Abstract
Candida auris is an emerging fungal pathogen of rising concern due to global spread, the ability to cause healthcare-associated outbreaks, and antifungal resistance. Genomic analyses revealed that early contemporaneously detected cases of C. auris were geographically stratified into four major clades. While Clades I, III, and IV are responsible for ongoing outbreaks of invasive and multidrug-resistant infections, Clade II, also termed the East Asian clade, consists primarily of cases of ear infection, is often susceptible to all antifungal drugs, and has not been associated with outbreaks. Here, we generate chromosome-level assemblies of twelve isolates representing the phylogenetic breadth of these four clades and the only isolate described to date from Clade V. This Clade V genome is highly syntenic with those of Clades I, III, and IV, although the sequence is highly divergent from the other clades. Clade II genomes appear highly rearranged, with translocations occurring near GC-poor regions, and large subtelomeric deletions in most chromosomes, resulting in a substantially different karyotype. Rearrangements and deletion lengths vary across Clade II isolates, including two from a single patient, supporting ongoing genome instability. Deleted subtelomeric regions are enriched in Hyr/Iff-like cell-surface proteins, novel candidate cell wall proteins, and an ALS-like adhesin. Cell wall proteins from these families and other drug-related genes show clade-specific signatures of selection in Clades I, III, and IV. Subtelomeric dynamics and the conservation of cell surface proteins in the clades responsible for global outbreaks causing invasive infections suggest an explanation for the different phenotypes observed between clades.
Keywords: Candida auris, fungal genome, chromosome rearrangement, karyotype variation, subtelomeric variation, cell wall proteins, selection
Introduction
The emerging fungal pathogen Candida auris has been reported in over 40 countries to date and has become a leading cause of invasive candidiasis in some hospitals, often in severely ill patients (van Schalkwyk et al. 2019). C. auris isolates are commonly resistant to one or more antifungal drugs and can survive for long periods both in the clinical environment and as a commensal on skin (Welsh et al. 2017). Initially identified in cases of ear infection in Japan and South Korea (Kim et al. 2009; Satoh et al. 2009), cases of systemic infection were soon after reported in India, South Africa, and Venezuela (Chowdhary et al. 2013; Magobo et al. 2014; Calvo et al. 2016). Initial genomic analysis of the global emergence identified four major genetic groups corresponding to these geographic regions or Clades I, II, III, and IV (Lockhart et al. 2017). Subsequent genomic analysis identified a single genetically diverse isolate from Iran representing a potential fifth clade (Chow et al. 2019). Clades I, III, and IV are responsible for the ongoing multidrug-resistant and difficult to control outbreaks in healthcare facilities worldwide (Chow et al. 2018, 2020). Clade II, also termed the East Asia clade, is predominantly associated with cases of ear infection and appears to be less resistant to antifungals than other clades (Welsh et al. 2019), though cases of Clade II infections have been reported on different continents (Chow et al. 2020).
Comparisons of global isolates have highlighted clade-specific variation in the levels of drug resistance and the mechanisms that contribute to resistance. While Clade II isolates are typically susceptible to azoles and other major antifungals, nearly all isolates in Clades I and III and roughly half of isolates in Clade IV are resistant to azoles. These resistant isolates contain one of three mutations (Y132F, K143R, and F126L) in the drug target lanosterol 14-α-demethylase (ERG11) (Lockhart et al. 2017; Chowdhary et al. 2018; Chow et al. 2020). Clades I and IV isolates also have mutations contributing to fluconazole resistance in the transcriptional factor TAC1b that regulates the efflux pump CDR1 (Rybak et al. 2020) and expression of CDR1 has been shown to be differentially regulated between isolates from Clades III and IV (Mayr et al. 2020). An average of seven percent of the isolates from Clades I, III, and IV are resistant to echinocandins and resistance has been associated to a single mutation at S639 (S639Y/F/P) in the hotspot1 of the drug target 1,3-beta-D-glucan synthase (FKS1) (Chowdhary et al. 2018; Chow et al. 2020). Although low susceptibility to amphotericin B is common in Clades I and IV, amphotericin B drug-resistant mutations have not been identified and linked to these clades. Together, this highlights interclade variation in drug resistance mechanisms.
Other than clade-specific drug resistance and correlated mutational profiles, little is known about the mechanisms responsible of the phenotypic differences between clades and the increased ability of C. auris to be transmitted and persist in healthcare settings. While genome comparisons with the well-studied species C. albicans suggest conservation of mechanisms of biofilm formation and host interaction, in addition to drug resistance, little is known about how variation within C. auris contributes to phenotypic differences other than drug resistance. Clade II isolates have primarily been isolated from cases of ear infection whereas Clades I, III and IV isolates are primarily linked to invasive infections (Welsh et al. 2019). In addition, Clade II isolates have increased susceptibility to disinfectants (Sexton et al. 2020), ultraviolet C light disinfection (Chatterjee et al. 2020), are more sensitive to environmental stressors (Heaney et al. 2020), and induce a lower cytokine production and have simpler mannans relative to Clades I, III, and IV (Bruno et al. 2020). Most of the clade-specific differences described thus far have been based on SNP-based analyses, and this reference-based analysis has not typically analyzed differences in genome structure and stability between C. auris isolates. We hypothesized that larger genomic differences could also be linked to phenotypic variation. In other haploid and asexual Candida species such as C. glabrata, chromosomal rearrangements are associated with antifungal drug resistance and host adaptation (Poláková et al. 2009), including differences in the repertoire of cell wall proteins (Carreté et al. 2018).
Previous studies have provided evidence of karyotypic variation in C. auris and of gene content differences between clades (Muñoz et al. 2018; Bravo Ruiz et al. 2019). Whole-genome alignment of an assembly of a Clade I isolate with that of a Clade III isolate identified an inversion and two translocations (Muñoz et al. 2018), and analysis of chromosomal sizes across a wider set of isolates suggested other major rearrangements between clades (Bravo Ruiz et al. 2019). Evidence for gene content variation includes the presence of a gene cluster involved in L-rhamnose utilization only in isolates of Clade III, which correlates with the ability of Clade III isolates to grow on L-rhamnose (Ambaraghassi et al. 2019). Here, to better understand the genomic variation between the clades linked to phenotypic differences, we leverage complete reference genomes for isolates from Clades I, II, III, and IV to better sample the diversity of these major clades and for the only isolate reported from Clade V. We find that the genomes of Clade II isolates are highly rearranged and are missing large subtelomeric regions that include candidate cell wall proteins conserved in all of the other clades. Many of these candidate cell wall proteins as well as several genes involved in drug resistance appear under selection in the three global clades causing outbreaks. These findings may help explain the differences in clinical presentation between isolates from this clade and those from the clades causing global outbreaks as well as the striking variation in vitro susceptibility to antifungal drugs and to disinfectants that impact patient treatment and infection control.
Methods
DNA purification
Twelve C. auris strains were selected for long-read sequencing, two from Clade I (B11205, B13916), four from Clade II (B11220, B12043, B11809, and B13463), three from Clade III (B12037, B12631, and B17721) two from Clade IV (B11245 and B12342) and the only isolate described to date from Clade V (B18474). High molecular weight DNA for long-read sequencing was obtained using the Epicentre MasterPure yeast DNA purification kit (MPY80200), and immediately followed by purification using Pacific Biosciences Ampure PB (100-265-900). DNA for Illumina sequencing was extracted using the ZYMO Research ZR Fungal/Bacterial DNA MiniPrep kit.
Genome sequencing and assembly
For B11220, B12043, B13463, B11245, and B18474 long-read sequencing was generated using an Oxford Nanopore Technology Ligation Sequencing Kit 1 D (SQK-LSK108), sequenced on a MinIon Flow Cell R9.4 or R9.4.1 (FLO-MIN106) and basecalled with Albacore v2.0.2. For B11205, B13916, B11809, B12037, B12631, B17721, and B12342 long reads were generated using Single-molecule real-time (SMRT) sequencing using the PacBio RS II or Sequel SMRT DNA sequencing system (Pacific Biosciences, Menlo Park, CA, USA). The total read depth ranged from 35.2X to 181X. Specific details of long-read sequencing are given in Supplementary Table S1.
