Abstract
Background
There is evidence regarding the role of two lncRNAs: MEG3 and H19 the pathomechanism of obesity and related disorders. Here, we aimed to evaluate the expression of MEG3 and H19 in visceral adipose tissues (VAT) and subcutaneous adipose tissues (SAT) of obese women (n = 18), as compared to normal‐weight women (n = 17). Moreover, we sought to identify the association of expression of MEG3 and H19 in SAT and VAT with obesity parameters, insulin resistance, and the mRNA expression of possible target genes involved in adipogenesis and lipogenesis including peroxisome proliferator‐activated receptor gamma (PPARγ), fatty acid synthase (FAS), and acetyl‐CoA carboxylase (ACC).
Methods
Real‐time PCR was performed to investigate the mRNA expression of the above‐mentioned genes in VAT and SAT from all participants.
Results
The results showed lower mRNA levels of H19 in SAT of obese women, compared to normal‐weight women, while MEG3 expression was significantly higher in the SAT of the obese group rather than controls. Correlation analysis indicated that the transcript level of H19 had an inverse correlation with obesity indices and HOMA‐IR values. However, MEG3 expression displayed a positive correlation with all the indicated parameters in all participants. Interestingly, a positive correlation was found between transcript level of MEG3 in SAT with FAS and PPARγ. However, there was an inverse correlation between SAT expression of H19 and FAS.
Conclusions
It appears that lncRNAs, MEG3 and H19, are involved in obesity‐related conditions. However, more clinical studies are still required to clarify the relationships between lncRNAs with obesity and related abnormalities.
Keywords: adipogenesis, lipogenesis, long non‐coding RNAs, obesity
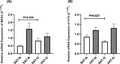
The expression level of MEG3 (a) and H19 (b) in VAT and SAT from obese and non‐obese groups. All data were expressed as an n‐fold difference relative to the calibrator sample (a mixture of the VAT and SAT tissues). SAT: subcutaneous adipose tissue; VAT: visceral adipose tissue; N: non‐obese; O: obese. Data were shown as the mean ± standard error of the mean (SEM).
1. INTRODUCTION
Obesity, one of the most devastating phenomena of the recent era, is a worldwide epidemic heterogeneous disorder with a high prevalence rate. 1 WHO defines obesity as a disease with the accumulation of excess fat in the body which has been linked to increased risk for metabolic diseases, including non‐alcoholic fatty liver diseases, insulin resistance, and cardiovascular diseases. 2 There is an anatomical difference in the distribution of the fat tissues in the body depending on various factors, such as age, sex, race, dietary regimen, physical activities, hormones, and drugs. 3 A concept that has emerged fairly recently is that abdominal obesity is more associated with diabetes and cardiovascular diseases compared to general obesity. 4 , 5 Abdominal fat is composed of abdominal subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT). 6 Unlike SAT, VAT is localized in mesentery and omentum and has metabolic properties. VAT and SAT are completely different from each other concerning cellular types, endocrine function, and the response to insulin and other hormones. 7 As compared to SAT, VAT has more metabolic activity, more sensitivity to lipolysis, more resistance to insulin, and more sensitivity to adrenergic stimulation. However, the SAT tends to absorb free fatty acids and triglycerides from blood circulation. 8 The amount of fat accumulation is dependent on the balance between anabolic processes (adipogenesis and lipogenesis) and catabolic processes (lipolysis, beta‐oxidation of fatty acids, and thermogenesis).
