Abstract
Background: Previous treatment guidelines have suggested that bacteria are associated with the severity of appendicitis, and the use of postoperative antibiotics should be guided according to the bacteria culture results derived from intraoperative samples. However, this approach has many limitations. Patients were commonly administrated antibiotics during the perioperative period, which can lead to inaccurate culture result. Aim: To assess the relationship between pathogenic bacteria and appendicitis and optimize the process of antibiotic selection. Methods: A nonconsecutive case series analysis was conducted from January to July 2017. Nineteen patients were divided into two groups according to their postoperative histological results (Non-perforated: phlegmonous/Perforated: gangrenous, n = 9/10) and postoperative bacterial culture results (Negative/Positive, n = 8/11). Patients were administrated same antibiotics during the perioperative period. During appendectomy, the diseased appendixes were collected, and whole metagenomic sequencing was used to identify the pathogenic bacteria in the specimens. Conventional technology was used to culture bacteria from appendix samples. Results: We identified 361 species in the appendix samples. Six species in the appendix samples had relative abundances > 5%. No significant differences were observed in the bacterial composition of the two assayed groups. In particular, according to the grouping of culture results, the sequencing analysis results were completely different from those of the culture-based method. Conclusion: In clinical practice, because patients are regularly administrated antibiotics during the perioperative period, these antibiotics inevitably affect the results of bacterial culture. Therefore, bacterial culture results are not suitable for exclusively guiding the use of antimicrobial agents after appendicitis. Next-generation sequencing has numerous advantages, such as precisely characterizing the profiles of microbiota and their antibiotic resistance in appendicitis patients. Based on the above results, we propose that a combination of bacterial culture and next-generation sequencing should be used to improve the efficacy of antibiotic therapy.
Keywords: Appendicitis, microbiology, metagenomics, sequencing, NGS
Introduction
Acute appendicitis (AA) is the most common surgical emergency, and after hundreds of years of research, surgeons have a considerable understanding of this disease. Appendectomy is recommended as the first choice for various types of appendicitis. However, it is difficult to explain why the severity and symptoms of AA patients vary despite exhibiting almost the same course of illness. A recent theory has suggested that the local microbiota is responsible for the occurrence and evolution of appendicitis [1-5]. Matthew B. Rogers reported that the appendixes collected from children with AA harbored populations of fusobacteria, which are generally absent in fecal samples from healthy adults and children and likely contribute to the pathogenesis of appendicitis [6]. In a 16S ribosomal RNA (rRNA) gene sequence analysis-based study, Zhong et al. observed that appendicitis samples contained an increased abundance of Fusobacterium spp. and other pathogens commonly detected in the oral cavity, while non-appendicitis samples contained a reduced abundance of Bacteroides spp. [7]. Alexander Swidsinski’s group applied rRNA-based fluorescence in situ hybridization to investigate sections of 70 appendixes, and their results supported the hypothesis that the local microbiota was responsible for the occurrence and development of appendicitis [8]. However, some scholars believe that there is no correlation between local microbiota and the grade of inflammation [9]. Moreover, it is difficult to explain why although the results of bacterial cultures were different, patients recovered similarly after treatment with appendectomy. At present, the relationship between the varying degrees of clinical symptoms and different intestinal microbial environments remains unclear, since the detection of appendicitis-causing pathogenic bacteria has been primarily based on bacterial culture methods. Patients were administrated antibiotics during the perioperative period commonly, which can lead to inaccurate culture result. However, the development and use of next-generation sequencing (NGS) approaches for the detection of pathogenic bacteria have obvious advantages and allow for the relationship between clinical manifestations and pathogenic bacteria to be studied in greater depth.
Currently, the methods used to detect pathogenic microorganisms include bacterial culture, 16S rRNA gene sequencing, fluorescence in situ hybridization (FISH) and metagenomic sequencing. All of these methods have certain limitations; specifically, culture-based approaches can only detect 1% of living bacteria, and 16S rRNA gene sequencing can identify pathogenic bacteria to the genus level but may not be able to identify species. Moreover, FISH cannot achieve 100% hybridization. As the cost of NGS technology continues to decrease, metagenomic sequencing can be increasingly used to detect pathogenic microorganisms. Metagenomic sequencing can detect 99% of the studied microbes and provide results that are accurate at the species level. Due to its many advantages, metagenomic sequencing shows promise as a sensitive and rapid method to investigate host microbiomes.