Reads were assembled using Canu v1.5 and v1.6 (genomeSize = 12000000; stopOnReadQuality=false; correctedErrorRate = 0.075) and Flye v 2.4.2 (genome-size = 12000000) (Koren et al. 2017; Kolmogorov et al. 2019; Supplementary Table S1). The most contiguous assembly was obtained with Canu for B11245, B11205, B13916, B12043, B11809, B13463, B12037, B17721, B12342, B18474, and with Flye for B11220 and B12631. A tandem motif (AGACACCACCTA{1,2}GAAA{1,2}CC{1,2}) was identified at contig ends; contig ends missing this motif were aligned to the unassembled contigs and manually extended. The genomes of B12043, B13463, and B18474 underwent initial polishing using Nanopore reads with Medaka version 0.8.1 using medaka_consensus with parameter -m r941_min_high (https://github.com/nanoporetech/medaka). All twelve assemblies had four or five iterations of Illumina read error correction using Pilon v1.23 (Walker et al. 2014). Assemblies were aligned to each other and to B8441 and B11221 (Muñoz et al. 2018) using NUCmer (MUMmer v3.22) (Kurtz et al. 2004), and rearrangement sites were visually inspected for support based on alignments of both long and short reads using Integrative Genomics Viewer (IGV) v2.3.72 (Robinson et al. 2011).
Genome annotation
Gene annotation was performed with BRAKER1 (Hoff et al. 2016) using RNA-Seq to improve gene structure predictions, as done for previously reported assemblies (Muñoz et al. 2018). Briefly, we mapped RNA-Seq reads to the genome assembly using Tophat2 v. 2.1.1 (Kim et al. 2013), and used the alignments to predict genes using BRAKER1 (Hoff et al. 2016), which combines GeneMark-ET (Lomsadze et al. 2014) and AUGUSTUS (Stanke et al. 2008), incorporating RNA-Seq data into unsupervised training and subsequently generates ab initio gene predictions. tRNAs were predicted using tRNAscan (Lowe and Eddy 1997) and rRNAs predicted using RNAmmer (Lagesen et al. 2007). Genes containing PFAM domains found in repetitive elements or overlapping tRNA/rRNA features were removed. Genes were named and numbered sequentially. For the protein-coding gene name assignment, we combined names from HMMER PFAM/TIGRFAM, Swissprot and Kegg products. For comparative analysis genes were functionally annotated by assigning PFAM domains, GO terms, and KEGG classification using KoalaBlast. HMMER3 (Eddy 2011) was used to identify PFAM domains using release 27. Orthologs were assigned using bidirectional blast and OrthoMCL v1.4 (Markov index 1.5; maximum e-value 1e-5) (Li et al. 2003). The predicted gene number was highly similar across all C. auris genomes, totaling 5,328 for B11220, 5,506 for B11245 and 5,294 for B18474. GPI anchored proteins were predicted with PredGPI using the general model and selecting proteins with high probability (> 99.90% specificity) (Pierleoni et al. 2008).
Genome alignments
Shared synteny regions of at least 10 kb were identified using NUCmer v3.22 (Kurtz et al. 2004). Chromosomal rearrangements (translocations, inversions, and deletions) were identified from the alignment blocks based on alignment length, chromosome mapping, and orientation. Both long read and Illumina read alignments were manually inspected in IGV v2.3.72 (Robinson et al. 2011) to confirm that the rearrangement junctions are well supported in each assembly. Illumina sequences of 17 Clade II isolates (B13463, B11220, B11808, B11809, B14308, B12081, B12043, B12040, B12082, ERR2300774, ERR2842675, TWCC13846, TWCC13847, TWCC13878, TWCC50952, TWCC58191, and TWCC58362) (Lockhart et al. 2017; Chow et al. 2018, 2020; Sekizuka et al. 2019; Wasi et al. 2019) were aligned to the B8441 and B11245 genomes using BWA mem v0.7.12 (Li 2013) and deleted regions identified using CNVnator v0.3 (1 kb windows; P-value < 0.01) (Abyzov et al. 2011) and visually inspected using IGV v2.3.72 (Robinson et al. 2011).
Variant identification
For variant identification and analysis, we included isolate sequences of Lockhart et al. (Lockhart et al. 2017) and five additional isolates from Clade II (B11808, B11809, B12043, B12081, and B14308) (Chow et al. 2018, 2020). We used FastQC and PRINSEQ (Schmieder and Edwards 2011) to assess the quality of reading data and perform read filtering low-quality sequences. Paired-end reads were aligned to the C. auris assembly strain B8441 [GenBank accession PEKT00000000.2; (Muñoz et al. 2018)] using BWA mem v0.7.12 (Li 2013). Variants were then identified using GATK v3.7 (McKenna et al. 2010) using the haploid mode and GATK tools RealignerTargetCreator, IndelRealigner, HaplotypeCaller for both SNPs and indels, CombineGVCFs, GenotypeGVCFs, GatherVCFs, SelectVariants, and Variant Filtration. Sites were filtered with Variant Filtration using “QD < 2.0 ‖ FS > 60.0 ‖ MQ < 40.0”. Genotypes were filtered if the minimum genotype quality < 50, percent alternate allele <0.8, or depth <10. Genomic variants were annotated and the functional effect was predicted using SnpEff v4.3T (Cingolani et al. 2012). The annotated VCF file was used to examine small polymorphisms (SNPs) and loss-of-function (LOF) mutations exclusively in isolates from Clade II adhesins.
Data and resource availability
The whole-genome sequence and assemblies in this study are available in NCBI as follows: B11205 (CP060353-CP060359), B13916 (CP060374-CP060380), B11220 (CP043531-CP043537), B12043 (CP050666-CP050672), B11809 (CP050659-CP050665), B13463 (CP050652-CP050658), B12037 (CP060367-CP060373), B12631 (CP060360-CP060366), B17721 (CP060353-CP060359), B11245 (CP043442-CP043448), B12342 (CP060346-CP060352), and B18474 (CP050673-CP050679). Illumina sequence of Clade II isolates is available in BioProject PRJNA328792. Isolates are available from the CDC and FDA Antimicrobial Resistance (AR) Isolate Bank, https://www.cdc.gov/drugresistance/resistance-bank/index.html. Supplemental Material available at figshare: https://doi.org/10.25386/genetics.13759276.
Results
Chromosomal level features of C. auris clades
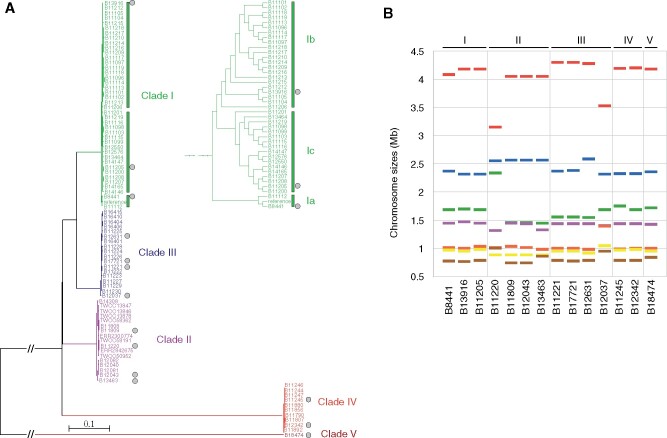
To investigate genomic changes that could explain the emergence and different phenotypes observed between C. auris clades, we generated complete chromosome scale assemblies using long-read sequencing for twelve isolates from each of the four major clades—two Clade I, four Clade II, three Clade III, and two Clade IV—and for the only isolate reported to date from Clade V (B18474; NG19339). These isolates were selected to expand the phylogenetic breadth of genome assemblies for C. auris (Figure 1A). These include two isolates representing the two major causing outbreak lineages in Clade I, which are diverged from a small subclade that includes the commonly used B8441 reference genome; we denominated these as Clade la for the B8441 subclade and Clade lb and lc for the outbreak subclades, which each contain a specific drug-resistant mutation in lanosterol 14-alpha-demethylase (ERG11), with Clade Ib corresponding to ERG11Y132F (isolate B13916) and Clade Ic corresponding to ERG11K143R (isolate B11205). Four divergent Clade II antifungal susceptible isolates from different countries were sequenced (B11220 from Japan, B11809 from South Korea, B12043 from the United States, and B13463 from Canada). For Clades III and IV, five isolates were selected for sequencing to represent different countries and drug resistance profiles, including two from Clade IV (B11245 ERG11Y132F from Venezuela and B12342 from Colombia) and three from Clade III (B17721 ERG11F126L and B12631 ERG11F126L; CNV from the United States and B12037 from Canada) (Table 1; Supplementary Table S1). These genomes were compared with the reference genomes of the isolates B8441 (Clade Ia) (Lockhart et al. 2017) and B11221 (Clade III; ERG11F126L) (Muñoz et al. 2018) (Figure 1A).
