Abstract
Diffuse capillary malformation with overgrowth (DCMO) is a clinical diagnosis describing patients with multiple, extensive capillary malformations (CMs) associated with overgrowth and foot anomalies. The purpose of the study was to identify somatic variants in DCMO. Skin containing CM and overgrown subcutaneous adipose tissue was collected from patients with DCMO. Exons from 447 cancer-related genes were sequenced using OncoPanel. Variant-specific droplet digital PCR (ddPCR) independently confirmed the variants and determined variant allele frequencies (VAF). One subject contained a somatic PIK3CA p.G106V variant. A second patient had a PIK3CA p.D350G variant. VAF was 27–29% in skin and 16–28% in subcutaneous adipose. Variants were enriched in endothelial cells (VAF 50–51%) compared to non-endothelial cells (1–8%). DCMO is associated with somatic PIK3CA variants and should be considered on the PIK3CA-related overgrowth spectrum (PROS). Variants are present in both skin and subcutaneous adipose and are enriched in endothelial cells.
Keywords: capillary malformation, phosphatidylinositol 3-kinases, PIK3CA-related overgrowth spectrum, vascular malformations, humans, mutation, genetic association studies
INTRODUCTION
Capillary malformation (CM) is the most common type of vascular malformation. The lesion can be localized or diffuse and part of vascular malformation syndromes. Somatic variants in GNAQ and GNA11 have been found in non-syndromic CMs as well as in Sturge-Weber syndrome.3–5 CMs may be present in conditions among the PIK3CA-related overgrowth spectrum (PROS), such as Klippel-Trenaunay syndrome and CLOVES (congenital lipomatous overgrowth, vascular malformations, epidermal nevi, scoliosis/skeletal/spinal).6,7 CMs also are associated with a clinical condition called “diffuse capillary malformation with overgrowth” (DCMO).8 Patients exhibit extensive and multiple CMs, facial asymmetry, limb overgrowth, and hand/foot anomalies.8 The purpose of this study was to identify somatic variants in patients with DCMO.
MATERIALS AND METHODS
The Committee on Clinical Investigation at Boston Children’s Hospital approved this study. The study conforms to recognized standards as described in the United States Federal Policy for the Protection of Human Subjects. Inclusion in the study was fully discussed with all participants and their parents/legal guardian when applicable. Proper informed consent was obtained and documented prior to inclusion in the study.
Skin containing CM and overgrown subcutaneous adipose tissue from two patients clinically diagnosed with DCMO were collected during an operative procedure to reduce the soft-tissue overgrowth of their trunk or leg.8 Both patient specimens previously tested negative for GNAQ and GNA11 variants by multiplex targeted sequencing using molecular inversion probe (MIP-seq) and droplet digital PCR (ddPCR). Because somatic variants in cancer-associated genes can cause vascular malformations, we sequenced exons from cancer-related genes in DNA from the two DCMO specimens. Samples were flash-frozen and stored at −80°C. Genomic DNA was extracted using standard methods to sequence exons from 447 cancer-related genes (OncoPanel, Dana Farber Cancer Institute, Boston, MA).9 Putative somatic variants were identified using MuTect v1.1.4 with pairing of each patient specimen to the project normal specimen, CEPH1408-H1. Non-coding sequence variants were excluded, as were variants having a population frequency of > 0.1% in the gnomAD database. Any variant that was present in the COSMIC database at least twice was flagged for manual review. Translocations were detected using CCGD in-house software called Breakmer. Likely pathogenic variants then were independently tested using variant-specific ddPCR assays on a separate DNA aliquot from the original specimen.
Skin was separated from subcutaneous adipose using 2.5x magnification immediately after excision in the operating room. Cell isolation was performed as previously described.4 Tissue was washed in phosphate-buffered saline to remove blood cell contaminants, digested with collagenase A (2.5 mg/mL) (Roche, Basel, Switzerland) for 1 hour at 37°C, then filtered through a 100 μm strainer to produce a single cell suspension. Cells were placed on fibronectin-coated (1 μg/cm2) tissue culture plates in endothelial growth medium-2 (EGM-2, Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA). After 5–7 days of expansion, cells were fractionated into two populations (endothelial and non-endothelial) using anti-human CD31 (endothelial cell marker) magnetic beads (DynaBeads™, Thermo Fisher Scientific, Waltham, MA). DNA was extracted from frozen tissue, cells, and blood (unaffected tissue) using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany).