To study the effects of pathogenic bacteria on appendicitis, we grouped patients according to postoperative pathology and postoperative routine bacterial culture results. First, perforated and non-perforated appendicitis may be of different types and different pathological processes [10], and the current treatment strategies used for these two types of appendicitis are different. Thus, it is necessary to analyze the bacterial composition of appendicitis for these two types of pathology. Second, appendicitis with positive and negative bacterial culture results may have different bacterial compositions. The results of bacterial culture have long been used as a guide for antibiotic use. However, the bacterial culture method has limitations, and it is necessary to group these results and use NGS methods to comprehensively compare the composition of bacteria. Therefore, in the present study, we analyzed the relationship between the classification of appendicitis and metagenome sequencing results to promote the use of metagenome sequencing to improve treatment strategies for appendicitis.
Methods
Study design and participants
We conducted an observational and nonconsecutive case series analysis from March 2017 to July 2017. Data were collected from the clinical database of the Fifth Affiliated Hospital of Southern Medical University. Preoperative diagnosis was based on medical history, physical examination, routine blood results and ultrasound. Postoperative pathology confirmed the preoperative diagnosis. After the surgeon suspected the diagnosis of AA, all patients received the same treatment, including second-generation cephalosporin therapy and a laparoscopic appendectomy. During the observation period, patients undergoing laparoscopic appendectomy for AA were preoperatively enrolled in the study. Patients with chronic diseases associated with other physiological systems, multiple infections, or low immune function were excluded from the study. We grouped the patients according to the results of postoperative pathology and routine bacterial culture. The details of the subjects are outlined in Tables 1 and 2. All laboratory tests were conducted in the BGI-Guangzhou Medical Laboratory. The Ethical Committee of the Fifth Affiliated Hospital of Southern Medical University reviewed the study details and confirmed that all methods were performed in accordance with the relevant guidelines and regulations (NYWY201804). Before any patient was enrolled in the study, we ensured that he or she sufficiently understood the study. All of the appendix specimens from our cases were examined by routine culture. According to the ethical approval guidelines, the subjects did not need to sign any written consent because all additional tests were conducted on the leftover specimens from routine microbiological examinations. The statistical analysts were blind to the identity information of the patients, and the sample information was grouped by specific identification codes.
Table 1.
Study population based on intraoperative exploration combined with postoperative histological examination results
| Histological type | Appendicitis categorized by histological type | |
|---|---|---|
|
| ||
| Non-perforated: phlegmonous (N = 9) | Perforated: gangrenous (N = 10) | |
| Age | 37 (14-64) | 36 (7-62) |
Table 2.
Study population on the basis of bacterial culture results
| Bacterial culture | Appendicitis categorized by bacterial culture | |
|---|---|---|
|
| ||
| Negative (N = 8) | Positive (N = 11) | |
| Age | 30 (7-64) | 41 (14-63) |
Table 2 Escherichia coli were detected in all bacterial culture samples, and Klebsiella pneumoniae was detected in one of them. Values are presented as the median (min-max) or as the absolute number of patients.
Sample acquisition
During the laparoscopic appendectomy, the appendix was removed through trocar holes, after which the appendix tissue was dissected into 3 subsamples and stored in aseptic tubes. One piece was used for bacterial culture, one piece was histopathologically examined, and the other piece was stored in a sterile tube at -80°C for use in subsequent whole metagenomic sequencing analyses.
Culturing of appendix sample
The appendix tissue was cut into small pieces and diluted with pre-reduced phosphate-buffered saline (PBS) to approximately 109 CFU/ml. The debris was precipitated briefly with 5 min standing at room temperature. A 100 µl of the supernatant was spread on agar plates of each medium, and the plates were incubated in the anaerobic glove box UM-017 Bugbox Plus at 37°C for 24 h. We selected 3 types of media (Columbia blood agar medium, Nutrient-Broth-Medium and Sabouraud plate) for bacterial culturing under 4 different conditions. For Sabouraud plate, the spread plates were incubated at 37°C with an Anaero Pack for microaerophilic culture at 37°C for 72 h and aerobic culture at 37°C for 24 h, respectively. GP identification card was used for gram-positive cocci and GN identification card was used for Gram-negative bacteria.
