Abstract
Mislocalization of the centromeric histone H3 variant (Cse4 in budding yeast, CID in flies, CENP-A in humans) to noncentromeric regions contributes to chromosomal instability (CIN) in yeast, fly, and human cells. Overexpression and mislocalization of CENP-A have been observed in cancers, however, the mechanisms that facilitate the mislocalization of overexpressed CENP-A have not been fully explored. Defects in proteolysis of overexpressed Cse4 (GALCSE4) lead to its mislocalization and synthetic dosage lethality (SDL) in mutants for E3 ubiquitin ligases (Psh1, Slx5, SCFMet30, and SCFCdc4), Doa1, Hir2, and Cdc7. In contrast, defects in sumoylation of overexpressed cse4K215/216/A/R prevent its mislocalization and do not cause SDL in a psh1Δ strain. Here, we used a genome-wide screen to identify factors that facilitate the mislocalization of overexpressed Cse4 by characterizing suppressors of the psh1Δ GALCSE4 SDL. Deletions of histone H4 alleles (HHF1 or HHF2), which were among the most prominent suppressors, also suppress slx5Δ, cdc4-1, doa1Δ, hir2Δ, and cdc7-4 GALCSE4 SDL. Reduced dosage of H4 leads to defects in sumoylation and reduced mislocalization of overexpressed Cse4, which contributes to suppression of CIN when Cse4 is overexpressed. We determined that the hhf1-20, cse4-102, and cse4-111 mutants, which are defective in the Cse4-H4 interaction, also exhibit reduced sumoylation of Cse4 and do not display psh1Δ GALCSE4 SDL. In summary, we have identified genes that contribute to the mislocalization of overexpressed Cse4 and defined a role for the gene dosage of H4 in facilitating Cse4 sumoylation and mislocalization to noncentromeric regions, leading to CIN when Cse4 is overexpressed.
Keywords: centromere, CENP-A, histone H4, CIN
Introduction
Centromeres are specialized chromosome loci that are essential for faithful chromosome segregation during mitosis and meiosis. The kinetochore (centromeric DNA and associated proteins) provides an attachment site for microtubules to promote proper segregation of sister chromatids during cell division (Allshire and Karpen 2008; Verdaasdonk and Bloom 2011; Burrack and Berman 2012; Choy et al. 2012; Maddox et al. 2012; McKinley and Cheeseman 2016). Despite the wide divergence of centromeric DNA sequence, establishment of centromeric chromatin is regulated by epigenetic mechanisms where incorporation of the essential and evolutionarily conserved centromeric histone H3 variant CENP-A (Cse4 in Saccharomyces cerevisiae, Cnp1 in Schizosaccharomyces pombe, CID in Drosophila melanogaster, and CENP-A in mammals) serves to nucleate kinetochore assembly (Kitagawa and Hieter 2001; Biggins 2013; McKinley and Cheeseman 2016).
The evolutionarily conserved CENP-A-specific histone chaperones (Scm3 in S. cerevisiae and S. pombe, CAL1 in D. melanogaster, Holliday Junction Recognition Protein HJURP in humans) mediate the centromeric localization of CENP-A (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007; Foltz et al. 2009; Pidoux et al. 2009; Williams et al. 2009; Shuaib et al. 2010; Chen et al. 2014). In budding yeast, other chaperones such as Chromatin Assembly Factor 1 (CAF-1), an evolutionarily conserved replication-coupled histone H3/H4 chaperone, can facilitate the deposition of overexpressed Cse4 when Scm3 is depleted (Hewawasam et al. 2018). The CAF-1 orthologues Mis16 in S. pombe and RbAp46/48 in humans and D. melanogaster also contribute to centromeric localization of CENP-A (Fujita et al. 2007; Pidoux et al. 2009; Williams et al. 2009; Boltengagen et al. 2016).
Restricting the localization of CENP-A to centromeres is essential for faithful chromosome segregation. However, overexpression of CENP-A leads to its mislocalization to noncentromeric chromatin and contributes to chromosomal instability (CIN) in yeast, flies, and humans (Collins et al. 2004; Heun et al. 2006; Moreno-Moreno et al. 2006; Au et al. 2008; Mishra et al. 2011; Lacoste et al. 2014; Athwal et al. 2015; Shrestha et al. 2017). Overexpression and mislocalization of CENP-A are observed in many cancers and are proposed to promote tumorigenesis (Tomonaga et al. 2003; Amato et al. 2009; Li et al. 2011; McGovern et al. 2012; Sun et al. 2016). Thus, defining the molecular mechanisms that promote and prevent mislocalization of CENP-A is an area of active investigation.
In budding yeast, post-translational modifications (PTMs) of Cse4, such as ubiquitination, sumoylation, and isomerization, are important for regulating steady-state levels of Cse4 and preventing its mislocalization to noncentromeric regions, thereby maintaining chromosome stability (Collins et al. 2004; Hewawasam et al. 2010; Ranjitkar et al. 2010; Ohkuni et al. 2014, 2016; Cheng et al. 2017; Au et al. 2020). Ubiquitin-mediated proteolysis of Cse4 by E3 ubiquitin ligases such as Psh1 (Hewawasam et al. 2010; Ranjitkar et al. 2010), SUMO-targeted ubiquitin ligase (STUbL) Slx5 (Ohkuni et al. 2016), SCFMet30/Cdc4 (Au et al. 2020), SCFRcy1 (Cheng et al. 2016), and Ubr1 (Cheng et al. 2017) and the proline isomerase Fpr3 (Ohkuni et al. 2014) regulate the cellular levels of Cse4. Psh1-mediated proteolysis of Cse4 has been well characterized and has been shown to be regulated by the FACT (Facilitates Chromatin Transcription/Transactions) complex (Deyter and Biggins 2014), CK2 (Casein Kinase 2) (Hewawasam et al. 2014), HIR (HIstone Regulation) histone chaperone complex (Ciftci-Yilmaz et al. 2018), and DDK (Dbf4-Dependent Kinase) complex (Eisenstatt et al. 2020). In general, mutation or deletion of factors that prevent Cse4 mislocalization show synthetic dosage lethality (SDL) when Cse4 is overexpressed from a galactose-inducible promoter (GALCSE4).
In contrast to the many studies that have characterized pathways that prevent mislocalization of CENP-A to noncentromeric regions, mechanisms that facilitate the mislocalization of overexpressed CENP-A have not been fully explored. Studies from our laboratory and those of others show that the transcription-coupled histone H3/H4 chaperone DAXX/ATRX promotes mislocalization of CENP-A to noncentromeric regions in human cells (Lacoste et al. 2014; Shrestha et al. 2017). In budding yeast, CAF-1 contributes to the mislocalization of overexpressed Cse4 to noncentromeric regions (Hewawasam et al. 2018). We have recently shown that sumoylation of Cse4K215/216 in the C-terminus of Cse4 facilitates its interaction with CAF-1 and this promotes the deposition of Cse4 to noncentromeric regions (Ohkuni et al. 2020). Notably, psh1Δ cac2Δ GALCSE4 strains and psh1Δ GALcse4K215/216R/A strains do not exhibit SDL due to reduced mislocalization of Cse4 (Hewawasam et al. 2018; Ohkuni et al. 2020).
Defining the mechanisms that facilitate the mislocalization of overexpressed Cse4 to noncentromeric regions is essential for understanding which pathways contributes to mislocalization of CENP-A in cancers with a poor prognosis. We performed a genome-wide screen using a synthetic genetic array (SGA) which combined mutants of essential genes and deletions of nonessential genes with psh1Δ GALCSE4 to identify suppressors of the psh1Δ GALCSE4 SDL. Deletion of the two alleles that encode histone H4 (HHF1 or HHF2) were among the most prominent suppressors of the psh1Δ GALCSE4 SDL and a role for the dosage of H4 in preventing mislocalization of Cse4 has not been previously examined. In this study, we focused on defining the molecular mechanisms that prevent the mislocalization of overexpressed Cse4 and suppress the psh1Δ GALCSE4 SDL when the gene dosage of H4 is reduced. We showed that deletion of HHF1 or HHF2 also suppresses the GALCSE4 SDL in slx5Δ, doa1Δ, hir2Δ, cdc4-1, and cdc7-4 strains. Deletion of HHF1 or HHF2 results in reduced Cse4 sumoylation and this correlates with reduced mislocalization to noncentromeric regions and rapid degradation of Cse4 in a psh1Δ strain. Moreover, cse4-102, cse4-111, and hhf1-20, which have mutations in their histone fold domains and are defective for the formation of the Cse4-H4 dimer (Smith et al. 1996; Glowczewski et al. 2000), show reduced Cse4 sumoylation and do not cause SDL in psh1Δ GALCSE4 strains. In summary, our genome-wide suppressor screen allowed us to identify genes that contribute to Cse4 mislocalization and to define a role for reduced gene dosage of H4 in preventing the mislocalization of Cse4 to noncentromeric regions and suppression of the psh1Δ GALCSE4 SDL and CIN when Cse4 is overexpressed.
Materials and methods
Strains and plasmids
Yeast strains used in this study are described in Supplementary Table S2 and plasmids in Supplementary Table S3. Yeast strains were grown in rich media (1% yeast extract, 2% bacto-peptone, and 2% glucose) or synthetic medium with glucose or raffinose and galactose (2% final concentration each) and supplements to allow for selection of the indicated plasmids. Double mutant strains were generated by mating wild-type or psh1Δ strains with empty vector or a plasmid containing GAL1-6His-3HA-CSE4 to mutant strains on rich medium at room temperature for 6 h followed by selection of diploid cells on medium selective for the plasmid and appropriate resistance markers. Diploids were sporulated for 5 days at 23°C and plated on selective medium without uracil, histidine, or arginine and with canavanine, clonNAT, and G418 to select for MATa double mutants. The SGA was performed as previously described (Costanzo et al. 2016).
Growth assays
Growth assays were performed as previously described (Eisenstatt et al. 2020). Wild-type and mutant strains were grown on medium selective for the plasmid, suspended in water to a concentration with an optical density of 1 measured at a wavelength of 600 nm (OD600, approximately 1.0 × 107 cells per ml), and plated in fivefold serial dilutions starting with 1 OD600 on synthetic growth medium containing glucose or galactose and raffinose (2% final concentration each) selecting for the plasmid. Strains were grown at the indicated temperatures for 3–5 days.
Protein stability assays
Protein stability assays were performed as previously described (Au et al. 2008). Briefly, logarithmically growing wild-type and mutant cells were grown for 3–4 h in media selective for the plasmid containing galactose/raffinose (2% final concentration each) at 30°C followed by addition of cycloheximide (CHX, 10 µg/ml) and glucose (2% final concentration). Protein extracts were prepared from cells collected 0, 30, 60, and 90 min after CHX addition with the TCA method as described previously (Kastenmayer et al. 2006). Equal amount of protein as determined by the Bio-Rad DC™ Protein Assay were analyzed by Western blot. Proteins were separated by SDS-PAGE on 4–12% Bis-TRIS SDS-polyacrylamide gels (Novex, NP0322BOX) and analysis was done against primary antibodies α-HA (1:1000, Roche, 12CA5) or α-Tub2 (1:4500, custom made for Basrai Laboratory) in TBS-T containing 5% (w/v) dried skim milk. HRP-conjugated sheep α-mouse IgG (Amersham Biosciences, NA931V) and HRP-conjugated donkey α-rabbit IgG (Amersham Biosciences, NA934V) were used as secondary antibodies. Stability of the Cse4 protein relative to the Tub2 loading control was measured as the percent remaining as determined with the Image Lab Software (BioRad).
Ubiquitination pull-down assay
Levels of ubiquitinated Cse4 were determined with ubiquitin pull-down assays as described previously (Au et al. 2013) with modifications. Cells were grown to logarithmic phase, induced in galactose-containing medium for 3 h at 30°C and pelleted. The cell pellet was resuspended in lysis buffer [20 mM Na2HPO4, 20 mM NAH2PO4, 50 mM NaF, 5 mM tetra-sodium pyrophosphate, 10 mM beta-glycerolphosphate, 2 mM EDTA, 1 mM DTT, 1% NP-40, 5 mM N-Ethylmaleimide, 1 mM PMSF, and protease inhibitor cocktail (Sigma, catalogue # P8215)] and equal volume of glass beads (lysing matrix C, MP Biomedicals). Cell lysates were generated by homogenizing cells with a FastPrep-24 5 G homogenizer (MP Biomedicals) and a fraction of the lysate was aliquoted for input. An equal concentration of lysates from wild type and mutant strains were incubated with tandem ubiquitin-binding entities (Agarose-TUBE1, Life Sensors, Inc., catalog # UM401) overnight at 4°C. Proteins bound to the beads were washed three times with TBS-T at room temperature and eluted in 2 × Laemmli buffer at 100°C for 10 min. The eluted protein was resolved on a 4–12% Bis-Tris gel (Novex, NP0322BOX) and ubiquitinated Cse4 was detected by Western blot using anti-HA antibody (Roche Inc., 12CA5). Levels of ubiquitinated Cse4 relative to the nonmodified Cse4 in the input were quantified using software provided by the Syngene imaging system. The percentage of ubiquitinated Cse4 levels is set to 100% in the wild-type strain.
