Abstract
Circular (circ) RNA expression vectors are used as a method of identifying and characterizing RNA sequences that harbor internal ribosome entry site (IRES) activity. During the course of developing a vector series tailored for IRES discovery, we found evidence for the occurrence of trans-spliced mRNAs arising when sequences with promoter activity were embedded between the upstream CTD and downstream NTD exons of the pre-mRNA. These trans-spliced products regenerate the same open reading frame expected from a circRNA and can lead to false-positive signals in screens relying on circRNA expression vectors for IRES discovery. Our results caution against interpretations of IRES activity solely based on results obtained from circRNA expression vectors.
Keywords: circRNA, trans-splicing, IRES, gene expression
INTRODUCTION
Internal ribosome entry sites (IRESes) were initially identified by positioning the 5′ leader regions from picornaviruses as intercistronic regions within a bicistronic mRNA configuration (Jang et al. 1988; Pelletier and Sonenberg 1988). The bicistronic system proved to be an easy and efficient way of identifying and characterizing novel viral and cellular sequences that harbor IRES activity, but is not without its caveats (Jackson 2013). The presence of cryptic promoter activity or splicing events, ribosome readthrough activity, and ribosome shunting are phenomena that can result in translation of the second cistron in the absence of a bona fide IRES. In order to bolster claims that bicistronic expression is truly attributed to IRES-driven translation, a number of additional complementary experiments are recommended, some of which include: (i) assessing the 5′ end- and cap-dependency of any sequence being tested as a putative IRES in the bicistronic context, (ii) RNAi-mediated targeting of the first cistron of a bicistronic mRNA (which should lead to a concomitant reduction in expression of the second cistron), (iii) undertaking transfection using bicistronic mRNA into cells rather than DNA transfection, and (iv) ensuring that polysome association of the bicistronic mRNA is maintained upon inhibition of 5′ end- or cap-dependent translation (Jackson 2013). However, many studies lack these critical authentication steps. An alternative method to verifying putative IRES activity assay is demonstrating translation of a circular mRNA (circRNA) template harboring said IRES sequences (Chen and Sarnow 1995). Due to the lack of a free 5′ end, validation experiments using circRNAs are not vulnerable to false positive signals stemming from cryptic promoter activity, ribosome readthrough, or ribosome shunting. However, generating circRNAs in vitro using enzymatic ligation is inefficient and as a result, circRNAs have been utilized far less widely compared to the bicistronic reporter.
Naturally occurring circRNAs are generated by a head-to-tail backsplicing mechanism whereby the splice donor (SD) of an exon is covalently joined to a splice acceptor (SA) of an upstream exon (Salzman et al. 2012; Jeck et al. 2013; Memczak et al. 2013). This backsplicing event is promoted by proximal positioning of the introns flanking the SA and SD sites, which can be mediated through sequence complementarity or via RNA binding proteins (Dubin et al. 1995; Jeck et al. 2013; Ashwal-Fluss et al. 2014; Liang and Wilusz 2014; Zhang et al. 2014; Conn et al. 2015; Ivanov et al. 2015). CircRNAs have been implicated in several facets of gene expression regulation including acting as microRNA sponges (Hansen et al. 2013; Memczak et al. 2013), regulating RNA-binding protein function (Ashwal-Fluss et al. 2014), and affecting transcription rates (Li et al. 2015), although the number of circRNAs with ascribed activity is quite low. CircRNAs are predominantly cytoplasmic, and some have been reported to be translated (Pamudurti et al. 2017), although the precise mechanism of ribosome recruitment has not been extensively dissected but presumably involves an IRES.
As an alternative to generating circRNAs in vitro via ligation, mammalian circRNA expression vectors have been developed. These vectors typically consist of split reporter exons flanked by two introns with complementary regions that promote backsplicing of the pre-mRNA. Upon successful backsplicing, a circular RNA containing the coding information of the full-length reporter protein is generated. This type of system has been adapted for a genetic screen in which a library of random 10-nt sequences were inserted upstream of a circRNA-encoded GFP expression vector. From this screen, 97 hexamers were found to have IRES activity (https://www.biorxiv.org/content/10.1101/473207v4). During our development and testing of circRNA expression vectors tailored for IRES discovery, we found that trans-spliced products arose from such expression systems if a promoter was situated between the upstream carboxy-terminal domain (CTD)- and the downstream amino-terminal domain (NTD)-encoded exons. Our results caution against the use of current configurations of circRNA expression vectors for IRES discovery unless complemented by rigorous downstream validation experiments.
