Abstract
Background
SARS‐CoV‐2 has become a global pandemic due to its capacity for rapid transmission. In this context, an early and rapid diagnosis of infected patients that do not require expensive equipment or highly trained personnel is crucial in order to reduce the contagious rate. The aim of this study was to evaluate a chromatographic immunoassay's performance for the rapid diagnosis of SARS‐CoV‐antigen.
Methods
A cross‐sectional study included 369 adults from Western México with diagnosis or suspicion of SARS‐CoV‐2 infection. Two samples were collected; a naso‐oropharyngeal was used for a molecular determination of SARS‐CoV‐2 RNA. The molecular analysis was carried out using DeCoV19 Kit Triplex (Genes2life S.A.P.I.) based on the CDC diagnostic panel for N1, N2, and N3 regions. The second sample was retrieved from a nasopharyngeal rub and used for the rapid diagnosis of SARS‐CoV‐2 antigen employing the commercial STANDARD™ Q COVID‐19 Ag Test (SD BIOSENSOR).
Results
Overall, in 28.2% of the patients was detected the SARS‐CoV‐2 RNA, and 21.4% were positive for antigen detection. The rapid antigen test showed a sensitivity and specificity of 75.9% and 100%, respectively, with a positive predictive and negative values of 100% and 91%. Symptoms as anosmia presented a high OR for the positive diagnosis for both test, reverse transcription‐polymerase chain reaction (RT‐PCR), and the rapid antigen test of 8.86 (CI = 4.91–16) and 6.09 (CI = 3.42–10.85), respectively.
Conclusion
SD BIOSENSOR is a useful assay, but some caveats must be considered before the general implementation.
Keywords: COVID‐19, rapid test, SARS‐CoV‐2
This cross‐sectional study included a population from western Mexico with a diagnosis or suspicion of SARS‐CoV‐2 infection. An antigen rapid‐test performance was evaluated against Reverse Transcription Polymerase Chain Reaction (RT‐PCR), resulting in a sensitivity of 75.9% and a sensitivity of 100%. Symptoms as anosmia, cough, and fatigue were highly related to positivity with both diagnosis‐methods. Antigen rapid‐test resulted in a useful essay for SARS‐CoV‐2 diagnosis.

1. INTRODUCTION
In December 2019, adults from Wuhan, China, began presenting at local hospitals with severe pneumonia of unknown etiology. On January 7, 2020, the disease‐causing agent was identified as a novel coronavirus that shared >95% homology with the bat coronavirus and >70% similarity to SARS‐CoV. This novel coronavirus was denominated Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2), and the underlying disease COVID‐19 (Coronavirus Disease 2019). 1 , 2 Despite multiple attempts to contain the disease in China, COVID‐19 has been spreading worldwide, with over 97,425,832 confirmed cases and 2,087,820 deaths on January 21. 3
The virus is highly transmissible in humans; it primarily spreads through the respiratory tract by droplets, respiratory secretions, and direct contact. Other transmission routes, such as feces and blood, have been proposed; however, this has not been completely elucidated. 4 , 5
The presentation of symptoms varies among the infected subjects; current data have shown that while cases may be asymptomatic (15.6–80%), other infected patients with SARS‐CoV‐2 present mild flu‐like symptoms and few patients develop critical conditions (acute respiratory distress syndrome, ARDS). The fatality rate increases with the severity of the illness, reaching up to 49% in critical patients. Therefore, early and accurate diagnosis is a current challenge for public health systems. 2
The RT‐PCR test is the gold standard for SARS‐CoV‐2 detection; however, it needs specialized staff and equipment, and the timeliness of the results is usually affected by the logistics of the test and the communication of the results, and the continuous increase in the demand. Consequently, the development and application of different diagnostic strategies, such as rapid tests, is an urgent need. SARS‐CoV‐2‐antigen detection using a rapid chromatographic immunoassay brings a perfect opportunity to reduce cost, time, and personnel in the SARS‐CoV2 diagnosis. However, their performance, mainly their specificity and sensitivity, are still on trial. In this matter, the validation of a SARS‐CoV‐2 antigen assay (STANDARD™ Q COVID‐19, SD BIOSENSOR) is of great importance and relevance for public health strategies that aid in earlier detection and isolation of confirmed cases.