In vivo sumoylation assay
Cell lysates were prepared from 50 ml culture of strains grown to logarithmic phase in raffinose/galactose (2% final concentration each) medium at 30°C for 4 h to induce expression of Cse4 from the galactose-inducible promoter. Cells were pelleted, rinsed with sterile water, and suspended in 0.5 ml of guanidine buffer (0.1 M Tris-HCl at pH 8.0, 6.0 M guanidine chloride, 0.5 M NaCl). Cells were homogenized with Matrix C (MP Biomedicals) using a bead beater (MP Biomedicals, FastPrep-24 5 G). Cell lysates were clarified by centrifugation at 6000 rpm for 7 min and protein concentration was determined using a DC protein assay kit (Bio-Rad). Samples containing equal amounts of protein were brought to a total volume of 1 ml with appropriate buffer.
In vivo sumoylation was assayed in crude yeast extracts using nickel-nitrilotriacetic acid (Ni-NTA) agarose beads to pull down His-HA-tagged Cse4 as described previously (Ohkuni et al. 2015) with modifications. Cell lysates were incubated with 100 µl of Ni-NTA superflow beads (Qiagen, 30430) overnight at 4°C. After being washed with guanidine buffer one time and with breaking buffer (0.1 M Tris-HCl at pH 8.0, 20% glycerol, 1 mM PMSF) five times, beads were incubated with 2x Laemmli buffer including imidazole at 100°C for 5 min. The protein samples were analyzed by SDS-PAGE and western blotting. Primary antibodies were anti-HA (12CA5) mouse (Roche, 11583816001), anti-Smt3 (y-84) rabbit (Santa Cruz Biotechnology, sc-28649), anti-c-Myc (A-14) rabbit (Santa Cruz Biotechnology, sc-789), anti-FLAG mouse (Sigma, F3165), and anti-Tub2 rabbit (Basrai laboratory). Secondary antibodies were ECL Mouse IgG, HRP-Linked Whole Ab (GE Healthcare Life Sciences, NA931V) or ECL Rabbit IgG, HRP-linked Whole Ab (GE Healthcare Life Sciences, NA934V). Protein levels were quantified using Image Lab software (version 6.0.0) from Bio-Rad Laboratories, Inc. (Hercules).
ChIP-qPCR
Chromatin immunoprecipitations were performed with two biological replicates per strain as previously described (Cole et al. 2014; Chereji et al. 2017; Eisenstatt et al. 2020) with modifications. Logarithmic phase cultures were grown in raffinose/galactose (2% final concentration each) media for 4 h and were treated with formaldehyde (1% final concentration) for 20 min at 30°C followed by the addition of 2.5 M glycine for 10 min at 30°C. Cell pellets were washed twice with 1 X PBS and resuspended in 2 mL FA Lysis Buffer (1 mM EDTA pH8.0, 50 mM HEPES-KOH pH7.5, 140 mM NaCl, 0.1% sodium deoxycholate, 1% Triton X-100) with 1 × protease inhibitors (Sigma) and 1 mM PMSF (final concentration). The cell suspension was split into four screw top tubes with glass beads (0.4–0.65 mm diameter) and lysed in a FastPrep-24 5 G (MP Biosciences) for 40 seconds three times, allowed to rest on ice for 5 min, and lysed two final times for 40 s each. The cell lysate was collected, and the chromatin pellet was washed in FA Lysis Buffer twice. Each pellet was resuspended in 600 µl of FA Lysis Buffer and combined into one 5 ml tube. The chromatin suspension was sonicated with a Branson digital sonifer 24 times at 20% amplitude with a repeated 15 s on/off cycle. After 3 min of centrifugation (13,000 rpm, 4°C), the supernatant was transferred to another tube. Input sample was removed (5%) and the average size of the DNA was analyzed. The remaining lysate was incubated with anti-HA-agarose beads (Sigma, A2095) or anti-H3 (Millipore 04-928) bound, or anti-H4 (Millipore 04-858) bound protein A magnetic beads overnight at 4–8°C. The beads were washed in 1 ml FA, FA-HS (500 mM NaCl), RIPA, and TE buffers for 5 min on a rotor two times each. The beads were suspended in ChIP Elution Buffer (25 mM Tris-HCl pH7.6, 100 mMNaCl, 0.5% SDS) and incubated at 65°C overnight. The beads were treated with proteinase K (0.5 mg/ml) and incubated at 55°C for 4 h followed by Phenol/Chloroform extraction and ethanol precipitation. The DNA pellet was resuspended in a total of 50 µl sterile water. Samples were analyzed by quantitative PCR (qPCR) performed with the 7500 Fast Real Time PCR System with Fast SYBR Green Master Mix (Applied Biosystems). qPCR conditions used: 95°C for 20 s; 40 cycles of 95°C for 3 s, 60° for 30 s. For Figure 3 and Supplementary Figure S4, the enrichment was measured as the percent input. For Supplementary Figure S5, relative protein occupancy was measured as the percent input where the relative occupancy of Cse4 and H3 at the indicated genomic locus were normalized against the relative occupancy of H4.
Figure 3.
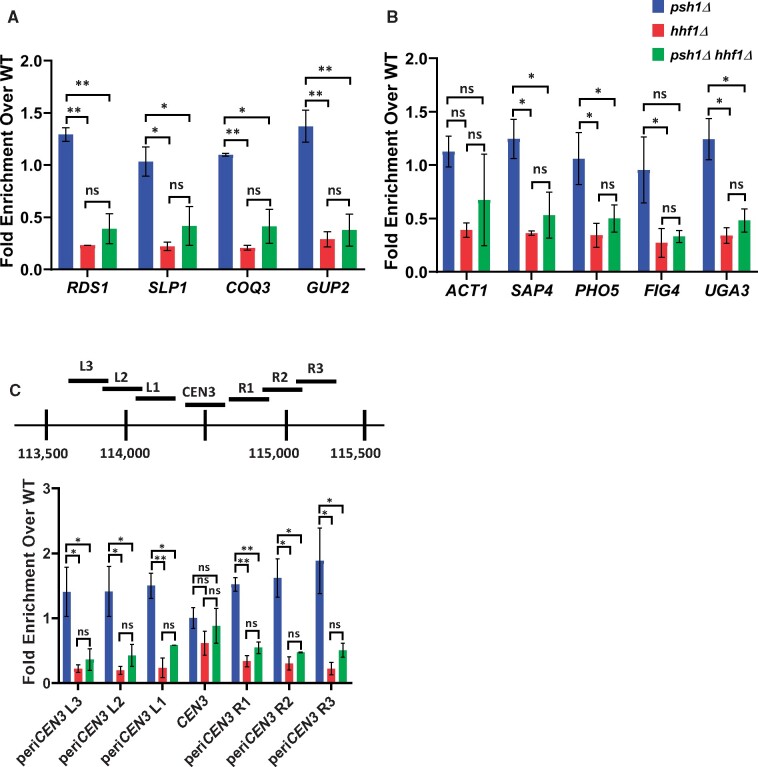
Deletion of the HHF1 allele reduces enrichment of Cse4 at peri-centromeric and noncentromeric regions. (A–C) ChIP-qPCR was performed on chromatin lysate from wild-type (YMB9804), psh1Δ (YMB10479), hhf1Δ (YMB10937), and psh1Δ hhf1Δ (YMB10822) strains transiently overexpressing GAL1-6His-3HA-CSE4 (pMB1458). Enrichment of 6His-3HA-Cse4 is shown as a fold over wild-type. Displayed are the mean of two independent experiments. Error bars represent standard deviation of the mean. **P < 0.0099, *P < 0.09, ns, not significant. (A and B) Levels of Cse4 enrichment are reduced at noncentromeric regions when HHF1 is deleted. qPCR at A: ACT1, SAP4, RDS1, SLP1, and PHO5 and B: FIG4, COQ3, GUP2, and UGA3. (C) Cse4 enrichment is reduced at peri-centromeric, but not the core centromere, regions in hhf1Δ strains. Top: A diagram of the peri-centromere and centromere of Chromosome III analyzed by ChIP-qPCR. Horizontal lines represent the regions amplified. Bottom: Enrichment of 6His-3HA-Cse4 at the core centromere and the left and right peri-centromeric regions on Chromosome III.
Primers used are listed in Supplementary Table S4.
Plasmid loss
Plasmid loss assays were performed for strains overexpressing Cse4 as in (Metzger et al. 2017) with minor modifications. Strains were grown in media selective for the plasmid (SC-Leu) with raffinose/galactose (2% final concentration each) for 24 h. Appropriate dilutions were plated on to rich media or selective media plates (0-h timepoint). Cultures were diluted into nonselective minimal media (SC+Leu) with raffinose/galactose (2% final concentration each) for 24 h, re-diluted into fresh nonselective media for an additional 24 h, and appropriate dilutions were plated on to rich media or selective media plates (48-h timepoint). Plates were incubated for two to three days and colonies were counted. Plasmid loss was determined as a percentage of colonies grown on selective media plates vs colonies grown on nonselective media plates at each timepoint.
Reverse transcription PCR
Cells were grown in 2% raffinose synthetic complete medium at 30°C to mid-logarithmic phase. Galactose was added to the media to a final concentration of 2% to induce CSE4 expression from the GAL promoter for 4 h at 30°C. Total RNAs were isolated from 1.5 OD600 equivalent cells using MasterPureTM Yeast RNA purification kit with DNase I treatment as indicated by the manufacturer (Epicentre). Total RNAs (100 ng for SIZ1 and SIZ2, 10 ng for SMT3, and 5 ng for UBI4) were analyzed by the AccessQuickTM RT-PCR system (Promega). M-MLV Reverse Transcriptase (Promega) or AMV Reverse Transcriptase (Promega) was used. Primer sets are listed in Supplementary Table S4. PCR conditions used are: 45°C for 45 min (Reverse Transcription); 95°C for 2 min (Initial denaturation); 28 cycles of 95°C for 30 s, 55°C for 1 min, 68°C for 1 min; and 68°C for 5 min (Final extension). PCR products were loaded onto Ethidium Bromide-stained 1.5% agarose gels in TBE (KD Medical) and band intensities were quantified with Image Lab software (version 6.0.0) from Bio-Rad Laboratories, Inc. Expression levels from two biological repeats were calculated based on the standard curve run on the same gel and relative values were determined with the levels in wild-type defined as 1.
Data availability
Strains and plasmids are available upon request. Supplementary Figures S1–S10 are available as JPG files. Supplementary Table S1 is an Excel file that describes mutations that suppress the psh1Δ GALCSE4 SDL, the gene systematic name, the gene name, the functional category, growth and colony scores, and validation information if applicable. Supplemental File S1 contains Supplementary Tables S2–S4 which describe the yeast strains, plasmids, and primers used in this study, respectively. Supporting information is available at figshare: https://doi.org/10.25386/genetics.13713937.
Results
A genome-wide screen identified suppressors of the SDL in a psh1Δ GALCSE4 strain
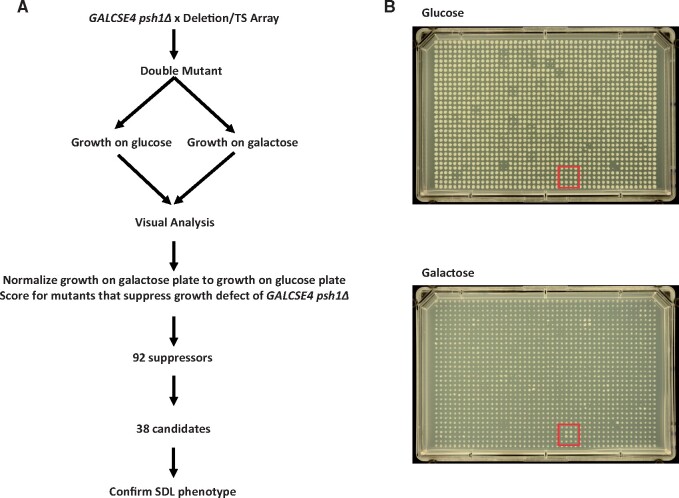
Identifying pathways that facilitate the deposition of overexpressed Cse4 to noncentromeric regions will provide insight into the mechanisms that promote CIN in CENP-A overexpressing cancers. Deletion of PSH1, which regulates ubiquitin-mediated proteolysis of overexpressed Cse4, results in SDL when Cse4 is overexpressed (GALCSE4) (Hewawasam et al. 2010; Ranjitkar et al. 2010). We reasoned that strains with deletions or mutations of factors that promote Cse4 mislocalization would rescue the SDL of a psh1Δ GALCSE4 strain. Therefore, we generated a psh1Δ query strain overexpressing CSE4 from a galactose-inducible plasmid and mated it to arrays of 3827 nonessential gene deletion strains and 786 conditional mutant alleles, encoding 560 essential genes, and 186 nonessential genes for internal controls (Costanzo et al. 2016). Growth of the haploid meiotic progeny plated in quadruplicate was visually scored on glucose-and galactose-containing media grown at 30°C for nonessential and 26°C for essential gene mutant strains (Figure 1A). Highlighted in the figure are all four replicates of deletion of histone H4 (hhf1Δ) showing better growth on galactose media compared to the control strains along the perimeter and other deletion strains on the plate (Figure 1B). Strains that suppress the psh1Δ GALCSE4 SDL on galactose-containing media were given a growth score of one (low suppression) to four (high suppression) (Supplementary Table S1). The number of replicates within the quadruplicate that displayed the same growth were given a colony score of one (one out of four replicates) to four (all four replicates). We identified 94 deletion and mutant alleles encoding 92 genes that suppressed the psh1Δ GALCSE4 SDL and the majority (81%) of quadruplicates had all four colonies displaying the same level of suppression, indicated by a colony score of four (Supplementary Table S1).
Figure 1.