Recent years have seen increased investigation into the role of long non‐coding RNA (lncRNAs) in the pathogenesis of obesity and related abnormalities. lncRNAs, a group of non‐coding RNAs with a length longer than 200 nucleotides, play substantial roles in epigenetic regulation, chromatin remodeling intracellular trafficking, transcription regulation, and post‐transcriptional processes. Given the contribution in the wide variety of intracellular processes, it is not surprising that any alteration in the expression pattern of lncRNAs contributes to the development of many diseases. 9 Among the long list of genes encoding lncRNA, MEG3 and H19 are two lncRNAs whose expression is restricted to adipose tissues. MEG3, a prominent gene located at chromosome 14 in humans and chromosome 12 in mice, encodes a lncRNA with about 700 nucleotides. 10 The data extracted from several studies shed light on the participation of lncRNA MEG3 in the pathogenesis of obesity. It has been indicated that while the down‐regulation of lncRNA MEG3 could inversely regulate the expression of lipogenesis‐related genes, the over‐expression of this gene may alleviate lipid over‐deposition. 11 Moreover, the up‐regulation of lncRNA MEG3 in high‐fat diet and ob/ob mice and the association between insulin resistance and the expression level of this lncRNA are all suggestive of the involvement of this non‐coding RNA in the etiology of obesity. 12 Apart from lncRNA MEG3, lncRNA H19 also seems to have a role in the regulation of obesity. lncRNA H19 owns its reputation due to its involvement in the pathogenesis of different human cancers. 13 Aside from carcinogenesis, this lncRNA could also inhibit adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modification. 14 Recent studies reported that H19 and MEG3 have interactions with an mRNA‐binding protein named PTBP1 and stabilize SERBP1c mRNAs, 15 , 16 which has a role in lipogenesis through regulating the expression level of acetyl‐CoA carboxylase (ACC) and fatty acid synthesis (FAS) and PPAR‐gamma target genes. 17 Data from in vitro studies and animal model surveys led us to investigate the clinical relevance of these lncRNAs in the context of obesity in humans. Hence, we intended to compare the expression pattern of H19 and MEG3 in VAT and SAT from obese women and normal‐weight ones. Next, we aimed to investigate the possible association of H19 and MEG3 with metabolic profile as well as the adipose tissue transcript levels of PPAR‐γ, FAS, and ACC.
2. PATIENTS AND METHODS
2.1. Study population
A total of 35 women were recruited and divided into obese (n = 18) and no non‐obese (n = 17) groups. Obese women (BMI ≥ 35 Kg/m2) were selected among ones referred to the Surgery Clinic of Imam Khomeini Hospital (Tehran University of Medical Sciences) and Irfan Hospital who were a candidate for bariatric surgery. The control group was recruited from normal‐weight women (BMI ≤ 25 kg/m2) who were undergoing elective surgery (cholecystectomy, abdominal hernia, appendectomy, diaphragm hernia) at Sina and Loqman Hakim hospitals, Tehran, Iran. The age of the participants was between 20 and 54 years old. Exclusion criteria for both obese and non‐obese groups were current smoking, type I diabetes, type II diabetes, chronic or acute inflammatory diseases, infectious diseases, cancer, and acute conditions causing steroid therapy, surgical intervention, and hospitalization. None of the patients had received drugs affecting metabolic profile (e.g. weight loss control drugs, metformin, statin medications, hypoglycemic and hypolipidemic drugs) in the past 6 months. The study protocol was in accordance with the Declaration of Helsinki and was approved by the ethics committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.RETECH. REC.1397.1175).
2.2. Sampling
After 12 h of fasting, 10 ml of venous blood samples were collected, two studied groups. The serum samples were separated and stored at −70°C until further experiments. Serum concentrations of fasting blood sugar (FBS), HDL‐C, LDL‐C, triglyceride (TG), and total cholesterol (TC) were measured by autoanalyzer using commercial kits (Pars Azmoon). Furthermore, the fasting insulin concentration was assessed using the ECL method in the Cobas6000 E601 auto analyzer. High‐sensitivity C‐reactive protein (hs‐CRP) was quantified by an immunoturbidometric method using the Roche Integra analyzer. Homeostasis model assessment of insulin resistance (HOMA‐IR) was estimated with the following equation: fasting blood glucose (mg/dL) × fasting blood insulin (µU/ml)/405.
The surgeon excised approximately 0.5 g of SAT and VAT from each participant at the beginning of the surgical procedure. VAT was obtained from the abdominal cavity. SAT was obtained after the surgeon used a scalpel to create a small incision 0.5 cm deep in the superficial skin. The adipose tissue samples were washed in the cold and sterile phosphate‐buffered saline and were aseptically divided into small sections and stored in RNase‐free tubes. Then, tissues were snap‐frozen in liquid nitrogen and immediately stored at −80°C until needed.
