Abstract
Cancer evolves through the accumulation of somatic mutations over time. Although several methods have been developed to characterize mutational processes in cancers, these have not been specifically designed to identify mutational patterns that predict patient prognosis. Here we present CLICnet, a method that utilizes mutational data to cluster patients by survival rate. CLICnet employs Restricted Boltzmann Machines, a type of generative neural network, which allows for the capture of complex mutational patterns associated with patient survival in different cancer types. For some cancer types, clustering produced by CLICnet also predicts benefit from anti-PD1 immune checkpoint blockade therapy, whereas for other cancer types, the mutational processes associated with survival are different from those associated with the improved anti-PD1 survival benefit. Thus, CLICnet has the ability to systematically identify and catalogue combinations of mutations that predict cancer survival, unveiling intricate associations between mutations, survival, and immunotherapy benefit.
INTRODUCTION
Cancer progression is a stochastic evolutionary process in which cells acquire somatic mutations that allow them to evade growth suppression, resist cell death signals, and enhance replication and immune suppression (1,2). Most cancers are caused by multiple somatic mutations that together lead to the cancer phenotype (1,3). The somatic mutations that cause cancer are often called driver mutations, or simply, drivers. The drivers can cause impairments in a variety of functional pathways, including DNA replication, DNA repair, cell cycle control, and programmed cell death (4,5). In addition, cancers are extremely heterogeneous, such that the driver mutations and affected genes vary greatly between patients, even within the same cancer type (6,7). Although some somatic mutations are indeed drivers that directly contribute to the cancer phenotype, the substantial majority are passengers, that is, mutations that are simply coincidentally present in tumors and have no discernible effect on the cancer phenotype (8,9,10), some of these mutations could result from DNA repair impairments in cancer. Thus, it is in general difficult to pinpoint mutations that are critical for tumor initiation and progression, and to identify clinically relevant combinations of mutations that could facilitate stratifying patients by survival rate and/or treatment response (11,12).
The rapid accumulation of cancer genomic data in recent years has enabled the creation of a comprehensive collection of somatic mutations in cancer and evaluation of their impact on tumor progression (13,14,15). In contrast to germline mutations, that is, predisposition variants detected in germline cells, somatic mutations that are the most common cause of cancer occur in diploid cells and are tissue-specific. To systematically characterize the mutational processes that promote cancer, mathematical methods have previously been used to decipher mutational signatures from somatic mutation catalogues (16). These approaches largely involve modelling specific mutation types in trinucleotides using Nonnegative Matrix Factorization (NMF) (16,17,18,19). Although these mutation signatures successfully characterize key mutational processes for numerous cancers (17,18,19), they are not optimized for the prediction of patient survival or treatment efficacy. The recent development of immune checkpoint blockade therapies, and particularly, anti-PD1 (programmed death-1) treatment has demonstrated durable responses in multiple cancer types, especially, melanoma, lung cancer and mismatch repair deficient gastrointestinal and endometrial cancers (20,21,22). However, not all patients respond to this treatment, which can incur severe side effects and costs (23,24), thus adding urgency to the need for the use of mutational data to predict treatment efficacy. Indeed, the first FDA-approved marker for anti-PD1 efficacy is based on high microsatellite instability (MSI-H) (25,26), which results from mismatch repair deficiency and is therefore linked to increased mutagenesis (27,28). More recently, high tumor mutational burden (TMB-H), has also been approved by the FDA as a marker for anti-PD1 efficacy based on similar research (29,30,31,32). However, the MSI-H marker is limited to gastrointestinal and endometrial cancers, where mismatch repair deficiency is observed almost exclusively (33). In addition, the predictive signal of TMB-H status can be confounded by disease subtype (34,35). When considered individually, some cancer types do not show association between TMB-H and survival with anti-PD1 immune checkpoint blockade treatment (31,34,35) although such association is strongly evident in non small cell lung cancers (30,34).
Several techniques have been developed to study the associations between cancer mutations and survival (36,37,38), and many studies have reported mutations in distinct genes that are associated with survival in particular cancer types. For example, mutations of TP53, KRAS and PIK3CA are associated with survival in colorectal cancers (39,40,41), mutations of BRCA1 and BRCA2 in breast and ovarian cancers (42,43,44), and mutations of BRCA2 in prostate cancers (45,46). ALK rearrangements are exploited for treatment and prognosis of non-small cell lung cancer (47), B2M mutations for multiple myeloma and leukemia prognoses (48), c-KIT mutations for gastrointestinal stromal tumors (49), BRAF V600E mutations are predictive of papillary thyroid cancer recurrence (50), and MYCN alterations are serves as a marker of spontaneous regression in neuroblastoma (51). In addition, some RNA based signatures are employed in the clinic, including the 21-gene signature to predict breast cancer recurrence (52), and a 17-gene signature to predict prostate cancer risk and recurrence (53,54). However, mutation-based studies usually focus on a single gene or cancer type, and do not include comprehensive analyses of potential gene interactions that could predict survival. Several methods have been developed to identify complex combinations of mutations that correspond to interaction networks (55,56,57) and/or can be used for cancer subtype clustering (9,58,59), but to our knowledge, these precise methods have not been directly harnessed towards survival prediction, despite the observations that subtype clustering often defines survival differences (60,61). We are unaware of efforts to systematically identify and catalogue combinations of mutations that would predict survival across different cancer types. There is therefore a pressing need for an approach that could uncover combinations of mutations that enable clustering cancer patients based on survival rates derived from mutational patterns, and could be used to systematically identify mutational patterns that predict survival in different cancer types.
Here, we present CLICnet (http://clicnet.pythonanywhere.com/), a computational method for Clinical Clustering of Cancer patients using neural NETworks, which includes a collection of independent predictors trained for different cancer types. To our knowledge, CLICnet is the first method to systematically identify and catalogue patterns of somatic mutations that are significantly predictive of survival in different cancer types, based on subsets of genes from the MSK-IMPACT panel. CLICnet relies on Restricted Boltzmann Machine (RBM) (62,63) neural networks to cluster cancer patients into high and low risk clusters, based on mutations in cancer type-specific sets of genes. We analyzed 10,141 tumors samples that represent 15 cancer types, from the Cancer Genome Atlas (TCGA) (64,65) and Memorial Sloan Kettering Cancer Center (MSKCC) cohorts (66). CLICnet was trained and validated on the TCGA and MSKCC mutation and survival data, respectively, for each cancer type, to cluster patients into two clusters with significantly different survival rates, based on mutation patterns. We catalogued the top 5 combinations of mutations for each cancer that are predictive of patient survival. In some cancer types, the CLICnet clusters were also predictive of the anti-PD1 immune checkpoint blockade therapy benefit. Thus, CLICnet allows the identification of combinations of mutations that predict survival, provides a catalogue of such combinations across different cancer types, and pinpoints mutation combinations that predict survival under anti-PD1 treatment in three cancer types.
MATERIALS AND METHODS
Training and validation sample collection and preprocessing
The TCGA (65) mutation data was downloaded from the Xena browser (67,68) and the corresponding clinical data was obtained (69); the two data sets were merged using the patient barcode. Survival was set to the maximum value between the ‘last_contact_days_to’ and ‘death_days_to’ columns. The MSKCC mutation and clinical data (66) were downloaded from the cBioPortal (70,71) (https://www.cbioportal.org) and merged using the patient ID. Survival was set to the ‘OS_MONTHS’ column. A large proportion of the samples in the MSK-IMPACT cohort was derived from distal metastases in different tissues (55%), which can bias the analysis, especially because the TCGA training data considered did not include samples from such sites. therefore, only the primary site tumor samples in the MSK-IMPACT cohort were included, by filtering for samples for which the ‘SAMPLE_TYPE’ column was set to ‘Primary’.