A genome-wide screen identified suppressors of the psh1Δ GALCSE4 SDL. (A) Schematic for the genome-wide screen. A psh1Δ strain (YMB8995) transformed with GAL1-6His-3HA-CSE4 (pMB1458) was mated to an array of nonessential gene deletions and an array of conditional alleles of essential genes. Growth of the haploid meiotic progeny plated in quadruplicate was visually scored on glucose- and galactose-containing media grown at 30°C for nonessential and 26°C for essential gene mutant strains. Ninety-two genes were identified as growing better on galactose-containing media than the psh1Δ GALCSE4 strain. Thirty-eight candidate genes were selected for confirmation of suppression of lethality. (B) Representative plates from the genome-wide screen. Shown is Plate 01 of the nonessential gene deletion array. The mutant strains were spotted in quadruplicate on selective media plates containing glucose (top) or galactose (bottom). The red box highlights psh1Δ hhf1Δ which displayed improved growth on galactose-containing plates compared to the psh1Δ GALCSE4 control strain (perimeter of plate) and did not show a growth defect or improved growth on the glucose plates.
Of the 94 alleles, we selected 38 candidate mutants (14 nonessential deletion strains and 24 conditional mutants) to confirm the suppression of the psh1Δ GALCSE4 SDL (Table 1). These candidates displayed a growth score of three or four where most of the replicates displayed high suppression and represent pathways involved in RNA processing and cleavage, DNA repair, chromatin remodeling, histone modifications, and DNA replication (Table 1). Secondary validation of the SDL suppressors was done by independently generating double mutant strains of psh1Δ GALCSE4 with candidate mutants. Growth assays were performed on media selective for the GALCSE4 plasmid and containing either glucose or raffinose and galactose. We used a hir2Δ psh1Δ strain as a negative control because hir2Δ psh1Δ GALCSE4 strains display SDL (Ciftci-Yilmaz et al. 2018). Of the 38 strains tested, 29 showed almost complete suppression, five strains showed a partial suppression, and four did not suppress the SDL on galactose media (Table 1 and Supplementary Table S1 and Supplementary Figure S1, A and B). We further tested a subset of the 38 genes to confirm overexpression of CSE4 and found that strains with mutations in genes involved in RNA processing and transcription do not show galactose-induced expression of CSE4 (Supplementary Table S1 and hchSupplementary Figure S1C), indicating that these are false positive hits. Through secondary validation, we confirmed that 89% of the candidate mutants tested suppressed the psh1Δ GALCSE4 SDL.
Table 1.
Candidate double mutant strains with the indicated mutant allele combined with psh1Δ GALCSE4 were generated and used for secondary validation using growth assays
| Allele | Systematic name | Gene name | Standard name | Growth score | Colony score | SDL suppression |
|---|---|---|---|---|---|---|
| Nonessential | ||||||
| hhf1Δ | YBR009C | HHF1 | Histone H4 | 3 | 4 | Y |
| hhf2Δ | YNL030W | HHF2 | Histone H4 | 3 | 4 | Y |
| ies2Δ | YNL215W | IES2 | Ino Eighty Subunit | 2 | 3 | N |
| arp8Δ | YOR141C | ARP8 | Actin-Related Protein | 3 | 4 | Y |
| swc5Δ | YBR231C | SWC5 | SWr Complex | 1 | 4 | N |
| eaf1Δ | YDR359C | EAF1 | Esa1p-Associated Factor | 2 | 3 | Partial |
| eap1Δ | YKL204W | EAP1 | EIF4E-Associated Protein | 2 | 4 | Y |
| cse2Δ | YNR010W | CSE2 | Chromosome SEgregation | 2 | 3 | Partial |
| cse2Δ_tsa | YNR010W | CSE2 | Chromosome SEgregation | 3 | 3 | Y |
| mrm2Δ | YGL136C | MRM2 | Mitochondrial rRNA Methyl transferase | 2 | 3 | N |
| hap3Δ | YBL021C | HAP3 | Heme Activator Protein | 3 | 4 | Partial |
| hap5Δ | YOR358W | HAP5 | Heme Activator Protein | 3 | 4 | Y |
| rpl6bΔ | YLR448W | RPL6B | Ribosomal Protein of the Large subunit | 2 | 3 | Y |
| rad4Δ | YER162C | RAD4 | RADiation sensitive | 2 | 3 | Partial |
| rad14Δ | YMR201C | RAD14 | RADiation sensitive | 2 | 2 | Y |
| Essential | ||||||
| act1-132 | YFL039C | ACT1 | ACTin | 3 | 4 | N |
| mob1-5001 | YIL106W | MOB1 | Mps One Binder | 4 | 4 | Y |
| tbf1-5001 | YPL128C | TBF1 | TTAGGG repeat-Binding Factor | 3 | 4 | Y |
| csl4-5001 | YNL232W | CSL4 | Cep1 Synthetic Lethal | 4 | 4 | Y |
| pop4-5001 | YBR257W | POP4 | Processing Of Precursor RNAs | 4 | 4 | Y |
| orc1-5001 | YML065W | ORC1 | Origin Recognition Complex | 4 | 4 | Y |
| orc6-5001 | YHR118C | ORC6 | Origin Recognition Complex | 4 | 4 | Y |
| cft2-1 | YLR115W | CFT2 | Cleavage Factor Two | 3 | 4 | Partial |
| cft2-5001 | YLR115W | CFT2 | Cleavage Factor Two | 4 | 4 | Y |
| clp1-5001 | YOR250C | CLP1 | CLeavage/Polyadenylation factor Ia subunit | 4 | 3 | Y |
| ipa1-5001 | YJR141W | IPA1 | Important for cleavage and PolyAdenylation | 3 | 4 | Y |
| hrp1-1 | YOL123W | HRP1 | Heterogenous nuclear RibonucleoProtein | 2 | 4 | Y |
| rpb5-5001 | YBR154C | RPB5 | RNA Polymerase B | 4 | 4 | Y |
| rpc17-5001 | YJL011C | RPC17 | RNA Polymerase C | 4 | 4 | Y |
| pol31-5001 | YJR006W | POL31 | POLymerase | 4 | 4 | Y |
| srp54-5001 | YPR088C | SRP54 | Signal Recognition Particle 54-kD subunit | 4 | 4 | Y |
| dbp6-5001 | YNR038W | DBP6 | Dead Box Protein | 4 | 4 | Y |
| dbp9-5001 | YLR276C | DBP9 | Dead Box Protein | 4 | 4 | Y |
| yef3-f650s | YLR249W | YEF3 | Yeast Elongation Factor | 2 | 4 | Y |
| cdc5-1 | YMR001C | CDC5 | Cell Division Cycle | 2 | 4 | Y |
| cdc31-1 | YOR257W | CDC31 | Cell Division Cycle | 2 | 4 | Y |
| hrr25-5001 | YPL204W | HRR25 | HO and Radiation Repair | 3 | 4 | Y |
| ost2-5001 | YOR103C | OST2 | OligoSaccharylTransferase | 4 | 4 | Y |
Indicated is the allele analyzed, systematic name, gene name, standard name, visual scoring from the primary screen for growth score (from 1 to 4) and colony score (from 1 to 4), and suppression of SDL (Y: SDL was suppressed; N: SDL was not suppressed; Partial: SDL was partially suppressed).
We initiated our studies with the INO80 chromatin remodeling complex as our screen identified deletion and mutant alleles corresponding to three components of the INO80 complex, Ies2, Arp8, and Act1 (Poch and Winsor 1997; Shen et al. 2000, 2003; Tosi et al. 2013). Secondary validation assays showed that arp8Δ did suppress the psh1Δ GALCSE4 SDL (Supplementary Figures S1A and S2A), however, the arp8Δ strain displayed polyploidy when analyzed by Fluorescent Activated Cell Sorting (FACS) (Supplementary Figure S2B) and we consequently did not pursue further studies with the INO80 complex.
Deletion of histone H4 alleles suppresses the SDL of a psh1Δ GALCSE4 strain
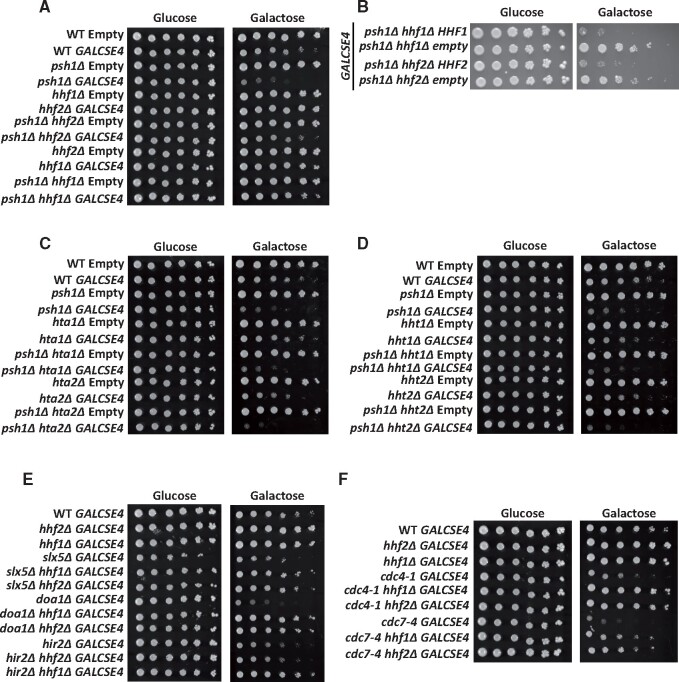
Two nonallelic loci, HHT1/HHF1 and HHT2/HHF2, encode identical H3 and H4 proteins in budding yeast. The screen identified the deletion of either one of the histone H4 alleles, HHT1/hhf1Δ (hhf1Δ) or HHT2/hhf2Δ (hhf2Δ), as among the most prominent suppressors of the psh1Δ GALCSE4 SDL. A role for the dosage of histone H4-encoding genes in mislocalization of Cse4 has not yet been reported. We confirmed that the hhf1Δ and hhf2Δ strains do not exhibit defects in ploidy or cell cycle by FACS analysis (Supplementary Figure S3). Growth assays confirmed that psh1Δ hhf1Δ GALCSE4 and psh1Δ hhf2Δ GALCSE4 strains plated on galactose media do not exhibit SDL (Figure 2A). We determined that the phenotype was linked to deletion of the H4 alleles because transformation of a plasmid with the respective wild type histone H4 gene into the psh1Δ hhf1Δ or psh1Δ hhf2Δ strains restored the SDL observed in the psh1Δ GALCSE4 strain (Figure 2B).
Figure 2.
Deletion of H4 genes suppresses GALCSE4 SDL. Three independent isolates for each strain were assayed and shown is a representative for each. (A) The psh1Δ GALCSE4 SDL is suppressed by deletion of HHF1 or HHF2. Growth assays of wild type, psh1Δ, hhf1Δ, hhf2Δ, psh1Δ hhf1Δ, and psh1Δ hhf2Δ strains with empty vector (pMB433; YMB9802, YMB10478, YMB10825, YMB11166, YMB10821, and YMB10823, respectively) or GAL1-6His-3HA-CSE4 (pMB1458; YMB9803, YMB10479, YMB10937, YMB10938, YMB10822, and YMB10824, respectively). Cells were spotted in fivefold serial dilutions on glucose (2% final concentration) or raffinose/galactose (2% final concentration each) media selective for the plasmid and grown at 30°C for 3–5 days. (B) The psh1Δ GALCSE4 SDL suppression is linked to the hhf1Δ and hhf2Δ alleles. Growth assays of psh1Δ hhf1Δ (YMB10824) and psh1Δ hhf2Δ (YMB10822) strains with GAL1-6His-3HA-CSE4 (pMB1458) transformed with empty vector (pRS425) or a plasmid containing wild type HHF1 (pMB1928) or HHF2 (pMB1929). Strains were assayed as described above in (A). (C) and (D) Deletion of genes encoding histones H2A (C) or H3 (D) does not suppress the SDL of a psh1Δ GALCSE4 strain. Growth assays of wild-type, psh1Δ, and (C) hta1Δ, hta2Δ, psh1Δ hta1Δ, psh1Δ hta2Δ, or (D) hht1Δ, hht2Δ, psh1Δ hht1Δ, and psh1Δ hht1Δ strains with empty vector (pMB433; YMB9802, YMB10478, YMB11258, YMB11266, YMB11260, YMB11268, YMB11274, YMB11282, YMB11276, and YMB11284, respectively) or GAL1-6His-3HA-CSE4 (pMB1458: YMB9803, YMB10479, YMB11262, YMB11270, YMB11264, YMB11272, YMB11278, YMB11286, YMB11280, and YMB11288, respectively). Strains were assayed as described above in (A). (E) Reduced gene dosage of H4 suppresses the SDL of slx5Δ, doa1Δ, and hir2Δ GALCSE4 strains. Growth assays of wild type (YMB9803), hhf1Δ (YMB10937), hhf2Δ (YMB10938), slx5Δ (YMB10963), slx5Δ hhf1Δ (YMB11046), slx5Δ hhf2Δ (YMB11047), doa1Δ (YMB11032), doa1Δ hhf1Δ (YMB11050), doa1Δ hhf2Δ (YMB11053), hir2Δ (YMB8332), hir2Δ hhf1Δ (YMB11107), hir2Δ hhf2Δ (YMB11105) strains expressing GAL1-6HIS-3HA-CSE4 (pMB1458). Strains were assayed as described above in (A) and grown at 30°C for 3–5 days. (F) Deletion of HHF1 or HHF2 suppresses the SDL of cdc4-1 and cdc7-4 GALCSE4 strains. Growth assays of wild-type (YMB9803), hhf1Δ (YMB10937), hhf2Δ (YMB10938), cdc4-1 (YMB9756), cdc4-1 hhf1Δ (YMB11051), cdc4-1 hhf2Δ (YMB11054), cdc7-4 (YMB9760), cdc7-4 hhf1Δ (YMB11052), and cdc7-4 hhf2Δ (YMB11055) with GAL1-6His-3HA-CSE4 (pMB1458). Strains were assayed as described above in (A) and grown at 23°C for 3–5 days.