2.3. RNA extraction and real‐time PCR
During the surgical procedure, approximately 0.5 g of VAT and SAT was obtained from the patients by the surgeon as described previously. Biopsy samples were rapidly washed in the cold and sterile phosphate‐buffered saline and were aseptically divided into small sections and aliquoted in RNase‐free tubes. Then, tissues were snap‐frozen in liquid nitrogen and immediately stored at −80°C until needed.
Tissue samples were homogenized in Qiazol lysis reagent (Qiagen) and total RNAs were extracted using RNeasy lipid tissue kit (Qiagen). Before reverse transcription, isolated RNAs were checked for quality and quantity with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific) and RNA integrity was checked by running RNA samples on a 1% agarose gel. Complementary DNA (cDNA) was synthesized from RNA (1 µg) using the PrimeScript 1st Strand cDNA Synthesis kit. Complementary DNA (cDNA) was synthesized from 1 µg of extracted RNA using the PrimeScript 1st Strand cDNA Synthesis kit.
Table S1 lists the used primer sequences: MEG3, H19, PPARγ, FAS, and ACC and the reference gene: β‐actin. 18 , 19 , 20 , 21 Each expression was quantified in duplicate. A melting curve analysis was done to check the specificity of all amplified products. The difference in Ct values (ΔCt) between the target gene and the reference gene was calculated for all samples.
The efficiency of amplification was calculated by the slope of the standard curve for all target genes and the reference gene. Since the value of efficiency was ranged from 90% to 100% in all assays, we used a 2−∆∆Ct method to perform mRNA analysis. A mixture of the SAT and VAT tissues was used as the calibrator sample and all results were expressed as an n‐fold difference relative to the calibrator sample.
2.4. Statistical analysis
The Shapiro‐Wilk test was used for evaluating the normality of continuous variables. While normal variables are expressed as means ± standard deviation (SD), those variables without normal distribution are shown as the interquartile range (IQR). Student t test and Mann‐Whitney U test were used for analyzing the comparison between groups. Moreover, for evaluating the association between the genes and between the genes and the clinical manifestations, we computed Pearson's rho correlation coefficients. All data with non‐normal distribution were logarithmically transformed before conducting correlation analysis. For evaluating the alteration of the genes at the mRNA level, we used Schmittgen and Livak method, which calculated 2−ΔCT (relative expression) from the measured ΔCT by real‐time PCR. Multiple stepwise regression analysis was performed to correct the effects of the covariates and to test independent factors. For all analyses, a p‐value of <.05 was considered to be statistically meaningful. All statistical analyses were performed using SPSS (version 16.0; SPP Inc.).
3. RESULT
3.1. Anthropometric, biochemical, and clinical characteristics of obese and non‐obese women
The comparison between biochemical parameters and metabolic characteristics of obese women and non‐obese counterparts are summarized in Table 1. The mean age of the obese group and non‐obese was 34.9 ± 6.6 and 39.7 ± 10.2 years, respectively (p‐value = .115). Notably, there was an obvious difference in terms of the obesity indicators, such as BMI, WC, and hip circumference between the two groups (p‐value = .000 for all parameters).
TABLE 1.
Clinical and laboratory characteristics of the obese and non‐obese groups
| Non‐obese group (n = 17) |
Obese group (n = 18) |
p‐value | |
|---|---|---|---|
| Age (years) | 39.7 ± 10.2 | 34.9 ± 6.6 | .115 |
| BMI (kg/m2) | 25.6 (24.9–27) | 41 (36–46) | .000 |
| WC (cm) | 89.6 ± 11.8 | 115.1 ± 10.2 | .000 |
| Hip circumference (cm) | 100.7 ± 10.8 | 129.2 ± 12.3 | .000 |
| WHR | 0.88 ± 0.05 | 0.89 ± 0.07 | .808 |
| SBP (mm Hg) | 117.2 (110–122.5) | 113.5 (107.5–122.5) | .973 |
| DBP (mm Hg) | 73.8 (70–80) | 72.8 (60–90) | .695 |
| FBS (mg/dl) | 88.05 ± 11.5 | 85.7 ± 10.1 | .525 |
| FINS | 8.7 ± 5.5 | 20.3 ± 8.1 | .000 |
| HOMA‐IR | 1.9 (1.04–2.19) | 4.3 (3.16–5.35) | .000 |
| hs‐CRP (mg/dl) | 4.9 (|0.94–2.84) | 6.5 (2.68–9.47) | .003 |
| HDL‐C (mg/dl) | 33.8 ± 9.9 | 44.8 ± 8 | .001 |
| LDL‐C (mg/dl) | 86.1 ± 28.1 | 114.9 ± 20.3 | .001 |
| VLDL (mg/dl) | 23.2 (11.5–29) | 22.2 (16.5–29) | .552 |
| TC (mg/dl) | 143 ± 38.5 | 182 ± 26.9 | .002 |
| TG (mg/dl) | 121.6 (63.1–125.5) | 130.9 (82.5–164.2) | .291 |
| Urea (mg/dl) | 22.7 ± 7.2 | 26.3 ± 6.2 | .124 |
| Creatinine (mg/dl) | 0.56 ± 0.16 | 0.74 ± 0.11 | .001 |
Data are reported as mean ± standard deviation (SD) or median (interquartile range [IQR]) as appropriate.
