Abstract
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a recently discovered coronavirus that causes severe and acute diarrhea and rapid weight loss in piglets. SADS-CoV was reported to be capable of infecting cell lines derived from diverse species, including bats, mice, hamsters, rats, chickens, pigs, nonhuman primates, and humans, implying its high risk of cross-species infection. However, its receptor is still unknown. In this study, the receptor-binding domain of the SADS-CoV spike (S) protein was purified and then subjected to affinity purification (AP)-coupled mass spectrometry (MS)-based proteomic analysis to identify the interactors of the SADS-CoV S protein. Forty-three host proteins were identified, and a Gene Ontology analysis indicated that these interactors can be grouped into categories such as “cell-cell adhesion”, “translation” “viral transcription”, suggesting that these processes may participate in the SADS-CoV life cycles. RNA interference-based screening of these interactors indicated that PPIB and vimentin can affect SADS-CoV replication. Our study provides an overarching view into the host interactome of the SADS-CoV S protein and highlights potential targets for the development of therapeutics against SADS-CoV.
Keywords: SADS-CoV, Spike protein, Virus-host interaction, PPIB, Vimentin
1. Introduction
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a recently discovered HKU2-related bat coronavirus that causes severe and acute diarrhea and rapid weight loss in piglets [1]. In 2017, an outbreak of SADS-CoV led to the death of almost 24,700 piglets in southern China and resulted in significant economic losses [1]. SADS-CoV belongs to the genus Alphacoronavirus within the subfamily Orthocoronavirinae of the family Coronaviridae. SADS-CoV shares 95% sequence identity of the overall genome with the bat coronavirus HKU2 and was reported to be capable of infecting cell lines derived from diverse species, including bats, mice, hamsters, rats, chickens, pigs, nonhuman primates, and humans [2], [3], [4]. The wide range of cell tropism of SADS-CoV in vitro implies its high risk of cross-species infection [4].
The coronavirus life cycle involves a number of stages, including entry into host cells, replication, and transcription of viral RNA, translation and processing of viral proteins, and assembly and budding of virions [5]. During infection, the spike (S) protein of CoV recognizes receptors in the host cell membrane for initial entry, and thus, the receptor is an important antiviral target. Recombinant human ACE2, which is the receptor for SARS-CoV and SARS-CoV-2 [6], [7], has been reported to significantly reduce SARS-CoV-2 entry into the human cell–derived organoids, presumably by acting as a decoy for virus binding [8]. Multiple receptors for CoVs have been identified, including ACE2, DPP4, and APN [6], [9], [10]. However, none of these proteins contributes to the infection of SADS-CoV [1], [4].
The identification of virus-host interactions has benefited greatly from the use of affinity purification (AP)-coupled mass spectrum (MS)-based proteomic analysis [11]. For example, by AP-MS analysis of S interactors, ACE2 and DPP4 were identified as receptors for SARS-CoV and MERS-CoV, respectively [6], [9]. Interactomics analysis of viral proteins of SARS-CoV-2 has led to the identification of multiple host proteins involved in SARS-CoV-2 infection [12]. However, the host protein partners involved in SADS-CoV infection have not yet been characterized. In this study, an AP-MS strategy was used to identify host proteins interacting with SADS S1, the receptor-binding protein of SADS-CoV, and proteins involved in “cell-cell adhesion”, “translation”, and “viral transcription” that were found to be enriched. Further RNA interfering (RNAi)-based functional screening of S1 interactors led to the identification of proteins that can affect SADS-CoV replication. Our study provides overarching views into the host interactome of the SADS-CoV S protein and highlights the potential targets for the development of therapeutics against SADS-CoV.
2. Materials and methods
2.1. Cells
A549, HEK 293T, and Vero cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% (v/v) fetal bovine serum (DMEM; Gibco) in 37 °C incubators with 5% CO2. FreeStyle 293-F (Gibco, R790-07) cells were maintained in FreeStyle 293 Expression Medium (Gibco, 12338026) in 37 °C incubators with 8% CO2 on an orbital shaker platform rotating at 150 rpm/min. For all the cells used, mycoplasma contamination was checked and excluded by MycoBlue Mycoplasma Detector (Vazyme, D101-02).
2.2. Plasmids
The mammalian expression plasmids pEVL9.3-SADS-CoV-S (aa22-532; humanized)-Fc (human IgG1) and pEVL9.3-Fc (human IgG1) were gifts from Professor Zhengli Shi (Wuhan Institute of Virology, Chinese Academy of Science). pMD2.G (Addgene, 12259), lentivirus package plasmids pCMV-dR8.91, and lentiCRISPR v2 were gifts from Professor Ke Peng (Wuhan Institute of Virology, Chinese Academy of Science). pLX304 plasmids encoding C-terminal V5-tagged ANXA2 and PRSS1 were obtained from CCSB-Broad Lentiviral Expression Library (Dharmacon). LentiCRISPR v2 plasmids with sgRNA targeting human PPIB/SUB1/VIM/CBR1, pEVL9.3-SADS-CoV-S1-ΔNTD (aa253-532)-Fc-strep, pEVL9.3-SADS-CoV-S1-SD1-CTD (aa253-460)-Fc-strep, pEVL9.3-SADS-CoV-S1-CTD (aa275-400)-Fc-strep and pEVL9.3-Fc-strep were constructed by standard molecular biology techniques. All these plasmids were confirmed by Sanger sequencing.