We next investigated if deletion of a single allele for either histone H3 or H2A genes could suppress the SDL of a psh1Δ GALCSE4 strain. Note that the two nonallelic loci, HTA1/HTB1 and HTA2/HTB2, encode almost identical H2A and H2B proteins. Deletion of HTA1 (hta1Δ/HTB1), HTA2 (hta2Δ/HTB2), HHT1 (hht1Δ/HHF1), or HHT2 (hht2Δ/HHF2) did not suppress the SDL of a psh1Δ GALCSE4 strain on galactose media (Figure 2, C and D and Table 2). Based on these results we conclude that the suppression of psh1Δ GALCSE4 SDL is specific to the reduced gene dosage of H4.
Table 2.
Summary of the SDL growth phenotypes of mutants that exhibit SDL with GALCSE4 and combined with hhf1Δ or hhf2Δ
| Protein function | Relevant strain genotype | Growth with GALCSE4 |
|---|---|---|
| WT | ++ | |
| Histone H4 | hhf1Δ | +++ |
| hhf2Δ | +++ | |
| Histone H2A | hta1Δ | ++ |
| hta2Δ | ++ | |
| Histone H3 | hht1Δ | ++ |
| hht2Δ | ++ | |
| E3 Ubiquitin Ligase | psh1Δ | — |
| psh1Δ hhf1Δ | +++ | |
| psh1Δ hhf2Δ | +++ | |
| psh1Δ hhf1Δ + HHF1 | — | |
| psh1Δ hhf2Δ + HHF2 | — | |
| psh1Δ hta1Δ | — | |
| psh1Δ hta2Δ | — | |
| psh1Δ hht1Δ | — | |
| psh1Δ hht2Δ | — | |
| SUMO-Targeted Ubiquitin Ligase | slx5Δ | — |
| slx5Δ hhf1Δ | +++ | |
| slx5Δ hhf2Δ | +++ | |
| Ubiquitin Binding | doa1Δ | — |
| doa1Δ hhf1Δ | +++ | |
| doa1Δ hhf2Δ | +++ | |
| HIR Nucleosome Binding Complex | hir2Δ | — |
| hir2Δ hhf1Δ | ++ | |
| hir2Δ hhf2Δ | — | |
| F-box of the SCF Complex | cdc4-1 | — |
| cdc4-1 hhf1Δ | +++ | |
| cdc4-1 hhf2Δ | +++ | |
| Dbf4-Dependent Kinase | cdc7-4 | — |
| cdc7-4 hhf1Δ | ++ | |
| cdc7-4 hhf2Δ | ++ |
Shown is the protein function, relevant strain genotype, and growth with GALCSE4. Wild type growth is indicated as ++; SDL as — and extent of suppression (++ or +++).
Reduced gene dosage of H4 suppresses the SDL of slx5Δ, doa1Δ, hir2Δ, cdc4-1, and cdc7-4 GALCSE4 strains
To determine if the SDL suppression by reduced H4 gene dosage is limited to the psh1Δ GALCSE4 strain, we deleted HHF1 or HHF2 in deletion or mutant strains encoding Slx5, Doa1, Hir2, Cdc4, and Cdc7 as deletion or mutation of these factors show SDL with GALCSE4 and mislocalization of transiently overexpressed Cse4 (Au et al. 2013, 2020; Ohkuni et al. 2016; Ciftci-Yilmaz et al. 2018; Eisenstatt et al. 2020). Growth on galactose media revealed that the SDL of doa1Δ, slx5Δ, cdc4-1, and cdc7-4 GALCSE4 strains is suppressed when either HHF1 or HHF2 is deleted (Figure 2, E and F and Table 2), while the SDL of hir2Δ GALCSE4 is suppressed only when HHF1 is deleted (Figure 2E and Table 2). These results suggest that the gene dosage of H4 contributes to the SDL of mutants that exhibit defects in Cse4 proteolysis and mislocalizes Cse4 to noncentromeric regions.
Reduced gene dosage of H4 reduces the mislocalization of Cse4 in psh1Δ strains
The SDL phenotype of psh1Δ GALCSE4 strains is correlated with the mislocalization of Cse4 to noncentromeric regions (Hewawasam et al. 2010; Ranjitkar et al. 2010). We examined if the suppression of SDL in the psh1Δ hhf1Δ GALCSE4 or psh1Δ hhf2Δ GALCSE4 strains is due to reduced mislocalization of Cse4. We performed ChIP-qPCR to assay the localization of Cse4 using chromatin from wild-type, psh1Δ, hhf1Δ, hhf2Δ, psh1Δ hhf1Δ, and psh1Δ hhf2Δ strains transiently overexpressing CSE4. In agreement with previously published data (Hildebrand and Biggins 2016; Hewawasam et al. 2018; Ohkuni et al. 2020), we found that Cse4 enrichment at noncentromeric regions such as the promoters of RDS1, SLP1, GUP2, and COQ3 is higher in the psh1Δ strain compared to the wild type strain (Figure 3, A and B; Supplementary Figure S4, A and B). In contrast, deletion of HHF1 in a wild type strain or when combined with psh1Δ showed reduced levels of Cse4 enrichment at these regions (Figure 3, A and B). Results for ChIP-qPCR with the hhf2Δ strain also showed reduced levels of Cse4 at noncentromeric loci (Supplementary Figure S4A and S4B). Consistent with previous studies (Hildebrand and Biggins 2016), we observed higher levels of Cse4 at peri-centromeric regions in a psh1Δ strain (Figure 3C and Supplementary Figure S4C). However, we observed reduced levels of Cse4 at peri-centromeric regions in psh1Δ hhf1Δ and psh1Δ hhf2Δ strains when compared to the psh1Δ strain (Figure 3C and Supplementary Figure S4C). Localization of Cse4 to the centromere was not significantly altered in hhf1Δ, hhf2Δ, psh1Δ hhf1Δ, and psh1Δ hhf2Δ strains (Figure 3C and Supplementary Figure S4C). We confirmed the reduced Cse4 occupancy at noncentromeric regions in the psh1Δ hhf1Δ strain when levels of Cse4 are normalized to levels of H4 (Supplementary Figure S5A). We next examined if the mislocalization of Cse4 affects the occupancy of H3-H4 nucleosomes at noncentromeric regions. ChIP-qPCR of H3 normalized to H4 at selected regions did not show a significant difference between psh1Δ and psh1Δ hhf1Δ strains (Supplementary Figure S5B, ACT1, SAP4, and RDS1). Taken together, our results show that reduced gene dosage of H4 contributes to reduced levels of Cse4 at noncentromeric and peri-centromeric regions in psh1Δ strains.
Scm3 is the primary chaperone for centromeric deposition of Cse4 and strains depleted for Scm3 are not viable (Camahort et al. 2007). However, overexpression of Cse4 can rescue the growth defect of Scm3-depleted cells, suggesting that nonScm3-based mechanisms can promote centromeric deposition of overexpressed Cse4 (Hewawasam et al. 2018). Our studies so far have shown that reduced gene dosage of H4 contributes to suppression of Cse4 mislocalization to noncentromeric regions. We next asked if the reduced gene dosage of H4 would affect the Scm3-independent centromeric deposition of Cse4 by assaying the growth of Scm3-depleted cells that overexpress CSE4. In these strains, expression of Scm3 is regulated by a galactose-inducible promoter and is only expressed when grown in galactose medium, but not in glucose medium. Overexpression of Cse4 from a copper-inducible promoter can suppress the growth defect caused by depletion of Scm3 on copper-containing medium (Hewawasam et al. 2018). We constructed hhf2Δ GAL-SCM3 Cu-CSE4 strains and performed Western blot analysis to confirm the induced overexpression of Cse4 in these strains when grown in copper-containing medium (Supplementary Figure S4D). Growth assays showed that deletion of HHF2 resulted in poor growth of cells when Cse4 is overexpressed in Scm3-depleted strains (Supplementary Figure S4E, glucose + 0.5 mM Cu). We conclude that physiological levels of histone H4 are required for centromeric association of Cse4 in cells depleted of Scm3 and for mislocalization of Cse4 to peri-centromeric and noncentromeric regions in psh1Δ strains.
Deletion of HHF1 contributes to reduced stability of Cse4 in a psh1Δ strain
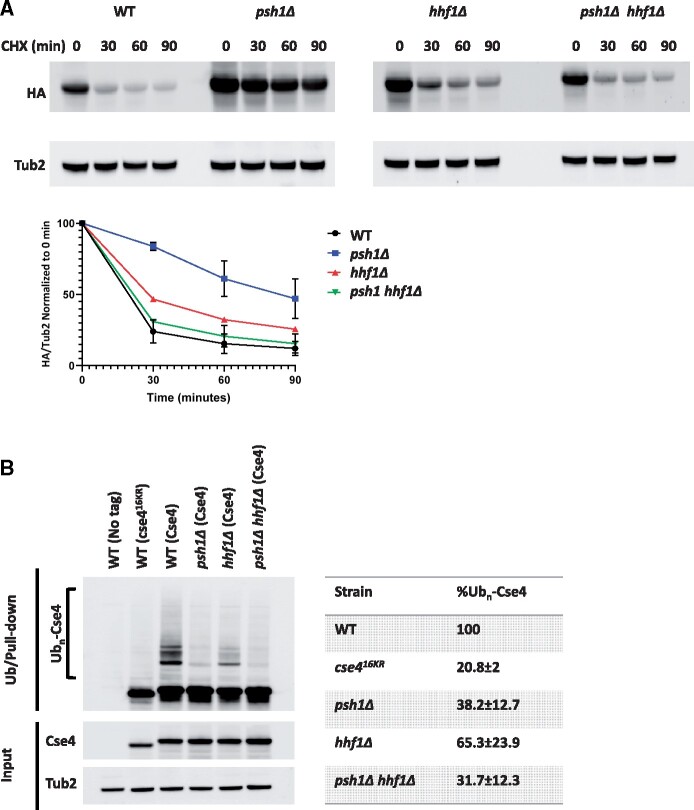
The SDL phenotype and mislocalization of Cse4 in a psh1Δ GALCSE4 strain is associated with a higher stability of Cse4 (Hewawasam et al. 2010; Ranjitkar et al. 2010). The suppression of the psh1Δ GALCSE4 SDL and the reduced mislocalization of Cse4 by hhf1Δ led us to hypothesize that the stability of Cse4 would be reduced in a psh1Δ hhf1Δ strain. Protein stability assays showed that, in agreement with previous studies (Hewawasam et al. 2010; Ranjitkar et al. 2010), transiently overexpressed Cse4 is highly stable in the psh1Δ strain when compared to that observed in a wild type strain. The stability of Cse4 was not significantly affected in the hhf1Δ strain when compared to the wild type strain. Consistent with our hypothesis, we observed reduced stability of Cse4 in the psh1Δ hhf1Δ strain compared to the psh1Δ strain (Figure 4A). These results show a correlation between suppression of SDL of a psh1Δ GALCSE4 strain, lower levels of mislocalized Cse4 at noncentromeric regions, and reduced stability of Cse4 due to reduced gene dosage of H4.
Figure 4.
Deletion of HHF1 contributes to reduced stability and ubiquitin-independent proteolysis of Cse4 in a psh1Δ strain. (A) hhf1Δ strains contribute to reduced stability of Cse4 in a psh1Δ strain. Western blot analysis of protein extracts from wild-type (YMB9803), psh1Δ (YMB10479), hhf1Δ (YMB10938), and psh1Δ hhf1Δ (YMB10824) strains transiently overexpressing GAL1-6His-3HA-CSE4 (pMB1458). Cells were grown to logarithmic phase in media selective for the plasmid and containing raffinose (2% final concentration) and induced with galactose (2% final concentration) for 4 h. Cultures were treated with cycloheximide (CHX, 10 µg/mL) and glucose (2%) and analyzed at the indicated time points. Extracts were analyzed by Western blot against HA (Cse4) and Tub2 as a loading control. Levels of 6His-3HA-Cse4 were normalized to Tub2 and the quantification of the percent remaining 6His-3HA-Cse4 after CHX treatment is shown in the graph. Error bars represent the SEM of two independent experiments. (B) Deletion of HHF1 does not increase ubiquitination of Cse4 in a psh1Δ strain. Ubiquitin-pull down assays were performed using protein extracts from wild type strains (BY4741) with no tag (pMB433) or overexpressing cse416KR (pMB1892) and from wild-type (YMB9803), psh1Δ (YMB10479), hhf1Δ (YMB10938), and psh1Δ hhf1Δ (YMB10824) strains overexpressing 6His-3HA-CSE4 (pMB1458). Lysates were incubated with Tandem Ubiquitin Binding Entity beads (LifeSensors) prior to analysis of ubiquitin-enriched samples by Western blot against HA and input samples against HA and Tub2 as a loading control. Poly-ubiquitinated Cse4 (Ubn-Cse4) is indicated by the bracket. HA levels in input samples were normalized to Tub2 levels and quantification of levels of Ubn-Cse4 were normalized to the levels of Cse4 in the input. The percentage of Ubn-Cse4 from two independent experiments with standard error is shown.