Figure 1.
Phylogenetic breadth and karyotypic variation in C. auris . (A) Phylogenetic tree of 87 C. auris whole-genome sequences clustering into four major clades and the basal isolate B18474; potential fifth clade. Maximum likelihood phylogeny using 63,674 SNPs based on 1,000 bootstrap replicates. Isolate labels are color-coded by clade and gray circles indicated those isolates selected for chromosomal-level sequencing and assembly. (B) Chromosome sizes of fourteen isolates based on chromosome scale contigs.
Table 1.
Chromosomal-level assembly statistics
| Isolate | Cladea | Country | Assembly size (Mb) | Chrs | Scaffolds | Genes | Telomeres | Accession |
|---|---|---|---|---|---|---|---|---|
| B8441 | 1a | Pakistan | 12.37 | 7 | 15 | 5,419 | NA | GCA_002759435.2 |
| B13916 | 1b | UAE | 12.4 | 7 | 7 | 5,305 | 12 | CP060374-CP060380 |
| B11205 | 1c | India | 12.4 | 7 | 7 | 5,316 | 11 | CP060353-CP060359 |
| B11220 | 2 | Japan | 12.2 | 7 | 7 | 5,227 | 13 | CP043531-CP043537 |
| B12043 | 2 | USA | 12.2 | 7 | 7 | 5,242 | 13 | CP050666-CP050672 |
| B11809 | 2 | South Korea | 12.2 | 7 | 7 | — | 13 | CP050659-CP050665 |
| B13463 | 2 | Canada | 12.2 | 7 | 7 | — | 13 | CP050652-CP050658 |
| B11221 | 3 | South Africa | 12.7 | 7 | 20 | 5,520 | NA | GCA_002775015.1 |
| B12037 | 3 | Canada | 12.3 | 7 | 7 | 5,304 | 12 | CP060367-CP060373 |
| B12631 | 3 | USA | 12.5 | 7 | 7 | — | 13 | CP060360-CP060366 |
| B17721 | 3 | USA | 12.4 | 7 | 7 | 5,309 | 12 | CP060353-CP060359 |
| B11245 | 4 | Venezuela | 12.4 | 7 | 7 | 5,506 | 13 | CP043442-CP043448 |
| B12342 | 4 | Colombia | 12.4 | 7 | 7 | — | 13 | CP060346-CP060352 |
| B18474 | 5 | Iran | 12.4 | 7 | 7 | 5,294 | 12 | CP050673-CP050679 |
1b = ERG11 Y132F; 1c = ERG11 K143R.
While seven chromosomes were assembled in each genome, substantial variation in chromosomes sizes was observed. While the three Clade I isolates contain similarly sized chromosomes, there was high variation across Clade II isolates, in particular B11220 (Figure 1B). Across the four Clade III assemblies, B12037 showed the most karyotypic variation. In the Clade III isolate B12631, a 220 kb increase in the size of chromosome 2 represents a segmental duplication and translocation of 267 kb of the region encompassing ERG11 from chromosome 3 (Supplementary Figure S3). This variation in chromosome sizes in genome assemblies is consistent with a prior description of extensive karyotypic variation across C. auris isolates (Bravo Ruiz et al. 2019).
All genome assemblies consisted of 7 nuclear contigs corresponding to nearly complete chromosomes with telomeres at both ends. The one or two uncaptured contig ends correspond to ribosomal DNA in each assembly (Figure 2; Supplementary Table S1). For all genomes, each chromosome contains one discrete region for which guanine-cytosine (GC) content is markedly reduced (average GC content 41%; average length 85 kb; Figure 2; Supplementary Figure S1). These regions in each chromosome represent candidate positions of centromeres, as similar GC-poor troughs in Pichia stipitis, Yarrowia lipolytica, and the closely related species Candida lusitaniae had been shown to coincide with experimentally identified centromeres (Lynch et al. 2010; Kapoor et al. 2015). In only one case, two GC-poor troughs were detected; these are located in tandem in B11220 Chromosome 6 and appear to have resulted from a translocation associated with a large tandem duplication (145 kb) encompassing the candidate centromere (Figure 2; Supplementary Figure S2). As large ribosomal DNA arrays play an important role in genome dynamics, we examined each assembly for rDNA arrays and identified at least one large rDNA array located at a chromosome end, ranging in size from 25 kb to 85 kb (Figure 2). We found that the rDNA arrays appear dynamic in C. auris as their location varies between isolates, even those from the same clade. In the Clade I isolates B11205 and B13916, the rDNA array is located at the end of different chromosomes, 4 or 5, respectively. While the rDNA position appears conserved in Clade II isolates, located at the end of chromosome 5, the location varies in isolates from Clade III (B17721 and B12037, chromosome 2 and 6, respectively) and is found at a different location in Clade IV (B11245, chromosome 3) and Clade V (B18474, chromosome 7) (Figure 2). This variation suggests that recombination may be frequently linked to the rDNA array or possibly to chromosome ends.
Figure 2.
Genome dynamics in C. auris . Guanine-Cytosine (GC) content 0.38 to 0.50. GC content is color-coded <0.42 blue or >0.42 gray. Ribosomal DNA array are depicted as green lines and telomeres as red lines.
Large chromosomal rearrangements in C. auris Clade II
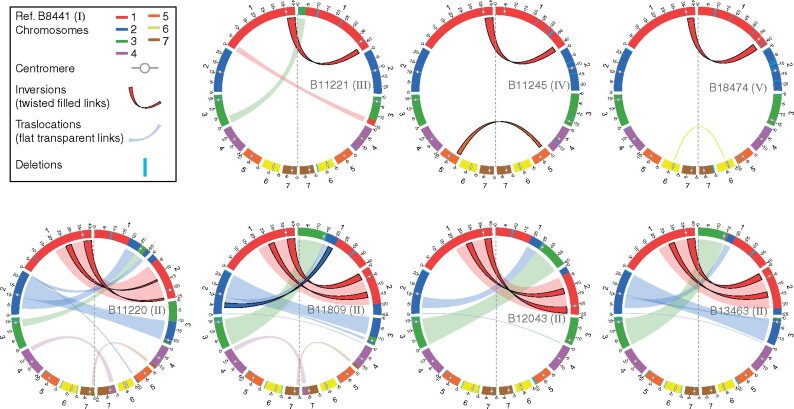
While the number of chromosomes is conserved across isolates and the total genome size is similar (ranging from 12.2 to 12.5 Mb), chromosome lengths can differ substantially. This is most pronounced in Clade II isolates, for which chromosome sizes which can vary by up to 1.1 Mb relative to those from other clades, even the distantly related Clade V isolate (B18474) (Figure 1B; Supplementary Table S1). Based on whole-genome alignments, we found that differences in chromosomes lengths are largely due to large (>10 kb) interchromosomal rearrangements between isolates (Figure 3). Each Clade II isolate has undergone 2 to 3 inversions and 4 to 8 translocations compared to B8441 (Clade Ia), resulting in large changes in chromosome size. While these translocations are commonly found in the largest chromosomes 1, 2, and 3, the translocated segments differ between isolates in size and location, suggesting recent recombination (Figure 3). Chromosomal breakpoints in Clade II isolates in chromosomes 2, 3, 4, and 7 are adjacent to candidate positions of centromeres (Figure 3, Supplementary Figure S1). Inter-chromosomal recombination near centromeres, which has resulted in a substantially different karyotype in Clade II, has previously been observed in Candida tropicalis (Chatterjee et al. 2016).