RESULTS
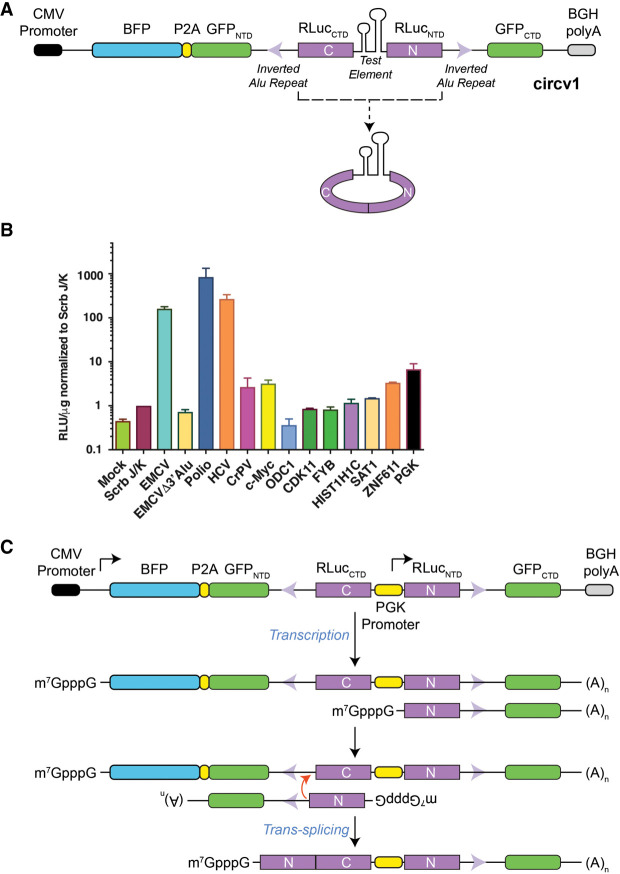
In designing a circRNA expression vector, we placed a fragment encoding the renilla (RLuc) luciferase NTD (RLucNTD) downstream from the RLuc CTD (RLucCTD) (Fig. 1A). We flanked these regions with introns derived from the PolR2A gene which harbor inverted Alu repeats since these have been previously shown to promote circRNA formation (Zhang et al. 2014). Our first-generation vector (circV1) also harbored a split GFP to report on linear splicing, as well as a BFP reporter to normalize for upstream transcriptional activity from the CMV promoter; however, these were not used in the current study given what emerged from our initial results (Fig. 1A).
FIGURE 1.
The PGK promoter, as test sequence in a circRNA expression vector context, leads to reporter expression. (A) Schematic diagram of circv1. The positions of the inverted Alu repeats with the two introns are denoted by violet arrowheads. (B) Relative RLuc expression obtained from 293T cells following transfection with circv1 harboring test elements from the indicated genes (Supplemental Table S1). Mock; mock transfection. Values are set relative to those obtained with EMCV Scrb J/K control sequences in circv1. n = 3 ± SEM. (C) Schematic illustrating potential mechanism by which a shortened RLucNTD-containing mRNA produced from the PGK promoter can participate in a trans-splicing reaction to generate a full-length, functional renilla ORF.