Because defects in the ubiquitin-proteasome mediated proteolysis of Cse4 contribute to its mislocalization and increased stability (Hewawasam et al. 2010; Ranjitkar et al. 2010), we investigated if deletion of HHF1 affects ubiquitination of Cse4 (Ubn-Cse4) in a psh1Δ strain. Ubiquitin pull-down assays were done to determine the levels of Ubn-Cse4 in wild type, psh1Δ, hhf1Δ, and psh1Δ hhf1Δ strains transiently overexpressing CSE4. Wild type strains expressing a nontagged Cse4 or a mutant form of Cse4 (cse416KR) that cannot be ubiquitinated, where the 16 lysine residues are mutated to arginine, were used as negative controls. As previously reported (Hewawasam et al. 2010; Ranjitkar et al. 2010), levels of Ubn-Cse4 were greatly reduced in the psh1Δ strain (38.2% ± 12.7) when compared to the wild type strain. The levels of Ubn-Cse4 in the psh1Δ hhf1Δ strain (31.7% ± 12.3) were similar to the psh1Δ strain, however, Ubn-Cse4 levels were decreased in the hhf1Δ strain (65.3% ± 23.9) compared to the levels in the wild type strain (Figure 4B). We propose that reduced mislocalization of Cse4 and ubiquitin-independent proteolysis of Cse4 contribute to reduced stability of Cse4 in a psh1Δ hhf1Δ GALCSE4 strain.
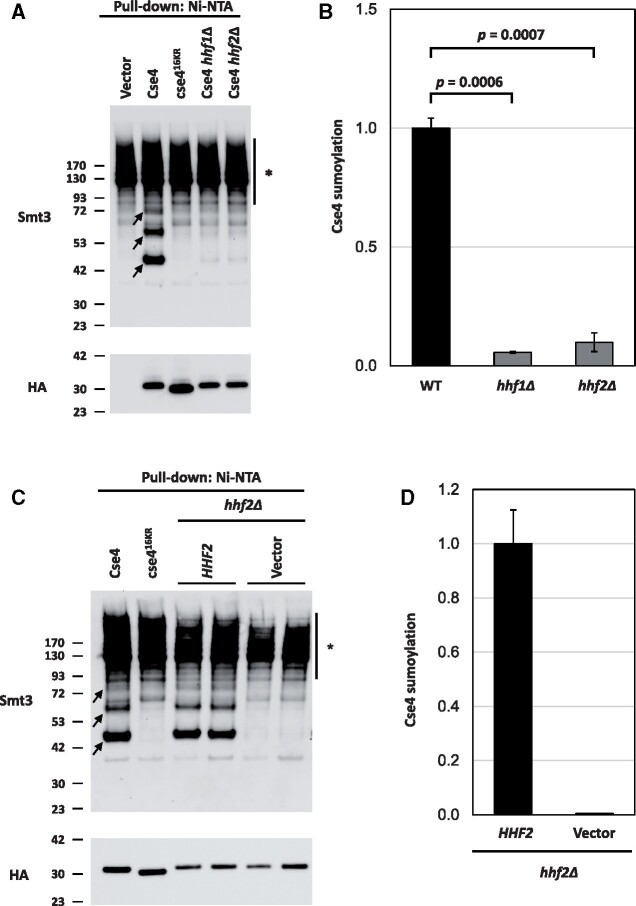
Reduced dosage of H4 is associated with defects in sumoylation of Cse4
We recently reported that Cse4 is sumoylated and that the sumoylation status of Cse4 at residues K215/216 correlates with the SDL of psh1Δ GALCSE4 strains (Ohkuni et al. 2020). Overexpression of the sumoylation-defective cse4K215/216R/A does not cause SDL in psh1Δ, slx5Δ, or hir2Δ strains; the lack of an SDL phenotype in the psh1Δ strain is due to reduced mislocalization and lower protein stability of cse4K215/216R/A. The phenotypic consequences related to defects in Cse4 sumoylation are similar to the ones we have observed due to reduced dosage of H4. We examined if sumoylation of Cse4 is affected due to reduced dosage of H4. Wild-type, hhf1Δ, and hhf2Δ GALCSE4 strains were assayed for Cse4 sumoylation. Consistent with previous results (Ohkuni et al. 2016, 2018, 2020), we detected sumoylated Cse4 as a pattern of three high molecular weight bands in wild type cells overexpressing wild type Cse4 but not in wild type cells expressing vector alone or overexpressing cse416KR (Figure 5A). Deletion of either histone H4 allele resulted in reduced levels of sumoylated Cse4 (Figure 5, A and B; P-value WT vs hhf1Δ = 0.0006, P-value WT vs hhf2Δ = 0.0007). To confirm that the reduction of sumoylated Cse4 is linked to deletion of the histone H4 allele, we assayed the levels of sumoylated Cse4 in hhf2Δ GALCSE4 strains transformed with an empty vector or with a plasmid borne HHF2. As expected, plasmid borne HHF2 restored the levels of sumoylated Cse4 to that observed in wild type cells (Figure 5, C and D). We conclude that physiological levels of histone H4 are required for Cse4 sumoylation.
Figure 5.
Histone H4 contributes to the sumoylation of Cse4. (A) Levels of sumoylated Cse4 are decreased in histone H4 deletion strains. Sumoylation levels were assayed on wild-type (BY4741) strains transformed with empty vector (pYES2), pGAL-8His-HA-CSE4 (pMB1345), or pGAL-8His-HA-cse416KR (pMB1344) and hhf1Δ (YMB10766) and hhf2Δ (YMB10767) strains transformed with pGAL-8His-HA-Cse4 (pMB1345). Sumoylated and nonmodified Cse4 were detected using cell lysates that were incubated with Ni-NTA beads followed by western blot analysis with antibodies against Smt3 and HA (Cse4), respectively. Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (B) Quantification of relative levels of sumoylated Cse4 in histone H4 deletion strains. Levels of sumoylated Cse4 were normalized to nonmodified Cse4 probed against HA in the pull-down sample. Statistical significance from two independent experiments was assessed by one-way ANOVA (P = 0.0004) followed by Tukey post-test (all pairwise comparisons of means). Error bars indicate average deviation from the mean. (C) The Cse4 sumoylation defect in a hhf2Δ strain is linked to the HHF2 allele. Sumoylation levels were determined from lysates from a hhf2Δ (YMB10767) strain with pGAL-8His-HA-CSE4 (pMB1345) transformed with vector (pRS425) or HHF2 (pMB1929) as described in (A). Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (D) Quantification of relative levels of sumoylated Cse4. Relative levels of sumoylated Cse4 were normalized to nonmodified Cse4 probed against HA in the pull-down sample. Error bars indicate average deviation from the mean from two biological replicates.
To eliminate the possibility that deletion of either histone H4 allele affects global sumoylation, we examined the sumoylation status of Ndc80, a kinetochore protein which has been characterized as a substrate for sumoylation (Montpetit et al. 2006; Ohkuni et al. 2015). Sumoylation levels of Ndc80 were examined in wild-type, hhf1Δ, and hhf2Δ strains expressing His-Flag-tagged Smt3 and Myc-tagged Ncd80 (Supplementary Figure S6A). His-Flag-Smt3 conjugates were purified using Ni-NTA beads, and the level of sumoylated proteins and sumoylated Ndc80 were determined using anti-Flag and anti-Myc antibodies, respectively. The levels of total sumoylated substrates were not significantly altered in the hhf1Δ and hhf2Δ strains compared to the wild-type (Supplementary Figure S6B). Consistent with previous results (Montpetit et al. 2006; Ohkuni et al. 2015), we observed sumoylated Ndc80 as a pattern of multiple bands in the wild-type strain (Supplementary Figure S6C). We did not observe a significant decrease in levels of sumoylated Ndc80 in hhf1Δ or hhf2Δ strains (Supplementary Figure S6, C and D). Reduced gene dosage of H4 specifically affects sumoylation of Cse4, but not of other substrates such as Ndc80.
We next performed reverse transcription PCR (RT-PCR) to examine the transcription level of genes associated with the SUMO pathway. Our results show that mRNA levels of the SUMO E3 ligases SIZ1 and SIZ2, which are responsible for sumoylating the majority of substrates, and the SUMO encoding gene SMT3 were not significantly altered in wild-type, psh1Δ, hhf1Δ, hhf2Δ, psh1Δ hhf1Δ, and psh1Δ hhf2Δ strains (Supplementary Figure S7, A–C). Given that overexpression of UBI4 partially suppresses the SDL of psh1Δ GALCSE4 strains (Au et al. 2013), we also examined the transcription of UBI4. We did not observe a significant difference in UBI4 transcription in the histone H4 deletion strains compared to the wild-type strain (Supplementary Figure S7D). Interestingly, increased levels of UBI4 mRNA were observed in the psh1Δ hhf1Δ strain compared to the wild-type strain (Supplementary Figure S7D, P < 0.01) despite the decreased Ubn-Cse4 in this strain (Figure 4B). These data are consistent with our conclusion that reduced mislocalization of Cse4 and ubiquitin-independent proteolysis of Cse4 contribute to reduced stability of Cse4 in a psh1Δ hhf1Δ GALCSE4 strain (Figure 4A). Taken together, these results show that defects in Cse4 sumoylation due to reduced dosage of H4 are not due to defects in the transcription of SUMO-pathway genes or sumoylation of nonCse4 substrates.
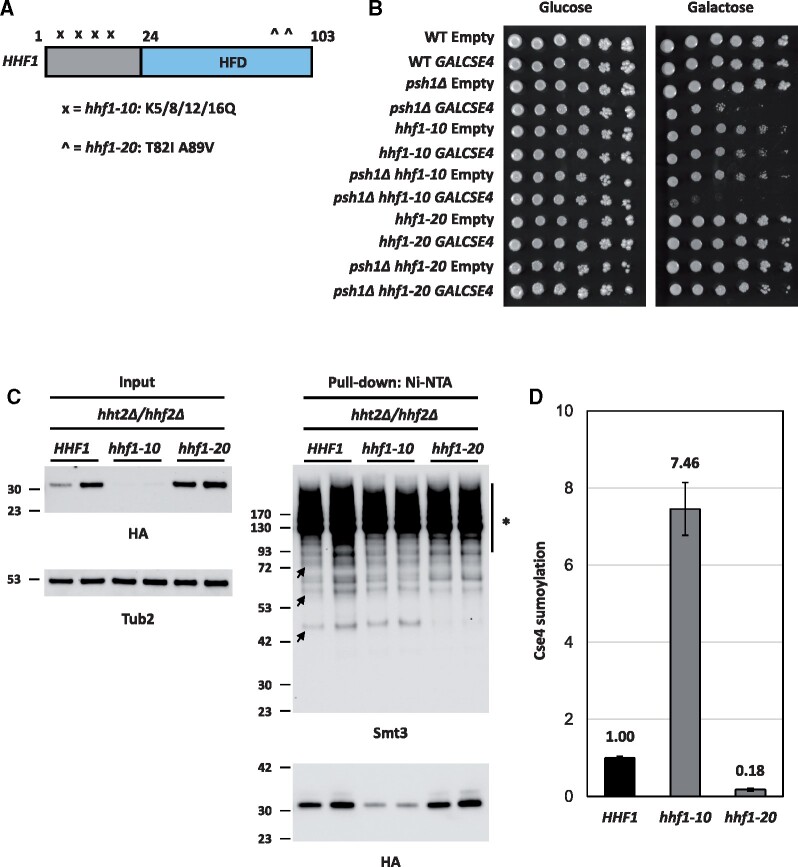
A histone H4 mutant defective for interaction with Cse4 suppresses the psh1Δ GALCSE4 SDL and shows defects in Cse4 sumoylation
Our results so far have shown that reduced gene dosage of H4 contributes to the suppression of the SDL phenotype, reduced stability of Cse4, decreased mislocalization of Cse4 in psh1Δ GALCSE4 strains, and defects in Cse4 sumoylation. We hypothesized that strains with defects in the interaction of H4 with Cse4 will display the same phenotypes that are observed due to reduced dosage of H4 in psh1Δ strains. To test our hypothesis, we used HHT1/hhf1 hht2Δ/hhf2Δ strains with mutations either in the N-terminal lysines (HHT1/hhf1-10) or in the histone fold domain (HHT1/hhf1-20) (Figure 6A) that have been well characterized by genetic and biochemical analysis (Smith et al. 1996; Glowczewski et al. 2000). The temperature sensitivity of the HHT1/hhf1-20 strain, but not the HHT1/hhf1-10 strain, is suppressed by overexpression of Cse4 and the HHT1/hhf1-20 strain is proposed to have defects in the formation of the Cse4-H4 dimer (Smith et al. 1996; Glowczewski et al. 2000). We deleted PSH1 in the same genetic background as the HHT1/HHF1, HHT1/hhf1-10, and HHT1/hhf1-20 strains, transformed these strains with CSE4 on a galactose-inducible plasmid, and performed growth assays. Compared to wild-type strains with a single copy of genes encoding histones H3/H4, HHT1/HHF1 psh1Δ strains display SDL on galactose medium when Cse4 is overexpressed, though to a less prominent degree compared to strains expressing both alleles encoding H3/H4 (compare Figure 6B to Figure 2A, psh1Δ GALCSE4). The relative decrease in SDL may be due to the expression of a single copy of the genes encoding histones H3/H4 in the strain background. The HHT1/hhf1-20 mutant suppresses the SDL of psh1Δ GALCSE4 strains while the HHT1/hhf1-10 mutant does not (Figure 6B). These findings suggest that the defect in the Cse4-H4 interaction contributes to the suppression of the psh1Δ GALCSE4 SDL in the HHT1/hhf1-20 strain.
