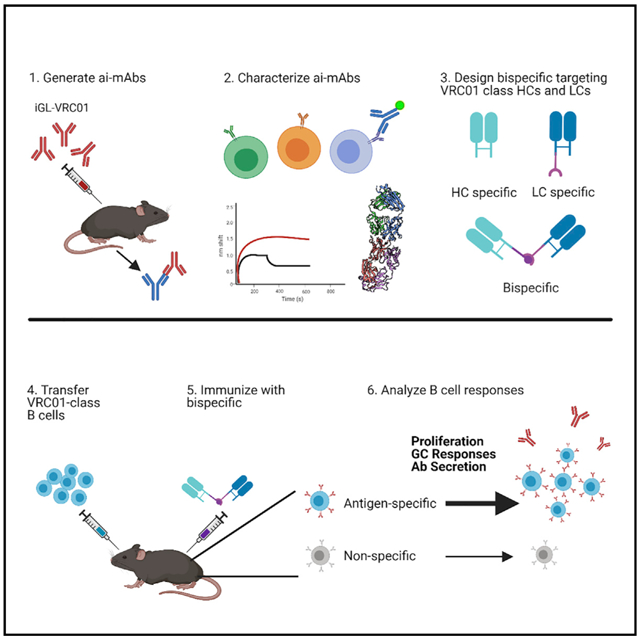
SUMMARY
An effective HIV-1 vaccine will likely need to elicit broadly neutralizing antibodies (bNAbs). Broad and potent VRC01-class bNAbs have been isolated from multiple infected individuals, suggesting that they could be reproducibly elicited by vaccination. Several HIV-1 envelope-derived germline-targeting immunogens have been designed to engage naive VRC01-class precursor B cells. However, they also present off-target epitopes that could hinder development of VRC01-class bNAbs. We characterize a panel of anti-idiotypic monoclonal antibodies (ai-mAbs) raised against inferred-germline (iGL) VRC01-class antibodies. By leveraging binding, structural, and B cell sorting data, we engineered a bispecific molecule derived from two aimAbs; one specific for VRC01-class heavy chains and one specific for VRC01-class light chains. The bispecific molecule preferentially activates iGL-VRC01 B cells in vitro and induces specific antibody responses in a murine adoptive transfer model with a diverse polyclonal B cell repertoire. This molecule represents an alternative non-envelope-derived germline-targeting immunogen that can selectively activate VRC01-class precursors in vivo.
Graphical abstract
In brief
Successful engagement of naive B cells that give rise to broadly neutralizing antibodies is thought to be key to a successful HIV-1 vaccine. In this study, Seydoux et al. present the characterization and assessment of anti-idiotypic antibodies targeting VRC01-class B cell precursors. Their results represent an alternative to non-envelope-derived immunogens.
INTRODUCTION
Numerous antibodies exhibiting remarkable breadth and potency in their ability to neutralize HIV-1 (broadly neutralizing antibodies [bNAbs]) have been isolated from HIV-1-infected individuals (Scheid et al., 2009; Walker et al., 2009; West et al., 2014; Burton et al., 2012; Falkowska et al., 2014; Doria-Rose et al., 2014; Huang et al., 2012, 2014, 2016; Wu et al., 2010; Corti et al., 2010; Sajadi et al., 2018; Gorman et al., 2020). Structural studies of bNAbs have provided critical information on their mechanisms of action and on the overall structure and function of the envelope protein Env and delineated critical neutralizing epitopes (Burton and Mascola, 2015; West et al., 2014; Burton and Hangartner, 2016; Kwong and Mascola, 2018). When passively delivered, or genetically expressed in animal models, bNAbs confer protection from experimental HIV-1 infection, indicating that if elicited by vaccination bNAbs should be protective in humans (Mascola et al., 1999, 2000; Shibata et al., 1999; Foresman et al., 1998; Hessell et al., 2007, 2009; Moldt et al., 2012; Balazs et al., 2011, 2014; Saunders et al., 2015; Pietzsch et al., 2012; Gruell et al., 2013; Baba et al., 2000; Parren et al., 2001).
VRC01-class antibodies are among the most broad and potent HIV-1 bNAbs known (Sajadi et al., 2018; Huang et al., 2016). They have been isolated from at least 10 different donors and all use the VH1–2*02 variable heavy (VH) gene, paired with light chains (LCs) expressing rare 5-aa CDRL3 domains (Wu et al., 2010, 2011; Scheid et al., 2011; Bonsignori et al., 2012; Zhou et al., 2013, 2015; Georgiev et al., 2013; Huang et al., 2016; Sajadi et al., 2018; Umotoy et al., 2019). Despite being highly somatically mutated and ~50% divergent in sequence, VRC01-class antibodies bind the CD4 binding site (CD4bs) on Env in a nearly identical manner predominantly through their CDHR2 regions (Zhou et al., 2010, 2013, 2015; Diskin et al., 2011; Huang et al., 2016). The three amino acids Trp50 (FWR2), Asn58 (FWR3), and Arg71 (FWR3) encoded by the VH1–2*02 gene are nearly invariable in VRC01-class bNAbs and make contacts with Env that are critical for binding and neutralization (Scharf et al., 2013, 2016; West et al., 2012; Zhou et al., 2015). Two additional amino acids, Glu96 in CDLR3 and Trp100b in CDRH3, are commonly found in VRC01-class antibodies. Trp100b hydrogen bonds with Asp279 or Asn279 on Env, and Glu96 hydrogen bonds with Gly459 on Env (Zhou et al., 2013; West et al., 2012). Since the CDRH3 and CDRL3 regions are generated through V(D)J recombination, they could be present on a naive B cell, or acquired through somatic mutation.
Inferred germline (iGL) versions of VRC01-class mAbs have been generated through homology-guided reversion of antibody variable (V) and joining (J) gene-encoded portions of the mutated heavy or LCs (as isolated from HIV-1-infected donors) to their chromosomally templated sequences to approximate VRC01-class precursors. These iGL-VRC01-class bNAbs display no reactivity to recombinant Env (rec Env) and they do not neutralize HIV-1, indicating that the inability to elicit bNAbs by rec Env immunization is in part due to a failure to engage and stimulate the appropriate B cells during this critical first step of antibody development (Hoot et al., 2013; Jardine et al., 2013; McGuire et al., 2013, 2014b; Zhou et al., 2010; Scheid et al., 2011; Klein et al., 2013). These observations led to the development of “germline-targeting” immunogens capable of engaging the iGL versions of VRC01-class antibodies (McGuire et al., 2013, 2016; Jardine et al., 2013; Medina-Ramírez et al., 2017; Jardine et al., 2016). These immunogens activate VRC01-class precursor B cells in vivo (McGuire et al., 2014a, 2016; Tian et al., 2016; Jardine et al., 2015; Medina-Ramírez et al., 2017; Briney et al., 2016; Dosenovic et al., 2015; Sok et al., 2016; Havenar-Daughton et al., 2018a). However, since they are Env-derived, they also present epitopes recognized by non-neutralizing or narrowly neutralizing antibodies (nNAbs) (McGuire et al., 2014a). Indeed, immunization of transgenic knockin (KI) animals expressing human VH and variable light (VL) genes with Env-derived germline-targeting immunogens leads to the activation and proliferation of a non-VRC01-class, off-target B cell response even in mice where the B cell repertoire is heavily skewed (Sok et al., 2016, Jardine et al., 2015; Tian et al., 2016; Dosenovic et al., 2015; Medina-Ramírez et al., 2017; McGuire et al., 2016; Briney et al., 2016). It is possible that these off-target B cell responses will be recalled and predominate the B cell responses upon subsequent Env immunizations. In line with this, adoptive transfer experiments where B cells expressing putative VRC01 precursors or intermediates with defined B cell receptor (BCR) specificity are introduced into a wild-type (WT) mouse at a controlled frequency demonstrated that immunogens with high affinity and/or avidity for the target BCR were required for successful inter-clonal competition of rare target B cells following a priming immunization (Dosenovic et al., 2018; Abbott et al., 2018; Huang et al., 2020; Kato et al., 2020).
We recently described an alternative approach to Env-derived immunogens; the use of anti-idiotypic monoclonal antibodies (ai-mAbs) to target unmutated BCRs with genetic features associated with bNAbs (Bancroft et al., 2019; Dosenovic et al., 2019). One ai-mAb, iv8, raised against the iGL-VRC01 mAb specifically activated VRC01-class target B cells in vivo, without eliciting Env-specific off-target responses (Dosenovic et al., 2019). A potential limitation, however, of the iv8 ai-mAb as an immunogen is that it preferentially recognizes VRC01-like LCs and may not selectively prime VRC01 precursors.
In this study, we describe the development and isolation of several novel ai-mAbs that can bind to not only one but multiple diverse, putative VRC01 precursors with high affinity. Despite their broader recognition properties, single B cell sorting from HIV-1-negative donors demonstrated that they selectively recognize BCRs with either VRC01-class LCs, which have a 5-aa CDRL3, or those with heavy chains (HCs) derived from VH1–2*02, but not both VH1–2*02 HCs paired with LCs with 5-aa CDRL3s. To overcome this limitation, we sought to combine the distinct binding activities of two such ai-mAbs in order to improve their ability to engage and activate B cells expressing VRC01 precursor-like BCRs. To this end we engineered a bispecific anti-idiotypic molecule in which one arm is derived from an ai-mAb, called iv9, that preferentially recognizes the VH1–2*02 HC, and a second arm derived from an ai-mAb, called iv4, that preferentially recognizes LCs with 5-aa CDRL3. The bispecific ai-mAb iv4/iv9 could specifically activate iGL-VRC01 B cells in vitro. Using a murine adoptive transfer model, we show that when used as an immunogen, the bispecific ai-mAb was more efficient at engaging and expanding putative VRC01 precursor B cells in vivo than either iv4 or iv9 antigen-binding fragments (Fabs). Our results are relevant not only to the development of an HIV-1 vaccine aimed at eliciting VRC01-class antibodies, but to a general effort to activate specific B cell lineages that produce protective antibodies, and they further suggest that ai-mAb-derived immunogens may have general utility as germline targeting immunogens against diverse B cell targets.
RESULTS
Generation of anti-idiotypic antibodies against germline VRC01
We previously described the isolation of the ai-mAbs iv1 and iv8 from mice immunized with iGL-VRC01 (Dosenovic et al., 2019). In the present study, we screened additional hybridomas from these mice. In addition, we generated additional hybridomas from mice immunized with a cocktail of iGL-12A21 and iGL-3BNC60, which are also inferred VRC01-class precursors. Our overall goal was to identify ai-mAbs that can specifically recognize BCRs comprised of a VH1–2*02-derived HC paired with a LC with a 5-aa CDRL3. Hybridomas were arrayed into 384-well plates at the Fred Hutchinson Antibody Technology Center. Culture supernatants from hybridoma-containing wells were screened using a high-throughput bead-based array against a small panel of antibodies, including the mAbs used to immunize, as well as control mAbs iGL-b12 (Hoot et al., 2013), iGL-FI6 (Corti et al., 2011), and iGL-pt1–695 (McGuire et al., 2014a), which were each conjugated to a different fluorescent microsphere. Wells containing hybridomas producing antibodies that bound the mAb used for immunization but not the control mAbs were re-arrayed and screened against a larger panel of mutated VRC01-class antibodies, which contain somatic mutations present in the mAbs isolated from HIV-1-infected donors and in which the CDRH3 regions are nearly identical to those found in the mutated bNAbs. The expanded panel also included additional iGL-VRC01-class antibodies, as well as other irrelevant control mAbs (data not shown). Using this approach, we identified seven hybridomas (iv1–iv6 and iv8) from animals immunized with iGL-VRC01. We note that the initial characterizations of iv1 and iv8 were previously described, but they are included herein for comparison (Dosenovic et al., 2019; Figure S1). We also isolated six hybridomas (iv9–iv14) from animals immunized with iGL-3BNC60 and iGL-12A21 that bound the iGL, but not the control, mAbs in the initial screening panel. From these, we established monoclonal hybridoma cell lines. Supernatants from monoclonal hybridomas were further screened against an expanded panel of mature and iGL-VRC01-class antibodies as well as other irrelevant control antibodies and HC/LC chimeras by biolayer interferometry (BLI) (Figures S1A and S1B).