The analyzed cancer types, which are included in both datasets are detailed in Figure 1 and Supplementary Table S4. Given the differences in the assignment of cancer types and the different cancer types that are included in each dataset, the samples were aggregated into a total of 15 types of cancer. We filtered the samples to retain those with one of the 15 cancer types included in both datasets. Colorectal adenocarcinomas (COAD, READ) were aggregated because they have similar mutations and clinical characteristics. Non-small cell lung cancers (LUSC, LUAD), and renal cell carcinomas (KIRC,KIRP) were aggregated by the tissue of origin to increase the sample size, as it has been done in previous studies (72,73,74,75,76). We verified that the identified CLICnet gene sets were also associated with the outcome independently in each of these cancer types (Supplementary Figure S6). In addition, to evaluate the predictive ability of CLICnet gene sets that were selected in non-small cell lung cancer and renal cell carcinoma specific subtypes, we individually trained CLICnet on LUAD, LUSC and KIRC (KIRP was excluded because the MSK data contains only 15 KIPR samples). We could not identify any gene set specifically for LUSC, likely, due either to the smaller sample size or the subtype discrepancy between the TCGA and MSK cohorts. We found five LUAD gene sets after five training iterations, all of which were significantly predictive on the MSK data (Supplementary Figure S7, Supplementary Table S5). In addition, we found five KIRC gene sets that were significantly predictive on the MSK data after seven iterations (Supplementary Figure S7, Supplementary Table S5). The LUAD specific gene sets show a slightly better predictive ability compared with the aggregated non-small cell lung cancer gene sets with fewer training iterations, and the KIRC specific gene sets show a comparable predictive ability to the aggregated renal cell carcinoma gene sets with a comparable number of iterations.
Figure 1.
The CLICnet dataset, and the training and validation pipeline. (A) Illustration of the training of CLICnet. The GA is used to select genes that are given as input to the RBM. The RBM produces two clusters of patients, given the mutations provided by the GA, and the survival difference between patients in the two clusters is evaluated (via log-rank P-value), and given back to the GA as the fitness function. (B) The datasets used for training and validation of CLICnet. The numbers in the table refer to the number of samples in each dataset.
Mutation values per gene were set to 1 if a non-synonymous mutation was present and to 0 otherwise. Gene level mutations were used rather than nucleotide level mutations to restrict the overall number of features, avoiding having many more features than samples, which would hinder application of machine learning methods. Two additional datasets were obtained for melanoma patients treated with anti-PD1 (77,78), for which the mutational and clinical data were obtained from the cBioPortal (70,71) (https://www.cbioportal.org).
RBM structure, training and assessment
RBMs are neural networks that are typically utilized for unsupervised learning tasks which involve automatically discovering and learning regularities and patterns in the input data such that the model learns to generate new examples (62,63). The choice of RBMs as the machine learning technique for this study was motivated by the following: (i) First, RBMs are specifically designed for applications to binary and sparse datasets (79). Given the sparse and binary nature of the mutation data, which is a severe bottleneck for the application of most machine learning techniques (80,81), we reasoned that the method of choice should mitigate these issues, by being well fitted to process and utilize this type of datasets. (ii) Second, RBMs are simple, shallow and unsupervised, thus allowing interrogation of the features and weights learned (and therefore, potential interpretability applications), and can learn a distribution over the data without explicitly optimizing a supervised classification task, which might lead to overfitting. (iii) Third, RBMs are generative models. Therefore, they can extract highly informative features from the input data to learn the hidden states, which then can be used for clustering. Because we were interested in clustering patients by mutations in sets of genes, which make a sparse and binary input data, and aimed for a simple method with potentially interpretable features, RBMs were considered to be the optimal choice for this study. Each CLICnet RBM is trained for a specific cancer type, on a specific set of genes. The RBMs used in this work were constructed with 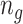 visible units and one hidden unit, where
visible units and one hidden unit, where  is the number of genes in the gene set. The number of epochs for training each RBM was set to 1000, with a 0.1 learning rate.
is the number of genes in the gene set. The number of epochs for training each RBM was set to 1000, with a 0.1 learning rate.
When a trained RBM is applied to mutation data from a new sample, the hidden unit activation can be either zero or one, defining the cluster assignment of the samples. To assess how each RBM clustering predicts patient survival rates, Cox's proportional hazard model was applied to the assigned clusters and the corresponding patient survival data. Patients were additionally stratified by sex, age and stage, to ensure that these were not confounders of the analysis. The subsets of patients with treatment information were additionally stratified by treatment, to ensure that these were not confounders of the analysis. The P-value was extracted, with a lower P-value indicating a stronger association between the defined clusters and overall survival.
Although RBMs are inherently stochastic in both training and application, the trained RBMs created for CLICnet include a deterministic procedure to define the clustering (or hidden states), and thus make the subsequent applications deterministic and reproducible. This was achieved by directly using the hidden probabilities to define the hidden state (where the median is set as cutoff), rather than randomly sampling a new distribution over these probabilities, to ensure that CLICnet always returns the same results for the same input once trained. In addition, for training of CLICnet, we set a constant random seed, to ensure that retraining CLICnet with the same input yields the same trained RBM. As a result, for any set of genes, there is one specific clustering of patients that is inferred by the CLICnet RBM.
Selecting sets of genes for CLICnet
The RBMs were incorporated with the GA feature selection to identify gene sets that yield RBM-inferred clusters with significantly different survival rates. The genes that were initially considered for training were those that are included in the MSK-IMPACT panel (82) and that, across the patients within each cancer, are mutated in the top 0.7 percentile frequency among all MSK-IMPACT panel genes.
From the set of genes that is used as initial input to CLICnet, three iterations of GA are applied to select the subset of genes that, when given as input to the RBM, optimizes the difference in survival rates between the two RBM clusters. Hence, the GA step depends on the RBM clustering to evaluate the fitness function, which is the survival difference between the two RBM-inferred clusters. The RBM step receives different solutions (sets of genes) from the GA, and evaluates each solution through the survival difference between the two inferred clusters (Figure 1).
The following steps of the GA were defined for each cancer type: (a) Initialization of a population of size 50, where 15% of the considered genes for the given cancer type was randomly selected for each instance in the initial population. (b) Evaluation of each instance in the population, where mutations in each gene set in the population were used to train an RBM, define two clusters of patients, and yield a Cox P-value which shows how well the clusters correspond to survival. This P-value was used to evaluate each of the gene sets. (c) The top half (25) instances in the population, that is, those with the lowest Cox P-values, were selected for reproduction, with randomly selected pairing. (d) Crossover was applied to the randomly selected pairs, until a population size of 50 was reached. Three iterations of steps (b)–(d) were repeated, and the best solution was retained, corresponding to the sets of genes that yielded the lowest P-values with the RBM clustering. These parameters (the population size and percentage of considered genes) were optimized for the TCGA training set via a grid search, with a 3-fold cross validation.
For each cancer type, the genetic algorithm was applied until five different sets of genes were found, such that each of them yielded an RBM clustering with a Cox's proportional hazard P-value ≤0.05 in the training (TCGA) and validation set (MSKCC). We used 100 iterations of this process as the upper bound, to reduce the risk of overfitting, where for all 15 cancer types, at least five sets were found in fewer than 100 iterations (the precise number of iterations required for each cancer type is shown in Figure 2). The number 5 was selected because it is the largest number of gene sets that were found for every cancer type under 100 iterations. Therefore, the top 5 gene sets in each cancer type are reported. The entire training of CLICnet was completed in less than 6 hours on a high performance computing cluster.
Figure 2.