Figure 6.
Mutation in the histone fold domain of histone H4 suppresses the SDL phenotype of a psh1Δ GALCSE4 strain and causes defects in Cse4 sumoylation. (A) Schematic of HHF1. Displayed is a cartoon of the HHF1 gene with mutations in hhf1-10 indicated by an “x” and hhf1-20 with a “^” in the histone fold domain (HFD, blue). The specific residues mutated in each allele are indicated below the schematic. (B) Mutations in the histone fold domain of histone H4 suppress the SDL phenotype of a psh1Δ GALCSE4 strain. Growth assays of wild-type (MSY559), psh1Δ (YMB11346), HHT1/hhf1-10 (MSY535), HHT1/hhf1-20 (MSY534), psh1Δ HHT1/hhf1-10 (YMB11347), and psh1Δ HHT1/hhf1-20 (YMB11348) with empty vector (pMB433) or expressing GAL1-6His-3HA-CSE4 (pMB1458). Cells were plated in fivefold serial dilutions on selective media plates containing either glucose (2% final concentration) or raffinose/galactose (2% final concentration each). Plates were incubated at 30°C for three to five days. Three independent transformants were tested and a representative image is shown. (C) Mutations in the histone fold domain of histone H4 decrease levels of sumoylated Cse4. The levels of sumoylated Cse4 were determined using lysates from HHT1/HHF1 (MSY559), HHT1/hhf1-10 (MSY535), and HHT1/hhf1-20 (MSY534) strains in the hht2Δ/hhf2Δ background, transformed with pGAL-8His-HA-CSE4 (pMB1345), as described in Figure 5A. Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (D) Quantification of the relative levels of sumoylated Cse4 in hhf1 strains. Levels of sumoylated Cse4 were normalized to nonmodified Cse4 probed against HA in the pull-down samples and levels in the HHT1/HHF1 strain were set to 1. Error bars indicate average deviation from the mean from two biological replicates.
We next examined the stability of Cse4 in HHT1/HHF1, HHT1/HHF1 psh1Δ, HHT1/hhf1-10, HHT1/hhf1-20, HHT1/hhf1-10 psh1Δ, and HHT1/hhf1-20 psh1Δ strains transiently overexpressing CSE4. In agreement with previous findings (Figure 4A), overexpressed Cse4 is rapidly degraded in HHT1/HHF1 cells and is stabilized in the HHT1/HHF1 psh1Δ strain (Hewawasam et al. 2010; Ranjitkar et al. 2010) (Supplementary Figure S8, top panels). Interestingly, degradation of overexpressed Cse4 in both HHT1/hhf1-10 and HHT1/hhf1-20 strains was faster compared to the HHT1/HHF1 strain. The HHT1/hhf1-20 psh1Δ strain showed rapid degradation of Cse4 when compared to the HHT1/HHF1 psh1Δ and HHT1/hhf1-10 psh1Δ strains (Supplementary Figure S8). The rapid degradation of overexpressed Cse4 in the HHT1/hhf1-20 psh1Δ GALCSE4 strain is consistent with our previous finding that the hhf1Δ psh1Δ GALCSE4 strain shows reduced stability of Cse4 (Figure 4A) and suggests that the histone fold domain of H4 contributes to rapid degradation of Cse4 due to the ubiquitin-independent proteolysis of Cse4.
To examine the effect of the HHT1/hhf1-20 and HHT1/hhf1-10 alleles on the levels of Cse4 sumoylation, we used HHT1/HHF1, HHT1/hhf1-10, and HHT1/hhf1-20 strains overexpressing CSE4 to examine the sumoylation status of Cse4. Western blot analysis was performed after equal amounts of protein (5 mg) for each strain were pulled down with Ni-NTA agarose beads and normalized to the levels of nonmodified Cse4 in the pull down (Figure 6, C and D). Sumoylated Cse4 was observed in the HHT1/HHF1 and the HHT1/hhf1-10 strains (Figure 6, C and D). Levels of sumoylated Cse4 were normalized to nonmodified Cse4 in the pull down samples. The low expression of Cse4 in the HHT1/hhf1-10 strain (Figure 6C, input) contributes to the higher levels of Cse4 sumoylation due to normalization to the low levels of nonmodified Cse4 in this strain (Figure 6D). In contrast, the levels of Cse4 sumoylation were barely detectable in the HHT1/hhf1-20 strain when compared to the HHT1/HHF1 strain (Figure 6, C and D). The reduced sumoylation of Cse4 in the HHT1/hhf1-20 strain is consistent with the rescue of SDL in the HHT1/hhf1-20 psh1Δ GALCSE4 strain. We conclude that defects in the interaction of hhf1-20 with Cse4 contributes to reduced Cse4 sumoylation and suppression of psh1Δ GALCSE4 SDL due to rapid degradation of Cse4.
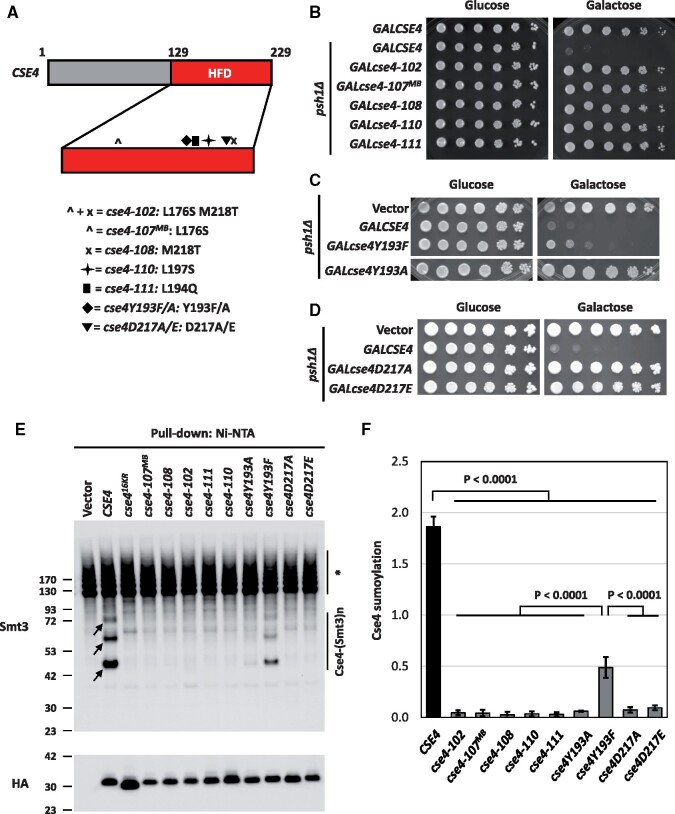
Cse4 mutants defective in the Cse4-H4 interaction do not cause SDL in a psh1Δ strain and exhibit defects in Cse4 sumoylation
To further confirm that the Cse4-H4 interaction contributes to SDL in a psh1Δ GALCSE4 strain and Cse4 sumoylation, we investigated if Cse4 residues that are essential for the Cse4-H4 dimer formation (Figure 7A) affect the SDL of a psh1Δ strain and sumoylation of Cse4. Like the HHT1/hhf1-20 mutant, the cse4 mutants cse4-102 (L176S M218T) and cse4-111 (L194Q) exhibit defects in the Cse4-H4 dimer formation, while cse4-110 (L197S) likely impairs formation of the (Cse4-H4)2 tetramer (Glowczewski et al. 2000). We hypothesized that overexpression of these cse4 mutants will not lead to SDL in a psh1Δ strain and these mutants will show defects in Cse4 sumoylation. To test these hypotheses, we generated galactose-inducible plasmids expressing cse4-102 (L176S M218T), cse4-107MB (L176S), cse4-108 (M218T), cse4-110 (L197S), and cse4-111 (L194Q). To test the effect of the cse4 mutants on SDL in a psh1Δ strain, we performed growth assays. We first determined that overexpression of mutant cse4 from these plasmids did not result in growth defects in a wild-type strain (Supplementary Figure S9). In agreement with our hypothesis, overexpression of all cse4 mutants did not cause SDL in a psh1Δ strain (Figure 7B). We conclude that the Cse4-H4 dimerization is essential for the SDL phenotype of a psh1Δ GALCSE4 strain.
Figure 7.
Cse4 mutants defective in the Cse4-H4 interaction do not cause SDL in a psh1Δ GALCSE4 strain and exhibit defects in Cse4 sumoylation. (A) Schematic of CSE4. Displayed is a cartoon of the CSE4 gene highlighting mutations in the histone fold domain (HFD, red). The HFD is expanded under the representation of CSE4. Below the gene schematic is a key describing the symbol that represents a specific mutant cse4 allele and the residues mutated. (B) Cse4-H4 assembly mutants in Cse4 do not cause SDL in a psh1Δ GALCSE4 strain. Growth assays of a psh1Δ (YMB8995) strain transformed with pGAL1-8His-HA-Cse4 (pMB1344), pGAL1-8His-HA-cse4-102 (pMB1984), pGAL1-8His-HA-cse4-107MB (pMB1985), pGAL1-8His-HA-cse4-108 (pMB1986), pGAL1-8His-HA-cse4-110 (pMB1987), or pGAL1-8His-HA-cse4-111 (pMB1988). Cells were plated in fivefold serial dilutions on selective media plates containing either glucose (2% final concentration) or raffinose/galactose (2% final concentration each). Plates were incubated at 30°C for three to five days. Three independent transformants were tested and a representative image is shown. (C) The Y193A mutation in Cse4 does not cause SDL in a psh1Δ GALCSE4 strain. Growth assays of a psh1Δ (YMB9034) strain transformed with empty vector (pYES2), pGAL1-8His-HA-cse4Y193A (pMB1766), or pGAL1-8His-HA-cse4Y193F (pMB1787). Fivefold serial dilutions of the indicated strains were plated on glucose (2% final concentration)- or galactose (2% final concentration)-containing medium selective for the plasmid. The plates were incubated at 30°C for 3 days. (D) The cse4D217A/E mutants do not cause SDL in a psh1Δ strain. Growth assays of a psh1Δ (YMB9034) strain transformed with empty vector (pYES2), pGAL1-8His-HA-cse4D217A (pMB1910), or pGAL1-8His-HA-cse4D217E (pMB1920). Strains were assayed as described in (C). (E) Cse4 sumoylation levels are decreased in Cse4-H4 assembly mutants. Levels of sumoylated Cse4 were assayed in a wild-type strain (BY4741) transformed with empty vector (pYES2), pGAL1-8His-HA-CSE4 (pMB1345), pGAL1-8His-HA-cse416KR (pMB1344), pGAL1-8His-HA-cse4-107MB (pMB1985), pGAL1-8His-HA-cse4-108 (pMB1986), pGAL1-8His-HA-cse4-102 (pMB1984), pGAL1-8His-HA-cse4-111 (pMB1988), pGAL1-8His-HA-cse4-110 (pMB1987), pGAL1-8His-HA-cse4Y193A (pMB1766), pGAL1-8His-HA-cse4Y193F (pMB1787), pGAL1-8His-HA-cse4D217A (pMB1910), or pGAL1-8His-HA-cse4D217E (pMB1920). Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (F) Quantification of the relative levels of sumoylated Cse4 in cse4 mutants. Levels of sumoylated Cse4 in arbitrary density units were normalized to nonmodified Cse4 probed against HA in the pull-down samples. Statistical significance from at least three biological repeats was assessed by one-way ANOVA (P < 0.0001) followed by Tukey post-test (all pairwise comparisons of means). Error bars indicate standard deviation from the mean.
Next, we generated galactose-inducible plasmids expressing cse4Y193A/F and cse4D217A/E for growth assays in a psh1Δ strain. The rationale for cse4Y193A/F is that Y193 is next to the mutated residue in cse4-111 (L194Q), is located at the center of the α2 helix of Cse4, and interacts with the α2 helix of H4 in the context of Scm3 (Zhou et al. 2011). For cse4D217A/E, the D217 residue is adjacent to the residue mutated in cse4-108 (M218T), is part of the K215/216 sumoylation consensus site, 214-MKKD-217 (Ψ-K-x-D/E), and is essential for dimerization of Cse4 (Camahort et al. 2009). Growth assays on galactose media showed that cse4Y193A and cse4D217A/E do not cause SDL in a psh1Δ strain (Figure 7, C and D). Note that cse4Y193F showed partial lethality in a psh1Δ strain when compared to CSE4 (Figure 7C). Taken together, these results show that overexpression of the cse4 mutants with defects in the formation of the Cse4-H4 dimer, do not lead to a SDL phenotype in a psh1Δ strain.
The lack of SDL in psh1Δ strains overexpressing cse4-102, cse4-107MB, cse4-108, cse4-110, cse4-111, cse4Y193A, and cse4D217A/E is similar to the suppression of psh1Δ GALCSE4 SDL when combined with hhf1Δ, hhf2Δ, and hhf1-20 strains. Defects in Cse4 sumoylation in hhf1Δ, hhf2Δ, and hhf1-20 strains led us to hypothesize that cse4-102, cse4-107MB, cse4-108, cse4-110, cse4-111, cse4Y193A, and cse4D217A/E strains will also show defects in Cse4 sumoylation. Thus, we examined the sumoylation status of the cse4 mutants used in the growth assays (Figure 7E). Consistent with our hypothesis, levels of Cse4 sumoylation were reduced in all cse4 mutants except cse4Y193F, which showed only a partial reduction of Cse4 sumoylation (Figure 7, E and F). The reduced sumoylation of cse4Y193F is consistent with the partial lethality observed in a psh1Δ strain expressing cse4Y193F. Our results demonstrate that overexpression of cse4 mutants defective for the Cse4-H4 dimer formation lead to defects in Cse4 sumoylation. We conclude that the Cse4-H4 dimer formation regulates Cse4 sumoylation and this contributes to psh1Δ GALCSE4 SDL.
