partially characterized, although their increasing relevance in various important biological functions is being gradually uncovered [1]. Indeed, ever-growing evidences are pointing to a significant role of this diverse family in mechanisms of protein degradation and other biological functions, hinting for several of them as potential players in cancer development or cancer prevention, as well as potential therapeutic targets in the treatment of tumor.
The KCTD family arose from a common ancestral gene by gene duplication and evolutive divergence and share a conserved domain, called BTB (Broad complex, Tramtrak and Bric‐a‐brac)/POZ (poxvirus zinc finger) domain [2, 3]. This domain (from now on BTB domain) is a simple protein motif (approximately 95 amino acids) which is functional for protein oligomerization [4] and to establish protein–protein interactions and is essential for the diverse biological functions of the KCTD family members [5].
Comparative analyses of the full aminoacidic sequences of all family members suggested that KCTD proteins can be clustered in seven clades, each composed of KCTD paralogs that often share similar biological functions [6]. However, a more recent classification from Teng et al., based on the minimal BTB domain sequences alignment and the N-terminal domain structures, subdivides the KCTD proteins into eight groups [1]. In this review, we will use this latter classification. We also refer to Teng et al.’s work for the available details on binding partners, BTB structures and additional protein domains of the KCTD family.
In a first attempt of characterization, KCTD family members can be classified according to the capability of binding to E3-ubiquitin ligases, through their BTB domain, and thus participate in degradative processes. Indeed, KCTD proteins of group B (KCASH1KCTD11, KCASH2KCTD21, KCASH3KCTD6), group C (KCTD10, -13, TNFAIP1), group D (KCTD3, SHKBP1), group E (KCTD2, -5, -9, -17) and group H (KCTD7, -14) can form a E3-ubiquitin ligase complex together with Cullin RING E3 Ligases (CRL, most frequently partnering with Cullin3 (Cul3) ligase).
Most KCTD proteins act therefore as adapters, recruiting selectively substrates for ubiquitination [4, 7–13].
On the other hand, CRL recruitment is not a property present in all KCTD proteins [14]: indeed, some KCTD, belonging to group A (KCTD1 and KCTD15), group F (KCTD8, -12, -16) and group G (KCTD20, BTBD10) seem to have Cullin-independent functions [6, 9, 15, 16].
So far, KCTD genes have been associated with several diseases, including neurodevelopmental, neuropsychiatric, and neurodegenerative disorders [1, 17]. More recently, several KCTD have also been associated to cancer (see below) although the identification of KCTD-containing players in cancer appears far from completed.
In this review, we aim to discuss the role of KCTD proteins in cancer, analyzing the published literature and adding further insights, by collecting evidences based on differential gene expression and presence of genic mutations in various tumor tissues obtained from two different databases (db), COSMIC (Catalogue Of Somatic Mutations In Cancer; https://cancer.sanger.ac.uk/cosmic [18, 19] and GENT2 (Gene Expression patterns across Normal and Tumor tissues; http://gent2.appex.kr/gent2/ [20].
COSMIC is the largest well annotated collection of somatic variants detected in tumor samples. A considerable fraction of COSMIC data is generated by whole genome screens, making them suitable for comparative analysis of mutability of human genes.
Gene expression data were analyzed through the Gene Expression database of Normal and Tumor tissues (GENT2) and based on the GEO public repository using the U133Plus2 (GPL570) platform and we focused on the information that was statistically significant.
Both db classify tumors according to their anatomic localization. While differential expression of a gene in a specific tumor type is generally consistent in published data (when available) and in db data, sometimes differences may occur; discrepancies are probably due to the existence of histological and molecular subgroups in the different tumor types, which are often not considered in these large databases. For this reason, we have taken in account in first approximation only data which are statistically significant (GENT) and consistent between the two db.
In the next sections we will first present the KCTD groups that have greater evidence of implication in tumorigenesis. Then, we will explore groups which, although do not appear yet to have strong experimental evidence, may still have a role in cancer, either because they are involved in signaling pathway known to lead to tumorigenesis or because of new hints obtained from our current databases observations.
An overview of the evidence on KCTD family’s role in cancer and the pathways involved is summarized in Table 1.
Table 1.
Differential expression of KCTD family members in human tumors
| Group | Gene | Cancer | Literature data | Cosmic | GENT2 | |||
|---|---|---|---|---|---|---|---|---|
| Expression | Molecular target | Gene expression | Frequency | FC (T/N tissue) | P value | |||
| Group C | KCTD10 | Burkitt's lymphoma | O/E [29] | |||||
| Lymphoblastic leukemia | O/E [29] | |||||||
| Colorectal adenocarcinoma | O/E [29] | |||||||
| Gastrointestinal stromal tumor | O/E [37] | |||||||
| Lung carcinoma | O/E [29] | |||||||
| Ovarian cancer | O/E [27] | |||||||
| Melanoma | O/E [29] | |||||||
| Breast cancer | O/E [25] | RhoB, Rac1 | ||||||
| Pancreatic cancer | O/E [28] | |||||||
| Hepatocellular carcinoma | U/E [38] | EIF3D | ||||||
| Adrenal gland cancer | O/E | 5/79 (6.3%) | 1.41 | < 0.001 | ||||
| TNFAIP1 | Osteosarcoma | O/E [43, 52] | NF- κB signaling | |||||
| Breast cancer | O/E [53] | |||||||
| Hepatocellular carcinoma | U/E [41] | CSNK2B | ||||||
| Non-small cell lung cancer | U/E [46, 47] | |||||||
| Uterus cancer | U/E [49] | |||||||
| Pancreatic cancer | U/E [48] | |||||||
| Gastric carcinoma | U/E [42–45] | |||||||
| Cervical carcinoma | U/E [49, 50] | RhoB | ||||||
| Adrenal gland cancer | U/E | 11/79 (13.9%) | 0.85 | < 0.05 | ||||
| KCTD13 | Breast cancer | O/E | 154/1104 (13.9%) | 1.85 | < 0.001 | |||
| Group B | KCTD11 | Medulloblastoma | U/E [59, 61, 62] | Hedgehog signaling | ||||
| Endometrium cancer | U/E [61] | |||||||
| Gallbladder cancer | U/E [61] | |||||||
| Urinary bladder cancer | U/E [61] | |||||||
| Colorectal cancer | U/E [61] | |||||||
| Stomach cancer | U/E [61] | |||||||
| Lung cancer | U/E [61] | |||||||
| Larynx cancer | U/E [61] | |||||||
| Breast cancer | U/E [61] | |||||||
| Esophageal cancer | U/E [61] | |||||||
| Prostate carcinoma | U/E [59, 61, 62] | |||||||
| Hepatocellular carcinoma | U/E [63] | MST1/GSK3β/p21 signaling | ||||||
| Ovarian cancer | U/E | 159/266 (59.8%) | 0.82 | |||||
| KCTD21 | Medulloblastoma | U/E [12, 15, 66] | Hedgehog signaling | |||||
| KCTD6 | Medulloblastoma | U/E [12, 15, 66] | Hedgehog signaling | |||||
| Group A | KCTD15 | Lymphoblastic leukemia | O/E [80] | |||||
| Myeloid leukemia | O/E [80, 81] | |||||||
| Medulloblastoma | U/E [15] | Hedgehog signaling | ||||||
| KCTD1 | Ovarian cancer | O/E | 37/266 (13.9%) | 1.47 | < 0.001 | |||
| Endometrium cancer | O/E | 59/600 (9.8%) | 1.43 | < 0.001 | ||||
| Pancreatic cancer | O/E | 20/179 (11.17%) | 1.32 | < 0.001 | ||||
| Lung cancer | O/E | 76/1019 (7.46%) | 1.21 | < 0.001 | ||||
| Group D | SHKBP1 | Small intestine neuroendocrine tumor | O/E [88] | |||||
| Osteosarcoma | O/E [86] | EGFR signaling | ||||||
| Myeloid leukemia | Mut.V89I [89] | FLT3 Tyrosine kinase | ||||||
| Cervical cancer | Mut [90] | O/E | 33/307 (10.8%) | 2.14 | < 0.05 | |||
| Pancreatic cancer | O/E | 24/179 (13.4%) | 2.05 | < 0.001 | ||||
| Nervous system tumor | Mutation | 7/130 (5.4%) | ||||||
| Large intestine tumor | Mutation | 113/2513 (4.5%) | ||||||
| KCTD3 | Stomach cancer | O/E | 37/285 (13.1%) | 1.83 | < 0.001 | |||
| Breast cancer | O/E | 157/1104 (14.2%) | 1.3 | < 0.001 | ||||
| Nervous system tumor | Mutation | 7/130 (3.9%) | ||||||
| Group F | KCTD12 | Cervical cancer | O/E [102, 103] | CDC25B, CDK1 | ||||
| Lung cancer | O/E [102, 103] | CDC25B, CDK1 | ||||||
| Gastrointestinal stromal tumor | U/E [96, 97] | |||||||
| Colorectal cancer | U/E [98, 102, 103] | CDK1 | ||||||
| Uveal melanoma | U/E [100] | |||||||
| Esophageal squamous cell carcinoma | U/E [101] | WNT, Notch signaling | ||||||
| Breast cancer | U/E [99] | FoxO/Akt signaling | ||||||
| KCTD8 | Cervical cancer | O/E | 17/307 (5.5%) | n.a | ||||
| Pancreatic cancer | O/E | 13/179 (7.3%) | n.a | |||||
| Esophageal cancer | O/E | 9/125 (7.2%) | n.a | |||||
| Thyroid cancer | O/E | 41/513 (8.0%) | n.a | |||||
| KCTD16 | Small cell lung cancer | O/E [104] | ||||||
| Thyroid cancer | O/E [105] | |||||||
| Hepatic cancer | Mutation | 296/2216 (13.6%) | ||||||
| Pancreatic cancer | Mutation | 216/1840 (11.7%) | ||||||
| Prostate carcinoma | Mutation | 177/1984 (8.9%) | ||||||
| Group E | KCTD5 | Melanonoma | O/E [110] | Rac1, Ca2 + signaling | ||||
| Breast cancer | O/E [110, 111] | TRPM4, Rac1, Ca2 + signaling | O/E | 140/1104 (12.5%) | 1.33 | < 0.001 | ||
| KCTD2 | Colonrectal cancer | O/E [115] | ||||||
| Glioma | U/E [8] | C-Myc | ||||||
| KCTD9 | Stomach cancer | O/E | 42/285 (14.7%) | 1.25 | < 0.05 | |||
| Group G | KCTD20 | Non-small cell lung cancer | O/E [113] | Akt signaling, E-cadherin | ||||
| BTBD10 | Glioma | U/E [120, 122] | Akt signaling | |||||
| Group H | KCTD7 | Glioblastoma | O/E [123] | |||||
| Lung adenocarcinoma | O/E [124] | |||||||
| Adrenal gland cancer | O/E | 11/79 (13.9%) | 1.61 | < 0.001 | ||||
| Skin cancer | O/E | 40/473 (8.46%) | 2.28 | < 0.001 | ||||
| Breast cancer | O/E | 68/1104 (6.16%) | 1.16 | < 0.001 | ||||
| Pancreatic cancer | Mutation | 9/165 (5.5%) | ||||||
| KCTD14 | Ovarian cancer | CNV gain | 31/684 (4.5%) | 1.53 | < 0.001 | |||
| N.C | KCNRG | Chronic lymphocitic leukemia | U/E [126] | |||||
| Multiple myelomas | U/E [126] | |||||||
| Hepatocellular carcinoma | Mut [127] | |||||||
| KCTD19 | Hematopoietic and lymphoid tumor | O/E | 28/221 (12.7%) | 1.14 | < 0.001 | |||
| Nervous system tumor | Mutation | 6/130 (4.7%) | ||||||
| Large intestine tumor | Mutation | 89/2502 (3.2%) | ||||||
| Skin cancer | Mutation | 76/1279 (4.2%) | ||||||
| KCTD4 | Lung cancer | O/E | 51/1019 (5.0%) | 1.32 | < 0.001 | |||
Summary of the KCTD family members and the evidence of their involvement (or differential expression) in tumors, as reported in literature. Where possible, data suggesting further roles for KCTD members in tumors, extrapolated from COSMIC db (in terms of gene expression and frequency of tumor samples with overexpression, underexpression or mutation of the gene of interest compared to total analysed tumor samples) and GENT2 db (in numerical terms of FC = fold change, calculated from the ratio of expression in tumor and normal tissues, and statistical ones of p-value) are also indicated. Numbers in brackets in the “Expression” column indicate the relative bibliographic references; O/E: overexpression; U/E: underexpression; n.a.: not available.