Evaluation of the performance and stability of CLICnet. (A) Bar plots show the number of iterations needed to obtain five gene sets that were significant on the TCGA training data, and subsequently significant on the MSKCC validation data, for each cancer type. The numbers within the bars show the percentage of validated gene sets, among those that were significant on the training data. Statistical significance (permutation P-value) is indicated with asterisks (* P < 0.01, ** P < 0.001). (B) Boxplots show the number of genes in the CLICnet gene set for each cancer type. (c) Boxplot with overlaid dot-plots showing the CLICnet log-rank P-values, when applied to randomly sampled 2/3 of the MSKCC validation set.
Predicting survival with anti-PD1 using the TMB-H status
The survival of MSKCC patients treated with anti-PD1 was predicted using the TMB-H status. To that end, we used different cut-offs of the TMB (ranging from 5 to 24), in order to define the TMB-H status and predict the survival of anti-PD1 treated patients. The prediction was performed separately for all anti-PD1 treated MSKCC samples, for primary samples (which were used for CLICnet), and for metastatic samples estimating the Cox proportional hazard ratio and P-values.
Mutational signatures analysis
To evaluate the mutational processes underlying the different CLICnet clusters, the mutational signatures reported by Alexandrov et al. were quantified for each TCGA sample (83). These signatures were compared between each of the high and low risk clusters defined by CLICnet, in every cancer type, through a two-sided rank sum P-value, and significant (P-value < 0.05) associations were identified. Whenever a significant association between a cancer type and a mutational signature was detected, at least three CLICnet clusters from the given cancer type were associated with that signature.
Statistical analyses
Survival analyses: Kaplan-Meier survival curves were plotted, where the two CLICnet clusters define the curves. Cox proportional hazard analyses were applied to estimate how the CLICnet clusters predict the survival time, through a hazard ratio (HR) and P-value.
Boxplots and comparisons: For all boxplots, center lines indicate medians, box edges represent the interquartile range, whiskers extend to the most extreme data points not considered outliers, and the outliers are plotted individually. Points are defined as outliers if they are greater than q3 + w × (q3–q1) or <q1–w × (q3 − q1), where w is the maximum whisker length, and q1 and q3 are the 25th and 75th percentiles of the sample data, respectively. All differential expression and distribution comparisons P-values are obtained via one-sided Rank-sum test.
Pathway enrichment analysis: Enrichment P-values were calculated using the hypergeometric enrichment test, using GO annotation pathway definitions.
RESULTS
Using CLICnet to identify combinations of mutations that cluster patients by survival rates
The fundamental idea behind CLICnet is the utilization of mutations to identify groups of genes that partition patients into high and low risk clusters with significantly different overall survival rates, using Restricted Boltzmann Machines (RBMs). Briefly, RBMs are stochastic, generative neural networks that are widely used for unsupervised learning tasks which involve automatically discovering and learning regularities and patterns in the input data (84). The RBMs are specifically designed to work with binary and sparse datasets (79), and are therefore a good fit for mutational data, given that mutational data tend to be sparse and, in this work, is represented with binary values. RBMs consist of a visible layer, which receives the input data (in our case, mutations in a set of genes), and a hidden layer, which consists of the evaluated hidden states for the input data (in this case, there is a single binary hidden state, which defines the clusters of patients). After the RBM is trained on a set of patients’ mutations, it can be applied to cluster new patients based on their mutations, and use the inferred clusters to predict patient survival. Because RBMs are unsupervised, the clustering itself is based solely on the input mutational data without any previous knowledge of patient survival.
When developing CLICnet, we sought to train RBMs that cluster patients based on combinations of somatic mutations, such that the resulting clusters predict the patients’ survival rates. Because the RBMs are unsupervised, we integrated this approach with a genetic algorithm (GA) feature selection step that actively selects sets of genes such that the patient clusters inferred with the RBM using these genes predict the probability of survival. The mutations in genes selected by the GA are used as input for the RBM, which partitions the patients into two clusters. The fitness function of the GA is the log-rank P-value, estimating the difference in survival rates between the two clusters. Therefore, the GA evaluates different solutions (i.e. combinations of mutations) by the difference in survival rates between the two clusters that are inferred by the RBM for each combination of mutations (Figure 1A). The best solution (i.e. the gene set with the lowest log-rank P-value) after three iterations of the GA is selected. By incorporating an unsupervised approach with only three GA iterations, we aimed to limit the fitting of the model to the survival objective and maintain a stochastic element in the training of CLICnet, to reduce the risk of overfitting.
In the input of CLICnet, each gene is assigned a zero or one value per patient sample, with zero denoting no non-synonymous mutations and one denoting at least one non-synonymous mutation (see Methods). The output of CLICnet is the cluster assignment (zero or one) for each patient. The training process was done using the TCGA (64,65) mutation data (henceforth the training set, Figure 1B), where gene sets are selected and used to train RBMs for each cancer type, such that the clustering predicts survival in TCGA samples. These are then applied to the MSK-IMPACT (66) data for validation (henceforth the validation set, Figure 1B), where the RBMs and the underlying gene sets that significantly predict survival in this additional set of tumors are kept.
We applied this procedure to 15 cancer types, aiming to identify sets of genes that yield CLICnet-inferred clusters with significantly different survival rates. For each cancer type, we selected five sets of genes (see Methods) that group patients into high versus low risk clusters, with significantly different survival rates in the training (TCGA) and validation (MSK-IMPACR) sets (log-rank P-value < 0.05).
Using CLICnet to predict cancer patient survival from combinations of mutations
For each cancer type, we catalogued the combinations of mutations that best predict patient survival. Hence, five gene sets were selected by CLICnet, leading to five different partitionings of the patients into clusters of high and low survival rates (see Materials and Methods for details). Given the mutation data for these genes, CLICnet can classify a new patient as either high or low risk, estimating the survival probability. To catalogue the combinations of mutations that were the best predictors of survival across different cancer types, we extracted the mutation sets that were highly and significantly predictive of survival in both TCGA and MSKCC. To assess the robustness of CLICnet in predicting survival across different cancer types, we recorded the number of iterations needed to obtain five gene sets that were significantly predictive of survival in the TCGA training data and also showed a significant performance on the MSKCC validation data, across the different cancer types. We found that for six of the cancer types (pancreatic, melanoma, renal, glioma, bladder, and head and neck cancer), 10 or fewer iterations were sufficient. For all but three cancer types, the majority of the gene sets that were significantly predictive of survival in the TCGA training data were also significantly predictive of survival in the MSKCC validation data. In four cancer types (pancreatic, melanoma, glioma and bladder cancer), 100% of the gene sets were significant for, Figure 2A).
For all cancer types, the percentage of CLICnet gene sets that were predictive on the MSKCC validation data was significantly higher than expected for a random gene set (using 1000 random gene sets, permutation P-value < 0.01, Figure 2A). In addition, the number of genes in selected CLICnet sets differed substantially across the cancer types. The cancer types associated with a high mutation load, such as melanoma, lung, gastrointestinal and endometrial cancers, tend to have more genes in the selected CLICnet sets than cancers with low mutation loads (Figure 2b). Moreover, by subsampling the MSKCC validation set to subsets of size 2/3 of the original validation size, 50 times for each cancer type, we showed that CLICnet clustering consistently produced significant survival predictions on these randomly sampled subsets of the validation data, for every cancer type (Figure 2C). For some cancer types, such as gliomas and pancreatic tumors, we consistently observed a higher than 3 hazard ratio (HR > 3) between the two clusters in the MSKCC validation cohort (Supplementary Table S1). Although some combinations of mutations identified by CLICnet generalized to more than one cancer type, such as head and neck, stomach, thyroid and prostate adenocarcinomas, others were predictive almost exclusively within the tumor type on which they were trained (such as those of gliomas and renal cancers, Figure 3A).
Figure 3.