Reduced gene dosage of H4 suppresses CIN due to overexpression of Cse4
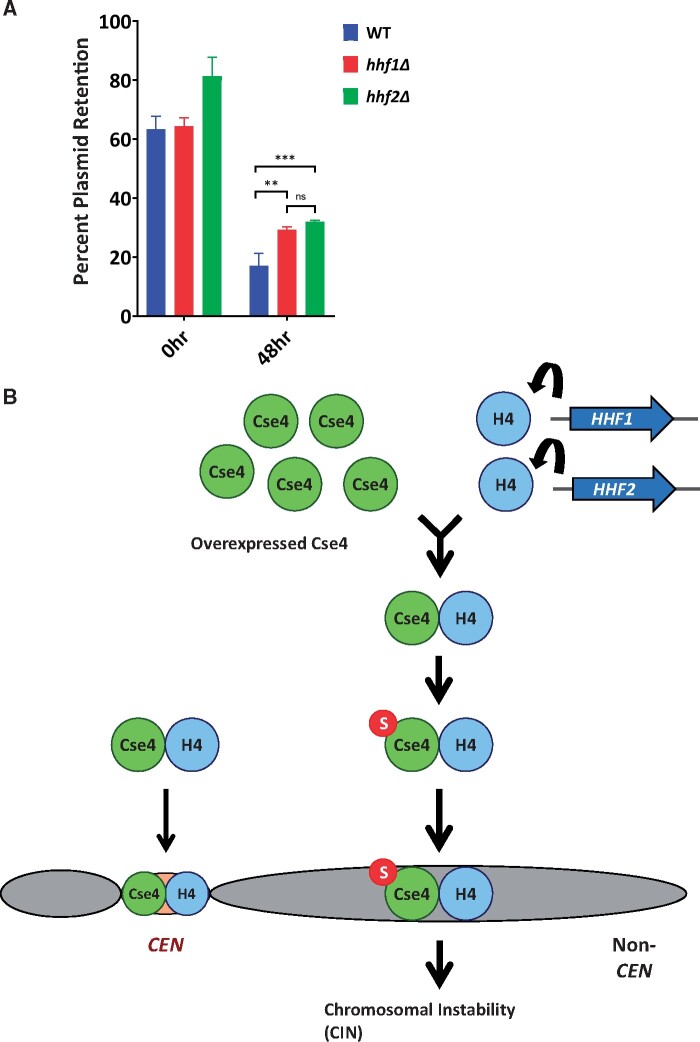
We next examined the physiological consequences of reduced gene dosage of H4 on CIN. A recent study has shown that overexpression of Cse4 (GALCSE4) contributes to CIN in a wild-type strain (Metzger et al. 2017). We have shown a correlation between CIN and mislocalization of Cse4 (Au et al. 2008, 2013, 2020). CIN was examined in wild-type, hhf1Δ, and hhf2Δ strains with GALCSE4 by quantifying the loss of a centromere (CEN)-containing plasmid after growth in nonselective medium (48 h) compared to growth in medium selective for the plasmid (0 h). Consistent with previous results (Metzger et al. 2017) we observed low CEN plasmid retention in the wild-type GALCSE4 strain after 48 h of nonselective growth (Figure 8A). Reduced dosage of H4 resulted in increased CEN plasmid retention at 48 h in hhf1Δ GALCSE4 and hhf2Δ GALCSE4 strains (Figure 8A, wt vs hhf1Δ P < 0.05; wt vs hhf2Δ P < 0.01). These results show that reduced gene dosage of H4 suppresses CIN due to overexpression of Cse4.
Figure 8.
The interaction of H4 with Cse4 promotes CIN caused by overexpressed Cse4 and Cse4 mislocalization. (A) Deletion of HHF1 or HHF2 increased CEN plasmid retention when Cse4 is overexpressed. Wild-type (SBY8904), hhf1Δ (YMB11603), and hhf2Δ (YMB11604) strains were transformed with a plasmid containing CEN LEU2 pFZO1-FZO1HA (pMM190). Cells were grown in media selective for the plasmid and containing 2% raffinose/2% galactose for 24 h prior (0 h) to shifting to nonselective media (48 h). Plasmid retention was calculated as the number of colonies that retain the plasmid as growth on selective plates vs colony number grown on nonselective plates. Error bars represent the SD of three replicates. ***P < 0.001, ** P < 0.01, ns = not significant. (B) Model for the interaction of Cse4 and H4 promoting incorporation into noncentromeric chromatin. The budding yeast genome possesses HHF1 and HHF2 which encode identical H4 proteins. Gene dosage of H4 and the Cse4-H4 interaction are key upstream events for the sumoylation of Cse4, which facilitates noncentromeric localization of overexpressed Cse4. The interaction of overexpressed Cse4 with H4 contributes to Cse4 sumoylation and this facilitates the mislocalization of overexpressed Cse4 to noncentromeric regions and CIN.
Discussion
Mislocalization of overexpressed CENP-A and its homologs contributes to CIN in yeast, fly, and human cells (Heun et al. 2006; Au et al. 2008; Mishra et al. 2011; Lacoste et al. 2014; Athwal et al. 2015; Shrestha et al. 2017) and overexpression and mislocalization of CENP-A are observed in many cancers (Tomonaga et al. 2003; Amato et al. 2009; Li et al. 2011; McGovern et al. 2012; Sun et al. 2016; Zhang et al. 2016). In this study, we performed the first genome-wide screen to identify deletion or temperature sensitive (ts) mutants that suppress the SDL due to mislocalization of overexpressed Cse4 in psh1Δ GALCSE4 strains. Deletion of either allele that encodes histone H4 (HHF1 and HHF2) were among the most prominent suppressors of psh1Δ GALCSE4 SDL. We determined that reduced gene dosage of H4 contributes to defects in Cse4 sumoylation and this prevents mislocalization of overexpressed Cse4 at peri-centromeric and noncentromeric regions, leading to suppression of the psh1Δ GALCSE4 SDL. We also determined that the Cse4-H4 interaction contributes to Cse4 sumoylation and psh1Δ GALCSE4 SDL as hhf1-20, cse4-102, and cse4-111 mutants, which are defective for the Cse4-H4 interaction, exhibit reduced sumoylation of Cse4 and do not exhibit psh1Δ GALCSE4 SDL. GALCSE4 contributes to CIN and reduced gene dosage of H4 suppresses the GALCSE4 CIN phenotype in a wild-type strain. Taken together, our genome-wide screen identified genes that contribute to Cse4 mislocalization and provides mechanistic insights into how reduced gene dosage of H4 prevents mislocalization of Cse4 into noncentromeric regions and CIN.
The suppressor screen was performed under a condition with high levels of Cse4 expression induced from a GAL1-6His-3HA-CSE4 plasmid, which contributes to mild growth sensitivity even in wild-type cells and this leads to lethality in psh1Δ strains (Figure 2). To reduce the number of false positive suppressors, we performed the screen with a psh1Δ GALCSE4 strain grown on 2% galactose medium to achieve maximum levels of Cse4 overexpression. These growth conditions limited us from identifying partial suppressors such as deletion of NHP10, which encodes a subunit of the INO80 chromatin remodeling complex and was previously shown to suppress the psh1Δ GALCSE4 SDL on medium with a lower concentration of galactose (0.1%) (Hildebrand and Biggins 2016). While our screen did not identify nhp10Δ, it did identify two deletions and one mutant allele for genes encoding INO80 subunits, Ies2, Arp8, and Act1, respectively, that are evolutionarily conserved between yeast and human cells (Poch and Winsor 1997; Shen et al. 2000, 2003; Tosi et al. 2013). Secondary growth validation showed that arp8Δ suppresses the psh1Δ GALCSE4 SDL. However, the polyploid nature of the arp8Δ strain precluded further study with this suppressor. The stringent growth conditions of the screen also prevented the identification of deletion of Cac2, a subunit of the CAF-1 complex, which promotes Cse4 incorporation at noncentromeric regions (Hewawasam et al. 2018). We determined that cac2Δ cannot suppress the psh1Δ GALCSE4 SDL under the conditions used in our screen (data not shown).
Previous studies have shown that mislocalization of Cse4 to noncentromeric regions contributes to the GALCSE4 SDL in psh1Δ, slx5Δ, doa1Δ, hir2Δ, cdc4-1, and cdc7-4 strains (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013, 2020; Ohkuni et al. 2016; Ciftci-Yilmaz et al. 2018; Eisenstatt et al. 2020). We sought to define mechanisms that prevent lethality due to mislocalization of overexpressed Cse4. The identification of both hhf1Δ and hhf2Δ as suppressors of psh1Δ GALCSE4 SDL led us to examine how reduced gene dosage of H4 contributes to preventing mislocalization of Cse4. A role for histone H4 in centromeric localization of Cse4 has been examined previously (Deyter et al. 2017), however, the effect of gene dosage of H4 in noncentromeric chromosome localization of Cse4 has not yet been explored. We determined that suppression of the GALCSE4 SDL phenotype by hhf1Δ and hhf2Δ is not restricted to psh1Δ strains and is also observed in slx5Δ, doa1Δ, cdc4-1, and cdc7-4 strains. The SDL phenotype of the hir2Δ GALCSE4 strain showed better suppression with hhf1Δ than with hhf2Δ. This may be due to the role of the HIR complex in histone gene expression (Prochasson et al. 2005; Fillingham et al. 2009; Kurat et al. 2014).
We used several approaches to understand the molecular mechanism for suppression of the psh1Δ GALCSE4 SDL phenotype by hhf1Δ and hhf2Δ. These include ChIP-qPCR at regions of known Cse4 association, protein stability assays, and determining the status of Cse4 ubiquitination and sumoylation. Genome-wide studies have shown that overexpressed Cse4 is significantly enriched at promoters and peri-centromeric regions in a psh1Δ strain (Hildebrand and Biggins 2016). Our ChIP-qPCR data showed reduced levels of Cse4 at peri-centromeric and noncentromeric regions in psh1Δ hhf1Δ and psh1Δ hhf2Δ strains when compared to the psh1Δ strain. The occupancy of H3 normalized to H4 was not significantly different in a psh1Δ hhf1Δ strain compared to that in the psh1Δ strain. This may be because reduced dosage of H4 may affect both H4-Cse4 and H4-H3 interactions or the high occupancy of H3-containing nucleosomes relative to those with Cse4 in a psh1Δ strain may limit our ability to discern differences in H3 association at noncentromeric regions in the psh1Δ hhf1Δ GALCSE4 strain with our ChIP-qPCR assay.
The mislocalization of overexpressed Cse4 to noncentromeric regions contributes to highly stable Cse4 in psh1Δ, slx5Δ, doa1Δ, hir2Δ, cdc4-1, and cdc7-4 strains (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013, 2020; Ohkuni et al. 2016; Ciftci-Yilmaz et al. 2018; Eisenstatt et al. 2020). We reasoned that reduced mislocalization of Cse4 to noncentromeric regions in psh1Δ hhf1Δ strains may contribute to faster degradation of Cse4 in these strains. Our results showed that the proteolysis of Cse4 was indeed faster in psh1Δ hhf1Δ strains when compared to the psh1Δ strain. Intriguingly, this was not due to increased ubiquitination of Cse4 (Ubn-Cse4) in psh1Δ hhf1Δ strains. These results suggest a ubiquitin-independent mechanism that may contribute to the proteolysis of Cse4 in hhf1Δ psh1Δ strains. Ubiquitin-independent proteolysis has also been reported previously as cse416KR, in which all lysine residues are mutated to arginine, is still degraded (Collins et al. 2004).
Our results showing that reduced dosage of H4 contributes to the suppression of GALCSE4 SDL in psh1Δ strains, reduced mislocalization of Cse4, and lower protein stability of Cse4 are similar to the phenotypes of the sumoylation-defective cse4K215/216R/A strains (Ohkuni et al. 2020). Consistent with these results, deletion of either histone H4 allele resulted in reduced levels of sumoylated Cse4. Reduced gene dosage of H4 did not affect sumoylation of Ndc80 or transcription of genes in the SUMO pathway. We therefore propose that physiological levels of H4 regulate the sumoylation of Cse4 and that this in turn facilitates mislocalization of overexpressed Cse4 to noncentromeric regions and GALCSE4 SDL in mutant such as psh1Δ. Importantly, in contrast to histone H4, reduced dosage of genes encoding other canonical histones such as histones H2A or H3 does not suppress the psh1Δ GALCSE4 SDL.
