Abstract
Prostate cancer is one of the most frequently diagnosed malignancies in developed countries and approximately 248,530 new cases of prostate cancer are likely to be diagnosed in the United States in 2021. During the late 1990s and 2000s, the prostate cancer-related death rate has decreased by 4% per year on average because of advancements in prostate-specific antigen (PSA) testing. However, the non-specificity of PSA to distinguish between benign and malignant forms of cancer is a major concern in the management of prostate cancer. Despite other risk factors in the pathogenesis of prostate cancer, recent advancement in molecular genetics suggests that genetic heredity plays a crucial role in prostate carcinogenesis. Approximately, 60% of heritability and more than 100 well-recognized single-nucleotide-polymorphisms (SNPs) have been found to be associated with prostate cancer and constitute a major risk factor in the development of prostate cancer. Recent findings revealed that a low to moderate effect on the progression of prostate cancer of individual SNPs was observed compared to a strong progressive effect when SNPs were in combination. Here, in this review, we made an attempt to critically analyze the role of SNPs and associated genes in the development of prostate cancer and their implications in diagnostics and therapeutics. A better understanding of the role of SNPs in prostate cancer susceptibility may improve risk prediction, enhance fine-mapping, and furnish new insights into the underlying pathophysiology of prostate cancer.
Keywords: Prostate cancer, polymorphism, single nucleotide polymorphisms, prostate specific antigen, tumorigenesis
Introduction
Prostate cancer is one of the major non-skin malignancies because of its health-associated costs, high prevalence rate, and mortality [1]. The increasing trend of incidence is seen more often in developed countries probably due to more advanced medical care facilities and PSA screening at early stages of disease development [2]. However, the highest mortality rate has been observed in the males of the African race than the white race [3]. The lowest rate of prostate cancer mortality has been recorded in Asian men [4]. Prostate cancer is mostly diagnosed in elderly males over young males and is expected that its incidence could increase in the coming future [5]. Owing to the presence of non-modifiable risk factors, such as ethnicity (race), age and genetic (BRCA2, BRCA1, HOXB13, NBS1, CHEK2 mutations, and SNPs) factors [6], it is extremely hard to reduce prostate cancer incidence, and thus gives more importance to early diagnostics and therapeutics [7]. Despite having clinically confined and dormant tumors at the time of diagnosis of prostate cancer, yet the malignancy of the prostate holds among the major cause of mortality worldwide [8]. Owing to have high mortality rate, prevalence, and socioeconomic-related issues, the field of prostate carcinogenesis and affected patients have a major challenge in both diagnostics and therapeutics [9]. The major reasons are the absence of specific cancer associated or prostate cancer patient-specific biomarkers, limitations, and non-specificity in existing diagnostics to distinguish between benign and aggressive tumors, and finally in the therapeutic modalities of prostate cancer due to over-treatment and development of drug resistance [10-12].
Prostate cancer is life threatening polygenetic disease, and many genes involved in the pathogenesis of this disease are imprecisely explored [13]. Therefore, there is a need to identify novel genetic markers which could be used as indicators to predict the most susceptible segments of the population to the disease or for the genes that are involved in prostate cancer pathogenesis. Single nucleotide polymorphisms (SNPs) are variations in a genome’s base pair in a DNA sequence and occur in nearly 1 out of 800 base pairs [14]. Conventionally, for a single nucleotide variation to be described as polymorphism it must occurs in the DNA of at least 1% of the population [15]. SNPs cause variations in genes which alters the protein and enzymatic machinery of the cell [16]. The inheritance of genes within families is strongly influenced by SNPs [17] and reports suggest that susceptibility to prostate cancer is associated with SNPs and the susceptibility of developing prostate cancer in certain individuals is higher than others [18]. It has become clear from genome-wide association studies (GWAS) and fine-mapping efforts that more than 100 common SNPs are associated with prostate cancer susceptibility [19]. For example, polymorphisms in gene 8q24 have shown strong links with prostate cancer susceptibility, signifying that 8q24 polymorphisms could be good markers in prostate cancer diagnosis and therapy [20]. Many other studies have shown the association of SNPs in candidate genes with increased susceptibility to prostate cancer [21]. The type of candidates that show association with increased susceptibility to prostate cancer include genes involved in steroid metabolism, oxidative stress, cell adhesion, angiogenesis, cell cycle and DNA repair as well as variants of other genes [22]. A recently conducted association analyses of more than 140,000 men have identified 63 new prostate cancer susceptibility loci [23].
Therefore, the current review outlines the role of common SNPs in prostate cancer development and how these SNPs could be utilized for the screening and management of prostate cancer. SNPs are predictors of aggressive prostate cancer and in this review the research findings highlighted that SNPs represents an important genetic biomarker that has strong association with susceptibility to prostate cancer. However, there are indications that research focused on genetic biomarkers is not complete and there is a need to identify clinically more relevant genetic biomarkers that could be used for screening, diagnosis, and prognosis of prostate cancer.
Prostate cancer progression
With the advancement in age, the enlargement of the prostate gland is common. At the age of around 40 years, benign prostate hyperplasia (BPH) develops from the transition zone of the prostate and implicates in urination complications [24]. Although there is no report that BPH is causally associated with prostate cancer, however recent evidence suggests that BPH might have a possible casual association with prostate cancer inflammation which is believed to be a key event in the progression of prostate cancer [25,26]. The other common complication in the prostate with age is prostatitis [27]. Prostate cancer is commonly adenocarcinoma and arises from epithelial tissue of the prostate gland [28]. Around 70%, 25% and 5% prostate cancer arise from peripheral, transition and central zone, respectively [29]. Prostate tumors are usually multifocal bearing multiple tumors and are believed to be advanced from the Prostatic Intraepithelial Neoplasia (PIN) [30] Figure 1. The advancement in disease progression affects neighboring organs such as the seminal vesicle, pelvis, urethra, urinary bladder via lymph nodes and finally spreads to bones through the pelvis [31]. A plethora of reports suggests that prostate cancer can also metastasis to lungs and other organs [32]. Recent reports suggest that prostate metastasis originates and spreads from a single independent clone present within the prostate gland [33,34]. This suggests the heterogeneity and aggressiveness of tumors within the prostate and implicates that future prostate cancer diagnostics and therapeutics will be a challenging task. Therefore, these studies suggest that prostate cancer is an elderly cancer in men which mainly arise from peripheral epithelial tissue of prostate. Additionally, it is a heterogenous malignancy that originate from PIN which further affects neighbouring organs and finally metastasis to various organs that includes bones and lungs.
Figure 1.
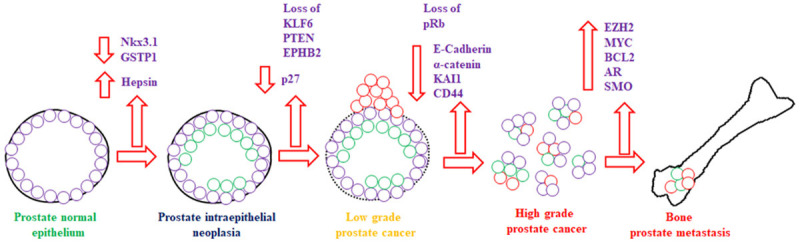
Prostate cancer initiation, progression, and metastasis to bone with upregulation and downregulation of tumor suppressor and proto-oncogenes involved in various signaling pathways.
Prostate cancer diagnosis and grading
The common mode of detection of prostate cancer is PSA testing or occasionally by clinical symptoms such as complications in urination to empty the urinary bladder [35]. At the initial stage of prostate malignancy when the tumor is confined to the prostate with no symptoms, the metastatic spread often emerges with pain in the hips, pelvis, and back portion of the skeleton [36]. The foremost step in the diagnostics of prostate malignancy is PSA testing, followed by digital rectal examination (DRE) [37]. The PSA testing was debatable for quite some time to fix the threshold level of PSA under various guidelines to limit the overdiagnosis of clinically invaluable malignancies associated with the prostate [38]. The Table 1 shows the PSA threshold level for the age group when the DRE is negative to be recommended for further investigations for the diagnosis of prostate malignancy. The follow-up for the suspected prostate cancer patients includes an examination of transrectal ultrasonography-guided needle biopsy with at least 12 cores of prostate samples followed by an examination of the specimens by histopathological means and the reporting should be performed as per the Gleason grading system [39]. The Gleason’s score ranges from 1 for well-characterized prostate glandular cells to 5 for poorly characterized glandular cells [40]. Therefore, the common mode to diagnose prostate cancer is detection of PSA. However, due to its non-specificity and sensitivity, the suspected prostate cancer patients should perform DRE, followed by transrectal ultrasonography-guided needle biopsy to characterize the stage of malignancy based on Gleason’s grading system.