2.3. Virus
SADS-CoV (GenBank: QNL24139.1) was a gift from Professor Zhengli Shi (Wuhan Institute of Virology, Chinese Academy of Science) [1], and was amplified in Vero cells. Briefly, Vero cells in DMEM with 2 μg/mL trypsin (Gibco, 15050065) were infected with viruses at an MOI of 0.01. After incubation for 1.5 h, viral supernatants were replaced with 10% tryptose phosphate broth (TPB) (Sigma-Aldrich, T9157) in DMEM with 2 μg/mL trypsin. Culture supernatants were harvested at 48 h postinfection and viral titers were determined by plaque assay.
2.4. Reagents and antibodies
MagStrep XT beads (IBA Lifesciences, 2-4090-002) and buffer BXT (IBA Lifesciences, 2-1041-250); n-Dodecyl-β-D-Maltopyranoside (Anatrace, D310A); mouse monoclonal antibodies against GAPDH (Proteintech, 60004-1-Ig) and V5 (Invitrogen, MA5-15253); rabbit monoclonal antibodies against CBR1 (Abcam, EPR9660), Vimentin (Cell Signaling Technology, 5741) and Annexin A2 (Cell Signaling Technology, 8235); rabbit polyclonal antibodies against α-Tubulin (Proteintech, 11224-1-AP), Human IgG Heavy Chain (Proteintech, 16402-1-AP) PPIB (Proteintech, 11607-1-AP), SUB1 (Proteintech, 10948-2-AP) and strep (Genscript, A00626-40) and PE anti-human IgG Fc (BioLegend, 409304) were purchased from the indicated companies.
2.5. Protein purification
Plasmids expressing truncated SADS-CoV S1-Fc-strep fusion proteins were transfected into FreeStyle-293F cells with polyethyleneimine (PEI) (Polysciences, 23966). From 4 to 6 h post-transfection, sodium valproate (Sigma, P4543) was added to control the overgrowth of cells. Cell culture supernatants were collected 7 days later, dialyzed with phosphate buffer (0.15 M NaCl, 20 mM Na2HPO4, pH 7.0), and then subjected to Protein A Resin (GenScript Biotech, TM0205) purification. Recombinant proteins were eluted with elution buffer (0.1 M glycine, pH 3.0) and immediately neutralized to pH 7.4 with neutralization Buffer (1 M Tris-HCl, pH 8.5). Then, concentrated proteins were subjected to gel filtration chromatography using Superdex 200 Increase 10/300 GL column and phosphate buffer (pH 7.4). Proteins were stored at −80 °C at a concentration of 2 mg/mL.
2.6. Protein inhibition of infection assay
Prechilled A549 cells (2.5 × 105) were co-incubated with prechilled DMEM containing the indicated concentrations of fusion proteins and viruses at an MOI of 0.1 at 4 °C for 1 h. With an extensively cold PBS wash, culture supernatants were replaced with 0.5 μg/mL trypsin and 10% TPB in DMEM containing the indicated concentrations of fusion proteins. Twenty-four hours later, total RNA was extracted for qRT-PCR analysis.
2.7. FACS analysis of fusion proteins binding to cells
A549 cells (1 × 106) were washed and blocked with 5% BSA at 4 °C for 1 h. Then, the cells were incubated with purified fusion proteins at 4 °C for 1 h, followed by extensive PBS washing. Cells were stained with PE-labeled human IgG-Fc antibody at 4 °C for 1 h. For FACS analysis, cells were detached with 0.5 mM EDTA in PBS, washed and resuspended in PBS containing 1% FBS, and analyzed with a FACS LSRII instrument (BD).