We initially inserted the EMCV IRES between the two RLuc synthetic exons and found expression of RLuc at levels 100-fold higher than obtained when using an EMCV IRES containing a scrambled J/K loop (Scrb), which leads to loss of IRES activity (Fig. 1B; Supplemental Table S1; Hoffman and Palmenberg 1995). RLuc expression from the EMCV IRES containing vector was Alu-dependent since removal of one of these repeats (from the intron downstream from RLuc NTD) reduced RLuc expression to levels comparable to those obtained with the Scrb J/K vector (Fig. 1B, see EMCVΔ3′Alu). Among other viral IRESes tested, polio and HCV sequences produced strong responses in this assay (Fig. 1B). The CrPV IRES was quite weak in the circv1 context, although it is possible that the spatial constraints of the circular RNA may have impeded proper folding of the IRES (Fig. 1B). We also tested nine different cellular 5′ leader regions previously reported to harbor IRES activity (Weingarten-Gabbay et al. 2016) and found that only two of these (c-Myc and ZNF611) showed activity above levels obtained with the Scrb J/K control (Fig. 1B; Supplemental Table S1). To assess whether the circv1 system was susceptible to false positive signals attributed to cryptic promoter activity, we inserted the PGK promoter into circv1. Surprisingly, the levels of RLuc obtained with circv1.PGK exceeded those obtained with any of the nine putative cellular 5′ leader regions. We hypothesize that our vector could produce two linear mRNAs: (i) the expected full-length mRNA driven from the CMV promoter, and (ii) a shorter mRNA arising from the PGK promoter harboring the RLuc NTD and downstream intron (Fig. 1C). Annealing of these two different mRNAs via their complementary Alu repeats would be predicted to favor a trans-splicing event with the resulting product being capped and capable of synthesizing RLuc protein (Fig. 1C). These results challenge the assumption that RLuc levels obtained with circRNA vectors, like circv1, solely arise from a circular RNA template.
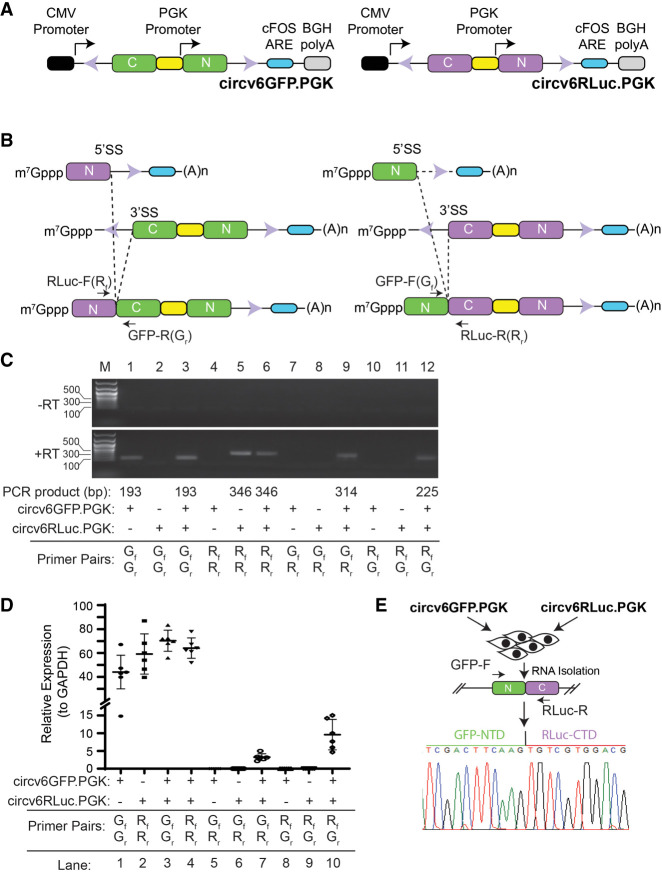
To test this possibility more directly, we simplified our vector design (circv6RLuc) and generated a second companion vector with split GFP exons, circv6GFP (Fig. 2A). If our hypothesis was correct, we reasoned that cotransfection of circv6RLuc.PGK and circv6GFP.PGK should produce not only circRNA products, but also trans-spliced GFP–RLuc and RLuc–GFP mRNA fusions which should be detectable using appropriately designed primer pairs (Fig. 2B). Following transfection of 293T cells with single or both plasmids, total RNA was isolated and the presence of spliced fusions probed by RT-PCR (Fig. 2C). GFP and RLuc primer pairs were designed to cross the NTD/CTD exon–exon junctions and successfully did so (Fig. 2C; compare lane 1 to 2 and 5 to 4). Using GFP/RLuc primer combinations, we detected PCR products only in RNA preparations from cells that had been cotransfected with both circv6RLuc.PGK and circv6GFP.PGK (Fig. 2C; compare lane 9 to 7–8 and 12 to 10–11). These results were independently confirmed in RT-qPCR experiments where the presence of GFP-RLuc and RLuc-GFP fusion mRNAs were detected in cells cotransfected with both circv6RLuc.PGK and circv6GFP.PGK (Fig. 2D). Sequencing of the fusion product obtained with primers GFP-F and RLuc-R confirmed the presence of a GFP(NTD)/RLuc(CTD) fusion junction in cells that had been cotransfected with circv6GFP.PGK and circv6RLuc.PGK (Fig. 2E).