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; FBS, fasting blood sugar; FINS, fasting insulin; HbA1c, hemoglobin A1c; HDL‐C, high‐density lipoprotein cholesterol; HOMA‐IR, homeostatic model assessment of insulin resistance; hs‐CRP, high‐sensitivity C‐reactive protein; LDL‐C, low‐density lipoprotein cholesterol; SBP, systolic blood pressure; TC, total cholesterol; TG, triglycerides; VLDL, very‐low‐density lipoprotein; WC, waist circumference; WHR, waist‐to‐hip ratio.
Continuous variables with normal and non‐normal distribution were described as the mean ± SD and median (IQR), respectively.
p‐value <.05 shows significant differences between the obese and normal‐weight groups.
Also, we observed that unlike the serum levels of TG and FBS, which displayed no difference between the obese and non‐obese groups, HOMA‐IR (p‐value = .000) and the serum levels LDL‐C (p‐value = .001), HDL‐C (p‐value = .001), TC (p‐value = .002), and creatinine (p‐value = .001) were significantly higher in the obese group compared with the control group.
3.2. Expression profile of lncRNAs: MEG3 and H19 and possible target genes: PPARγ, FAS, and ACC
The expression pattern of MEG3 and H19 in the SAT and VAT was evaluated by qRT‐PCR analysis in two studied groups. Our findings revealed that the expression of MEG3 in the SAT from the obese patient was higher than the control group (p‐value = .036), while there was a reduction in the expression of H19 in the SAT from the obese group (p‐value = .027; Figure 1). However, the transcript level of MEG3 and H19 in VAT was not statistically significant between the obese and non‐obese groups.
FIGURE 1.

The expression level of MEG3 (A) and H19 (B) in VAT and SAT from obese and non‐obese groups. All data were expressed as an n‐fold difference relative to the calibrator sample (a mixture of the VAT and SAT tissues). SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue; N, non‐obese; O, obese. Data were shown as the mean ± standard error of the mean (SEM)
Furthermore, the mRNA level of PPARγ, a master regulator of adipocyte differentiation, as well as FAS and ACC in SAT and VAT, was analyzed between two studied groups (Figure 2). Our data showed that the expression of PPARγ, FAS, and ACC was elevated in the SAT obtained from the obese women (p‐value = .000, p‐value = .013, and p‐value = .033, respectively) as compared with the non‐obese counterparts. However, we found no statistically significant differences between the two study groups regarding VAT transcript levels of PPARγ, FAS, and ACC.
FIGURE 2.

The expression level of PPARγ (A), FAS (B), and ACC (C) in VAT and SAT from obese and non‐obese groups. All data were expressed as an n‐fold difference relative to the calibrator sample (a mixture of the VAT and SAT tissues). SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue; N, non‐obese; O, obese. Data were shown as the mean ± standard error of the mean (SEM)
3.3. The Correlation of MEG3 and H19 Expression with Anthropometric and Biochemical Measurements
The results of Pearson's correlation analysis (Table 2) showed that MEG3 expression in SAT had a significant correlation with both obesity indices: BMI (r = .422, p = .006), WC (r = .468, p = .002), hip circumference (r = .398, p = .009), insulin levels (r = .504, p = .001), and HOMA‐IR (r = .483, p = .002). However, we failed to find any significant correlation between MEG3 expression in VAT and the indicated parameters. Moreover, mRNA expression of H19 in SAT inversely correlated with obesity indices: BMI (r = −.384, p = .011), WC (r = −.316, p = .032), hip circumference (r = −.359, p = .017), insulin levels (r = −.373, p = .014), and HOMA‐IR (r = −.320, p = .031).