2.8. Coimmunoprecipitation and immunoblot analysis
For transient transfection and coimmunoprecipitation experiments, A549 cells (1 × 107) were lysed in 1 mL of lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 0.3% β-DDM, pH 7.4, complete protease inhibitor cocktail (Roche, 11697498001)). For each immunoprecipitation, purified S1 ΔNTD-Fc-strep or Fc-strep proteins (20 μg/mL) were incubated with 50 μL of precleared MagStrep XT beads at 4 °C for 2 h. After extensive PBS-T (0.02% Tween-20) washing, a 1 mL aliquot of the lysate was incubated with protein-bound beads at 4 °C for 4 h or overnight. The beads were washed once with 1 mL of lysis buffer, twice with 1 mL of PBS-T (0.02% Tween-20), and three times with PBS. Proteins specifically interacting with beads were sent for mass spectrometry identification or immunoblot analysis. For immunoblot analysis, proteins were eluted and denatured by adding 20 μL of 1× Laemmli SDS-PAGE buffer (containing DL-dithiothreitol) followed by heating for 10 min at 100 °C. The proteins were then separated on SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes by a Trans-Blot Turbo rapid transfer system (Bio-Rad) according to the manufacturer’s instructions. The membranes were blocked in 5% defatted milk (dissolved in Tris-buffered saline (TBS)) for 1 h at room temperature and then incubated with a primary antibody for 1 h at room temperature or overnight at 4 °C. The membranes were then washed extensively in wash buffer (TBS containing 0.1% Tween 20) three times (for 5 min each time) with agitation and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Proteintech) for 1 h at room temperature according to the species source of the primary antibody. The membranes were washed three times in wash buffer and imaged using an enhanced chemiluminescence substrate solution (Millipore, P90720) to visualize the protein bands. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or ɑ-Tubulin was utilized as a loading control by stripping the membranes with stripping buffer (Beyotime, China) and re-probing with an anti-GAPDH or anti-ɑ-Tubulin antibody according to the same procedures.
2.9. Mass spectrometry and data analysis
An innovative in-bead digestion strategy was used to obtain the highest yield. Briefly, the beads obtained from the aforementioned immunoprecipitation steps were washed additionally with PBS to remove the remaining detergent. Washed beads were resuspended in 100 μL of denaturing buffer (2 M urea, 50 mM Tris-HCl, pH 8.0) and then reduced in 2 mM dithiothreitol (Sigma-Aldrich) at 56 °C for 30 min, followed by alkylation in 5 mM iodoacetamide (Sigma-Aldrich) for 30 min at room temperature in the dark. Then, 0.5 μg of sequencing-grade modified trypsin (Promega) was added, and the mix was incubated overnight at 37 °C. The resulting peptides were separated from the beads by a magnetic rack and subjected to desalination and concentration by C18 bonded solid-phase extraction discs (Empore). Purified peptides were then subjected to electrospray ionization, followed by liquid chromatography-mass spectrometry (LC-MS) detection, as described previously [13]. The obtained raw MS spectra were subjected to analysis by ProteinPilot version 5.0 for peptide sequence identification against the Swiss-Prot database (downloaded in 2017), which was set to human species restricted. The threshold value of the false discovery rate (FDR) was set to 0.05. S1-ΔNTD-interacting proteins were compared with Fc-interacting proteins in volcano plot using the ggplot2 package in R. S1-ΔNTD specifically bound proteins (fold change > 2, p value < 0.05) were subjected to Gene Ontology analysis using DAVID Bioinformatics Resources 6.8. For each analysis, only the top 10 GO terms are displayed. The N-glycosylation and O-glycosylation sites of interacting proteins were calculated by referring to the UniProt database (https://www.uniprot.org/).
2.10. RNAi experiments
siRNAs corresponding to the target sequences were chemically synthesized by GenePharma (China). For each gene, at least 2 pairs of siRNAs were designed to avoid off-target effects. siRNA was transfected into HEK 293T cells using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher, 13778075) in a final concentration of 40 nM. Culture supernatants were replaced with maintenance medium at 24 h post-transfection. Another 24 h later, the cells were infected with viruses. Forty-eight hours later, the cells were lysed for RNA extraction. siRNA without human mRNA targets (NC) was used as a control for RNAi-related experiments. All siRNA sequences are listed in Supplementary Table 1.
2.11. qRT-PCR
Total RNA was isolated by RNAiso Plus TRIzol reagent (TaKaRa, 9109). RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit with gDNA eraser (TaKaRa, RR047A). One microliter of cDNA was used for real-time PCR assay using TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa, RR820A). The relative quantity was determined by performing a comparative Ct (ΔΔCt) experiment using GAPDH as an endogenous control. Gene-specific primer sequences are listed in Supplementary Table 2.
2.12. Establishment of CRISPR/Cas9 based specific gene knockout in A549 cells
sgRNA sequences targeting exons of the human STAT3 were designed by using the Millipore&Sigma CRISPR design tool (https://www.milliporesigmabioinfo.com/bioinfo_tools/faces/informatics.xhtml). The predesigned sequence with the fewest predicted off-target cleavage sites was selected. Oligos were synthesized, annealed, and ligated to a FastDigest BsmBI (Thermo, FD0454) digested lentiCRISPR v2 plasmid by using T4 DNA ligase (Thermo, EL0014). LentiCRISPR v2 plasmid containing sgRNA with no human genome target sequence (NT) was used as a control.