2. METHODS
2.1. Study population and study design
We performed a cross‐sectional study where the study population consisted of 369 adult people visiting the Laboratorio de Diagnóstico de Enfermedades Emergentes y Reemergentes (LaDEER) in the Universidad de Guadalajara, Jalisco from October to November 2020 (Ethical committee approval number: CI 06220). The patients received for the antigen determination manifested symptoms such as headache, fever, fatigue, other respiratory signs, or gastrointestinal symptoms that suggested COVID‐19. This information (the severity of the disease, age, and the onset of symptoms) was retrieved during the visit. Moreover, people diagnosed by RT‐PCR for infection of SARS‐CoV‐2 and individuals in contact with those in the last 3–5 days, with or without symptoms, were included. Exclusion criteria for this research were applied when a sample was unavailable, degraded, or insufficient. All subjects participated voluntarily and signed an informed consent.
2.2. Sample collection
Two nasopharyngeal swab specimens were collected from the population by trained staff. A sterile nasopharyngeal specimen collection swab (NEST Scientific, USA) was introduced in one nostril until resistance was felt at the nasopharynx, rotated 180°, and then withdrawn. A sterile oropharyngeal specimen collection swab was introduced in the mouth, and until reaching the oropharynx, then the swab was rub on the posterior wall for 10–15 seconds. 6 Both swab applicators were placed into 3 ml of viral transport media. The samples for RT‐PCR were immediately shipped to the laboratory for SARS‐CoV‐2 RNA determination, while SARS‐CoV‐2 antigen analysis was carried out in the place.
2.3. Test methods for diagnosis challenge
2.3.1. RT‐PCR for SARS‐CoV‐2 diagnosis
For the molecular analysis, samples were processed in a BSL‐2 laboratory. First, for viral inactivation; the sample in the medium was stirred for 20 minutes, then 140 μl was transferred to a tube with 560 μl of AVL buffer (Qiagen). RNA extraction was performed with the QIAamp Viral RNA Mini Kit (Qiagen). For the RT‐PCR assay, the samples were examined using DeCoV19 Kit Triplex (Genes2life S.A.P.I de C.V., Mexico), which is based on the CDC diagnostic panel for SARS‐CoV2 detection. It is commercialized in México and has received the Instituto de Diagnóstico y Referencia Epidemiológicos (InDRE) emergency use authorization as well as the approval from the Comisión Federal para la Protección contra Riesgos Sanitarios (COFEPRIS). The assay target genes are regions of the virus nucleocapsid (N), it also includes a primer mix to detect the human RNase P gene (RP). Samples with Ct‐values below 35 with an exponential growth curve of 2 or more genes were considered as positive.
2.3.2. SARS‐CoV‐2 antigen determination
Another nasopharyngeal swab specimen was collected with the diagnosis test kit's material following the same procedure before mentioned. Afterward, the swab was placed in the extraction buffer supplemented in the kit and gently rotated at least ten times to resuspend the sample. Three drops from this medium were deposited in the cassette well STANDARD™ Q COVID‐19 Ag Test (SD BIOSENSOR), which detects the SARS‐CoV‐2 C‐terminal‐nucleocapsid (N) antigen in respiratory specimens, and read in 15 minutes. The test was considered positive when marks were observed in the control and test positions. Negative results were interpreted when only a mark in the control position was observed. The test was invalidated when no marks were detected.
2.4. Statistical analysis
The quantitative (continuous) variables were summarized using mean ± standard deviation or as median and percentiles 25 and 75 according to the data distribution, evaluated by a Kolmogorov‐Smirnov test. Frequencies and percentages were used for the description of the qualitative (categorical) variables. Furthermore, the specificity and sensitivity were calculated according to the following formulas: Specificity (%) = 100 × [Negative/(Negative + Positive)] and Sensitivity (%) = 100 × [Positive/(Positive + Negative)], respectively, based on the final diagnosis. Additionally, positive/negative predictive values and positive/negative plausibility values were estimated. All analyses were performed using the software IBM SPSS statistics, version 25 for Windows (IBM Corp, Inc.).
3. RESULTS
3.1. Demographic and clinical data
As shown in Table 1, samples consisted mostly of female population, with an age average of 36.6 ± 13.16. From the symptoms registered in our study, headache, fever, and cough were the most common among patients. In contrast, symptoms such as arthralgia, breath shortness, and diarrhea were less frequent in the study population. In 28.2% of the overall patients, the viral genome was detected by RT‐PCR, indicating an active infection with SARS‐CoV‐2; likewise, 21.4% resulted positive for the rapid test viral‐antigen diagnosis.
TABLE 1.