FIGURE 2.
CircRNA expression vectors can generate trans-spliced products. (A) Schematic representation of circv6GFP.PGK and circv6RLuc.PGK expression vectors. (B) Trans-spliced products expected from circv6RLuc.PGK and circv6GFP.PGK vectors. Only trans-spliced products formed from the CMV (full-length mRNA) and PGK (shorter mRNAs) promoters are shown due to space limitations, but we envisage homotypic (GFP and RLuc) products also being produced, as well as products from trans-splicing of two full-length, CMV-synthesized mRNAs. The relative location of PCR primers (RLuc-F; Rf, RLuc-R; Rr, GFP-F; Gf, and GFP-R; Gr) used for product detection are shown. (C) Endpoint RT-PCR analysis of RNA isolated from 293T cells transfected with the indicated expression plasmids. RNA was isolated 48 h following transfection. (Top panel) PCRs performed on RNA samples that had not been converted to cDNA (−RT). (Bottom panel) RT-PCR products obtained after 25 cycles. (D) RT-qPCR analysis of RNA from 293T cells transfected with the indicated plasmids. The combination of plasmids used in the transfection, as well as primer pairs used in the qPCR, are shown below the graph. The relative abundance of each PCR product was calculated using the ΔΔCt method with GFP or RLuc signal calculated relative to GAPDH. n = 6 ± SD. (E) Sequencing chromatogram of the PCR product obtained from RNA of cells cotransfected with circv6GFP.PGK and circv6RLuc.PGK. PCR primers crossed the GFP(NTD)/RLuc(CTD) exon junction.
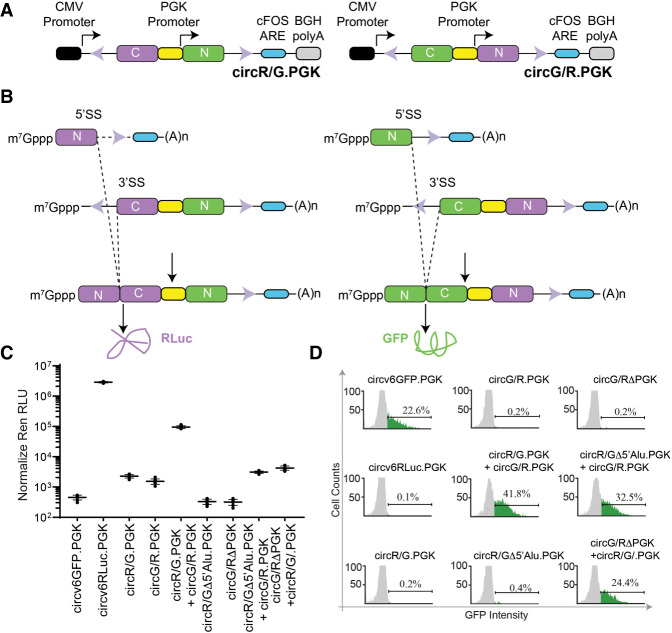
Our results indicate that trans-spliced products can be readily generated from circRNA expression vectors. To determine if the levels of trans-spliced mRNA products would be sufficient to generate detectable RLuc or GFP protein levels, we created two chimeric circRNA vectors containing either: (i) an upstream RLuc CTD, the PGK promoter, and a downstream GFP NTD (circR/G.PGK); or (ii) an upstream GFP CTD, the PGK promoter, and a downstream RLuc NTD (circG/R.PGK) (Fig. 3A). In this scenario, only trans-splicing can lead to functional RLuc or GFP proteins when these two vectors are cointroduced into cells (Fig. 3A,B). We also generated two control vectors: (i) circG/R.ΔPGK in which the PGK promoter was deleted, and (ii) circR/GΔ5′Alu.PGK in which the upstream Alu sequence was deleted (Supplemental Fig. S1a,b). circG/R.ΔPGK should not produce the shorter RenNTD-containing mRNA hence no RLuc protein is expected upon cotransfection with circR/G.PGK—only GFP should be produced (Supplemental Fig. S1a). The longer, CMV-driven transcript from circR/GΔ5′Alu.PGK will be unable to participate in a trans-splicing reaction with the shorter, PKG-driven mRNA from circG/R.PGK, and thus we also expect the production of only GFP, and no RLuc, when both of these vectors are cotransfected (Supplemental Fig. S1b).