The KCTD proteins of group C: oncogenes or tumor suppressors?
KCTD10, TNFAIP1 (Tumor Necrosis Factor Induced Protein 1, also known as B12 or Bacurd2) and KCTD13 (also known as Bacurd1) proteins, belong to Group C. In addition to the capability of binding Cul3 ubiquitin ligase and thus inducing degradation of some interacting partners, this group is characterized by a proliferating cell nuclear antigen (PCNA)-binding motif at the C-terminus [1, 21]. Indeed, they are TNF-α/IL-6 -inducible proteins which are able to stimulate the activity of DNA polymerase δ in presence of PCNA, playing a role in DNA replication, repair and cell cycle control [22]. Given the pleiotropic role of TNF in immune response and inflammation [23] and the panoply of biological processes in which these KCTD proteins may be involved, their differential expression between normal and cancer tissues is not surprising.
KCTD10 gene maps to chromosome 12q24.11 and is strongly conserved from zebrafish to human [24]. Its expression is detectable in all human tissues with higher levels in heart, skeletal muscle, and placenta. In HER2‐positive breast cancers, one of the most aggressive subtypes of breast cancer, KCTD10 induces RhoB degradation and activation of Rac1 [25], which promotes progression of tumors, and resistance to therapy [26]. Similarly, KCTD10 expression has been associated to unfavorable prognosis in patients with early-stage clear-cell ovarian carcinoma [27] suggesting that in these contexts KCTD10 may be a therapeutic target.
Moreover, KCTD10 expression results upregulated, and it has been indicated as key to the pancreatic carcinogenesis [28].
High KCTD10 expression levels have also been detected in lymphoblastic leukemia, Burkitt’s lymphoma, colorectal adenocarcinoma, lung carcinoma and melanoma tumor lines [29]; nevertheless, the KCTD10 involvement in these tumors has yet to be investigated in detail.
Analysis of the COSMIC and GENT db confirm observations in melanoma, but also suggests KCTD10 overexpression in adrenal gland tumors, in a 6.3% of cancer tissues analyzed. Similarly, GENT2 db indicates a gene expression ratio of KCTD10 in adrenal tumor vs normal tissues [FC (fold change) = 1.41 (p value < 0.001)], suggesting the involvement of KCTD10 in this tumor type.
Ren and colleagues, through the generation of a KCTD10KO mouse model, have demonstrated that KCTD10 could be involved in embryonic angiogenesis and heart development by negatively regulating the Notch signaling pathway, via Notch1 proteolytic degradation [30]. The role of KCTD10 on Notch signaling may underlie its putative involvement not only in cardiac diseases but also in tumor types induced by Notch deregulation, among whom are hematopoietic tumors [31, 32].
KCTD10, complexing with Cul3, also mediates the degradation of CEP97 protein, which is involved in blocking unscheduled ciliogenesis in proliferating cells [33, 34]. This observation suggests that KCTD10 defects may lead to a plethora of developmental defects, ciliary diseases and tumors caused by deregulation of pathways in which the primary cilium plays a role [35], including the potentially oncogenic Hedgehog (Hh) signaling pathway [36].
On the other hand, it has to be noted that KCTD10 expression has been suggested as a favorable prognostic marker in patients affected by gastrointestinal stromal tumor [37], and in hepatocellular carcinoma (HCC) cells. Indeed, Cul3/KCTD10 binds to and leads to degradation of Eukaryotic Translation Initiation Factor 3 subunit D (EIF3D), thus inhibiting cell growth [38].
Taken together, the available information seems to indicate a context-dependent function of KCTD10 in tumorigenesis, probably associated with the wide range of potential targets, and their relative concentration and activity in different cellular and tissue contexts.
TNFAIP1 is differentially regulated in a tissue-specific manner during mouse embryos development: particularly, it presents a high expression in brain, liver and heart tissues [39]. The gene is highly expressed in normal cell lines and is downregulated in cancer cell lines [40]. Indeed, in HCC cell lines, TNFAIP1 suppresses cell proliferation, metastasis, angiogenesis [41]. Several microRNAs have been described to negatively regulate TNFAIP1 in a large number of tumors, confirming its pivotal role of tumor suppressor: miR-372 and miR-373 in gastric carcinoma [42–45], miR-224 and miR-424 in non-small cell lung cancer (NSCLC) [46, 47], miR-181a in pancreatic cancer [48].
TNFAIP1 is frequently downregulated in uterine cancer tissues, and upregulation of TNFAIP1 expression inhibits tumorigenicity and cell growth in human cervical carcinoma (HeLa) and endometrial carcinoma cell line [49].
Analysis of COSMIC db suggest that lower levels of TNFAIP1 are also present in 14% of adrenal gland, data confirmed by analysis of GENT2 db (FC = 0.85; p value < 0.05).
In HeLa cells, TNFAIP1 overexpression and interaction with RhoB induces cell apoptosis via SAPK/JNK-mediated signal pathway [50]. Therefore, the capability to bind RhoB appears to be shared between KCTD10 and TNFAIP1, although the effects of this interaction may be different, suggesting the possibility of either a competition between the two proteins, or a partial redundancy of their functions.
Interestingly, KCTD10 and TNFAIP1 are also able to heterodimerize and TNFAIP1 has been shown to promote the proteasomal degradation of KCTD10 and inhibition of the transcriptional activities of NF-κB and AP-1 [51].
On the other hand, knockdown of TNFAIP1 in osteosarcoma (OS) cells repressed cell proliferation and invasion, and induced cell apoptosis, together with the downregulation of NF-κB signaling [43]. In the OS context, TNFAIP1 expression was significantly increased compared with adjacent non-cancerous tissues and positively correlated with metastasis [43]; targeting the 3’-UTR of TNFAIP1 mRNA by miR-15 has been suggested as a novel therapeutic strategy in this context [52].
Moreover, it was found that TNFAIP1 overexpression was related to an unfavorable prognosis in breast cancer [53] and TNFAIP1 has been suggested as a valid therapeutic target for the treatment of cancer with paclitaxel resistance, since it confers acquired resistance to this potent antitumor agent in various human malignancies [42].
KCTD13 is the least studied member of the clade C and its role remains largely elusive. Although it shares common features with its paralogs, it has been mainly described in neuropsychiatric and autism spectrum disorders, rather than pathological and cancerous status [54, 55].
KCTD13 has been recently demonstrated to target adenylosuccinate synthetase (ADSS) for ubiquitination and degradation [56]. ADSS is an enzyme that catalyzes the first step in adenosine monophosphate (AMP) synthesis. Of note, ADSS deletion has been previously highlighted in lung adenocarcinoma [57]. Based on these observations it can be hypothesized that ADSS reduction following overexpression of KCTD13 may favor cancerogenesis. Indeed, data extrapolated by us from COSMIC database suggest a positive role for KCTD13 in breast tumor, since its overexpression is found in 14% of samples analyzed and GENT2 db analysis confirmed this data (FC = 1.85; p value < 0.001).
Group B: suppressors of Hh-dependent tumorigenesis
Group B is composed of KCTD11, KCTD21 and KCTD6 proteins. These proteins seem to have a more straightforward role in tumor suppression, since they share structural and functional features in control of the developmental and tumorigenic processes driven by Hh signalling. For this reason, the group B of KCTD proteins has recently been renamed as KCASH (KCTD Containing-Cul3 Adaptors, Suppressors of Hedgehog) family of proteins: in particular, KCTD11 (also known as REN) has become KCASH1, KCTD21 is now KCASH2 and KCTD6 as KCASH3 [12]. Indeed, these proteins may act in concert to negatively regulate the Hh signalling: binding to Cul3, they promote the ubiquitination and degradation of the Histone Deacetylase 1 (HDAC1), leading to hyperacetylation of Gli1 (the main transcription factor of the Hh signalling), and thus blocking its transcriptional activity [12, 13].
KCASH1KCTD11, the first member of KCASH family, maps on the chromosome 17p13.2 and its expression promotes growth arrest, differentiation, apoptosis and antagonizes Hh signalling in cerebellar granule cell progenitors [58, 59].
Interestingly, among the genetic alterations observed in Hh-dependent medulloblastoma (MB, cerebellar tumor), the most frequent one is represented by allelic deletion on chromosome 17p, occurring in up to 50% of tumors and frequently restricted to 17p13.2–13.3.19 region, where KCASH1KCTD11 maps [60]. Furthermore, KCASH1KCTD11 has been shown to be frequently down-regulated also by epigenetic modifications both in MB and prostate carcinoma [59, 61, 62].
A significant reduction of KCASH1KCTD11 expression has also been observed in other different types of human cancers: larynx, esophagus, stomach, colon-rectum, urinary bladder, lung, breast, gallbladder and endometrium [61]. Finally, in HCC tumors KCASH1KCTD11 inhibits cell proliferation and migration in vitro and tumor growth and metastasis in vivo [63]: indeed, it induces the G1/S cell cycle arrest through the p21 activation and repression of cycle-related proteins expression and inhibits tumor metastasis, by repressing EMT (epithelium-mesenchymal transition).