The CLICnet-derived clusters. (A) Heatmap showing the log10 Cox hazard ratio (HR) obtained with the five CLICnet clustering applied to the MSKCC validation set, for each cancer type (vertical axes), when the trained RBMs are applied to data from each of the 15 cancer types (horizontal axes). Significant Cox P-values are denoted by a red or blue border, where red corresponds to negative HR (where the majority of the mutations are observed in the high-risk cluster) and blue corresponds to positive HR (where the majority of the mutations are observed in the low-risk cluster). (B) The survival curves corresponding to one selected CLICnet clustering in five cancer types (where blue curves denote CLICnet cluster 0, and red denotes cluster 1), for the training and validation cohorts. (C) The heatmaps showing the mutations in the selected gene sets and cancer types in panel (C), for the two CLICnet clusters (cluster 0 in blue and cluster 1 in red).
CLICnet derives non-trivial combinations of mutations to construct the patient clusters, which are not simply defined by the total number of mutations in a set of genes, but rather, by mapping presence-absence pattern of mutations in a set of genes, to clusters of patients. Therefore, CLICnet identifies mutations that are not significantly associated with survival by themselves, but only when co-occurring with other mutations (Supplementary Figure S1). Nonetheless, we found that, for most cancer types, all high risk clusters were associated with either an increased or a decreased number of mutations across the respective selected gene sets (Figure 3A, Supplementary Figures S2 and S3). For example, in lung, thyroid and pancreatic adenocarcinomas, the high-risk clusters are characterized by increased numbers of mutations in the CLICnet sets of genes, implying that these mutations might be drivers. By contrast, in stomach and endometrial cancers, the high-risk clusters are associated with reduced numbers of mutations in the respective CLICnet gene sets, suggesting that in these cancers the increased mutation rates could be linked to impaired DNA repair (and therefore, would enhance the responses to DNA damage inducing therapeutics), or could enhance neoantigen presentation (and therefore, would increase the immune infiltration). These combinations of mutations also included mutations in the TP53 gene (Figure 3B, C, Supplementary Figure S2 and S3) that are associated with high risk; conceivably, the negative impact of the other mutations on the cancer cell fitness overrode the effect of the TP53 mutations. One exception are head and neck carcinomas, where in four CLICnet gene sets, the clusters with higher mutation rates were associated with low risk and improved survival, whereas in one gene set (gene set 2), it was the cluster with the lower mutation rate that was associated with low risk. Notably, TP53 was selected for all of the head and neck tumors gene sets, and the TP53 mutations were always associated with the high-risk clusters.
We investigated the genes that were selected by CLICnet for significant clustering by survival rate. As expected, many genes known to harbor driver mutations were frequently selected (Figure 4A). These were enriched for functions and pathways involved in cell cycle and cell death regulation, response to radiation, and several developmental processes (Figure 4B). The most frequently selected genes across all tumor types were well known pan-cancer drivers (Figure 4C). Overall, TP53 was most frequently selected across cancers, with only five tumor types where it was never selected (Figure 4A). In every gene set selected by CLICnet that included TP53, TP53 mutations were associated with decreased survival rate (Supplementary Figure S2 and S3). Nevertheless, there were pronounced differences between the selected genes among the tumor types. Some genes were frequently selected in a single tumor type but never in other types, such as NF2 in renal cancer, SMO in stomach cancer, IDH1 in glioma, and IRS2 in pancreatic cancer (Figure 4A, Supplementary Tables S2 and S3). Notably, in gliomas, where higher mutation rates are associated with the high-risk clusters, IDH1 mutations are exclusively linked with all low-risk clusters (Supplementary Figure S2 and S3). Examining the pairwise correlations between the selected gene sets in different tumor types, we found that lung, stomach and endometrial tumors shared the largest fraction of selected genes with other types of tumors, whereas renal, prostate, melanoma and breast tumors share the lowest fraction (Figure 4D). This is likely to be the case because, in the former group of tumors, the CLICnet-selected gene sets included pan-cancer drivers, such as TP53, MTOR, PTEN and POLE, whereas in the latter cancer types, the CLICnet gene sets were more cancer type-specific (Figure 4A, Supplementary Tables S2, S3).
Figure 4.
Genes selected by CLICnet and the associated pathways. (A) Heatmap showing the top genes selected by CLICnet for each cancer type, where the color intensity ranges from 0 to 1 and denotes the frequency that each gene was selected in each cancer type (the complete data is available in Supplementary Table S2). (B) Heatmap showing the GO pathway enrichment analysis (log transformed enrichment P-value) of the CLICnet selected genes for each cancer type. (C) Structure of the network connecting cancer types to genes that were selected by CLICnet, with the most frequently selected genes marked in the middle. (D) Correlation heatmap correlating the frequency that each gene was selected by CLICnet across all pairs of cancer types. Significant correlations are circled.
Predicting survival of anti-PD1 treated patients with CLICnet risk clusters
The CLICnet mutational clusters predict overall survival in different tumor types, without considering the particular treatments given to different patients. To evaluate whether some of these mutational clusters could also predict survival of patients treated with immune checkpoint blockade, we applied CLICnet to the primary samples of MSKCC patients that were treated with anti-PD1 (31), to cluster these patients into high and low risk groups. The purpose of this analysis was not to identify the strongest or most informative predictors of anti-PD1 benefit, which would require both training and validation datasets of anti-PD1 treated samples that are not available for the majority of cancer types. Rather, we aimed to examine whether in some of the cancer types, the mutational processes governing treatment-general survival were also linked with anti-PD1 benefit. We found that in melanoma, bladder cancer and gliomas, the high-risk clusters were significantly associated with poor survival in the subsets of patients treated with anti-PD1 (Figure 5A–C). When focusing on primary tumor samples, the TMB-H (high tumor mutation burden) status was not predictive of survival in the anti-PD1 treated patients, emphasizing the relevance of CLICnet derived clusters, which capture non-trivial mutational patterns, for predicting anti-PD1 survival in these types of tumors (Figure 5A, Supplementary Figure S4). Moreover, we applied CLICnet to two additional mutation datasets of melanoma patients treated with anti-PD1 (77,78) and found that three of the five CLICnet clustering predicted survival in the Liu et al. (78) dataset (144 patients), one of which is also significantly associated with survival in the Riaz et al. (77) dataset (where the relatively small size of 68 samples might diminish the effect, Figure 5D, Supplementary Figure S5). These results mark CLICnet melanoma gene combination 3 as a potentially strong prognostic marker, which predicted survival after anti-PD1 treatment in 3 independent datasets (MSKCC anti-PD1, Liu et al. and Riaz et al.; P-values: 4e–2, 3e–2 and 1.7e–2, respectively; hazard ratios: 2.6, 1.6 and 3.2, respectively). By contrast, the high risk clusters inferred for lung, esophagus, renal and colorectal tumors, were not significantly associated with poor survival in the anti-PD1 treated patients, and some were even associated with improved survival (in particular, in colorectal, lung and renal tumors, Figure 5A, Supplementary Figure S5).
Figure 5.
Association between CLICnet clustering and survival under anti-PD1 treatment. (A) Heatmap showing the log10 transformed Cox HR resulting from application of each of the five CLICnet RBMs trained for each cancer type, to the original training data (top panel), the validation data (second top panel), the anti-PD1 treated MSKCC samples (third top panel), and when predicting survival of the anti-PD1 treated MSKCC samples using TMB-H status (bottom panel). Red colors correspond to negative HR (where the majority of mutations are observed in the high-risk CLICnet cluster) and blue colors correspond to positive HR (where the majority of mutations are observed in the low-risk CLICnet cluster). The significant P-values (P < 0.05) are shown with a bold border. (B) The survival curves corresponding to selected CLICnet clustering of bladder cancer and glioma, for the training data (left panels), validation data (middle panels), and the subset of MSKCC samples treated with anti-PD1 (right panels). The blue curve corresponds to the high-risk CLICnet cluster and the red curve corresponds to the low-risk cluster (as defined on the TCGA training data). (C) The survival curves corresponding to three CLICnet clustering of melanoma, for the training data (left panels), validation data (middle panels) and the subset of MSKCC samples treated with anti-PD1 (right panels). The blue curve corresponds to the high-risk CLICnet cluster and the red curve corresponds to the low-risk CLICnet cluster. (D) The survival curves corresponding to three CLICnet clustering of melanoma when the trained CLICnet RBMs are applied to two additional melanoma datasets of patients treated with anti-PD1 (77,78).