FIGURE 3.
Protein expression from trans-spliced mRNA products. (A) Schematic representation of circR/G.PGK and circG/R.PGK expression vectors. (B) Schematic diagram illustrating how trans-splicing of the indicated cotransfected vectors can lead to the production of full-length RLuc and GFP proteins. Due to space constraints, not all possible trans-splicing reactions are shown. (C) Luciferase values obtained from 293T cells cotransfected with the indicated expression vectors. RLuc RLU values were normalized to total protein levels in the extracts. n = 4 ± SD. (D) Flow cytometry showing representative GFP expression obtained from 293T cells transfected with the indicated individual or combinations of plasmids.
As expected, introduction of circv6GFP.PGK into 293T cells did not produce significant levels of RLuc (Fig. 3C), but did produce quantifiable GFP levels (Fig. 3D). Introduction of circv6RLuc.PGK produced significant levels of RLuc, but no GFP (Fig. 3C,D). Transfection of only circR/G.PGK or circG/R.PGK alone failed to produce significant levels of RLuc or GFP (Fig. 3C,D). However, cotransfection of circR/G.PGK and circG/R.PGK generated robust levels of both RLuc and GFP proteins. On their own, neither control vector (circR/GΔ5′Alu.PGK nor circG/R.ΔPGK) produced significant RLuc or GFP products. Cotransfection of circR/GΔ5′Alu.PGK with circG/R.PGK produced GFP, and significantly reduced levels of RLuc (Fig. 3C,D). Similarly, when circG/R.ΔPGK was cotransfected with circR/G.PGK, significant levels of GFP were apparent and low levels of RLuc were obtained (Fig. 3C,D). Taken together, our results highlight that trans-splicing is a source of protein products arising from circRNA expression vectors.
DISCUSSION
The phenomenon of trans-splicing was initially documented by Konarska et al. (1985) who showed that this process is favored when intervening sequences between two target mRNAs can base-pair. This phenomenon is well described and has even been explored as a potential therapeutic route by which germline mutations could be eliminated from endogenous pre-mRNAs if an appropriate wild-type donor RNA template is available (Berger et al. 2016). Trans-splicing events have also been found in cancers and postulated to precede and facilitate chromosome translocations (Zaphiropoulos 2011).
Although our data emerged from studying only the PGK promoter, the amount of trans-spliced mRNA product arising from our circRNA expression vector was quite significant. In our experience with circv6GFP.PKG and circv6RLuc.PGK, chimeric mRNA levels reach ∼2.5–9 times the levels of GAPDH in transfected 293T cells (Fig. 2D). As well, RLuc activity was easily detected in cells cotransfected with circR/G.PGK and circG/R.PGK, where its production could only have arisen from trans-splicing (Fig. 3B,C). We note that levels were ∼25-fold lower than obtained with circv6RLuc.PGK, and this may reflect that trans-splicing is more efficient when transcripts are generated in proximity of each other from the same expression vector or locus versus from two different plasmids at geographically distinct nuclear locations. Although we have highlighted in our schematic diagrams trans-spliced reactions involving a longer, CMV-derived mRNA with a shorter, PGK-derived mRNA to explain how RLuc activity could have been attained from circv1.PGK (Fig. 1B), we are cognizant that two longer CMV originating transcripts could also trans-splice to each other. For example, in Supplemental Figure S1a, one could expect a functional RLuc protein being generated from trans-splicing of the CMV-originating mRNA from circR/G.PGK with the CMV-originating mRNA from circG/R.ΔPGK. It may be that circularization in cis by the circG/R.ΔPGK full-length mRNA is more efficient since splicing generally occurs during transcription (Herzel et al. 2017), minimizing the amount of unspliced full-length RNA available for trans-splicing. Since the shorter, PGK-originating transcript cannot participate in cis-splicing reactions (Fig. 2B), its accumulation may then permit it to participate in trans-splicing reactions.
