Abstract
SARS-CoV-2 has surged across the globe causing the ongoing COVID-19 pandemic. Systematic testing to facilitate index case isolation and contact tracing is needed for efficient containment of viral spread. The major bottleneck in leveraging testing capacity has been the lack of diagnostic resources. Pooled testing is a potential approach that could reduce cost and usage of test kits. This method involves pooling individual samples and testing them ‘en bloc’. Only if the pool tests positive, retesting of individual samples is performed. Upon reviewing recent articles on this strategy employed in various SARS-CoV-2 testing scenarios, we found substantial diversity emphasizing the requirement of a common protocol. In this article, we review various theoretically simulated and clinically validated pooled testing models and propose practical guidelines on applying this strategy for large scale screening. If implemented properly, the proposed approach could contribute to proper utilization of testing resources and flattening of infection curve.
Keywords: SARS-CoV-2, COVID-19, Pooled testing, RT-PCR, Diagnosis, Sensitivity
1. Introduction
A novel coronavirus was recognized as the pathogen responsible for the chronic cases of pneumonia that were reported in Wuhan, China in late December 2019 (Andersen et al., 2020; Zhou et al., 2020). This virus was identified as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) (Lu et al., 2020) causing coronavirus disease 2019 (COVID‐19). Due to its extensive transmission across the globe, COVID-19 was declared as a pandemic by the World Health Organization on 11 March 2020 (Cucinotta and Vanelli, 2020). As of 9 February 2021, the WHO has reported 105.4 million cases and 2.3 million deaths worldwide (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). Owing to limited monitoring capacity, diagnostic testing is often limited to symptomatic individuals or people with underlying risk factors for chronic diseases and healthcare workers. A significant proportion of potentially infectious carriers in the community remain undiagnosed, since a majority of population are asymptomatic or present with mild symptoms (Wu and McGoogan, 2020). However, it has been documented that asymptomatic individuals can very well transmit SARS-CoV-2 (Rothe et al., 2020; Ye et al., 2020). With gradual evolution of the pandemic over the last few months, diagnostic centres encounter new challenges in performing surveillance screening of asymptomatic population, driving test numbers even beyond those executed during the peak of the pandemic. Therefore, it is vital to amplify the screening capacity for index case isolation and contact tracing to prevent further spread. The primary diagnostic test that has been employed worldwide to screen COVID-19 is the Real‐Time Reverse Transcriptase‐Polymerase Chain reaction (RT‐PCR) with acceptable sensitivity and specificity levels (Corman et al., 2020). But even in developed countries, testing by RT-PCR is highly constrained due to shortage of reagents and the limited tests that can be performed in a single run. In addition to this, low-income countries with inadequate resources cannot afford the test. Therefore, a rapid, widespread and cost-effective testing strategy is required to curb further spread. A promising strategy to escalate screening capacity is to pool multiple samples in a single test.
Pooled testing (pooling or group testing) involves the screening of a sample pool comprising of multiple individual samples, followed by individual testing (pool deconvolution) only of pools that test positive. A pool that tests negative implies a negative result for all individual samples in the pool. This potentially leads to a much larger number of people being tested when compared to individual testing, while keeping the number of tests the same thereby accelerating the throughput of the existing testing infrastructure (Hanel and Thurner, 2020). Pooling bears the advantage of estimating the positivity rate in a population with fewer tests. It also reduces the testing time. This strategy of grouping samples was pioneered by Robert Dorfman in 1943 to screen men with syphilis in the US military (Dorfman, 1943). Thenceforth, pooling of samples has been used as a screening strategy in multiple infectious diseases like HIV (Emmanuel et al., 1988; Litvak et al., 1994; Sherlock et al., 2007; Stramer et al., 2004), Influenza (Hourfar et al., 2007; Van et al., 2012), Malaria (Taylor et al., 2010) and various bacterial diseases (Currie et al., 2004; Edouard et al., 2015; Singer et al., 2006).
In this review article, we present multiple pooled testing approaches reported so far for the diagnosis of COVID-19. We evaluate the efficiency of these methods and provide insights on their practical relevance. We also provide a meta-analysis of various clinical studies that successfully employed pooled testing. Using the practical guidelines furnished, pooling protocols can be framed by testing centres to increase their diagnostic capacity.
2. Pooled testing strategies
Effective application of pooled testing requires baseline parameters like the prevalence rate of the population, sensitivity and specificity of the test and limit of detection. A key step in this approach is to ascertain an ideal pool size that conserves maximum resources with consistent testing performance (Abdalhamid et al., 2020b). In multiple recent articles and preprints, ideal batch sizes for different ranges of prevalence along with the expected reduction in fraction of tests and costs are put forth based on various algorithms (Aragón-Caqueo et al., 2020; Brynildsrud, 2020; Gu et al., 2020; Rai et al., 2020; Regen et al., 2020; Tan et al., 2020)
Pooled testing works on two paradigms, adaptive and non-adaptive testing. Adaptive pooling is a sequential multi-stage grouping design. In this method, the pool results of the former stage impact the pooling strategy of the consecutive stages. Non-adaptive pooling methods involve parallel testing of multiple pools in a single stage.
2.1. Adaptive testing
In this approach, the tests are performed sequentially. A group is randomly selected based on predicted prevalence rate and tested. The outcome of this test determines the next group to test and so on. Thus, the size and samples of a pool are chosen adaptively based on a previous pool and its test outcome (Mentus et al., 2020).
2.1.1. Dorfman pooling
Dorfman Pooling is the simplest form of pooled testing wherein a set of individual samples are clustered into a pool. If the pool tests positive, it is deconvoluted to decrypt the positive individuals. If the pool is tested negative, all the individuals in the group are interpreted as negative (Dorfman, 1943). Dorfman testing is the most preferred pooling method in clinical scenarios although it usually utilises the highest number of tests among all pooling approaches.
2.1.2. Binary splitting
In Binary splitting, instead of performing individual tests of a positive group as in Dorfman's method, the pool is further divided into two equal sized sub-pools and tested again. The sub-pool that tests positive is split in the next stage until all the negative pools are excluded (Litvak et al., 1994). Binary method is feasible with a maximum of four steps since several dividing steps can be time consuming.
2.1.3. Multistage testing
Multistage model involves sampling different pool sizes at different stages contrary to the binary split method, which uses just two pools in every stage. The number of tests and stages are also substantially minimized. A three-stage testing scheme with pool size of 16 samples in a prevalence rate of 5% has an efficiency gain of 3. A lower prevalence rate favours large pool size and multiple stages while smaller pool sizes with lesser number of stages are to be chosen in higher prevalence settings (Eberhardt et al., n.d.). Gajpal et al employed dynamic programming and have furnished details on the optimum stages required for different sample sizes (Gajpal et al., 2020).