Sample processing and sequencing
DNA was extracted from the appendix samples using a TIANamp Micro DNA kit (DP316, TIANGEN Biotech, Beijing, China) and a DNeasy® Blood & Tissue kit (69504, Qiagen, Hilden, Germany) according to the manufacturers’ recommendations. DNA libraries were subsequently constructed through fragmentation, end repair, adapter ligation and PCR amplification. An Agilent 2100 bioanalyzer (Agilent, Santa Clara, USA) was used to assess the quality of the DNA libraries, and libraries of sufficient quality were sequenced using the BGISEQ-500 platform (BGI, Shenzhen, China) [11,12]. We used conventional microbiological techniques to culture bacteria from the appendix samples [13,14].
Identification of bacterial sequences
An average of 46,439,586 sequence reads per sample was generated. High-quality sequencing data were obtained by removing low-quality and short (length < 35 bp) reads, followed by the subtraction of human host sequences mapped to the indicated human reference genome (hg19) using the Burrows-Wheeler Aligner (BWA) [15]. Reads with low complexity were identified using PRINSEQ [16] with the parameters “-derep 14 -derep_min 100 dust-threshold = 7” and then removed. The BWA was used to search for putative bacterial matches for the remaining reads against the bacterial and microbial genome databases, which were downloaded from the National Center for Biotechnology Information (NCBI) RefSeq database [17]. This database contains 1,494 bacterial genomes or scaffolds associated with human diseases. Reads with unique bacterial hits were retained for the next comparative analysis.
Statistical analyses
To increase the reliability of the results, species with less than 10 reads per 20,000,000 reads within each sample were filtered. To account for potential variations in sequencing efficacy, we transformed read abundances into percentages based on the total number of high-quality mapped sequences within each sample at all taxonomic levels of classification, namely, the phylum, genus and species levels. These normalized percentages were used in all subsequent data and statistical analyses. All statistical analyses were performed using the R software environment (R 3.5.2 for Windows). The evenness index was calculated using the formula E = S/log(R), where S is the Shannon diversity index and R is the number of operational taxonomic units (OTUs) in the sample (richness) [18]. In the boxplots, the black central lines represent the median, and the box edges represent the first and third quartiles. Differences between groups were calculated with the Wilcoxon rank-sum test. Principal coordinate analyses (PCoAs) were conducted using species abundance profiles. These results were confirmed with a permutational multivariate analysis of variance (PERMANOVA) that resulted in increased pseudo-F and R2 values. These analyses were performed in R using the vegan library with 999 permutations and the Euclidean distance method. Significant differences between the case and control groups in genus and species abundances were assessed using the Wilcoxon rank-sum test with a significant p-value threshold of less than 0.05.
Results
Participants
Twenty-one patients diagnosed with appendicitis were included in the study, and appendix samples were collected from 19 subjects. Detailed information for the included patients is provided in Table 1 (grouped by postoperative histological results) and Table 2 (grouped by bacterial culture results).
Taxonomic profiles of appendix microbiomes
Genus level
One hundred ninety-four genera were identified in the appendix samples, with six genera exhibiting relative abundances > 5%, including Bacteroides (30.8%), Odoribacter (8.0%), Escherichia (6.4%), Porphyromonas (6.1%), Tannerella (5.3%) and Shigella (5.1%) (Figure 1).
Figure 1.
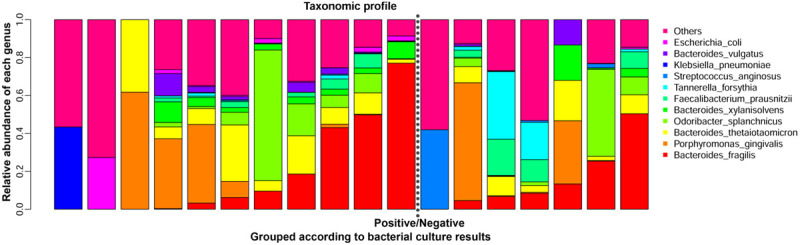
Genus-level taxonomic profiles of the microbiomes from the appendix samples.
The bar graph was generated by the script developed by BGI in the R software environment. Each vertical bar represents a unique sample. Samples were ordered by different groups shown below the figure. The y-axis represents the relative abundance of each genus. The vertical bars of the graph are grouped according to bacterial culture results and divided into Positive (case) and Negative (control). Only the top 11 genera were plotted. After filtering, no bacteria were detected in one of the negative group; therefore, this sample is not shown.
