Abstract
The detection of fecal viral pathogens in water is hampered by their great variety and complex analysis. As traditional bacterial indicators are poor viral indicators, there is a need for alternative methods, such as the use of somatic coliphages, which have been included in water safety regulations in recent years. Some researchers have also recommended the use of reference viral pathogens such as noroviruses or other enteric viruses to improve the prediction of fecal viral pollution of human origin. In this work, phages previously tested in microbial source tracking studies were compared with norovirus and adenovirus for their suitability as indicators of human fecal viruses. The phages, namely those infecting human-associated Bacteroides thetaiotaomicron strain GA17 (GA17PH) and porcine-associated Bacteroides strain PG76 (PGPH), and the human-associated crAssphage marker (crAssPH), were evaluated in sewage samples and fecal mixtures obtained from different animals in five European countries, along with norovirus GI + GII (NoV) and human adenovirus (HAdV). GA17PH had an overall sensitivity of ≥83% and the highest specificity (>88%) for human pollution source detection. crAssPH showed the highest sensitivity (100%) and specificity (100%) in northern European countries but a much lower specificity in Spain and Portugal (10 and 30%, respectively), being detected in animal wastewater samples with a high concentration of fecal indicators. The correlations between GA17PH, crAssPH, or the sum of both (BACPH) and HAdV or NoV were higher than between the two human viruses, indicating that bacteriophages are feasible indicators of human viral pathogens of fecal origin and constitute a promising, easy to use and affordable alternative to human viruses for routine water safety monitoring.
Keywords: Bacteroides, bacteriophages, indicators, fecal pollution, crAssphage, enteric viruses, norovirus, adenovirus
Introduction
To date, over 150 human enteric viruses have been detected in water bodies (Rodríguez-Lázaro et al., 2012; Tran et al., 2015; Farkas et al., 2019), including adenoviruses, noroviruses, enteroviruses, sapoviruses, rotaviruses, and polyomaviruses, among others. As the assessment of all pathogenic viral strains in water is not feasible, indicators have been traditionally used as a proxy. The most used fecal indicators worldwide are Escherichia coli and enterococci, but they are less resistant to environmental stresses than viruses such as temperature, pH and sun irradiation and behave differently in the environment (Gerba et al., 1979; Wyer et al., 1995; Baggi et al., 2001; Borchardt et al., 2004; Harwood et al., 2005; Costán-Longares et al., 2008). As an alternative, the monitoring of reference viruses such as NoV and human adenovirus (HAdV) has been proposed (Hundesa et al., 2006; Ahmed et al., 2010; Rusiñol et al., 2014; Mayer et al., 2016). Enteroviruses are the most easily detectable enteric viruses by cell culture, and have been considered as potential indicators of the human enteric viruses (Grabow, 2007). Enteroviruses are recommended in the few water quality regulations that include virus-based criteria (EEC, 1976; USEPA, 1992). However, cell culture is expensive, time-consuming and needs specialized staff and equipment. Since the advent of genomic techniques, genome fragments of enteric viruses excreted by humans and animals have also been proposed as potential viral indicators (Hot et al., 2003; Albinana-Gimenez et al., 2009; Hunt et al., 2010; Silva et al., 2011; Wong et al., 2012). Additionally, viruses that exclusively infect humans, such as HAdV, have been postulated as microbial source tracking (MST) markers (Hundesa et al., 2006; Ahmed et al., 2010; Rusiñol et al., 2014).
However, human infectious viruses and human viral genomes both present serious drawbacks as indicators of waterborne enteric viruses. Firstly, the concentration of human viruses is variable in sewage and low in other types of water, requiring cumbersome, time-consuming and costly, concentration procedures (Hewitt et al., 2013; Haramoto et al., 2015). Secondly, there is no evidence that the presence and concentration of a given human virus in a water matrix unequivocally predicts the presence of other viruses (Irving and Smith, 1981; Chapron et al., 2000; Jiang et al., 2001). Finally, genomic methods have the inconvenience of not being able to distinguish between infectious and non-infectious viruses without additional unwieldy steps (Nuanualsuwan and Cliver, 2002; Jofre and Blanch, 2010). Such a distinction is essential when evaluating water treatments and natural inactivation and risk to human health.
Bacteriophages infecting bacteria have been proposed as efficient indicators of human viruses transmitted in water (IAWPRC, 1991; Grabow, 2004), and concurrence between enteroviruses and human-associated bacteriophages infecting Bacteroides fragilis has been demonstrated in a range of matrices including bivalve molluscs, treated wastewater, etc. (Tartera et al., 1988; Ebdon et al., 2007; Costán-Longares et al., 2008). Bacteriophages have been incorporated in recently developed guidelines for water quality monitoring (NHMRC, 2011; European Commission, 2018; Health Canada, 2019). In particular, somatic bacteriophages and F-RNA phages have proved to be reliable indicators of fecal viral pollution and suitable for use in water treatment processes (Jofre et al., 2016; Jebri et al., 2017). MST studies have used genogroups of F-specific RNA phages (Jofre et al., 2011) and bacteriophages infecting selected strains of Bacteroides (Tartera et al., 1989; Gómez-Dóñate et al., 2011; Jofre et al., 2014) to detect the source of fecal pollution in water environments. Recent studies have identified crAssphage (crAssPH) as the most abundant bacteriophage family in sewage, with Bacteroides as the putative host (Dutilh et al., 2014; Shkoporov et al., 2018), and its potential as a human-associated MST molecular marker is being evaluated (García-Aljaro et al., 2017; Ahmed et al., 2018; Ballesté et al., 2019). A study comparing the efficacy of human viruses with that of the molecular marker crAssPH and phages infecting the human-associated Bacteroides GA17 strain (GA17PH) to predict the origin of fecal pollution is therefore timely and of interest.