Figure 3.
Chromosomal rearrangements in C. auris . Circos plots showing syntenic chromosomes by color and links for inversions (twisted filled links) and translocations (flat transparent links) using B8441 (Clade I) as reference compared to B11221 (Clade III), B11245 (Clade IV), and B18474 (Clade V) in the top row. The lower row of Circos plots depicts B8441 compared to four Clade II isolates (B11220, B11809, B12043, and B13463). Scaffolds/contigs to chromosome mapping for these genome assemblies is included in Supplementary Table S1.
In B11220, in addition to the translocation close to a tandem duplication of the candidate centromere, there was one rearrangement that resulted in an internal telomeric repeat array. This region is located on Chromosome 4, 22.7 kb from the chromosome end (Supplementary Figure S4). As telomeric repeats are otherwise only found at chromosome ends, the presence of this unusual structure in B11220 suggests an additional sign of increased genome instability. Comparison of B11220 with a nearly complete assembly of JCM 15448 (Sekizuka et al. 2019), a clonally related isolate with only 73 SNP differences, suggests a recent origin of the Chromosome 7 centromeric duplication, which is not present the JCM 15448 genome (Supplementary Figure S2). Inspection of read support for a potential fusion of B11220 contigs1 and 2 in JCM 15448 contig1 (BGOX01000001.1) suggests this is a mis-join in JCM 15448, not supported by read alignments (Supplementary Figure S5). These differences between closely related isolates in Clade II further support increased genome instability in this clade relative to Clades I, III, IV, and V.
Outbreak-causing clades have highly syntenic stable genomes
While isolates from each C. auris clade associated with global outbreaks (I, III, and IV) have limited genetic diversity, we found evidence that intra-chromosomal rearrangements, particularly inversions, could occur between closely related isolates. By comparing Clade I isolates representing each of the subclades (Ia, Ib, and Ic), we identified three inversions. One inversion was found in chromosome 1 (scaffold01: 2720075-3096521) in B8441 (subclade 1a); the genes adjacent to the upstream breakpoint included B9J08_003891 (a small predicted protein-coding gene with similarity to the transcription factor MRR1) and B9J08_003892 (ORC4; a subunit of the origin recognition complex). The second breakpoint was located adjacent to the 3’ end of the transcriptional regulator MRR1 (B9J08_004061), producing a truncated coding region relative to the full-length copy of MRR1 in Clade III (CJI97_003963). An inversion in B11205 (subclade lc, ERG11K143R) is located in chromosome 1 (768159-904752) and the breakpoints are both adjacent to predicted proteins similar to the C. albicans Zorro3 retrotransposon gene FGR14 (B9J08_003706 and B9J08_004248). Lastly, an inversion in B13916 (subclade l b, ERG11Y132F) is located in chromosome 5 (364491-767207); the breakpoints are most closely linked to the genes B9J08_004620 (RNA15)—B9J08_004798 (ARG3) and B9J08_004621 (PTI1)—B9J08_004799. These subclade-specific rearrangements, likely mediated by sequence similarity between the breakpoints, may result in differences in gene function, such as MRR1 for subclade la, or possibly in gene regulation for other inversions.
Whole-genome alignments were also used to explore genome structure differences in Clade III. Most isolates from Clade III are azole-resistant and carry the ERG11F126L (represented by B11221 from South Africa; B17721 and B12631 from the United States); genomes of these isolates appear highly syntenic. However, a divergent and drug susceptible isolate (B12037 from Canada, Figure 1A) showed higher karyotypic variation, largely due to four translocations (Supplementary Figure S6). In addition, this genome has undergone four large deletions at subtelomeric regions, at the beginning of chromosomes 6 and 7 and the end of chromosomes 4 and 5. In addition, we closely examined rearrangements associated with copy number variation in ERG11, one of the mechanisms contributing to azole-resistance in C. auris (Muñoz et al. 2018; Bhattacharya et al. 2019) predominantly observed in Clade III isolates (Chow et al. 2020). We found using both short read mapping and chromosomal assembly that B12631 has two copies of a ∼200kb region encompassing ERG11. Notably, one of the duplications resulted in a translocation of the ERG11 region to the beginning of chromosome 2 (Supplementary Figure S3).
Two isolates from different countries in Clade IV were also examined. Comparison of chromosomal assemblies of the two Clade IV isolates, B11245 (Venezuela; ERG11Y132F) and B12342 (Colombia), revealed a large inversion in chromosome 1 (2481613–2925007). Both breakpoints are located in intergenic regions; the adjacent genes include B9J08_002936 (RMS1)—B9J08_002933 and the efflux transporter BOR1 (B9J08_002715) and a predicted protein with similarity to retrotransposons (B9J08_002717).
These intraclade chromosomal changes, including in closely related isolates, suggests that the C. auris genome can rapidly undergo rearrangements. Some of these events appear to be associated with retroelements, suggesting nonallelic homologous recombination can occur between these small elements. While this appears to be a frequent mechanism of variation between isolates, isolates in this study were selected to maximize phylogenetic representation, and the wider frequency and impact on clinical phenotypes need to be established. In addition, these alterations in C. auris karyotype, most dramatically of Clade II, likely serve as a barrier to the production of viable progeny following mating and recombination.
Large subtelomeric deletions and decay in C. auris Clade II
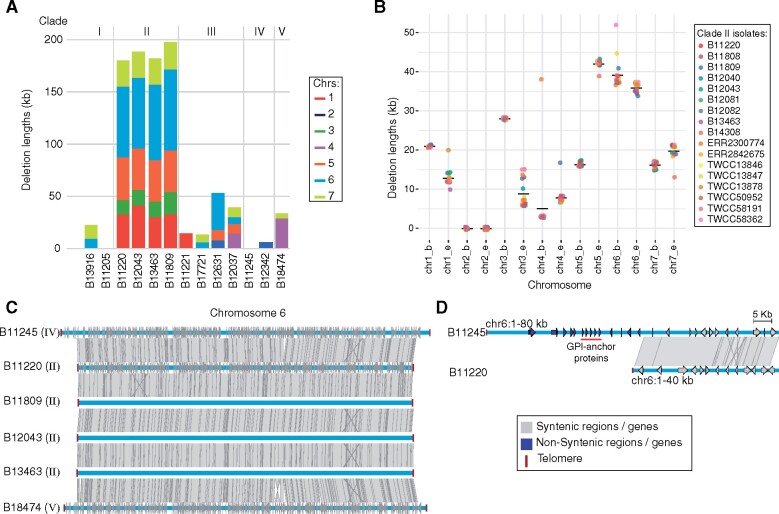
In addition to rearrangements, we identified large genomic regions that were missing in one or more clades, predominantly subtelomeric regions that are absent in Clade II isolates relative to Clades I, III, IV, and V. Comparing the Clade I (B8441) and Clade II (B11220) genomes, we identified 11 large regions (>5 kb) absent in Clade II that encompassed 226 kb and 74 genes; 10 of these 11 regions are subtelomeric in B8441 (Figure 4A; Supplementary Table S2). Comparing B8441 and B11220 with assemblies of Clade III (B11221) and Clade IV (B11245), we confirmed that these regions were also subtelomeric and only absent in Clade II isolates. These subtelomeric deletions are a common feature of isolates from Clade II, as these regions are also absent in the genome assemblies of other Clade II isolates from Canada, South Korea, and United States (Figure 4A). This is also apparent across 13 additional Clade II isolates from Japan (Sekizuka et al. 2019; Wasi et al. 2019), South Korea (Chow et al. 2020), Malaysia (ERR2300774), and the United States (Chow et al. 2018, 2020), based on alignment of Illumina sequence to the Clade I and IV references (Figure 4B). The length of the telomeric deletions varies by a few kilobases between Clade II isolates (Figure 4B). In addition, further decay might be impeded by the presence of homologs of genes shown to be essential in C. albicans at most Clade II chromosome ends (Supplementary Table S3). In addition, we observed differences in the length of subtelomeric regions even between samples from the same patient (Supplementary Figure S7). Three Clade II samples collected from a colonized patient over the course of one month (B12040, B12043, and B12082) (Chow et al. 2018) are highly clonal based on genomic analysis, with less than seven SNP differences between each pair. While these samples displayed subtelomeric deletions typical of Clade II, B12040 displayed a larger deletion at the end of chromosome 3; a region of 10 kb was deleted whereas only 6 kb was deleted in the two other clonal isolates from this same patient. The additional deleted region includes a putative alpha-1,3-mannosyltransferase (MNN1; B9J08_001516; Supplementary Figure S7). Mannosyltransferases play an important role in cell wall composition and immune recognition in C. albicans (Hall and Gow 2013) and C. auris (Bruno et al. 2020). This suggests that structural variation can contribute to intrahost variation of C. auris and may occur over a short period of time.