Table 1.
Threshold level of PSA and age group of patients with digital rectal examination
| S. No | Digital Rectal Examination | PSA level | Age |
|---|---|---|---|
| 01 | Negative | ≥ 2 µg/l | < 50 years |
| 02 | Negative | ≥ 3 µg/l | 50-70 years |
| 03 | Negative | ≥ 5 µg/l | 70-80 years |
| 04 | Negative | ≥ 7 µg/l | > 80 years |
Primary risk factors and their causal association with prostate cancer
The primary risk factor for the development of prostate cancer is familial history, age, and ethnicity [41]. Recent studies of epidemiology suggest that prostate cancer is one of the prominent heritable malignancy and suggests a strong causal association between genetic factors and the development of prostate cancer [42,43]. A person having a heritable link with a person who had diagnosed with prostate cancer has a 2-3-fold higher risk of developing prostate malignancy as compared with the family without any familial history of prostate cancer [44]. A Nordic twin study revealed that around 60% of prostate cancer patients had familial history to develop prostate cancer [45].
With the advancement in technology, recent studies suggest that molecular genetics plays a crucial role in the development of prostate cancer [46]. The human genome contains nearly 30 million base-pairs of nucleotides, of which 99% of these nucleotide base pairs are in the same sequence in all human beings [47]. During the evolutionary process, the sequence sites where the mutations have happened and dispersed to more than 1% of the population are called polymorphic [48]. The human genome has various types of variation called polymorphisms and has been reported to be associated with the progression of cancer [49]. One of the common polymorphisms is SNPs. The SNP is a variation in the sequence of DNA at a single position of a nucleotide (A, T, C, or G) in the genome that varies from the regularly expected nucleotide as shown in Figure 2 [15]. Various genetic sequence programs at the international level have revealed the identification of millions of SNPs in the human genome [15]. SNPs does not directly influence the protein-coding function because they are localized within the introns and intergeneric regions of the genome [50]. The SNPs act as an interesting genetic marker to differentiate the susceptibility of a person to disease [51]. The determination of SNPs is quite easy, simple, and performed only once, these features make them appealing biomarkers in the field of molecular genetics to analyze the disease status [52]. The growing demand for the reputation of SNPs in prostate cancer pathogenesis is verified by the numerous studies conducted recently and suggests a key role of SNPs in the field of prostate cancer diagnostics [42,53].
Figure 2.
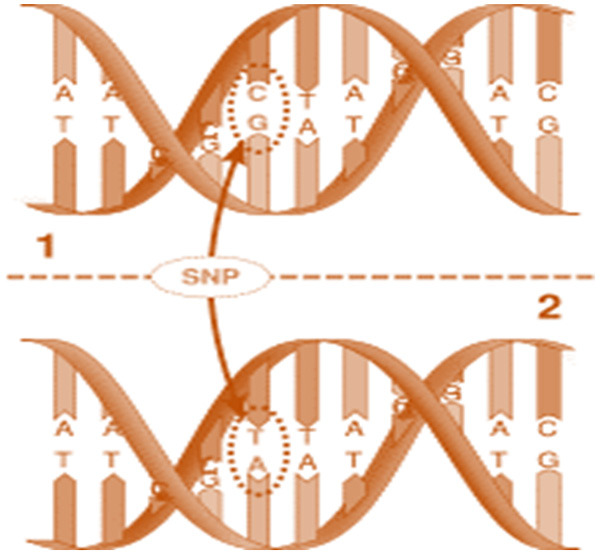
A small change in nucleotide sequence results in the formation of single nucleotide polymorphism.
Genome-wide association studies (GWAS) have demonstrated that more than 100 SNPs have been identified with low to moderate association with prostate cancer Table 2 [54]. Interestingly, studies reveal that a moderate effect of prostate cancer risk was observed when SNPs were single. However, the effect of prostate cancer risk increases significantly when the SNPs were in combination [54]. This can be explained by approximately more than 39% of prostate cancer risk is associated with familial history. Further, analysis of GWAS in present and future, to add some more genetic variants which could help the scientific community to better explain the familial risk in prostate cancer. However, the interesting question raised here is, does the identification of more SNPs and their association with increased prostate cancer risk has the worth over the established SNPs.
Table 2.
List of SNPs along with their endpoint conclusion discussed in table with the appropriate references
| High risk SNPs involved in prostate cancer | Investigated gene(s)/loci | Number of cases in the study | Endpoint Conclusion | References |
|---|---|---|---|---|
| rs17021918, rs7679673, rs1447295, rs721048, rs9364554, rs16901979, rs2928679, rs4430796, rs1512268, rs6983267, rs16902094, rs12621278, rs920861, rs1855962, rs4962416, rs2660753, rs10934853, rs7127900, rs10486567, rs12418451, rs10993994, rs8102476, rs10896449, rs2735839, rs5945619, rs5759167, rs1465618 | - | 4621 | Risk prediction models at genetic level lead the identification of a subset of high-risk suspected prostate cancer patients at early curable stage | Sun et al. 2011 [74] |
| rs7841060, rs620861, rs6983267, rs1447295, rs721048, rs4242382, rs12621278, rs7837688, rs4857841, rs16902094, rs12500426, rs1571801, rs9364554, rs10993994, rs6465657, rs4962416, rs1512268, rs7127900, rs1016343, rs12418451, rs16901979, rs7931342, rs1465618, rs10896449, rs2660753, rs11649743, rs4430796, rs7501939, rs1859962, rs17021918, rs266849, rs7679673, rs2735839, rs10486567, rs5759167, rs6465657, rs5945572, rs2928679, rs5945619, rs4961199 | SLC25A37, CPNE3, EHBP1, MSMB, BIK, SLC22A3, ITGA6, TCF2, PDLIM5, EEFSEC, CTBP2, KLK3, TET2, THADA, CNGB3, JAZF1, LMTK2, NUDT11, NKX3-1 | 15161 | Addition of familial history and genetic information apart from PSA screening in predictive risk models of prostate cancer could be beneficial for young suspected prostate cancer patients | Lindstrom et al. 2012 [75] |
| rs721048, rs1465618, rs12621278, rs2660753, rs17021918, rs12500426, rs7679673, rs9364554, rs10486567, rs6465657, rs10505483, rs6983267, rs1447295, rs2928679, rs1512268, rs10086908, rs620861, rs10993994, rs4962416, rs7931342, rs7127900, rs4430796, rs1859962, rs2735839, rs5759167, rs5945619 | - | 2885 | Development of algorithm to predict prostate cancer using familial history and genotypes of more than 25 SNPs. This can be used to assess the pedigrees of an arbitrary population size or structure. | Macinnis et al. 2011 [76] |
| rs2736098, rs11067228, rs10788160, rs17632542 | - | 964 | Combination of genotyping and PSA detection more specifically detect suspected prostate cancer and delay or prevent needless prostate biopsies | Helfand et al. 2013 [88] |
| rs10486567, rs17632542, rs11228565, rs5759167 | JAZF1, IGF2, TPCN2, HNF1B, DPF1, MYC, PPP1R14A, NCOA4, SPINT2, TH, KLK3, INS, TTLL1, MYEOV, BIK, MSMB, NUDT11 | 3772 | Using SNPs for assessing prostate cancer risk is less accurate than PSA detection and there is no advancement in prostate cancer diagnostics using both SNPs and PSA. | Klein et al. 2012 [80] |
| rs721048, rs10993994, rs12621278, rs7127900, rs10086908, rs10896449, rs13252298, rs11649743, rs16901979, rs4430796, rs6983267, rs1859962, rs1016343, rs8102476, rs6983561, rs2735839, rs7679673, rs5759167, rs16902094, rs5945619, rs1447295 | EEFSEC, CTBP2, THADA, HNF1B, ITGA6, PPP1R14A, PDLIM5, KLK3, FLJ20032, TNRC6B, JAZF1, BIK, LMTK2, NUDT11, MSMB, 8q24.21, EHBP1, 11q15.5, SLC22A3, 11q13.2, NKX3-1, 17q24.3 | 5241 | Implementation of risk-prediction and genetic risk score decreases biopsies by 22.7% and missing diagnosis cost of prostate cancer by 3% | Aly et al. 2011 [131] |
| rs8844019 | EGFR | 212 | After radical prostatectomy, the authors observed a strong association between prostate biochemical recurrence and the SNPs | Perez et al. 2010 [132] |
| rs9282861, rs198977, rs11536889 | SULT1A1, KLK2, TLR4 | 703 | After radical prostatectomy predicting biochemical recurrence in prostate cancer were significantly improved by the inclusion of patient genetic information and clinicopathological data interpretation. | Morote et al. 2010 [133] |