Species level
Three hundred sixty-one species were identified in the appendix samples, six of which had relative abundances > 5%, including Bacteroides fragilis (17.1%), Bacteroides thetaiotaomicron (10.3%), Odoribacter splanchnicus (8.0%), Porphyromonas gingivalis (6.1%) and Tannerella forsythia (5.3%) (Figures 2 and 3).
Figure 2.
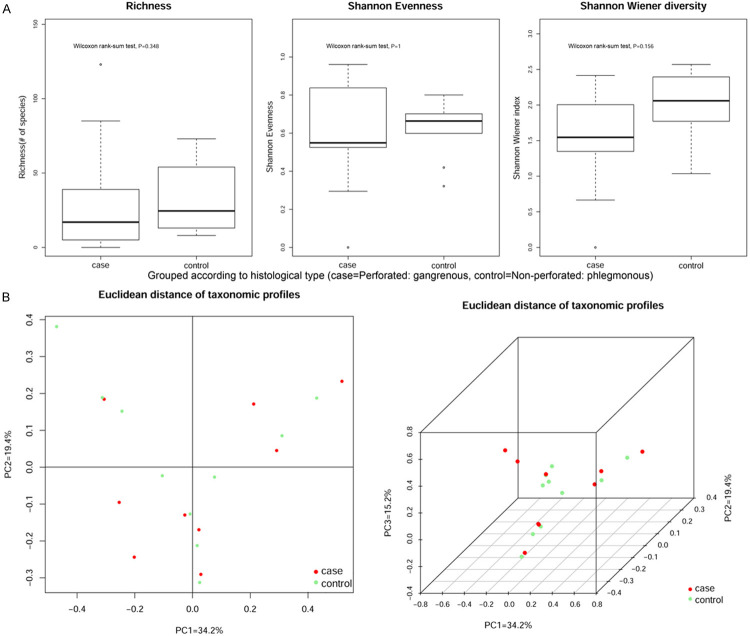
A. According to postoperative histological type, the samples were divided into Perforated: gangrenous group (n = 10) and Non-perforated: phlegmonous group (n = 9). Although it is greater than 0.05 for the P values of richness, Shannon evenness and Shannon Wiener diversity, no significant differences were observed in the bacterial composition. B. Alpha- and beta-diversity comparison of the samples grouped by histological type. Red and green represent patients with perforated (case) and non-perforated (control) appendicitis, respectively.
Figure 3.
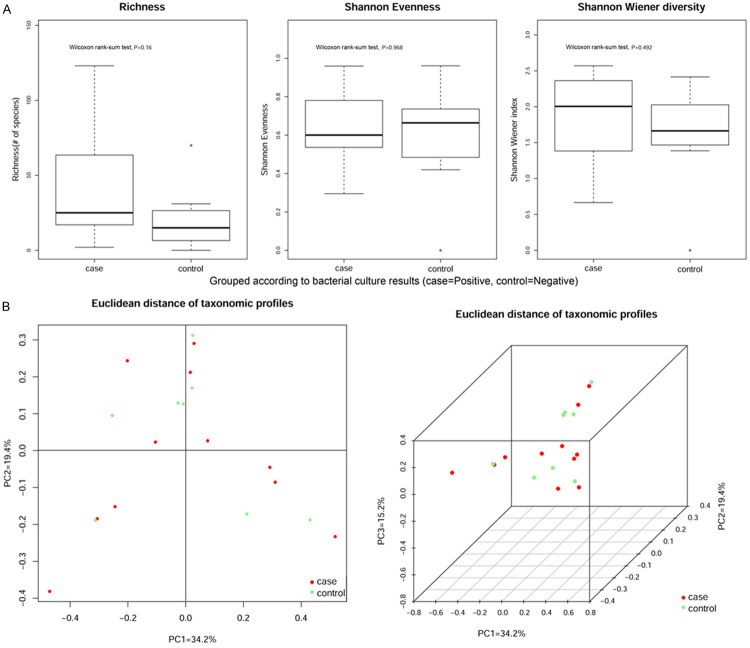
A. According to bacterial culture results, the samples were divided into Positive group (n = 11) and Negative group (n = 8). Although it is greater than 0.05 for the P values of richness, Shannon evenness and Shannon Wiener diversity, no significant differences were observed in the bacterial composition. B. Alpha- and beta-diversity comparison of the samples grouped by bacterial culture results. Red and green represent patients who were positive (case) and negative (control) for appendicitis, respectively.