We sequenced the VH and VL transcripts of ai-mAb-secreting hybridomas by RT-PCR and used the cDNA to produce recombinant mAbs with human constant regions (Tiller et al., 2009; Siegel, 2009). The sequence analysis revealed that iv4 and iv5 were identical, and iv6 was a closely related clonal variant; therefore, we only produced iv4 as a recombinant mAb. BLI was used to determine the affinity of the recombinant ai-mAbs to the iGL-VRC01-class mAb they were raised against (Figure 1A; Table S1). iv1–iv4 and iv9 bound exclusively to iGL-VRC01 with high (0.8–6 nM) affinity. In contrast, iv13 bound weakly to iGL-VRC01, above the μM range, but strongly to iGL-3BNC60 and iGL-12A21 (0.6–60 nM), while binding of iv12 to iGL-3BNC60 and iGL-12A21 was ~100 nM. From this initial screen, we concluded that we had isolated ai-mAbs that recognize iGL-VRC01-class mAbs differently. To further evaluate the binding specificity of these ai-mAbs we used a multiplex array in which VRC01-class and control antibodies were immobilized onto individual microspheres and incubated with biotinylated ai-mAbs followed by BV421-labeled streptavidin (Figure 1B). iv1–iv4 displayed reactivity to the iGL-VRC01 mAb and the iGL-NIH45–46 mAb, which differs from iGL-VRC01 by 6 aa in the CDRH3 (McGuire et al., 2013). iv1–iv4 did not bind to other iGL-VRC01-class antibodies, which have diverse CDRH3 regions and different LCs (Figure 1B, “iGL-VRC01 Abs”), or to non-VRC01 antibodies used as controls (Figure 1B, “non-iGL-VRC01 control Abs”). The VH1–2*02, *03, and *04 alleles encode Trp50, which hydrogen bonds with Asn280 on Env and is unmutated in all VRC01-class antibodies. The *01, *05, and *06 alleles encode Arg50. iv1–iv3 did not bind to an iGL-VRC01 variant derived from the VH1–2*01 allele, indicating that Trp50 is important for their ability to recognize iGL-VRC01. In agreement with our previous report, iv1 bound weakly to mature VRC01.
Figure 1. Binding analyses of ai-mAbs raised against iGL-VRC01-class antibodies.

(A) Binding affinities of recombinant ai-mAbs to iGL-VRC01-class Fabs was measured using biolayer interferometry (BLI). Symbols on the dotted line indicate detectable binding but that it was too low to accurately determine at the highest concentration tested (700 nM). Symbols on the abscissa indicate that no binding was detected at the highest concentration tested.
(B) Biotinylated ai-mAbs were incubated with the indicated mAbs immobilized on magnetic microspheres, washed, and then incubated with streptavidin coupled to BV421. The mean BV421 fluorescence on each bead is shown as indicated. The fluorescence scale for each mAb is shown on the far right. iGL-VRC01-class mAbs are inferred germline-reverted variants of VRC01-class antibodies; control mAbs include inferred germline-reverted variants of non-VRC01-class antibodies, as well as mutated VRC01-class antibodies; chimeric mAbs include antibodies comprised of either an iGL-VRC01-class heavy chain (HC) or light chain (LC) paired with a non-VRC01-class LC or HC, respectively; naturally paired mAbs include putative VRC01-class antibodies isolated from naive donors by total paired VH/VL sequencing (unselected), or sorted with eOD-GT8 (eOD-GT8 sorted).
(C) A B cell line engineered to stably express the iGL-VRC01 B cell receptor was loaded with Fluo4 calcium indicator dye. The Fluo4 fluorescence signal was measured on a cytometer for 30 s. The indicated ai-mAb was added to the B cells to a final concentration of 125 nM and the Fluo4 fluorescence signal was measured for an additional 150 s.
To ascertain whether these ai-mAbs recognize the VH and/or the VL regions of iGL-VRC01 antibodies, we evaluated their binding to chimeric mAbs composed of iGL-VRC01 HCs paired with non-VRC01-class LCs, or non-VRC01-class HCs paired with iGL-VRC01-class LCs (Figure 1B, “chimeric mAbs”). iv2 and iv3 did not bind any chimeric antibodies, indicating that their epitopes span both the VH and the VL of iGL-VRC01 and iGL-NIH45–46. iv1 bound weakly to the iGL-3BNC60/iGL-VRC01 LC chimera but no other chimeras with an iGL-VRC01 LC. iv4 bound to several chimeric antibodies with the iGL-VRC01 LC, including iGL-b12-HC/iGL-VRC01-LC and iGL-1NC9-HC/iGL-VRC01-LC chimeras, which have HCs derived from VH1–3 and VH1–46, respectively, indicating that iv4 preferentially recognizes the iGL-VRC01 LC.
One ai-mAb, iv9, bound all of the iGL-VRC01 mAbs and all chimeric mAbs with a VH1–2*02 HC, irrespective of their LCs. It also bound to iGL-IOMA, which is a non-VRC01-class mAb that also utilizes a VH1–2 HC (Gristick et al., 2016), indicating that the epitope recognized by iv9 is entirely encoded by the VH1–2*02 gene.
The iGL-VRC01-class mAbs were generated through germ-line reversion based on sequence homology to the mutated bNAbs. We therefore sought to determine whether the ai-mAbs could bind to naturally paired, putative VRC01-class precursors; antibodies with a VH1–2*02 HC paired with a CDRL3 of 5 aa (Figure 1B, “naturally paired mAbs”). Such naturally paired putative VRC01 precursors have been isolated from human peripheral blood mononuclear cell (PBMC) samples collected from HIV-1 uninfected donors through unbiased high-throughput paired VH/VL sequencing (DeKosky et al., 2016; Figure 1B, “unselected”), or by using fluorescence-activated cell sorting (FACS) with eOD-GT8 as bait (Jardine et al., 2016; Figure 1B, “eOD-GT8 sorted”). Again, iv9 bound all of the putative VRC01 precursors tested (Figure 1B), while iv4 and iv12 bound to some but not all putative VRC01 precursors. In sum, the ai-mAbs displayed a differential ability to recognize diverse inferred and putative VRC01-class precursors, indicating that they recognize distinct idiotypes that are not universally conserved across putative VRC01-class precursors. The exception is iv9, which recognizes an epitope that is encoded by the VH1–2*02 gene and is therefore present on VRC01-class precursors, but also present on non-VRC01-class mAbs that are also derived from this VH gene.
ai-mAbs do not identify VRC01-class BCRs in diverse naive B cell pools
The observation that ai-mAbs could recognize putative VRC01-class precursors suggests that they could potentially be used as immunogens to activate the corresponding B cells. To test this, we used a calcium flux assay based on BCR engagement and B cell activation (Hoot et al., 2013; McGuire et al., 2013) to evaluate the ability of the ai-mAbs to recognize cell surface-expressed iGL-VRC01 BCRs and activate B cells (Figure 1C). All ai-mAbs except iv12 and the anti-iGL-b12 idiotype ib3 (Bancroft et al., 2019), which was used as a negative control, activated iGL-VRC01 B cells. Although iv12 was unable to activate iGL-VRC01 B cells, it could readily activate B cells expressing iGL-3BNC60 and iGL-12A21 BCRs (Figure S2), for which it has a much higher affinity (Figure 1A).
To ascertain whether ai-mAbs could recognize bona fide putative VRC01 precursors, they were fluorescently labeled and used as baits to single-cell sort B cells from PBMCs drawn from HIV-1-negative, healthy donors (Figure 2A). Using RT-PCR (Tiller et al., 2008; Figure 2B; Table S2), we recovered VH and VL transcripts from ~1,100 ai-mAb+ B cells and compared the frequency of B cells expressing VH1–2 HC and the frequency of those that express a 5-aa CDRL3 with the frequencies of these two features determined by unbiased paired sequencing using the chromium platform from 10x Genomics (Figure 2B; Table S2). iv9 and iv12 showed a significant enrichment for B cells expressing the VH1–2*02 HC (80%–99% versus 1%–5% from total unselected B cells) (Figure 2B; Table S2). B cells sorted with iv4 showed a strong (85-fold) enrichment for LCs with a 5-aa CDRL3 (Figure 2B; Table S2). These data indicate that iv9 and iv12 bind preferentially to BCRs with VH1–2*02-derived HCs and iv4 efficiently engages BCRs with a 5-aa CDRL3.
Figure 2. ai-mAb-specific B cell sorting and sequencing.

(A) ai-mAbs fluorescently labeled with either phycoerythrin (PE) or allophycocyanin (APC) were used as bait to single-sort CD19+CD20+IgD+IgM+CD27− B cells from PBMCs obtained from healthy, HIV-1-negative donors. VH and VL transcripts were recovered using RT-PCR, and amplicons were Sanger sequenced.
(B and C) Frequencies of B cells expressing VH1–2 HC (B) and 5-aa CDRL3 (C) were compared between the different ai-mAbs. Frequencies in total unselected B cells were assessed as control.
Data in (B) and (C) are presented as mean ± SD. All frequency data are given in Table S2.
However, from all sorted B cells, we isolated only one putative VRC01-class precursor (defined as having a VH1–2*02 HC and 5-aa CDRL3; PHVP2) using iv12 as bait. This mAb bound to recombinant iv12, but not to germline-targeting Env derivatives 426c.TM4ΔV1–3 or eOD-GT8 (Figure S3). The absence of reactivity of PHVP2 to these two germline-targeting immunogens is not surprising, as the two immunogens are not expected to bind all available VH1–2 HC/5-aa LC BCRs (Umotoy et al., 2019; Havenar-Daughton et al., 2018b; Lin et al., 2020). Considering that we screened approximately 3.7 million naive B cells (Table S2), and estimates of the frequency of VRC01 precursors range from ~1 in 300,000 based on eOD-GT8 sorting to ~1 in 10,000 based on unbiased high-throughput sorting (Havenar-Daughton et al., 2018b), these data indicate that none of the ai-mAbs tested was capable of efficiently engaging VRC01-class precursors in HIV-1-negative PBMC samples.
ai-mAbs show distinct modes of interaction with iGL-VRC01-class mAbs
To understand the molecular basis of the differences in the ability of ai-mAbs to bind different VRC01 precursors, we determined crystal structures of iGL-VRC01-class mAb/ai-mAb complexes. We obtained well-diffracting crystals of iv1, iv4, and iv9 in complex with iGL-VRC01 and were able to solve their structures at 2.0, 2.5, and 2.6 Å, respectively (Table S3). We also obtained the crystal structure of iv12 in complex with iGL-12A21 at 2.6 Å (Table S3). These structures were compared to the recently reported structure of the iv8-iGL-VRC01 complex (PDB: 6OL7) in which iv8 makes most of its contacts with the iGL-VRC01 LC (Dosenovic et al., 2019; Figure 3).
Figure 3. Crystal structures of ai-mAbs in complex with iGL-VRC01-class mAbs.

(A–E) iv8 (PDB: 6OL7) (A), iv1 (B), iv4 (C), and iv9 Fab (D) in complex with iGL-VRC01, and iv12 in complex with iGL-12A21 (E) are shown in cartoon and surface representations. HCs and LCs are labeled and color coded. Only the Fv is depicted for clarity.