2.1.4. Household grouping
In case of a homogenous population like a locality or neighbourhood, an exposed individual has high chances of spreading the disease immediately to families, neighbours, etc., In this approach, the first step involves forming optimum sized pools of a homogenous population followed by swab tests for all members of the pool. In a second step, positively tested pools will be retested individually. In a population of 150 000, this method can lead to reduction of existing tests by around 56% in moderate prevalence settings with pool sizes up to 25 (Deckert et al., 2020; Takyi-Williams, 2020).
2.1.5. Challenges in adaptive testing
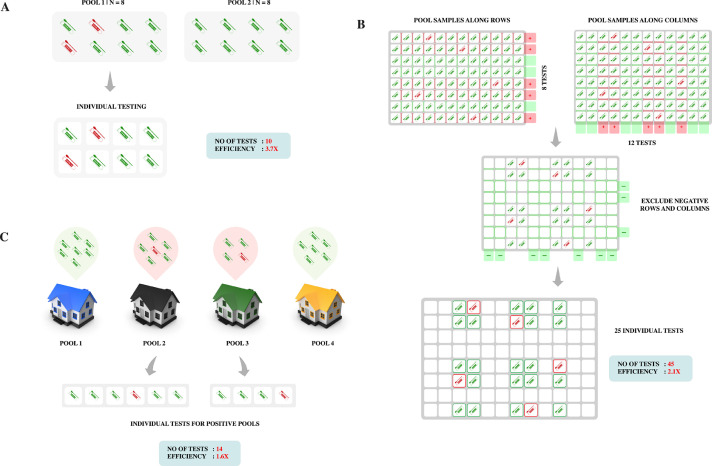
The storing of multiple samples and rerunning of RT-PCR makes lab organization for adaptive approach a resource intensive process. The diagnosis process is liable to delays because of the follow-up loop. Fig.1A-C represents various adaptive pooled testing models that are implemented for COVID-19 diagnosis.
Fig. 1.
Adaptive pooled testing strategies: (A) Dorfman Pooling: Population is divided into two pools of optimum size 8 and tested. The positively tested pool is then deconvoluted to identify the individual infected sample. (B) Column and row pooling: On a 96-well plate, rows and columns are pooled separately. The negative rows and columns are excluded. The individuals from positive rows and columns are then tested individually. In the above illustration, 5 positive samples are identified with 45 tests, compared to 96 tests if tested individually. (C) Household grouping: Pools are formed from each household in a locality and tested. Positive groups undergo individual testing.
2.2. Non-Adaptive Testing
In the non-adaptive approach, each sample is distributed over multiple pools, and all the tests are simultaneously run in parallel yielding reliable results after one round of testing. With increase in pool size and sample multiplicity, a gradual rise in the number of false positives is expected. But initial observation of the prevalence rate and calculation of the explicit error bounds of the false positives leads to streamlined testing and reduced detection times for mass screening (Täufer, 2020).
2.2.1. Matrix testing
Matrix or array testing is a high throughput non-adaptive screening procedure originally proposed by Phatarfod and Sudbury (Phatarfod and Sudbury, 1994). This approach involves constructing a matrix-like grid of samples and grouping samples within rows and columns of the matrix. Unlike adaptive testing, where individuals are consigned to one pool, here each sample will be distributed in two or more individual pools. Samples that traverse the positive rows and columns are tested individually to diagnose the positive cases whereas samples present outside the row-column intersections are declared negative.
2.2.2. Hypercube algorithm
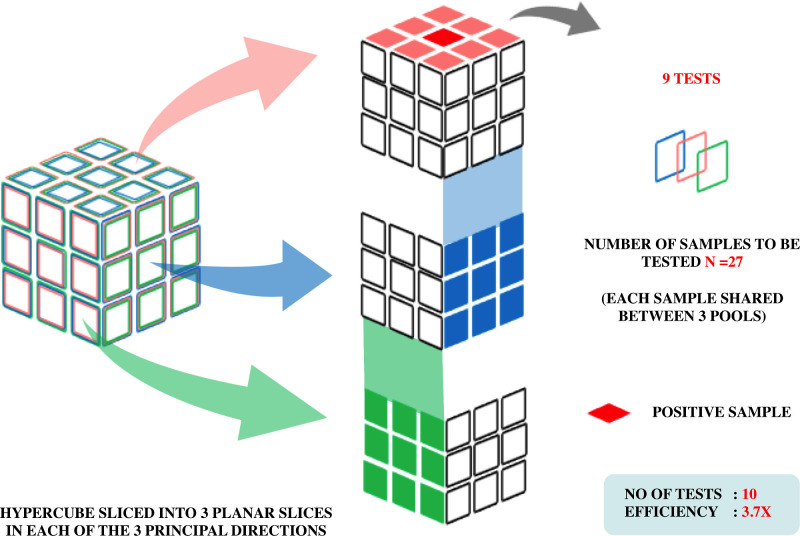
A recent article proposes to arrange samples in the geometry of a hypercube by grouping along its hyperplanes thereby allocating each member in pool sizes of three or more. Based on the viral prevalence, a pool size N = 3D (D= dimension) is determined initially. With decrease in prevalence, D rises, leading to reduced number of tests. Fig. 2 depicts pooling based on hypercube algorithm. This algorithm was validated in oropharyngeal swabs in Rwanda, with precise diagnostic accuracy even after a 100-fold dilution (adding 99 negative samples to one positive sample). In a prevalence setting of less than 0.01%, an improvement of 400 times efficiency compared to regular testing was observed (Mutesa et al., 2021). But an intersection of any two hyperplanes in more than one point might occur in dimensions above three. This might impair testing efficiency by creating unnecessary correlations between different pools.
Fig. 2.
Adaptive pooled testing strategy: Sample pooling based on Hypercube algorithm. For this example, the three sets of slices are shown in blue, red and green. If one infected individual is present, tests on each set of slices identify their coordinate in that direction. Hence only nine tests would uniquely identify them. (Colour version of figure is available online)
2.2.3. Compressed sensing
In compressed sensing, the result is not just a binary reading (positive or negative), but also reveals the viral loads (copies/mL) of individuals who test positive (Ghosh et al., 2020a; Yi et al., 2020).
2.2.3.1. P-BEST(Pooling-Based Efficient SARS-CoV-2 Testing)algorithm
In this approach, samples are stratified on the basis of Reed- Solomon error correction. P-BEST was tested using four sets of 384 samples, each containing positive cases ranging from two to five. This method was able to correctly identify all positive cases using only 48 tests with a testing efficiency gain of 8. Modelling algorithms report very less false positive and negative numbers (<2.75 and <0.33 respectively) utilising P-BEST decoding (Shental et al., 2020).
2.2.3.2. Tapestry
Tapestry pooling is a single round testing where each individual is assigned to three different groups. The algorithm takes ct values from the pooled RT-PCR tests as input and yields the output result. If the sample is tested positive, an estimated viral titre is also provided. An app has also been developed to facilitate implementation in clinical laboratory settings. The method has been shown to work in vitro with zero false positives and false negatives. Validation in clinical samples is ongoing (Ghosh et al., 2020b; Gopalkrishnan and Krishna, 2020).