We next sought to investigate why in some tumor types, namely, melanoma, glioma and bladder cancers, there was a clear, direct link between the CLICnet-inferred mutational clusters and the survival rates of patients treated with anti-PD1, whereas in other tumor types, weak and even inverse associations were found. We reasoned that some of the mutations captured with CLICnet (especially, those affecting DNA repair) could increase the incidence of mutational processes and thus could promote the emergence of specific mutation signatures. To evaluate this, we used the mutation signatures previously reported by Alexandrov et al., which derive distinct patterns of substitutions to define nucleotide biases in subsets of cancers, including some with known environmental triggers (16,18,83). By using the quantified measure of each mutational signature in every cancer type, we aimed to identify mutational signatures that were significantly increased or decreased in CLICnet patients with a low risk and favorable survival. We hypothesized that such mutational signature could uncover oncogenic processes that underlie the complex relationships between the overall survival rates with and without anti-PD1 treatment in different cancer types.
To this end, we investigated the association between the CLICnet-inferred clusters and previously published mutational signatures (16,18,83). We compared the levels of previously reported mutational signatures (83) in the CLICnet-defined high and low-risk patients, for the seven tumor types where anti-PD1 treatment data was available. Surprisingly, the association between CLICnet risk clusters and different mutational signatures was found to be cancer type-specific, so that five of the seven tumor types showed a unique association, that is, the signatures associated with the CLICnet risk clusters were unique to that particular type of cancer. The one signature that was associated with the CLICnet risk clusters in more than one cancer type was signature 1, which consists of genes that are related to endogenous mutational processes in most cancer types (Figure 6A). Specifically, we found that signatures 2 and 13, which have been attributed to the activity of AID/APOBEC family of cytidine deaminases (85), were significantly associated with CLICnet risk clusters in bladder tumors. Signature 3, linked with failure of DNA double-strand break-repair by homologous recombination (HR (86)), was significantly associated with CLICnet risk clusters in renal tumors. Signature 4 that is linked to smoking and tobacco mutagens (87) is significantly associated with CLICnet risk clusters in lung tumors. Signature 6 that is linked with defective DNA mismatch repair and is found in microsatellite unstable tumors is significantly associated with CLICnet risk clusters in colorectal tumors, and signature 7 that is linked with ultraviolet (UV) radiation is significantly associated with CLICnet risk clusters in melanoma.
Figure 6.
Associations between the measures of mutational signatures and CLICnet clusters. (A) A map showing the significant associations between mutational signature and either high or low risk CLICnet clusters in each of the seven cancer types with anti-PD1 treatment data. (B) The average quantification of each mutational signature in each cancer type. (C and D) Boxplots showing the quantification of mutational signatures that are significantly increased in low risk clusters (C) or in high risk clusters (D) of each cancer type, for the five CLICnet selected high (red) and low (blue) risk clusters in each cancer type. (E) The survival curves corresponding to selected CLICnet clustering of lung, colorectal and renal cancers, for the training data (left panels), validation data (middle panels) and the subset of MSKCC samples treated with anti-PD1 (right panels). The blue curve corresponds to the high-risk cluster and the red curve corresponds to the low risk cluster (as defined on the TCGA training data). The mutational signatures were from Alexandrov et al. (16,17,18,19).
These cancer-specific associations are in accord with the type of intrinsic mutagenesis that is characteristic of each of these cancer types (Figure 6B). Indeed, we found that in melanoma, increased UV mutational signature was associated with the low risk clusters and improved survival (signature 7, Figure 6C), in agreement with previous reports (88). The increased UV signature was also weakly associated with a better immunotherapy response (89). Thus, the link between the low risk cluster and improved immunotherapy survival in melanoma could be mediated through UV mutagenesis. Similarly, increased activation of AID/APOBEC cytidine deaminases and the increased signatures 2 and 13 coupled with it were linked with tissue inflammation and immunity as well as viral infection (90), potentially, explaining why these signatures were associated with improved immunotherapy survival (91,92). Because the higher levels of AID/APOBEC signatures 2 and 13 were associated with the low-risk clusters in bladder tumors, the improved survival benefit from anti-PD1 in patients that are clustered by CLICnet as low-risk might be mediated through activation of AID/APOBEC mutagenesis.
By contrast, the increased smoking-associated mutagenesis (signature 4) is significantly associated with the high risk CLICnet clusters for lung cancers (Figure 6D), in agreement with the well-known association between smoking and poor lung cancer outcome (93,94). However, within the subset of lung cancer patients treated with anti-PD1, the patients matching the high-risk CLICnet clusters showed similar or even improved survival compared with those matching the low-risk clusters (Figure 6E, Supplementary Figure S5), in accordance with previous findings of improved immunotherapy responses and higher PD-L1 in smoking lung cancer patients (95,96,97). Additionally, the high-risk CLICnet clusters in colorectal cancer are significantly associated with increased mutagenesis of defective mismatch repair (MMR) genes and the MSI status (signature 6, Figure 6E). Thus, MSI-H, which is an established marker of immunotherapy response, could underlie the improved survival of anti-PD1 treated patients matching the high-risk CLICnet clusters in colorectal tumors. Finally, a mild increase of mutagenesis related to DNA double-strand break-repair by homologous recombination (HR) is associated with the high-risk renal cancer clusters (Figure 6E). Thus, although this connection has not been previously reported, this observation made with the CLICnet clusters suggests that HR mutagenesis could be associated with poor outcome in renal cancer patients, and possibly, to improved survival in patients treated with anti-PD1. However, this association appears weak compared to the other associations detected (Figure 6E). Overall, these findings demonstrate that, in tumors where high mutation burden is associated with low risk and improved survival, such as melanoma and bladder, the low-risk CLICnet clusters also predict improved survival with anti-PD-1 treatment. In contrast, tumors where increased mutagenesis is linked to poor outcome, such as lung and colorectal tumors, have a complex relation between CLICnet prognostic clusters and immunotherapy benefit, which is likely mediated through the differential impact of mutagenesis on survival in anti-PD1 treated patients compared to patients undergoing other treatment regimes.
DISCUSSION
In this work, we present CLICnet, to our knowledge, the first approach that harnesses mutational patterns to cluster cancer patients by survival, using subsets of genes from the MSK-IMPACT panel (82). We limited the search to genes within this panel to harmonize the discovery set between the training (TCGA) and validation (MSKCC) datasets, where the latter only included MSK-IMPACT panel mutations. When additional pan-cancer studies with comprehensive mutations and clinical data become available, it will be possible to apply CLICnet to perform a broader search for combinations of mutations that predict clinical outcomes, which is expected to reveal new mutations with context-specific clinical relevance. CLICnet captures stochastic mutational processes that are predictive of survival in different cancer types, and partitions patients in each cancer type into high and low risk clusters. By utilizing RBMs for clustering, CLICnet can infer non-trivial combinations of mutations that predict survival, and capture the signal arising from combinations of mutations that are associated with improved and poor survival, or mutations that only predict survival in the context of other mutations. We applied CLICnet to 15 cancer types with data from the TCGA (64,65) and MSKCC (66) cohorts and identified gene sets for each cancer type that were significantly predictive of survival rates in both datasets.