RESULTS
One subject (Figure 1) at birth had diffuse, multifocal capillary malformations of her trunk and extremities and was diagnosed with DCMO. She developed significant posterior trunk subcutaneous adipose growth during puberty that was treated with suction-assisted lipectomy. The second individual (Figure 2) had overgrowth of the left side of the body (face, arm, trunk, and leg), as well as a foot anomaly (syndactyly of the 2nd and 3rd toes, macrodactyly, sandal gap) at birth and was diagnosed with DCMO. During adolescence she was diagnosed with lymphedema of the left leg. She underwent pulsed dye laser of the capillary malformation of her face as well as epiphysiodesis and suction-assisted lipectomy to reduce the size of the left lower extremity.
Figure 1.
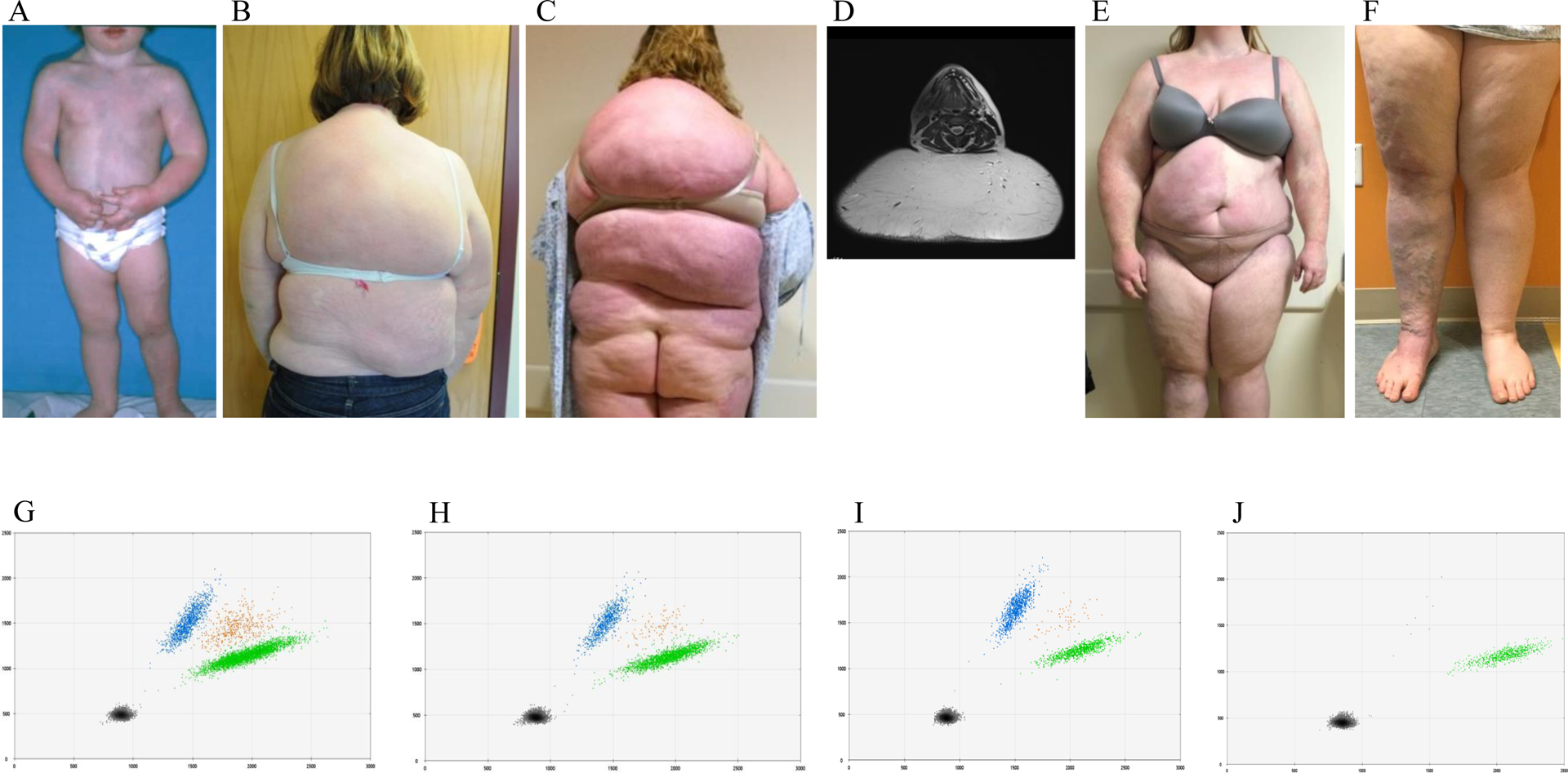
Female with a somatic PIK3CA p.G106V variant. (A) Early childhood illustrates diffuse CMs (capillary malformations) involving her trunk and extremities and a diagnosis of DCMO (diffuse capillary malformation with overgrowth). (B) Appearance of posterior trunk at age 17 years. (C) Significant posterior trunk overgrowth 4 years later at age 21. (D) MR (magnetic resonance) image shows subcutaneous adipose hypertrophy. Anterior trunk (E) and lower extremities (F). ddPCR (droplet digital polymerase chain reaction) graphs of skin (G) VAF (variant allele frequency) 27%, subcutaneous adipose (H) VAF 28%, endothelial cells isolated from subcutaneous adipose (I) VAF 50%, non-endothelial cells isolated from subcutaneous adipose (J) VAF 1%. Left upper blue droplets contain variant alleles. Right middle orange droplets have variant and wild-type alleles. Right lower green droplets contain wild-type alleles. Left lower black droplets are empty.