3. Validation of Pooled Testing in clinical samples
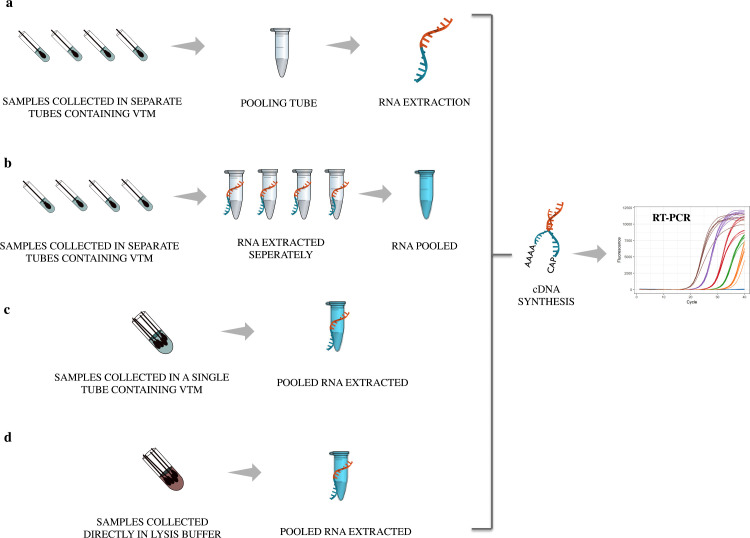
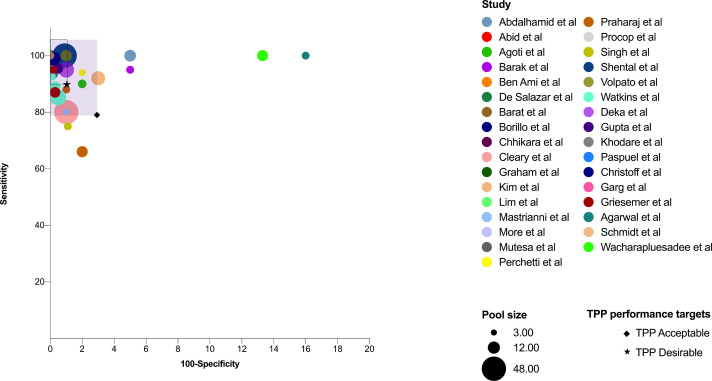
A foremost requisite of any modelling algorithm is the successful translation of theoretical simulations into clinically relevant applications. There are effective feasibility studies of various pooling methods in SARS-CoV-2 diagnosis. Studies have also compared pooled testing with naïve testing to ensure that detection accuracy is not compromised. Table 1 summarizes different studies worldwide that have validated pooled testing in various pool sizes and prevalence rates. Four different pooling strategies have been employed so far in clinical setups (Fig. 3 ). The most common method is to collect the samples and pool them just before extracting RNA. Few studies have reported pooling the extracted RNA and then proceeding for RT-PCR. Gupta et al have reported 95.4% sensitivity in detecting positive samples while pooling RNA keeping individual testing as reference (Gupta et al., 2020). Though the turn-around time is swift in this method, it does not save many resources. Swab pooling in a single tube at the time of collection is another practice. This is found to be as sensitive as individual testing (Christoff et al., 2021) but if a positive pool is encountered, samples have to be collected again. Hence double swabs have to be collected from every individual in the beginning (Chen et al., 2020). A couple of studies have directly collected the samples in lysis buffer instead of the viral transport media (VTM) to avoid dilution. This direct pooling also inactivates the virus easing the extraction process (Schmidt et al., 2020; Wacharapluesadee et al., 2020). Owing to the non-invasive nature of sample collection, the feasibility of utilising pooled saliva has also been tested and found that detection is possible without much compromise in sensitivity (Barat et al., 2020; Fogarty et al., 2020; Pasomsub et al., 2020). Studies have reported improved test turn- around time and conservation of laboratory resources while adapting pooled testing (Abdalhamid et al., 2020a; Agoti et al., 2020). The diagnostic accuracy while adapting pooled testing is found to be highly efficient in most of the studies (Fig. 4 ). The WHO has furnished an acceptable level of ≥80% sensitivity and ≥97% specificity for COVID-19 diagnostics in the Target Product Profile (TPP).
Table 1.
Summary of global pooled testing studies-Validation of pooled testing in different clinical settings. Pooling works with maximum efficiency in prevalence <3% with pool size <10)
| Study setting | Location | Sample size | Sample type | Prevalence (%) | Optimum Pool size | Reduction in tests (%) |
|---|---|---|---|---|---|---|
| Pooling samples before RNA extraction | ||||||
| (Abdalhamid et al., 2020b) | Nebraska, US | 60 | Nasopharyngeal swabs | 1 | 11 | 80 |
| (Abid et al., 2020) | Tunisia, North Africa | 800 | Nasopharyngeal swabs | <1 | 10 | 80 |
| (Agoti et al., 2020) | Kilifi, Kenya | 1500 | Nasopharyngeal swabs | <4 | 6 | 53.3 |
| (Barak et al., 2020) | Jerusalem, Israel | 133816 | Nasopharyngeal swabs | 1-6 | 5,8 | 76 |
| (Lohse et al., 2020) | Homburg, Germany |
1191 | Nasopharyngeal and Oropharyngeal swabs | 1.93 | 30 | 78 |
| (Hogan et al., 2020) | San Francisco Bay Area, California, US | 2888 | Nasopharyngeal and bronchoalveolar lavage swabs | 0.07 | 9, 10 | 77 |
| (Ben-Ami et al., 2020) | Jerusalem, Israel | 26576 | Nasopharyngeal swabs | 0.23 | 8 | 85 |
| (de Salazar et al., 2020) | Spain | 3519 | Nasopharyngeal swabs | 6.86 | 10 | 61.57 |
| (Barat et al., 2020) | Maryland, Washington D.C., US | 449 | Saliva | 5 |
5 | 40 |
| (Ho et al., 2021) | Hong Kong, China | 55 | Deep throat saliva | 0.39 | 5 | - |
| (Borillo et al., 2020) | California, US |
101 | Nasopharyngeal, midturbinate swabs | ≤10 | 4 |
- |
| (Chen et al., 2020) | Wuhan, China | 23 | Oropharyngeal swabs | - | 6,10 | - |
| (Chhikara et al., 2021) | Chandigarh, India | 500 | Nasopharyngeal swabs | <2 | 10 | 69 |
| (Chong et al., 2020) | Melbourne, Australia | 29000 | Nasopharyngeal swabs | <0.5 | 8 | 87.5 |
| (Cleary et al., 2020) | Massachusetts, US | 960 | Nasopharyngeal swabs | 3.1 | 48 | 65 |
| (Das et al., 2020) | Maryland, US | 7000 | Nasal mid-turbinate and nasopharyngeal swabs | 0.11 | 10 | 89 |
| (Graham et al., 2021) | Melbourne, Australia | 31 | Nasopharyngeal swabs | <5 | 4,6 | >50 |
| (Hirotsu et al., 2020) | Tokyo, Japan | 555 | Nasopharyngeal swabs | 3.6 | 5,10 |
47 |
| (Kim et al., 2020) | Seoul, Jeonju, South Korea | 350 | Nasopharyngeal and oropharyngeal swabs | - | 2-16 | - |
| (Li et al., 2020) | Hainan, China | 944 | Nasopharyngeal swabs | Airport | 10 | 87.6 |
| (Lim et al., 2020) | Malaysia | 1745 | Nasopharyngeal and oropharyngeal swabs | <5 | 10 | 57 |
| (Mastrianni et al., 2020) | New York, US | 530 | Nasopharyngeal swabs | 1-2 | 3 | 67 |
| (Mohanty et al., 2020) | Odisha, India | 7228 | Nasopharyngeal and oropharyngeal swabs | 3.5 | 4 | 64.2 |
| (More et al., 2021) | Oklahoma, US | 630 | Nasopharyngeal swabs | <8 | 5 | 52.5 |