For lentivirus package, HEK 293T cells (1.5 × 106) were transfected with STAT3-sgRNA LentiCRISPR v2 plasmid (800 ng) or NT-sgRNA LentiCRISPR v2 plasmid (800 ng) and two packaging plasmids (pCMV-dR8.91 (1200 ng) coding HIV-Gag and HIV-Pol proteins and pMD2.G (400 ng) coding VSV-G protein) by Lipofectamine 2000 transfection reagent. The culture medium was replaced with a maintenance medium at 12 h after transfection. After an additional 36 h, the recombinant virus-containing supernatants were filtered by 0.22-μm PES filter and used to infect A549 cells three times in the presence of polybrene (8 μg/mL) for higher transduction efficiency. 4 days after the first transduction, cells were selected by incubating with 1 μg/mL puromycin for another 10 days, and then the single cell was isolated by serial dilutions and allowed to expand for 2 to 3 weeks without puromycin. Genomic DNA from the cell lines was extracted by using a TIANamp genomic DNA kit (Tiangen, DP304). The modified regions were amplified using specific genomic cleavage detection primers. PCR production was ligated to T-vector by using 5 × TA/Blunt-Zero Cloning Kit (Vazyme, C601-01), followed by transforming into NEB stable bacteria. At least 10 bacteria clones of each transforming were defined by Sanger sequencing and A549 cell lines exhibiting frameshift mutations at the corresponding sites were selected and further confirmed by Western blotting. Two isogenic cell lines bearing the desired gene editing outcomes and one control cell line (NT) were selected for indicated assay. sgRNA sequences and specific genomic cleavage detection primers were listed in Supplementary Table 3.
2.13. Virus-cell binding assay
A549 cells (2.5 × 105) were prechilled at 4 °C for 15 min. Then cells were incubated with viruses at an MOI of 0.1 at 4 °C for 1 h, followed by extensively cold PBS wash. Cells were lysed in Trizol reagent. Cell-attached viral RdRp genes were measured by qRT-PCR and normalized to the level of GAPDH mRNA.
3. Results
3.1. Expression of truncated S1 proteins revealed that S1-ΔNTD accounted for the binding of SADS-CoV
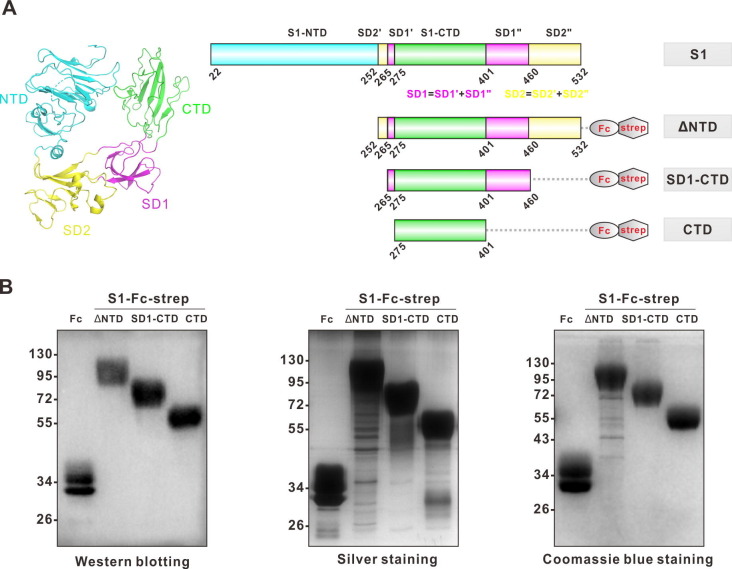
The S1 subunit of the SADS-CoV S protein, which contributes to the binding of the host cell receptor, comprises two major domains, the N-terminal domain (NTD) and the C-terminal domain (CTD), which are followed by subdomains 1 and 2 (SD1 and SD2) [14] (Fig. 1 A). It has been proposed that the CTD of S1 may account for binding to host cells [14], [15], and we then expressed truncated S1 proteins without NTD (ΔNTD), without both NTD and SD2 (SD1-CTD), or with CTD only (Fig. 1A). Briefly, the C-terminus of truncated S1 proteins was fused to the Fc region of human IgG and Strep tag and expressed in FreeStyle-293F cells. Then, cultured supernatants were collected, and truncated S1 proteins were purified with protein A agarose. The purification of the truncated S1 proteins was confirmed by Western blotting, silver staining, and Coomassie blue staining (Fig. 1B).
Fig. 1.
Expression and purification of truncated SADS-CoV S1 fusion proteins. (A) The structure of the SADS-CoV S1 (PDB code: 6 M39) monomer was displayed by PyMOL software (left). The NTD, CTD, SD1 and SD2 domains of S1 are labeled cyan, green, magenta and yellow, respectively. NTD, N-terminal domain; CTD, C-terminal domain; and SD1 to SD2, subdomains 1 to 2. The DNA sequences of CTD with distinct subdomains fused with Fc and a strep tag (right) were cloned into a pcDNA3.1 eukaryotic protein expression vector. The DNA sequence of the IFNA1 signal peptide was added between the Kozaka sequence and the CTD sequence to promote protein secretion. (B) Truncated S1 fusion proteins were expressed on Freestyle 293F cells. Proteins purified from supernatants were analyzed by Western blotting (left), silver staining (middle) and Coomassie blue staining (right) respectively. Fc-strep fusion protein was used as the negative control.