TABLE 2.
Pearson correlation of Meg3 and H19 genes expression in subcutaneous and visceral adipose tissue of whole participants with metabolic‐related indices
| Variable | MEG3 in SAT | Meg3 in VAT | H19 in SAT | H19 in VAT |
|---|---|---|---|---|
| BMI (Kg/m2) |
r = .422 p = .006 |
r = −.125 p = .237 |
r = −.384 p = .011 |
r = −.061 p = .364 |
| WC (cm) |
r = .468 p = .002 |
r = −.073 p = .339 |
r = −.316 p = .032 |
r = −.037 p = .417 |
| Hip circumference (cm) |
r = .398 p = .009 |
r = −.108 p = .269 |
r = −.359 p = .017 |
r = −.029 p = .435 |
| FINS |
r = .504 p = .001 |
r = −.036 p = .419 |
r = −.373 p = .014 |
r = −.111 p = .263 |
| HOMA‐IR |
r = .483 p = .002 |
r = .026 p = .441 |
r = −.320 p = .031 |
r = −.063 p = .360 |
Abbreviations: BMI, body mass index; FINS, fasting insulin; HOMA‐IR, homeostatic model assessment of insulin resistance; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue; WC, waist circumference.
Multivariate stepwise regression analysis included MEG3 and H19 transcript levels in SAT as dependent variables and, on one hand, the factors which p < .05 from the bivariate correlation. These are BMI, WC, hip, HOMA‐IR, and insulin for MEG3 and H19 gene expression. Each model was controlled for age. In multiple stepwise regression analysis, HOMA‐IR was positively associated with MEG3 gene expression in SAT (β = 0.485; adjusted R 2 change: .211; p = .004, CI: 0.001–0.003) and remained unchanged after control for age. However, multivariate stepwise regression analysis indicated that BMI was significant covariates associated with the H19 transcript level in SAT (β = −0.397; adjusted R 2 change: .158; p = .02, CI: −0.006–0.001).
3.4. Correlations of mRNA expression of MEG3, H19 with PPARγ, FAS, and ACC Transcript Levels
The results of Pearson's correlation analysis (Table 3) showed MEG3 expression in SAT had a significant correlation with the transcript level of PPARγ (r = .333, p = .025) and FAS (r = .293, p = .044) in the SAT. Moreover, the H19 mRNA level in SAT inversely correlated with the transcript level of FAS (r = −.396, p = .009) in the SAT. We failed to find any significant correlation of MEG3 and H19 expression in VAT with PPARγ, FAS, and ACC transcript levels.
TABLE 3.
Pearson correlation of MEG3 and H19 genes expression in subcutaneous and visceral adipose tissue of whole participants with metabolic‐related genes
| Variable | MEG3 in SAT | Meg3 in VAT | H19 in SAT | H19 in VAT |
|---|---|---|---|---|
| PPARγ |
r = .333 p = .025 |
r = −.217 p = .106 |
r = −.03 p = .403 |
r = −.232 p = .090 |
| FAS |
r = .293 p = .044 |
r = −.174 p = .159 |
r = −.396 p = .009 |
r = −.143 p = .207 |
| ACC |
r = .047 p = .394 |
r = −.126 p = .236 |
r = −.209 p = .114 |
r = −.077 p = .331 |
Abbreviations: ACC, acetyl‐CoA carboxylase; FAS, fatty acid synthase; PPARγ, peroxisome proliferator‐activated receptor gamma.
4. DISCUSSION
The possible role of lncRNAs in underlying mechanism pertinent to obesity and associated metabolic disorders have started to emerge in recent years. However, the clinical relevance and functional roles of these molecules in the context of obesity are still poorly understood. 22
Among the long list of genes involved in metabolic disorders, MEG3 and H19 are notorious for their participation in several diseases, where the alteration in their expression is coupled with the metabolic homeostasis disturbance. To unravel the role of lncRNAs in the pathogenesis of obesity, we for the first time investigate the gene expression of MEG3 and H19 concerning obesity‐associated metabolic parameters in VAT and SAT from obese and non‐obese women.