Figure 2.
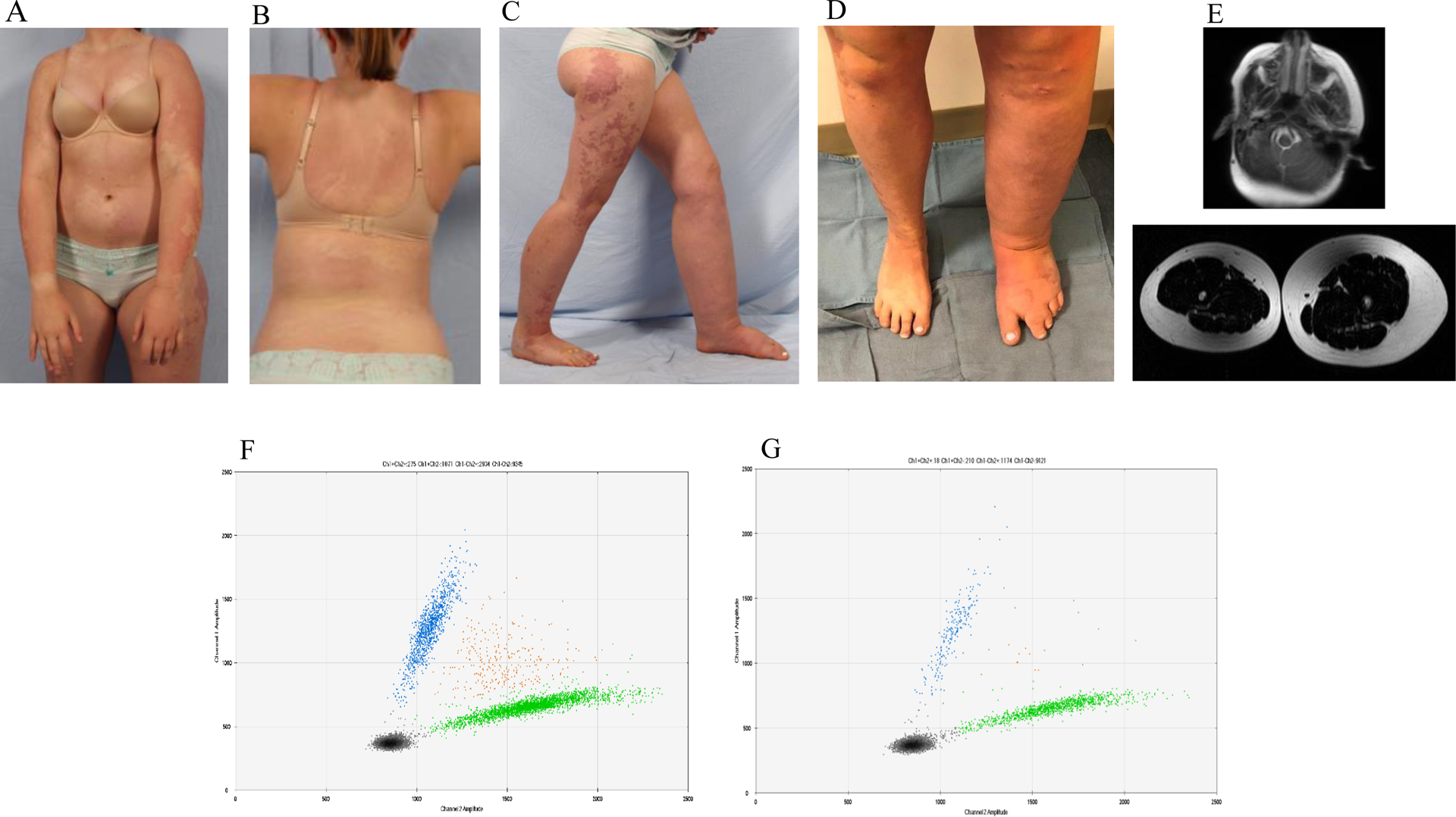
Female subject with a somatic PIK3CA p.D350G variant. Age 15 years with diffuse CMs (capillary malformations) involving her trunk (A, B) and extremities (C, D) and a diagnosis of DCMO (diffuse capillary malformation with overgrowth). She had overgrowth of the left side of her body, as well as syndactyly and lymphedema of the left lower extremity. (E) MR (magnetic resonance) of the face illustrates overgrowth and subcutaneous adipose excess of the left side. MR image of the legs exhibits subcutaneous fat overgrowth of the left lower extremity. ddPCR (droplet digital polymerase chain reaction) graphs of skin (F) VAF (variant allele frequency) 29% and subcutaneous adipose (G) VAF 16%. Left upper blue droplets contain variant alleles. Right middle orange droplets have variant and wild-type alleles. Right lower green droplets contain wild-type alleles. Left lower black droplets are empty.
Each sample achieved at least 30-fold coverage for 80% of the targeted bases; the mean target coverage was 265x (range 65–822x). We detected a PIK3CA variant in both patients: p.G106V (NM_006218.4(PIK3CA_v001):c.317G>T) and p.D350G (NM_006218.4(PIK3CA_v001):c.1049A>G). Variants were independently tested and confirmed using ddPCR. Variant allele frequencies (VAF) were: skin (27%–29%), subcutaneous adipose tissue (16%–28%), endothelial cells isolated from skin (51%) or subcutaneous adipose tissue (50%), non-endothelial cells isolated from skin (8%) or subcutaneous adipose tissue (1%). Variants were not present in unaffected tissue (white blood cell DNA).