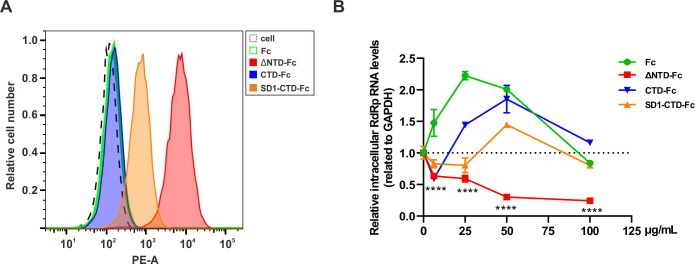
We then examined whether truncated S1 proteins can bind susceptible cells. A549 cells, which are permissive to SADS-CoV infection [2], [3], were incubated with truncated S1 proteins and then washed with PBS to remove unbound proteins, while bound proteins were detected with flow cytometry. As shown in Fig. 2 A, compared to CTD or SD1-CTD, S1-ΔNTD can effectively bind A549 cells. We also explored whether the truncated S1 protein can block SADS-CoV infection, presuming it may competitively bind the host cell receptor. A549 cells were incubated with truncated S1 proteins or Fc and then infected with SADS-CoV. At twenty-four hours post-infection, the intracellular level of SADS-CoV RNA was measured with quantitative real-time PCR (RT-PCR). We found that CTD and SD1-CTD did not inhibit SADS-CoV replication effectively, while S1-ΔNTD blocked SADS-CoV infection significantly (Fig. 2B), which is in accordance with its ability to bind A549 cells (Fig. 2A), suggesting that S1-ΔNTD can bind to receptors in host cells and thus inhibit SADS-CoV infection.
Fig. 2.
Functional analysis of truncated SADS-CoV S1 proteins. (A) Protein binding activity to A549 cell surface. ΔNTD, SD1-CTD, CTD or Fc fusion proteins were incubated with adherent A549 cells at a concentration of 20 μg/mL at 4 °C. One hour later, the cells were extensively washed with PBS to remove unbound protein. Following incubation with PE anti-human IgG secondary antibody and washing, the cells were detached using 0.5 mM EDTA in PBS for FACS analysis. (B) Protein inhibition activity of SADS-CoV infection. SADS-CoV mixed with indicated concentrations of ΔNTD, SD1-CTD, CTD or Fc fusion proteins, and then added to A549 cells at a multiplicity of infection (MOI) of 0.01. Twenty-four hours later, intracellular mRNA was extracted. The relative SADS-CoV genome level was measured using quantitative RT-PCR. ****p < 0.0001, unpaired t-test.
3.2. Proteomic analysis led to the identification of host proteins that can interact with SADS-CoV S1
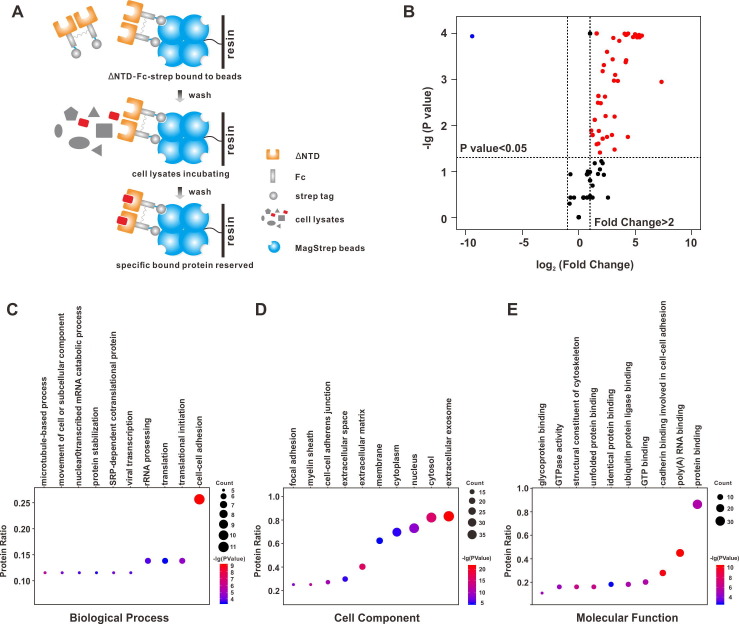
To identify host proteins that may interact with S1-ΔNTD, purified S1-ΔNTD was incubated with host cell lysate, and then, protein enrichment was determined by pulling down the Strep tag. Purified Fc was used as a control. Then, the enriched proteins were digested with trypsin and identified by MS analysis (Fig. 3 A). After normalizing the proteins in the control group (Fc enriched), 43 proteins were identified as S1-ΔNTD interactors (Fig. 3B, enrichment fold > 2, p value < 0.05; the adjusted p values of these interactors are indicated in Fig. S2). Using Gene Ontology (GO) analysis, we found that S1-ΔNTD interactors were mainly involved in “cell-cell adhesion”, “translation”, “viral transcription” processes (Fig. 3C-E), suggesting that these biological processes may be involved in SADS-CoV infection by enriched proteins interacting with S1. GO analysis based on cellular components indicated that the majority of interactors were enriched in the exosome, and these proteins may be released from cells and existed in the extracellular space (Fig. 3D). We then analyzed the glycosylation of these interactors and found that the average numbers of N-glycosylation and O-glycosylation are 0.23 and 0.28, respectively. Among these interactors, vimentin has four O-glycosylation sites, while PPIB has one O-glycosylation site.