Figure 4.
Subtelomeric decay in C. auris Clade II. (A) Length of the deleted sequences in subtelomeric regions in thirteen C. auris genomes. (B) Length of the deleted sequences in subtelomeric regions in seventeen isolates from Clade II. (C) Chromosome wide synteny between B11245 (Clade IV) and B11220 (Clade II). Chromosome 6 includes telomeres at both ends in both isolates (dark red square). Shared synteny regions based on genome alignment (blastn) are depicted in gray vertical blocks connecting the chromosome regions. Depicted genes in blue and light gray arrows showing the direction of transcription are color-coded according to the location in conserved (gray) or nonconserved (blue) regions. (D) Comparison corresponds to a zoom in of the subtelomeric region depleted in Clade II (B11220), which encompasses auris-clade specific adhesins in tandem (red line).
To search for genetic variation that could explain these dramatic changes in genome integrity in Clade II, we identified mutations found exclusively in Clade II isolates (see Section Methods). These included 37 genes with loss-of-function mutations only found in Clade II, and only one of these impacted a gene involved in chromosome or DNA stability, a nonsense mutation near the start of DCC1 (B9J08_000232) (amino acid change=Y10*; codon change=taC/taG; Supplementary Table S4). DCC1 is a member of an alternate RFC complex involved in sister chromatid cohesion and telomere length maintenance (Askree et al. 2004). While the role of DCC1 in C. auris has not been tested, gene deletion in Saccharomyces cerevisiae DCC1 results in shorter telomeres (Askree et al. 2004) and genome instability (Yuen et al. 2007); this profile differs from that observed in hypermutator isolates with defects in DNA repair. This suggests that this naturally occurring loss of function mutation in DCC1, found in all Clade II isolates, might contribute to instability of heterochromatin, leading to shorter telomeres and genome rearrangements close to centromeres in this clade.
Basally branching Clade V isolate is highly syntenic with outbreaks causing clades
The initial description of an isolate from Iran (B18474) corresponding to a fifth clade found that it appeared highly diverged from isolates from each of the other four clades (Chow et al. 2019). This drug-sensitive isolate came from a case of ear infection, properties shared with Clade II isolates (Chow et al. 2019). Using SNP-based analysis and whole-genome alignment of a de novo assembly, we confirmed that B18474 is distantly related to isolate from all other clades, yet the genome is highly syntenic with those of isolates from Clades I, III, and IV but not Clade II. Relative to B8441 (Clade I), B18474 (Clade V) only displayed two inversions (Figure 3) and one large deletion of 35 kb at the beginning of chromosome 4 (Figure 4A). This region is not deleted in any Clade II isolate examined, supporting an independent origin. This deleted region encompassed eleven genes, including three tandem copies of putative nicotinic acid transporters (TNA1), an acylglycerol lipase (MGL2) and the SKS1 putative ser/thr kinase involved in glucose transport.
As B18474 appeared at a basally branching position compared to the other C. auris clades, we also examined the relationship of this isolate to those of species from the Candida haemulonii complex using a phylogenetic analysis of six genes (Supplementary Figure S8). In this phylogenetic tree, B18474 appears closely related to C. auris Clades I, II, III, and IV and highly divergent from the C. haemulonii species complex, supporting this isolate as a basally branching clade within C. auris. As hybrids have been reported for other Candida species, we measured the allele sharing between B18474 and all isolates from each of the other clades to evaluate if it could be a hybrid or potentially a mixed sample. The number of fixed alleles between B18474 and isolates from Clades I, II, III, and IV is 255,014, 263,799, 259,647, and 275,146, respectively. The roughly equal number of clade diagnostic alleles present in B18474 supports that it is similarly diverged from each of the four other clades; hybrid isolates would be expected to share a higher proportion of diagnostic alleles with parental lineages, as has been demonstrated in Cryptococcus neoformans for example (Rhodes et al. 2017). This is fivefold times higher than the number of fixed alleles between Clades I and II, which are the closest clades, and 1.5-fold times higher than the number of fixed alleles between Clades I and IV. In addition, B18474 has 176 K singleton SNPs, which is sevenfold times higher than the next isolate with the most unique SNPs. Although there is a single isolate from Clade V, the high number of private SNPs and similar number of fixed alleles shared with each of the four other clades supports that B18474 diverged before the separation of the four clades.
Based on read mapping and whole-genome alignment B18474 appeared heterothallic with mating type a similar to Clades I and IV isolates (Supplementary Figure S8). This isolate conserved the L-rhamnose cluster similar to Clade III isolates. Clade III isolates are able to assimilate L-rhamnose, but not Clades I, II, and IV isolates, which have deleted the L-rhamnose cluster (Ambaraghassi et al. 2019). The fact that this cluster is conserved in Clade V supports that in C. auris the absence of this cluster in Clades I, II, and IV is a loss rather than a gain in Clade III.