DISCUSSION
Most CMs contain pathogenic variants in GNAQ.3,4 Previously we found GNA11 variants in patients with diffuse lesions involving an extremity that tested negative for GNAQ variants.5 In this study we show two patients with diffuse, multifocal CMs that did not have GNAQ or GNA11 variants, but contained PIK3CA variants. These subjects previously were diagnosed with “diffuse capillary malformation with overgrowth (DCMO).”8 Both patients had extensive CMs involving multiple anatomic areas, including the trunk and extremities. They exhibited subcutaneous adipose overgrowth underneath the area of the cutaneous birthmark that commonly occurs with sporadic and syndromic CMs.10,11
CMs also are a component of vascular malformation overgrowth syndromes associated with PIK3CA variants, including M-CM (megalencephaly-capillary malformation), CLOVES, and Klippel-Trenaunay.6,7,12–14 The patients in this study were diagnosed with DCMO prior to genetic testing and were not thought to be on the PIK3CA-related overgrowth spectrum (PROS) because they did not exhibit features associated with M-CM, CLOVES, or Klippel-Trenaunay syndromes such as brain malformations, macrocephaly, epidermal nevi, or other vascular anomalies.14 Consequently, they were not tested for PIK3CA mutations and instead underwent broad exome sequencing. After identifying PIK3CA mutations, however, we realized in retrospect that both patients exhibited features that are also associated with PROS. One individual had rapid subcutaneous adipose overgrowth of her trunk during puberty, and the other exhibited lymphedema and a foot malformation.