It has been reported that circRNAs can be translated to give rise to protein products (Legnini et al. 2017; Pamudurti et al. 2017). The mechanism by which ribosomes would be recruited to circRNAs should involve an IRES, and the m6A reader, YTHDF3, and eIF4G2 have been implicated in this process (Yang et al. 2017; Zhao et al. 2019; Di Timoteo et al. 2020). Our results do not challenge the existence of circRNAs or the fact that polypeptide products arise from some of these (Begum et al. 2018; Zhang et al. 2018a,b; Zheng et al. 2019), they simply raise caution concerning interpretations of translation initiation mechanisms of circRNAs when using expression vectors of the type used herein. Indeed, similar concerns were recently raised by Ho-Xuan et al. (2020), who undertook a detailed analysis of the protein products arising from circZNF609 and concluded that they were unlikely to be the consequence of translation from the circular mRNA template, but more likely to arise from trans-splicing events (although the latter was not formally investigated).
The results presented here raise concerns over the use of circRNA expression vectors as a facile way of identifying IRESes. Cryptic promoter activity from an embedded sequence can lead to the production of transcripts harboring an intron with an ALU repeat that will be able to participate in trans-splicing reaction with transcripts made from the upstream CMV promoter on the same vector. A validation assay for any IRESs discovered using circRNA expression vectors could be to position the putative IRES sequence upstream of a reporter ORF, generate ApppG-capped mRNA in vitro, and transfect the transcribed reporter into cells. The presence of an ApppG-cap will effectively block any 5′-dependent initiation events. Alternatively, if the IRES requires a nuclear experience to function (Semler and Waterman 2008), one could isolate circRNA from transfected cells, demonstrate their circular nature in vitro (e.g., using RNase R resistance), followed by reintroduction into cells. Our results caution against solely the use of circRNA expression vectors for IRES discovery, unless accompanied by experimental approaches to eliminate trans-splicing interference.
MATERIALS AND METHODS
Cell culture conditions
HEK293T cells were maintained in DMEM supplemented with 10% FBS (Wisent), 100 U/mL penicillin/streptomycin, and 2 mM l-glutamine at 37°C and 5% CO2. For transfection, HEK293T cells were seeded to 70% confluency in 24 well plates and transfected with 1 µg plasmid using polyethylenimine (PEI) mediated transfection. Media was refreshed the next day and 2 d following transfection (Fukumoto et al. 2010), GFP expression was measured using flow cytometry (Guava EasyCyte), and renilla values were determined on a Berthold luminometer (Lumat LB 9507) (Dyer et al. 2000).
CircRNA plasmid design
The expression cassette shown in Figure 1A was custom synthesized in pUC57 by GenScript and then subcloned into pCMV6-XL6 using EcoRI/XbaI sites to produce circV1.EMCV. To generate different reporter plasmids containing different sequences between the Ren CTD and NTD, the circV1 vector was digested using MfeI and MluI restriction sites and the sequence of interest (produced as a gBlock [IDT]) was cloned into the digested vector using Gibson Assembly (NEB). The more streamlined circV6 vectors were also custom synthesized by GenScript and subcloned in pCMV6-XL6.