Finally, analysis of the COSMIC db shows a previously undescribed reduction of KCASH1KCTD11 expression in 59.8% of ovary cancer and GENT2 db confirmed this observation (FC = 0.82; p value < 0.05). Since the Hh pathway plays a role in the development and differentiation of the internal organs and its activation has been found in ovarian tumorigenesis [64], it would not be surprising if KCASH1KCTD11 had a role, direct or not, in ovarian maturation and malignancy. Hitherto, evidence of KCASH1KCTD11’s involvement in ovarian differentiation comes from studies in rat where the authors have observed that KCASH1 induction may be important for theca and granulosa cell differentiation into luteal cells [65].
KCASH2KCTD21 and KCASH3KCTD6 analogously to KCASH1KCTD11, present high expression in cerebellum. The expression levels of KCASH2KCTD21 and KCASH3KCTD6 genes are significantly downregulated in the group of MB which are characterized by increased Hh signalling (Group SHH; [12, 15, 66]).
In addition to still unidentified epigenetic silencing events, KCASH2KCTD21 allelic deletion was also observed to contribute to the reduced expression of this gene [12]. Of note, chromosome 11q, where KCASH2KCTD21 gene is localized, can be lost in several sporadic tumors, including MB [67, 68], neuroblastoma [69], leukemia [70] and prostate cancer [71].
The role of KCASH3KCTD6 in Hh tumorigenesis seems to be less critical. Unlike its paralogs, KCASH3KCTD6 needs to heterodimerize with KCASH1 to downregulate the Hh signalling, since it is not able to directly bind HDAC1 [12].
KCASH3KCTD6 appears also to play other functions, being able to bind and lead to mono-ubiquitination the centrosome-associated deubiquitylase USP21. It has been demonstrated that USP21 recruits and stabilizes phosphorylated-Gli1 at the centrosome, where it could pilot the entry into or the exit of Gli1 from the primary cilium [72]. However, the mechanism underlying the role of KCASH3KCTD6 -USP21 complex on the Hh signalling regulation needs to be further explored.
Although it is likely that the KCASH family members present not only redundant functions, but also specific functions not yet identified, KCASH1KCTD11 and KCASH2KCTD21 appear as good candidates for novel biomarkers, and the design of methods to reactivate or increase their expression in tumor cells may be useful for treating different type of cancers.
To this end, the recent discovery of KCTD15 as a partner of KCASH2KCTD21, which stabilizes the protein and increases its activity, seems to be a promising result ([15] and see below). Furthermore a recent work has demonstrated the potential transcriptional modulation of KCASH2 by Sp1 and p53 [73].
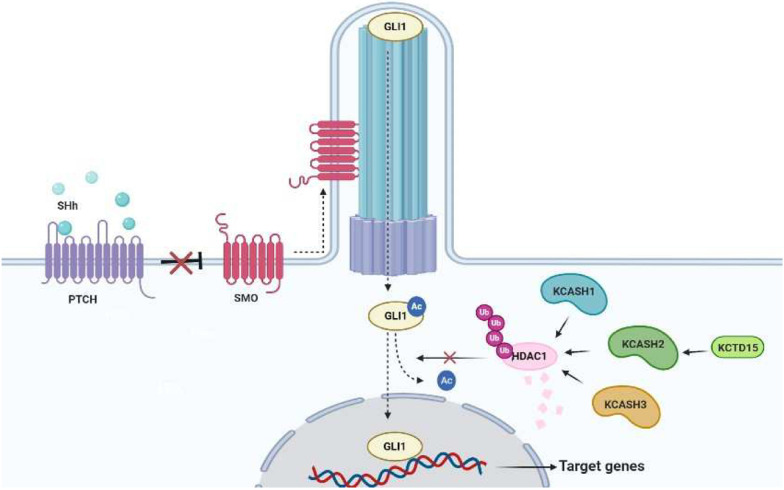
The role of the group B family members in the control of the Hh pathway is depicted in Fig. 1.
Fig. 1.
Group B KCTDs (KCASH family) suppress Hh/Gli1-dependent tumorigenesis. An example of cooperation between KCTD members. The Hh pathway is activated in the presence of SHh ligand, which binding to PTCH abolishes PTCH-mediated SMO inhibition allowing SMO to translocate into the primary cilium. Subsequently, ciliary trafficking of Hh pathway components leads to activation and transport of the transcription factor Gli1 towards the nucleus, where it transcribes target genes involved in proliferation and tumorigenesis. Along the way, Gli1 is kept transcriptionally active by HDAC1 deacetylation. The three members of the KCASH family act as negative regulators, promoting the ubiquitination and proteasomal degradation of HDAC1, thereby inhibiting transcriptional activity of Gli1. In this context, another KCTD protein, KCTD15, positively regulates KCASH2KCTD21 protein levels, enhancing its inhibitory activity. SHh: Sonic Hedgehog. PTCH: Patched
Group A
Group A is composed by KCTD15 and KCTD1.
The KCTD15 gene maps to chromosome 19q13.11 and is expressed in several tissues, including spleen, kidney, adult brain, and cerebellum [74, 75].
KCTD15 protein has been attributed to non-protein degradation functions. In fact, it was observed that KCTD15 is not able to interact with Cul3 and does not appear to be able to directly ubiquitinate and degrade its interactors [14].
KCTD15 has been previously suggested, by genome wide association studies (GWAS), to play a role in obesity and eating disorders [76, 77], although other groups did not confirm this association [78, 79]. On the other side, interaction between GRP78, a fundamental player in adipogenesis, and KCTD15 has been described [80].
Recently, a role for KCTD15 in cancer has been unveiled, based on its interaction with and stabilization of KCASH2KCTD21 protein [15]. In this way, KCTD15 enhances KCASH2KCTD21 inhibitory effect in MB cells, reducing tumor cell proliferation. Coherently, KCTD15 expression is reduced in a percentage of human sporadic MB which present Hh hyperactivation [15].
On the other hand, Smaldone and colleagues have shown that KCTD15 is upregulated in B-cell type acute lymphoblastic leukemia (B-ALL) patients [80] and acute myeloid leukemia (AML) patients [80, 81].
KCTD1 is highly expressed in mammary glands, kidney, brain, and ovary [82].
KCTD1 was first identified as a transcriptional repressor, which repressed AP-2alpha-mediated transactivation through the BTB domain [82, 83]) and had the capability of binding to AP2α and removing it from the nucleus [84]. Subsequently KCTD1 was identified as the cause of Scalp-ear-nipple (SEN) syndrome, a rare autosomal-dominant disorder [17].
Although KCTD1 shares several features and biological functions with KCTD15, including the inability to bind to Cul3 [9], its role in cancer has still not been intensively investigated. However, unlike KCTD15, it has been shown that KCTD1 is involved in proteasomal degradation processes, by complexing with other E3-ubiquitine ligases, through which it participates in negative regulation of pathways which are aberrantly activated in many human cancers [85]. Indeed, KCTD1 has been proposed as an inhibitor of the WNT/β-catenin developmental pathway [85]: KCTD1 binds, through BTB domain, β-catenin, triggering its cytoplasmic accumulation. Here, KCTD1 contributes to an overall decrease of β-catenin protein levels, via β-TrCP-mediated proteasome pathway, dependent on the phosphorylation of the Ser45 and Ser33/Thr41 sites mediated by CK1 and GSK-3β kinases. Interestingly, ectopic expression of KCTD1 in cervix tumor HeLa cells inhibits the expression of β-Catenin downstream target genes and while APC counteracts the KCTD1-mediated downregulation of β-catenin, p53 enhances KCTD1 inhibitory role [85].
Therefore, it is likely that KCTD1 may play a oncosuppressive role by promoting the degradation of β-catenin, a core player in canonical WNT/β-catenin signaling pathway [85].
Intriguingly, evidence from COSMIC and GENT2 db, suggest a potential pro-tumorigenic role of KCTD1, which is overexpressed in subsets of ovary tumors (13.9% of tumor analyzed, FC = 1.47, p value < 0.001), pancreatic cancer (11.17% of samples; FC = 1.32, p value < 0.001), endometrium tumor (9.8%; FC = 1.43, p value < 0.001), lung tumor (7.46%; FC = 1.21, p value < 0.001).
The high homology of sequence between KCTD1 and KCTD15, both at level of the BTB domain and of the full sequence, would have suggested similar mechanisms of action. Surprisingly while KCTD1 enhances β-catenin degradation by the β-TrCP-mediated proteasome pathway [85], KCTD15 is involved in KCASH2KCTD21 protein stabilization. Of course, even though KCTD15 cannot bind Cul3, [14, 15] we cannot exclude the possibility that also KCTD15 may bind other E3 ligases in different contexts. On the other hand, it cannot be ruled out that also KCTD1 may stabilize specific proteins modulating tumorigenesis.
Group D
Group D comprises SHKBP1 (also known as SETA binding protein 1, SB1) and KCTD3 proteins.
SHKBP1 biological function has not been completely characterized, although several hints suggest a significant role in tumorigenesis.
SHKBP1 presents a 55% aa identity to the renal tumor antigen NY-REN-45 and was initially identified as a protein that binds to SH3 domains of the SETA adapter protein, whose hyperexpression has been associated with adult brain tumors [84]. In this context, it is interesting that our analysis of COSMIC db shows that 5.4% of nervous system tumors presents SHKBP1 mutations.
SHKBP1 expression levels are also increased in osteosarcoma (OS) samples compared to their normal counterparts [86]. In this context, SHKBP1 may promote EGFR signaling pathway by interrupting c-Cbl-CIN85 complex and inhibiting EGFR degradation [87].
Furthermore, serum protein profiling identified SHKBP1 as a candidate in earlier detection of small intestine neuroendocrine tumor (WD-SI-NETs) tumor [88].
Moreover, whole genome sequencing identified a missense mutation of SHKBP1 in acute myeloid leukemia (AML), which characterizes SHKBP1 as a putative proto-oncogene. In particular, the V891I mutation is suggested to interfere with binding to SETA, disturbing the degradation of the FLT3 tyrosine kinase [89]. Recently, through a comprehensive genomic study of cervical cancer, SHKBP1 has been also identified as a mutated gene in the squamous subtype [90].
Finally, analysis of COSMIC db suggests SHKBP1 overexpression in pancreatic tumor (13.4%; FC = 2.05, p value < 0.001 extrapolated by GENT2 db) and cervical cancer (10.8%; FC = 2.14, p value < 0.05 by GENT2 analysis). It is also interesting to note that 4.5% of large intestine cancer present SHKBP1 mutations.
While KCTD3 has been associated with neurogenetic and neurodevelopmental disorders [1, 91], no correlation with cancer has been reported so far.
Nevertheless, KCTD3 has been demonstrated to interact and increase stability and cell surface expression of hyperpolarization-activated cyclic nucleotide-gated channel 3 (HCN3) [92]. Intriguingly, HCN3 has been found overexpressed in neuroblastoma tumors and has been indicated to protect cells from apoptosis driven by HIF-1a and p53 [93]. Furthermore, analysis of COSMIC db indicate that 4% of nervous system tumor samples present KCTD3 mutations, suggesting the need to verify KCTD3 role in this context.