In the present study, we found that unlike MEG3, which disclosed an elevating expression pattern, the expression level of H19 was lower in the obese group as compared with the non‐obese counterparts. This association was further substantiated by correlation analysis, as revealed that while transcript level of H19 had an inverse correlation with BMI, WC, hip circumference, and HOMA‐IR values. However, MEG3 expression displayed a positive correlation with all the indicated parameters in all participants. Of great interest, the involvement of lncRNAs: MEG3 and H19 has been reported in different metabolic disorders performed in animal models and in vitro surveys albeit with conflicting results. For instance, the down‐regulation of MEG3 was inversely associated with lipogenesis‐related genes expression, 23 while its over‐expression alleviates lipid over‐deposition in vitro and in vivo models of NAFLD. lncRNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modification 14 ; indicating this lncRNA can be regarded as a potential therapeutic target for bone marrow adiposity. The up‐regulation of lncRNA MEG3 in high‐fat diet and ob/ob mice, induction of insulin resistance following over‐expression of lncRNA MEG3, and protection against dietary obesity after H19 over‐expression in brown fat provide other information regarding the involvement of these lncRNAs in the etiology of obesity. 24 , 25
While in several studies up‐regulation of MEG3 has been introduced to be a mechanism through which both adipogenesis and lipogenesis could be halted in different metabolic syndromes, 11 , 26 , 27 others reported a positive relationship between the expression level of this gene and obesity development.
A similar conflict has been related to H19 and its association with metabolic disorders. Zhang et al. have reported that the expression level of H19 is up‐regulated in hyperglycemic mice, introducing H19 long non‐coding RNA as a molecule responsible for the progression of diabetes. 28 Likewise, it has been declared that over‐expressed H19 could induce adipogenesis and inflammatory response in Raw264.7 cells through down‐regulating miR‐130b. 29 On contrarily, it has been suggested that H19 expression declined with ascending BMIs in obese patients, suggesting that probably the expression of this gene may prevent the evolution of dietary obesity through suppressing the expression of monoallelic genes in brown fat. 30 It is worth mentioning that the majority of the literature is performed on the animal models and it can be considered as one possible reason for these discordant findings.
Interestingly, a positive correlation was found between transcript level of MEG3 in SAT with FAS and PPARγ. However, there was an inverse correlation between SAT expression of H19 and FAS. In a mechanistic view, MEG3 and H19 can affect the expression level of PPARγ. Huang et al. have shown that the suppression of H19 expression was coupled with the induction of adipogenesis, as revealed by the up‐regulation of PPARγ, C/EBPα, and FABP4. 31 Moreover, in other studies, it has been claimed that MEG3 could regulate apoptosis in diverse types of cells by increasing the expression level of PPARγ. 32 , 33 Our results also showed that the expression level of both acetyl‐CoA carboxylase (ACC) and fatty acid synthesis (FAS), two critical molecules required for fatty acid biogenesis were elevated to a greater extent in the obese patients as compared to the control group, suggesting that presumably, the alteration of MEG3 and H19 expression in adipocyte tissues may provide signaling which increases the fatty biogenesis through promoting ACC and FAS expression.
Although the current study cannot determine the exact mechanism by which alteration in the pattern of these lncRNAs is linked to the pathogenesis of obesity, a possible candidate would be PPARγ, a key nuclear receptor that engages in unique cross talk with ACC and fatty acid trafficking. However, more experiment with a mechanistic view is warranted to establish. It should be noted that we failed to find any correlation between the expression of these genes and lncRNA MEG3/H19 in VAT, suggesting that probably the expression of these genes might be controlled by other regulators. Differences in gene expression between subcutaneous and visceral human adipose tissue have been reported previously. 34 , 35 VAT is associated with insulin resistance, diabetes, hypertension, atherosclerosis, and hepatic steatosis, while, SAT responds better to insulin and secretes more adiponectin and less inflammatory cytokines. 36 These differences may be due to this notion that subcutaneous and visceral adipocytes have arisen from different progenitor cells that exhibit different gene expression patterns.