| 10 | 45 | |||||
| (Mutesa et al., 2021) | Rwanda, East Africa | 1280 | Oropharyngeal swabs | 2 | 20 | - |
| (Perchetti et al., 2020) | Seattle, US | 160 | Nasopharyngeal swabs | <8 | 4 | - |
| (Praharaj et al., 2020) | India | 1000 | Nasopharyngeal and oropharyngeal swabs | - | 5 10 |
- |
| (Procop et al., 2021) | Ohio, US | 20 | Nasopharyngeal swabs | 0.5 | 10 | |
| (Salimnia et al., 2021) | Detroit, US | 15 | Nasopharyngeal swabs | 2 | 6 | 70 |
| (Singh et al., 2020) | Madhya Pradesh, India | 545 | Nasopharyngeal and oropharyngeal swabs | 4.8 | 5 | 70 |
| (Shental et al., 2020) | Beer-Sheva, Israel | 1115 | Nasopharyngeal and oropharyngeal swabs | 1 | 48 | 87.1 |
| (Thanh et al., 2021) | Da Nang, Vietnam | 96123 | Nasopharyngeal swabs | <1 | 5 | 77 |
| (Torres et al., 2020) | Valencia, Spain | 40 | Nasopharyngeal swabs | - | 5, 10 | - |
| (Volpato et al., 2020) | Porto Alegre, Brazil | 220 | Nasopharyngeal and oropharyngeal swabs | - | 10 | - |
| (Wang et al., 2020) | San Francisco Bay, California, US | 1648 | Nasopharyngeal and oropharyngeal swabs | 19.1 | 4, 8 |
47 |
| (Watkins et al., 2020) | New Haven, US | Saliva | >3 <1 |
5, 10, 20 | - | |
| (Yelin et al., 2020) | Haifa, Israel | 388 | Nasopharyngeal and oropharyngeal swabs | - | 32 | - |
| (Denny et al., 2020) | Duke University, Durham, North Carolina | 10265 | Nasopharyngeal swabs | ≤0.1 | 5 | 90 |
| (Schneitler et al., 2020) | Homburg, Germany | 25978 | Nasopharyngeal and oropharyngeal swabs | 0.9 | 10 | 91 |
| Pooling extracted RNA | ||||||
| (Pasomsub et al., 2020) | Bangkok, Thailand | 200 | Saliva | 9 | 5 | 47.5 |
| (Deka et al., 2020) | Uttarakhand, India | 102 | - | 4.1 | 20 | 40 |
| (Farfan et al., 2020) | Chile, South America | 63 | Nasopharyngeal swabs | 10 | 5 | - |
| (Gupta et al., 2020) | New Delhi, India | 280 | Nasopharyngeal and oropharyngeal swabs | - |
8 |
85 |
| (Eis-Hübinger et al., 2020) | Freiburg, Bonn, Leipzig, Regensburg, Frankfurt (Germany) | 700 | Pharyngeal swabs, sputum, broncho-alveolar lavage fluid | - | 10 | 99.89 |
| (Cabrera Alvargonzalez et al., 2020) | Spain | 100 60 |
Nasopharyngeal swabs | 2 1.7 |
20 | 77 |
| Sub Pool 5 | 80 | |||||
| (Khodare et al., 2020) | New Delhi, India |
55 | Nasopharyngeal and oropharyngeal swabs | 4 | 6 | 63 |
| (Freire-Paspuel et al., 2020) | Galapagos, Equador | 114 | Nasopharyngeal swabs | <5 | 3 | - |
| Pooling at the time of collection in a single VTM tube | ||||||
| (Christoff et al., 2021) | Brazil | 19535 | Nasopharyngeal swabs | 1.26 | 16 | 77 |
| (Alcoba-Florez et al., 2020) | Tenerif, Spain | 4475 | Nasopharyngeal swabs | 5 | 5 | 62 |
| (Garg et al., 2020) | Lucknow, India | 19570 | Nasopharyngeal and Oropharyngeal swabs | <5 | 5 | 76 |
| 10 | 93 | |||||
| (Griesemer et al., 2021) | New York, US | 20 | Upper respiratory swab | <1.5 | 5, 9 | 75 |
| (Agarwal et al., 2021) | Delhi, India |
230 | Nasopharyngeal swabs | - | 5 | 70 |
| Pooling directly into lysis buffer | ||||||
| (Schmidt et al., 2020) | Frankfort, Germany | 100 | Nasopharyngeal and oropharyngeal swabs | 2 | 5, 10 | 40 |
| (Wacharapluesadee et al., 2020) | Bangkok, Thailand | 99 | Nasopharyngeal and oropharyngeal swabs | 0.1-10 | 10 | 80 |
‘-’ indicates that studies that did not report prevalence rate at the time of testing and the resources saved
Fig. 3.
Pooling methods employed in clinical setups: (A) Sample pooling before RNA extraction (B) Pooling of extracted RNA (C) Sample pooling at the time of collection (D) Sample collected directly into the lysis buffer; VTM= viral transport media.
Fig. 4.
Diagnostic accuracy utilising pooled testing strategy: The size of the bubble depicts pool size, while the different colours code for different studies. The purple-shaded area represents areas with sensitivity and specificity combinations that meet the acceptable levels of Target Product Profile (TPP) put forth by WHO for COVID-19 diagnostics. The dashed square box within the purple-shaded area portrays the desirable level. Study bubbles clustered within the acceptable limits infer efficient diagnostic capability in detecting SARS-CoV-2 by pooling samples. (Colour version of figure is available online)
Other works over the last few months have suggested refined models, from a more theoretical viewpoint with real world data as example. In a 2% prevalence rate, a matrix- based 96 well pooling is predicted to reduce tests up to four-fold while an eight-fold reduction is expected at 0.5% prevalence (Sinnott-Armstrong et al., 2020). A simulation study based on the available positivity rate data proposes an optimum pool size of up to 32 for a population of 10 000 (Bukhari et al., 2020). With the underlying prevalence rate of 2% in South Korea, sample stratification in pools up to 16 using a multi- stage scheme allows a five -fold efficiency gain relative to individual testing (Eberhardt et al., n.d.). Other simulation models and algorithms have also been proposed (Chow and Chow, 2021; Nalbantoglu, 2020; Pilcher et al., 2005; Polage et al., 2020; Vukičević and Polašek, 2020).