Similarly to its paralog SHKBP1, analyses performed on COSMIC database point out KCTD3 overexpression in other tumor types, such as breast cancer (14.2%; FC = 1.3, p value < 0.001 in the GENT2 db) and stomach (13%; FC = 1.83, p value < 0.001 by GENT2 analysis).
Group F: not only neurodegenerative and neuropsychiatric disorders
KCTD12, KCTD8 and KCTD16 proteins belong to the F clade of the KCTD family and are known as auxiliary subunits of the GABAB1/2 receptor (G-protein coupled receptors for GABA, the main inhibitory neurotransmitter in the central nervous system [94]).
Given the role they play in regulating GABA receptor signalling, mutations in KCTD12, KCTD8 and KCTD16 genes have been implicated in neurodegenerative and neuropsychiatric disorders [1].
Among these proteins, KCTD12 has also been linked to tumorigenesis, in some contexts as a potential oncosuppressor, in other contexts as oncogene. KCTD12 (also known as Pfetin) was identified as a gene predominantly expressed in fetal tissues [95] and suggested as a biomarker for the diagnosis and prognosis of gastrointestinal stromal tumors (GIST). In particular, a decreased pfetin expression was observed in a subset of GISTs with poor clinical outcomes and pfetin expression significantly affected the disease-free and overall survival of the patients [96, 97].
In colorectal cancer (CRC) cells down-regulation of KCTD12 leads to an increase of staminality markers [98]. The KCTD12 inhibitory role on stemness has also been confirmed in vivo through xenotransplantation experiments [98].
KCTD12 displays a reduced expression in breast cancer tissues and cells, correlated with patients’ overall poorer survival. Indeed, downregulation of KCTD12 significantly promotes cancer cell proliferation and G1/S transition through the AKT/FOXO1 signaling [99]. Consistently, ectopic expression of KCTD12 in human uveal melanoma cells causes a retention in the G2/M phase, an inhibition of proliferation and an increase of apoptosis, which has been confirmed in vivo through xenograft experiments [100]. Moreover, KCTD12 acts as a tumor suppressor also in esophageal squamous cell carcinoma (ESCC), downregulating WNT and NOTCH signaling [101].
In other contexts, KCTD12 may promote cell proliferation: increased expression of KCTD12 has been observed in cervical, colon and lung cancers and its high levels have been correlated with poor prognosis [102, 103]. In fact, KCTD12 interacts with CDK1 and CDC25B, forming a complex that supports CDK1 phosphorylation and cell cycle progression [103], while pharmacological inhibition of KCTD12-CDK1 interaction suppresses growth of colon cancer cells in vitro and in vivo [102].
KCTD16 is a transcriptional target of ASCL1, one of the master transcription factors of small cell lung carcinoma, (SCLC) [104]. Comparative RNA-seq studies and immunohistochemical staining revealed a strong KCTD16 expression in H69 cells (ASCL1-positive, classical type SCLC) and in ASCL1-transfected A549 adenocarcinoma cell lines, while in H69AR (ASCL1-negative, variant type SCLC) and A549 (control) cell lines the level of KCTD16 protein does not result upregulated [104]. The definition of the biological significance of KCTD16 upregulation in neoplastic development requires further investigation.
KCTD16 expression has been suggested as a negative prognostic marker in thyroid cancers [105]. On the other side, analysis of the COSMIC db indicates that KCTD16 presents mutations in hepatic tumors (13.6% of samples), pancreas (11.7%) and prostate (8.9%), suggesting the need to evaluate if these mutations are inactivating or hyperactivating the protein.
Almost unexplored is the potential role of KCTD8 in cancer. Hypermethylation and transcriptional repression of KCTD8 was observed in breast cancer samples [106]. On the other hand, COSMIC db analysis suggests high KCTD8 expression in thyroid cancer (8% of samples analyzed), pancreas (7.3%), esophagus (7.2%) and cervix (5.5%).
Insides into the role of E, G, H groups and other KCTD proteins in cancer
KCTD2, -5, -9, -17 proteins belong to group E. For all these proteins, there is still little information and their role in physiological and pathological contexts and even less is known about their involvement in the onset or progression of cancer.
The most studied protein of this clade is undoubtedly KCTD5 the only member of the KCTD family whose three-dimensional structure has been resolved. Indeed, KCTD5 presents a pentameric association of both the BTB and the C-terminal domains and a central spanning cavity [107]. KCTD5 and KCASH3KCTD6 share a high sequence identity (> 40%) at BTB domain [108]. Like KCASH3KCTD6, KCTD5 is able to bind Cul3 [109].
KCTD5 has been suggested to act as a negative regulator of cell migration by modulating cell spreading and focal adhesion dynamics. Indeed, KCTD5 is a negative regulator for the migration of melanoma and breast cancer cells, by affecting Rac1 activity and Ca2+ signaling [110]. Given these data, KCTD5 could play a role in inhibition of processes associated with cancer metastasis.
On the other side, KCTD5 positively regulates the Transient Receptor Potential Melastatin 4 (TRPM4) [111]. Since TRPM4 activity has been proposed to contribute to the pathophysiology of different cancers, and TRPM4 and KCTD5 expression are increased in breast cancer (in particular in the most aggressive subtype, the triple negative one), study of the TRPM4-KCTD5 protein interaction could be useful to develop drugs that modulate their activity with therapeutics purposes [111].
Confirming these data, COSMIC and GENT2 db indicate KCTD5 overexpression in 12.5% of breast cancer samples (FC = 1.33, p-value < 0.001). It would be interesting to further evaluate if KCTD5 is restricted to non metastatic breast tumors, as it could be suggested by its role on TRPM4.
Further involvement of KCTD5 in other tumor types has not yet been described, but Brockmann and colleagues demonstrated that KCTD5 may switch off the Akt pathway, which regulates cell survival, cell cycle progression and cellular growth [112]. Furthermore, since the AKT pathway interplays with Hh signaling through the non-canonical pathway, it may be possible that KCTD5 alterations affect the Hh signaling-dependent tumorigenesis [113, 114].
Evidence of KCTD2 involvement in cancer derive from studies conducted on glioblastoma tumor cells and CRC cancer patients. Indeed, KCTD2 promotes ubiquitination and degradation of the oncogene c-Myc, and reduced KCTD2 mRNA levels are present in glioma cells, promoting tumor growth in vivo [8]. On the other hand, microarray analysis suggests that KCTD2 may be upregulated in colorectal cancer [115].
KCTD17 protein controls ciliogenesis through polyubiquitylation and subsequent degradation of trichoplein, a keratin-binding protein that controls the recruitment of microtubules to centrioles, therefore participating in the initial step of axonemal extension during ciliogenesis [116].
Knockdown of KCTD17 stabilizes trichoplein which in turn binds Aurora A and elevates its kinase activity. In turn Aurora A destabilizes axonemal microtubules, resulting in the deciliation at the cell cycle reentry or the inhibition of unscheduled ciliary assembly, affecting ciliary dynamics in development and tissue homeostasis. [117]. KCTD17 is therefore likely to play a role in tumorigenesis, given the role of cilia in transmission of environmental and intracellular signals, both repressive and promoter of proliferation and control of cell cycle [118].
Indeed, recently has been demonstrated that depletion of Trichoplein expression results in chromosome mis-segregation, DNA damage and chromosomal instability in cancer cells [119].
KCTD9 is the only human protein containing five pentapeptide repeats (presumably a DNA mimicking structure) [6], which is described to play a role in NK cell lineage commitment and maturation, mediating by direct or indirect interaction-dependent degradation of transcription factors or chromatin regulators a marked increase in CD69 expression, cytotoxicity, IFN‐γ secretion and a significant decrease in NKG2A receptor expression (120).
No information has been published yet on KCTD9 and cancer, although we would expect a role for KCTD9 in tumorigenesis, given that NK cells are a key immune constituent in the protective antitumor immune response [121]. Of note, COSMIC and GENT2 db analysis indicate KCTD9 overexpression in 14.7% of gastric tumor samples and FC = 1.25 (p value < 0.05) respectively.
KCTD20 and BTBD10 belong to group G. The amino acid sequences of these two proteins present a high homology, especially in correspondence of the C-terminal region, which suggests that BTBD10 and KCTD20 share functional characteristics and can play similar biological role. Indeed, both proteins are involved in the activation of Akt signaling [122, 123] and it is therefore possible that aberrant expression of these genes can be implicated in tumorigenic processes.
A recent study correlates high KCTD20 expression with advanced non-small cell lung cancer (NSCLC), characterized by positive regional lymph node metastasis and poor prognosis [113]. Indeed, it has been observed that KCTD20, through activation of the Akt signaling, increases cell proliferation and facilitate the invasion of NSCLC cells by inhibiting E-cadherin expression (113).
Similarly, BTBD10, also known as glucose metabolism-related protein 1 (GMRP1), inhibits apoptosis of neuronal and islet β-cells via Akt pathway. On the other hand, another group indicates that BTBD10 is downregulated in glioma tumor, suggesting that it might play a negative role in the proliferation and progression of glioma cells [120, 123].
KCTD7 and KCTD14 have been clustered in group H, according to their sequence homology, but have not been characterized yet.
KCTD7 alternative splicing has been suggested to have prognostic value in glioblastoma [124] and lung adenocarcinoma [125]. Analysis of GENT2 and COSMIC Db indicate that KCTD7 is upregulated in tumors affecting the following tissues: adrenal gland (13.9%; FC = 1.61, p-value < 0.001 by GENT2 db), skin (8.46%, coherent with GENT2 FC = 2.28, p value < 0.001), breast (6.16%; FC = 1.16, p value < 0.001 by GENT2 analysis). Moreover, 5.5% of the pancreatic tumors analyzed (COSMIC) present a KCTD7 mutation.
KCTD14 has not so far been investigated, and involvement in the onset and progression of cancer has not been documented, DB analysis suggests a potential protumor role in ovary cancers: COSMIC reports a CNV gain in 4.5% of ovary cancers and GENT2 a FC = 1.5 (p value < 0.001) in expression.
The KCTD proteins that are not classified in any group, since they do not share homologies with other KCTD members, are in general not characterized.
Among them, KCNRG is the only one whose role in cancer is known, albeit partially.
The human KCNRG is located on 13q14.3 and was described as a K+ channel regulator [126].
KCNRG overexpression in leukemia cell lines exerts growth suppression, promotes apoptosis, leads to changes in size and shape, and causes the decrease of cell migration capacity [126]. Interestingly, deletion of 13q14 is the most frequent chromosomal anomaly in chronic lymphocytic leukemia (CLL). Rearrangements and/or deletions in the 13q14.3 region were found also in other types of hematopoietic neoplasms, including 38% of mantle cell lymphomas and approximately 54% of multiple myelomas (MM). In most of these non-CLL cases, 13q14 deletions were associated with a poor prognosis [127]. These data suggest that KCNRG can act as tumor suppressor and may play a relevant role in the pathogenesis of CLL and MM tumors.