Depleted Hyr/Iff and species-specific cell wall protein families in C. auris Clade II
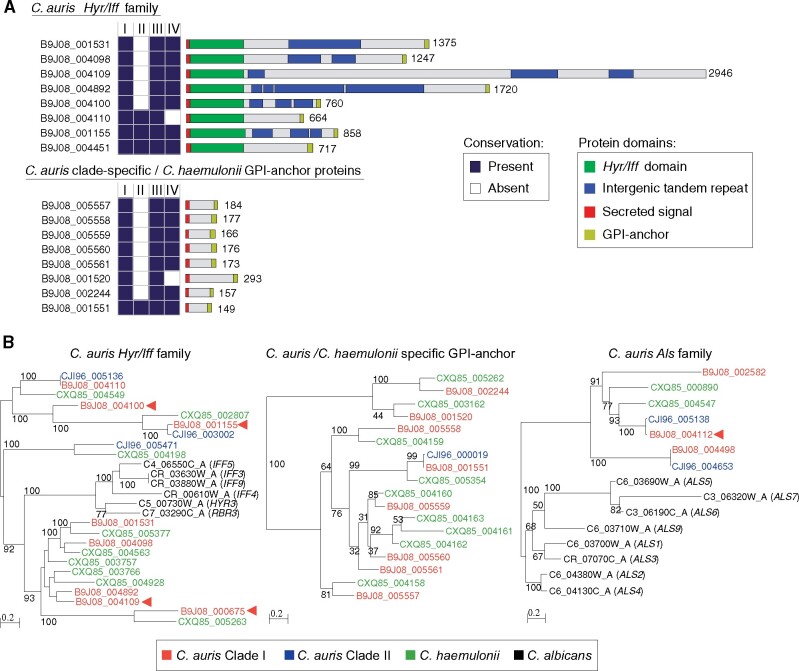
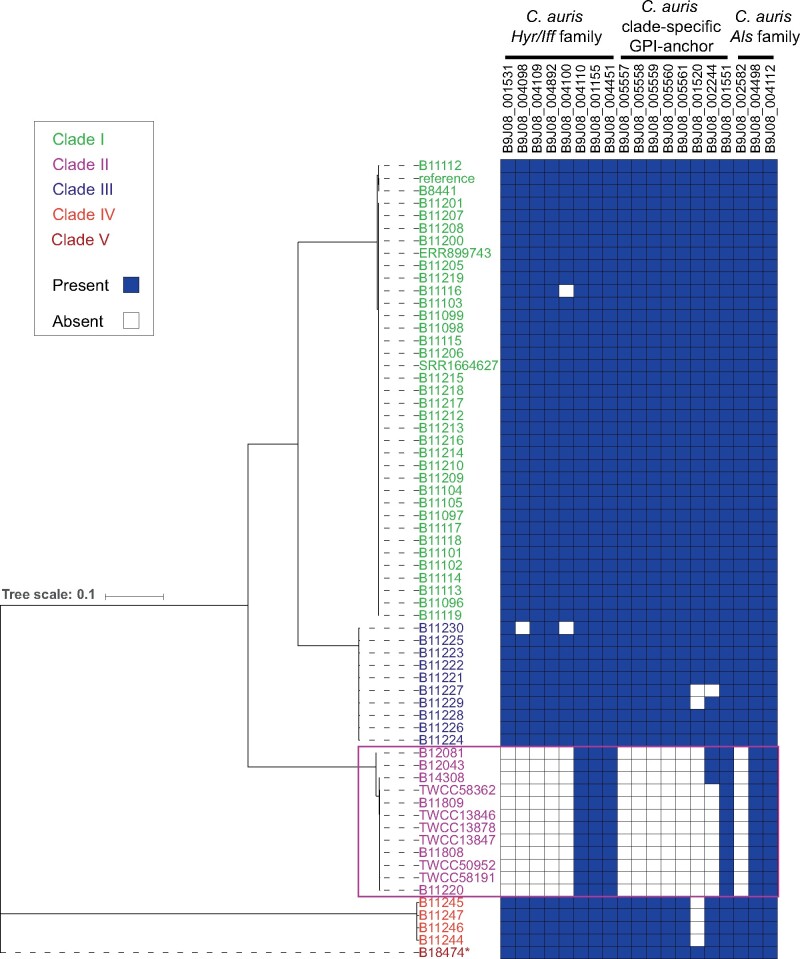
The subtelomeric regions deleted in Clade II likely contribute to the phenotypic differences of this clade. Notably, these include the loss of fourteen candidate adhesins present in Clades I, III, IV, and V (Figure 5 and 6). These candidate adhesins are divided into two sets of genes that encode predicted GPI anchors and secretion signals, one set sharing sequence similarity to C. albicans adhesins from the Hyr/Iff family and a second set of tandemly arrayed genes only found in C. auris and the closely related species C. haemulonii and C. duobushaemulonii (Figure 5; Supplementary Table S5). The Hyr/Iff gene family was previously noted to be the most highly enriched family in pathogenic Candida species and has been associated with cell wall organization and virulence (Butler et al. 2009; de Groot et al. 2013). Six of eight Hyr/Iff proteins found in C. auris contain intergenic tandem repeats, which can vary in copy number and modulate adhesion and virulence (Bates et al. 2007; de Groot et al. 2013); five Hyr/Iff genes are deleted in Clade II isolates (Figure 5A; Supplementary Table S5). The second set of candidate adhesins are small proteins (149–293 amino acids) with serine/threonine-rich regions (9.0–17.3% Serine; 17.5–22.8% Threonine), and are tandemly located in subtelomeric regions conserved in Clades I, III, IV, and V, but absent in Clade II (Figure 2, 5A, and B; Supplementary Table S5). These proteins are conserved in species from the C. haemulonii complex but not more distantly related species (Figure 5B). The subtelomeric location and serine/threonine-rich region are properties shared with C. glabrata EPA adhesins (De Las Peñas et al. 2003), and the expansion of EPA adhesins has been linked to the emergence of the ability to infect humans in the C. glabrata lineage (Gabaldón et al. 2013). Several of the genes in deleted regions of Clade II [three of the C. auris-clade specific adhesins, one HYR-like gene, and other cell wall associated proteins (ALS4, CSA1, and RBR3)] were transcriptionally up-regulated in developing C. auris biofilms (Kean et al. 2018; Supplementary Table S2), suggesting they play a role in biofilm formation. Other changes associated with cell wall and mannose modifications include the loss of another putative alpha-1,3-mannosyltransferase (MNT4; B9J08_004891) and a loss of function mutation in all Clade II isolates in the N-acetylglucosamine (GlcNAc) kinase (HXK1; B9J08_003836; amino acid change=Q101*; codon change=Caa/Taa; Supplementary Table S4). The loss of adhesin-like genes, as well as genes involved in cell wall modifications in Clade II isolates, could help explain epidemiological differences between this clade and the three other clades that are more commonly observed.
Figure 5.
Differences in the repertoire GPI-anchor proteins in C. auris . (A) Conserved domains in two clusters of GPI-anchor found in C. auris B8441. (Top) Hyr/Iff GPI-anchor family and (bottom) C. auris clade-specific GPI-anchor protein. Conservation across C. auris isolates representing Clades I, II, III, and IV is color-coded indicating whether the gene is present (dark blue) or absent (white). (B) Phylogenetic analysis of the Hyr/Iff GPI-anchor family (left), the C. auris clade-specific GPI-anchor family (middle) and Als GPI-anchor family (right) using one representative isolate for C. auris Clades I and II, C. haemulonii and C albicans. Gene IDs are color-coded by species/isolate.
Figure 6.
Conservation of GPI-anchor proteins across C. auris clades. Phylogenetic tree of 64 C. auris isolates from Clades I, II, III, IV, and V. Right panel depicts the conservation (present: blue; absent: white) of three families of adhesins. *B18474 a.k.a. NG19339.
Clade-specific cell wall proteins and drug-related genes have signatures of selection
We examined the genomes of C. auris for signatures of selection using the composite likelihood ratio (CLR) test (Nielsen et al. 2005) and scanned genes for signatures of selective sweeps in C. auris Clades I, III, and IV. We did not carry out this analysis in Clades II and V due to the limited number of isolates from these clades. Overall, the genes in the top 5% of CLR values in Clades I, III, and IV fall into two categories. One group of genes associated with drug resistance, including the drug targets ERG11 and FKS1, the transcriptional regulator TAC1b and the drug efflux pump CDR1 (Table 2). The second group of genes related to invasion and biofilm formation, including the cell wall proteins RBR3, HYR3, IFF6, and ALS4 (Figure 5), the secreted yeast wall protein YWP1 and the transcriptional regulators WOR2, ZCF32, and OPI1 (Table 2). Notably, some of these genes display signatures of selection in all three clades or in two of the three (Table 2), suggesting shared and clade-specific mechanism of host and drug adaptation. The drug targets ERG11 and FKS1 and the cell wall proteins HYR3 (B9J08_000675) and RBR3 (B9J08_004100) displayed signatures of selection in all three clades. Drug resistance mutations in ERG11 (Lockhart et al. 2017; Chowdhary et al. 2018 p. 350; Chow et al. 2020) and FKS1 (Chowdhary et al. 2018; Chow et al. 2020) have been reported in Clades I, III, and IV. In addition, TAC1b and CDR1 appear under selection in Clades I and IV, but not Clade III, suggesting a stronger contribution of variants in these genes to azole-resistance in these clades. This is supported by recent work that demonstrated that mutations in TAC1b contribute to fluconazole resistance (Rybak et al. 2020) and that expression of CDR1 is differentially regulated between isolates from Clades III and IV (Mayr et al. 2020). Together, this suggests shared and clade-specific evolution of both drug-resistance and cell wall proteins that may impact host interaction. These candidate cell wall proteins are strong candidates for experimental validation to better understand the high rate of transmission of the C. auris outbreak clades within health care facilities and the ability to persist in colonized patients and on plastic surfaces.