From the research perspective, this work provides the first systematic approach to identify and catalogue sets of mutations that are jointly associated with survival in different cancer types. From the clinical standpoint, CLICnet provides a way to cluster patients based on multiple mutations, in order to construct clinically relevant clusters of patients. Although numerous outcome-associated clinical and molecular parameters have been identified for many of the tumor types analyzed here, for some tumor types, such as pancreatic adenocarcinoma, there are few clinically relevant somatic mutations. Moreover, even for cancer types with many outcome markers in clinical use, additional parameters are likely to be helpful, to increase the number of patients that can benefit from these, especially, in the case of rare mutations. By identifying combinations of multiple mutations, CLICnet can utilize rare mutations to predict survival of many patients simultaneously.
By using RBMs with a single hidden node, the training of CLICnet becomes similar to estimating the posterior probabilities of the true labels (clustering of samples), making the training process and application of CLICnet straightforward and interpretable. By incorporating three iterations of a GA within the unsupervised RBM framework, we aimed to focus on simply inferred combinations that are not truly optimized for the objective and thus reduce the risk of overfitting. Future studies are warranted to develop more complex techniques for this purpose, for example, by employing deep, supervised neural networks, or incorporating additional data types and treatment information. Additionally, given that the ultimate goal of this study is to uncover and catalogue the complex relations between groups of genes that produce similar survival rates, the validation data is used to filter sets of genes and identify combinations of mutations that are reproducibly predictive of the overall survival across two large cancer cohorts, with the exception of the anti-PD1 analysis. Future studies based on this work could incorporate additional testing steps to evaluate the clinical utility of this technique for patient stratification without considering treatment regimes.
We applied survival profiling with CLICnet to the subset of primary MSKCC tumor samples from seven cancer types that were treated with anti-PD1 (31), and found that for three cancer types, namely, melanoma, bladder cancer and glioma, the high-risk clusters constructed by CLICnet also predict poor survival with anti-PD-1 treatment. Furthermore, for melanoma, we showed that CLICnet predicts survival rates after anti-PD-1 treatment in additional datasets (78,98), suggesting that these clusters can be developed as strong markers of survival from primary site sequencing of melanoma patients under the anti-PD-1 treatment. Although CLICnet mutation sets predict treatment-independent overall survival and not direct treatment responses, in melanoma, bladder cancer and glioma, all five CLICnet gene sets were significantly predictive of the anti-PD1 benefit. When more mutational and clinical data becomes available for anti-PD1 treated patients, allowing CLICnet training and validation, we expect that for the majority of cancer types, it becomes possible to derive stronger, treatment-specific mutational clusters. We demonstrated that, although the TBM-H status was highly predictive of survival of anti-PD1 treated patients when different cancer types were aggregated, the signal originated primarily from metastatic samples in specific cancer types and therefore might not be predictive across all individual cancer types or when applied to non-metastatic samples (Supplementary Figure S4). Indeed, we found that TMB-H was not predictive of survival with anti-PD1 for primary site samples, when considering most cancer types individually, and is never predictive of anti-PD1 survival in glioma patients (34,35) (Supplementary Figure S4). Therefore, the combinations of mutations that are identified by CLICnet and predict anti-PD1 survival in glioma could be especially clinically important. Notwithstanding that these observations might be partially due to the small sample size for some cancer types, the CLICnet clusters for melanoma, bladder cancer and glioma show a clear predictive signal for anti-PD1 treated patients and indicate that CLICnet mutational clusters potentially could be developed as an alternative marker for anti-PD1 efficacy, which is specifically predictive for primary site tumors.
The mutational processes captured with CLICnet reveal intricate relationships between overall survival rates, and specifically, with immune checkpoint blockade therapy. In some tumor types, the mutational patterns that characterize the high-risk, poor survival clusters are paradoxically associated with improved survival after anti-PD-1 treatment, i.e. the same clusters that predict poor survival when considering all samples predict improved survival when considering only anti-PD1 treated samples. This connection could be due to increased mutagenesis, which likely contributes to tumor aggressiveness and simultaneously induces immune infiltration and neoantigen presentation. For cancers in which increased mutagenesis could be linked to a particular type of DNA damage, whether exogenous or endogenous, increased mutagenesis was also associated with better survival under anti-PD1 treatment. This observation is recapitulated in cancer types where increased mutagenesis was originally linked with poor survival, such as mutational processes associated with smoking in lung cancer, emphasizing the complex relations between patients’ prognoses with and without immune checkpoint blockade therapy.
In this study, we examined both homogenous and heterogenous cancer types (those that include several tumor subtypes aggregated together by the tissue of origin), and in both cases, CLICnet demonstrated a strong survival prediction on the training and validation sets. However, in heterogenous cancer types (such as colorectal, renal and non-small cell lung cancers), CLICnet clustering did not predict the anti-PD1 benefit. The heterogenous cancer types might have varying response rates to different treatments, confounding the generalization of survival prediction when examining a specific treatment regime. It is therefore advisable to apply CLICnet within tumor subtypes when aiming to derive mutational predictors of treatment response. In addition, in this work, we focused on somatic mutations data, as a proof of concept for the approach. Integration of other data types in follow-up studies is highly desirable and could reveal complex relationships between different types of alterations that affect clinical outcomes. In particular, incorporating RBMs based on germline mutations could uncover links between genetic and environmental mutagenesis and cancer survival, and provide means for early diagnosis. This would probably require developing an ensemble technique to integrate RBMs based on different data types, to allow investigation and interoperation of the associations between different alterations. Because mutated genes are often not expressed and therefore difficult to target, reducing their physiological relevance, incorporation of other data types, such as copy number variations, fusion events and epigenetic alterations, that are not considered in this work, is expected to allow for more complete inference of the factors governing patients’ outcomes, and reveal targetable combinations of events.
In conclusion, this work introduces CLICnet, an RBM-based method that identifies combinations of mutations that cluster cancer patients by survival rates. CLICnet does not depend on arbitrary, user-determined thresholds and is deterministic once trained (albeit depending on the initial gene set selection), thus, directly facilitating patient clustering. As more data becomes available, CLICnet can be easily adapted for clustering based on combinations of mutations that specifically predict responses to various cancer treatments from mutational data in selected panels of genes. If carefully validated with additional data, CLICnet can be used as a predictor of anti-PD1 immunotherapy efficacy in particular cancers through analysis of primary site tumor samples, aiding clinicians in the selection of patients that are most likely to benefit from this treatment.