Demographic and clinical characteristics of patients
| N (%) | ||
|---|---|---|
| Age (mean/SD) | — | 36.6 ± 13.13 |
| Gender (M/F) | — | 154/215, (41.7/58.3) |
| Clinical features | Fever | 92, (24.9) |
| Anosmia | 66, (17.9) | |
| Breath shortness | 25, (6.8) | |
| Myalgia | 79, (21.4) | |
| Cough | 86, (23.3) | |
| Fatigue | 60, (16.3) | |
| Diarrhea | 38, (10.3) | |
| Arthralgia | 16, (4.3) | |
| Headache | 154, (41.7) | |
| RT‐PCR Positive | — | 104, (28.2) |
| Ag‐Positive | — | 79, (21.4) |
Abbreviations: Ag, Antigen; F, Female; M, Male.
3.2. Antigen test, sensitivity, specificity, and accuracy
Based on the results obtained previously, 21.4% of the patients were classified as true‐positive, and 71.82% were true‐negative. Also, there were no patients with false‐positive results by the rapid antigen test, and only 6.8% showed a false‐negative result (Table 2). The rapid test performance analysis is summarized in Table 3; the antigen test showed a sensitivity performance of 75.9% and a specificity of 100%; similarly, this test exhibited a positive predictive value of 100% and a negative predictive value of 91%.
TABLE 2.
Performance comparison between antigen test and RT‐PCR for SARS‐CoV‐2 diagnosis
| Gold standard (RT‐PCR) | |||
|---|---|---|---|
| Positive | Negative | Total | |
| Rapid diagnostic antigen test (Standard Q COVID−19 Ag) | |||
| Positive | TP = 79 | FP = 0 | 79 |
| Negative | FN = 25 | TN = 265 | 290 |
| Total | 104 | 265 | 369 |
Abbreviations: FN, False‐negative; FP, False‐positive; TN, True‐negative; TP, True‐positive.
TABLE 3.
Performance comparison of RT‐PCR and rapid antigen test
| Estimated values (%) | C.I. (%) | |
|---|---|---|
| Sensitivity | 75.9 | 66.5–83.8 |
| Specificity | 100 | 98.6–100 |
| Positive Predictive Value | 100 | NA |
| Negative Predictive Value | 91 | 88.2–93.7 |
When the antigen test results were evaluated according to the Ct‐values, we observed significant differences between positives and negatives (Figure 1). In specimens with a high viral load (Ct <25), the sensitivity increased to 88%. Nevertheless, 10.30% of our population had a medium‐low viral load (Ct >25), affecting significantly (70.3%) of the assay. Ct‐values from those patients positive to the antigen test presented a Ct‐mean of 23.74 ± 4.34, 23.59 ± 4.37, and 22.03 ± 4.26 in N1, N2, and N3 regions, respectively. By contrast, patients with negative results to the antigen test exhibited Ct‐values in 27.91 ± 4.50, 28.02 ± 4.43, and 26.43 ± 4.26 in N1, N2, and N3, respectively, showing significant differences with p‐values <0.001.
FIGURE 1.
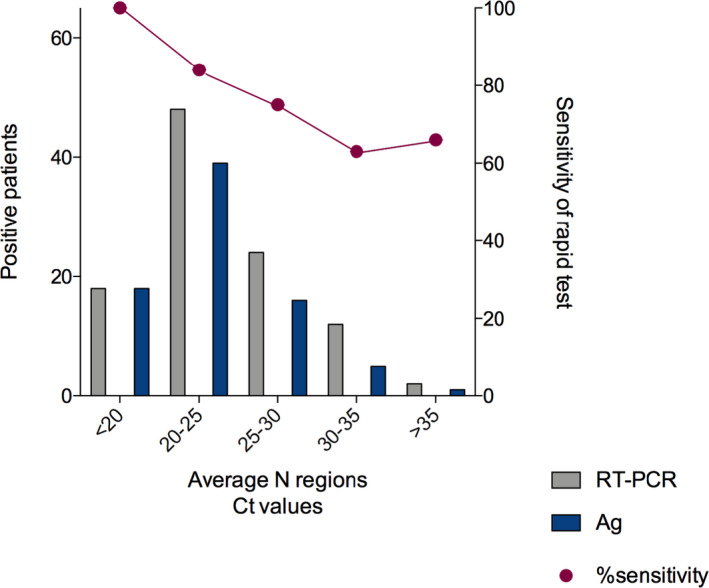
Ct‐values are directly related to the antigen test results. The Ct‐values from positive patients to SARS‐CoV‐2 diagnosed by reverse transcription‐polymerase chain reaction (RT‐PCR) were significantly higher than the negatives. Moreover, we observed that the sensitivity decreased
3.3. Anosmia is related to SARS‐CoV‐2 infection
Analysis of the symptomatology reported by the study population demonstrated that anosmia was the most frequent in the individual's diagnosed positive by both tests, showing an OR of 8.86 (CI = 4.91–16) and 6.09 (CI = 3.42–10.85) for RT‐PCR and antigen rapid test, respectively, followed by fatigue and cough. By contrast, symptoms such as diarrhea and arthralgia were not associated with the positive viral diagnosis (Figure 2).