In this work, we determined the presence and abundance of genome fragments of HAdV, noroviruses GI and GII (NoV), crAssPH, and cultured phages infecting Bacteroides thetaiotaomicron strain GA17 (GA17PH), B. fragilis strain PG76 for porcine pollution (PGPH), and the sum of these two host-associated Bacteroides phages (BACPH) in human and animal wastewaters and fecal slurries in five European countries. The aim was to evaluate their sensitivity and specificity as MST markers for human pollution source detection and to assess the potential of the bacteriophages as indicators of the two human viruses (HAdV and NoV).
Materials and Methods
Samples and Sampling Campaigns
A total of 120 sewage and wastewater samples and animal fecal slurries were collected from municipal wastewater treatment plants, abattoirs and farms in five different countries (Austria, Finland, Germany, Portugal, and Spain) from 2013 to 2014 within the framework of the European Project AQUAVALENS. These samples were of different origins: human (35), porcine (24), and other animals (61) [bovine (23), poultry (24), horse (7), dog (2), cat (2), goat (1), bird (1), and rabbit (1)]. The sewage samples came from communities with 2,100 to 4.0 million inhabitants. Wastewater was taken from abattoirs processing about 400 pigs, 8,000 ruminant animals and 100,000 poultry per day. Animal fecal slurries were obtained by mixing feces from at least 10 individual animals with sterile water. Details of each sample can be found in Supplementary Material. All the countries took samples of the different origins throughout the year to avoid any seasonality effect. They were collected in sterile containers and kept at 4°C while in transit to the laboratory. One hundred ml of each sample was sent to the other partner institutions at 4°C to perform the assigned parameter analysis (NoV, Finland; HAdV, Portugal; PGPH, crAssPH and GA17PH, Spain) and immediately processed.
Detection and Enumeration of MST Markers
Detection of Host-Associated Bacteroides Phages
Phages infecting human- and porcine-associated Bacteroides species were enumerated according to the ISO standard method 10705-4 (ISO, 2001). Plaque-forming units (PFU) of host-associated Bacteroides phages were enumerated by the double-agar-layer technique using the B. thetaiotaomicron strain GA17 to detect human pollution and B. fragilis strain PG76 for porcine pollution (Payan et al., 2005; Gómez-Dóñate et al., 2011). One ml of fresh samples was directly analyzed in triplicate with the corresponding host strain.
Detection of Human Viruses: Adenovirus and Norovirus
To detect HAdV, 200 μl of sewage samples were extracted directly using a QIAamp DNA Blood Mini Kit (Qiagen). HAdV sequences were amplified following a previously described protocol (Hernroth et al., 2002). Each run included the original sample, 10- and 100-fold dilutions of each sample, a standard curve and positive and negative controls.
Norovirus GI and GII (NoV) sequences were amplified following ISO/TS 15216-1 (ISO, 2013) with some modifications. Briefly, a sample volume of 250 μl was used for RNA extraction, which was performed using a NucliSens® Magnetic Extraction Kit and NucliSens® MiniMag® instrument (Biomerieux, Boxtel, Netherlands) according to the manufacturer’s instructions. The sample was spiked with mengovirus to be used as a positive process control. Samples were amplified in a 20 μl-reaction using the QuantiTect Probe RT-PCR Kit (Qiagen, Hilden, Germany) and Rotorgene PCR cycler (Corbett) as described by Oristo et al. (2018), except using primer-probe sets as mentioned in ISO 15216: QNIF2 (FW) ATGTTCAGRTGGATGAGRTTCTCWGA, COG2R (REV) TCGACGCCATCTTCATTCACA, and QNIFs (PROBE) AGCACGTGGGAGGGCGATCG. Reverse transcrip tion- PCR runs included sample RNAs undiluted and diluted 1:10. For every set of samples, a negative extraction control, positive external RNA controls, and dilutions of purified plasmid dsDNA for construction of a standard curve were added.
Detection of Human-Associated crAssphage
DNA was extracted directly from 1 ml using the QIAamp DNA Blood Mini Kit (Qiagen). The abundance of crAssPH was analyzed with TaqMan Environmental Master Mix 2.0 (Applied Biosystems) using ABI StepOne Real-Time qPCR as previously described (García-Aljaro et al., 2017). DNA extraction controls were run together with the samples.
Statistical Analyses
Specificity and sensitivity of the different markers in detecting fecal pollution of known human or non-human origin were calculated as follows: specificity was defined as the proportion of negative samples in which the marker was not detected (true negatives/[true negatives + false positives]), whereas sensitivity was defined as the proportion of positive samples in which the marker was detected (true positives/[true positives + false negatives]). Spearman’s correlation coefficients were used to study the relationship between the different markers. Traditional multidimensional scaling was carried out in R using the library stats (R Core Team, 2019). Receiver operating characteristic (ROC) curve analysis was performed using OptimalCutpoints R library (López-Ratón et al., 2014). ROC curve analysis was performed to define the cut-off levels for the predictors of NoV and HAdV. The criteria for obtaining the optimal cut-off point was based on the Youden index. The optimal cut-off point was defined as the point on the ROC curve nearest to the point where both the sensitivity and specificity were 1. Positive predictive values (PPV) for NoV and HAdV were calculated as the proportion of true positive results out of the number of samples with a positive result (true positives/[true positives + false positives]). Negative predictive values (NPV) were calculated as the proportion of true negative results out of the number of samples with a negative result (true negatives/[true negatives + false negatives]). The best cut-off points defined the thresholds, and thresholds were included in PPV and NPV calculations.
All the results were log transformed and the mean used for the calculations.