4. Discussion
The diagnostic screening of SARS-CoV-2 can be performed in a lesser time span by pooling samples. It is also evident that testing in pools substantially reduces the number of reagents, labour costs and resources. However, pooled testing is efficient only in low prevalence settings with least number of positive pools. Larger number of positive pools might lead to exhaustive deconvolution for retesting individual samples. Though several novel algorithms have been simulated as a strategy for pooled testing, Dorfman's Pooling method with certain improvements remain the gold standard for pooled testing. A major challenge with pooled testing is the increase in false negative rates since grouping multiple negative specimens with few positives could lead to undetectable levels of the viral load. But studies investigating the accuracy of pooled testing in wide ranges of dilutions have revealed false-negative rates below 10%, suggesting that the dilution introduces minimal error (Bateman et al., 2020; Smalley et al., 2021; Yelin et al., 2020). The general documented challenges during individual testing include low viral load in some patients, errors during RT-PCR testing, and RNA degradation due to improper handling. These effects might be augmented in pooled testing procedures, so it is even more crucial to carefully monitor the testing process. Further inevitable practical difficulties are time constraints, sample conservations for multi-stage pooling, tracking and retesting individuals of positive pools. Many studies have reported models based on the assumption of the presence of one positive sample per pool. It is important to tailor algorithms for more than one positive case. These are some points to ponder to implement pooled testing strategy over the ongoing individual testing method.
5. Practical guidelines for employing Pooled Testing for COVID-19 diagnosis
With gradual relaxation in lockdown rules, reopening of Universities and schools and resumption of transport, it is critical to ramp up the numbers of testing of asymptomatic population. By comparing various pooled testing strategies implemented so far, we put forth the optimal choice for various scenarios.
Select Pooling method
For community surveillance in resource limited settings with inadequate testing facilities, a door-to-door approach is suitable. Asymptomatic people in households and localities can be tested using this approach. Individual samples from houses in close vicinity can be pooled together considering the spatial clustering of the outbreak in a particular area. This can be organized by mapping the region, plotting the households in the locale and pooling samples from adjoining houses.
Binary splitting algorithms can be employed to test market vendors, bank and supermarket agents and other frontline workers who were actively exposed during the lockdown enforced by the Government.
Routine screening of health care workers and hospital staff can be done with Matrix Pooling since it involves a single round of tests.
When the testing speed is a primary factor, it may be feasible to employ Multistage testing as it enables us to perform parallel group tests
Hypercube algorithm can find application in regular testing of players and staff of sports teams.
Estimate the Positivity rate
At the beginning of each test, it is vital to estimate the positivity rate. Based on the empirical results received from a specific laboratory in a population, the rate of positive tests should be updated on a daily basis.
Define the maximal Pool size
Medical councils from across the globe have recommended to use a pool size of 5. But in areas with prevalence lesser than 1%, the pool size can go up to 10 based on clinical validation studies around the world. It is always important to restrict pooled testing approach for prevalence up to 5%. As prevalence increases, the efficiency of the pooled testing flattens.
Web based applications like shiny app (https://bilder.shinyapps.io/PooledTesting/) (Hou et al., 2020) can also be used to determine the efficient pool size based on parameters like prevalence, sensitivity and specificity of the assay.
Proceed with RT-PCR of each pool, neglect negative pools, deconvolute positive pools.
6. Conclusion
Testing in pools has the potential to substantially expand the testing capacity and lower the limitations present in the current diagnostic tests for SARS-CoV-2. Studies have reported considerable increase in testing throughput and detection efficiency with consistent levels of sensitivity and specificity. Thus, pooled testing is a feasible method to circumvent the bottlenecks in the existing testing methods of COVID-19. Given the successful implementation of this strategy in different countries, the approach may find applicability in routine community surveys.
Authors’ Contribution
Evangeline Ann Daniel: Conceptualization, Design and Methodology Validation, Data collection, curation and Analysis, Writing Original Draft
Bennett Henzeler Esakialraj L: Conceptualization, Design and Methodology Validation, Data collection, curation and Analysis, Writing Original Draft, Visualization
Anbalagan S: Data collection, curation and Analysis
Kannan Muthuramalingam: Data collection, curation and Analysis
Ramesh Karunaianantham: Data collection, curation and Analysis
Lucia Precilla Karunakaran: Data collection, curation and Analysis; Reviewing and editing draft
Manohar Nesakumar: Data collection, curation and Analysis; Reviewing and editing draft
Murugesan Selvachithiram: Data curation and analysis
Sathyamurthi Pattabiraman: Data curation and analysis
Sudhakar Natarajan: Data curation and analysis; Reviewing and editing draft
Srikanth Prasad Tripathy: Supervision; Reviewing and editing draft
Luke Elizabeth Hanna: Conceptualization; Supervision; Reviewing and editing draft
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115432.