13q14.3 deletions are also common in prostate cancer and gastrointestinal stromal tumors. Sequencing analysis of 77 Hepatocellular carcinomas (HCCs) showed that KCNRG can be mutated in HCC tumors. This missense mutation is caused by Arg to His substitution at codon 92 in exon 1 encoding the KCNRG BTB domain. Similarly to leukemia cells, KCNRG overexpression suppresses the growth activity of Hep3B hepatoma cells while its inactivation, through mutations or allelic losses, contributes to the development or progression of HCC [128].
KCTD19, so far observed mutated only in 44% of patients affected by Cerebral Visual Impairment [129], is found upregulated in hematopoietic and lymphoid district tumors (12.7%; FC = 1.14, p-value < 0.001). Similarly, KCTD19 is mutated in cancer tissues from nervous system (4.7%), large intestine (3.2%) and skin (4.2%).
KCTD4 has not been characterized. Db analysis indicate is overexpressed in 5% of sample of analyzed tumor associated to lung cancer (GENT2 db: FC = 1.32, p-value < 0.001).
Finally, KCTD18 has been only potentially associated to Restless Legs Syndrome [130] and no information are available that link KCTD18 to tumor development. Since the protein structure has not been defined, no inference can be done on its function. Also, analysis of the db did not provide univocal indications or correlations with tumor types.
Conclusions
The family of KCTD proteins is formed by 25 members, with most of them still poorly characterized.
Over time, some of the KCTD have been associated with neurodevelopmental and neuropsychiatric disorders, obesity, or modulation of signal transduction pathways, while information of their role in cancer is scattered within the literature.
Furthermore, analysis of the KCTD family is complex, since homology between these proteins is often limited to the conserved BTB domain, and these proteins, even when members of the same groups, may be involved in rather different pathological processes. This can in part due to the roles played by some of these proteins in ubiquitination and degradation of a wide plethora of different targets. Furthermore, some of the characterized (and most of the uncharacterized) members of the family are likely involved in other basic processes that are yet to be identified, including fundamental biological processes that can play a role also in tumorigenesis.
Cancer is considered the leading cause of death worldwide. Genetic and histological heterogeneity of cancers, the ability of cancer cells to evade the immune response and to remodel pleiotropic systems contribute to the difficulty of understanding how different cancer develops, grows, metastasizes and identifying valid therapeutic strategies that do not lead to resistance and relapse. In the era of precision medicine, the knowledge of all perturbations of molecular pathways and identification of all the genes playing a role in tumorigenesis has become more necessary than ever, both for therapeutic purposes and for diagnosis and prognosis.
Few of the KCTD groups have been identified as key players in the modulation of specific tumorigenic pathway (e.g., the KCASH family; see Fig. 1), but the collection of all the evidence available up to now allow us to hypothesize that the number of KCTD family member involved in tumorigenesis (either as positive or negative modulator) may be far bigger than so far demonstrated. Figure 2 summarizes the available data on the role of KCTDs in modulation of other potentially tumorigenetic pathways. Homology between family members, capability to participate in ubiquitination and degradation of different targets and to take part to the signal transduction of developmental and tumorigenic pathways, ability to heterodimerize between members, bioinformatic analysis of available databases in the search for positive or negative correlations with cancers, are all factors that need to be considered in the search for new functions in this family. In this review we referred to COSMIC and GENT2 db, two user-friendly search platform for gene expression patterns across different normal and tumor tissues. The gene expression profiles generated by a large number of samples (68,000 for GENT2 [20]; and genome-wide analysis of over 37,000 genomes for COSMIC2) may provide an extensive coverage of the cancer genomic landscape.
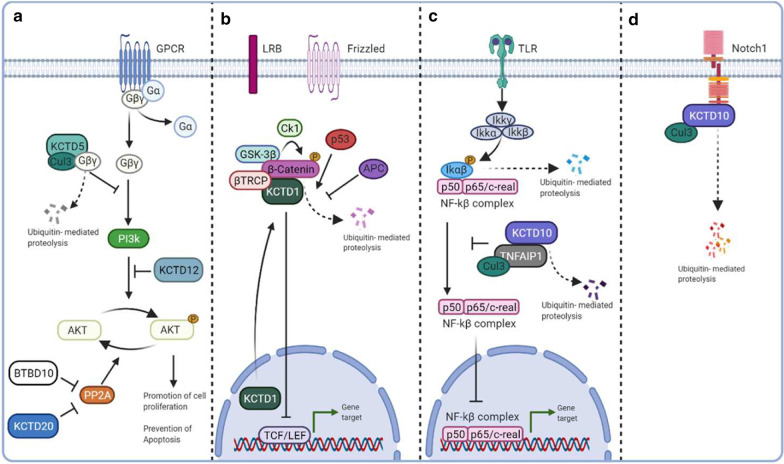
Fig. 2.
Schematic representation of the role of KCTDs in modulation of potentially tumorigenetic pathways. a: PI3K/AKT pathway. The activation of the GPCR receptor leads to the dissociation of the heterotrimer G into Gα and Gβγ. Subsequently, Gβγ activates PI3K that phosphorylates AKT. KCTD5 and KCTD12 negatively regulate the AKT pathway by triggering proteolysis of Gβγ heterodimer and inhibiting the phosphorylation of AKT, respectively. KCTD20 and BTBD10 suppress the PP2A-mediated dephosphorylation of AKT, acting as positive regulators of the pathway. b: WNT pathway. In the absence of Wnt ligand, β-catenin is degraded through the “destruction complex” (composed of CK1, axin, GSK-3β and APC), which phosphorylates β-catenin for ubiquitin-mediated proteasomal degradation. KCTD1 negatively regulates Wnt signalling by Enhancing β-catenin degradation. c: NF-κB pathway. Binding of inflammatory cytokines to TLR leads to the activation of the IKK complex, which phosphorylates IκBα. IκBα phosphorylation induces its ubiquitination and degradation by the proteasome, leading to nuclear translocation of the NF-κB transcriptional complex. TNFAIP1 interacts with KCTD10 inducing its degradation and inhibits the transcriptional activity of NF-κB. d: Notch pathway. KCT10 interacts with Cullin3 and the intracellular domain of Notch1, mediating Notch1 ubiquitination and proteolytic degradation
In order to provide new hints to the scientific community we have summarized in this review most of the published material integrated with data extrapolated from COSMIC and GENT2 databases (summarized in Fig. 3), whose knowledge could be a good starting point for further necessary research.
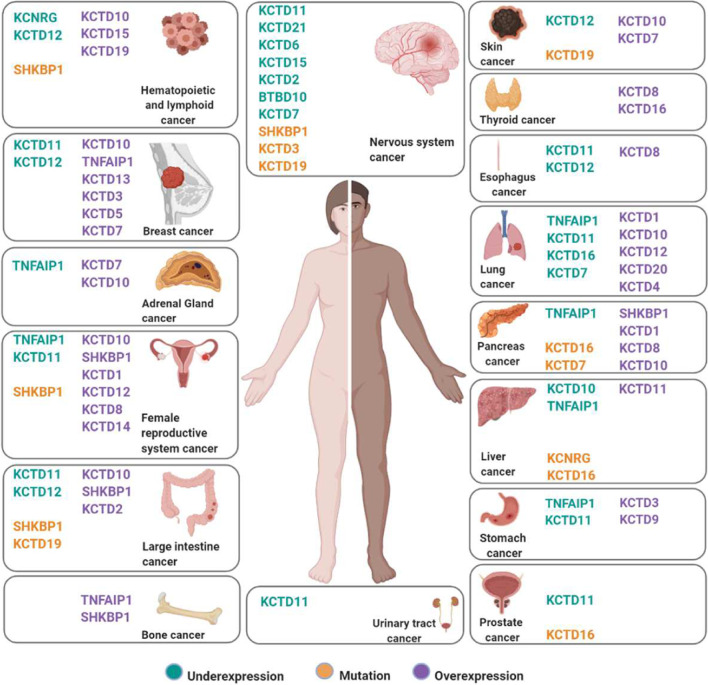
Fig. 3.
The KCTD family in human cancers. Synthesis of the current knowledge on the role of KCTD proteins in different cancer types. KCTDs whose expression is downregulated in cancer are in green; mutated KCTDs are in orange; KCTDs which are overexpressed in violet. All information is the result of data extrapolated by literature and analysis of COSMIC-GENT2 db, details are in the text
Acknowledgements
Figures were Created with BioRender.com program.
Abbreviations
- AML
Acute Myeloid Leukemia
- BTB/POZ
Bric-a-brac, Tramtrak, Broad complex/ Poxvirus zinc finger
- CLL
Chronic lymphocytic Leukemia
- CNV
Copy Number Variant
- COSMIC
Catalogue Of Somatic Mutations In Cancer
- CRC
Colon-Rectal Cancer
- CRL
Cullin Ring Ligase
- Cul3
Cullin 3
- Db
Database
- EIF3D
Eukaryotic Translation Initiation Factor 3 subunit 3D
- EMT
Epithelial–Mesenchymal Transition
- ESCC
Esophageal Squamous Cell Carcinoma
- FC
Fold Change
- GENT2
Gene Expression patterns across Normal and Tumor tissues
- GIST
Gastro Intestinal Stromal Tumors
- GMRP1
Glucose Metabolism-Related Protein 1
- GWAS
Genome Wide Associations Studies
- HCC
HepatoCellular Carcinoma
- Hh
Hedgehog
- KCASH
KCTD containing Cul3 Adaptors Suppressor of Hedgehog
- KCTD
Potassium (K+) Channel Tetramerization Domain
- MB
Medulloblastoma
- MM
Multiple Myelomas
- NSCLC
No Small Cell Lung Cancer
- OS
Osteosarcoma
- PCNA
Proliferating Cell Nuclear Antigen
- PDIP1
Polymerase Delta-Interacting Protein 1
- SCLC
Small Cell Lung Carcinoma
- SNV
Single Nucleotide Variant
- TNFAIP1
Tumor Necrosis Factor Induced Protein 1
- TRPM4
Transient Receptor Potential Melastatin 4
- Ub
Ubiquitination
Authors' contributions
A.A, A.D.F, E.D.S. and M.M. conceived this study. A.A, A.D.F., M.M. and E.D.S contributed to writing and revising the manuscript. A.A. and A.D.F collected the data and generated the pictures. All authors read and approved the final version of the manuscript.