| rs2208532, rs4952197, rs12470143, rs518673, rs523349, rs12470143 | SRD5A2, SRD5A1 | 846 | Multiple variations in SRD5A2 and SRD5A1 are associated with either decreased or increased rates of BCR after radical prostatectomy. | Audet-Walsh et al. 2011 [134] |
| rs10895304 | MMP7 | 212 | Predictive analysis of A/G genotype in clinically localized prostate cancer patients reduces recurrence | Jaboin et al. 2011 [135] |
| rs2279115 | bcl2 | 290 | Biochemical recurrence frequency was more observed in -938 A/A genotype carriers than -938 CC + C/A carriers | Bachmann et al. 2011 [136] |
| rs3846716 | APC, CTNNB1 | 307 | After radical prostatectomy, the SNP genotype of AA/GA has promising prognostic role | Huang et al. 2010 [137] |
| rs2016347, rs2946834 | IGF1R, IGF1 | 320 | Post radical prostatectomy associated with biochemical recurrence has a strong genetic association between IGF1R rs2016347 and IGF1 rs2946834 | Chang et al. 2013 [138] |
| rs2569733, rs9282861, rs198977, rs1800247 | KLK3, SULT1A1, KLK2, BGLAP | 670 | In post radical prostatectomy Genetic testing such as SNPs and clinicopathological data allows improvement of preoperative prediction of early biochemical recurrence | Borque et al. 2013 [139] |
| rs25489 | XRCC1 | 603 | In post radiotherapy polymorphism of XRCC1 Arg280His could be protective against the high-grade late toxicity | Langsenlehner et al. 2011 [140] |
| rs1982073, rs1800469 | TGF β1 | 322 | After radical prostatectomy followed by radiotherapy with IMRT, the genetic variants of TGF β1 in codon 10 T > C and codon -509 C > T are involved in nocturia induced by radiations | De langhe et al. 2013 [141] |
| rs1799794 | ERCC2, MLH1, ATM, XRCC3, LIG4 | 698 | Development of gastrointestinal toxicity by the mean dose and SNP in post 3D-CRT | Fachal et al. 2012 [142] |
| None | TGF β1 | 413 | No association was observed between the risk of late toxicity and haplotypes or investigated SNPs | Fachal et al. 2012 [143] |
| None | XRCC6, ABCA1, MRE11A, ALAD, MSH2, APEX1, NEIL3, BAX, NFE2L2, ATM, NOS3, CDKN1A, PAH, DCLRE1C, PRKDC, EPDR1, PTTG1, ERCC2, RAD17, ERCC4, RAD21, GSTA1, RAD9A, LIG4, REV3L, HIF1A, SART1, LIG3, SH3GL1, MED2L2, SOD2, TGFB1, MLH1, TGFB3, MAP3K7, TP53, MAT1A, XPC, CD44, XRCC1, IL12RB2, XRCC3, GSTP1, XRCC5, MPO | 637 | No association was confirmed in the current study after evaluating previous reports. The study further evaluates the SNPs P value distribution against the toxicity score. | Barnett et al. 2012 [144] |
| rs12422149, rs1077858, rs1789693, | SLCO1B3, SLCO2B1 | 538 | Patients on ADT possess three SNPs in SLCO2B1 and have a strong associated with time to progression. However, SLCO1B3 and SLCO2B1genotype patients on ADT translocate androgens efficiently and displayed median 2-year shorter TTP | Yang et al. 2011 [99] |
| - | TGFBR2 | 1765 | The post ADT patients have more risk of early relapse when TGFBR2-875GG are in homozygous condition. Combination of genetic interpretation and clinicopathological data revealed high ability to envisage the risk of failure of ADT | Teixeira et al. 2009 [145] |
| rs6900796, rs1268121 | TRMT11, WBSCR22, PRMT2, SRD5A1, PRMT7, SRD5A2, HSD17B1, UGT2B10, PRMT6, HSD17B12, HSD11B1, PRMT3, THBS1, UGT2B7, SULT2A1, CYP3A4, UGT2B4, SULT2B1, PRMT5, CYP11B2, PRMT8, METTL6, HSD3B2, CARM1, UGT1A4, HEMK1, ARSE, METTL2B, UGT1A8, UGT2B11, UGT1A5, UGT1A10, CYP19A1, ESR2, ESR1, LCMT2, UGT2A3, UGT1A9, SERPINE1, AR, UGT1A6, UGT1A7 AKR1D1, AKR1C4, UGT2A1, STS, SULT1E, HSD17B8, UGT2B28, ARSD, HSD17B3, HSD17B2, LCMT1, HSD17B7 CYP11B1, UGT1A1, HSD3B1, UGT1A3 | 304 | The study demonstrated that a strong association was observed between TRMT11 and time to ADT failure. Further, the study revealed that two out of 4 TRMT11 tag SNPs are strongly associated with time to ADT failure. | Kohli et al. 2012 [101] |
| rs6728684, rs1071738, rs3737336, rs998754, rs1045747, rs4351800 | KIF3C, PALLD, ACSL1, CDON, GABRA1, IFI30, SYT9, ETS1, ZDHHC7, has-mir-423, MTRR | 601 | In multivariate models that includes clinicopathological predictors, the genotypes IFI30 RS1045747, CDON rs3737336 and KIF3C rs6728684 emerge significant predictors for the progression of disease. More number of unfavourable type of genotypes was found to be associated with quick disease progression and lessens the prostate cancer-specific survival time after ADT | Bao et al. 2011 [102] |
| rs2051778, rs16934641, rs3763763, rs3763763 | SKAP2, GNPDA2, TACC2, BNC2, SKAP1, ZNF507, KLHL14, ZNF521, NR4A2, SPRED2, FBXO32, ALPK1, AATF | 601 | In post ADT, the clinical outcome is associated with genetic variant in TACC2, BNC2 and ALPK1. Further, the effect is cumulative when combination of ACM and genotypes of ADT of two loci of interest were investigated | Huang et al. 2012 [103] |
| rs9508016, rs2939244, rs7830622, rs9508016, rs6504145 | FLT1, ACTN2, PSMD7, ARRDC3, SKAP1, XRCC6BP1, FBXO32, FLRT3, NR2F1 | 601 | For PCSM the genetic variants in FLT1, ARRDC3, and SKAP1 are significant predictors. However, for ACM the genetic variants that are significant predictors are in FLT1 and FBXO32. Together, there was a strong combined effect on ACM and PCSM. | Huang et al. 2012 [104] |
| rs4862396, rs7986346, rs3734444 | BMP5, RXRA, NCOR2, ERG, IRS2, BMPR1A, MAP2K6 | 601 | The study suggests a strong association of genetic variants in BMP5, CASP3 and IRS2 with ACM. However, a significant association was observed in genetic variants in IRS2 and BMP5 with PCSM. In ADT patients, the greater number of unfavourable genotypes at interested loci have less time to PCSM and ACM. | Huang et al. 2012 [146] |
| - | IGF-1 | 251 | The study suggests that considering sum of all the genetic risk factors in each LD block, there is reduction in cancer-specific survival significantly when compared to 0-2 risk factors. | Tsuchiya et al. 2013 [105] |
| rs1056836 | CYP1B1 | 60 | The clinical outcome in CPRC patients to docetaxel and predictive marker is polymorphism. | Pastina et al. 2010 [120] |
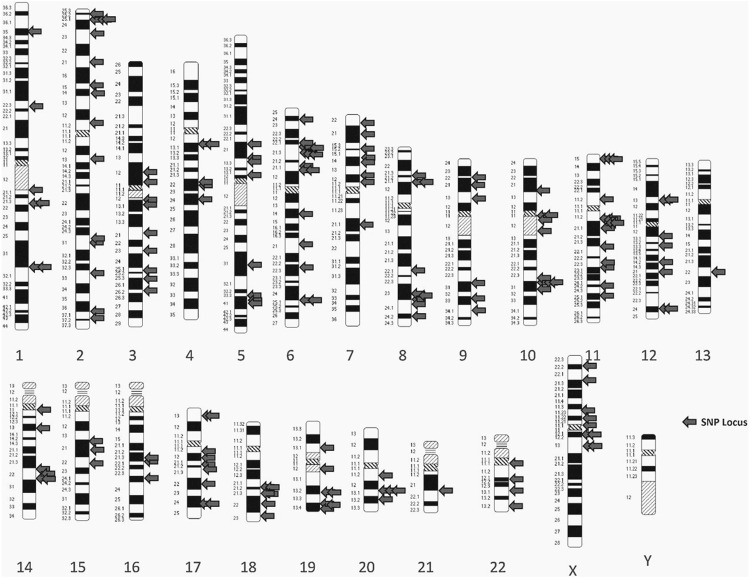
Various SNPs are associated with each other via “linkage disequilibrium”. It is an association (non-random) of alleles at two loci or more than two loci projected from a distinct inherited chromosome [55]. Nevertheless, the identification of SNPs through GWAS studies are mainly “index SNPs” that are different from SNPs connected through linkage disequilibrium [56]. Reports suggest that index SNPs are not associated with the increase in prostate cancer risk [57]. Therefore, the identification of causative linkage-type SNPs and their underlying molecular analysis is warranted. Some SNPs that are located within an open reading frame of an exon can modify the stability of messenger RNA (mRNA) or efficiency of translation of proteins as well as modulations in the activity or structure of translated proteins [58]. However, most SNPs lie outside the exonic region of the functional gene and could modulate both transcriptions as well as the level of genome organization [59] Figure 3. Therefore, the recent findings revealed that genetic factors have a major contribution in prostate malignancy. Among them SNPs which acts as an appealing genetic marker to differentiate the susceptibility of a person to prostate cancer progression. Thus, the need to identify and investigate the role of different SNPs in the progression of prostate cancer as biomarkers is deeply needed for early prostate cancer diagnosis.