Table 2.
Genes in the top 5% of composite likelihood ratio (CLR) test values of selection
| Clade | Gene ID | Category | Gene | Description |
|---|---|---|---|---|
| Clades I, III, and IV | B9J08_001448 | Drug target | ERG11 | Lanosterol 14-alpha-demethylase |
| B9J08_000964 | Drug target | FKS1 | Beta-1,3-glucan synthase | |
| B9J08_000675 | Cell wall | RBR3 | Cell wall adhesin-like protein | |
| B9J08_004100 | Cell wall | HYR3 | Putative GPI-anchored adhesin-like protein | |
| Clades I and III | B9J08_000336 | Cell wall | — | Mucin-like protein |
| B9J08_004109 | Cell wall | RBR3 | Cell wall adhesin-like protein | |
| B9J08_000045 | Chromatin remodeling | ISW2 | Translocase involved in chromatin remodeling | |
| B9J08_000092 | Intracellular transport | DYN1 | Dynein; microtubule motor protein | |
| B9J08_004769 | Protein degradation | UBI4 | Ubiquitin | |
| B9J08_001715 | Phosphorylation | URK1 | Uridine/cytidine kinase | |
| B9J08_004988 | Transcription factor | — | Predicted transcription factor | |
| B9J08_005188 | Transcription factor | TIF4631 | Putative translation initiation factor eIF4G | |
| B9J08_001729 | Transcription factor | ZCF32 | Zn(II)2Cys6 transcription factor (biofilm) | |
| B9J08_002715 | Transporter | BOR1 | Borate efflux transmembrane transporter | |
| B9J08_002864 | Transporter | NGT1 | N-acetylglucosamine (GlcNAc)-specific transporter | |
| B9J08_003323 | Transporter | YOR1 | Plasma membrane ABC transporter | |
| Clades I and IV | B9J08_000164 | Transporter | CDR1 | Multidrug transporter of ABC superfamily |
| B9J08_004718 | Transporter | OSH2 | OxySterol binding protein Homolog | |
| B9J08_004820 | Transcription factor | TAC1b | Zn(2)-Cys(6) transcriptional activator (efflux pumps) | |
| B9J08_002136 | Transcription factor | WOR2 | Zn(II)2Cys6 transcription factor (white-opaque switching) | |
| Clades III and IV | B9J08_003550 | Cell wall | YWP1 | Secreted yeast wall protein |
| B9J08_001155 | Cell wall | IFF6 | Putative GPI-anchored adhesin-like protein | |
| B9J08_004112 | Cell wall | ALS4 | Putative GPI-anchored adhesin-like protein | |
| B9J08_001181 | Transcription factor | OPI1 | Leucine zipper transcriptional regulator |
Discussion
In this study, we generated and analyzed 12 complete, telomere-to-telomere, C. auris genomes representing the phylogenetic breadth of the five phylogenetic clades. By comparing these genomes, we characterized a major genetic difference between isolates from Clade II compared to those from Clades I, III, IV, and V. We discovered that the genomes of Clade II isolates, which are often drug susceptible and associated with ear infections, have increased chromosomal rearrangements, genome instability, and telomere shortening. We found that C. auris subtelomeric regions are enriched in cell wall proteins, which are largely lost in Clade II isolates. These cell wall proteins fall into three families that are also targets of selection in Clades I, III, and IV, along with other predicted cell wall proteins and genes involved in drug resistance. The loss of these cell wall proteins in Clade II potentially impacts adhesion and biofilm formation, and may help explain the lower virulence and potential for transmission in Clade II isolates. While the isolates from the outbreak-causing Clades I, III, and IV have very low intra-clade genetic diversity, comparison of complete genomes identified large genomic changes that will be important to further evaluate for an impact on host interaction and drug resistance. This analysis underscores the importance of generating genome assemblies of diverse isolates to understand the impact of genomic plasticity across a population.
A major challenge in managing C. auris infections is that unlike other Candida this species can be easily transmitted within health care facilities. This may be accelerated by the ability to persist on plastic surfaces common in health care settings (Welsh et al. 2017). In addition, the ability of C. auris to form biofilms might enhance the capacity of C. auris to colonize patients’ skin for long periods of time and survive in healthcare setting even after cleaning, further increasing transmission and potentiating outbreaks. These are common features of isolates from Clades I, III, and IV that are the primary cause of invasive infections and have each caused outbreaks in healthcare settings (Welsh et al. 2019). The identification of candidate cell wall proteins and mannosyltransferases that are absent in the highly rearranged genome of Clade II, highlights the major differences that can occur between otherwise closely related isolates of a species. Understanding how C. auris survives in healthcare settings and on patients’ skin, and how different clades respond to different stressors, is essential to understand its pathobiology, however, studies to date have only tested small numbers of samples for each of the four major clades. Overall, C. auris isolates showed salt tolerance and thermotolerance up to 42°C, with a subset showing an aggregating phenotype (Borman et al. 2016; Sherry et al. 2017). A recent study of sixteen C. auris isolates found they were generally resistant to the cationic, oxidative, nitrosative and cell wall stresses, however, the one Clade II isolate tested was highly susceptible to Calcofluor White and Congo red (Heaney et al. 2020), suggesting that Clade II isolates are more susceptible to cell wall stress. A pair of recent studies showed that Clade II isolates from Japan and South Korea were more sensitive to UV-C killing than Clades I, III and IV isolates originating from Venezuela, Spain and India (de Groot et al. 2019; Chatterjee et al. 2020). Although most Clade II isolates are drug susceptible isolated from ear infection, some isolates appear to cause fungemia and acquire azole resistance (Lee et al. 2011), suggesting that, even with a highly rearranged genome and large subtelomeric losses, this clade has the potential to cause severe, drug-resistant infections.
Our study highlights the importance of more widely evaluating phenotypic variation across C. auris clades. It will be important to understand how genomic variation including clade-specific differences may affect properties such as biofilm formation, adhesion, invasion and damage to elucidate the basis of infection, colonization, and transmission of C. auris. Future studies are also needed to test the direct role of subtelomeric gene families of potential adhesins in human colonization and virulence. Recent studies have found that some predicted cell wall protein genes are differentially regulated under physiologically-relevant conditions, including the candidate adhesins in subtelomeric regions deleted in Clade II (Kean et al. 2018; Yue et al. 2018). One adhesin homolog of C. albicans ALS4 (B9J08_004112) is differentially expressed during C. auris filamentous growth (Yue et al. 2018), and other GPI-anchored cell wall genes (IFF4, PGA26, PGA52, and HYR3) and potential ALS adhesin (B9J08_002582) were upregulated during C. auris in vitro biofilm formation compared to planktonic cells (Kean et al. 2018). In addition, two ALS family orthologs, were expressed in biofilms (Kean et al. 2018). In C. albicans, ALS3 was used to generate a vaccine against C. albicans that cross-protects against C. auris (Singh et al. 2019); ALS3 (B9J08_004498) is conserved in all clades, and is even found in Clade II and the only isolate described to date from potential Clade V, suggesting this vaccine may be broadly effective.