Results
Bacteriophages as Human MST Trackers
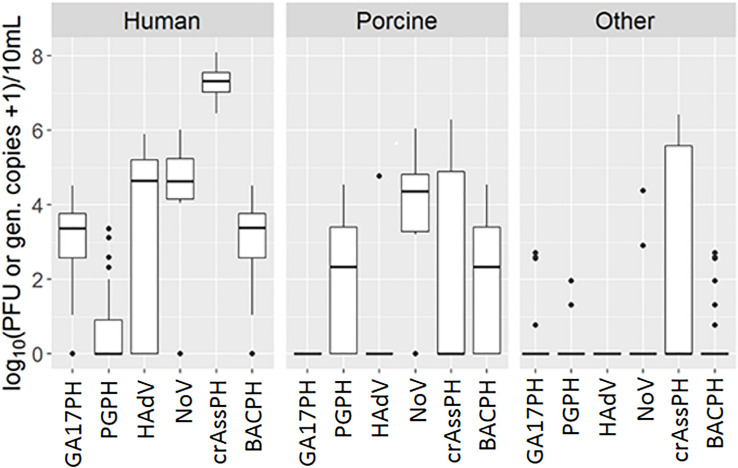
Most samples of human origin tested positive for NoV, as did a similar proportion of porcine samples. Briefly, a total of 30 out of 35 human samples (85.7%) tested positive for norovirus by RT-PCR, compared to19 out of 24 porcine samples (79.2%) and only 2 out of 61 samples (3.3%) of other origins, including poultry and cow. The concentrations found in positive samples of different origin ranged from 4.05 to 6.01 log10 GC/10 ml (human), 3.20 to 6.04 log10 GC/10 ml (porcine), and 2.90 to 4.37 log10 GC/10 ml (other) (Figure 1 and Supplementary Table 1). Important geographical differences were observed for the specificity and sensitivity for human source detection. Whereas the NoV marker presented a sensitivity ≥80% for all the countries except Portugal, its specificity was low (71, 63, and 55% in Germany, Portugal, and Spain, respectively), mainly due to its detection in pig samples (Table 1); in contrast, specificity in Austria was 100%.
FIGURE 1.
Boxplot representation of GA17PH, phages infecting human-associated Bacteroides thetaiotaomicron strain GA17; PGPH, phages infecting porcine-associated B. fragilis strain PG76; HAdV, human Adenoviruses; NoV, GI + GII Noroviruses, crAssPH, crAssphage, BACPH, sum of GA17PH, and PGPH. Outliers are shown as black dots.
TABLE 1.
Specificity (sp) and sensitivity (se) of the different markers at the different geographical locations in detecting human pollution (GA17PH, crAssPH, HAdV, and NoV) and porcine pollution (PGPH).
| GA17PH |
PGPH |
crAssPH |
HAdV |
NoV |
||||||
| sp | se | sp | se | sp | se | sp | se | sp | se | |
| Austria | 1.00 | 0.88 | 0.79 | 0.00 | 1.00 | 1.00 | 1.00 | 0.88 | 1.00 | 1.00 |
| Germany | 0.93 | 1.00 | 0.93 | 0.00 | 1.00 | 1.00 | 0.93 | 0.60 | 0.71 | 0.80 |
| Finland | 1.00 | 0.86 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 | 0.71 | 0.82 | 1.00 |
| Portugal | 0.88 | 1.00 | 0.94 | 1.00 | 0.38 | 1.00 | 1.00 | 0.14 | 0.63 | 0.57 |
| Spain | 0.95 | 0.83 | 0.65 | 1.00 | 0.10 | 1.00 | 1.00 | 0.67 | 0.55 | 1.00 |
GA17PH, phages infecting human-associated Bacteroides thetaiotaomicron strain GA17; PGPH, phages infecting porcine-associated B. fragilis strain PG76; BACPH, sum of GA17PH and PGPH; crAssPH, crAssphage; HAdV, human Adenoviruses; and NoV, GI + GII Noroviruses.
Human adenovirus was detected almost exclusively in samples of human origin, with the exception of 1 positive porcine sample out of 24. Nevertheless, only 20 out of 33 (60.6%) human samples were positive by qPCR (the remaining 2 samples up to 35 were not analyzed due to technical problems). The concentrations in the positive samples were high, ranging from 4.29 to 5.90 log10 GC/10 ml (Figure 1 and Supplementary Table 1). Once again, geographical differences were observed. In this case, although the specificity of the HAdV marker for human source detection was very high in all the countries (≥90%), its sensitivity was low, being ≤80% in all the countries except Austria (Table 1).
The performance of two human-related bacteriophages (GA17PH and crAssPH) was also assessed in terms of specificity and sensitivity for human source detection. The GA17PH marker displayed a higher specificity and sensitivity for human sources than either NoV or HAdV. Accordingly, GA17PH was detected in most of the human samples (32 out of 35, 91.4%), not detected in any porcine sample, and rarely detected in the samples of mixed animal origin (4 out of 61). The concentration in the positive human source samples ranged from 1.00 to 4.52 log10 PFU/10 ml, whereas in the other animal wastewater samples it was 2 log10 units lower in those with a higher fecal load (Figure 1 and Supplementary Table 1). No geographical differences were observed for GA17PH, with specificity ≥88% and sensitivity ≥83% for human source detection (Table 1). On the other hand, crAssPH was present at higher levels in human samples, which ranged from 6.45 to 8.10 log10 GC/10 ml, although it was also detected in 11 out of 24 samples of porcine origin and in 17 out of 61 samples of other animals (Figure 1 and Supplementary Table 1). This marker exhibited a notable geographical variability in specificity, which was 100% for the Northern European countries, yet very low in Portugal and Spain (38 and 10%, respectively; Table 1).
As stated above, most false positives in the NoV assay were due to interference from NoV of porcine origin. These viruses may also infect humans and therefore, it could be advantageous to have a suite of markers able to predict the presence of NoV of both origins. In this study, the sum of the human-associated (GA17PH) and porcine-associated (PGPH) Bacteroides phages was assessed as an indicator for the presence of NoV of either human or porcine origin. It should be noted that the results of the PGPH marker showed considerable geographical variability (Table 1), with a sensitivity of 0% in Austria and Germany, where it was not detected, and a specificity of 65% in Spain. The concentration of PGPH in positive samples of wastewater and slurries ranged from 0.70 to 4.55 log10 PFU/10 ml.