(F) Sequences of iGL-VRC01 and iGL-12A21 are shown with interacting residues within 5 Å of iv1, iv4, iv8, iv9, and iv12 marked with a filled star under the sequences. Data collection and refinement statistics for the crystal structures are provided in Table S3. Details on the interactions of the ai-mAbs in complex with iGL-VRC01 precursors are provided in Tables S4–S7.
iv1 binds iGL-VRC01 head-on and makes extensive contacts with both the VH (~48% of the total buried surface area [BSA]) and VL (~52% of the total BSA) of iGL-VRC01, including the CDRH3 (27.5% of the BSA from the HC [or ~13% of the total BSA]) and CDRL3 (~22% of the LC BSA [or ~11% of the total BSA]) (Figure 3B; Table S4). Tyr98 on the iv1 HC makes a hydrogen bond with Trp50 on iGL-VRC01, which may explain the lack of iv1 binding to iGL-VRC01*01 (Figure 2B). Since the CDRH3 and CDRL3 are the product of recombination, and are unique among putative VRC01-class precursors, this mode of interaction likely explains how iv1 achieves a high-affinity interaction with iGL-VRC01 and the closely related iGL-NIH45–46, but fails to bind to other putative glVRC01-class precursors (Figures 1A and 1C). The contacts between iv1 and the CDRH3 and CDRL3 of iGL-VRC01 explain the weak binding observed to mutated VRC01, which shares nearly identical CDRH3 and CDLR3 regions with iGL-VRC01.
Similar to iv8 (Dosenovic et al., 2019), iv4 primarily contacts the iGL-VRC01 LC (~84% of the total BSA; Figure 3B; Table S5). The iv4 CDRL1 reaches across the iGL-VRC01 CDRL3 and makes additional contacts with the iGL-VRC01 CDRH3. This mode of interaction would restrict binding to antibodies with longer CDRL3 regions, providing a structural basis for the enrichment of antibodies with 5-aa CDRL3s (Figure 2C; Table S2), a rare but key feature of VRC01-class antibodies. The CDRH3 and CDRL1 of iv4 make direct contacts with the CDRL3 region, including the QQYE motif formed by the junction of the V and J genes, and in particular with Glu96, which hydrogen bonds with Gly459 on Env (Table S5). Only 5 of 93 5-aa CDRL3s identified by unbiased 10x sequencing encoded Glu96, while 2 of 23 5-aa CDRL3s sorted by iv4 encode Glu96 (Figure S4).
iv9 binds to HC framework regions 1 and 3, which are encoded entirely by the VH1–2*02 gene, and makes no contact with the iGL-VRC01 LC (Figures 3D and 3F; Table S6). The mode framework region-restricted mode of iv9 binding is consistent with the ability of this antibody to bind all mAbs tested that have a VH1–2*02-derived HC (Figure 1B).
We note that Arg82b of the iGL-VRC01HC (Kabat numbering) makes three salt bridges and a hydrogen bond with Asp95 iv9HC as well as three salt bridges and a hydrogen bond with Asp91 iv9LC (Table S6). VH1–2 is the only member of the VH1 gene family with an arginine at position 82b; all of the others have a serine (Figure S5), which would be unable to form the salt bridges and hydrogen with Asp95 iv9HC and Asp91 iv9LC. The extensive contacts between iv9 and Arg82b rationalize the specificity of iv9 for B cells expressing VH1–2-derived HCs and not HCs derived from other VH1 family genes (Figure 2B). Moreover, the framework-directed mode of binding utilized by iv9 is consistent with the ability of this ai-mAb to bind the iGL-VRC01*01 allele.
Similar to iv9, iv12 predominantly interacts with the VH1–2*02 HC of iGL-12A21 (~88.5% of the total BSA) (Figures 3E and 3F; Table S7). Most contacts with iv12 are made with CDRH regions (~23%, ~24%, and ~35% of the total BSA for CDRH1, CDRH2, and CDRH3, respectively) and a few contacts with FW3 (~7% of the total BSA), CDRL1 (~8% of the total BSA), and CDRL2 (3% of the total BSA) of iGL-12A21. The CDR-dominated mode of HC recognition utilized by iv12, which includes several contact residues in the CDRH3, could account for the more limited ability of iv12 to bind mAbs and BCRs with VH1–2-derived HCs compared to iv9.
Collectively, the HC-dominated mode of recognition utilized by iv9 and iv12 explains why these two mAbs predominately recognize B cells with VH1–2 HCs paired with diverse LCs in B cell sorting experiments (Figure 2B; Table S1). The LC-focused mode of iv4 binding to iGL-VRC01 helps to rationalize the strong preference this mAb has for engaging naive BCRs with 5-aa CDRL3, without selecting for BCRs that also have a VH1–2*02 HC. The extensive contacts between the iGL-VRC01 CDRs and iv1 demonstrate the exquisite binding specificity of this ai-mAb for iGL-VRC01 and likely explain the lack of binding to other iGL-VRC01-class mAbs tested (Figure 1B). Since we would expect BCRs that closely mimic the iGL-VRC01 sequence to be rare in the naive repertoire, this mode of interaction explains the inefficiency of iv1 to recognize B cells expressing VRC01-class precursors (Figures 2B and 2C; Table S2).
Generation of a bispecific iv4/iv9 molecule
Based on the results of our ai-mAb binding, B cell sorting, and structural analyses, none of the ai-mAbs would be predicted to specifically activate and expand diverse VRC01-class precursors when used as single immunogens. Thus, we sought to engineer a bispecific molecule that combines the properties of two ai-mAbs with distinct recognition properties: one that recognizes VH1–2-encoded HCs and another that recognizes LCs with 5-aa CDRL3s. We chose iv9 and iv4 as being highly specific for VRC01-class HCs and LCs, respectively. To do this we expressed and purified a recombinant Fab variant of iv4 fused to SpyCatcher (iv4-SC) and a recombinant Fab variant of iv9 fused to SpyTag (iv9-ST) (Zakeri et al., 2012; Figure 4A). Mixing of Spy-Catcher-tagged and SpyTag-tagged Fabs results in a covalent linkage with 1:1 stoichiometry through the spontaneous isoamide bond formation between SpyCatcher and SpyTag. Using this approach, we can dissect the binding properties of each Fab component individually and in complex following conjugation. This design also permits the production of a bispecific molecule without an Fc region, which we have previously shown to be detrimental to an ai-mAb-induced B cell response (Dosenovic et al., 2019).
Figure 4. Generation of a bispecific iv4/iv9 molecule.

(A) Schematic of iv9-Fab with a SpyTag (iv9-ST), iv4-Fab fused to SpyCatcher (iv4-SC), and a divalent iv4/iv9 bispecific molecule generated though covalent SpyCatcher-SpyTag linkage.
(B) Chromatograms of iv9-ST, iv4-SC, or iv4-SC incubated with an excess of iv9-ST (iv4/iv9) run on a size-exclusion column (SEC).
(C) iv4-SC, iv9-ST, and SEC-purified iv4/iv9 were subjected to non-reducing SDS-PAGE and stained with Coomassie blue.
(D–F) The binding of iv4-SC, iv9-ST, or SEC-purified iv4/iv9 to a chimeric antibody comprised of the iGL-NIH45–46-HC paired with the iGL-8AN131-LC (D), iGL-b12-HC paired with the iGL-NIH45–46-LC (E), or iGL-NIH45–46 (F) was assessed by BLI.
(G) iv4/iv9 was directly conjugated to DL550 and used as bait to single-cell sort iv4/iv9+ B cells from healthy, HIV-1-negative donors. Subsequently, VH and VL transcripts were recovered using RT-PCR and amplicons were Sanger sequenced. Frequencies of iv4/iv9+ B cells expressing VH1–2*02 HC and 5-aa CDRL3 were assessed. Data in (G) are presented as mean ± SD. All frequency data are provided in Table S2.
(H and I) Putative VRC01-class HC/LC pairs PHVP6 (H) and PHVP7 (I) were produced as recombinant IgG and their binding to various proteins was assessed by BLI.
Mixing of iv4-SC with an excess of iv9-ST produced a complex that eluted earlier than the individual components during size-exclusion chromatography (SEC) (Figure 4B). Fractions from the larger peak migrated more slowly than iv4-SC or iv9-ST alone under non-reducing SDS-PAGE (Figure 4C), consistent with the formation of a covalent iv4-SC/iv9-ST complex (hereafter referred to as iv4/iv9). To verify the antigenicity of iv4/iv9, we measured binding of iv4-SC, iv9-ST, and iv4/iv9 to chimeric antibodies, including an iGL-b12-HC/iGL-VRC01-LC chimera that binds to iv4 but not iv9 and to a iGL-NIH45–46/iGL-8ANC131 chimera that binds iv9 but not iv4 (Figure 1B). We did not observe binding of iv4-SC to glNIH45–46/iGL-8ANC131, which has a 9-aa CDRL3 (Figure 4D), but measured weak binding of iv4-SC to iGL-b12-HC/iGL-VRC01-LC (Figure 4E). The binding profile of iv9-ST was opposite, as it bound to iGL-NIH45–46/iGL-8ANC131 but not iGL-b12-HC/iGL-VRC01-LC (Figures 4D and 4E). The iv4/iv9 complex bound to both chimeric antibodies (Figures 4D and 4E), demonstrating that the bispecific molecule maintained the binding properties of both individual components. We also evaluated the ability of the iv4-SC, iv9-ST, and the iv4/iv9 to bind to iGL-NIH45–46. Compared to the individual Fab components, the bispecific molecule dissociated more slowly from iGL-NIH45–46, consistent with an increase in avidity afforded by bivalent binding (Figure 4F).
iv4/iv9 identifies putative VRC01 precursor B cells
We next evaluated whether iv4/iv9 was more efficient than iv4 or iv9 at sorting putative VRC01-class precursors from pools of HIV-1 naive PBMCs. In four separate experiments we single-cell sorted 488 B cells using fluorescently labeled iv4/iv9 as bait (Table S2). 95%–100% of recovered HC sequences were derived from VH1–2 and 0%–7% of the LC sequences had a 5-aa CDRL3 (Figure 4G; Table S2). In total, we recovered three putative VRC01-class heavy LC pairs using iv4/iv9 as bait. We produced these as recombinant immunoglobulin (Ig)Gs (PHVP6, PHVP7, and PHVP8). PHVP6 and PHVP7 both bound to iv4 and iv9 (Figures 4H and 4I), PHVP8 only bound iv9 (Figure S6). We assessed whether these could bind to two multimeric Env-derived germline-targeting immunogens, 426cTM4ΔV1–3-ferritin and eOD-GT8-ferritin. PHVP6 bound to eOD-GT8-ferritin but not to 426cTM4ΔV1–3-ferritin (Figure 4H), and it was specific for the CD4-BS, as it did not bind to eOD-GT8-KO-ferritin, which contains mutations that abrogate VRC01-class B cell binding (Figure 4H). In contrast, PHVP7 and PHVP8 did not bind either 426cTM4ΔV1–3 or eOD-GT8 (Figures 4I and S6).
iv4/iv9 selectively activates iGL-VRC01 B cells in vitro
Based on the observations that the frequency of B cells that stained with iv4/iv9 were comparable to the frequency that stained with iv9, and that only a modest enrichment for 5-aa CDRL3 was achieved, we concluded that iv4/iv9 can bind to any B cell expressing a VH1–2*02 HC through the iv9 arm of the bispecific molecule. However, we hypothesized that avid binding to an adjacent BCR through the iv4 arm of iv4/iv9 would lead to preferential B cell activation of putative VRC01-class precursor B cells. To test this directly, we evaluated the ability of iv4/iv9 to bind to and activate primary B cells from knockin mice homozygous for either the iGL-VRC01 HC (Jardine et al., 2015), the iGL-VRC01 LC (Abbott et al., 2018), or both using flow cytometry and calcium flux as readouts. Consistent with the notion that the iv9 arm of the bispecific molecule could bind to any BCR with a VH1–2-derived HC, iv4/iv9 directly conjugated to DL550 readily stained all of the B cells from the iGL-VRC01 HC knockin and iGL-VRC01 HC/LC double knockin mice (Figure 5A). iv4/iv9 stained ~40% of the iGL-VRC01 LC knockin B cells, which may be due to the relatively weak binding to BCRs with VRC01 LCs, paired with irrelevant HCs, as observed in Figures 1B and 4E.
Figure 5. The bispecific iv4/iv9 molecule binds to and activates iGL-VRC01 B cells.