Figure 3.
Idiogram showing SNPs locus associated with prostate cancer risk by GWAS studies.
Early diagnostics in prostate cancer
The only diagnostic test for the suspected prostate cancer patients at the early stage of the disease is the detection of serum PSA level which acts as a marker for prostate cancer [60]. Owing to its low specificity and sensitivity issues, PSA screening at the mass level has been immensely debated [61]. Two primary screening trail studies: The Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial and The European Randomized Study of Screening for Prostate Cancer (ERSPC) has discussed and concluded that there is no significant difference in prostate cancer patient mortality who are screened or not [62]. Based on the data from the ERSPC study which concludes that detection of prostate cancer by PSA-over diagnosis prevents one prostate cancer-associated death, while screening as many as 1410 suspected men to treat additional 48 prostate cancer cases [63]. Based on these studies, the US Preventive Services Task Force has reduced the mass detection of PSA and ignored approximately 50% of prostate cancer-associated mortality since the introduction of PSA [64]. However, without PSA screening in most cases, diagnosis of prostate cancer was detected late with prominent symptoms and often disease has been progressed to advanced stages with limited therapeutic response. Therefore, the currently followed European association of urology (EAU) guidelines urge for opportunistic screening for suspected prostate cancer patients. This includes PSA screening, genetic analysis for the detection of SNPs, and searching for novel biomarkers which is the need of an hour for prostate cancer diagnostics [65]. To conclude, recent findings suggest that in addition to PSA screening detection of SNPs and other associated markers are extremely important for early diagnosis of prostate cancer.
Using opportunistic screening strategy to improve prostate cancer diagnostics
Although overdiagnosis of PSA screening remains a concern for both clinicians and suspected patients of prostate cancer [66]. However, as per new EAU guidelines, opportunistic PSA testing would be performed in high-risk suspected patients apart from genetic screening [67]. Recent studies suggest that molecular genetics play a crucial role in the development of prostate cancer. Therefore, apart from performing opportunistic screening of PSA, analysis of genetic modifications could be a deciding factor to predict the high-risk patient groups which could be susceptible to develop prostate cancer [68]. Interestingly, this could have a great advantage in determining which suspected patients would be benefited from PSA testing that in turn will decide the suspected patient’s group go for prostate biopsies. The present genetic variations were analyzed by GWAS studies to correlate the risk associations. However, the only limitation in clinics is the characterization and prediction of risk in suspected patients is based on establishing the categorizing people into various groups. This categorization of suspected patients allows the stratification of the high-risk group which has greater chances of developing prostate cancer than other groups in the population. This stratification provides a platform for clinicians to better understand and calculate the absolute prostate cancer risk for patients after assessing an individual’s medical information.
Numerous epidemiological models developed by clinicians and scientists reveal that using various risk factors such as age, history and genetic modification could have a great impact on predicting the risk and analyzing the development of prostate cancer [69] Figure 4. Zheng et al. demonstrate that good efficiency was achieved in prostate cancer detection when a comparison of risk factors was combined with a level of PSA cut-off of 4.1 ng/mL [70]. Owing to the low sensitivity and specificity of PSA screening, initially, the analysis looks like irrelevant, but the efficiency of detection was good. The genetic analysis reveals that it is economically cost-effective as well as determined once in a lifetime, however, determination of PSA level in serum varies and needs to fix by performing many times. Unfortunately, once the PSA level, age and familial history were well recorded, the cumulative predictive analysis of genetic screening and PSA screening for the diagnosis of prostate cancer did not have improved the prostate cancer diagnostics [70,71].
Figure 4.
Various risk factors involved in prostate cancer progression.
The debate is still going on whether SNPs are good enough to use them to predict prostate cancer risk. A recent report suggests that SNPs are not able to differentiate cases within the control groups but could be especially useful in the analysis of determining the suspected prostate cancer patients [72]. To predict the absolute risk for the development of prostate cancer risk, Xu et al. predict an absolute model based on the cumulation of 14 SNPs, familial history, and a specific age group [73]. This model was used in the Swedish and USA population to detect a small subset of suspected patients (0.5-1%) who are at extremely high risk (52% and 41%) for developing prostate cancer between the age of 55-74 years. A similar study by Sun et al. demonstrated the absolute risk of three sets of SNPs in predicting and developing prostate cancer. The model efficiently differentiates the suspected patients who are significantly at higher risk (2-3-fold increased risk) of developing prostate cancer than the median population [74]. The significant impact of these risk factors predicted from various models is with the younger suspected patients with a familial history of developing prostate cancer. The logic behind this is that a suspected person with familial history has some inherent, specific subset of SNPs from parents and remain throughout one’s life without any modification. Thus, the outcome of these inheritable SNPs could be from the initial stages of one’s life [75]. A model developed by Macinnis et al. predicts prostate cancer risk algorithm using 26 common variants for familial risk of prostate cancer. The model predicts collective prostate cancer risk based on familial history (incidental prostate cancer to exceedingly burdened prostate cancer) and the number of inheritable SNPs (expressed in percentage) [76].
In general, the diagnostic role of SNP genotyping in prostate cancer is to analyze the suspected patients who are at high risk. The genotyping of SNPs for high-risk prostate cancer should be performed frequently at an early stage of life. Additionally, the high-risk group with familial history should also take preventive measures in lifestyle, diet, and chemoprevention [77,78]. With respect to age-specific SNPs screening, the personal screening results in a 16% reduction in suspected patients to categorize them to be eligible for screening and a 3% reduction in cost for eligible cases [78]. The determination of SNPs can be performed at any age with low cost and makes them appealing candidates for predicting high risk suspected patients to develop prostate cancer [79]. Therefore, these findings suggest that SNPs underlie differences in the susceptibilities of subsections of population to diseases, signifying the role of SNPs in diagnosis and therapeutic intervention of different diseases.