Bacteriophages as Indicators of HAdV and NoV
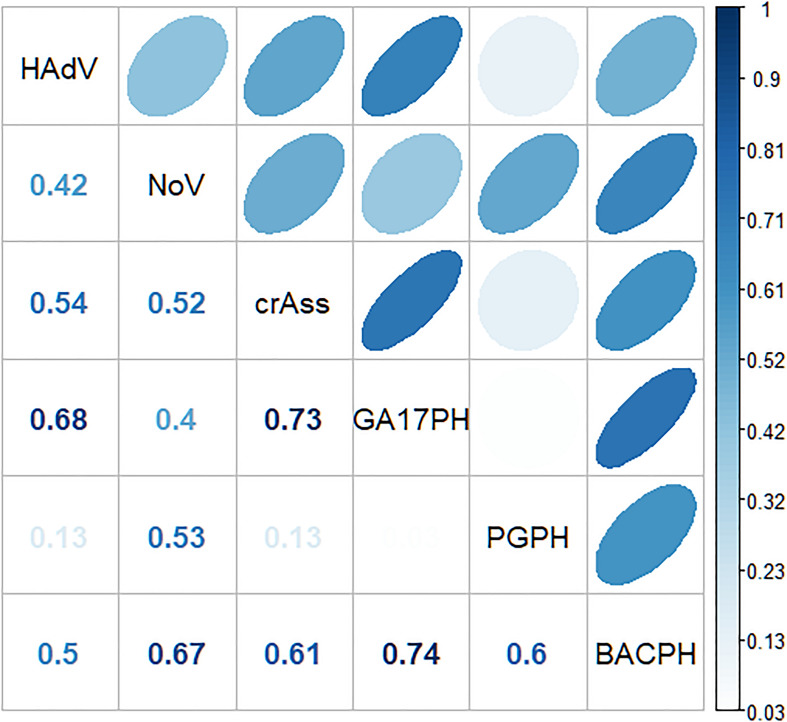
A significant correlation was observed between GA17PH and HAdV (p < 0.01, Spearman correlation coefficient rho = 0.66), as well as between crAssPH and both human viruses (p < 0.01, rho = 0.55, and 0.54 for HAdV and NoV, respectively; Figure 2). The sum of the two Bacteroides markers (GA17PH + PGPH, indicated as BACPH) gave a higher correlation with NoV (rho = 0.7) but lower with HAdV (rho = 0.44). NoV correlated more strongly with BACPH than with HAdV (rho = 0.44), indicating a weak correlation between the viruses.
FIGURE 2.
Spearman’s correlation coefficients between the different markers (GA17PH, phages infecting human-associated Bacteroides thetaiotaomicron strain GA17; PGPH, phages infecting porcine-associated B. fragilis strain PG76; BACPH, sum of GA17PH and PGPH; crAssPH, crAssphage; HAdv, human Adenoviruses; and NoV, GI + GII Noroviruses).
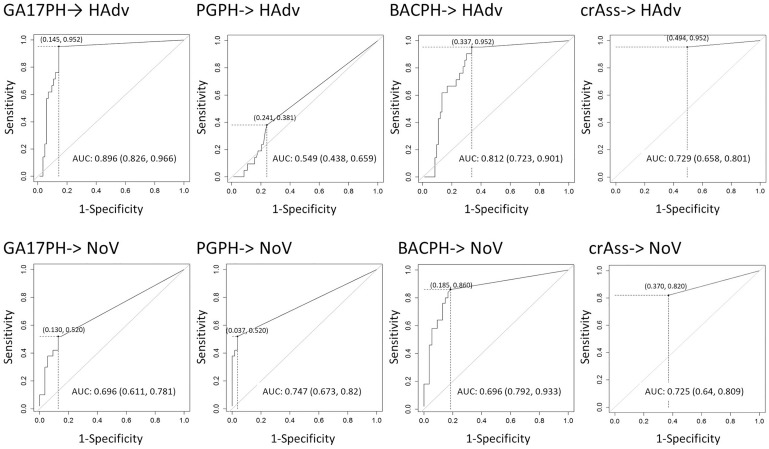
In view of these results, the concentration limits of the different bacteriophage markers that would be required to predict the presence of the enteric viruses were assessed statistically with the analysis of ROC curves (Figure 3). Based on the ROC curve analyses, the minimum concentration of crAssPH and GA17PH needed to predict HAdV was 1.00 and 1.54 log10 units, respectively, which was slightly higher than the respective 1.00 and 1.04 log10 units required to predict NoV. GA17PH showed a PPV of 63% and NPV of 99% for HAdV; for NoV a higher PPV of 79% and a lower NPV of 66% were obtained (Table 2). The sum of the markers PGPH and GA17PH (BACPH) increased the PPV and NPV for NoV to 81 and 86%, respectively, but gave a lower PPV of 42% for HAdV. In the case of crAssPH, a higher NPV of 79% was obtained for NoV, but the PPV was lower for both HAdV and NoV, being 33 and 67%, respectively. In summary, the highest PPV and NPV for HAdV were obtained with GA17PH (63 and 99%, respectively) followed by crAssPH (33 and98%, respectively), whereas for NoV the most acceptable PPV and NPV were obtained with BACPH (81 and 86%, respectively).
FIGURE 3.
ROC curve analysis with the prediction of HAdV and NoV by the different markers (GA17PH, phages infecting human-associated Bacteroides thetaiotaomicron strain GA17; PGPH, phages infecting porcine-associated B. fragilis strain PG76; BACPH, sum of GA17PH and PGPH; crAssPH, crAssphage; HAdv, human Adenoviruses; and NoV, GI + GII Noroviruses). AUC with the log of the minimal concentration (in brackets) needed to predict HAdV and NoV are indicated in the table below.