(A) B cells from wild-type C57BL/6 mice and mice homozygous for either the iGL-VRC01 LC, HC, or HC/LC were stained with DL550-labeled iv4/iv9. Histogram of iv4/iv9 from stained knockin (KI) B cells (blue) is overlaid on histogram from stained B cells from C57BL/6 mice (gray) as indicated.
(B) B cells from mice homozygous for either the iGL-VRC01 LC, HC, or HC/LC were loaded with Fluo4 calcium indicator dye. The Fluo4 fluorescence signal was measured on a cytometer for 30 s. Either iv4 IgG, iv9 IgG, an isotype control (top panels), iv4-SC, iv9-ST, or iv4/iv9 (bottom panels) was then added to the B cells to a final concentration of 100 nM as indicated, and the Fluo4 fluorescence signal was measured for an additional 210 s.
As expected, iv9 IgG, which can engage two BCRs through its two Fab arms, activated B cells from the iGL-VRC01 HC but not the iGL-VRC01 LC knockin mice, while iv4 IgG activated B cells from the iGL-VRC01 LC but not the iGL-VRC01 HC knockin mice (Figure 5B, top panels), and both iv4 IgG and iv9 IgG activated B cells from the iGL-VRC01 HC/LC double knockin mice (Figure 5B, top panels). In contrast to the divalent IgG, the monovalent iv9-ST or iv4-SC Fab fragments generated no, or weak, Ca2+ flux responses in any of the single or double knockin cells (Figure 5B, bottom panels). The bifunctional iv4/iv9 generated a robust Ca2+ flux response in VRC01 HC/LC double knockin cells but failed to activate iGL-VRC01 HC or iGL-VRC01 LC knockin B cells (Figure 5B, bottom panels). Collectively, these data demonstrate that iv4/iv9 can bind BCRs comprised of an iGL-VRC01-like HC or LC, but that it preferentially crosslinks and activates B cells expressing both iGL-VRC01 HCs and LCs in vitro.
iv4/iv9 selectively activates iGL-VRC01 B cells in vivo
The specific in vitro activation of iGL-VRC01 B cells with iv4/iv9, but not with each of the individual components, indicates that the bispecific antibody may have a selective advantage when used as an immunogen. However, in vitro assays do not necessarily reflect B cell activation in vivo where antigens can be presented to B cells on follicular dendritic cells or as parts of immune complexes. Moreover, the calcium flux assay does not inform on how an activated iGL-VRC01 B cell would compete in a polyclonal B cell response. Therefore, we set up adoptive transfer experiments in which B cells from a mouse homozygous for the iGL-VRC01 BCR were transferred into a WT mouse (Abbott et al., 2018). In this system, congenic marking allows for identification and tracking of the iGL-VRC01 B cells in a polyclonal background, with the iGL-VRC01 B cells being CD45.2+ and the B cells from the recipient WT mice being CD45.1+. In this study, no B cells from CD45.1+ recipient mice stained positive for iv4/iv9 (Figure S7A), while almost all B cells from the donor iGL-VRC01 mice were recognized by fluorescently labeled iv4/iv9 (Figure S7B).
CD45.1+ mice each received 500,000 purified, CellTrace Violet (CTV)-labeled B cells from homozygous CD45.2+ iGL-VRC01 mice. The precursor frequency of iGL-VRC01 B cells 24 h after injection was approximately 0.005% of all B cells corresponding to approximately 1 in 20,000 B cells (Figure 6A). Additional CD45.1+ mice each received 500,000 purified, CTV-labeled B cells from the iGL-VRC01 mice and were immunized intraperitoneally with adjuvanted PBS, iv4-SC, iv9-ST, or iv4/iv9. 5 or 14 days later the splenic B cell responses were analyzed by flow cytometry, and IgG titers were measured in the serum.
Figure 6. The bispecific iv4/iv9 molecule activates iGL-VRC01 B cells in vivo.

500,000 purified, CellTrace Violet (CTV)-labeled B cells from homozygous iGL-VRC01 CD45.2+ mice were transferred into wild-type recipient CD45.1+ mice. The next day, mice were immunized with adjuvant plus 10 μg of iv4-SC, iv9-ST, iv4/iv9, or with adjuvant alone mixed in PBS. Splenocytes and serum were harvested 5 or 14 days after immunization, and the B cell response was analyzed by flow cytometry and ELISA.
(A) The precursor frequency of live iGL-VRC01 B cells at the time of immunization (24 h after injection) was assessed in two separate experiments represented by white and black dots.
(B and C) The proliferation of antigen-specific (iv4/iv9+) B cells upon immunization was assessed by analyzing the intensity of CTV staining to determine the percentage of cells that had divided 5 days after immunization.
(D and E) Frequencies of total CD45.2+ B cells after 5 days (D) and 14 days (E).
(F) Frequency of antigen-specific (iv4/iv9+) CD45.2+ B cells in germinal centers (GC; CD45.2+iv4/iv9+, Bcl6+GL7+) at day 14.
(G) Gating strategy to identify antigen-specific GC (Bcl6+GL7+) B cells that are endogenous (CD45.1+) or transferred (CD45.2+).
(H) Frequency of antigen-specific GC B cells that were recruited from CD45.1+ or CD45.2+ B cells at day 14.
(I and J) Total IgG binding to eOD-GT8 (solid circles) and eOD-GT8 knockout (KO) (open circles, dashed lines) in serum of animals 14 days after immunization as measured by ELISA. (I) PBS immunized. (J) iv4/iv9 immunized.
Results are shown as mean ± SD, with each circle representing a single animal. The p values were determined by one-way ANOVA and a Tukey’s multiple comparison test (day 5) or Student’s t test (day 14). *p < 0.05, ***p < 0.001, ****p < 0.0001.
Five days after immunization, we observed that significantly more iv4/iv9-specific CD45.2+ B cells had divided in mice immunized with iv4/iv9 compared to mice immunized with monovalent iv4, iv9, or adjuvant alone (Figures 6B and 6C). These results indicate that although monomeric iv4-SC and iv9-ST proteins are immunogenic in vivo, iv4/iv9 is superior in targeting and successfully activating the desired iGL-VRC01 B cells even in a polyclonal B cell background. This increased proliferation, however, did not correlate with an increased frequency of CD45.2+ splenic B cells in recipient mice in any of the groups, at 5 or 14 days (Figures 6D and 6E). We observed that the frequency of antigen-specific CD45.2+ B cells in germinal centers (GCs) was significantly higher in animals immunized with iv4/iv9 compared to the other groups at day 14 (Figure 6F).
We next sought to confirm the on-target activation of B cells and analyzed whether antigen-specific GC B cells (Bcl6+GL7+iv4/iv9+) were of CD45.1+ (endogenous) or CD45.2+ (transferred) origin (Figure 6G). We observed that compared to the iv4-SC and iv9-ST groups, immunization with iv4/iv9 resulted in a larger percentage of antigen-specific GC B cells. Among these cells almost all originated from the iGL-VRC01 donor CD45.2+ mice, indicating that iv4/iv9 inducedminimal off-target activation (Figure 6H). In contrast, immunization with iv4-SC and iv9-ST elicited a greaterproportionof off-target responses (Figure 6H). We also tested serum reactivity with the germline-targeting immunogen eOD-GT8 (Figures 6I and 6J). Animals immunized with iv9-ST or iv4/iv9 showed detectable binding to eOD-GT8 14 days after immunization (Figure 6J) whereas the PBS or iv4-SC-immunized controls did not (Figure 6I). This binding is due to VRC01-class antibody responses, asno binding to the eOD-GT8 KO was detected (Figure 6I and 6J).
DISCUSSION
As an alternative to Env-derived germline-targeting immunogens, we recently reported that an ai-mAb raised against iGL-VRC01 can activate B cells expressing iGL-VRC01-class B cells in vivo (Dosenovic et al., 2019). A key observation from this study and our previous work on germline-targeting ai-mAbs is that identifying a single ai-mAb capable of engaging diverse VRC01-class precursor antibodies is very difficult. Indeed, most ai-mAbs recognize one unique feature of VRC01-class precursors, i.e., a VH1–2*02-derived HC or a rare 5-aa CDRL3, limiting their use as general priming immunogens, as they would activate off-target, non-germline VRC01-class antibodies (Figures 7A and 7B). These findings underscore the requirement for a comprehensive approach to screen and characterize ai-mAbs capable of recognizing the specific features of VRC01-class antibodies. However, we note that ai-mAbs may be particularly well suited to target B cells with HC-restricted features such as apex-precursor bNAbs that have rare, long CDRH3s.
Figure 7. Model representing how different divalent or multivalent ai-mAbs bind and activate B cells that have only VRC01-class HCs (left column), only VRC01-class LCs (middle column), or both (right column).

(A–D) Activation due to BCR cross-linking is depicted by the orange lightning bolt. BCR cross-linking with the bispecific molecule iv4/iv9 could lead to preferential activation of precursor B cells that possess the VRC01-class HC and LC features (VH1–2*2 and 5-aa CDRL3, respectively).
By integrating data from binding, B cell sorting, and structural studies we engineered a bispecific molecule derived from a HC-specific ai-mAb and a LC-specific ai-mAb. Based on its framework-directed mode of binding, the iv9 arm of the bispecific molecule would be expected to bind to any VH1–2-expressing B cell, including putative VRC01-class precursors with high (nanomolar) affinity (Figure 7C). Indeed, we observe that iv4/iv9 recognizes VH1–2-expressing B cells in pools of naive polyclonal B cells, and it readily stains murine B cells expressing knockin BCRs with a fixed iGL-VRC01 HC paired with diverse LCs (or vice versa). However, the bispecific design is intended to enforce specific BCR cross-linking and activation of VRC01-class precursor B cells through engagement of a VRC01 LC through the iv4 arm on an adjacent BCR, which would further improve the interaction strength through avidity (Figure 7C). The ability of the bispecific antibody to only activate B cells with the correct heavy/light pairing supports this design principle, and it suggests that not all of the B cells that were sorted by iv4/iv9 are representative of those that would be activated upon immunization. A similar “immunosubversion” concept has been applied to the design of mosaic nanoparticles that co-display diverse influenza hemagglutinin (HA) trimers in order to favor the cross-linking of BCRs that recognize conserved epitopes on diverse HAs, while avoiding cross-linking of BCRs specific for strain-specific epitopes (Kanekiyo et al., 2019).
We previously demonstrated that immunization with a multimeric variant of the iv8 ai-mAb more readily activated B cells compared to iv8 IgG or iv8 Fab in a 3BNC60SI knockin mouse (Dosenovic et al., 2019). The observation that divalent iv9 or iv4 IgG can activate B cells with only iGL-VRC01 HCs or only VRC01 LCs suggests that multimerization of the bispecific molecule described herein would lead to cross-linking of off-target BCRs (Figure 7D).
An advantage of using ai-mAbs as germline-targeting immunogens is that even if the ai-mAb activates off-target B cells, they will not be boosted by antigenically distinct Env-derived boosting immunogens. In this study, we show that this bispecific antibody is superior to the individual components in activating and expanding target B cells despite the presence of a diverse polyclonal repertoire. Moreover, most antigen-specific B cells and serum responses elicited by iv4/iv9 were on target.
We note that the VRC01 B cells in our adoptive transfer experiments were present at the high end of the estimated frequency in humans (Havenar-Daughton et al., 2018b), and they expressed a fixed inferred-germline BCR. However, our data indicate that the bispecific ai-mAb was superior at identifying VRC01-class B cell precursors among polyclonal human B cells compared to the individual components. Indeed, iv4/iv9 identified at least one putative VRC01 precursor B cell that cross-reacted with eOD-GT8. Assuming that such B cells would respond to immunization with iv4/iv9, they could potentially be boosted with a specifically designed germline-targeting Env. Future studies that include various prime-boost regimens with iv4/iv9 and Env derivatives, ideally in a polyclonal setting where multiple, diverse VRC01 precursors are present, will be necessary to fully assess the potential of iv4/iv9 as an effective prime immunogen.