Funding
This work was supported by grants from Italian Ministry of University and Research, PRIN projects (to E.D.S.); La Sapienza Research Grant years 2015 and 2017 (to E.D.S); La Sapienza Research Grant 2019 (to A.A.).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the following repository: COSMIC (Catalogue Of Somatic Mutations In Cancer; https://cancer.sanger.ac.uk/cosmic and GENT2 (Gene Expression patterns across Normal and Tumor tissues; http://gent2.appex.kr/gent2/.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Annapaola Angrisani and Annamaria Di Fiore have contributed equally (co-first authors)
Enrico De Smaele and Marta Moretti have contributed equally (co-last authors)
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Teng X, Aouacheria A, Lionnard L, Metz KA, Soane L, Kamiya A, et al. KCTD: A new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci Ther. 2019;25(7):887–902. doi: 10.1111/cns.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Xiang Y, Sun G. The KCTD family of proteins: structure, function, disease relevance. Cell Biosci. 2013;3(1):45. doi: 10.1186/2045-3701-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinkas DM, Sanvitale CE, Bufton JC, Sorrell FJ, Solcan N, Chalk R, et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem J. 2017;474(22):3747–3761. doi: 10.1042/BCJ20170527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. BioEssays. 2006;28(12):1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 6.Skoblov M, Marakhonov A, Marakasova E, Guskova A, Chandhoke V, Birerdinc A, et al. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. BioEssays. 2013;35(7):586–596. doi: 10.1002/bies.201300002. [DOI] [PubMed] [Google Scholar]
- 7.Kovačević I, Sakaue T, Majoleé J, Pronk MC, Maekawa M, Geerts D, et al. The Cullin-3-Rbx1-KCTD10 complex controls endothelial barrier function via K63 ubiquitination of RhoB. J Cell Biol. 2018;217(3):1015–1032. doi: 10.1083/jcb.201606055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim EJ, Kim SH, Jin X, Kim H. KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ. 2017;24(4):649–659. doi: 10.1038/cdd.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji AX, Chu A, Nielsen TK, Benlekbir S, Rubinstein JL, Privé GG. Structural Insights into KCTD Protein Assembly and Cullin3 Recognition. J Mol Biol. 2016;428(1):92–107. doi: 10.1016/j.jmb.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Rutz N, Heilbronn R, Weger S. Interactions of cullin3/KCTD5 complexes with both cytoplasmic and nuclear proteins: Evidence for a role in protein stabilization. Biochem Biophys Res Commun. 2015;464(3):922–928. doi: 10.1016/j.bbrc.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 11.Azizieh R, Orduz D, Van Bogaert P, Bouschet T, Rodriguez W, Schiffmann SN, et al. Progressive myoclonic epilepsy-associated gene KCTD7 is a regulator of potassium conductance in neurons. Mol Neurobiol. 2011;44(1):111–121. doi: 10.1007/s12035-011-8194-0. [DOI] [PubMed] [Google Scholar]
- 12.De Smaele E, Di Marcotullio L, Moretti M, Pelloni M, Occhione MA, Infante P, et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia. 2011;13(4):374–385. doi: 10.1593/neo.101630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 14.Smaldone G, Pirone L, Balasco N, Di Gaetano S, Pedone EM, Vitagliano L. Cullin 3 Recognition Is Not a Universal Property among KCTD Proteins. PLoS One. 2015;10(5):e0126808. [DOI] [PMC free article] [PubMed]
- 15.Spiombi E, Angrisani A, Fonte S, De Feudis G, Fabretti F, Cucchi D, et al. KCTD15 inhibits the Hedgehog pathway in Medulloblastoma cells by increasing protein levels of the oncosuppressor KCASH2. Oncogenesis. 2019;8(11):64. doi: 10.1038/s41389-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng S, Abreu N, Levitz J, Kruse AC. Structural basis for KCTD-mediated rapid desensitization of GABA. Nature. 2019;567(7746):127–131. doi: 10.1038/s41586-019-0990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marneros AG, Beck AE, Turner EH, McMillin MJ, Edwards MJ, Field M, et al. Mutations in KCTD1 cause scalp-ear-nipple syndrome. Am J Hum Genet. 2013;92(4):621–626. doi: 10.1016/j.ajhg.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–7. [DOI] [PMC free article] [PubMed]
- 19.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12(Suppl 5):101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He H, Tan CK, Downey KM, So AG. A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2001;98(21):11979–11984. doi: 10.1073/pnas.221452098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Ren K, Liu X, Xiong X, Hu X, Zhang J. A novel PDIP1-related protein, KCTD10, that interacts with proliferating cell nuclear antigen and DNA polymerase delta. Biochim Biophys Acta. 2005;1729(3):200–203. doi: 10.1016/j.bbaexp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Wajant H. The role of TNF in cancer. Results Probl Cell Differ. 2009;49:1–15. doi: 10.1007/400_2008_26. [DOI] [PubMed] [Google Scholar]
- 24.Hu X, Gan S, Xie G, Li L, Chen C, Ding X, et al. KCTD10 is critical for heart and blood vessel development of zebrafish. Acta Biochim Biophys Sin (Shanghai) 2014;46(5):377–386. doi: 10.1093/abbs/gmu017. [DOI] [PubMed] [Google Scholar]
- 25.Murakami A, Maekawa M, Kawai K, Nakayama J, Araki N, Semba K, et al. Cullin-3/KCTD10 E3 complex is essential for Rac1 activation through RhoB degradation in human epidermal growth factor receptor 2-positive breast cancer cells. Cancer Sci. 2019;110(2):650–661. doi: 10.1111/cas.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De P, Rozeboom BJ, Aske JC, Dey N. Active RAC1 Promotes Tumorigenic Phenotypes and Therapy Resistance in Solid Tumors. Cancers (Basel). 2020;12(6). [DOI] [PMC free article] [PubMed]
- 27.Engqvist H, Parris TZ, Kovács A, Rönnerman EW, Sundfeldt K, Karlsson P, et al. Validation of Novel Prognostic Biomarkers for Early-Stage Clear-Cell, Endometrioid and Mucinous Ovarian Carcinomas Using Immunohistochemistry. Front Oncol. 2020;10:162. doi: 10.3389/fonc.2020.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu B, Shi C, Jiang HX, Qin SY. Identification of novel therapeutic target genes and pathway in pancreatic cancer by integrative analysis. Medicine (Baltimore). 2017;96(42):e8261. [DOI] [PMC free article] [PubMed]
- 29.Wang Y, Zheng Y, Luo F, Fan X, Chen J, Zhang C, et al. KCTD10 interacts with proliferating cell nuclear antigen and its down-regulation could inhibit cell proliferation. J Cell Biochem. 2009;106(3):409–413. doi: 10.1002/jcb.22026. [DOI] [PubMed] [Google Scholar]
- 30.Ren K, Yuan J, Yang M, Gao X, Ding X, Zhou J, et al. KCTD10 is involved in the cardiovascular system and Notch signaling during early embryonic development. PLoS One. 2014;9(11):e112275. [DOI] [PMC free article] [PubMed]
- 31.Pelullo M, Zema S, Nardozza F, Checquolo S, Screpanti I, Bellavia D. Wnt, Notch, and TGF-β Pathways Impinge on Hedgehog Signaling Complexity: An Open Window on Cancer. Front Genet. 2019;10:711. doi: 10.3389/fgene.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsaouli G, Barbarulo A, Vacca A, Screpanti I, Felli MP. Molecular Mechanisms of Notch Signaling in Lymphoid Cell Lineages Development: NF-κB and Beyond. Adv Exp Med Biol. 2020;1227:145–164. doi: 10.1007/978-3-030-36422-9_10. [DOI] [PubMed] [Google Scholar]
- 33.Nagai T, Mukoyama S, Kagiwada H, Goshima N, Mizuno K. Cullin-3-KCTD10-mediated CEP97 degradation promotes primary cilium formation. J Cell Sci. 2018;131(24). [DOI] [PubMed]
- 34.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130(4):678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Dynlacht BD. The regulation of cilium assembly and disassembly in development and disease. Development. 2018;145(18). [DOI] [PMC free article] [PubMed]
- 36.Liu H, Kiseleva AA, Golemis EA. Ciliary signalling in cancer. Nat Rev Cancer. 2018;18(8):511–524. doi: 10.1038/s41568-018-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota D, Yoshida A, Tsuda H, Suehara Y, Okubo T, Saito T, et al. Gene expression network analysis of ETV1 reveals KCTD10 as a novel prognostic biomarker in gastrointestinal stromal tumor. PLoS One. 2013;8(8):e73896. [DOI] [PMC free article] [PubMed]
- 38.Maekawa M, Hiyoshi H, Nakayama J, Kido K, Sawasaki T, Semba K, et al. Cullin-3/KCTD10 complex is essential for K27-polyubiquitination of EIF3D in human hepatocellular carcinoma HepG2 cells. Biochem Biophys Res Commun. 2019;516(4):1116–1122. doi: 10.1016/j.bbrc.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Wolf FW, Marks RM, Sarma V, Byers MG, Katz RW, Shows TB, et al. Characterization of a novel tumor necrosis factor-alpha-induced endothelial primary response gene. J Biol Chem. 1992;267(2):1317–1326. doi: 10.1016/S0021-9258(18)48432-3. [DOI] [PubMed] [Google Scholar]
- 40.Liu XW, Lu FG, Zhang GS, Wu XP, You Y, Ouyang CH, et al. Proteomics to display tissue repair opposing injury response to LPS-induced liver injury. World J Gastroenterol. 2004;10(18):2701–2705. doi: 10.3748/wjg.v10.i18.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, Huang S, Qiu F, Ding X, Sun Y, Wei C, et al. Tumor necrosis factor α-induced protein 1 as a novel tumor suppressor through selective downregulation of CSNK2B blocks nuclear factor-κB activation in hepatocellular carcinoma. EBioMedicine. 2020;51:102603. [DOI] [PMC free article] [PubMed]
- 42.Zhu Y, Yao Z, Wu Z, Mei Y, Wu M. Role of tumor necrosis factor alpha-induced protein 1 in paclitaxel resistance. Oncogene. 2014;33(25):3246–3255. doi: 10.1038/onc.2013.299. [DOI] [PubMed] [Google Scholar]
- 43.Zhang CL, Wang C, Yan WJ, Gao R, Li YH, Zhou XH. Knockdown of TNFAIP1 inhibits growth and induces apoptosis in osteosarcoma cells through inhibition of the nuclear factor-κB pathway. Oncol Rep. 2014;32(3):1149–1155. doi: 10.3892/or.2014.3291. [DOI] [PubMed] [Google Scholar]