TABLE 2.
Predictive capacity of the different markers for HAdV and NoV.
| Markers | GA17PH |
PGPH |
BACPH |
crAssPH |
||||
| PPV | NPV | PPV | NPV | PPV | NPV | PPV | NPV | |
| HAdV | 0.63 | 0.99 | 0.29 | 0.83 | 0.42 | 0.98 | 0.33 | 0.98 |
| NoV | 0.79 | 0.66 | 0.93 | 0.68 | 0.81 | 0.86 | 0.67 | 0.79 |
Positive predictive value (PPV), negative predictive value (NPV). GA17PH, phages infecting human-associated Bacteroides thetaiotaomicron strain GA17; PGPH, phages infecting porcine-associated B. fragilis strain PG76; BACPH, sum of GA17PH and PGPH; crAssPH, crAssphage; HAdV, human Adenoviruses; and NoV, GI + GII Noroviruses.
Discussion
In the current work, we assessed the suitability of human viruses (NoV and HadV) as markers of fecal viral pollution (Albinana-Gimenez et al., 2009; Rusiñol et al., 2014), and compared their performance with that of different source-associated Bacteroides phages (Gómez-Dóñate et al., 2011), including the recently reported human bacteriophage crAssPH (García-Aljaro et al., 2017; Stachler et al., 2017; Ballesté et al., 2019).
In this study, HAdV was detected in 60% of the human sewage samples, NoV in 80%, GA17PH in 91%, and crAssPH in 100%, indicating that the bacteriophage markers were more sensitive than the enteric viruses. Comparable with our results, studies in Japan and New Zealand did not detect NoV in 10–40% of raw sewage samples, and also report a variable detection (Wolf et al., 2010; Hewitt et al., 2013; Haramoto et al., 2015). On the other hand, high sensitivity and specificity for human source detection has been generally observed for HAdV (Albinana-Gimenez et al., 2009; Ahmed et al., 2010; Rodríguez-Lázaro et al., 2012; Hewitt et al., 2013; Rusiñol et al., 2014; Haramoto et al., 2015), leading to its proposal as a human MST marker. Here, although HAdV specificity was high, sensitivity was only 60%, with variability among geographical areas. Another study has also reported lower specificity in individual human feces or septic tanks and sewage, which could be attributed to the variable prevalence of HAdV in the population pool, and the variety of HAdV species associated with outbreaks (Wolf et al., 2010). Another explanation for the differences among studies could be the different volumes and concentrating methods used. In our study, HAdV was measured directly from a low volume (0.2 ml) of sewage, but as no concentration method was required, DNA loss due to concentration was avoided. It has to be pointed out that in our study GA17PH and PGPH were detected by culture-based methods, whereas NoV and AdV were detected by qPCR. In theory, qPCR analysis of the two Bacteroides phages would provide useful data, however, it was not possible to perform because there are not available qPCR methods for these phages since they are a very heterogeneous group and difficult to analyze them by this technique.
The arithmetic mean of NoV GI and GII combined in samples was 4.79 log10 GC/10 ml. Values reported in the literature are highly variable, ranging from 1 to 7 log10 GC/10 ml (Kitajima et al., 2012; Hewitt et al., 2013; Haramoto et al., 2015; Katayama and Vinjé, 2017). Such variability hampers NoV detection in the environment and its application as an MST marker, another disadvantage being its presence in pig feces in some areas in this study. The mean of HAdV in raw sewage (5.13 log10 GC/10 ml) was found to be higher than in other studies (about 3–4 log10 GC/1,000 ml; Hewitt et al., 2013; Rusiñol et al., 2014; Haramoto et al., 2015), although a considerable variability (2–7 log10 GC/10 ml) between geographical areas has also been observed (Allard and Vantarakis, 2017). As a marker, HAdV showed good specificity for human source detection, being found mainly in human fecal samples, but its low sensitivity limits its use for MST unless combined with more sensitive markers (Blanch et al., 2006). To overcome the limitations of viruses as MST markers, host-associated bacteriophages have been proposed as MST tools, such as human-associated Bacteroides strains GA17 and GB124, the animal-associated PG76 (Ebdon et al., 2007; Gómez-Dóñate et al., 2011), or crAssPH (García-Aljaro et al., 2017), recently related to human pollution. In this study, we assessed the capacity of human phages infecting the human B. thetaiotaomicron GA17 strain as well as the porcine strain PG76 to predict the presence of human viruses. The GA17PH marker displayed the highest sensitivity and specificity, giving values from 1 to 4.52 log10 PFU/10 ml, in accordance with previous reports of 1.69 to 5.84 PFU/100 ml in Spain, Sweden, France, United Kingdom, Tunisia, and Colombia (Payan et al., 2005; Blanch et al., 2006; Venegas et al., 2015; Yahya et al., 2015). CrAssPH was sensitive, although its specificity varied according to location. The concentration of crAssPH found in our study was around 107 GC/10 ml, which was 2.5 log10 units higher compared to NoV. These results are in accordance with those of Farkas and co-workers, who compared crAssPH with several enteric viruses (NoVGI, NoVGII, SaV, AdV, and JCV; Farkas et al., 2019). However, their crAssPH numbers only correlated with JCV and not with any of the human viruses analyzed this study. The low concentration of GA17PH in raw sewage may be a problem for the analysis of environmental samples where fecal pollution is diluted. However, as the concentration of crAssPH in river water is 2–3 log10 units higher than GA17PH, the two markers could be used in combination in certain geographical locations or in catchments not containing large numbers of pigs or abattoirs (Ballesté et al., 2019).