Affinity maturation toward an ai-mAb could potentially reshape the BCRs following immunization. Since the iv9 arm of the bispecific molecule binds exclusively to the framework region, away from the Env-binding paratope (Borst et al., 2018; Scharf et al., 2013), it is unlikely that mutations that would improve affinity for iv9 would affect regions that interact with Env.
iv4 makes extensive contacts with the iGL-VRC01 CDRL3. Although iv4 selects for short 5-aa CDRL3s, only a minority of these contain Glu96. Glu96 has been demonstrated to be important for binding of putative VRC01 precursors to germline-targeting Env (Lin et al., 2020). Of the threeputative VRC01 precursorswe isolated in this study, PHVP6 was the only putative VRC01 precursor that bound a heterologous germline-targeting immunogen, and the only one with Glu96E. Since Tyr27 on the iv4 LC makes a hydrogen bond with Glu96 on iGL-VRC01, it is possible that B cells that are activated by iv4/iv9 may select for Glu96E through affinity maturation.
Our study highlights the potential ai-mAbs, and derivatives, as immunogens capable of initiating the production of VRC01-class antibodies and further suggests that the basic principle described herein is applicable to other classes of HIV-1 bNAbs. Similarly, ai-mAbs against non-HIV B cell targets with well-defined genetic features could be generated, such as those that target unmutated VH3–53-derived B cells, which recognize the receptor-binding domain of SARS-CoV-2 and potently neutralize the virus (Barnes et al., 2020; Yuan et al., 2020; Hurlburt et al., 2020).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew McGuire (amcguire@fredhutch.org).
Materials availability
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact. Materials described herein can be provided upon request and with execution of an MTA with The Fred Hutchinson Cancer Research Center. The use of pTT3-derived plasmids requires a license from the National Research Council of Canada.
Data and code availability
The accession number for the sequences of the variable regions of the ai-mAbs reported in this paper is GenBank: MT561048 - MT561059. The accession numbers for the structures of ai-mAbs in complex wiht iGL-VRC01 class antibodies reported in this paper are PDB: 7JLK (iv1), 6XOC (iv4), 7JLN (iv9), 6VRQ (iv12).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
All cell lines were incubated at 37°C in the presence of 5% CO2 and were not tested for mycoplasma contamination. 293-F and 293E (human female) were maintained in Freestyle 293 media with gentle shaking. DG-75 B cells (human male) were maintained in suspension in complete RPMI media.
Human subjects
Peripheral blood mononuclear cells (PBMCs) and serum were collected from HIV-uninfected adults recruited at the Seattle HIV Vaccine Trials Unit (Seattle, Washington, USA) as part of the study “Establishing Immunologic Assays for Determining HIV-1 Prevention and Control.” All participants signed informed consent, and the Fred Hutchinson Cancer Research Center institutional review committee (Seattle, Washington, USA) approved the protocol prior to study initiation. PBMC samples from donors were blindly selected at random with no considerations made for age or sex.
Mice
Mice homozygous for the iGL-VRC01 heavy and light chains (iGL-VRC01-Homo) were generated by crossing iGL-VRC01 HC mice (Jardine et al., 2015) with iGL-VRC01 LC mice (Abbott et al., 2018) to generate mice that were heterozygous for the iGL-VRC01 heavy and light chain. Heterozygous mice were interbred and genotyped to identify animals that were homozygous for the iGL-VRC01 heavy and light chains, which were used to set up and maintain a breeding colony. C57BL/6J and B6 CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) mice were purchased from Jackson Laboratory or bred in house. The mouse husbandry and experiments were approved and supervised by Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee.
METHOD DETAILS
Hybridoma generation
Wild-type C57BL/6J mice were injected three times with germline VRC01 which has a VH1–2*02 heavy chain and a VK3–11 light chain. Another set of mice were immunized with iGL-12A21 and iGL-3BNC60, which are also derived from VH1–2*02 heavy chain and have differing CDRH3 regions but utilize the VK1–33 light chain. 3 days after the final injection spleens were harvested and used to generate hybridomas at the Fred Hutchinson Antibody Technology Center. Hybridoma supernatants were initially screened against iGL-VRC01 or iGL-3BNC60 and iGL-12A21 to identify antigen-specific hybridomas. Supernatants from positive wells were then screened against a small panel of monoclonal antibodies that included iGL-VRC01 or iGL-3BNC60 and iGL-12A21 as well as other germline non-VRC01-class antibodies that served as isotype controls by ELISA. Wells containing hybridomas that displayed positive binding for iGL-VRC01, iGL-3BNC60 and iGL-12A21, but negative binding to isotype controls, were sub-cloned by limiting dilution and screened for binding to a larger panel of antibodies using biolayer interferometry (BLI).
Biolayer Interferometry (BLI)
BLI assays were performed on the Octet Red instrument at 30°C with shaking at 1,000 RPM.
ai-mAb binding screen
secreted ai-mAbs were captured using Anti-Mouse IgG Fc capture (AMC) biosensors by immersing sensors directly into hybridoma culture supernatants for 600 s. A baseline signal was recorded for 1 min in kinetics buffer (KB: 1X PBS, 0.01% BSA, 0.02% Tween 20, and 0.005% NaN3,) at pH 7.4. After loading, the baseline signal was then recorded for 60 s in KB. The sensors were then immersed in KB containing 20 μg/mL of purified human antibody for a 100 s association step. The maximum response (Rmax) was determined by averaging the nm shift over the last 5 s of the association step after subtracting the background signal from each analyte-containing well using empty AMC sensors at each time point. The binding screen included germline and mature VRC01-class mAbs, non-VRC01-class mAbs, and chimeric mAbs comprised of either a VRC01-class IgH paired with a non-VRC01-class IgL, or a non-VRC01-class IgH paired with a VRC01-class IgL.
Kinetic analysis
Anti-Human IgG Fc capture (AHC) sensors were immersed in KB containing 10 μg/ml of purified ai-mAb for 200 s. After loading, the baseline signal was then recorded for 60 s in KB. The sensors were then immersed into wells containing serial dilutions of purified recombinant iGL-VRC01-class Fabs diluted in KB for 200 s (association phase), followed by immersion in KB for an additional 600 s (dissociation phase). The background signal from each analyte-containing well was measured using empty reference sensors and subtracted from the signal obtained with each corresponding ai-mAb loaded sensor. The background signal of ligand-coupled sensors in KB was subtracted from each sensor at each time-point. Kinetic analyses were performed at least twice with an independently prepared analyte dilution series. Curve fitting was performed using a 1:1 binding model and the ForteBio data analysis software. Mean kon, koff values were determined by averaging all binding curves that matched the theoretical fit with an R2 value of ≥ 0.99.
Binding screens with ai-mAbs and putative human VRC01-like precursors (PHVPs)
Streptavidin (SA) biosensors were immersed in KB containing 10 μg/ml of biotinylated PHVPs for 180 s. After loading, the baseline signal was then recorded for 60 s in KB. The sensors were then immersed into wells containing various proteins (ai-mAb at 250nM, Env derivatives at 1μM) diluted in KB for 200 s (association phase), followed by immersion in KB for an additional 600 s (dissociation phase). The background signal of ligand-coupled sensors in KB was subtracted from each sensor at each time-point.
Multiplex bead array
Purified recombinant human mAbs were directly conjugated to one of five intensities of EDC activated fluorescent QuantumPlex M Carobxyl magnetic beads using EDC according to the manufacturer’s instructions. Following conjugation excess mAb was removed by immobilizing the beads on a magnet and subsequent washing with PBS. The beads were then stored in PBS containing 1% goat serum and 0.02% sodium azide until use. To verify conjugation efficiency each bead was incubated with an anti-human IgG FITC antibody. Beads that showed an MFI > 250,000 were used in further analyses.
Beads were pooled into sets of 5 based on the internal (starfire red) fluorescence intensity of the bead. Bead sets were arrayed on a 96-well plate at a density of 100,000 beads per well (20,000 of each fluorescence intensity). 2.5 ng of biotinylated recombinant ai-mAb was added to wells containing each bead set in a total volume of 20 μl. After 5 minutes incubation at room temperature, 5 μl of BV421-streptavidin (1:200) was added to each well. Samples were then run on an advanced high throughput flow cytometry instrument iQue to measure the BV421 MFI for each bead. The MFI of ai-mAb incubated with beads conjugated to the control anti-EBV mAb AMMO1 were subtracted from every other bead.
Plasmids
The VH and VL sequences of the anti iGL-VRC01 class hybridomas were recovered using the mouse iG primer set using the protocol outlined in Siegel (2009), and Sanger sequenced (Genewiz). The VH/VL sequences were codon-optimized and cloned into full-length pTT3 derived IgG1 and IgL kappa expression vectors containing human constant regions using Gibson assembly (Snijder et al., 2018).
The mAb leader sequences and VH regions were also cloned in frame with AA 1–106 of the IgG1 constant region encompassing the C1 fused to AA 24–121 of spycatcher (Zakeri et al., 2012), followed by a gly-gly linker and a 6X His tag in pTT3 (pTT3-iv4-Fab-HC-SC).
Alternatively, mAb leader sequences and VH regions were also cloned in frame with AA 1–106 of the IgG1 constant region encompassing the C1 region followed by a gly-ser linker, 6X His tag and spytag (Zakeri et al., 2012) (pTT3-iv9-Fab-HC-ST).
The plasmid encoding the pTT3 derived IgG1 heavy chain of iGL-VRC01 was modified to express a 6X His tag and a stop codon downstream of the C1 region using site-directed mutagenesis to create pTT3-iGL-VRC01-HC-Fab.
pTT3-iv1-scFv (single chain variable fragment) was modified with a (GGGGS)3 linker from the C terminus of the light chain variable region to the N terminus of the heavy chain variable region followed by a 6X His tag.
pTT3-iv4 was cloned as a Fab with a 6X His tag on the C terminus of the heavy chain. To improve stability for protein crystallization, an elbow region mutation was inserted in the heavy chain at residues (114SSASTKG120) by deleting a serine and mutating (115SASTKG120) into (111FNQIKG118) (Bailey et al., 2018; Henderson et al., 2019).
iGL-VRC01 (VK3-1-derived kappa chain with C98S mutation in the heavy chain (Borst et al., 2018) and iGL-12A21 expression plasmids were prepared as previously described (Hoot et al., 2013; Scheid et al., 2011; McGuire et al., 2013).
pTT3 plasmids expressing eOD-GT8 and eOD-GT8-KO which contains the D276N, W277F, R278T, D279A, and D368R mutations have been described previously (Jardine et al., 2015; Dosenovic et al., 2019).
Recombinant protein expression and purification
All recombinant proteins were produced in 293E cells in Freestyle 293 media using the 293Free transfection reagent according to the manufacturer’s instructions. Cells were transfected at a density of 106 cells/mL. For Fabs and IgGs, 250 μg heavy and 250 μg light chain encoding plasmids were co-transfected per liter of suspended culture. For scFv, eOD-GT8, or eOD-GT8-KO 250 μg of DNA was transfected per liter of suspended culture.
Expression was carried out for 6 days after which cells and cellular debris were removed by centrifugation at 4,000 × g followed by filtration through a 0.22 μm filter. Clarified cell supernatant containing recombinant antibodies was passed over Protein A Agarose resin. Protein A resin was extensively washed with PBS and eluted with IgG elution buffer. iv9 IgG and iv12 IgG were digested into Fab using Endoproteinase Lys-C by adding 1 μg of LysC per 10 mg of IgG and the mixture incubated at 37°C overnight. Following digestion, the protein was incubated with Protein A agarose resin for 1 hour to remove any Fc or undigested IgG from the preparation. iv4 and iGL-12A21 Fabs and iv1 scFv were purified over His60 Ni-Superflow resin. Ni resin was washed with 150 mM NaCl, 5 mM HEPES pH 7.5, 20 mM Imidazole; and eluted with 150 mM NaCl, 5 mM HEPES pH 7.5, and 300 mM imidazole. Fabs and scFv were further purified by size-exclusion chromatography (SEC) using a HiLoad 16/600 Superdex 200 pg column in 150 mM NaCl, 5 mM HEPES pH 7.5. Clarified cell supernatant containing eOD-GT8-his-avi and eOD-GT8-his-avi, was passed over Nickel nitrilotriacetic acid (Ni-NTA) resin pre-equilibrated with Ni-NTA binding buffer (0.3 M NaCl, 20 mM Tris,10mM imidazole, pH 8.0), followed by extensive washing with Ni-NTA binding buffer, and then eluted with 250 mM imidazole, 0.3 M NaCl, 20 mM Tris, pH 8.0 (Ni-NTA elution buffer). All affinity purified proteins were further purified using a using a HiLoad 16/600 Superdex 200 pg column pre-equilibrated in PBS. Fractions containing purified proteins were pooled, aliquoted, frozen in liquid nitrogen and stored at −20°C.