Appendix. Supplementary materials
References
- Abdalhamid B, Bilder CR, Garrett JL, Iwen PC. Cost effectiveness of sample pooling to test for SARS-CoV-2. J Infect Dev Ctries. 2020;14:1136–1137. doi: 10.3855/jidc.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid S, Ferjani S, El Moussi A, Ferjani A, Nasr M, Landolsi I, et al. Assessment of sample pooling for SARS-CoV-2 molecular testing for screening of asymptomatic persons in Tunisia. Diagn Microbiol Infect Dis. 2020;98 doi: 10.1016/j.diagmicrobio.2020.115125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Gupta E, Dubey S, Padhi A, Khodare A, Kumar G, et al. Pooled nasopharyngeal swab collection in a single vial for the diagnosis of SARS CoV-2 infection: An effective cost saving method. Indian J Med Microbiol. 2021 doi: 10.1016/j.ijmmb.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoti CN, Mutunga M, Lambisia AW, Kimani D, Cheruiyot R, Kiyuka P, et al. Pooled testing conserves SARS-CoV-2 laboratory resources and improves test turn-around time: experience on the Kenyan Coast. Wellcome Open Res. 2020;5:186. doi: 10.12688/wellcomeopenres.16113.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoba-Florez J, Gil-Campesino H, García-Martínez de Artola D, Díez-Gil O, Valenzuela-Fernández A, González-Montelongo R, et al. Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int J Infect Dis. 2020;103:19–22. doi: 10.1016/j.ijid.2020.11.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón-Caqueo D, Fernández-Salinas J, Laroze D. Optimization of group size in pool testing strategy for SARS-CoV-2: A simple mathematical model. J Med Virol. 2020;92:1988–1994. doi: 10.1002/jmv.25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak N, Ben-Ami R, Sido T, Perri A, Shtoyer A, Rivkin M, et al. Lessons from applied large-scale pooling of 133,816 SARS-CoV-2 RT-PCR tests. Infect Dis (except HIV/AIDS) 2020 doi: 10.1101/2020.10.16.20213405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat B, Das S, De Giorgi V, Henderson DK, Kopka S, Lau AF, et al. Pooled Saliva Specimens for SARS-CoV-2 Testing. J Clin Microbiol. 2020 doi: 10.1128/JCM.02486-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman AC, Mueller S, Guenther K, Shult P. Assessing the dilution effect of specimen pooling on the sensitivity of SARS-CoV-2 PCR tests. J Med Virol. 2020 doi: 10.1002/jmv.26519. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R, Klochendler A, Seidel M, Sido T, Gurel-Gurevich O, Yassour M, et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect. 2020;26:1248–1253. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borillo GA, Kagan RM, Baumann RE, Fainstein BM, Umaru L, Li H-R, et al. Pooling of upper respiratory specimens using a SARS-CoV-2 Real-time RT-PCR assay authorized for emergency use in low-prevalence populations for high-throughput testing. Open Forum Infect Dis. 2020;7 doi: 10.1093/ofid/ofaa466. ofaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O. COVID-19 prevalence estimation by random sampling in population - optimal sample pooling under varying assumptions about true prevalence. BMC Med Res Methodol. 2020;20:196. doi: 10.1186/s12874-020-01081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SUK, Khalid SS, Syed A, Shah SSH. Smart pooled sample testing for COVID-19: A possible solution for sparsity of test kits. MedRxiv. 2020 doi: 10.1101/2020.04.21.20044594. 2020.04.21.20044594. [DOI] [Google Scholar]
- Cabrera Alvargonzalez JJ, Rey Cao S, Pérez Castro S, Martinez Lamas L, Cores Calvo O, Torres Piñon J, et al. Pooling for SARS-CoV-2 control in care institutions. BMC Infect Dis. 2020;20:745. doi: 10.1186/s12879-020-05446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Geng Z, Wang J, Liuchang W, Huang D, Xu Y, et al. Comparing two sample pooling strategies for SARS-CoV-2 RNA detection for efficient screening of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26632. [DOI] [PubMed] [Google Scholar]
- Chhikara K, Kanta P, Ghosh A, Prakash RC, Goyal K, Singh MP. Validation of SARS CoV-2 detection by real-time PCR in matched pooled and deconvoluted clinical samples before and after nucleic acid extraction: a study in tertiary care hospital of North India. Diagn Microbiol Infect Dis. 2021;99 doi: 10.1016/j.diagmicrobio.2020.115206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong BSW, Tran T, Druce J, Ballard SA, Simpson JA, Catton M. Sample pooling is a viable strategy for SARS-CoV-2 detection in low-prevalence settings. Pathology. 2020;52:796–800. doi: 10.1016/j.pathol.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WK, Chow CL. A discussion on implementing pooling detection tests of novel coronavirus (SARS-CoV-2) for a large population. Epidemiol Infect. 2021;149:e17. doi: 10.1017/S0950268820003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff AP, Cruz GNF, Sereia AFR, Boberg DR, de Bastiani DC, Yamanaka LE, et al. Swab pooling: A new method for large-scale RT-qPCR screening of SARS-CoV-2 avoiding sample dilution. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary B, Hay JA, Blumenstiel B, Gabriel S, Regev A, Mina MJ. Efficient prevalence estimation and infected sample identification with group testing for SARS-CoV-2. MedRxiv. 2020 doi: 10.1101/2020.05.01.20086801. [DOI] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio Medica Atenei Parmensis. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie MJ, McNiven M, Yee T, Schiemer U, Bowden FJ. Pooling of clinical specimens prior to testing for Chlamydia trachomatis by PCR is accurate and cost saving. J Clin Microbiol. 2004;42:4866–4867. doi: 10.1128/JCM.42.10.4866-4867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Lau AF, Youn J-H, Khil PP, Zelazny AM, Frank KM. Pooled testing for surveillance of SARS-CoV-2 in asymptomatic individuals. J Clin Virol. 2020;132 doi: 10.1016/j.jcv.2020.104619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert A, Bärnighausen T, Kyei NN. Simulation of pooled-sample analysis strategies for COVID-19 mass testing. Bull World Health Organ. 2020;98:590–598. doi: 10.2471/BLT.20.257188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka S, Kalita D, Mangla A, Shankar R. Analysis of multi-sample pools in the detection of SARS-CoV-2 RNA for mass screening: An Indian perspective. Indian J Med Microbiol. 2020;38:451–456. doi: 10.4103/ijmm.IJMM_20_273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny TN, Andrews L, Bonsignori M, Cavanaugh K, Datto MB, Deckard A, et al. Implementation of a pooled surveillance testing program for asymptomatic SARS-CoV-2 infections on a college campus - duke university, Durham, North Carolina, August 2-October 11, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1743–1747. doi: 10.15585/mmwr.mm6946e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R. The detection of defective members of large populations. Ann Math Statist. 1943;14:436–440. doi: 10.1214/aoms/1177731363. [DOI] [Google Scholar]
- Eberhardt JN, Breuckmann NP, Eberhardt CS. Multi-stage group testing improves efficiency of large-scale COVID-19 screening. J Clin Virol n.d. https://doi.org/10.1016/j.jcv.2020.104382. [DOI] [PMC free article] [PubMed]