SNPs and their interpretation for possible Novel Biomarkers
Determination of SNPs by genotyping indicates its role in predicting the risk for the development of prostate cancer. Despite recent reports suggest that SNPs are not to be considered as true diagnostic markers for prostate cancer. However, Klein et al. demonstrated in a recent study that SNPs could be useful in clinics in other ways but hypothetically, SNPs in combination with PSA screening could have critical importance in predicting the risk of developing prostate cancer in high-risk groups [80]. In addition, SNPs could be of much importance in modulating some novel biomarkers [81]. Their analysis and interpretation with reference to other risk factors are of great importance in prostate cancer diagnostics [82]. Previous studies revealed that numerous SNPs play an important role in modulating the expression and functions of a plethora of genes including transmembrane serine protease 2 (TMPRSS2) [83], hexokinase-2 (hK2) [84], and beta microseminoprotein (β-MSP) [85]. This modulation of genes by SNPs could have a promising causal association and its interpretation could have a great impact on the diagnostics of prostate cancer. Recent reports of epidemiological studies suggest that genetic modulations could occur in PSA as well. This can be explained by a recent report which suggests that around 40-45% PSA level could be modulated by genetic variations in the general population [86]. Owing to this genetic variation the specificity and sensitivity of PSA screening are low. This explains why in general there is no PSA threshold level of suspected patients to undergo prostate biopsies. To explain the variability of PSA level Gudmundson et al. demonstrated the deletion of six loci that modulates the PSA levels. Among them, four loci had a cumulative effect on the variation of PSA level. The PSA variability was supported by different groups which suggest that genetic modification has a great impact on the risk of developing prostate cancer by changing the abnormal PSA level in men, thus prevents approximately 20% of biopsies that were supposed to undergo prostate biopsies [87,88]. This helps in improving diagnostics, quality of life, and more importantly decreases the complications and cost which remains one of the important factors in developing nations for prostate cancer diagnostics. Therefore, with greater interest in the development of novel biomarkers in the diagnostics of prostate cancer, SNPs are the potential candidates that could modify the novel biomarkers and plays a critical role in the correct interpretation and analysis for the proper diagnosis of prostate cancer. It is believed that if SNPs are not taken into consideration for the identification of novel biomarkers, we might expect comparable hindrances as we are facing in PSA screening for prostate cancer diagnostics.
Predictive analysis of genetic polymorphism and androgen deprivation therapy (ADT) in metastatic prostate cancer
Hormonal therapy such as ADT showed a significant effect with promising efficacy in metastatic prostate cancer patients [89]. However, the main debacle of ADT to metastatic prostate cancer is an elevated relapse rate that initiates the development of castration-resistant prostate cancer (CRPC) [90]. To evaluate the patient response to ADT, the clinicopathological parameter monitored by clinicians such as the Gleason score, the kinetics of PSA screening are the crucial factors in predicting the patient’s response to ADT and prognosis [91]. Despite providing information with respect to ADT response and improving prognosis in metastatic prostate cancer patients, the clinical parameters evaluated by clinicians are still insufficient to predict the complete picture of patients who develop CRPC [92]. Thus, as per EAU guidelines, the agonists of luteinizing hormone-releasing hormone (LHRH) have been recommended for those patients which are in state or have developed CRPC [93]. Considering, to improve risk stratification and predict the metastatic prostate cancer patient response to ADT and the outcome of prognosis, the identification of genetic markers might play a significant role in determining the response to hormonal therapy and prognosis outcome in CRPC. A recent study by Ross et al. revealed that pharmacogenomics plays a crucial role in determining the patient’s response to hormonal therapy such as ADT [94]. The study further demonstrated that three SNPs which are strongly associated with genes involved in prostate cancer progression such as hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (HSD3B1) (prostate cancer susceptibility), cytochrome P450 family 19 subfamilies A member 1 (CYP19A1) (encodes aromatase enzyme which synthesizes estrogen from testosterone), and hydroxysteroid 17-beta dehydrogenase (HSD17B4) (associated with Gleason score) [95,96]. Additionally, these three SNPs not only help in the progression of prostate cancer individually, but they have a profound cumulative effect on the progression of prostate cancer.
Besides the involvement of SNPs which are close to genes and play an important role in cancer progression, SNPs from various other loci have been documented to play a significant role in prostate cancer patients relapse that are on hormonal therapy such as ADT [97]. The SNPs of various loci are located within the genes such as epidermal growth factor (EGF) which plays a critical role in the activation of many prooncogenic signaling cascades [98]. The other SNPs loci are within genes are solute carrier organic anion transport family member 2B1 (SLCO2B1) and solute carrier organic anion transport family member 2B3 (SCCO2B3) which encodes for androgen transporter proteins and tumor-growth factor beta receptor 2 (TGFβR2) gene which encodes for receptor protein of TGFβ signaling cascade [99,100]. These signaling pathways play a crucial role in the development of tumorigenesis. Apart from these, other SNPs have a key role in the relapse of prostate cancer hormonal therapy ADT and their location is still debatable and unknown [101].
A recent study from Taiwan has established a DNA library of 601 advanced prostate cancer patients treated with hormonal therapy ADT. The study further revealed that 5 SNPs detected from the established DNA library had a causal association with prostate cancer progression, whereas 14 other SNPs have a strong causal association with prostate cancer-specific mortality who undergo ADT hormonal therapy. A similar study by Bao et al. demonstrated that 4 SNPs that were detected within the nucleotide sequence of mRNA and miRNAs were observed to have a causal association with disease progression in prostate cancer [102]. Additionally, 49 and 55 SNPs of estrogen and androgen-receptor binding sites were thoroughly evaluated by Huang et al. 5 SNPs which were localized on arrestin domain-containing protein 3 (ARRDC3), transforming acidic coiled-coil-containing protein 2 (TACC2), Src-kinase associated phosphoprotein 1 (SKAP1), Fms related receptor tyrosine kinase 1 (FLT1), and basonuclin 2 (BNC2) were observed to have a strong correlation with prostate cancer mortality specifically [103]. Interestingly, a single SNP was found to be associated with insulin receptor substrate 2 (IRS2) and bone morphogenetic protein 5 (BMP5) genes and are observed to have a role in prostate cancer metastatic survival [104]. The IRS2 is an especially important gene that encodes for various proteins which act as adaptor proteins for intracellular signaling of various key physiological processes within the cell such as IGF1 [105,106]. This example nicely explains the intricacy and interwoven of genetic modulations and their outcome in the form of clinical effects.
Besides the interesting findings revealed from the genetic variations in the form of SNPs, it is worth to mentioning that majority of evaluated prostate cancer cases are heterogeneous [107]. The tumor characterization of advanced prostate cancer cases has 33% Gleason score varies from score 2 to 6, around 32% of prostate cancer cases are in T1/T2 stage [108]. Additionally, the proper protocol which was followed while giving ADT hormonal therapy was also in a heterogeneous state, which varies from ADT for biochemical failure to ADT in neoadjuvant setting after post-radical prostatectomy [101]. This heterogeneity makes things more complex and as the result gives a poor interpretation of genetic variation in these types of clinical settings.
Considering all the facts associated with metastatic prostate cancer, SNPs, and ADT, it is necessary to perform SNP genotyping that might be useful in multi-ways for patients who are supposed to undergo ADT hormonal therapy. Firstly, it could identify the patient group which is at elevated risk of treatment failure to the current regiment of ADT hormonal therapy, thus might have a prognostic role [101]. Therefore, predictive analysis of genetic polymorphism assists in the identification of a sub-group of patients who need more aggressive combinatorial treatment initially with novel drugs. Secondly, the implications of these SNPs on the functional analysis of enzymes could be targeted with the new therapeutic regiment to improve the efficacy of ADT hormonal therapy.
Role of genetic polymorphism in castration-resistant prostate cancer (CRPC)
To date, the only therapeutic option for CRPC is docetaxel which is derived from taxane a well-known anticancer agent [109]. Based on randomized phase III clinical trials from the two multicentre, there is a moderate increased effect in overall survival time of CRPC patients after docetaxel treatment [110]. Over the last decade, various novel small molecules and their derivatives for prostate cancer therapeutics have been developed [111-116]. However, there are equally many questions to be addressed regarding their optimization and long-term side effects. Although clinical evidence suggests that significant effects on CRPC were observed with one therapeutic regimen, the same regimen has deleterious effects from other side, this makes the job tough for clinicians to optimize the therapeutic regimen which has less long-term adverse effects [117]. Docetaxel treatment showed extreme variability in the clinical settings. Thus, to predict this clinical response before the docetaxel treatment would be of great interest [118]. Based on these observations, clinicians have a second line thought whether to continue with docetaxel as a first-line therapeutic regimen or to choose another therapeutic option [119].