It should be noted that most of the false positives of NoV as an MST marker of human fecal contamination were due to the cross-reaction of the assay with NoV of porcine origin. A possible explanation is that NoV GII real-time RT-PCR primers and probes could detect porcine GII NoV strains, since primers and probes used in the RT-qPCR assay are highly similar to the sequences available in GeneBank for porcine NoV (LC509111, AB126329, HQ392821, and AY823305). In fact, strong homologies between swine NoVs genomic sequences and human primer sequences have been reported previously (L’Homme et al., 2009). Although human infections with porcine GII NoV have not been documented (Villabruna et al., 2019), they cannot be excluded. On the other hand, pigs may be susceptible to infection with human NoV strains, which have been detected in porcine feces (Mattison et al., 2007; Sisay et al., 2016), although how commonly this occurs is not known. Given the risk of these viruses infecting humans, it would be of interest to have a suite of markers able to predict the presence of NoV of both origins (Mattison et al., 2007). In this case, the combination of GA17PH and PGPH (BACPH) could be used as an indicator for the presence of NoV of either human or porcine origin. Nevertheless, it should be noted that the PGPH marker showed geographical variability, as reported for other Bacteroides phages, and thus a Bacteroides pig-associated strain may be isolated in each targeted area (Payan et al., 2005) or other MST marker capable of identifying porcine sources such as Bacteroidales PG markers.
In summary, the results presented here show that bacteriophages infecting human-associated Bacteroides strains are a promising alternative for the prediction of human viruses of fecal origin as they outperform human viruses such as AdV and NoV, and provide an easier and more affordable technique for routine monitoring, avoiding the need to look for pathogenic viruses. Moreover, they also provide useful information on infectiousness/viability of the phages which is useful when establishing risk to human health.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
EB, LS-C, CG-A, SM, and LM participated in the sampling and carried out the experiments. JM performed the statistical analyses. SM, LM, AB, AF, and AT conceived and supervised the sampling experiments in the different laboratories. EB, JJ, and CG-A contributed to the data analysis and writing of the manuscript. All authors revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer, JL, declared a past collaboration with one of the authors, LM.
Acknowledgments
The authors are very grateful to Satu Oristo and Kirsi Söderberg for their support.
Footnotes
Funding. This project was supported by the European Union AQUAVALENS project, European Union (FP7-KBBE.2012.2.5–01, grant agreement No.: 311846), and the Catalan government, Spain (2017 SGR 170). AF was additionally supported by the Austrian Science Fund (FWF) as part of the Vienna Doctoral Program on Water Resource Systems (W1219-N22) and the Project AQUASAFE (SC15-016) funded by the Niederösterreichische Forschungs- und Bildungsgesellschaft (NFB).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.619495/full#supplementary-material
References
- Ahmed W., Goonetilleke A., Gardner T. (2010). Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Res. 44 4662–4673. 10.1016/j.watres.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Ahmed W., Lobos A., Senkbeil J., Peraud J., Gallard J., Harwood V. J. (2018). Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res. 131 142–150. 10.1016/j.watres.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Albinana-Gimenez N., Miagostovich M. P., Calgua B., Huguet J. M., Matia L., Girones R. (2009). Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 43 2011–2019. 10.1016/j.watres.2009.01.025 [DOI] [PubMed] [Google Scholar]
- Allard A., Vantarakis A. (2017). “Adenoviruses,” in Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project). (J.S Meschke, and R. Girones (eds), Part 3: Specific Excreted Pathogens: Environmental and Epidemiology Aspects - Section 1: Viruses) Michigan State University, eds Rose J. B., Jiménez-Cisneros B. (E. Lansing, MI: UNESCO; ). 10.14321/waterpathogens.11 [DOI] [Google Scholar]
- Baggi F., Demarta A., Peduzzi R. (2001). Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res. Microbiol. 152 743–751. 10.1016/S0923-2508(01)01255-4 [DOI] [PubMed] [Google Scholar]
- Ballesté E., Pascual-Benito M., Martín-Díaz J., Blanch A. R., Lucena F., Muniesa M., et al. (2019). Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 155 233–244. 10.1016/j.watres.2019.02.042 [DOI] [PubMed] [Google Scholar]
- Blanch A. R., Belanche-Munoz L., Bonjoch X., Ebdon J., Gantzer C., Lucena F., et al. (2006). Integrated analysis of established and novel microbial and chemical methods for microbial source tracking. Appl. Environ. Microbiol. 72 5915–5926. 10.1128/aem.02453-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M. A., Haas N. L., Hunt R. J. (2004). Vulnerability of drinking-water wells in La Crosse, Wisconsin, to enteric-virus contamination from surface water contributions. Appl. Environ. Microbiol. 70 5937–5946. 10.1128/AEM.70.10.5937-5946.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapron C. D., Ballester N. A., Fontaine J. H., Frades C. N., Margolin A. B. (2000). Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66 2520–2525. 10.1128/AEM.66.6.2520-2525.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costán-Longares A., Montemayor M., Payán A., Méndez J., Jofre J., Mujeriego R., et al. (2008). Microbial indicators and pathogens: removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 42 4439–4448. 10.1016/j.watres.2008.07.037 [DOI] [PubMed] [Google Scholar]
- EEC (1976). The Council of European Economic Communities Directive of 8 December 1975 Concerning the Quality of Bathing Waters. Official J. the European Communities, Directive no. 76/160/EEC. Brussels: Council of European Economic Communities. [Google Scholar]