- Edouard S, Prudent E, Gautret P, Memish ZA, Raoult D. Cost-effective pooling of DNA from nasopharyngeal swab samples for large-scale detection of bacteria by real-time PCR. J Clin Microbiol. 2015;53:1002–1004. doi: 10.1128/JCM.03609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis-Hübinger AM, Hönemann M, Wenzel JJ, Berger A, Widera M, Schmidt B, et al. Ad hoc laboratory-based surveillance of SARS-CoV-2 by real-time RT-PCR using minipools of RNA prepared from routine respiratory samples. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel JC, Bassett MT, Smith HJ, Jacobs JA. Pooling of sera for human immunodeficiency virus (HIV) testing: an economical method for use in developing countries. J Clin Pathol. 1988;41:582–585. doi: 10.1136/jcp.41.5.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan MJ, Torres JP, O'Ryan M, Olivares M, Gallardo P, Lastra J, et al. Optimizing RT-PCR detection of SARS-CoV-2 for developing countries using pool testing. Rev Chilena Infectol. 2020;37:276–280. doi: 10.4067/s0716-10182020000300276. [DOI] [PubMed] [Google Scholar]
- Fogarty A, Joseph A, Shaw D. Pooled saliva samples for COVID-19 surveillance programme. Lancet Respir Med. 2020;8:1078–1080. doi: 10.1016/S2213-2600(20)30444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Paspuel B, Vega-Mariño P, Velez A, Cruz M, Garcia-Bereguiain MA. Sample pooling of RNA extracts to speed up SARS-CoV-2 diagnosis using CDC FDA EUA RT-qPCR kit. Virus Res. 2020;290 doi: 10.1016/j.virusres.2020.198173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajpal Y, Appadoo SS, Shi V, Liao Y. Optimal multi-stage group partition for efficient coronavirus screening. Rochester, NY. Social Sci Res Network. 2020 doi: 10.2139/ssrn.3591961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Ghoshal U, Patel SS, Singh DV, Arya AK, Vasanth S, et al. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. 2020 doi: 10.1002/jmv.26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Agarwal R, Rehan MA, Pathak S, Agrawal P, Gupta Y, et al. A compressed sensing approach to group-testing for COVID-19 detection. ArXiv:200507895 [q-Bio] 2020 [Google Scholar]
- Ghosh S, Rajwade A, Srikar Krishna, Gopalkrishnan N, Schaus TE, Chakravarthy A, et al. Tapestry: A single-round smart pooling technique for COVID-19 testing. MedRxiv. 2020 doi: 10.1101/2020.04.23.20077727. 2020.04.23.20077727. [DOI] [Google Scholar]
- Gopalkrishnan M, Krishna S. Pooling Samples to Increase SARS-CoV-2 Testing. J Indian Inst Sci. 2020;100:787–792. doi: 10.1007/s41745-020-00204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M, Williams E, Isles N, Buadromo E, Toatu T, Druce J, et al. Sample pooling on the Cepheid Xpert Xpress SARS-CoV-2 assay. Diagn Microbiol Infect Dis. 2021;99 doi: 10.1016/j.diagmicrobio.2020.115238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesemer SB, Slyke GV, George KS. Assessment of sample pooling for clinical SARS-CoV-2 testing. J Clin Microbiol. 2021 doi: 10.1128/JCM.01261-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Yao L, Meng X, Graff JC, Thomason D, Li J, et al. A cost-effective plan for global testing - an infection rate stratified, algorithm guided, multiple-level, continuously pooled testing strategy. Sci Total Environ. 2020;765 doi: 10.1016/j.scitotenv.2020.144251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta E, Padhi A, Khodare A, Agarwal R, Ramachandran K, Mehta V, et al. Pooled RNA sample reverse transcriptase real time PCR assay for SARS CoV-2 infection: A reliable, faster and economical method. PLOS ONE. 2020;15 doi: 10.1371/journal.pone.0236859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel R, Thurner S. Boosting test-efficiency by pooled testing for SARS-CoV-2—Formula for optimal pool size. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci Rep. 2020;10:18899. doi: 10.1038/s41598-020-76043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YI, Wong AH, Tang KPS, Wong RCW, Leung ECM, Lai RWM. Comparison of three commercial SARS-CoV-2 assays for pooled testing of deep throat saliva for surveillance of patients attending general outpatient clinics. J Med Virol. 2021 doi: 10.1002/jmv.26764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan CA, Sahoo MK, Pinsky BA. Sample Pooling as a Strategy to Detect Community Transmission of SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Tebbs JM, Wang D, McMahan CS, Bilder CR. Array testing for multiplex assays. Biostatistics. 2020;21:417–431. doi: 10.1093/biostatistics/kxy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourfar MK, Themann A, Eickmann M, Puthavathana P, Laue T, Seifried E, et al. Blood Screening Influenza. Emerg Infect Dis. 2007;13:1081–1083. doi: 10.3201/eid1307.060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodare A, Padhi A, Gupta E, Agarwal R, Dubey S, Sarin SK. Optimal size of sample pooling for RNA pool testing: An avant-garde for scaling up severe acute respiratory syndrome coronavirus-2 testing. Indian J Med Microbiol. 2020;38:18–23. doi: 10.4103/ijmm.IJMM_20_260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee J, Sung H, Lee H, Han MG, Yoo CK, et al. Pooling upper respiratory specimens for rapid mass screening of COVID-19 by Real-Time RT-PCR. Emerg Infect Dis. 2020;26:2469–2472. doi: 10.3201/eid2610.201955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sun K, Persing DH, Tang Y-W, Shen D. Real-time Screening of Specimen Pools for Coronavirus Disease 2019 (COVID-19) Infection at Sanya Airport, Hainan Island, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Johari NA, Wong ST, Khaw LT, Tan BK, Chan KK, et al. A novel strategy for community screening of SARS-CoV-2 (COVID-19): Sample pooling method. PLOS ONE. 2020;15 doi: 10.1371/journal.pone.0238417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak E, Tu XM, Pagano M. Screening for the Presence of a Disease by Pooling Sera Samples. J Am Statis Associat. 1994;89:424–434. doi: 10.1080/01621459.1994.10476764. [DOI] [Google Scholar]
- Lohse S, Pfuhl T, Berkó-Göttel B, Rissland J, Geißler T, Gärtner B, et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. The Lancet Infectious Diseases. 2020;0 doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrianni D, Falivena R, Brooks T, McDermott B, Tan J, Vandell R, et al. Pooled testing for SARS-CoV-2 in hospitalized patients. J Hosp Med. 2020;15:538–539. doi: 10.12788/jhm.3501. [DOI] [PubMed] [Google Scholar]
- Mentus C, Romeo M, DiPaola C. Analysis and applications of adaptive group testing methods for COVID-19. MedRxiv. 2020 doi: 10.1101/2020.04.05.20050245. 2020.04.05.20050245. [DOI] [Google Scholar]
- Mohanty S, Ravindra A, Gupta K, Hallur V, Behera B, Mahaptra A, et al. Intricacies in characterizing positivity in pooled sample testing for SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.26618. [DOI] [PubMed] [Google Scholar]
- More S, Narayanan S, Patil G, Ghosh P, Pushparaj S, Cooper E, et al. Pooling of Nasopharyngeal (NP) swab samples to overcome global shortage of rRT-PCR COVID-19 Test Kits. J Clin Microbiol. 2021 doi: 10.1128/JCM.01295-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutesa L, Ndishimye P, Butera Y, Souopgui J, Uwineza A, Rutayisire R, et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. 2021;589:276–280. doi: 10.1038/s41586-020-2885-5. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu OU. Group testing performance evaluation for SARS-CoV-2 massive scale screening and testing. BMC Med Res Methodol. 2020;20:176. doi: 10.1186/s12874-020-01048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasomsub E, Watcharananan SP, Watthanachockchai T, Rakmanee K, Tassaneetrithep B, Kiertiburanakul S, et al. Saliva sample pooling for the detection of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchetti GA, Sullivan K-W, Pepper G, Huang M-L, Breit N, Mathias P, et al. Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatarfod RM, Sudbury A. The use of a square array scheme in blood testing. Stat Med. 1994;13:2337–2343. doi: 10.1002/sim.4780132205. [DOI] [PubMed] [Google Scholar]