The predicted outcome of docetaxel treatment in CRPC patients depends upon the genetic variability of cytochrome P450 family 1 subfamily B member 1 (CYP1B1) gene. A recent study by Pastina et al. demonstrated that the CRPC patients who carry the GG genotype of CYP1B1 gene at 4326 positions had significantly decreased in overall survival period than those CRPC patients who had CC genotype of CYP1B1 gene at 4326 positions. Further, the study reveals that after normalizing every other risk factor such as pathological, demographic, or biochemical parameters, the genotypic factor remains unchanged independent of the strong predictive parameter with an elevated risk of death in CRPC patients. This explains the genotypic parameter such as the GG at 4326 positions in CYP1B1 might act as an appealing pharmacogenetic marker for the CRPC patient’s prevalent response to docetaxel treatment. Reports suggest that GG genotype of CYP1B1 at 4326 positions plays a crucial role in modulating the therapeutic effect of docetaxel via its major metabolite called 4-hydroxyestradiol. Mechanistically, the presence of GG genotype of CYP1B1 at 4326 position has elevated levels of 4-hydroxyestradiol, that induces structural modulation in docetaxel thereby interfere with the docetaxel functions by negatively regulates the stabilization of microtubules [120,121]. Another interesting example is the ATP binding cassette subfamily B member 1 (ABCB1) gene. The important function of the ABCB1 gene is the efflux of docetaxel. The presence of three different types of genotypes 2677, 1236, and 3435 within the ABCB1 gene has a strong causal association with CRPC patient’s survival who receive docetaxel chemotherapy [122]. Docetaxel in combination with thalidomide to CRPC patients who had these three types of genotypes has a strong cumulative effect to developed long-term neuropathy in patients receiving this combination therapy. With the advancement in developing novel therapeutic modalities, the outcome of these chemotherapeutics is of great interest in the future particularly in clinical settings [123]. Based on the patient’s genotype and role of SNPs, clinicians have an option to use these novel therapeutics to predict the treatment response in advance. The therapeutic efficacy outcome of these novel modalities like enzalutamide and abiraterone could be assessed in the future using the same strategy to predict the outcome [124]. Therefore, the genetic variability of patients could play a crucial role in modifying drug metabolism, and associated genes to predict the outcome of chemotherapy in advance.
Limitations of SNPs in clinic
Besides, utilization of SNPs for population-based risk stratification to carry prostate cancer screening in susceptible men [125]. However, there are certain limitations associated with SNPs that needs great attention. Albeit several SNPs have been well documented with prostate cancer development, the single gene SNPs are known to provide little contribution to prostate cancer risk, therefore the utilization of multiple gene SNPs panels is often recommended [126,127]. Moreover, many SNPs are located outside of the genes and are suspected to influence gene expression and genome organization, thus, warranting the need to determine the role of these SNPs in prostate cancer development [123,128]. Public database of SNPs is often used to choose SNPs to study their association with complex disorders [129]. However, due to incomplete and uneven representation of the candidate SNPs in the public databases of different ethnic population groups, SNPs fail to explain the pathogenesis of complex disorders [130]. Therefore, there is an imperative need to accurately characterize SNPs in different ethnic groups at large-scale.
Conclusion
Genetic variability apparently plays an important role in assessing the susceptibility of individuals to various diseases. GWAS studies suggest that SNPs (single or multiple) groups could predict the elevated risk for prostate cancer. Besides, useful in identification and understanding the novel targets for therapeutic intervention, SNPs in combination with somatic tumor profile of patient could make personalized treatment strategy and work towards developing true precision medicine for managing prostate cancer.
However, there are limitations using SNPs in clinics for diagnosis and prognosis. First a small insignificant heterogeneous population of subjects might have false negative and positive results. Secondly, the causal association of SNPs and risk to develop prostate cancer are mostly conducted through GWAS. The genetic variability analysis to evaluate the phenotypic prostate cancer that varies from low penetrance to high has chances to be independent on GWAS and SNPs studies. These studies might not be sufficient to explain the underlying mechanism of these variations within response to the progression of prostate cancer. Additionally, Some SNPs are suspected to regulate transcriptional activity and genome organization, therefore necessitating to evaluate their involvement in prostate cancer development. Furthermore, due to incomplete and uneven representation of the candidate SNPs in the public databases of different ethnic population groups, SNPs fail to explain the pathogenesis of complex disorders. Therefore, there is an imperative need to accurately characterize SNPs in different ethnic groups at large-scale.
Future perspectives
It becomes clear throughout the review that there are still major challenges to evaluate the translational role of SNPs in clinics for prostate cancer. The future studies should thrust on the following approaches, firstly, there is a need to perform well-powered clinical analysis of prostate cancer on SNPs that should explain all underlying queries in present as well as in the future with conclusive results. Secondly, more thrust should be given to basic research to validate the role of SNPs in the progression of prostate cancer. Hopefully, in near the future we could reveal the comprehensive role of SNPs not only as predictive markers for prostate cancer but could also help in finding the other novel biomarkers. Their causal association with signaling pathways in the development of prostate cancer could possibly be the new therapeutic targets in prostate cancer.
Acknowledgements
Researchers would like to thank the Deanship of Scientific Research, Qassim University for funding publication of this project.
Disclosure of conflict of interest
None.
References
- 1.Berglund A, Garmo H, Robinson D, Tishelman C, Holmberg L, Bratt O, Adolfsson J, Stattin P, Lambe M. Differences according to socioeconomic status in the management and mortality in men with high risk prostate cancer. Eur J Cancer. 2012;48:75–84. doi: 10.1016/j.ejca.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Taitt HE. Global trends and prostate cancer: a review of incidence, detection, and mortality as influenced by race, ethnicity, and geographic location. Am J Mens Health. 2018;12:1807–1823. doi: 10.1177/1557988318798279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly SP, Rosenberg PS, Anderson WF, Andreotti G, Younes N, Cleary SD, Cook MB. Trends in the incidence of fatal prostate cancer in the United States by race. Eur Urol. 2017;71:195–201. doi: 10.1016/j.eururo.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung BH, Horie S, Chiong E. The incidence, mortality, and risk factors of prostate cancer in Asian men. Prostate Int. 2019;7:1–8. doi: 10.1016/j.prnil.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou CK, Check DP, Lortet-Tieulent J, Laversanne M, Jemal A, Ferlay J, Bray F, Cook MB, Devesa SS. Prostate cancer incidence in 43 populations worldwide: an analysis of time trends overall and by age group. Int J Cancer. 2016;138:1388–1400. doi: 10.1002/ijc.29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuzick J, Thorat MA, Andriole G, Brawley OW, Brown PH, Culig Z, Eeles RA, Ford LG, Hamdy FC, Holmberg L. Prevention and early detection of prostate cancer. Lancet Oncol. 2014;15:e484–e492. doi: 10.1016/S1470-2045(14)70211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adjakly M, Ngollo M, Dagdemir A, Judes G, Pajon A, Karsli-Ceppioglu S, Penault-Llorca F, Boiteux JP, Bignon YJ, Guy L. Prostate cancer: the main risk and protective factors-Epigenetic modifications. Ann Endocrinol. 2015;76:25–41. doi: 10.1016/j.ando.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Sharma S, Zapatero-Rodriguez J, O’Kennedy R. Prostate cancer diagnostics: clinical challenges and the ongoing need for disruptive and effective diagnostic tools. Biotechnol Adv. 2017;35:135–149. doi: 10.1016/j.biotechadv.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 9.de Dieuleveult M, Marchal C, Jouinot A, Letessier A, Miotto B. Molecular and clinical relevance of ZBTB38 expression levels in prostate cancer. Cancers. 2020;12:1106. doi: 10.3390/cancers12051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okarvi SM. Recent developments of prostate-specific membrane antigen (PSMA)-specific radiopharmaceuticals for precise imaging and therapy of prostate cancer: an overview. Clin Transl Imaging. 2019;7:189–208. [Google Scholar]