Figures 2A and 3A show Alpha-diversity (Shannon-Wiener) indices in the different groups. Each p-value obtained by the Wilcoxon rank-sum test is reported in the boxes denote the interquartile range (IQR) between the 25th and the 75th percentile (first and third quartiles), and the central line represents the median. Figures 2B and 3B show Euclidean distance principal coordinate analysis (PCoA) of species-level taxonomic profiles. The proportion of variance explained by each principal component is denoted in the corresponding axis label.
The top 10 most abundant bacterial species detected by the whole metagenomic sequencing-based approach are shown in Table 3. Since no significant difference was observed between the culture positive and negative groups, these data were combined. These data may allow pharmacists to make better decisions.
Table 3.
Top 10 most abundant bacterial species detected by NGS
| Species | Culture positive | Culture negative |
|---|---|---|
| Shigella dysenteriae | 0.00028 | 0.024359 |
| Escherichia fergusonii | 0.000362 | 0.024747 |
| Escherichia coli | 0.001924 | 0.037056 |
| Streptococcus constellatus | 0.02913 | 0.034143 |
| Klebsiella pneumoniae | 0 | 0.039488 |
| Klebsiella variicola | 0 | 0.024776 |
| Morganella morganii | 0.047511 | 0.047511 |
| Bacteroides xylanisolvens | 0.03085 | 0.067361 |
| Tannerella forsythia | 0.07353 | 0.078547 |
| Odoribacter splanchnicus | 0.077448 | 0.180031 |
| Streptococcus anginosus | 0.056816 | 0.058706 |
| Bacteroides thetaiotaomicron | 0.069636 | 0.18821 |
| Faecalibacterium prausnitzii | 0.054137 | 0.073353 |
| Porphyromonas gingivalis | 0.120998 | 0.257946 |
| Bacteroides fragilis | 0.135933 | 0.324893 |
Discussion
Appendicitis is an acute abdomen disease caused by bacterial infection. The treatment of appendicitis consists of surgical removal of the appendix and antibiotic administration. For non-perforated appendicitis, antibiotic treatment plays an important role in preventing postoperative wound infection and intraperitoneal abscess and avoiding chronic appendicitis [19,20]. It has been reported that patients who have received an emergency appendectomy should accept preventive antibiotic therapy [21,22]. Several studies have proposed guidelines for the selection of antibiotic therapy for appendicitis patients [22-24]. Berríos-Torres et al. suggested that it is not necessary to administer appendicitis patients with antibiotic treatment postoperatively [25]. However, for perforated appendicitis, antibiotic therapy is necessary. Empirical antibiotic therapy should treat gram-negative bacilli and anaerobic bacteria, and the antibiotics used should be adjusted according to the results of bacterial culture, which is in accord with most reports. However, due to the limitation of technology, only 1% bacteria can be successfully cultured. As shown in Table 3, Escherichia coli could be cultured in all samples and was not the most abundant bacterium in either the culture positive or negative groups. Furthermore, we are skeptical of the negative culture results because they may result from limitations of the culture method or the use of antibiotics. Even a positive culture result may not fully reflect the bacterial composition because, according to the guideline based on the results of bacterial culture, antibiotics were used before surgery. Finally, bacterial culture requires a great deal of time, longer than what is acceptable in some instances. Currently, antibiotics are chosen based on culture results for postoperative patients, which is probably not appropriate. From our perspective, surgeons should monitor the changes in bacterial composition and abundance so that better decisions can be made for patients. Therefore, in the present study, we used NGS to examine bacteria in the guts of patients to completely characterize appendicitis-associated pathogenic bacteria. Our results showed that the NGS results were not in accord with those of bacterial culture, further suggesting that bacterial culture has limits in guiding the use of antibiotics in clinical practice.
By using NGS, we did not observe any significant difference between the bacterial culture negative and positive groups or between the phlegmonous and gangrenous groups. Jackson et al. extracted DNA from the microbiota of non-perforated and perforated appendixes for use in 16S rRNA gene sequencing analysis and observed a significant difference between non-perforated and perforated samples [26]. Salö et al. conducted a study of 3 control samples, 11 phlegmonous samples, 4 gangrenous samples and 4 perforated samples. They also utilized 16S rRNA gene sequencing but did not observe a significant difference among the different groups [9]. Our results demonstrated that there was not significant difference in the microbiota between the non-perforated and perforated appendicitis groups, suggesting that antibiotic selection does not need to be altered based on bacterial culture results. However, we advise that medical institutions should use NGS to regularly reanalyze the microbiota spectrum of appendicitis patients and assess the presence of drug resistance genes to allow for antibiotic treatments to be adjusted in a timely manner.