- Pilcher CD, Fiscus SA, Nguyen TQ, Foust E, Wolf L, Williams D, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352:1873. doi: 10.1056/NEJMoa042291. –83. [DOI] [PubMed] [Google Scholar]
- Polage CR, Lee MJ, Hubbard C, Rehder C, Cardona D, Denny T, et al. Assessment of an online tool to simulate the effect of pooled testing for SARS-CoV-2 detection in asymptomatic and symptomatic populations. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.31517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praharaj I, Jain A, Singh M, Balakrishnan A, Dhodapkar R, Borkakoty B, et al. Pooled testing for COVID-19 diagnosis by real-time RT-PCR: A multi-site comparative evaluation of 5- & 10-sample pooling. Indian J Med Res. 2020;152:88–94. doi: 10.4103/ijmr.IJMR_2304_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procop GW, Tuohy M, Ramsey C, Rhoads DD, Rubin BP, Figler R. Asymptomatic patient testing after 10:1 pooling using the xpert xpress SARS-CoV-2 Assay. Am J Clin Pathol. 2021 doi: 10.1093/ajcp/aqaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai B, Shukla A, Choudhary G, Singh A. Pool testing for COVID-19: suitable splitting procedure and pool size for India. Disaster Med Public Health Prep. 2020:1–17. doi: 10.1017/dmp.2020.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regen F, Eren N, Heuser I, Hellmann-Regen J. A simple approach to optimum pool size for pooled SARS-CoV-2 testing. Int J Infect Dis. 2020;100:324–326. doi: 10.1016/j.ijid.2020.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Eng J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Salazar A, Aguilera A, Trastoy R, Fuentes A, Alados JC, Causse M, et al. Sample pooling for SARS-CoV-2 RT-PCR screening. Clin Microbiol Infect. 2020;26:1687.e1–1687.e5. doi: 10.1016/j.cmi.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimnia H, Mitchell R, Gundel A, Cambell A, Gammou F, Chopra T, et al. Pooling samples: a testing option for SARS-CoV-2 during a supply shortage. Diagn Microbiol Infect Dis. 2021;99 doi: 10.1016/j.diagmicrobio.2020.115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Hoehl S, Berger A, Zeichhardt H, Hourfar K, Ciesek S, et al. Novel multiple swab method enables high efficiency in SARS-CoV-2 screenings without loss of sensitivity for screening of a complete population. Transfusion. 2020;60:2441–2447. doi: 10.1111/trf.15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitler S, Jung P, Bub F, Alhussein F, Benthien S, Berger FK, et al. Simple questionnaires to improve pooling strategies for SARS-CoV-2 laboratory testing. Ann Glob Health. 2020;86:148. doi: 10.5334/aogh.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shental N, Levy S, Wuvshet V, Skorniakov S, Shalem B, Ottolenghi A, et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Science Advances. 2020;6:eabc5961. doi: 10.1126/sciadv.abc5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock M, Zetola NM, Klausner JD. Routine detection of acute HIV infection through RNA pooling: survey of current practice in the United States. Sex Transm Dis. 2007;34:314–316. doi: 10.1097/01.olq.0000263262.00273.9c. [DOI] [PubMed] [Google Scholar]
- Singer RS, Cooke CL, Maddox CW, Isaacson RE, Wallace RL. Use of pooled samples for the detection of Salmonella in feces by polymerase chain reaction. J Vet Diagn Invest. 2006;18:319–325. doi: 10.1177/104063870601800401. [DOI] [PubMed] [Google Scholar]
- Singh AK, Nema RK, Joshi A, Shankar P, Nema S, Raghuwanshi A, et al. Evaluation of pooled sample analysis strategy in expediting case detection in areas with emerging outbreaks of COVID-19: A pilot study. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott-Armstrong N, Klein D, Hickey B. Evaluation of group testing for SARS-CoV-2 RNA. MedRxiv. 2020 doi: 10.1101/2020.03.27.20043968. 2020.03.27.20043968. [DOI] [Google Scholar]
- Smalley DL, Cisarik PM, Grantham J, Cloud W, Neil RB, DePriest P. Impact of pool testing in detection of asymptomatic patients with COVID-19. Lab Med. 2021;52:e15–e16. doi: 10.1093/labmed/lmaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, Wright DJ, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- Takyi-Williams J. Household representative sample strategy for COVID-19 large-scale population screening. Med Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JG, Omar A, Lee WB, Wong MS. Considerations for group testing: a practical approach for the clinical laboratory. Clin Biochem Rev. 2020;41:79–92. doi: 10.33176/AACB-20-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Täufer M. Rapid, large-scale, and effective detection of COVID-19 via non-adaptive testing. J Theor Biol. 2020;506 doi: 10.1016/j.jtbi.2020.110450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Juliano JJ, Trottman PA, Griffin JB, Landis SH, Kitsa P, et al. High-throughput pooling and real-time PCR-based strategy for malaria detection. J Clin Microbiol. 2010;48:512–519. doi: 10.1128/JCM.01800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh TT, Nhan NTT, Mai HK, Trieu NB, Huy LX, Thuy HTT, et al. The application of sample pooling for mass screening of SARS-CoV-2 in an outbreak of COVID-19 in Vietnam. Am J Trop Med Hyg. 2021;1 doi: 10.4269/ajtmh.20-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I, Albert E, Navarro D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J Med Virol. 2020;92:2306–2307. doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van TT, Miller J, Warshauer DM, Reisdorf E, Jernigan D, Humes R, et al. Pooling nasopharyngeal/throat swab specimens to increase testing capacity for influenza viruses by PCR. J Clin Microbiol. 2012;50:891–896. doi: 10.1128/JCM.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpato F, Lima-Morales D, Wink PL, Willig J, de-Paris F, Ashton-Prolla P, et al. Pooling of samples to optimize SARS-CoV-2 diagnosis by RT-qPCR: comparative analysis of two protocols. Eur J Clin Microbiol Infect Dis. 2020 doi: 10.1007/s10096-020-04071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukičević D, Polašek O. Optimizing the diagnostic capacity for COVID-19 PCR testing for low resource and high demand settings: The development of information-dependent pooling protocol. J Glob Health. 2020;10 doi: 10.7189/jogh.10.020515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Kaewpom T, Ampoot W, Ghai S, Khamhang W, Worachotsueptrakun K, et al. Evaluating the efficiency of specimen pooling for PCR-based detection of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AE, Fenichel EP, Weinberger DM, Vogels CBF, Brackney DE, Casanovas-Massana A, et al. Pooling saliva to increase SARS-CoV-2 testing capacity. MedRxiv. 2020 doi: 10.1101/2020.09.02.20183830. [DOI] [Google Scholar]
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirusdisease 2019 (COVID-19) outbreak in China: Summary of a Report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Ye F, Xu S, Rong Z, Xu R, Liu X, Deng P, et al. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. International Journal of Infectious Diseases. 2020;94:133–138. doi: 10.1016/j.ijid.2020.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin I, Aharony N, Tamar ES, Argoetti A, Messer E, Berenbaum D, et al. Evaluation of COVID-19 RT-qPCR test in multi sample pools. Clin Infect Dis. 2020;71:2073–2078. doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Mudumbai R, Xu W. Low-cost and high-throughput testing of COVID-19 viruses and antibodies via compressed sensing: system concepts and computational experiments. ArXiv:200405759 [Cs, Eess, Math, q-Bio] 2020 [Google Scholar]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.