- 11.Matin F, Jeet V, Moya L, Selth LA, Chambers S, Clements JA, Batra J. A plasma biomarker panel of four microRNAs for the diagnosis of prostate cancer. Sci Rep. 2018;8:1–15. doi: 10.1038/s41598-018-24424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salehi B, Fokou PVT, Yamthe LRT, Tali BT, Adetunji CO, Rahavian A, Mudau FN, Martorell M, Setzer WN, Rodrigues CF, Martins N, Cho WC, Sharifi-Rad J. Phytochemicals in prostate cancer: from bioactive molecules to upcoming therapeutic agents. Nutrients. 2019;11:1483. doi: 10.3390/nu11071483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoury MJ, Janssens AC, Ransohoff DF. How can polygenic inheritance be used in population screening for common diseases? Genet Med. 2013;15:437–443. doi: 10.1038/gim.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan EY. Single Nucleotide Polymorphisms. Springer; 2009. Next-generation sequencing methods: impact of sequencing accuracy on SNP discovery; pp. 95–111. [DOI] [PubMed] [Google Scholar]
- 15.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–934. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 16.Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 17.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 18.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benafif S, Kote-Jarai Z, Eeles RA. A review of prostate cancer genome-wide association studies (GWAS) Cancer Epidemiol Prev Biomark. 2018;27:845–857. doi: 10.1158/1055-9965.EPI-16-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matejcic M, Saunders EJ, Dadaev T, Brook MN, Wang K, Sheng X, Al Olama AA, Schumacher FR, Ingles SA, Govindasami K. Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-018-06863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thibodeau SN, French AJ, McDonnell SK, Cheville J, Middha S, Tillmans L, Riska S, Baheti S, Larson MC, Fogarty Z. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat Commun. 2015;6:1–10. doi: 10.1038/ncomms9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beuten J, Gelfond JA, Franke JL, Weldon KS, Crandall AC, Johnson-Pais TL, Thompson IM, Leach RJ. Single and multigenic analysis of the association between variants in 12 steroid hormone metabolism genes and risk of prostate cancer. Cancer Epidemiol Prev Biomark. 2009;18:1869–1880. doi: 10.1158/1055-9965.EPI-09-0076. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D, Anokian E, Cieza-Borrella C. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu S, Tsounapi P, Shimizu T, Honda M, Inoue K, Dimitriadis F, Saito M. Lower urinary tract symptoms, benign prostatic hyperplasia/benign prostatic enlargement and erectile dysfunction: are these conditions related to vascular dysfunction? Int J Urol. 2014;21:856–864. doi: 10.1111/iju.12501. [DOI] [PubMed] [Google Scholar]
- 25.De Nunzio C, Kramer G, Marberger M, Montironi R, Nelson W, Schröder F, Sciarra A, Tubaro A. The controversial relationship between benign prostatic hyperplasia and prostate cancer: the role of inflammation. Eur Urol. 2011;60:106–117. doi: 10.1016/j.eururo.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 26.Ørsted DD, Bojesen SE. The link between benign prostatic hyperplasia and prostate cancer. Nat Rev Urol. 2013;10:49. doi: 10.1038/nrurol.2012.192. [DOI] [PubMed] [Google Scholar]
- 27.Sarwar S, Adil MA, Nyamath P, Ishaq M. Biomarkers of prostatic cancer: an attempt to categorize patients into prostatic carcinoma, benign prostatic hyperplasia, or prostatitis based on serum prostate specific antigen, prostatic acid phosphatase, calcium, and phosphorus. Prostate Cancer. 2017;2017:5687212. doi: 10.1155/2017/5687212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, Zhang H, Kohn DB, Huang J, Witte ON. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci. 2013;110:20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol. 1988;12:897–906. doi: 10.1097/00000478-198812000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev. 2018;32:1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moghanaki D, Turkbey B, Vapiwala N, Ehdaie B, Frank SJ, McLaughlin PW, Harisinghani M. Advances in prostate cancer magnetic resonance imaging and positron emission tomography-computed tomography for staging and radiotherapy treatment planning. Semin Radiat Oncol. 2017;27:21–33. doi: 10.1016/j.semradonc.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drake JM, Gabriel CL, Henry MD. Assessing tumor growth and distribution in a model of prostate cancer metastasis using bioluminescence imaging. Clin Exp Metastasis. 2005;22:674–684. doi: 10.1007/s10585-006-9011-4. [DOI] [PubMed] [Google Scholar]
- 33.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Högnäs G, Annala M. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gratzke C, Bachmann A, Descazeaud A, Drake MJ, Madersbacher S, Mamoulakis C, Oelke M, Tikkinen KA, Gravas S. EAU guidelines on the assessment of non-neurogenic male lower urinary tract symptoms including benign prostatic obstruction. Eur Urol. 2015;67:1099–1109. doi: 10.1016/j.eururo.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 36.Logothetis CJ, Kim J, Davis JW, Chapin BF, Kuban D, Efstathiou E, Aparicio A. Neoplasms of the prostate. Holl Cancer Med. 2016:1–38. [Google Scholar]
- 37.Saba K, Wettstein MS, Lieger L, Hötker AM, Donati OF, Moch H, Ankerst DP, Poyet C, Sulser T, Eberli D. External validation and comparison of prostate cancer risk calculators incorporating multiparametric magnetic resonance imaging for prediction of clinically significant prostate cancer. J Urol. 2020;203:719–726. doi: 10.1097/JU.0000000000000622. [DOI] [PubMed] [Google Scholar]
- 38.Filella X, Fernández-Galan E, Bonifacio RF, Foj L. Emerging biomarkers in the diagnosis of prostate cancer. Pharmacogenomics Pers Med. 2018;11:83. doi: 10.2147/PGPM.S136026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG, Freeman A, Kirkham AP, Oldroyd R, Parker C, Emberton M PROMIS study group. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey PA. Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol. 2004;17:292–306. doi: 10.1038/modpathol.3800054. [DOI] [PubMed] [Google Scholar]
- 41.Bostwick DG, Burke HB, Djakiew D, Euling S, Ho S, Landolph J, Morrison H, Sonawane B, Shifflett T, Waters DJ. Human prostate cancer risk factors. Cancer Interdiscip Int J Am Cancer Soc. 2004;101:2371–2490. doi: 10.1002/cncr.20408. [DOI] [PubMed] [Google Scholar]
- 42.Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, Easton D, Kote-Jarai Z. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11:18. doi: 10.1038/nrurol.2013.266. [DOI] [PubMed] [Google Scholar]
- 43.Pernar CH, Ebot EM, Wilson KM, Mucci LA. The epidemiology of prostate cancer. Cold Spring Harb Perspect Med. 2018;8:a030361. doi: 10.1101/cshperspect.a030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johns LE, Houlston RS. A systematic review and meta-analysis of familial prostate cancer risk. BJU Int. 2003;91:789–794. doi: 10.1046/j.1464-410x.2003.04232.x. [DOI] [PubMed] [Google Scholar]
- 45.Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, Pukkala E, Christensen K, Adami HO, Holm NV. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Prev Biomark. 2014;23:2303–2310. doi: 10.1158/1055-9965.EPI-13-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhi J, Afridi A, Vatsia S, Joshi G, Joshi G, Kaplan SA, Smith NL, Khan SA. The molecular biology of prostate cancer: current understanding and clinical implications. Prostate Cancer Prostatic Dis. 2018;21:22–36. doi: 10.1038/s41391-017-0023-8. [DOI] [PubMed] [Google Scholar]
- 47.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 48.Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet. 2013;14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ku CS, Loy EY, Salim A, Pawitan Y, Chia KS. The discovery of human genetic variations and their use as disease markers: past, present and future. J Hum Genet. 2010;55:403–415. doi: 10.1038/jhg.2010.55. [DOI] [PubMed] [Google Scholar]