- 44.Zhou C, Li X, Zhang X, Liu X, Tan Z, Yang C, et al. microRNA-372 maintains oncogene characteristics by targeting TNFAIP1 and affects NFκB signaling in human gastric carcinoma cells. Int J Oncol. 2013;42(2):635–642. doi: 10.3892/ijo.2012.1737. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Li X, Tan Z, Liu X, Yang C, Ding X, et al. MicroRNA-373 is upregulated and targets TNFAIP1 in human gastric cancer, contributing to tumorigenesis. Oncol Lett. 2013;6(5):1427–1434. doi: 10.3892/ol.2013.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui R, Kim T, Fassan M, Meng W, Sun HL, Jeon YJ, et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget. 2015;6(26):21802–21815. doi: 10.18632/oncotarget.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y, et al. MiR-424 Promotes Non-Small Cell Lung Cancer Progression and Metastasis through Regulating the Tumor Suppressor Gene TNFAIP1. Cell Physiol Biochem. 2017;42(1):211–221. doi: 10.1159/000477314. [DOI] [PubMed] [Google Scholar]
- 48.Zhang P, Guo Z, Hu R, He X, Jiao X, Zhu X. Interaction between microRNA-181a and TNFAIP1 regulates pancreatic cancer proliferation and migration. Tumour Biol. 2015;36(12):9693–9701. doi: 10.1007/s13277-015-3704-8. [DOI] [PubMed] [Google Scholar]
- 49.Tan ZW, Xie S, Hu SY, Liao T, Liu P, Peng KH, et al. Caudatin targets TNFAIP1/NF-κB and cytochrome c/caspase signaling to suppress tumor progression in human uterine cancer. Int J Oncol. 2016;49(4):1638–1650. doi: 10.3892/ijo.2016.3662. [DOI] [PubMed] [Google Scholar]
- 50.Kim DM, Chung KS, Choi SJ, Jung YJ, Park SK, Han GH, et al. RhoB induces apoptosis via direct interaction with TNFAIP1 in HeLa cells. Int J Cancer. 2009;125(11):2520–2527. doi: 10.1002/ijc.24617. [DOI] [PubMed] [Google Scholar]
- 51.Hu X, Yan F, Wang F, Yang Z, Xiao L, Li L, et al. TNFAIP1 interacts with KCTD10 to promote the degradation of KCTD10 proteins and inhibit the transcriptional activities of NF-κB and AP-1. Mol Biol Rep. 2012;39(11):9911–9919. doi: 10.1007/s11033-012-1858-7. [DOI] [PubMed] [Google Scholar]
- 52.Tian X, Zhang J, Yan L, Dong JM, Guo Q. MiRNA-15a inhibits proliferation, migration and invasion by targeting TNFAIP1 in human osteosarcoma cells. Int J Clin Exp Pathol. 2015;8(6):6442–6449. [PMC free article] [PubMed] [Google Scholar]
- 53.Grinchuk OV, Motakis E, Kuznetsov VA. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. 2010;11 Suppl 1:S9. [DOI] [PMC free article] [PubMed]
- 54.Escamilla CO, Filonova I, Walker AK, Xuan ZX, Holehonnur R, Espinosa F, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551(7679):227–231. doi: 10.1038/nature24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485(7398):363–7. [DOI] [PMC free article] [PubMed]
- 56.Madison JM, Duong K, Vieux EF, Udeshi ND, Iqbal S, Requadt E, et al. Regulation of purine metabolism connects KCTD13 to a metabolic disorder with autistic features. iScience. 2021;24(1):101935. [DOI] [PMC free article] [PubMed]
- 57.Miller JC, Blake DC, Herzog CR. Adenylosuccinate synthetase 1 gene is a novel target of deletion in lung adenocarcinoma. Mol Carcinog. 2009;48(12):1116–1122. doi: 10.1002/mc.20563. [DOI] [PubMed] [Google Scholar]
- 58.Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci. 2005;25(36):8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101(29):10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Smaele E, Di Marcotullio L, Ferretti E, Screpanti I, Alesse E, Gulino A. Chromosome 17p deletion in human medulloblastoma - A missing checkpoint in the Hedgehog pathway. Cell Cycle. 2004;3(10):1263–1266. doi: 10.4161/cc.3.10.1200. [DOI] [PubMed] [Google Scholar]
- 61.Mancarelli MM, Zazzeroni F, Ciccocioppo L, Capece D, Po A, Murgo S, et al. The tumor suppressor gene KCTD11REN is regulated by Sp1 and methylation and its expression is reduced in tumors. Mol Cancer. 2010;9:172. doi: 10.1186/1476-4598-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zazzeroni F, Nicosia D, Tessitore A, Gallo R, Verzella D, Fischietti M, et al. KCTD11 tumor suppressor gene expression is reduced in prostate adenocarcinoma. Biomed Res Int. 2014;2014:380398. [DOI] [PMC free article] [PubMed]
- 63.Tong R, Yang B, Xiao H, Peng C, Hu W, Weng X, et al. KCTD11 inhibits growth and metastasis of hepatocellular carcinoma through activating Hippo signaling. Oncotarget. 2017;8(23):37717–37729. doi: 10.18632/oncotarget.17145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szkandera J, Kiesslich T, Haybaeck J, Gerger A, Pichler M. Hedgehog signaling pathway in ovarian cancer. Int J Mol Sci. 2013;14(1):1179–1196. doi: 10.3390/ijms14011179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Li HR, Yu JJ, Li FX. Induction of tumor suppressor KCTD11 during periovulatory period in rat ovary. Reprod Biol. 2019;19(2):173–178. doi: 10.1016/j.repbio.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 66.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reardon DA, Jenkins JJ, Sublett JE, Burger PC, Kun LK. Multiple genomic alterations including N-myc amplification in a primary large cell medulloblastoma. Pediatr Neurosurg. 2000;32(4):187–191. doi: 10.1159/000028932. [DOI] [PubMed] [Google Scholar]
- 68.Lescop S, Lellouch-Tubiana A, Vassal G, Besnard-Guerin C. Molecular genetic studies of chromosome 11 and chromosome 22q DNA sequences in pediatric medulloblastomas. J Neurooncol. 1999;44(2):119–127. doi: 10.1023/A:1006387518100. [DOI] [PubMed] [Google Scholar]
- 69.Luttikhuis ME, Powell JE, Rees SA, Genus T, Chughtai S, Ramani P, et al. Neuroblastomas with chromosome 11q loss and single copy MYCN comprise a biologically distinct group of tumours with adverse prognosis. Br J Cancer. 2001;85(4):531–537. doi: 10.1054/bjoc.2001.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stankovic T, Skowronska A. The role of ATM mutations and 11q deletions in disease progression in chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55(6):1227–1239. doi: 10.3109/10428194.2013.829919. [DOI] [PubMed] [Google Scholar]
- 71.Schleutker J, Baffoe-Bonnie AB, Gillanders E, Kainu T, Jones MP, Freas-Lutz D, et al. Genome-wide scan for linkage in finnish hereditary prostate cancer (HPC) families identifies novel susceptibility loci at 11q14 and 3p25-26. Prostate. 2003;57(4):280–289. doi: 10.1002/pros.10302. [DOI] [PubMed] [Google Scholar]
- 72.Heride C, Rigden DJ, Bertsoulaki E, Cucchi D, De Smaele E, Clague MJ, et al. The centrosomal deubiquitylase USP21 regulates Gli1 transcriptional activity and stability. J Cell Sci. 2016;129(21):4001–4013. doi: 10.1242/jcs.188516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Angrisani A, Di Fiore A, Di Trani CA, Fonte S, Petroni M, Lospinoso Severini L, et al. Sp1 and p53 interplay modulates the expression of the oncosuppressor KCASH2. Front Cell Dev Biol. 2021. [DOI] [PMC free article] [PubMed]
- 74.Elks CE, Loos RJ, Sharp SJ, Langenberg C, Ring SM, Timpson NJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010;7(5):e1000284. [DOI] [PMC free article] [PubMed]
- 75.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gamero-Villarroel C, González LM, Rodríguez-López R, Albuquerque D, Carrillo JA, García-Herráiz A, et al. Influence of. Brain Behav. 2017;7(9):e00784. [DOI] [PMC free article] [PubMed]
- 77.Waalen J. The genetics of human obesity. Transl Res. 2014;164(4):293–301. doi: 10.1016/j.trsl.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Takeuchi F, Yamamoto K, Katsuya T, Nabika T, Sugiyama T, Fujioka A, et al. Association of genetic variants for susceptibility to obesity with type 2 diabetes in Japanese individuals. Diabetologia. 2011;54(6):1350–1359. doi: 10.1007/s00125-011-2086-8. [DOI] [PubMed] [Google Scholar]
- 79.Renström F, Payne F, Nordström A, Brito EC, Rolandsson O, Hallmans G, et al. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet. 2009;18(8):1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smaldone G, Pirone L, Capolupo A, Vitagliano L, Monti MC, Di Gaetano S, et al. The essential player in adipogenesis GRP78 is a novel KCTD15 interactor. Int J Biol Macromol. 2018;115:469–475. doi: 10.1016/j.ijbiomac.2018.04.078. [DOI] [PubMed] [Google Scholar]
- 81.Smaldone G, Coppola L, Incoronato M, Parasole R, Ripaldi M, Vitagliano L, et al. KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia. Diagnostics (Basel). 2020;10(6). [DOI] [PMC free article] [PubMed]
- 82.Ding XF, Luo C, Ren KQ, Zhang J, Zhou JL, Hu X, et al. Characterization and expression of a human KCTD1 gene containing the BTB domain, which mediates transcriptional repression and homomeric interactions. DNA Cell Biol. 2008;27(5):257–265. doi: 10.1089/dna.2007.0662. [DOI] [PubMed] [Google Scholar]
- 83.Ding X, Luo C, Zhou J, Zhong Y, Hu X, Zhou F, et al. The interaction of KCTD1 with transcription factor AP-2alpha inhibits its transactivation. J Cell Biochem. 2009;106(2):285–295. doi: 10.1002/jcb.22002. [DOI] [PubMed] [Google Scholar]
- 84.Borinstein SC, Hyatt MA, Sykes VW, Straub RE, Lipkowitz S, Boulter J, et al. SETA is a multifunctional adapter protein with three SH3 domains that binds Grb2, Cbl, and the novel SB1 proteins. Cell Signal. 2000;12(11–12):769–779. doi: 10.1016/S0898-6568(00)00129-7. [DOI] [PubMed] [Google Scholar]
- 85.Li X, Chen C, Wang F, Huang W, Liang Z, Xiao Y, et al. KCTD1 suppresses canonical Wnt signaling pathway by enhancing β-catenin degradation. PLoS One. 2014;9(4):e94343. [DOI] [PMC free article] [PubMed] [Retracted]
- 86.Wang T, Wang D, Zhang L, Yang P, Wang J, Liu Q, et al. The TGFβ-miR-499a-SHKBP1 pathway induces resistance to EGFR inhibitors in osteosarcoma cancer stem cell-like cells. J Exp Clin Cancer Res. 2019;38(1):226. doi: 10.1186/s13046-019-1195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feng L, Wang JT, Jin H, Qian K, Geng JG. SH3KBP1-binding protein 1 prevents epidermal growth factor receptor degradation by the interruption of c-Cbl-CIN85 complex. Cell Biochem Funct. 2011;29(7):589–596. doi: 10.1002/cbf.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darmanis S, Cui T, Drobin K, Li SC, Öberg K, Nilsson P, et al. Identification of candidate serum proteins for classifying well-differentiated small intestinal neuroendocrine tumors. PLoS One. 2013;8(11):e81712. [DOI] [PMC free article] [PubMed]
- 89.Greif PA, Eck SH, Konstandin NP, Benet-Pagès A, Ksienzyk B, Dufour A, et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25(5):821–827. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- 90.Network CGAR, Medicine AECo, Services AB, Hospital BC, Medicine BCo, Hope BRIoCo, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–84. [DOI] [PMC free article] [PubMed]