Specifically, completely excising lesions of the appendix and eliminating all the intraperitoneal effusion are the most important tasks in the treatment of appendicitis. Different acute abdomen infections harbor different bacterial species. Because of the similar bacterial species, appendicitis has similar pathogenesis among individuals [27]. In the light of all the above deficiencies, it is difficult to make an optimal decision on antibiotic usage based on bacterial culture results alone. Moreover, bacterial species and drug resistance genes change over time. Antibiotic resistance determinants are encoded by several genes, many of which can be transferred between bacteria [28,29]. Thus, NGS should be used to examine drug resistance genes at a regular interval to adjust the choice of antibiotics. However, the cost of NGS is high, making it difficult to use in all patients.
Based on the results of the present study and those of previous studies, we propose that a combination of NGS and bacterial culture technology should be used to guide treatment of appendicitis. NGS can be used to examine the bacterial species present in the appendix and analyze their drug resistance profiles. Before empirical antibiotic treatment of appendicitis patients, bacterial culture should be performed to assess refractory infections and evaluate the species of bacteria present throughout the treatment period. The combination of NGS and bacterial culture will improve the effects of treatments and the use of medical resources. From the perspective of pharmacists, we suggest regular monitoring of the top 10 abundant bacteria to provide evidence that allows them to make antibiotic prescription recommendations.
Our research has several limitations. First, the sample size of our study was small. We collected samples from 22 patients, which may affect the reliability of our results. However, we utilized NGS in our research, rather than 16S rRNA gene sequencing, and we performed metagenomic sequencing to evaluate all the genes present to the fullest extent possible. The second limitation of our research was the use of antibiotics. Preoperative use of antibiotics is in line with the current clinical situation. Before we collected the samples, all patients had already accepted antibiotic treatment, which may also have affected the results of our study. Thus, samples without antibiotic treatment should be evaluated to avoid alterations in the composition of the gut microbiota of appendicitis patients. However, performing such an analysis would be impossible, since antibiotic treatment is an important part of the standard treatment of appendicitis patients. Moreover, our analysis was based on DNA, which is stable enough to resist the influence of antibiotics. After bacteria are killed by antibiotics, their DNA can still be examined, although its copy number would decrease over time. The interval between antibiotic administration and surgery was quite short, which guaranteed the integrity of DNA and the reliability of our results. Since the time between antibiotic administration and surgery was short, it is unlikely that antibiotic administration would have substantially altered our results.
Conclusion
In our present study, we divided appendicitis patients into non-perforated and perforated appendicitis groups. Surgical excision is still the gold standard in appendicitis treatment, and antibiotic administration plays an important role in preventing preoperative wound infection and intraperitoneal abscess. Traditionally, suitable antibiotics are typically chosen according to the results of bacterial culture. However, because bacterial culture has several disadvantages, such as being time-consuming and having a low positive rate, and because patients were regularly administrated antibiotics during the perioperative period, which inevitably affected bacterial culture results, we believe that bacterial culture is not suitable as the sole guideline for antibiotic administration. Our findings demonstrated that compared with bacterial culture, NGS can characterize the profiles of microbiota of appendicitis patients more precisely and can also analyze drug resistance. Furthermore, the results can be rationally applied based on current antibiotic administration analysis. Since the cost of bacterial culture is relatively low, we apply it to all the appendicitis patients to analyze the refractory infection and the retrospective analysis of empirical antibiotic treatment throughout the hospitalization. The combination of two technologies will optimize the treatment of appendicitis. Furthermore, in a future study, we should increase our sample size and further verify our findings.
Acknowledgements
We acknowledge the work of microbiologist involved in the sequencing and bioinformatics analysis in these individuals (Dr. Huayong Liu and Zhuo Xiao). Supported by Guangdong Medical Science and Technology Research Foundation (A2018096). The Ethical Committee of the Fifth Affiliated Hospital of Southern Medical University reviewed the study details and confirmed that all methods were performed in accordance with the relevant guidelines and regulations (NYWY201804). Before any patient was enrolled in the study, it was confirmed that they had a sufficient understanding of the study. All of the appendix specimens from our cases were examined by routine culture. According to the ethical approval guidelines, the subjects did not need to sign a written consent form because all additional tests were run on specimens left over from routine microbiological examinations. Statistical analysts were blind to the identity information of the patients, and the sample information available to analysts was grouped by specific identification codes.