DATA AVAILABILITY
CLICnet is freely available with a Python package (https://github.com/gussow/clicnet) and a webtool that allows application and visualization of the different CLICnet profiles (http://clicnet.pythonanywhere.com/).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eytan Ruppin and Sushant Patkar for helpful discussion and comments on the manuscript. This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Contributor Information
Ayal B Gussow, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Eugene V Koonin, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Noam Auslander, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
Intramural Research Program of the National Library of Medicine at the National Institutes of Health. Funding for open access charge: U.S. National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Yates L.R., Campbell P.J. Evolution of the cancer genome. Nat. Rev. Genet. 2012; 13:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 3. Loeb K.R., Loeb L.A. Significance of multiple mutations in cancer. Carcinogenesis. 2000; 21:379–385. [DOI] [PubMed] [Google Scholar]
- 4. Vandin F., Upfal E., Raphael B.J. De novo discovery of mutated driver pathways in cancer. 2012; 22:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matthew Bailey A.H., Tokheim C., Porta-Pardo E., Mills G.B., Karchin R., Ding L., Bailey M.H., Sengupta S., Bertrand D., Weerasinghe A. et al. Comprehensive characterization of cancer driver genes and mutations article comprehensive characterization of cancer driver genes and mutations. Cell. 2018; 173:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013; 501:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lawrence M.S., Stojanov P., Polak P., Kryukov G.V, Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.a et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013; 499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burrell R.A., McGranahan N., Bartek J., Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013; 501:338–345. [DOI] [PubMed] [Google Scholar]
- 9. Hofree M., Shen J.P., Carter H., Gross A., Ideker T. Network-based stratification of tumor mutations. Nat. Methods. 2013; 10:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bielas J.H., Loeb L.A. Quantification of random genomic mutations. Nat. Methods. 2005; 2:285–290. [DOI] [PubMed] [Google Scholar]
- 11. Nussinov R., Jang H., Tsai C.-J., Cheng F. Precision medicine and driver mutations: computational methods, functional assays and conformational principles for interpreting cancer drivers. PLoS Comput. Biol. 2019; 15:e1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dugger S.A., Platt A., Goldstein D.B. Drug development in the era of precision medicine. Nat. Rev. Drug Discov. 2018; 17:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al. COSMIC: the catalogue of somatic mutations In cancer. Nucleic Acids Res. 2018; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A. et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011; 39:D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., Stratton M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013; 3:P246–P259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alexandrov L.B., Ju Y.S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T. et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016; 354:618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L. et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N. et al. The repertoire of mutational signatures in human cancer. Nature. 2020; 578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Postow M.A., Callahan M.K., Wolchok J.D. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 2015; 33:1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei S.C., Duffy C.R., Allison J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018; 8:1069–1086. [DOI] [PubMed] [Google Scholar]
- 22. Galluzzi L., Chan T.A., Kroemer G., Wolchok J.D., López-Soto A. The hallmarks of successful anticancer immunotherapy. Sci. Transl. Med. 2018; 10:eaat7807. [DOI] [PubMed] [Google Scholar]
- 23. Zimmer L., Goldinger S.M., Hofmann L., Loquai C., Ugurel S., Thomas I., Schmidgen M.I., Gutzmer R., Utikal J.S., Göppner D. et al. Neurological, respiratory, musculoskeletal, cardiac and ocular side-effects of anti-PD-1 therapy. Eur. J. Cancer. 2016; 60:210–225. [DOI] [PubMed] [Google Scholar]
- 24. Hofmann L., Forschner A., Loquai C., Goldinger S.M., Zimmer L., Ugurel S., Schmidgen M.I., Gutzmer R., Utikal J.S., Göppner D. et al. Cutaneous, gastrointestinal, hepatic, endocrine, and renal side-effects of anti-PD-1 therapy. Eur. J. Cancer. 2016; 60:190–209. [DOI] [PubMed] [Google Scholar]
- 25. Dudley J.C., Lin M.-T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for PD-1 blockade. Clin. Cancer Res. 2016; 22:813–820. [DOI] [PubMed] [Google Scholar]
- 26. Food and Drug Administration FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication | FDA. 2017; (24 October 2017, date last accessed)www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm560040.htm. [Google Scholar]
- 27. Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010; 138:2073–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015; 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A., Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019; 30:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019; 51:202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Food and Drug Administration FDA approves pembrolizumab for adults and children with TMB-H solid tumors | FDA. 2020; (17 June 2020, date last accessed)https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors. [Google Scholar]
- 33. Hause R.J., Pritchard C.C., Shendure J., Salipante S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016; 22:1342–1350. [DOI] [PubMed] [Google Scholar]
- 34. Gurjao C., Tsukrov D., Imakaev M., Luquette L.J., Mirny L.A. Limited evidence of tumour mutational burden as a biomarker of response to immunotherapy. 2020; bioRxiv doi:04 September 2020, preprint: not peer reviewed 10.1101/2020.09.03.260265. [DOI]
- 35. Wood M.A., Weeder B.R., David J.K., Nellore A., Thompson R.F. Burden of tumor mutations, neoepitopes, and other variants are weak predictors of cancer immunotherapy response and overall survival. Genome Med. 2020; 12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith J.C., Sheltzer J.M. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. Elife. 2018; 7:e39217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macintyre G., Goranova T.E., De Silva D., Ennis D., Piskorz A.M., Eldridge M., Sie D., Lewsley L.A., Hanif A., Wilson C. et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018; 50:1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu D., Abbosh P., Keliher D., Reardon B., Miao D., Mouw K., Weiner-Taylor A., Wankowicz S., Han G., Teo M.Y. et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat. Commun. 2017; 8:2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phipps A.I., Buchanan D.D., Makar K.W., Win A.K., Baron J.A., Lindor N.M., Potter J.D., Newcomb P.A. KRAS-mutation status in relation to colorectal cancer survival: The joint impact of correlated tumour markers. Br. J. Cancer. 2013; 108:1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato S., Iida S., Higuchi T., Ishikawa T., Takagi Y., Yasuno M., Enomoto M., Uetake H., Sugihara K. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int. J. Cancer. 2007; 121:1771–1778. [DOI] [PubMed] [Google Scholar]
- 41. Hamelin R., Laurent-Puig P., Olschwang S., Jego N., Asselain B., Remvikos Y., Girodet J., Salmon R.J., Thomas G. Association of p53 mutations with short survival in colorectal cancer. Gastroenterology. 1994; 106:42–48. [DOI] [PubMed] [Google Scholar]
- 42. McLaughlin J.R., Rosen B., Moody J., Pal T., Fan I., Shaw P.A., Risch H.A., Sellers T.A., Sun P., Narod S.A. Long-term ovarian cancer survival associated with mutation in BRCA1 or BRCA2. J. Natl. Cancer Inst. 2013; 105:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kurian A.W., Sigal B.M., Plevritis S.K. Survival analysis of cancer risk reduction strategies for BRCA1/2 mutation carriers. J. Clin. Oncol. 2010; 28:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robson M.E., Chappuis P.O., Satagopan J., Wong N., Boyd J., Goffin J.R., Hudis C., Roberge D., Norton L., Bégin L.R. et al. A combined analysis of outcome following breast cancer: Differences in survival based on BRCA1/BRCA2 mutation status and administration of adjuvant treatment. Breast Cancer Res. 2003; 6:R8–R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tryggvadóttir L., Vidarsdóttir L., Thorgeirsson T., Jonasson J.G., Ólafsdóttir E.J., Ólafsdóttir G.H., Rafnar T., Thorlacius S., Jonsson E., Eyfjord J.E. et al. Prostate cancer progression and survival in BRCA2 mutation carriers. J. Natl. Cancer Inst. 2007; 99:929–935. [DOI] [PubMed] [Google Scholar]
- 46. Thorne H., Willems A.J., Niedermayr E., Hoh I.M.Y., Li J., Clouston D., Mitchell G., Fox S., Hopper J.L., Bolton D. Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev. Res. 2011; 4:1002–1010. [DOI] [PubMed] [Google Scholar]
- 47. Du X., Shao Y., Qin H.F., Tai Y.H., Gao H.J. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac. Cancer. 2018; 9:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li L., Dong M., Wang X.G. The implication and significance of beta 2 microglobulin: a conservative multifunctional regulator. Chin. Med. J. (Engl). 2016; 129:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Willmore-Payne C., Layfield L.J., Holden J.A. c-KIT mutation analysis for diagnosis of gastrointestinal stromal tumors in fine needle aspiration specimens. Cancer. 2005; 105:165–170. [DOI] [PubMed] [Google Scholar]