Fig. 3.
Interactome analysis of S1-ΔNTD. (A) Fc-strep or S1-ΔNTD-Fc-strep protein (20 μg/mL) was bound to MagStrep XT beads at 4 °C. Unbound proteins were washed with PBST (0.02% Tween-20). Then, precleared A549 cell lysates were incubated with beads at 4 °C for 4 h. After another washing, the beads were digested with trypsin and subjected to MS. Fc-strep bound proteins were used as controls. (B) S1-ΔNTD-interacting proteins were compared with Fc-interacting proteins in volcano plot using the ggplot2 package in R. Dots with a p value < 0.05 were labeled. ΔNTD specifically bound proteins (fold change > 2) are labeled in red. (C-E) S1-ΔNTD specifically bound proteins were subjected to Gene Ontology analysis using DAVID Bioinformatics Resources 6.8. For each analysis, only top 10 GO terms are displayed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. RNA interference-based screening highlighted PPIB and vimentin that can affect SADS-CoV infection
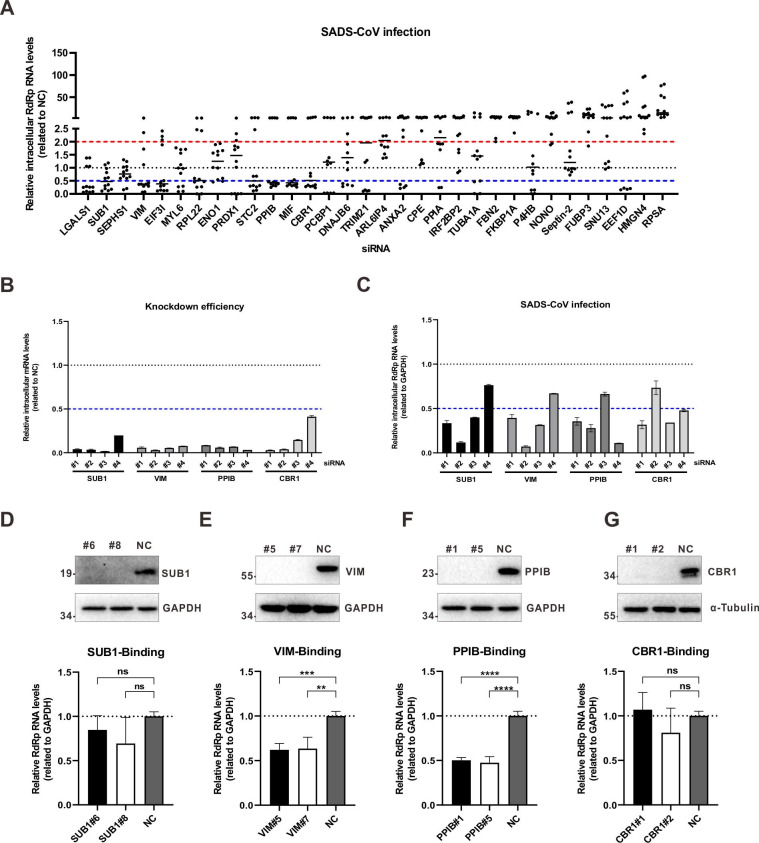
We then used RNAi to explored whether S1-ΔNTD interactors can affect SADS-CoV infection. Only proteins identified with a least two peptides were investigated. To this end, siRNAs targeting S1-ΔNTD interactors were synthesized, and their knockdown efficiency was examined using quantitative RT-PCR. Only siRNAs that could effectively decrease targeting genes without causing cytotoxicity were used for further analysis. Then, A549 cells were transfected with these siRNAs, and 48 h later, these cells were infected with SADS-CoV at a multiplicity of infection (MOI) of 0.01. At forty-eight hours post-infection, the cells were collected, and total RNAs were extracted. The intracellular level of SADS-CoV RNA was measured with quantitative RT-PCR. As shown in Fig. 4 A-C, among the 32 interactors identified with at least two peptides (high confidence), knockdown of CBR1, PPIB, SUB1, and vimentin effectively inhibited SADS-CoV infection. To confirm the effect of these four proteins on SADS-CoV infection, CRISPR-based knockout of these four proteins was performed (Fig. S1). As shown in Fig. 4D-G, knocking out PPIB or vimentin inhibited SADS-CoV infection, confirming the roles of these two proteins in SADS-CoV infection.
Fig. 4.