The single isolate from Iran that represents a potential fifth clade was hypothesized to be related to Clade II, based on shared drug susceptibility and causing ear infections. We confirmed B18474 is a basally branching isolate relative to the four major clades and is not closely related to the C. haemulonii species complex. As the genome was found to be highly syntenic with those of isolates of Clades I, III, and IV, this supports that the high rate of genome rearrangements and subtelomeric loss are unique features of Clade II isolates. Once more isolates are reported from this potential fifth clade, it would be possible to confirm if the ear infection and drug susceptibility are general patterns of this clade; isolates could also be sequenced to measure genetic diversity and allele sharing for Clade V compared to the other clades.
These chromosomal assemblies have highlighted the genomic instability and gene loss that has occurred in Clade II isolates as well as how structural variation contributes to diversity in other clades. The lost cell wall proteins identified in our study are not well studied to date, even the Iff/Hyr family shown to be fast evolving in other Candida species. These cell wall proteins could have implications for the observed differences in susceptibility to disinfectants, ability to persist outside human host, and transmission dynamics. Future experimental testing of these genes may help explain differences in the clinically important properties of these clades, including the severity of infection and propensity to cause outbreaks, which will help develop strategies to control this emerging fungal threat.
Disclaimer
The use of product names in this manuscript does not imply their endorsement by the US Department of Health and Human Services. The finding and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Acknowledgments
The authors thank Johanna Rhodes, Matthew Fisher, Andrew Borman, and Elizabeth Johnson for use of the unpublished Malaysian Clade II isolate sequence (SRA accession ERR2300774).
Funding
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under award U19AI110818 to the Broad Institute. CAC is a CIFAR fellow in the Fungal Kingdom Program.
Contributor Information
José F Muñoz, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Rory M Welsh, Mycotic Diseases Branch, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Terrance Shea, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Dhwani Batra, Division of Scientific Resources, Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Lalitha Gade, Mycotic Diseases Branch, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Dakota Howard, Division of Scientific Resources, Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Lori A Rowe, Division of Scientific Resources, Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Jacques F Meis, Department of Medical Microbiology and Infectious Diseases, Canisius Wilhelmina Hospital, Center of Expertise in Mycology Radboudumc/CWZ, Nijmegen, The Netherlands.
Anastasia P Litvintseva, Mycotic Diseases Branch, U.S. Department of Health and Human Services, Atlanta, GA, USA.
Christina A Cuomo, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Literature cited
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambaraghassi G, Dufresne PJ, Dufresne SF, Vallières É, Muñoz JF, et al. 2019. Identification of Candida auris by use of the updated Vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J Clin Microbiol. 57:e00884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, et al. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 101:8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, de la Rosa JM, MacCallum DM, Brown AJ, Gow NA, et al. 2007. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect Immun. 75:2922–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Holowka T, Orner EP, Fries BC. 2019. Gene duplication associated with increased fluconazole tolerance in Candida auris cells of. Sci Rep. 9:5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Szekely A, Johnson EM. 2016. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere 1:e00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Ruiz G, Ross ZK, Holmes E, Schelenz S, Gow NAR, et al. 2019. Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr Genet. 65:1217–1228. https://doi.org/10.1007/s00294-019-00976-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Kersten S, Bain JM, Jaeger M, Rosati D, et al. 2020. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 5:1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo B, Melo ASA, Perozo-Mena A, Hernandez M, Francisco EC, et al. 2016. First report of Candida auris in America: Clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 73:369–374. [DOI] [PubMed] [Google Scholar]
- Carreté L, Ksiezopolska E, Pegueroles C, Gómez-Molero E, Saus E, et al. 2018. Patterns of genomic variation in the opportunistic pathogen Candida glabrata suggest the existence of mating and a secondary association with humans. Curr Biol. 28:15–27.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee G, Sankaranarayanan SR, Guin K, Thattikota Y, Padmanabhan S, et al. 2016. Repeat-associated fission yeast-lie regional centromeres in the ascomycetous budding yeast Candida tropicalis. PLOS Genet. 12:e1005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Choi H, Ochoa B, Garmon G, Coppin JD, et al. 2020. Clade-specific variation in susceptibility of Candida auris to broad-spectrum ultraviolet C light (UV-C). Infect Control Hosp Epidemiol. 41:1–4. 10.1017/ice.2020.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, et al. 2019. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 25:1780–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, et al. 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 18:1377–1384. https://doi.org/10.1016/S1473-3099(18)30597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, et al. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, et al. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 73:891–899. [DOI] [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, et al. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 19:1670–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff Fly (Austin). 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Peñas A, Pan S-J, Castaño I, Alder J, Cregg R, et al. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, et al. 2013. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics 14:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot PWJD, Bader O, de Boer AD, Weig M, Chauhan N. 2013. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell 12:470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot TD, Chowdhary A, Meis JF, Voss A. 2019. Killing of Candida auris by UV‐C: importance of exposure time and distance. Mycoses 62:408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Gow NAR. 2013. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol. 90:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney H, Laing J, Paterson L, Walker AW, Gow NAR, et al. 2020. The environmental stress sensitivities of pathogenic Candida species, including Candida auris, and implications for their spread in the hospital setting. Med Mycol. 58:744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Zhu L, Froyd C, Liu T, Rusche LN. 2015. Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc Natl Acad Sci USA. 112:12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean R, Delaney C, Sherry L, Borman A, Johnson EM, et al. 2018. Transcriptome assembly and profiling of Candida auris reveals novel insights into Biofilm-Mediated resistance. mSphere 3:e00334–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-N, Shin JH, Sung H, Lee K, Kim E-C, et al. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 48:e57–e61. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37:540–546. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, et al. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 49:3139–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, et al. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 64:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Burns PD, Borodovsky M. 2014. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DB, Logue ME, Butler G, Wolfe KH. 2010. Chromosomal G + C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol. 2:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia. South Africa Emerging Infect Dis. 20:1250–1251. 10.3201/eid2007.131765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E-M, Ramírez-Zavala B, Krüger I, Morschhäuser J. 2020. A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere 5:e00279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz JF, Gade L, Chow NA, Loparev VN, Juieng P, et al. 2018. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 9:5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, et al. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15:1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. 2008. PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poláková S, Blume C, Zárate JA, Mentel M, Jørck-Ramberg D, et al. 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA. 106:2688–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Desjardins CA, Sykes SM, Beale MA, Vanhove M, et al. 2017. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics 207:327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak JM, Muñoz JF, Barker KS, Parker JE, Esquivel BD, et al. 2020. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11:e00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, et al. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 53:41–44. [DOI] [PubMed] [Google Scholar]
- Schalkwyk EV, Mpembe RS, Thomas J, Shuping L, Ismail H, et al. 2019. Epidemiologic shift in candidemia driven by Candida auris, South Africa, 2016–2017. Emerg Infect Dis. 25:1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizuka T, Iguchi S, Umeyama T, Inamine Y, Makimura K, et al. 2019. Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS ONE 14:e0223433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton DJ, Welsh RM, Bentz ML, Forsberg K, Jackson B, et al. 2020. Evaluation of nine surface disinfectants against Candida auris using a quantitative disk carrier method: EPA SOP-MB-35. Infect Control Hosp Epidemiol. 41:1219–1221. 10.1017/ice.2020.278 [DOI] [PubMed] [Google Scholar]
- Sherry L, Ramage G, Kean R, Borman A, Johnson EM, et al. 2017. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 23:328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, et al. 2019. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 15:e1007460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasi M, Khandelwal NK, Moorhouse AJ, Nair R, Vishwakarma P, et al. 2019. ABC transporter genes show upregulated expression in drug-resistant clinical isolates of Candida auris: a genome-wide characterization of ATP-Binding Cassette (ABC) Transporter Genes. Front Microbiol. 10:1445. 10.3389/fmicb.2019.01445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, et al. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 55:2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. 2019. Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 57:e00007–e00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Bing J, Zheng Q, Zhang Y, Hu T, et al. 2018. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KWY, Warren CD, Chen O, Kwok T, Hieter P, et al. 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci USA. 104:3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]