SpyCatcher/SpyTag Fab purification
pTT3-iv4-Fab-HC-SC plus pTT3-iv4 LC, or pTT3-iv9-Fab-HC-ST plus pTT3-iv9 LC were transfected into 293E cells at a density of 106 cells/mL in Freestyle 293 media using the 293Free transfection reagent according to the manufacturer’s instructions. Expression was carried out for 6 days after which cells and cellular debris were removed by centrifugation at 4,000 × g followed by filtration through a 0.22 μm filter. Clarified cell supernatant containing iv4-SC or iv9-ST was passed over Ni-NTA resin, pre-equilibrated with Ni-NTA binding buffer (0.3 M NaCl, 20 mM Tris,10mM imidazole, pH 8.0), followed by extensive washing with Ni-NTA binding buffer, and then eluted with 250 mM imidazole, 0.3 M NaCl, 20 mM Tris, pH 8.0 (Ni-NTA elution buffer). Proteins were further purified by SEC using a Enrich SEC 650 10×300 column equilibrated in PBS.
To generate a bispecific iv4/iv9, iv4-Fab-SC was incubated with a 1.5-fold excess of iv9-Fab-ST for 1 hour at room temperature. The complex was then separated from excess iv9-Fab ST using a Enrich SEC 650 10×300 column equilibrated in PBS. iv4SC/iv9ST (referred to iv4/iv9) was either conjugated to DyLight 550-NHS ester and then flash frozen, or was flash frozen without a fluorescent label and stored at −20°C.
Crystallization and data collection
Complexes of iv1 scFv/iGL-VRC01 Fab, iv4-HL Fab/iGL-VRC01 Fab, iv9 Fab/iGL-VRC01 Fab and iv12 Fab/iGL-12A21Fab were concentrated to ~4.5, ~9, ~10, and ~10 mg/mL, respectively (Table S3). Complexes were screened using commercially available screens: Rigaku Wizard Precipitant Synergy block #2, Molecular Dimensions Proplex HT-96, Clear Strategy screens #1, and Hampton Research Crystal Screen HT; using vapor diffusion method. An NT8 dispensing robot, and Rock Imager was used to set up and screen crystallization conditions. Crystallization conditions were as follows: 0.1 M HEPES pH 7.0, PEG 8000 for iv1 scFv/iGL-VRC01 Fab; 0.1 M Tris pH 7.5, 8, %w/v PEG 20K, 8%w/v PEG MME 550, 0.2 M KSCN for iv4-HL Fab/iGL-VRC01 Fab; 0.08% Sodium Acetate pH 3.5, 24% PEG 4000, 0.16M Ammonium Acetate for iv9 Fab/iGL-VRC01 Fab and 0.1M KCL, 0.1 M Tris pH8, and 15%w/v PEG MME 2 K for iv12 Fab/iGL-12A21 Fab.
Data collection, structure solution, and refinement
Diffraction data was collected at SSRL beamline 41–1, and ALS beamlines 5.0.1 and 5.0.2. Data was processed using HKL2000 (Otwinowski and Minor, 1997). The structures were solved by molecular replacement using Phaser in CCP4 (McCoy et al., 2007) using the iGL-VRC01 PDB ID: 6MFT (Dosenovic et al., 2019; Borst et al., 2018) as a starting model. Structure building and refinement was done in COOT (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010). Data collection and refinement statistics for the crystal structures are provided in Table S3.
Preparation of zenon mAb labeling decoys
Zenon APC-DL755 was generated by conjugating the Zenon APC Human IgG labeling reagent, from the Zenon Allophycocyanin Human IgG Labeling Kit to DyLight 755 NHS Ester according to manufacturers’ instructions. Zenon PE-DL650, was generated by conjugating the Zenon PE Human IgG labeling reagent from the Zenon R-Phycoerythrin Human IgG Labeling Kit with DyLight 650 NHS Ester according to the manufacturers’ instructions. To create decoy reagents, Zenon APC-DL755 and Zenon PE-DL650 were incubated with Zenon blocking reagent at a 1:1 ratio.
Human B cell sorting
Cryopreserved PBMCs were thawed and resuspended in 200 μl of FACS buffer (PBS, 2% heat inactivated fetal bovine serum, 1 mM EDTA). B cells were isolated using the StemCell Human B Cell Enrichment Kit according to the manufacturers’ instructions. Enriched B cells were resuspended in 200 μl of FACS buffer and incubated with 10 μl rat serum, 10 μl mouse serum, 10 μl mouse anti-human CD32, 10 μl mouse anti-human CD23 and 10 μl mouse anti-human CD16. Cells were then washed with FACS buffer and resuspended in 200 μl FACS buffer and incubated with 5 μl of Zenon APC-DL755 and 5 μl of Zenon PE-DL650 and incubated on ice for 10 minutes. Next, B cells were co-stained with ai-mAbs conjugated to either Zenon-PE or Zenon-APC and incubated for an additional 10 minutes on ice. B cells were then stained with CD19-BV711 (1:200 dilution), CD27-PE-Cy7 (1:600 dilution), CD14- FITC (1:60 dilution), CD3-FITC (1:60 dilution), CD20-AF700 (1:300 dilution), IgD-PerCP-Cy5.5 (1:120 dilution), IgM-BV605 (1:120 dilution), and Fixable Viability Dye-eFluor 506 (1:300 dilution). Cells were then washed with 5 mL of FACS buffer and resuspended in 0.5 mL of FACS buffer. Cells were sorted by flow cytometry using a BD FACS Aria II. Naive ai-mAb+ B cells were defined as live, CD14−, CD3−, CD19+, CD20+, IgM+, IgD+, CD27−, APC-DL755−, PE-DL650−, PE+, APC+.
Naive ai-mAb+ B cells were single-cell sorted into 96 well plates. cDNA was generated using iScript and the VH and VL sequences were recovered using gene specific primers and cycling conditions previously described (Tiller et al., 2008). VH or VL amplicons were sanger sequenced (Genewiz). The antibody gene usage was assigned using IMGT V-quest (Brochet et al., 2008).
An estimation of the number of naive B cells analyzed for each sort was calculated based on the frequency of naive B cells that stained positive for each ai-mAb and the number of cells sorted (number of naive B cells analyzed = the number of cells sorted divided by the frequency of naive B cells that stained, × 100).
Engineering B cells that stably express Env-specific B cell receptors
Lentiviral delivery plasmids to express the iGL-12A21 and iGL-BNC60 BCRs were created by replacing the iGL-3BNC60 and iGL-12A21 VH and VL regions in the pRRL.EuB29.VRC01.BCR.GFP.WPRE plasmid (McGuire et al., 2014a). Lentiviruses were produced by co-transfecting the pRRL EuB29, psPAX2 and pMD2.G plasmids at a 4:2:1 ratio (6, 3, and 1.5 μg, respectively) into 293T in DMEM + 10% FBS using the GeneJuice Transfection reagent according to the manufacturer’s instructions. Transfected cells were cultured at 37°C in the presence of 5% CO2 for 72 hours. The supernatant was then passed through a 0.22 μm filter and then added directly to DG-75 B cells at a density of 1×106 cells/ml in cRPMI + 4 μg/ml polybrene. 18–24 hours later the cells were diluted to 0.5 ×106 cells/ml and maintained in culture. 7–10 days post-transduction GFP positive cells (i.e., BCR-transduced cells) were sorted by FACS on a Beckman FACS Aria II. Sorted cells were expanded and cultured indefinitely. Expression of exogenous BCRs was verified by staining aliquots of sorted cells with APC-conjugated mouse monoclonal anti-human IgG.
Mouse B cell binding
B cells were enriched from single cell suspensions of splenocytes harvested from mice homozygous for either the iGL-VRC01 LC or HC using the Mouse B Cell Isolation Kit. Retro-orbital bleeds were performed to obtain PMBCs from a HC/LC double knock-in mouse and a C57BL/6 WT mouse; no enrichment for B cells was performed for the RO blood samples. B cells purified from splenocytes or PMBCs from whole blood were stained with: anti-CD19 BUV395 (1:200 dilution), anti-CD3 BV711 (1:100 dilution), anti-Ly6G BV711 (1:100 dilution), anti-F4/80 BV711 (1:100 dilution), CD45R/B220 BV786 (1:250 dilution), Fixable Viability Dye eFluor 506 (1:200 dilution) and iv4/iv9 conjugated directly to DyLight 550. The percentage of live, CD3− GR1− F4/80− CD19+ B220+ B cells positive for iv4/iv9 was assessed on a Fortessa X50 flow cytometer and analyzed using FlowJo software.
Calcium flux assays
DG-75 derived cell lines
Calcium mobilization in B cells stably expressing BCRs upon stimulation with different ai-mAbs was monitored as previously described (Hoot et al., 2013; McGuire et al., 2013, 2014a, 2014b). Briefly, prior to antigenic challenge, cells were loaded with Fluo-4 Direct calcium indicator, mixed 1:1 with complete RPMI at 37°C for 45 min. Cells were pelleted and stained with APC-conjugated mouse monoclonal anti-human IgG (1:10 dilution in 100 μl complete RPMI plus Fluo-4 Direct) for 15 min. The cells were washed with 5 mL of complete RPMI and resuspended at ~1×106 cells/ml in complete RPMI and subjected to Ca2+ flux analysis at a medium flow rate on a Fortessa X50 cytometer. In all cases BCR transduced cells were mixed with un-transduced cells as an internal control at a 7:3 ratio.
Levels of background fluorescence (MinFL) were determined by averaging the background Fluo-4 absorbance in cells for 30 s. After that, activation of B cells expressing exogenous BCRs by ai-mAbs was determined by monitoring changes in Fluo-4 fluorescence associated with cells expressing the exogenous BCRs (APC positive cells) for 210 s. Ionomycin was added to a final concentration of 6.5 nM for 60 s and maximum Fluo-4 fluorescence (MaxFL) was established by averaging changes in Fluo-4 fluorescence recorded during the last 10 s. The percent of maximum Fluo-4 fluorescence at each time point t was determined using the formula: (Fluorescence at t-MinFL) / (MaxFL-MinFL) × 100. This analysis was performed on both the BCR positive (anti-IgG-APC positive) and BCR negative cells (anti-IgG-APC negative) simultaneously. The background Fluo-4 fluorescence signal from the BCR negative cells was subtracted from that of the BCR positive population at each time point.
Primary murine cells
B cells were isolated from single cell splenocyte suspensions of mice homozygous for either the iGL-VRC01 LC, HC, or HC/LC using the Mouse B cell isolation kit. Prior to antigenic challenge, purified B cells were loaded with Fluo-4 Direct calcium indicator, mixed 1:1 with complete RPMI at 37°C for 45 min. The cells were washed with 5 mL of complete RPMI and resuspended at ~1×106 cells/ml in complete RPMI and subjected to Ca2+ flux analysis as described above using a Fortessa X50 cytometer. The following antigens were tested on the mouse B cells at a 100 nM concentration: iv4 IgG, iv9 IgG, iv4-spycatcher, iv9-spytag, iv4/iv9, and an isotype control.