Disclosure of conflict of interest
None.
Abbreviations
- AA
Acute appendicitis
- rRNA
ribosomal RNA
- FISH
fluorescence in situ hybridization
- BWA
Burrows-Wheeler Aligner
- NCBI
National Center for Biotechnology Information
- OTUs
operational taxonomic units
- PERMANOVA
permutational multivariate analysis of variance
- NGS
Next-generation sequencing
References
- 1.Swidsinski A, Dörffel Y, Loening-Baucke V, Tertychnyy A, Biche-Ool S, Stonogin S, Guo Y, Sun ND. Mucosal invasion by fusobacteria is a common feature of acute appendicitis in Germany, Russia, and China. Saudi J Gastroenterol. 2012;18:55–58. doi: 10.4103/1319-3767.91734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guinane CM, Tadrous A, Fouhy F, Ryan CA, Dempsey EM, Murphy B, Andrews E, Cotter PD, Stanton C, Ross RP. Microbial composition of human appendices from patients following appendectomy. mBio. 2013;4:e00366–12. doi: 10.1128/mBio.00366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt A, Bharti R, Ilves I, Häsler R, Miettinen P, Paajanen H, Brunke G, Ellrichmann M, Rehman A, Hauser C, Egberts JH, Ott SJ, Schreiber S, Rosenstiel P, Herzig KH. Characteristic changes in microbial community composition and expression of innate immune genes in acute appendicitis. Innate Immun. 2015;21:30–41. doi: 10.1177/1753425913515033. [DOI] [PubMed] [Google Scholar]
- 4.Peeters T, Penders J, Smeekens SP, Galazzo G, Houben B, Netea MG, Savelkoul PH, Gyssens IC. The fecal and mucosal microbiome in acute appendicitis patients: an observational study. Future Microbiol. 2019;14:111–127. doi: 10.2217/fmb-2018-0203. [DOI] [PubMed] [Google Scholar]
- 5.Vanhatalo S, Munukka E, Sippola S, Jalkanen S, Grönroos J, Marttila H, Eerola E, Hurme S, Hakanen AJ, Salminen P. Prospective multicentre cohort trial on acute appendicitis and microbiota, aetiology and effects of antimicrobial treatment: study protocol for the MAPPAC (Microbiology APPendicitis ACuta) trial. BMJ Open. 2019;9:e031137. doi: 10.1136/bmjopen-2019-031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers MB, Brower-Sinning R, Firek B, Zhong D, Morowitz MJ. Acute appendicitis in children is associated with a local expansion of fusobacteria. Clin Infect Dis. 2016;63:71–78. doi: 10.1093/cid/ciw208. [DOI] [PubMed] [Google Scholar]
- 7.Zhong D, Brower-Sinning R, Firek B, Morowitz MJ. Acute appendicitis in children is associated with an abundance of bacteria from the phylum Fusobacteria. J Pediatr Surg. 2014;49:441–446. doi: 10.1016/j.jpedsurg.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Swidsinski A, Dörffel Y, Loening-Baucke V, Theissig F, Rückert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kühn S, Schilling J, Dörffel WV. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 9.Salö M, Marungruang N, Roth B, Sundberg T, Stenström P, Arnbjörnsson E, Fåk F, Ohlsson B. Evaluation of the microbiome in children’s appendicitis. Int J Colorectal Dis. 2017;32:19–28. doi: 10.1007/s00384-016-2639-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson R, Hugander A, Thulin A, Nystrom PO, Olaison G. Indications for operation in suspected appendicitis and incidence of perforation. BMJ. 1994;308:107–110. doi: 10.1136/bmj.308.6921.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 12.Long Y, Zhang Y, Gong Y, Sun R, Su L, Lin X, Shen A, Zhou J, Caiji Z, Wang X, Li D, Wu H, Tan H. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch Med Res. 2016;47:365–371. doi: 10.1016/j.arcmed.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Humphries RM, Linscott AJ. Practical guidance for clinical microbiology laboratories: diagnosis of bacterial gastroenteritis. Clin Microbiol Rev. 2015;28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain JM, Fournier PE, Raoult D. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacci G, Paganin P, Lopez L, Vanni C, Dalmastri C, Cantale C, Daddiego L, Perrotta G, Dolce D, Morelli P, Tuccio V, De Alessandri A, Fiscarelli EV, Taccetti G, Lucidi V, Bevivino A, Mengoni A. Pyrosequencing unveils cystic fibrosis lung microbiome differences associated with a severe lung function decline. PLoS One. 2016;11:e0156807. doi: 10.1371/journal.pone.0156807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu AM, Huang L, Li TJ. Single-incision versus three-port laparoscopic appendectomy for acute appendicitis: systematic review and meta-analysis of randomized controlled trials. Surg Endosc. 2015;29:822–843. doi: 10.1007/s00464-014-3735-z. [DOI] [PubMed] [Google Scholar]
- 21.Fry DE. Surgical site infections and the surgical care improvement project (SCIP): evolution of national quality measures. Surg Infect (Larchmt) 2008;9:579–584. doi: 10.1089/sur.2008.9951. [DOI] [PubMed] [Google Scholar]
- 22.Flum DR, Davidson GH, Monsell SE, Shapiro NI, Odom SR, Sanchez SE, Drake FT, Fischkoff K, Johnson J, Patton JH, Evans H, Cuschieri J, Sabbatini AK, Faine BA, Skeete DA, Liang MK, Sohn V, McGrane K, Kutcher ME, Chung B, Carter DW, Ayoung-Chee P, Chiang W, Rushing A, Steinberg S, Foster CS, Schaetzel SM, Price TP, Mandell KA, Ferrigno L, Salzberg M, DeUgarte DA, Kaji AH, Moran GJ, Saltzman D, Alam HB, Park PK, Kao LS, Thompson CM, Self WH, Yu JT, Wiebusch A, Winchell RJ, Clark S, Krishnadasan A, Fannon E, Lavallee DC, Comstock BA, Bizzell B, Heagerty PJ, Kessler LG, Talan DA. A Randomized trial comparing antibiotics with appendectomy for appendicitis. N Engl J Med. 2020;383:1907–1919. doi: 10.1056/NEJMoa2014320. [DOI] [PubMed] [Google Scholar]
- 23.Slomski A. Antibiotics on par with surgery for appendicitis. JAMA. 2020;324:2020. doi: 10.1001/jama.2020.22576. [DOI] [PubMed] [Google Scholar]
- 24.Mahase E. Antibiotics are as good as surgery for appendicitis, study reports. BMJ. 2020;371:m3870. doi: 10.1136/bmj.m3870. [DOI] [PubMed] [Google Scholar]
- 25.Berríos-Torres SI, Umscheid CA, Bratzler DW, Leas B, Stone EC, Kelz RR, Reinke CE, Morgan S, Solomkin JS, Mazuski JE, Dellinger EP, Itani KMF, Berbari EF, Segreti J, Parvizi J, Blanchard J, Allen G, Kluytmans J, Donlan R, Schecter WP. Centers for disease control and prevention guideline for the prevention of surgical site infection, 2017. JAMA Surg. 2017;152:784–791. doi: 10.1001/jamasurg.2017.0904. [DOI] [PubMed] [Google Scholar]
- 26.Jackson HT, Mongodin EF, Davenport KP, Fraser CM, Sandler AD, Zeichner SL. Culture-independent evaluation of the appendix and rectum microbiomes in children with and without appendicitis. PloS one. 2014;9:e95414. doi: 10.1371/journal.pone.0095414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu SC, Rau CS, Liu HT, Kuo PJ, Chien PC, Hsieh TM, Tsai CH, Chuang JF, Huang CY, Hsieh HY, Hsieh CH. Metagenome analysis as a tool to study bacterial infection associated with acute surgical abdomen. J Clin Med. 2018;7:346. doi: 10.3390/jcm7100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chon JW, Lee UJ, Bensen R, West S, Paredes A, Lim J, Khan S, Hart ME, Phillips KS, Sung K. Virulence characteristics of mecA-positive multidrug-resistant clinical coagulase-negative staphylococci. Microorganisms. 2020;8:659. doi: 10.3390/microorganisms8050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marathe NP, Salvà-Serra F, Karlsson R, Larsson DGJ, Moore ERB, Svensson-Stadler L, Jakobsson HE. Scandinavium goeteborgense gen. nov., sp. nov., a new member of the family enterobacteriaceae isolated from a wound infection, carries a novel quinolone resistance gene variant. Front Microbiol. 2019;10:2511. doi: 10.3389/fmicb.2019.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]