- 50. Xing M., Alzahrani A.S., Carson K.A., Shong Y.K., Kim T.Y., Viola D., Elisei R., Bendlová B., Yip L., Mian C. et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J. Clin. Oncol. 2015; 33:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Westermann F., Muth D., Benner A., Bauer T., Henrich K.O., Oberthuer A., Brors B., Beissbarth T., Vandesompele J., Pattyn F. et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008; 9:R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004; 351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 53. Cullen J., Rosner I.L., Brand T.C., Zhang N., Tsiatis A.C., Moncur J., Ali A., Chen Y., Knezevic D., Maddala T. et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur. Urol. 2015; 68:123–131. [DOI] [PubMed] [Google Scholar]
- 54. Klein E.A., Cooperberg M.R., Magi-Galluzzi C., Simko J.P., Falzarano S.M., Maddala T., Chan J.M., Li J., Cowan J.E., Tsiatis A.C. et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 2014; 66:550–560. [DOI] [PubMed] [Google Scholar]
- 55. Leiserson M.D.M., Vandin F., Wu H.T., Dobson J.R., Eldridge J.V., Thomas J.L., Papoutsaki A., Kim Y., Niu B., McLellan M. et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015; 47:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shrestha R., Hodzic E., Sauerwald T., Dao P., Wang K., Yeung J., Anderson S., Vandin F., Haffari G., Collins C.C. et al. HIT’nDRIVE: patient-specific multidriver gene prioritization for precision oncology. Genome Res. 2017; 27:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leiserson M.D.M., Blokh D., Sharan R., Raphael B.J. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput. Biol. 2013; 9:e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vandin F., Clay P., Upfal E., Raphael B.J. Discovery of mutated subnetworks associated with clinical data in cancer. Pacific Symposium on Biocomputing. 2012; 55–66. [PubMed] [Google Scholar]
- 59. Altieri F., Hansen T.V., Vandin F. NoMAS: a computational approach to find mutated subnetworks associated with survival in genome-wide cancer studies. Front. Genet. 2019; 10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hovestadt V., Ayrault O., Swartling F.J., Robinson G.W., Pfister S.M., Northcott P.A. Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat. Rev. Cancer. 2020; 20:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang C., Armasu S.M., Kalli K.R., Maurer M.J., Heinzen E.P., Keeney G.L., Cliby W.A., Oberg A.L., Kaufmann S.H., Goode E.L. Pooled clustering of high-grade serous ovarian cancer gene expression leads to novel consensus subtypes associated with survival and surgical outcomes. Clin. Cancer Res. 2017; 23:4077–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006; 313:504–507. [DOI] [PubMed] [Google Scholar]
- 63. Smolensky P. Information processing in dynamical systems: foundations of harmony theory; CU-CS-321-86. Parallel Distributed Processing: Explorations in the Microstructure of Cognition. 1986; 1:194–281. [Google Scholar]
- 64. Hoadley K.A., Yau C., Hinoue T., Stuart J.M., Benz C.C., Laird P.W., Hoadley K.A., Yau C., Hinoue T., Wolf D.M. et al. Cell-of-origin patterns dominate the molecular article cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018; 173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nat. Med. 2017; 23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goldman M., Craft B., Swatloski T., Cline M., Morozova O., Diekhans M., Haussler D., Zhu J. The UCSC cancer genomics browser: Update 2015. Nucleic Acids Res. 2015; 43:D812–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goldman M., Craft B., Kamath A., Brooks A.N., Zhu J., Haussler D. The UCSC Xena Platform for cancer genomics data visualization and interpretation. 2018; bioRxiv doi:18 May 2018, preprint: not peer reviewed 10.1101/326470. [DOI]
- 69. Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cline M.S., Craft B., Swatloski T., Goldman M., Ma S., Haussler D., Zhu J. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci. Rep. 2013; 3:2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dhanasekaran S.M., Balbin O.A., Chen G., Nadal E., Kalyana-Sundaram S., Pan J., Veeneman B., Cao X., Malik R., Vats P. et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat. Commun. 2014; 5:5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016; 48:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen F., Zhang Y., Şenbabaoğlu Y., Ciriello G., Yang L., Reznik E., Shuch B., Micevic G., De Velasco G., Shinbrot E. et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016; 14:2476–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ricketts C.J., De Cubas A.A., Fan H., Smith C.C., Lang M., Reznik E., Bowlby R., Gibb E.A., Akbani R., Beroukhim R. et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018; 23:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martín-Algarra S., Mandal R., Sharfman W.H. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017; 171:934–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Amon L., Zimmer L., Gutzmer R., Satzger I., Loquai C. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019; 25:1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hinton G.E. A practical guide to training restricted Boltzmann machines. Momentum. 2012; 9:926. [Google Scholar]
- 80. Shepperd M., Cartwright M. Predicting with sparse data. IEEE Trans. Softw. Eng. 2001; 11:987–998. [Google Scholar]
- 81. Demiriz A. Enhancing product recommender systems on sparse binary data. Data Min. Knowl. Discov. 2004; 9:485–502. [Google Scholar]
- 82. Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). J. Mol. Diagnostics. 2015; 17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Huang P.J., Chiu L.Y., Lee C.C., Yeh Y.M., Huang K.Y., Chiu C.H., Tang P. MSignatureDB: a database for deciphering mutational signatures in human cancers. Nucleic Acids Res. 2018; 46:D964–D970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fischer A., Igel C. An introduction to restricted Boltzmann machines. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2012; 7441:Berlin, Heidelberg: Springer; 14–36. [Google Scholar]
- 85. Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E. et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014; 46:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Riaz N., Blecua P., Lim R.S., Shen R., Higginson D.S., Weinhold N., Norton L., Weigelt B., Powell S.N., Reis-Filho J.S. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 2017; 8:857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Alexandrov L.B., Ju Y.S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T. et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016; 354:618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Trucco L.D., Mundra P.A., Hogan K., Garcia-Martinez P., Viros A., Mandal A.K., Macagno N., Gaudy-Marqueste C., Allan D., Baenke F. et al. Ultraviolet radiation–induced DNA damage is prognostic for outcome in melanoma. Nat. Med. 2019; 25:221–224. [DOI] [PubMed] [Google Scholar]
- 89. Miao D., Margolis C.A., Vokes N.I., Liu D., Taylor-Weiner A., Wankowicz S.M., Adeegbe D., Keliher D., Schilling B., Tracy A. et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018; 50:1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Conticello S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008; 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang S., Jia M., He Z., Liu X.S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018; 37:3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Boichard A., Pham T.V., Yeerna H., Goodman A., Tamayo P., Lippman S., Framton G.M., Tsigelny I.F., Kurzrock R. APOBEC-related mutagenesis and neo-peptide hydrophobicity: implications for response to immunotherapy. Oncoimmunology. 2019; 8:1550341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sun S., Schiller J.H., Gazdar A.F. Lung cancer in never smokers - a different disease. Nat. Rev. Cancer. 2007; 7:778–790. [DOI] [PubMed] [Google Scholar]
- 94. Tammemagi C.M., Neslund-Dudas C., Simoff M., Kvale P. Smoking and lung cancer survival: the role of comorbidity and treatment. Chest. 2004; 125:27–37. [DOI] [PubMed] [Google Scholar]
- 95. Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., Chow L.Q., Vokes E.E., Felip E., Holgado E. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015; 373:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Garon E.B., Hellmann M.D., Rizvi N.A., Carcereny E., Leighl N.B., Ahn M.J., Eder J.P., Balmanoukian A.S., Aggarwal C., Horn L. et al. Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: results from the phase i KEYNOTE-001 study. J. Clin. Oncol. 2019; 37:2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gainor J.F., Shaw A.T., Sequist L.V., Fu X., Azzoli C.G., Piotrowska Z., Huynh T.G., Zhao L., Fulton L., Schultz K.R. et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin. Cancer Res. 2016; 22:4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martín-Algarra S., Mandal R., Sharfman W.H. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017; 171:934–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CLICnet is freely available with a Python package (https://github.com/gussow/clicnet) and a webtool that allows application and visualization of the different CLICnet profiles (http://clicnet.pythonanywhere.com/).