Taken together, it seems that our findings along with other studies can unravel the possible role of lncRNAs MEG3 and H19 in the pathomechanism of obesity and related metabolic abnormalities. However, some limitations need to be considered. Firstly, the cross‐sectional design of this study limits the extrapolation of the results. Secondly, there is evidence regarding the possible effect of gender on the pattern of epigenetic signatures. Hence, more studies with a higher sample size are needed to evaluate the lncRNAs in both obese women and men. Finally, mechanistic studies should be conducted to unravel the exact role of lncRNAs in the pathogenesis of obesity. Moreover, we found that the alteration in the expression of genes, such as FAS, and PPARγ was statistically meaningful in the SAT, but not in VAT.
5. CONCLUSION
Here, we showed that while the expression level of MEG3 was higher in obese women than the normal‐weight group, the expression level of H19 disclosed a down‐regulation pattern. Moreover, correlation analysis indicated that the transcript level of H19 had an inverse correlation with obesity indices and HOMA‐IR values. However, MEG3 expression displayed a positive correlation with all the indicated parameters in all participants. Interestingly, a positive correlation was found between transcript level of MEG3 in SAT with FAS and PPARγ. However, there was an inverse correlation between SAT expression of H19 and FAS. Hence, it appears that lincRNAs; MEG3 and H19 have an important role in obesity‐related conditions. However, more clinical studies are still required to clarify the relationships between lncRNAs with obesity and related abnormalities.
CONFLICT OF INTEREST
We have no conflict of interest to declare.
Supporting information
Table S1
ACKNOWLEDGMENT
This study was supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant number: 16189).
Solaleh Emamgholipour and Mehrnoosh shanaki contributed equally to this study.
Contributor Information
Solaleh Emamgholipour, Email: shanaki_m@sbmu.ac.ir.
Mehrnoosh shanaki, Email: semamgholipour@sina.tums.ac.ir.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author.
REFERENCES
- 1. Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Investig. 2017;127(1):1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim SY. The definition of obesity. Korean J Fam Med. 2016;37(6):309‐309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fruh SM. Obesity: risk factors, complications, and strategies for sustainable long‐term weight management. J Am Assoc Nurse Pract. 2017;29(S1):S3‐S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jain M, Singh C, Agarwal P. Association of food patterns, central obesity measures and metabolic risk factors for coronary heart disease in adult men. Indian J Public Health Res Develop. 2018;9(9):71‐76. [Google Scholar]
- 5. Despres J. Intra‐abdominal obesity: an untreated risk factor for Type 2 diabetes and cardiovascular disease. J Endocrinol Invest. 2006;29(3):77. [PubMed] [Google Scholar]
- 6. Ellis K, et al. Abdominal Subcutaneous Adipose Tissue (SAT) and Visceral Adipose Tissue (VAT) Measurements in HIV+ Adults: Influences of Measurement Site. In Antiviral Therapy. London EC3R 5DD, England: Int Medical Press Ltd 2–4 Idol Lane; 2004. [Google Scholar]
- 7. Lee M‐J, Wu Y, Fried SK. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol Aspects Med. 2013;34(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev. 2010;11(1):11‐18. [DOI] [PubMed] [Google Scholar]
- 9. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354‐361.21550244 [Google Scholar]
- 10. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45‐R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang P, Huang FZ, Liu HZ, Zhang TY, Yang MS, Sun C. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR‐21 with LRP6. Metabolism. 2019;94:1‐8. [DOI] [PubMed] [Google Scholar]
- 12. Schmidt E, Dhaouadi I, Gaziano I, et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat Commun. 2018;9(1):3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matouk IJ, DeGroot N, Mezan S, et al. The H19 non‐coding RNA is essential for human tumor growth. PLoS ONE. 2007;2(9):e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non‐coding RNA H19 Inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep. 2016;6(1):28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017;65(2):604‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu C, Yang Z, Wu J, et al. Long noncoding RNA H19 interacts with polypyrimidine tract‐binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67(5):1768‐1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moldes M, Boizard M, Liepvre XLE, Fève B, Dugail I, Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element‐binding protein‐1c (ADD1/SREBP‐1c) trans‐factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem J. 1999;344(3):873‐880. [PMC free article] [PubMed] [Google Scholar]