FIGURE 2.
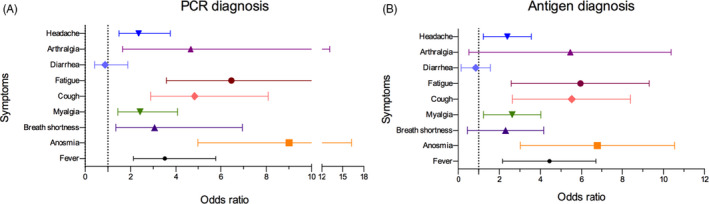
Symptoms with risk factors associated with the SARS‐CoV‐2 positive diagnosis. (A) Represents the symptoms risk related to polymerase chain reaction (PCR) and (B) to the rapid antigen test. Both diagnostic tests showed a better performance when anosmia symptoms were present. Diarrhea was the least relevant symptom
4. DISCUSSION
The current COVID‐19 pandemic has proven the relevance of an early and rapid viral diagnosis in order to reduce the SARS‐CoV‐2 infection rate. New tests that improve the results timelines have been introduced in the general market; nonetheless, their validity and reliability must be evaluated thoroughly.
STANDARD™ Q COVID‐19 Ag Test (SD BIOSENSOR) is a novel diagnostic tool based on chromatographic immunoassay that detects specific viral antigens from SARS‐CoV‐2 recovered from nasopharynx samples. Its use and interpretation are simple, the use of a safety cabinet or highly trained personnel are not needed, and it renders results within 30 minutes, collectively improving the procedure costs and helping in the resolution of the public health problem.
In the study population, we observed a tolerable sensitivity (75.9%) and very high specificity (100%); both results are in line with previous reports, which described sensitivity of 70.6% and specificity of 100% 7 and 70.7%, and 96%, respectively. 8 Meanwhile, other studies in a different population showed a higher sensitivity using antigen‐based detection test (98.33%); nevertheless, this study used clinical specimens mixed in viral transport media, whence a pre‐treatment was performed to the sample previous to the antigen analysis, 9 probably increasing the efficiency of the procedure, but also the cost and the need for highly trained personnel. These values indicate the effectiveness of a test compared with the gold standard (RT‐PCR). This test approaches the acceptable limit based on the Target Product Profiles 10 and reaches the qualification of a useful test by the estimation of sensitivity + specificity (1.75 value), according to Power, et al., 2020. 11 Therefore, the SD BIOSENSOR Ag has demonstrated to be a useful diagnostic test for SARS‐CoV‐2.
Similar to previous studies, 7 , 8 the antigen detection assay's sensitivity and specificity demonstrated to be highly Ct‐dependent, decreasing according to the viral load. Nonetheless, a positive molecular test may not imply transmissible live virus 12 as it has been demonstrated that low viral load samples rarely allow culturing of the virus. 8 , 13 , 14 Thus, the test may compromise the diagnosis of people during the early or late phase of the infection, and clinical data must be taken into consideration as well as a second test for confirmation, especially when we consider that in the clinical field, disease‐prevalence might influence in the diagnostic‐test performance. 10 In this context, a significant highlight is the differences in sensitivity and specificity of 93.12% and 100%, respectively, published by manufacturers against our results. This marked difference can be due to our samples' clinical nature, which include a wide range of viral load, showing low, medium, and high Ct´s values, in contrast with those reported by manufacturers, including limited samples with low viral load.
Even though the measures mentioned above are of great interest, other values such as Positive Predictive Value (PPV) and Negative Predictive Value (NPV) must be integrated to evaluate a test thoroughly. Both values strongly depend on the prevalence of the studied population, which in our study was 28.2%. Therefore, the PPV (100%) and the NPV (91%) that we obtained are classified as high and acceptable, respectively. 10
Moreover, symptoms with their respective risk factor were associated with a SARS‐CoV‐2 positive outcome either by PCR or the Ag test. Despite limited Mexican clinical reports, we found that anosmia was the most related symptom to both diagnosis techniques; it was followed by fatigue, cough, and fever. On the other hand, the symptoms with lower risk in both tests (diarrhea, breath shortness, myalgia, and headache) reflect that our studied population was not severely affected by the disease, which may be in part due to the mean reported age (36.6 ± 13.13).