The effects of knockdown of S1-ΔNTD interactors on SADS-CoV infection. (A) For RNAi-based screening, three different siRNAs were designed for each gene. Each siRNAs were transfected into A549 cells. Forty-eight hours later, the cells were infected with SADS-CoV at an MOI of 0.01. Another 48 h later, intracellular mRNA was extracted. The relative SADS-CoV RNA level was measured using quantitative RT-PCR. Scramble siRNAs without target genes were used as negative controls (NCs). Data are representative of two independent experiments and three different siRNAs for each gene. (B, C) Four siRNAs were designed for SUB1, VIM, PPIB and CBR1 and used as (A). Relative levels of targeted genes (B) and the SADS-CoV RNA (C) were measured using quantitative RT-PCR. The data are representative of three independent experiments with similar results. Graphs show the means ± SD. (D, E, F, G) For CRISPR-Cas9-based knockout of the SUB1, VIM, PPIB and CBR1 genes, Cas9 and gene-specific sgRNAs were packaged into lentiviruses. A549 cells were transduced with lentivirus and selected under puromycin at least 1 week. Then, single cells were separated for expansion. Following Sanger sequencing and Western blotting (upper), the cell lines with frameshift mutations and merely protein expression were selected. For each gene, 2 cell lines were chosen to perform the virus-binding assay (below). SADS-CoV was incubated with A549 cells at an MOI of 0.1 at 4 °C for 1 h. Then, the cells were extensively washed with PBS to remove unbound viruses. Total RNA was extracted, and SADS-CoV genome level was measured with quantitative RT-PCR. A549 cells transduced with scramble sgRNA without target gene were used as negative control. The data are representative of two independent experiments with similar results. The graphs show means ± SD; n = 2. *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant, unpaired t-test.
4. Discussion
SADS-CoV is a newly identified CoV that causes high mortality in piglets. Recent studies have indicated that SADS-CoV can infect cell lines derived from diverse species [2], [3], [4], implying its high risk of cross-species infection [4]. Multiple CoV receptors have been identified, including ACE2, DDP4, and APN [6], [9], [10]; however, none of these proteins contribute to the entry of SADS-CoV.
Although it has been proposed that the CTD of SADS-CoV may contribute to its binding to the host cell receptor, we found that S1-ΔNTD but not CTD can bind permissive cells and block SADS-CoV infection effectively, suggesting that in addition to CTD, both SD1 and SD2 may play roles in the binding of SADS-CoV to the host cell receptor. In this study, to identify host proteins that can interact with the receptor-binding domain of S1, an AP-MS strategy was used to analyze S1-ΔNTD interactors. Forty-three proteins were identified, which can be grouped into “cell-cell adhesion”, “translation” “viral transcription” and other process categories. Further functional analysis indicated that, among the enriched proteins, only PPIB and vimentin affected SADS-CoV infection.
PPIB is a PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore facilitate protein folding [16]. PPIB can interact with the HCV NS5B protein to stimulate the RNA-binding activity of NS5B and promote HCV replication [17]. It has been reported that PPIA and PPIB can interact with the HIV-1 Gag protein, and these interactions may be important for the HIV-1 life cycle [18]. A genome-wide yeast two-hybrid interaction screen study indicated that PPIH can interact with the SARS-CoV S protein and Nsp1, while PPIA, PPIB, and PPIG can interact with Nsp1, and inhibition of immunophilins (PPIA, PPIB, PPIH, and PPIG) by CspA can inhibit SARS-CoV replication [19]. In this study, we found that PPIB can interact with the receptor-binding domain of the SADS-CoV S protein, and knockdown of PPIB decreased SADS-CoV infectivity, suggesting that PPIB can affect SADS-CoV replication. However, further studies are needed to explore the mechanism underlining these phenomena.
Vimentin is a class-III intermediate filament that plays key roles in the integration of cytoskeletal functions, and therefore in basic cellular processes such as cell division and migration [20]. Although vimentin is a predominantly cytoplasmic protein, it can also be transported to the cell surface or secreted to the extracellular location. Extracellular vimentin has been reported to act as a receptor or coreceptor for several viruses, including severe acute respiratory syndrome-related coronavirus (SARS-CoV) [21], dengue virus [22], and Japanese encephalitis virus [23]. Here, we find that vimentin can interact with SADS-CoV S1, and further functional analysis indicated that knockdown of vimentin can inhibit SADS-CoV replication. However, only a slight decrease in SADS-CoV (~2-fold) was observed after the knockdown of vimentin, suggesting that vimentin may partially contribute to the replication of SADS-CoV.
5. Conclusions
Overall, in this study, a truncated S1 protein that maintains the receptor-binding activity of the SADS-CoV S protein was purified and subjected to AP-MS analysis and used to identify host proteins that may participate in the life cycles of SADS-CoV, especially in the entry stage of SADS-CoV. After RNAi-based functional screening of these interactors, we found that PPIB and vimentin can affect the replication of SADS-CoV. Our studies to further explore the mechanism of these two proteins on SADS-CoV replication are ongoing. We did not identify the receptor for SADS-CoV, and this may be due to the proteins with low abundance are not easy to be detected by MS. Genome-wide CRISPR screening may be an alternative strategy for further receptor identification of SADS-CoV.