Mouse adoptive transfers and immunization scheme
Single cell suspensions of splenocytes were prepared from CD45.2+ mice homozygous for the iGL-VRC01 heavy and light chains by mechanical disruption of the spleens through a 70 μm filter and incubating with 2 mL of RBC Lysing Buffer. B cells were enriched using the Mouse B cell isolation kit according to the manufacturer’s instructions and resuspended at 5 × 107 cell/mL in warm 1x DPBS. CellTrace Violet was added to a final concentration of 5 μM and incubated at 37C for 12 min. Labeling was quenched by diluting cells in PBS + 10% FBS + 1 mM EDTA. 5×105 labeled cells were resuspended in 100 μL of warm 1x DPBS per mouse and transferred into B6 CD45.1+ recipient hosts via retro-orbital injection. One day after cell transfer, 10 μg of recombinant ai-mAbs (iv4-spycatcher, iv9-spytag or iv4/iv9) resuspended in 50 μL PBS were mixed with 50 μL of adjuvant and injected intraperitoneally. Mice were euthanized 5 or 14 days after immunization, where spleens were harvested and whole blood was collected by terminal cardiac bleed. Splenocyte isolation and B cell enrichment using the Mouse B cell isolation kit was performed prepared as described above. Serum was obtained by subsequent centrifugation at 10,000 rpm for 5 min.
The isolated B cells were surface stained with anti-GL7 PerCP-Cy5.5 (1:200 dilution), anti-CD45.2 Alexa Fluor 647 (1:100 dilution), anti-CD38 Alexa Fluor 700 (1:100 dilution), anti-CD45.1 PE-eFluor 610 (1:100 dilution), anti-CD19 BUV395 (1:200 dilution), anti-CD45R/B220 BV786 (1:250 dilution), anti-CD3 BV711 (1:100 dilution), anti-Ly6G BV711 (1:100 dilution), anti-F4/80 BV711 (1:100 dilution), and Fixable Viability Dye eFluor 506 (1:200 dilution) in FACS buffer (PBS + 2% FBS + 1 mM EDTA) at 4°C for 30 minutes. Cells were washed once with FACS buffer and subsequently fixed and permeabilized in Foxp3/Transcription factor Fix and Perm buffer for 1 hour on ice and then intracellularly stained overnight at 4°C with Bcl6-FITC (1:100 dilution). Cells were washed once with FACS buffer and resuspended in FACS buffer for acquisition. Staining was measured on a Fortessa X50 cytometer and analyzed with FlowJo. For proliferation analysis, cells were gated as singlets > lymphocytes > live, CD3− Ly6G− F4/80− > CD19+ B220+ B cells > CD45.2+ > iv4/iv9+ > CTV+. For GC analysis, cells were gated as singlets > lymphocytes > live, CD3− Ly6G− F4/80− > CD19+ B220+ B cells > CD45.2+ > iv4/iv9+ > Bcl6+ GL7+. For on- and off-target activation analysis, cells were gated as singlets > lymphocytes > live, CD3− Ly6G− F4/80− > CD19+ B220+ B cells > iv4/iv9+ > Bcl6+ GL7+ > CD45.1 or CD45.2.
Serum IgG ELISA
Serum titers against eOD-GT8 and eOD-GT8 KO were evaluated by Ab capture ELISA. Briefly, clear flat-bottom immuno 384-well plates were coated with 30 μL eOD-GT8 or eOD-GT8 KO at 2 μg/mL in 0.1M sodium bicarbonate coating buffer overnight at room temperature. Plates were washed 4x with wash buffer (PBS + 0.02% Tween 20) and subsequently blocked with 100 μL of blocking buffer (PBS + 10% non-fat milk + 0.03% Tween 20) for 1.5 hours at 37C. After washing 4x with wash buffer, serum was prepared at a starting dilution of 1:10 in blocking buffer, serially diluted three-fold in plate and incubated for 1 hour at 37C. The secondary antibody HRP goat anti-mouse IgG was added after 4x wash and plates were incubated for 1 hour at 37C. After a final 4x wash, 30 μL of SureBlue Reserve TMB peroxidase substrate was added and incubated for 5 min, followed by addition of 30 μL of 1N H2SO4. Plates were analyzed at 450 nm using a SpectraMax M2 plate reader. All wash steps were performed using a BioTek 405 Select Microplate Washer.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as mean ± SD. Data were analyzed using GraphPad Prism 8.4.1 software by one-way ANOVA and Tukey multiple comparisons post-test or Student’s t test, as indicated. Values were considered statistically different with *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD32 | BD Biosciences | Cat# 551900; RRID: AB_394290 |
| Anti-human CD16 | BD Biosciences | Cat# 550383; RRID: AB_393649 |
| Anti-human CD23 | BD Biosciences | Cat# 555707; RRID: AB_396053 |
| Anti-human CD19-BV711 | BioLegend | Cat# 302246; RRID: AB_2562065 |
| Anti-human CD27-PE-Cy7 | eBioscience | Cat# 25-0271-82; RRID: AB_1724035 |
| Anti-human CD14-FITC | BD Biosciences | Cat# 557153; RRID: AB_396589 |
| Anti-human CD3-FITC | BD Biosciences | Cat# 556611; RRID: AB_396484 |
| Anti-human CD20-Alexa Fluor 700 | BioLegend | Cat# 302322; RRID: AB_493753 |
| Anti-human IgD-PerCP-Cy5.5 | eBioscience | Cat# 46-9868-42; RRID: AB_2573920 |
| Anti-human IgM-BV605 | BioLegend | Cat# 314524; RRID:AB_2562374 |
| Anti-human IgG-APC | BD Biosciences | Cat# 550931; RRID: AB_398478 |
| Anti-human IgG-FITC | Jackson ImmunoResearch | Cat# 109-095-008; RRID: AB_2337651 |
| Anti-mouse CD19-BUV395 | BD Biosciences | Cat# 563557; RRID: AB_2722495 |
| Anti-mouse CD3-BV711 | BioLegend | Cat# 100349; RRID: AB_2565841 |
| Anti-mouse Ly6G-BV711 | BioLegend | Cat# 127643; RRID: AB_2565971 |
| Anti-mouse F4/80-BV711 | BioLegend | Cat# 123147; RRID: AB_2564588 |
| Anti-mouse B220-BV786 | BD Biosciences | Cat# 563894; RRID: AB_2738472 |
| Anti-mouse GL7-PerCP-Cy5.5 | BioLegend | Cat# 144609; RRID: AB_2562978 |
| Anti-mouse CD45.2-Alexa Fluor 647 | BioLegend | Cat# 109818; RRID: AB_492870 |
| Anti-mouse CD38-Alexa Fluor 700 | eBioscience | Cat# 56-0381-82; RRID: AB_657740 |
| Anti-mouse CD45.1-PE-eFluor 610 | eBioscience | Cat# 61-0453-82; RRID: AB_2574560 |
| Anti-mouse Bcl6-FITC | BioLegend | Cat# 358513; RRID: AB_2860942 |
| HRP-conjugated goat anti-mouse IgG | BioLegend | Cat# 405306; RRID: AB_315009 |
| Biological Samples | ||
| PMBC from HIV-uninfected donors (Establishing Immunologic Assays for Determining HIV-1 Prevention and Control) | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Streptavidin-BV421 | BioLegend | Cat# 405226 |
| Fixable Viability dye-eFluor 506 | eBioscience | Cat# 65-0866-14 |
| Protein A agarose resin | Thermo Scientific | Cat# 20333 |
| IgG elution buffer | Thermo Scientific | Cat# 21004 |
| Endoproteinase LysC | New England BioLabs | Cat# P8109S |
| His60 Ni superflow resin | Takara | Cat# 635664 |
| HiLoad 16/600 Superdex 200 pg column | GE Healthcare | Cat# 28-9893-35 |
| Ni-NTA agarose resin | QIAGEN | Cat# 30230 |
| ENrich SEC 650 10 × 300 column | Biorad | Cat# 7801650 |
| DyLight 550 NHS ester | Thermo Scientific | Cat# 62262 |
| DyLight 650 NHS ester | Thermo Scientific | Cat# 62266 |
| Zenon Allophycocyanin human IgG labeling kit | Thermo Scientific | Cat# Z25451 |
| Zenon R-Phycoerythrin human IgG labeling kit | Thermo Scientific | Cat# Z25455 |
| Fluo-4 direct calcium indicator | Thermo Scientific | Cat# F10471 |
| Ionomycin | Millipore-Sigma | Cat# I9657 |
| SureBlue Reserve TMB Peroxidase Substrate | Seracare KPL | Cat# 5120-0080 |
| CellTrace Violet (CTV) | Thermo Scientific | Cat# C34557 |
| Sigma Adjuvant System | Sigma | Cat# S6322 |
| FoxP3/Transcription factor Fix and Perm buffer | eBioscience | Cat# 00-5523-00 |
| RBC lysing buffer | Sigma | Cat# R7757 |
| QuantumPlex M Carobxyl magnetic beads | Bangs Laboratories | Cat# 250 |
| Mouse Ig-Primer set | Millipore-Sigma | Cat# 69831 |
| GeneJuice | Millipore-Sigma | Cat# 70967 |
| 293 Freestyle media | Thermo Scientific | Cat# 12-338-018 |
| 293 Free transfection reagent | Millipore-Sigma | Cat# 72181 |
| Critical commercial assays | ||
| Mouse B cell isolation kit | StemCell Technologies, Inc. | Cat# 19854 |
| Human B cell enrichment kit | StemCell Technologies, Inc. | Cat# 19054 |
| iScript | Bio-Rad | Cat# 1708891 |
| Deposited data | ||
| ai-mAb VH and VL sequences | This paper | GenBank: MT561048 - MT561059 |
| iv1-iGL-VRC01 complex | This paper | PDB: 7JLK |
| iv4-iGL-VRC01 complex | This paper | PDB: 6XOC |
| Iv9-iGL-VRC01 complex | This paper | PDB: 7JLN |
| Iv12-iGL-12A21 complex | This paper | PDB: 6VRQ |
| Experimental models: Cell lines | ||
| 293-6E | National Research Council, Canada | N/A |
| HEK293T | ATCC | Cat#C RL-3216 |
| DG-75 B cells | ATCC | Cat# CRL-2625 |
| Experimental models: Organisms/strains | ||
| iGL-VRC01 HC homozygous mice | This paper | N/A |
| iGL-VRC01 LC homozygous mice | This paper | N/A |
| iGL-VRC01 HC+LC homozygous mice | This paper | N/A |
| B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | 002014 |
| C57BL/6J | The Jackson Laboratory | 000664 |
| Oligonucleotides | ||
| Primers for antibody nested PCR and sequencing | Tiller et al., 2008 | N/A |
| Spycatcher (AA 24-121) | Zakeri et al., 2012 | GenBank: AFD50637.1 |
| Spytag: GGSGGSGHHHHHHGSAHIVMVDAYKPTK | Zakeri et al., 2012 | N/A |
| Recombinant DNA | ||
| pTT3-iv4-Fab-HC-SC | This paper | N/A |
| pTT3-iv9-Fab-HC-ST | This paper | N/A |
| pTT3-iv1-scFv | This paper | N/A |
| pTT3-iGL-VRC01-HC-Fab | This paper | N/A |
| pTT3-iv4 LC | This paper | N/A |
| pTT3-iv9 LC | This paper | N/A |
| pTT3-eOD-GT8 | Jardine et al., 2015 | N/A |
| pTT3-eOD-GT8-KO | Dosenovic et al., 2019 | N/A |
| pRRL.EuB29.VRC01.BCR.GFP.WPRE | McGuire et al., 2014a | N/A |
| psPAX2 | Addgene | Cat# 12260 |
| pMD2.G | Addgene | Cat# 12259 |
| Software and algorithms | ||
| Flow Jo, version 9.9.4 | Tree Star | https://www.flowjo.com |
| Prism, version 8.4.1 | GraphPad Software | https://www.graphpad.com |
| ForteBio data analysis software | ForteBio | N/A |
| HKL2000 | Otwinowski and Minor, 1997 | https://www.hkl-xray.com/how-reference-hkl-hkl-2000-and-hkl-3000 |
| Phaser | McCoy et al., 2007 | https://www.phenix-online.org/documentation/reference/phaser.html |
| Phenix.refine | Afonine et al., 2012 | https://www.phenix-online.org/documentation/reference/refinement.html |
| Coot | Emsley et al., 2010 | RRID:SCR_014222 |
| Pymol | Delano, 2002 | https://pymol.org/2/ |
| Other | ||
| SpectraMax M2 plate reader | Molecular Devices | https://www.moleculardevices.com |
| BioTek 405 select microplate washer | BioTek | https://www.biotek.com |
| FACS Aria II | Becton, Dickinson and Company | N/A |
| Symphony X50 flow cytometer | Becton, Dickinson and Company | N/A |
| IQue3 high throughput flow cytometer | Intellicyt | N/A |
| Octet RED96e | ForteBio | N/A |
| Anti-mouse IgG Fc capture (AMC) biosensors | ForteBio | Cat# 18-5088 |
| Anti-human IgG Fc capture (AHC) biosensors | ForteBio | Cat# 18-5060 |
| Streptavidin biosensors | ForteBio | Cat# 18-5019 |
| Rock Imager | Formulatrix | N/A |
Highlights.