Overall, these results reveal that the SD Biosensor Ag test is useful under particular scrutiny. First, an optimal sample is of preeminence, as specimen quality and collection method directly impact the test results. 15 , 16 Here, a combination of NP and OP swabs was used for molecular detection, while only NP was employed for the Ag detection; thus, affecting the assay's sensitivity. Second, the viral load influences the result; low Ct‐values render the nadir sensitivity percentage values; hence, a retest may be suggested for confirmation. Lastly, symptomatology may be related to the risk factor of a positive diagnosis, linking clinical information with the test interpretation and follow‐up.
In conclusion, there is an urgent need for rapid diagnosis so that the transmission burden is dampened. Several challenges must be overcome in the timely detection of SARS‐CoV‐2. The SD BIOSENSOR is a useful assay for this; nonetheless, important caveats must be considered before the community implementation of this diagnostic test.
CONFLICT OF INTEREST
Bustillo‐Armendaríz Gustavo and García‐Cedillo Fernanda declared that the rapid antigen test was supplied by P.M.I 12.10, S.A.P.I. de C.V company.
ACKNOWLEDGEMENTS
We appreciate all the personnel from LaDEER who are fighting the COVID‐19 epidemic. We also recognize Diaz‐Bribiesca Carlos A. for the collaboration on population database management.
[Correction added on 22 March 2021, after first online publication: The first author's last name has been corrected from, "Peña‐Rodrígez" to "Peña‐Rodríguez" and the eighth author's last name has been corrected from "Bustillo‐Armendaríz" to "Bustillo‐Armendáriz".]
DATA AVAILABILITY STATEMENT
All the data related to this work are available at the corresponding author. The data used to support the findings of this study are included in the article.
REFERENCES
- 1. Zhou P, Yang X‐L, Wang X‐G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johns Hopkins University . COVID‐19 Map. Johns Hopkins Coronavirus Resource Center. [cited November 30, 2020]. Available in: https://coronavirus.jhu.edu/map.html
- 4. Kutti‐Sridharan G, Vegunta R, Vegunta R, Mohan BP, Rokkam VRP. SARS‐CoV2 in different body fluids, risks of transmission, and preventing COVID‐19: a comprehensive evidence‐based review. Int J Prev Med. 2020;11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meselson M. Droplets and aerosols in the transmission of SARS‐CoV‐2. N Engl J Med. 2020;382(21):2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coden E, Russo F, Arosio AD, Castelnuovo P, Karligkiotis A, Volpi L. Optimum naso‐oropharyngeal swab procedure for COVID‐19: step‐by‐step preparation and technical hints. Laryngoscope. 2020;130(11):2564‐2567. [DOI] [PubMed] [Google Scholar]
- 7. Krüttgen A, Cornelissen CG, Dreher M, Hornef MW, Imöhl M, Kleines M. Comparison of the SARS‐CoV‐2 rapid antigen test to the real star Sars‐CoV‐2 RT PCR kit. J Virol Methods. 2020;288:e114024. [Google Scholar]
- 8. Cerutti F, Burdino E, Milia MG, et al. Urgent need of rapid tests for SARS CoV‐2 antigen detection: evaluation of the SD‐Biosensor antigen test for SARS‐CoV‐2. J Clin Virol. 2020;132:e104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chutikarn C, Bualan K, Nattaya T, Niracha A, Rujipas S, Methee C. Rapid SARS‐CoV‐2 antigen detection assay in comparison with real‐time RT‐PCR assay for laboratory diagnosis of COVID‐19 in Thailand. Virol J. 2020;17(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. WHO . COVID‐19 target product profiles for priority diagnostics to support response to the COVID‐19 pandemic v.1.0. WHO. [cited December 1, 2020]. Available in: https://www.who.int/publications/m/item/covid‐19‐target‐product‐profiles‐for‐priority‐diagnostics‐to‐support‐response‐to‐the‐covid‐19‐pandemic‐v.0.1
- 11. Power M, Fell G, Wright M. Principles for high‐quality, high‐value testing. Evid Based Med. 2013;18(1):5‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walsh KA, Jordan K, Clyne B, et al. SARS‐CoV‐2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinez RM. Clinical samples for SARS‐CoV‐2 detection: review of the early literature. Clin Microbiol Newsl. 2020;42(15):121‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LeBlanc JJ, Heinstein C, MacDonald J, Pettipas J, Hatchette TF, Patriquin G. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS‐CoV‐2. J Clin Virol. 2020;128:e104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data related to this work are available at the corresponding author. The data used to support the findings of this study are included in the article.