Acknowledgements
We thank Juan Min in the Center for Instrumental Analysis and Metrology of the Wuhan Institute of Virology and Xi Chen in the SpecAlly Life Technology for their technical assistance. The study was supported by National Natural Science Foundation of China (31830096), the Open Research Fund Program of Wuhan National Bio-Safety Level 4 Lab of CAS (2020ACCP-MS01), and the Youth Innovation Promotion Association CAS (grants 2018367 to L.-K.Z).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Author contributions
Qingxing Wang: Investigation, Methodology, Validation, Visualization, Writing - Original Draft, Writing - Review & Editing. Yun Luo: Resources, Investigation. Weijuan Shang: Validation. Zhengli Shi: Conceptualization, Funding Acquisition, Resources. Gengfu Xiao: Conceptualization, Supervision, Funding Acquisition. Leike Zhang: Conceptualization, Funding Acquisition, Project Administration, Supervision, Writing - Original Draft, Writing - Review & Editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bsheal.2021.05.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y.L., Qin P., Wang B., Liu Y., Xu G.H., Peng L., Zhou J., Zhu S.J., Huang Y.W. Broad Cross-Species Infection of Cultured Cells by Bat HKU2-Related Swine Acute Diarrhea Syndrome Coronavirus and Identification of Its Replication in Murine Dendritic Cells In Vivo Highlight Its Potential for Diverse Interspecies Transmission. J. Virol. 2019;93 doi: 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y., Chen Y., Geng R., Li B., Chen J., Zhao K., Zheng X.S., Zhang W., Zhou P., Yang X.L., Shi Z.L. Broad Cell Tropism of SADS-CoV In Vitro Implies Its Potential Cross-Species Infection Risk. Virol. Sin. 2020;35:1–5. doi: 10.1007/s12250-020-00321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards C.E., Yount B.L., Graham R.L., Leist S.R., Hou Y.J., Dinnon K.H., 3rd, Sims A.C., Swanstrom J., Gully K., Scobey T.D., Cooley M.R., Currie C.G., Randell S.H., Baric R.S. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. U.S.A. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greco T.M., Diner B.A., Cristea I.M. The impact of mass spectrometry-based proteomics on fundamental discoveries in virology. Annu. Rev. Virol. 2014;1:581–604. doi: 10.1146/annurev-virology-031413-085527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D.E. Gordon, J. Hiatt, M. Bouhaddou, V.V. Rezelj, S. Ulferts, H. Braberg, A.S. Jureka, K. Obernier, J.Z. Guo, J. Batra, et al., Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms, Science 370 (2020), eabe9403. https://doi.org/10.1126/science.abe9403. [DOI] [PMC free article] [PubMed]
- 13.Zhu S., Wan W., Zhang Y., Shang W., Pan X., Zhang L.K., Xiao G. Comprehensive Interactome Analysis Reveals that STT3B is Required for the N-Glycosylation of Lassa Virus Glycoprotein. J. Virol. 2019;93 doi: 10.1128/JVI.01443-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan H., Wang Y., Perculija V., Saeed A., Liu Y., Li J., Jan S.S., Li Y., Zhu P., Ouyang S. Cryo-electron microscopy structure of the swine acute diarrhea syndrome coronavirus spike glycoprotein provides insights into evolution of unique coronavirus spike proteins. J. Virol. 2020;94 doi: 10.1128/JVI.01301-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J., Qiao S., Guo R., Wang X. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat. Commun. 2020;11:3070. doi: 10.1038/s41467-020-16876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis T.L., Walker J.R., Campagna-Slater V., Finerty P.J., Paramanathan R., Bernstein G., MacKenzie F., Tempel W., Ouyang H., Lee W.H., Eisenmesser E.Z., Dhe-Paganon S. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watashi K., Ishii N., Hijikata M., Inoue D., Murata T., Miyanari Y., Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Luban J., Bossolt K.L., Franke E.K., Kalpana G.V., Goff S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 19.Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos I., Stamatakis K., Oeste C.L., Perez-Sala D. Vimentin as a multifaceted player and potential therapeutic target in viral infections. Int. J. Mol. Sci. 2020;21:4675. doi: 10.3390/ijms21134675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y.T., Chien S.C., Chen I.Y., Lai C.T., Tsay Y.G., Chang S.C., Chang M.F. Surface vimentin is critical for the cell entry of SARS-CoV. J. Biomed. Sci. 2016;23:14. doi: 10.1186/s12929-016-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Zou L., Yang Y., Yuan J., Hu Z., Liu H., Peng H., Shang W., Zhang X., Zhu J., Rao X. Superficial vimentin mediates DENV-2 infection of vascular endothelial cells. Sci. Rep. 2016;6:38372. doi: 10.1038/srep38372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J.J., Yu C.Y., Liao C.L., Lin Y.L. Vimentin binding is critical for infection by the virulent strain of Japanese encephalitis virus. Cell. Microbiol. 2011;13:1358–1370. doi: 10.1111/j.1462-5822.2011.01624.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.