- Dutilh B. E., Cassman N., McNair K., Sanchez S. E., Silva G. G. Z., Boling L., et al. (2014). A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5 37–43. 10.1038/ncomms5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebdon J., Muniesa M., Taylor H. (2007). The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of faecal pollution in a temperate river catchment. Water Res. 41 3683–3690. 10.1016/j.watres.2006.12.020 [DOI] [PubMed] [Google Scholar]
- European Commission (2018). Directive of the European Parliament and of the Council on the Quality of Water Intended for Human Consumption. COM/2017/0753 final – 2017/0332 (COD). Brussels: European Commission. [Google Scholar]
- Farkas K., Adriaenssens E. M., Walker D. I., McDonald J. E., Malham S. K., Jones D. L. (2019). Critical evaluation of crAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ. Virol. 11 113–119. 10.1007/s12560-019-09369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Aljaro C., Ballesté E., Muniesa M., Jofre J. (2017). Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 10 1775–1780. 10.1111/1751-7915.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerba C. P., Goyal S. M., LaBelle R. L., Cech I., Bodgan G. F. (1979). Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Public Health 69 1116–1119. 10.2105/AJPH.69.11.1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Dóñate M., Payán A., Cortés I., Blanch A. R., Lucena F., Jofre J., et al. (2011). Isolation of bacteriophage host strains of Bacteroides species suitable for tracking sources of animal faecal pollution in water. Environ. Microbiol. 13 1622–1631. 10.1111/j.1462-2920.2011.02474.x [DOI] [PubMed] [Google Scholar]
- Grabow W. (2004). Bacteriophages: update on application as models for viruses in water. Water SA 27 251–268. 10.4314/wsa.v27i2.4999 [DOI] [Google Scholar]
- Grabow W. O. K. (2007). “Chapter 1 Overview of health-related water virology,” in Human Viruses in Water: Perspectives in Medical Virology, ed. A. Bosch (Amsterdam: Elsevier B.V.), 1–25. 10.1016/S0168-7069(07)17001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Fujino S., Otagiri M. (2015). Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci. Total Environ. 520 32–38. 10.1016/j.scitotenv.2015.03.034 [DOI] [PubMed] [Google Scholar]
- Harwood V. J., Levine A. D., Scott T. M., Chivukula V., Lukasik J., Farrah S. R., et al. (2005). Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163. 10.1128/AEM.71.6.3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada (2019). Guidelines for Canadian Drinking Water Quality: Guideline Technical Document – Enteric Viruses. Ottawa, ON: Health Canada. [Google Scholar]
- Hernroth B. E., Conden-Hansson A.-C., Rehnstam-Holm A.-S., Girones R., Allard A. K. (2002). Environmental factors influencing human viral pathogens and their potential indicator organisms in the Blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 68 4523–4533. 10.1128/AEM.68.9.4523-4533.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J., Greening G. E., Leonard M., Lewis G. D. (2013). Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res. 47 6750–6761. 10.1016/j.watres.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Hot D., Legeay O., Jacques J., Gantzer C., Caudrelier Y., Guyard K., et al. (2003). Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res. 37 4703–4710. 10.1016/S0043-1354(03)00439-1 [DOI] [PubMed] [Google Scholar]
- Hundesa A., Maluquer de M. C., Bofill-Mas S., Binana-Gimenez N., Girones R. (2006). Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 72 7886–7893. 10.1128/aem.01090-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt R. J., Borchardt M. A., Richards K. D., Spencer S. K. (2010). Assessment of sewer source contamination of drinking water wells using tracers and human enteric viruses. Environ. Sci. Technol. 44 7956–7963. 10.1021/es100698m [DOI] [PubMed] [Google Scholar]
- IAWPRC (1991). Bacteriophages as model viruses in water quality control. Water Res. 25 529–545. 10.1016/0043-1354(91)90126-B [DOI] [Google Scholar]
- Irving L. G., Smith F. A. (1981). One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41 51–59. 10.1128/AEM.41.1.51-59.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISO (2013). Microbiology of Food and Animal Feed – Horizontal method for Determination of Hepatitis A Virus and Norovirus in Food Using Real-Time RT-PCR – Part 1: Method for Quantification. Geneva: ISO. [Google Scholar]
- ISO (2001). ISO 10705-4: Water Quality – Detection and Enumeration of Bacteriophages. Part 4: Enumeration of Bacteriophages Infecting Bacteroides fragilis. Geneva: ISO. [Google Scholar]
- Jebri S., Muniesa M., Jofre J. (2017). “General and host-associated bacteriophage indicators of faecal pollution,” in Global Water Pathogen Project. http://www.waterpathogens.org (Farnleitner, A, Blanch, A. (eds) Part 2 Indicators and Microbial Source Tracking Markers) http://www.waterpathogens.org/book/coliphage Michigan State University, eds Rose J. B., Jiménez-Cisneros B. (E. Lansing, MI: UNESCO; ), 10.14321/waterpathogens [DOI] [Google Scholar]
- Jiang S., Noble R., Chu W. (2001). Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67 179–184. 10.1128/AEM.67.1.179-184.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre J., Lucena F., Blanch A. R., Muniesa M. (2016). Coliphages as model organisms in the characterization and management of water resources. Water (Switzerland) 8:199. 10.3390/w8050199 [DOI] [Google Scholar]
- Jofre J., Blanch A. R. (2010). Feasibility of methods based on nucleic acid amplification techniques to fulfil the requirements for microbiological analysis of water quality. J. Appl. Microbiol. 109 1853–1867. 10.1111/j.1365-2672.2010.04830.x [DOI] [PubMed] [Google Scholar]
- Jofre J., Blanch A. R., Lucena F., Muniesa M. (2014). Bacteriophages infecting Bacteroides as a marker for microbial source tracking. Water Res. 55 1–11. 10.1016/j.watres.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Jofre J., Stewart J. R., Grabow W. (2011). “Phage methods,” in Microbial Source Tracking: Methods, Applications, and Case Studies, eds Hagedorn C., Blanch A. R., Harwood V. J. (New York, NY: Springer New York; ), 137–156. 10.1007/978-1-4419-9386-1_6 [DOI] [Google Scholar]