Many patients previously diagnosed with DCMO are on the PIK3CA-related overgrowth spectrum, similar to the paradigm of its related condition M-CM. DCMO has a milder phenotype compared to M-CM which exhibits macrocephaly/megalencephaly, hypotonia, hydrocephalus, and developmental delay.8 DCMO, like M-CM, shares several features seen in other PROS conditions: extensive and multiple CMs, facial asymmetry, limb overgrowth, and hand/foot anomalies (e.g., macrodactyly and syndactyly).8 Individuals with a diffuse CM involving an extremity without any other features associated with PROS are most likely to have a GNAQ or GNA11 variant.3–5
One PIK3CA variant (G106V) we identified in DCMO has been reported in an individual with CLOVES13; the other variant (D350G) has not previously been documented in a patient with a PROS condition. The G106V PIK3CA variant is considered pathogenic by American College of Medical Genetics guidelines (Functional Analysis through Hidden Markov Models (FATHMM) prediction score 0.98) and has been reported in carcinomas of the gastrointestinal tract, bladder, oropharynx, uterus, breast, and brain.15–18 The D350G PIK3CA variant is considered pathogenic and has a FATHMM prediction score of 0.99. It is associated with tumors of the breast, colon, uterus, pancreas, salivary gland, and brain glioma.15,19,20 The most common somatic PIK3CA variants (E542K, E545K, H1047R) account for 80% of identified variants in the gene21,22 and are most commonly found in patients with a PROS condition.6,13,23–25 These mutations have been shown to be activating and stimulate AKT signaling.1,6 It is likely that the G106V and D350G variants also are activating. The G106V variant affects the adaptor-binding domain which can activate PI3 kinase due to altered interactions with the inhibitory p85 subunit.26 The D350G variant lies within the C2 domain of the p110α subunit; mutations at residues 345 and 453 occur in cancers and also may alter the regulatory effect of p85.26
We believe that continuing to diagnose patients clinically with DCMO is useful because it differentiates them from other PROS conditions. PROS has significant phenotypic variability with a wide range of morbidities and treatments. Separating patients with DCMO from individuals with CLOVES or Klippel-Trenaunay syndrome, for example, is important because DCMO is very different (e.g, no risk of having other types of vascular malformations).
This study extends the PIK3CA-related overgrowth spectrum by including patients with the clinical diagnosis of DCMO. Similar to other vascular malformations such as capillary malformation, Sturge-Weber syndrome, and arteriovenous malformation, we found that variants were enriched in endothelial cells suggesting that this cell-type is driving the pathophysiology of DCMO.4,27 Individuals with DCMO might benefit from pharmacotherapy that has been described for patients with other PROS conditions.28,29
Acknowledgements:
Research reported in this publication was supported by the National Heart Lung and Blood Institute under Award Number R01 HL127030 (AKG and JB), Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number R01 HD093735 (AKG), and the Translational Research Program Boston Children’s Hospital (AKG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement:
The authors have no conflicts of interest to declare.