Anti-idiotypic mAbs (ai-mAbs) were raised against germline VRC01-class mAbs
One is specific for unmutated VRC01-class heavy chains, another for light chains
These were used to develop a bispecific germline-targeting immunogen
The bispecific molecule selectively activates germline VRC01-class B cells in mice
ACKNOWLEDGMENTS
We thank Dr. M. Juliana McElrath for providing PBMC samples, Dr. David Nemazee for providing the iGL-VRC01 HC and iGL-VRC01 LC knockin mice, Dr. Pia Dosenovic for advice on adoptive transfer experiments, Jane Haass as well as the Fred Hutch Flow Cytometry Shared Resource for assistance with sorting, Ben Hoffstrom and Norman Boiani from the Fred Hutch Antibody Technology Facility for assistance with hybridoma generation and isolation, and the Pendleton Charitable Trust for its generous support of Formulatrix robotic instruments. Structural results shown in this study were collected at Structural Biology beamlines 5.0.1 and 5.0.2, which are supported in part by the National Institute of General Medical Sciences, National Institutes of Health. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the United States Department of Energy under contract no. DE-AC02-05CH11231. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). Research reported in this publication was supported by funding from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under award numbers R21 AI127249 (to A.T.M.), 1P01AI138212 (to L.S., A.T.M., and M.P.), R01 AI081625 (to L.S.), and R01 AI122912 (to J.J.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109084.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, et al. (2000). Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med 6, 200–206. [DOI] [PubMed] [Google Scholar]
- Bailey LJ, Sheehy KM, Dominik PK, Liang WG, Rui H, Clark M, Jaskolowski M, Kim Y, Deneka D, Tang WJ, and Kossiakoff AA (2018). Locking the elbow: Improved antibody Fab fragments as chaperones for structure determination. J. Mol. Biol 430, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs AB, Chen J, Hong CM, Rao DS, Yang L, and Baltimore D (2011). Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs AB, Ouyang Y, Hong CM, Chen J, Nguyen SM, Rao DS, An DS, and Baltimore D (2014). Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med 20, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft T, DeBuysscher BL, Weidle C, Schwartz A, Wall A, Gray MD, Feng J, Steach HR, Fitzpatrick KS, Gewe MM, et al. (2019). Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes. J. Exp. Med 216, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CO, West AP Jr., Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, et al. (2020). Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, Kozink DM, Parks RJ, Tomaras GD, Crump JA, et al. (2012). Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: Implications for vaccine design. J. Virol 86, 4688–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst AJ, Weidle CE, Gray MD, Frenz B, Snijder J, Joyce MG, Georgiev IS, Stewart-Jones GB, Kwong PD, McGuire AT, et al. (2018). Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core. eLife 7, e37688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, et al. (2016). Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166, 1459–1470.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, and Giudicelli V (2008). IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, and Hangartner L (2016). Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol 34, 635–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, and Mascola JR (2015). Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol 16, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, et al. (2012). A blueprint for HIV vaccine discovery. Cell Host Microbe 12, 396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, et al. (2010). Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 5, e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, and Georgiou G (2016). Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA 113, E2636–E2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2002). The PyMOL Molecular Graphics System (DeLano Scientific).
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. ; NISC Comparative Sequencing Program (2014). Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. (2015). Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, Kara EE, Pettersson AK, McGuire AT, Gray M, Hartweger H, Thientosapol ES, Stamatatos L, and Nussenzweig MC (2018). Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. USA 115, 4743–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, Pettersson A-K, Wall A, Thientosapol ES, Feng J, Weidle C, Bhullar K, Kara EE, Hartweger H, Pai JA, et al. (2019). Anti-idiotypic antibodies elicit anti-HIV-1-specific B cell responses. J. Exp. Med 216, 2316–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP Jr., Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, and Bjorkman PJ (2011). Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334, 1289–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. (2014). Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresman L, Jia F, Li Z, Wang C, Stephens EB, Sahni M, Narayan O, and Joag SV (1998). Neutralizing antibodies administered before, but not after, virulent SHIV prevent infection in macaques. AIDS Res. Hum. Retroviruses 14, 1035–1043. [DOI] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. (2013). Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756. [DOI] [PubMed] [Google Scholar]
- Gorman J, Chuang GY, Lai YT, Shen CH, Boyington JC, Druz A, Geng H, Louder MK, McKee K, Rawi R, et al. (2020). Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep. 31, 107488. [DOI] [PubMed] [Google Scholar]
- Gristick HB, von Boehmer L, West AP Jr., Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, and Bjorkman PJ (2016). Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol 23, 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H, Bournazos S, Ravetch JV, Ploss A, Nussenzweig MC, and Pietzsch J (2013). Antibody and antiretroviral preexposure prophylaxis prevent cervicovaginal HIV-1 infection in a transgenic mouse model. J. Virol 87, 8535–8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Abbott RK, Schief WR, and Crotty S (2018a). When designing vaccines, consider the starting material: The human B cell repertoire. Curr. Opin. Immunol 53, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Sarkar A, Kulp DW, Toy L, Hu X, Deresa I, Kalyuzhniy O, Kaushik K, Upadhyay AA, Menis S, et al. (2018b). The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med 10, eaat0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Watts BE, Ergin HN, Anasti K, Parks R, Xia SM, Trama A, Liao HX, Saunders KO, Bonsignori M, et al. (2019). Selection of immunoglobulin elbow region mutations impacts interdomain conformational flexibility in HIV-1 broadly neutralizing antibodies. Nat. Commun 10, 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, et al. (2007). Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449, 101–104. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, and Burton DR (2009). Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med 15, 951–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, and Stamatatos L (2013). Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 9, e1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, et al. (2014). Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Abbott RK, Havenar-Daughton C, Skog PD, Al-Kolla R, Groschel B, Blane TR, Menis S, Tran JT, Thinnes TC, et al. (2020). B cells expressing authentic naive human VRC01-class BCRs can be recruited to germinal centers and affinity mature in multiple independent mouse models. Proc. Natl. Acad. Sci. USA 117, 22920–22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt NK, Wan YH, Stuart AB, Feng J, Mcguire AT, Stamatatos L, and Pancera M (2020). Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. (2015). Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereño-Orbea J, Kalyuzhniy O, Deresa I, et al. (2016). HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol 20, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Abbott RK, Freeman BL, Haupt S, Groschel B, Silva M, Menis S, Irvine DJ, Schief WR, and Crotty S (2020). Multifaceted effects of antigen valency on B cell response composition and differentiation in vivo. Immunity 53, 548–563.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. (2013). Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871. [DOI] [PubMed] [Google Scholar]
- Lin YR, Parks KR, Weidle C, Naidu AS, Khechaduri A, Riker AO, Takushi B, Chun JH, Borst AJ, Veesler D, et al. (2020). HIV-1 VRC01 germline-targeting immunogens select distinct epitope-specific B cell receptors. Immunity 53, 840–851.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, et al. (1999). Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol 73, 4009–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, and Lewis MG (2000). Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med 6, 207–210. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. (2013). Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med 210, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, and Stamatatos L (2014a). Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science 346, 1380–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Glenn JA, Lippy A, and Stamatatos L (2014b). Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447–52D. J. Virol 88, 2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Gray MD, Dosenovic P, Gitlin AD, Freund NT, Petersen J, Correnti C, Johnsen W, Kegel R, Stuart AB, et al. (2016). Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun 7, 10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramírez M, Garces F, Escolano A, Skog P, de Taeye SW, Del Moral-Sanchez I, McGuire AT, Yasmeen A, Behrens AJ, Ozorowski G, et al. (2017). Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med 214, 2573–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, and Burton DR (2012). Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA 109, 18921–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, and Burton DR (2001). Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol 75, 8340–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J, Gruell H, Bournazos S, Donovan BM, Klein F, Diskin R, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, and Nussenzweig MC (2012). A mouse model for HIV-1 entry. Proc. Natl. Acad. Sci. USA 109, 15859–15864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi MM, Dashti A, Rikhtegaran Tehrani Z, Tolbert WD, Seaman MS, Ouyang X, Gohain N, Pazgier M, Kim D, Cavet G, et al. (2018). Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Pegu A, Georgiev IS, Zeng M, Joyce MG, Yang ZY, Ko SY, Chen X, Schmidt SD, Haase AT, et al. (2015). Sustained delivery of a broadly neutralizing antibody in nonhuman primates confers long-term protection against simian/human immunodeficiency virus infection. J. Virol 89, 5895–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, West AP Jr., Gao H, Lee T, Scheid JF, Nussenzweig MC, Bjorkman PJ, and Diskin R (2013). Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. USA 110, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, West AP, Sievers SA, Chen C, Jiang S, Gao H, Gray MD, McGuire AT, Scheid JF, Nussenzweig MC, et al. (2016). Structural basis for germline antibody recognition of HIV-1 immunogens. eLife 5, e13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. (2009). Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. (2011). Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho MW, and Martin MA (1999). Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med 5, 204–210. [DOI] [PubMed] [Google Scholar]
- Siegel RW (2009). Antibody affinity optimization using yeast cell surface display. Methods Mol. Biol 504, 351–383. [DOI] [PubMed] [Google Scholar]
- Snijder J, Ortego MS, Weidle C, Stuart AB, Gray MD, McElrath MJ, Pancera M, Veesler D, and McGuire AT (2018). An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein-Barr virus. Immunity 48, 799–811.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, Wood A, Lee EC, Le KM, Jones M, et al. (2016). Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Cheng C, Chen X, Duan H, Cheng HL, Dao M, Sheng Z, Kimble M, Wang L, Lin S, et al. (2016). Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires. Cell 166, 1471–1484.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H (2008). Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Busse CE, and Wardemann H (2009). Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350, 183–193. [DOI] [PubMed] [Google Scholar]
- Umotoy J, Bagaya BS, Joyce C, Schiffner T, Menis S, Saye-Francisco KL, Biddle T, Mohan S, Vollbrecht T, Kalyuzhniy O, et al. ; IAVI Protocol C Investigators; IAVI African HIV Research Network (2019). Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51, 141–154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. ; Protocol G Principal Investigators (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP Jr., Diskin R, Nussenzweig MC, and Bjorkman PJ (2012). Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. USA 109, E2083–E2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP Jr., Scharf L, Scheid JF, Klein F, Bjorkman PJ, and Nussenzweig MC (2014). Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. ; NISC Comparative Sequencing Program (2011). Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Liu H, Wu NC, Lee CD, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, et al. (2020). Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, and Howarth M (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 109, E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. (2010). Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. ; NISC Comparative Sequencing Program (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. ; NISC Comparative Sequencing Program (2015). Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequences of the variable regions of the ai-mAbs reported in this paper is GenBank: MT561048 - MT561059. The accession numbers for the structures of ai-mAbs in complex wiht iGL-VRC01 class antibodies reported in this paper are PDB: 7JLK (iv1), 6XOC (iv4), 7JLN (iv9), 6VRQ (iv12).