To further examine the role of H4 in regulating the mislocalization of Cse4, we pursued studies using well-characterized separation of function alleles of H4 (hhf1-20) and CSE4 (cse4-102 and cse4-111) with defects in the Cse4-H4 interaction (Smith et al. 1996; Glowczewski et al. 2000). Consistent with a role of H4 for its interaction with Cse4, we observed suppression of the psh1Δ GALCSE4 SDL in a hhf1-20 strain and lack of SDL when cse4-102 or cse4-111 were overexpressed in a psh1Δ GALCSE4 strain. The hhf1 mutant strains lack the HHT2/HHF2 allele and express only a single copy of H3/H4 (HHT1/HHF1). In this strain background, the psh1Δ GALCSE4 SDL was less severe compared to results in our strains with wild-type copies of both HHT1/HHF1 and HHT2/HHF2 (Figure 2). Despite this, we were able to unambiguously establish that HHT1/hhf1-20, but not HHT1/hhf1-10, suppresses the psh1Δ GALCSE4 SDL. Interestingly, the hhf1-10 psh1Δ GALCSE4 strain displayed a more lethal phenotype than the wild-type HHT1/HHF1 psh1Δ GALCSE4 strain. The N-terminal lysine residues on histone H4 (K5, 8, 12, 16) are acetylated and the HHT1/hhf1-10 mutations mimic the hyperacetylated state of the lysine residues (K to Q). We have previously shown that levels of acetylated H4 are low at centromeres and that the maintenance of hypoacetylated H4 at the centromere is essential for kinetochore function and faithful chromosome segregation (Choy et al. 2011). We propose that the hyperacetylated state of H4 in the HHT1/hhf1-10 strain contributes to the more severe SDL that we observed. A recent study showed that strains with a mutation of histone H4 arginine 36 to alanine (H4R36A) display SDL when Cse4 is overexpressed and that this is due to defects in the interaction of H4R36A with Psh1, thereby leading to enrichment of Cse4 and Psh1 at noncentromeric regions in these cells (Deyter et al. 2017).
Consistent with our previous studies (Ohkuni et al. 2020), we observed a correlation between the suppression of GALCSE4 SDL and reduced sumoylation of Cse4 in HHT1/hhf1-20, cse4-102, and cse4-111 strains. Similar results were observed with cse4Y193A/F, which is adjacent to the mutated site in cse4-111 (L194Q), and with cse4D217D/E, which is adjacent to the residue mutated in cse4-108 (M218T) and a part of the K215/216 sumoylation consensus site (Camahort et al. 2009). Accordingly, low levels of sumoylated cse4Y193F correlate with a partial lethality of a psh1Δ GALcse4Y193F strain and severe defects in sumoylated cse4Y193A correlate with a lack of SDL in a psh1Δ GALcse4Y193A strain. Phenylalanine (F) is identical to tyrosine (Y) except for the hydroxyl group present on Y. It is possible that the structural similarity between Y and F allows at least partial formation of the Cse4-H4 dimer, resulting in partial sumoylation of cse4Y193F. In contrast, we observed a reduction of Cse4 sumoylation of both cse4D217A and cse4D217E mutants compared to wild-type. The D217 residue of Cse4 is essential for growth and is important for the Cse4 dimerization. Since the cse4D217E mutant, which is part of the intact sumoylation consensus site, shows reduction of Cse4 sumoylation and does not complement the null mutation (Supplementary Figure S10), we propose that D217 has a role besides regulating sumoylation of Cse4K215/216. Sumoylation of Cse4 is not essential for centromeric localization of Cse4 because a cse416KR strain with all 16 lysine (K) residues mutated to arginine (R) is viable in the context of the wild-type centromeric chaperone Scm3 (Au et al. 2008). Sumoylation of Cse4K215/216 or physiological levels of H4 are indispensable only when Scm3 is not expressed (Ohkuni et al. 2020). Our results show that defects in Cse4 sumoylation contribute to reduced levels of noncentromere associated Cse4 with no significant effect on levels of centromere associated Cse4 in psh1Δ hhf1Δ and psh1Δ hhf2Δ strains. We propose that reduced dosage of H4 serves to protect the cells from the detrimental effects of overexpressed Cse4 due to defects in Psh1, SCFCdc4, Cdc7, Slx5/8, HIR, and Doa1-mediated proteolysis of Cse4. We define a previously undefined role for histone H4 gene dosage and the Cse4-H4 interaction as key upstream events for the sumoylation of Cse4, which facilitates noncentromeric localization of overexpressed Cse4 and SDL in a psh1Δ GALCSE4 strain.
We have previously shown that mislocalization of Cse4 contributes to CIN (Au et al. 2008, 2013, 2020). A recent report showed that overexpression of Cse4 contributes to CIN in a wild-type strain and that the plasmid loss in psh1Δ strains is independent of Cse4 overexpression (Metzger et al. 2017), suggesting that Psh1 has additional roles in chromosome stability. We observed that the CIN phenotype due to GALCSE4 was suppressed in the hhf1Δ and hhf2Δ strains. We propose a model in which the interaction of overexpressed Cse4 with histone H4 facilitates Cse4 sumoylation and this promotes the mislocalization of Cse4 to noncentromeric regions and CIN (Figure 8B). Reduced gene dosage of H4 or mutants defective for the Cse4-H4 interaction exhibit reduced Cse4 sumoylation, which contributes to the reduced mislocalization and suppression of CIN due to overexpression of Cse4.
In summary, our genome-wide screen identified suppressors of psh1Δ GALCSE4 SDL with deletions of either allele that encodes histone H4 (HHF1 and HHF2) as among the most prominent suppressors. We present several experimental evidences to support our conclusions that reduced gene dosage of H4 contributes to defects in Cse4 sumoylation and reduced mislocalization of overexpressed Cse4 at peri-centromeric and noncentromeric regions, which in turn results in faster degradation of Cse4, suppression of the psh1Δ GALCSE4 SDL, and CIN due to overexpressed Cse4. The suppression of SDL by hhf1Δ and hhf2Δ is not limited to a psh1Δ GALCSE4 background but is also observed in other mutants that exhibit GALCSE4 associated SDL. Most importantly, our results with the hhf1-20, cse4-102, and cse4-111 mutants, which are defective in the Cse4-H4 interaction, showed that the Cse4-H4 interaction is essential for noncentromeric association of Cse4. These studies are important from a clinical standpoint given the poor prognosis of CENP-A overexpressing cancers (Tomonaga et al. 2003; Amato et al. 2009; Li et al. 2011; McGovern et al. 2012; Sun et al. 2016; Zhang et al. 2016). Future studies with histone H4 and other mutants identified in our screen will provide insights into mechanisms that promote mislocalization of overexpressed Cse4 and how defects in these mechanisms may safeguard the cell from the lethal effect due to mislocalization of overexpressed Cse4.
Acknowledgments
We gratefully acknowledge Jennifer Gerton and Mitch Smith for reagents, Kathy McKinnon of the National Cancer Institute Vaccine Branch FACS Core for assistance with FACS analysis, Anthony Dawson for strain construction, and the members of the Basrai laboratory for helpful discussions and comments on the manuscript.
Funding
M.A.B. is supported by the National Institutes of Health Intramural Research Program at the National Cancer Institute. This research was also supported by grants from the National Institutes of Health to C.B. and M.C. (R01HG005853) and from the Canadian Institute of Health Research to C.B. (FDN-143264). C.B. is a fellow in the Canadian Institute for Advanced Research (CIFAR, https://www.cifar.ca/) Fungal Kingdom: Threats and Opportunities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
None declared.
Literature cited
- Allshire RC, Karpen GH.. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 9:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Schillaci T, Lentini L, Leonardo AD.. 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer 8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, et al. 2015. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA.. 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, et al. 2013. A novel role of the N-terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Zhang T, Mishra PK, Eisenstatt JR, Walker RL, et al. 2020. Skp, Cullin, F-box (SCF)-Met30 and SCF-Cdc4-Mediated Proteolysis of CENP-A prevents Mislocalization of CENP-A for chromosomal stability in budding yeast. PLoS Genet. 16:e1008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S. 2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltengagen M, Huang A, Boltengagen A, Trixl L, Lindner H, et al. 2016. A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. Nucleic Acids Res. 44:2145–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J.. 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, et al. 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26:853–865. [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, et al. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell 35:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, et al. 2014. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 204:313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Bao X, Gan X, Luo S, Rao H.. 2017. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci Rep. 7:8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Bao X, Rao H.. 2016. The F-box protein Rcy1 is involved in the degradation of histone H3 variant Cse4 and genome maintenance. J Biol Chem. 291:10372–10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ocampo J, Clark DJ.. 2017. MNase-sensitive complexes in yeast: nucleosomes and non-histone barriers. Mol Cell 65:565–577.e563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Acuna R, Au WC, Basrai MA.. 2011. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics 189:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Mishra PK, Au WC, Basrai MA.. 2012. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim Biophys Acta 1819:776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Au WC, Mishra PK, Eisenstatt JR, Chang J, et al. 2018. A genome-wide screen reveals a role for the HIR histone chaperone complex in preventing mislocalization of budding yeast CENP-A. Genetics 210:203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ.. 2014. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res. 42:12512–12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S.. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 14:1968–1972. [DOI] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, et al. 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353:aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter GM, Biggins S.. 2014. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 28:1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter GM, Hildebrand EM, Barber AD, Biggins S.. 2017. Histone H4 facilitates the proteolysis of the budding yeast CENP-ACse4 centromeric histone variant. Genetics 205:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstatt JR, Boeckmann L, Au WC, Garcia V, Bursch L, et al. 2020. Dbf4-Dependent Kinase (DDK)-mediated proteolysis of CENP-a prevents mislocalization of CENP-A in Saccharomyces cerevisiae. G3 (Bethesda). 10:2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Kainth P, Lambert J-P, van Bakel H, Tsui K, et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell 35:340–351. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, et al. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, et al. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12:17–30. [DOI] [PubMed] [Google Scholar]
- Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM.. 2000. Histone-histone interactions and centromere function. Mol Cell Biol. 20:5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, et al. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, et al. 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam GS, Dhatchinamoorthy K, Mattingly M, Seidel C, Gerton JL.. 2018. Chromatin assembly factor-1 (CAF-1) chaperone regulates Cse4 deposition into chromatin in budding yeast. Nucleic Acids Res. 46:4440–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam GS, Mattingly M, Venkatesh S, Zhang Y, Florens L, et al. 2014. Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J Biol Chem. 289:29297–29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand EM, Biggins S.. 2016. Regulation of budding yeast CENP-a levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 12:e1005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, et al. 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16:365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P.. 2001. Evolutionary concervation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2:678–687. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Recht J, Radovani E, Durbic T, Andrews B, et al. 2014. Regulation of histone gene transcription in yeast. Cell Mol Life Sci. 71:599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, et al. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 53:631–644. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu Z, Zhang S, Yu D, Yu H, et al. 2011. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE 6:e17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Corbett KD, Desai A.. 2012. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 22:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA.. 2012. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 14:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM.. 2016. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 17:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Scales JL, Dunklebarger MF, Weissman AM.. 2017. The Ubiquitin Ligase (E3) Psh1p is required for proper segregation of both centromeric and two-micron plasmids in Saccharomyces cerevisiae. G3 (Bethesda). 7:3731–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, et al. 2011. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 7:e1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C.. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P.. 2006. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 174:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Torras-Llort M, Azorin F.. 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34:6247–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Abdulle R, Kitagawa K.. 2014. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl Cis-Trans isomerase. Genetics 196:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Levy-Myers R, Warren J, Au WC, Takahashi Y, et al. 2018. N-terminal sumoylation of centromeric Histone H3 variant Cse4 regulates its proteolysis to prevent mislocalization to non-centromeric Chromatin. G3 (Bethesda). 8:1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Suva E, Au WC, Walker RL, Levy-Myers R, et al. 2020. Deposition of centromeric histone H3 variant CENP-A/Cse4 into chromatin is facilitated by its C-terminal sumoylation. Genetics 214:839–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Takahashi Y, Basrai MA.. 2015. Protein purification technique that allows detection of sumoylation and ubiquitination of budding yeast kinetochore proteins Ndc10 and Ndc80. J Vis Exp. 99:e52482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Takahashi Y, Fulp A, Lawrimore J, Au WC, et al. 2016. SUMO-Targeted Ubiquitin Ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol Biol Cell. 27:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, et al. 2009. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O, Winsor B.. 1997. Who's Who among the Saccharomyces cerevisiae actin-related proteins? a classification and nomenclature proposal for a large family. Yeast 13:1053–1058. [DOI] [PubMed] [Google Scholar]
- Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL.. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, et al. 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C.. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541–544. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C.. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12:147–155. [DOI] [PubMed] [Google Scholar]
- Shrestha RL, Ahn GS, Staples MI, Sathyan KM, Karpova TS, et al. 2017. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8:46781–46800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A.. 2010. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 107:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Yang H, Santisteban MS, Boone PW, Goldstein AT, et al. 1996. A Novel Histone H4 Mutant Defective in Nuclear Division and Mitotic Chromosome Transmission. Mol Cell Biol. 16:1017–1026., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, et al. 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 104:10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Clermont PL, Jiao W, Helgason CD, Gout PW, et al. 2016. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer 139:899–907. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, et al. 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63:3511–3516. [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, et al. 2013. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154:1207–1219. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K.. 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 12:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P.. 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mao JH, Zhu W, Jain AK, Liu K, et al. 2016. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 7:12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou B-R, Ghirlando R, Hu K, et al. 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472:234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supplementary Figures S1–S10 are available as JPG files. Supplementary Table S1 is an Excel file that describes mutations that suppress the psh1Δ GALCSE4 SDL, the gene systematic name, the gene name, the functional category, growth and colony scores, and validation information if applicable. Supplemental File S1 contains Supplementary Tables S2–S4 which describe the yeast strains, plasmids, and primers used in this study, respectively. Supporting information is available at figshare: https://doi.org/10.25386/genetics.13713937.