Data Availability Statement:
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/ with accession numbers MN256643 and MN256644.1,2
REFERENCES
- 1.Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2018;46(D1):D41–D47. doi: 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GenBank [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1982] - [cited 2019 Aug 05]. Available from: https://www.ncbi.nlm.nih.gov/nucleotide/ [Google Scholar]
- 3.Shirley MD, Tang H, Gallione CJ, et al. Sturge–Weber Syndrome and Port-Wine Stains Caused by Somatic Mutation in GNAQ. N Engl J Med. 2013;368(21):1971–1979. doi: 10.1056/nejmoa1213507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Couto JA, Huang L, Vivero MP, et al. Endothelial cells from capillary malformations are enriched for somatic GNAQ mutations. Plast Reconstr Surg. 2016;137(1):77e–82e. doi: 10.1097/PRS.0000000000001868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couto JA, Ayturk UM, Konczyk DJ, et al. A somatic GNA11 mutation is associated with extremity capillary malformation and overgrowth. Angiogenesis. 2017;20(3):303–306. doi: 10.1007/s10456-016-9538-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurek KC, Luks VL, Ayturk UM, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90(6):1108–1115. doi: 10.1016/j.ajhg.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166(4):1048–1054.e5. doi: 10.1016/j.jpeds.2014.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MS, Liang MG, Mulliken JB. Diffuse capillary malformation with overgrowth: A clinical subtype of vascular anomalies with hypertrophy. J Am Acad Dermatol. 2013;69(4):589–594. doi: 10.1016/j.jaad.2013.05.030 [DOI] [PubMed] [Google Scholar]
- 9.Garcia EP, Minkovsky A, Jia Y, et al. Validation of oncopanel a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141(6):751–758. doi: 10.5858/arpa.2016-0527-OA [DOI] [PubMed] [Google Scholar]
- 10.Greene AK, Taber SF, Ball KL, Padwa BL, Mulliken JB. Sturge-weber syndrome: Soft-tissue and skeletal overgrowth. J Craniofac Surg. 2009;20(SUPPL. 1):617–621. doi: 10.1097/SCS.0b013e318192988e [DOI] [PubMed] [Google Scholar]
- 11.Couto JA, Maclellan RA, Greene AK. Management of Vascular Anomalies and Related Conditions Using Suction-Assisted Tissue Removal. Plast Reconstr Surg. 2015;136(4):511e–4e. doi: 10.1097/PRS.0000000000001558 [DOI] [PubMed] [Google Scholar]
- 12.Rivière JB, Mirzaa GM, O’Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuentz P, St-Onge J, Duffourd Y, et al. Molecular diagnosis of PIK3CA-related overgrowth spectrum (PROS) in 162 patients and recommendations for genetic testing. Genet Med. 2017;19(9):989–997. doi: 10.1038/gim.2016.220 [DOI] [PubMed] [Google Scholar]
- 14.Keppler-Noreuil KM, Rios JJ, Parker VER, et al. PIK3CA-related overgrowth spectrum (PROS): Diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet Part A. 2015;167(2):287–295. doi: 10.1002/ajmg.a.36836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Dios DA, Lambrechts D, Coenegrachts L, et al. High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma. Gynecol Oncol. 2013;128(2):327–334. doi: 10.1016/j.ygyno.2012.11.037 [DOI] [PubMed] [Google Scholar]
- 17.Ali SM, Yao M, Yao J, et al. Comprehensive genomic profiling of different subtypes of nasopharyngeal carcinoma reveals similarities and differences to guide targeted therapy. Cancer. 2017;123(18):3628–3637. doi: 10.1002/cncr.30781 [DOI] [PubMed] [Google Scholar]
- 18.Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122(10):2255–2259. doi: 10.1002/ijc.23388 [DOI] [PubMed] [Google Scholar]
- 19.Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Tsutsumi S, Kawaguchi T, et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012;22(2):208–219. doi: 10.1101/gr.123109.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. https://www.nature.com/articles/nature11412?report=reader. Accessed October 31, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie M, Zhang Q, Mcmichael JF, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634.Mutational [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindhurst MJ, Parker VER, Payne F, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet. 2012;44(8):928–933. doi: 10.1038/ng.2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keppler-Noreuil KM, Sapp JC, Lindhurst MJ, et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am J Med Genet Part A. 2014;164(7):1713–1733. doi: 10.1002/ajmg.a.36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hucthagowder V, Shenoy A, Corliss M, et al. Utility of clinical high-depth next generation sequencing for somatic variant detection in the PIK3CA-related overgrowth spectrum. Clin Genet. 2017;91(1):79–85. doi: 10.1111/cge.12819 [DOI] [PubMed] [Google Scholar]
- 26.Huang CH, Mandelker D, Schmidt-Kittler O, et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science (80-). 2007;318(5857):1744–1748. doi: 10.1126/science.1150799 [DOI] [PubMed] [Google Scholar]
- 27.Couto JA, Huang AY, Konczyk DJ, et al. Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation. Am J Hum Genet. 2017;100(3):546–554. doi: 10.1016/j.ajhg.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker VER, Keppler-Noreuil KM, Faivre L, et al. Safety and efficacy of low-dose sirolimus in the PIK3CA-related overgrowth spectrum. Genet Med. 2019;21(5):1189–1198. doi: 10.1038/s41436-018-0297-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venot Q, Blanc T, Rabia SH, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558(7711):540–546. doi: 10.1038/s41586-018-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]