- 91.Faqeih EA, Almannai M, Saleh MM, AlWadei AH, Samman MM, Alkuraya FS. Phenotypic characterization of KCTD3-related developmental epileptic encephalopathy. Clin Genet. 2018;93(5):1081–1086. doi: 10.1111/cge.13227. [DOI] [PubMed] [Google Scholar]
- 92.Cao-Ehlker X, Zong X, Hammelmann V, Gruner C, Fenske S, Michalakis S, et al. Up-regulation of hyperpolarization-activated cyclic nucleotide-gated channel 3 (HCN3) by specific interaction with K+ channel tetramerization domain-containing protein 3 (KCTD3) J Biol Chem. 2013;288(11):7580–7589. doi: 10.1074/jbc.M112.434803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang W, Dong K, Li K, Dong R, Zheng S. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1α and p53 pathways. Sci Rep. 2016;6:36268. doi: 10.1038/srep36268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ivankova K, Turecek R, Fritzius T, Seddik R, Prezeau L, Comps-Agrar L, et al. Up-regulation of GABA(B) receptor signaling by constitutive assembly with the K+ channel tetramerization domain-containing protein 12 (KCTD12) J Biol Chem. 2013;288(34):24848–24856. doi: 10.1074/jbc.M113.476770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Resendes BL, Kuo SF, Robertson NG, Giersch AB, Honrubia D, Ohara O, et al. Isolation from cochlea of a novel human intronless gene with predominant fetal expression. J Assoc Res Otolaryngol. 2004;5(2):185–202. doi: 10.1007/s10162-003-4042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suehara Y, Kondo T, Seki K, Shibata T, Fujii K, Gotoh M, et al. Pfetin as a prognostic biomarker of gastrointestinal stromal tumors revealed by proteomics. Clin Cancer Res. 2008;14(6):1707–1717. doi: 10.1158/1078-0432.CCR-07-1478. [DOI] [PubMed] [Google Scholar]
- 97.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22(18):3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 98.Li L, Duan T, Wang X, Zhang RH, Zhang M, Wang S, et al. KCTD12 Regulates Colorectal Cancer Cell Stemness through the ERK Pathway. Sci Rep. 2016;6:20460. doi: 10.1038/srep20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye RY, Kuang XY, Zeng HJ, Shao N, Lin Y, Wang SM. KCTD12 promotes G1/S transition of breast cancer cell through activating the AKT/FOXO1 signaling. J Clin Lab Anal. 2020;34(8):e23315. [DOI] [PMC free article] [PubMed]
- 100.Luo L, Cui J, Feng Z, Li Y, Wang M, Cai Y, et al. Lentiviral-mediated overexpression of KCTD12 inhibits the proliferation of human uveal melanoma OCM-1 cells. Oncol Rep. 2017;37(2):871–878. doi: 10.3892/or.2016.5325. [DOI] [PubMed] [Google Scholar]
- 101.Abbaszadegan MR, Taghehchian N, Li L, Aarabi A, Moghbeli M. Contribution of KCTD12 to esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):853. doi: 10.1186/s12885-018-4765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang J, Xu WW, Hong P, Ye F, Huang XH, Hu HF, et al. Adefovir dipivoxil sensitizes colon cancer cells to vemurafenib by disrupting the KCTD12-CDK1 interaction. Cancer Lett. 2019;451:79–91. doi: 10.1016/j.canlet.2019.02.050. [DOI] [PubMed] [Google Scholar]
- 103.Zhong Y, Yang J, Xu WW, Wang Y, Zheng CC, Li B, et al. KCTD12 promotes tumorigenesis by facilitating CDC25B/CDK1/Aurora A-dependent G2/M transition. Oncogene. 2017;36(44):6177–6189. doi: 10.1038/onc.2017.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kudoh S, Tenjin Y, Kameyama H, Ichimura T, Yamada T, Matsuo A, et al. Significance of achaete-scute complex homologue 1 (ASCL1) in pulmonary neuroendocrine carcinomas; RNA sequence analyses using small cell lung cancer cells and Ascl1-induced pulmonary neuroendocrine carcinoma cells. Histochem Cell Biol. 2020;153(6):443–456. doi: 10.1007/s00418-020-01863-z. [DOI] [PubMed] [Google Scholar]
- 105.Cai WY, Chen X, Chen LP, Li Q, Du XJ, Zhou YY. Role of differentially expressed genes and long non-coding RNAs in papillary thyroid carcinoma diagnosis, progression, and prognosis. J Cell Biochem. 2018;119(10):8249–8259. doi: 10.1002/jcb.26836. [DOI] [PubMed] [Google Scholar]
- 106.Faryna M, Konermann C, Aulmann S, Bermejo JL, Brugger M, Diederichs S, et al. Genome-wide methylation screen in low-grade breast cancer identifies novel epigenetically altered genes as potential biomarkers for tumor diagnosis. Faseb j. 2012;26(12):4937–4950. doi: 10.1096/fj.12-209502. [DOI] [PubMed] [Google Scholar]
- 107.Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, et al. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387(1):175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pirone L, Esposito C, Correale S, Graziano G, Di Gaetano S, Vitagliano L, et al. Thermal and chemical stability of two homologous POZ/BTB domains of KCTD proteins characterized by a different oligomeric organization. Biomed Res Int. 2013;2013:162674. [DOI] [PMC free article] [PubMed]
- 109.Balasco N, Pirone L, Smaldone G, Di Gaetano S, Esposito L, Pedone EM, et al. Molecular recognition of Cullin3 by KCTDs: insights from experimental and computational investigations. Biochim Biophys Acta. 2014;1844(7):1289–1298. doi: 10.1016/j.bbapap.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 110.Canales J, Cruz P, Díaz N, Riquelme D, Leiva-Salcedo E, Cerda O. K + Channel Tetramerization Domain 5 (KCTD5) Protein Regulates Cell Migration, Focal Adhesion Dynamics and Spreading through Modulation of Ca 2+ Signaling and Rac1 Activity. Cells. 2020;9(10). [DOI] [PMC free article] [PubMed]
- 111.Rivas J, Díaz N, Silva I, Morales D, Lavanderos B, Álvarez A, et al. KCTD5, a novel TRPM4-regulatory protein required for cell migration as a new predictor for breast cancer prognosis. FASEB J. 2020. [DOI] [PubMed]
- 112.Brockmann M, Blomen VA, Nieuwenhuis J, Stickel E, Raaben M, Bleijerveld OB, et al. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature. 2017;546(7657):307–311. doi: 10.1038/nature22376. [DOI] [PubMed] [Google Scholar]
- 113.Zhang X, Zhou H, Cai L, Fan C, Liu Y, Wang L, et al. Kctd20 promotes the development of non-small cell lung cancer through activating Fak/AKT pathway and predicts poor overall survival of patients. Mol Carcinog. 2017;56(9):2058–2065. doi: 10.1002/mc.22660. [DOI] [PubMed] [Google Scholar]
- 114.Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370(2):313–323. doi: 10.1016/j.canlet.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 115.Huang MY, Wang HM, Chang HJ, Hsiao CP, Wang JY, Lin SR. Overexpression of S100B, TM4SF4, and OLFM4 genes is correlated with liver metastasis in Taiwanese colorectal cancer patients. DNA Cell Biol. 2012;31(1):43–49. doi: 10.1089/dna.2011.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kasahara K, Kawakami Y, Kiyono T, Yonemura S, Kawamura Y, Era S, et al. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat Commun. 2014;5:5081. doi: 10.1038/ncomms6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Inaba H, Goto H, Kasahara K, Kumamoto K, Yonemura S, Inoko A, et al. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol. 2016;212(4):409–423. doi: 10.1083/jcb.201507046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68(7):2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lauriola A, Martello A, Fantini S, Marverti G, Zanocco-Marani T, Davalli P, et al. Depletion of Trichoplein (TpMs) Causes Chromosome Mis-Segregation, DNA Damage and Chromosome Instability in Cancer Cells. Cancers (Basel). 2020;12(4). [DOI] [PMC free article] [PubMed]
- 120.Zheng M, Zhu H, Gong Y, Wang D, Xie Q, Tang H, et al. Involvement of GMRP1, a novel mediator of Akt pathway, in brain damage after intracerebral hemorrhage. Int J Clin Exp Pathol. 2013;6(2):224–229. [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang C, Hu Y, Shi C. Targeting Natural Killer Cells for Tumor Immunotherapy. Front Immunol. 2020;11:60. doi: 10.3389/fimmu.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nawa M, Matsuoka M. KCTD20, a relative of BTBD10, is a positive regulator of Akt. BMC Biochem. 2013;14:27. doi: 10.1186/1471-2091-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J, Xu J, Ying K, Cao G, Hu G, Wang L, et al. Molecular cloning and characterization of a novel human BTB domain-containing gene, BTBD10, which is down-regulated in glioma. Gene. 2004;340(1):61–69. doi: 10.1016/j.gene.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 124.Xie ZC, Wu HY, Dang YW, Chen G. Role of alternative splicing signatures in the prognosis of glioblastoma. Cancer Med. 2019;8(18):7623–7636. doi: 10.1002/cam4.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chang YS, Tu SJ, Chiang HS, Yen JC, Lee YT, Fang HY, et al. Genome-Wide Analysis of Prognostic Alternative Splicing Signature and Splicing Factors in Lung Adenocarcinoma. Genes (Basel). 2020;11(11). [DOI] [PMC free article] [PubMed]
- 126.Birerdinc A, Nohelty E, Marakhonov A, Manyam G, Panov I, Coon S, et al. Pro-apoptotic and antiproliferative activity of human KCNRG, a putative tumor suppressor in 13q14 region. Tumour Biol. 2010;31(1):33–45. doi: 10.1007/s13277-009-0005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ivanov DV, Tyazhelova TV, Lemonnier L, Kononenko N, Pestova AA, Nikitin EA, et al. A new human gene KCNRG encoding potassium channel regulating protein is a cancer suppressor gene candidate located in 13q14.3. FEBS Lett. 2003;539(1–3):156–60. [DOI] [PubMed]
- 128.Cho YG, Kim CJ, Song JH, Rhie DJ, Park YK, Kim SY, et al. Genetic and expression analysis of the KCNRG gene in hepatocellular carcinomas. Exp Mol Med. 2006;38(3):247–255. doi: 10.1038/emm.2006.30. [DOI] [PubMed] [Google Scholar]
- 129.Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, de Ligt J, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet. 2016;24(5):660–665. doi: 10.1038/ejhg.2015.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pichler I, Schwienbacher C, Zanon A, Fuchsberger C, Serafin A, Facheris MF, et al. Fine-mapping of restless legs locus 4 (RLS4) identifies a haplotype over the SPATS2L and KCTD18 genes. J Mol Neurosci. 2013;49(3):600–605. doi: 10.1007/s12031-012-9891-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the following repository: COSMIC (Catalogue Of Somatic Mutations In Cancer; https://cancer.sanger.ac.uk/cosmic and GENT2 (Gene Expression patterns across Normal and Tumor tissues; http://gent2.appex.kr/gent2/.