- 18. Feng J, Xu H, Pan F, et al. An integrated analysis of mRNA and lncRNA expression profiles indicates their potential contribution to brown fat dysfunction with aging. Front Endocrinol. 2020;11(46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Li K, Zhang X, Chen J, Li M, Liu L. The novel long noncoding RNA lncRNA‐Adi regulates adipogenesis. Stem Cells Transl Med. 2020;9(9):1053‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao L, Ma Z, Guo Z, Zheng M, Li K, Yang X,. Analysis of long non‐coding RNA and mRNA profiles in epicardial adipose tissue of patients with atrial fibrillation. Biomed Pharmacother. 2020;121:109634. [DOI] [PubMed] [Google Scholar]
- 21. Ruan Y, Lin N, Ma Q, et al. Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet β‐cell function. Cell Physiol Biochem. 2018;46(1):335‐350. [DOI] [PubMed] [Google Scholar]
- 22. Ebrahimi R, Toolabi K, Ali Pour NJ, et al. Adipose tissue gene expression of long non‐coding RNAs; MALAT1, TUG1 in obesity: is it associated with metabolic profile and lipid homeostasis‐related genes expression? Diabetol Metab Syndr. 2020;12:36‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng L, Jin C, Chen S, et al. Long non‐coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose‐derived mesenchymal stem cells via miR‐140‐5p. Mol Cell Biochem. 2017;433. [DOI] [PubMed] [Google Scholar]
- 24. Schmidt E, Dhaouadi I, Gaziano I, et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. 2018. [DOI] [PMC free article] [PubMed]
- 25. Zhang Y, Fu Y, Zheng Y, Wen Z, Wang C. Identification of differentially expressed mRNA and the Hub mRNAs modulated by lncRNA Meg3 as a competing endogenous RNA in brown adipose tissue of mice on a high‐fat diet. Adipocyte. 2020;9(1):346‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z, Jin C, Chen S, et al. Long non‐coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose‐derived mesenchymal stem cells via miR‐140‐5p. Mol Cell Biochem. 2017;433(1–2):51‐60. [DOI] [PubMed] [Google Scholar]
- 27. Liu H, Wang QY, Zhang Y, et al. Pioglitazone up‐regulates long non‐coding RNA MEG3 to protect endothelial progenitor cells via increasing HDAC7 expression in metabolic syndrome. Biomed Pharmacother. 2016;78:101‐109. [DOI] [PubMed] [Google Scholar]
- 28. Zhang N, Geng T, Wang Z, et al. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI insight. 2018;3(10):e120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han Y, Ma J, Wang J, Wang L. Silencing of H19 inhibits the adipogenesis and inflammation response in ox‐LDL‐treated Raw264. 7 cells by up‐regulating miR‐130b. Mol Immunol. 2018;93:107‐114. [DOI] [PubMed] [Google Scholar]
- 30. Schmidt E, Dhauuadi I, Gaziano I, et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat Commun. 2018;9(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non‐coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep. 2016;6:28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu H, Zhu H, Zhuang Y, et al. LncRNA ACART protects cardiomyocytes from apoptosis by activating PPAR‐γ/Bcl‐2 pathway. J Cell Mol Med. 2020;24(1):737‐746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Gao Y. Long non‐coding RNA MEG3 inhibits cervical cancer cell growth by promoting degradation of P‐STAT3 protein via ubiquitination. Cancer Cell Int. 2019;19(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lefebvre AM, Laville M, Vega N, et al. Depot‐specific differences in adipose tissue gene expression in lean and obese subjects. Diabetes. 1998;47(1):98‐103. [DOI] [PubMed] [Google Scholar]
- 35. Modesitt SC, Hsu JY, Chowbina SR, Lawrence RT, Hoehn KL. Not all fat is equal: differential gene expression and potential therapeutic targets in subcutaneous adipose, visceral adipose, and endometrium of obese women with and without endometrial cancer. Int J Gynecol Cancer. 2012;22(5):732‐741. [DOI] [PubMed] [Google Scholar]
- 36. Gil A, Olza J, Gil‐Campos M, Gomez‐Llorente C, Aguilera CM. Is adipose tissue metabolically different at different sites? Int J Pediatr Obes. 2011;6(Suppl 1):13‐20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author.