RNA isolation and RT-qPCR
RNA isolation was performed 48 h post-transfection (as described above but scaled up into six-well plates using 3 µg of DNA) using TRIzol (Thermo Fisher Scientific) following the manufacturer's recommendations. Reverse-transcription reactions were performed using M-MuLV-reverse transcriptase (New England Biolab) and random primer mix (New England Biolab) according to the manufacturer's protocol. qPCR experiments were performed on cDNA diluted 1/10 using the following oligonucleotides (GFP-F: 5′ACGACGGCAACTACAAGACC3′; GFP-R: 5′TTGAAGTTCACCTTGATGCC3′; Ren-F: 5′CCCAGATCTGATCGGAATGG3′; Ren-R: 5′GATCTTGCTTGGGAGCATGG3′; GAPDH-F: 5′GGTATCGTGGAAGGACTCAT3′; GAPDH-R: 5′GCAGGGATGATGTTCTGGAG3′) in a CFX96 PCR System (Bio-Rad) using SsoFast Evagreen Supermix (Bio-Rad). Gene expression data analysis was performed using the Bio-Rad CFX Manager 3.1 software.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by research funding to J.P. (Canadian Institutes of Health Research [#FDN-148366]).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078261.120.
REFERENCES
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56: 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Begum S, Yiu A, Stebbing J, Castellano L. 2018. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene 37: 4055–4057. 10.1038/s41388-018-0230-3 [DOI] [PubMed] [Google Scholar]
- Berger A, Maire S, Gaillard MC, Sahel JA, Hantraye P, Bemelmans AP. 2016. mRNA trans-splicing in gene therapy for genetic diseases. Wiley Interdiscip Rev RNA 7: 487–498. 10.1002/wrna.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Di Timoteo G, Dattilo D, Centron-Broco A, Colantoni A, Guarnacci M, Rossi F, Incarnato D, Oliviero S, Fatica A, Morlando M, et al. 2020. Modulation of circRNA metabolism by m6A modification. Cell Rep 31: 107641. 10.1016/j.celrep.2020.107641 [DOI] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. 1995. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167: 245–248. 10.1016/0378-1119(95)00639-7 [DOI] [PubMed] [Google Scholar]
- Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. 2000. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem 282: 158–161. 10.1006/abio.2000.4605 [DOI] [PubMed] [Google Scholar]
- Fukumoto Y, Obata Y, Ishibashi K, Tamura N, Kikuchi I, Aoyama K, Hattori Y, Tsuda K, Nakayama Y, Yamaguchi N. 2010. Cost-effective gene transfection by DNA compaction at pH 4.0 using acidified, long shelf-life polyethylenimine. Cytotechnology 62: 73–82. 10.1007/s10616-010-9259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Herzel L, Ottoz DSM, Alpert T, Neugebauer KM. 2017. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol 18: 637–650. 10.1038/nrm.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MA, Palmenberg AC. 1995. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol 69: 4399–4406. 10.1128/JVI.69.7.4399-4406.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho-Xuan H, Glazar P, Latini C, Heizler K, Haase J, Hett R, Anders M, Weichmann F, Bruckmann A, Van den Berg D, et al. 2020. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res 48: 10368–10382. 10.1093/nar/gkaa704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10: 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Jackson RJ. 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 5: a011569. 10.1101/cshperspect.a011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643. 10.1128/JVI.62.8.2636-2643.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA. 1985. Trans splicing of mRNA precursors in vitro. Cell 42: 165–171. 10.1016/S0092-8674(85)80112-4 [DOI] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. 2017. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66: 22–37. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. 2017. Translation of circRNAs. Mol Cell 66: 9–21.e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler BL, Waterman ML. 2008. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol 16: 1–5. 10.1016/j.tim.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351: aad4939. 10.1126/science.aad4939 [DOI] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG. 2011. Trans-splicing in higher eukaryotes: implications for cancer development? Front Genet 2: 92. 10.3389/fgene.2011.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. 2014. Complementary sequence-mediated exon circularization. Cell 159: 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al. 2018a. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37: 1805–1814. 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, et al. 2018b. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9: 4475. 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al. 2019. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun 10: 2300. 10.1038/s41467-019-10246-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, Wu C, Zhou Q, Hu W, Wu C, et al. 2019. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer 18: 47. 10.1186/s12943-019-1010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.