- Katayama H., Vinjé J. (2017). “Norovirus and other caliciviurses,” in Global Water Pathogens Project. Michigan State University, eds Rose J. B., Jimenez-Cisneros B. (East Lansing, MI: UNESCO; ). [Google Scholar]
- Kitajima M., Haramoto E., Phanuwan C., Katayama H., Furumai H. (2012). Molecular detection and genotyping of human noroviruses in influent and effluent water at a wastewater treatment plant in Japan. J. Appl. Microbiol. 112 605–613. 10.1111/j.1365-2672.2012.05231.x [DOI] [PubMed] [Google Scholar]
- L’Homme Y., Sansregret R., Simard C. (2009). Broad range RT-PCR assays targeting human noroviruses also detect swine noroviruses. Food Microbiol. 26 552–555. 10.1016/j.fm.2009.03.011 [DOI] [PubMed] [Google Scholar]
- López-Ratón M., Rodríguez-Álvarez M. X., Suárez C. C., Sampedro F. G. (2014). OptimalCutpoints: an R package for selecting optimal cutpoints in diagnostic tests. J. Stat. Softw. 61 1–36. 10.18637/jss.v061.i08 [DOI] [Google Scholar]
- Mattison K., Shukla A., Cook A., Pollari F., Friendship R., Kelton D., et al. (2007). Human noroviruses in swine and cattle. Emerg. Infect. Dis. 13 1184–1188. 10.3201/eid1308.070005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R. E., Bofill-Mas S., Egle L., Reischer G. H., Schade M., Fernandez-Cassi X., et al. (2016). Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 90 265–276. 10.1016/j.watres.2015.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHMRC (2011). Australian Drinking water Guidelines Paper 6 National Water Quality Management Strategy. Canberra, NSW: NHMRC. [Google Scholar]
- Nuanualsuwan S., Cliver D. O. (2002). Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 104 217–225. 10.1016/S0166-0934(02)00089-7 [DOI] [PubMed] [Google Scholar]
- Oristo S., Lee H.-J., Maunula L. (2018). Performance of pre-RT-qPCR treatments to discriminate infectious human rotaviruses and noroviruses from heat-inactivated viruses: applications of PMA/PMAxx, benzonase and RNase. J. Appl. Microbiol. 124 1008–1016. 10.1111/jam.13737 [DOI] [PubMed] [Google Scholar]
- Payan A., Ebdon J., Taylor H., Gantzer C., Ottoson J., Papageorgiou G. T., et al. (2005). Method for isolation of Bacteroides bacteriophage host strains suitable for tracking sources of fecal pollution in water. Appl. Environ. Microbiol. 71 5659–5662. 10.1128/AEM.71.9.5659-5662.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ [Google Scholar]
- Rodríguez-Lázaro D., Cook N., Ruggeri F. M., Sellwood J., Nasser A., Nascimento M. S. J., et al. (2012). Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 36 786–814. 10.1111/j.1574-6976.2011.00306.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Fernandez-Cassi X., Hundesa A., Vieira C., Kern A., Eriksson I., et al. (2014). Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 59 119–129. 10.1016/j.watres.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Shkoporov A. N., Khokhlova E. V., Fitzgerald C. B., Stockdale S. R., Draper L. A., Ross R. P., et al. (2018). ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9:4781. 10.1038/s41467-018-07225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva H. D., García-Zapata M. T. A., Anunciação C. E. (2011). Why the use of adenoviruses as water quality virologic marker? Food Environ. Virol. 3 138–140. 10.1007/s12560-011-9069-2 [DOI] [Google Scholar]
- Sisay Z., Djikeng A., Berhe N., Belay G., Abegaz W. E., Wang Q. H., et al. (2016). First detection and molecular characterization of sapoviruses and noroviruses with zoonotic potential in swine in Ethiopia. Arch. Virol. 161 2739–2747. 10.1007/s00705-016-2974-9 [DOI] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O. C. (2017). Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 51 9146–9154. 10.1021/acs.est.7b02703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartera C., Jofre J., Lucena F. (1988). Relationship between numbers of enteroviruses and bacteriophages infecting bacterowes fragilis in different environmental samples. Environ. Technol. Lett. 9 407–410. 10.1080/09593338809384584 [DOI] [Google Scholar]
- Tartera C., Lucena F., Jofre J. (1989). Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl. Environ. Microbiol. 55 2696–2701. 10.1128/AEM.55.10.2696-2701.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N. H., Gin K. Y.-H., Ngo H. H. (2015). Fecal pollution source tracking toolbox for identification, evaluation and characterization of fecal contamination in receiving urban surface waters and groundwater. Sci. Total Environ. 538 38–57. 10.1016/j.scitotenv.2015.07.155 [DOI] [PubMed] [Google Scholar]
- USEPA (1992). Environmental Regulations and Technology: Control of Pathogens and Vector Attraction in Sewage Sludge Under 40 CFR Part 503. Washington, DC: U.S. Environmental Protection Agency. [Google Scholar]
- Venegas C., Diez H., Blanch A. R., Jofre J., Campos C. (2015). Microbial source markers assessment in the Bogotá River basin (Colombia). J. Water Health 13 801–810. 10.2166/wh.2015.240 [DOI] [PubMed] [Google Scholar]
- Villabruna N., Koopmans M. P. G., de Graaf M. (2019). Animals as Reservoir for human norovirus. Viruses 11:478. 10.3390/v11050478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Greening G. E. (2010). Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 76 1388–1394. 10.1128/AEM.02249-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Fong T.-T., Bibby K., Molina M. (2012). Application of enteric viruses for fecal pollution source tracking in environmental waters. Environ. Int. 45 151–164. 10.1016/j.envint.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Wyer M. D., Fleisher J. M., Gough J., Kay D., Merrett H. (1995). An investigation into parametric relationships between enterovirus and faecal indicator organisms in the coastal waters of England and Wales. Water Res. 29 1863–1868. 10.1016/0043-1354(95)00003-4 [DOI] [Google Scholar]
- Yahya M., Hmaied F., Jebri S., Jofre J., Hamdi M. (2015). Bacteriophages as indicators of human and animal faecal contamination in raw and treated wastewaters from Tunisia. J. Appl. Microbiol. 118 1217–1225. 10.1111/jam.12774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.