- 50.Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almal SH, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet. 2012;57:6–13. doi: 10.1038/jhg.2011.108. [DOI] [PubMed] [Google Scholar]
- 52.Kwok PY, Chen X. Detection of single nucleotide polymorphisms. Curr Issues Mol Biol. 2003;5:43–60. [PubMed] [Google Scholar]
- 53.Zhen JT, Syed J, Nguyen KA, Leapman MS, Agarwal N, Brierley K, Llor X, Hofstatter E, Shuch B. Genetic testing for hereditary prostate cancer: current status and limitations. Cancer. 2018;124:3105–3117. doi: 10.1002/cncr.31316. [DOI] [PubMed] [Google Scholar]
- 54.Eeles RA, Al Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 57.Babic A, Bao Y, Qian ZR, Yuan C, Giovannucci EL, Aschard H, Kraft P, Amundadottir LT, Stolzenberg-Solomon R, Morales-Oyarvide V. Pancreatic cancer risk associated with prediagnostic plasma levels of leptin and leptin receptor genetic polymorphisms. Cancer Res. 2016;76:7160–7167. doi: 10.1158/0008-5472.CAN-16-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steri M, Idda ML, Whalen MB, Orrù V. Genetic variants in mRNA untranslated regions. Wiley Interdiscip Rev RNA. 2018;9:e1474. doi: 10.1002/wrna.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatarinova TV, Chekalin E, Nikolsky Y, Bruskin S, Chebotarov D, McNally KL, Alexandrov N. Nucleotide diversity analysis highlights functionally important genomic regions. Sci Rep. 2016;6:1–12. doi: 10.1038/srep35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 61.Stefancu A, Moisoiu V, Couti R, Andras I, Rahota R, Crisan D, Pavel IE, Socaciu C, Leopold N, Crisan N. Combining SERS analysis of serum with PSA levels for improving the detection of prostate cancer. Nanomed. 2018;13:2455–2467. doi: 10.2217/nnm-2018-0127. [DOI] [PubMed] [Google Scholar]
- 62.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 63.Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 64.Fleshner K, Carlsson SV, Roobol MJ. The effect of the USPSTF PSA screening recommendation on prostate cancer incidence patterns in the USA. Nat Rev Urol. 2017;14:26. doi: 10.1038/nrurol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heidenreich A, Bolla M, Joniau S, Mason MD, Matveev V, Mottet N, Schmid HP, van der Kwast TH, Wiegel T, Zattoni F. Guidelines on. Update 2007 [Google Scholar]
- 66.Tikkinen KAO, Dahm P, Lytvyn L, Heen AF, Vernooij RWM, Siemieniuk RAC, Wheeler R, Vaughan B, Fobuzi AC, Blanker MH, Junod N, Sommer J, Stirnemann J, Yoshimura M, Auer R, MacDonald H, Guyatt G, Vandvik PO, Agoritsas T. Prostate cancer screening with prostate-specific antigen (PSA) test: a clinical practice guideline. BMJ. 2018;362:k3581. doi: 10.1136/bmj.k3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K, Bangma CH, Roobol MJ. Prostate cancer screening in Europe and Asia. Asian J Urol. 2017;4:86–95. doi: 10.1016/j.ajur.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mottet N, van den Bergh RC, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, Fanti S, Fossati N, Gandaglia G, Gillessen S. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2021;79:243–262. doi: 10.1016/j.eururo.2020.09.042. [DOI] [PubMed] [Google Scholar]
- 69.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10:10. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng SL, Sun J, Wiklund F, Gao Z, Stattin P, Purcell LD, Adami HO, Hsu FC, Zhu Y, Adolfsson J. Genetic variants and family history predict prostate cancer similar to prostate-specific antigen. Clin Cancer Res. 2009;15:1105–1111. doi: 10.1158/1078-0432.CCR-08-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salinas CA, Koopmeiners JS, Kwon EM, FitzGerald L, Lin DW, Ostrander EA, Feng Z, Stanford JL. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–372. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Sun J, Kader AK, Lindström S, Wiklund F, Hsu FC, Johansson JE, Zheng SL, Thomas G, Hayes RB. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1565–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun J, Kader AK, Hsu FC, Kim ST, Zhu Y, Turner AR, Jin T, Zhang Z, Adolfsson J, Wiklund F. Inherited genetic markers discovered to date are able to identify a significant number of men at considerably elevated risk for prostate cancer. Prostate. 2011;71:421–430. doi: 10.1002/pros.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindström S, Schumacher FR, Cox D, Travis RC, Albanes D, Allen NE, Andriole G, Berndt SI, Boeing H, Bueno-de-Mesquita HB. Common genetic variants in prostate cancer risk prediction-results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Prev Biomark. 2012;21:437–444. doi: 10.1158/1055-9965.EPI-11-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacInnis RJ, Antoniou AC, Eeles RA, Severi G, Al Olama AA, McGuffog L, Kote-Jarai Z, Guy M, O’Brien LT, Hall AL. A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol. 2011;35:549–556. doi: 10.1002/gepi.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dadaev T, Saunders EJ, Newcombe PJ, Anokian E, Leongamornlert DA, Brook MN, Cieza-Borrella C, Mijuskovic M, Wakerell S, Al Olama AA. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat Commun. 2018;9:1–19. doi: 10.1038/s41467-018-04109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pashayan N, Duffy SW, Chowdhury S, Dent T, Burton H, Neal DE, Easton DF, Eeles R, Pharoah P. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104:1656–1663. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu X, Albertsen PC, Andriole GL, Roobol MJ, Schröder FH, Vickers AJ. Risk-based prostate cancer screening. Eur Urol. 2012;61:652–661. doi: 10.1016/j.eururo.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klein RJ, Hallden C, Gupta A, Savage CJ, Dahlin A, Bjartell A, Manjer J, Scardino PT, Ulmert D, Wallström P. Evaluation of multiple risk-associated single nucleotide polymorphisms versus prostate-specific antigen at baseline to predict prostate cancer in unscreened men. Eur Urol. 2012;61:471–477. doi: 10.1016/j.eururo.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vatandoost N, Ghanbari J, Mojaver M, Avan A, Ghayour-Mobarhan M, Nedaeinia R, Salehi R. Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol. 2016;142:341–351. doi: 10.1007/s00432-015-1928-z. [DOI] [PubMed] [Google Scholar]
- 82.Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M, Fossati N, Gross T, Henry AM, Joniau S. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 83.Clinckemalie L, Spans L, Dubois V, Laurent M, Helsen C, Joniau S, Claessens F. Androgen regulation of the TMPRSS2 gene and the effect of a SNP in an androgen response element. Mol Endocrinol. 2013;27:2028–2040. doi: 10.1210/me.2013-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein RJ, Halldén C, Cronin AM, Ploner A, Wiklund F, Bjartell AS, Stattin P, Xu J, Scardino PT, Offit K. Blood biomarker levels to aid discovery of cancer-related single-nucleotide polymorphisms: kallikreins and prostate cancer. Cancer Prev Res (Phila Pa) 2010;3:611–619. doi: 10.1158/1940-6207.CAPR-09-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu X, Valtonen-André C, Sävblom C, Halldén C, Lilja H, Klein RJ. Polymorphisms at the microseminoprotein-β locus associated with physiologic variation in β-microseminoprotein and prostate-specific antigen levels. Cancer Epidemiol Prev Biomark. 2010;19:2035–2042. doi: 10.1158/1055-9965.EPI-10-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bansal A, Murray DK, Wu JT, Stephenson RA, Middleton RG, Meikle AW. Heritability of prostate-specific antigen and relationship with zonal prostate volumes in aging twins. J Clin Endocrinol Metab. 2000;85:1272–1276. doi: 10.1210/jcem.85.3.6399. [DOI] [PubMed] [Google Scholar]
- 87.Walsh PC. Re: genetic correction of PSA values using sequence variants associated with PSA levels. J Urol. 2011;186:103–104. [Google Scholar]
- 88.Helfand BT, Loeb S, Hu Q, Cooper PR, Roehl KA, McGuire BB, Baumann NA, Catalona WJ. Personalized prostate specific antigen testing using genetic variants may reduce unnecessary prostate biopsies. J Urol. 2013;189:1697–1701. doi: 10.1016/j.juro.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shore ND, Abrahamsson PA, Anderson J, Crawford ED, Lange P. New considerations for ADT in advanced prostate cancer and the emerging role of GnRH antagonists. Prostate Cancer Prostatic Dis. 2013;16:7–15. doi: 10.1038/pcan.2012.25. [DOI] [PubMed] [Google Scholar]
- 90.Ojo D, Lin X, Wong N, Gu Y, Tang D. Prostate cancer stem-like cells contribute to the development of castration-resistant prostate cancer. Cancers. 2015;7:2290–2308. doi: 10.3390/cancers7040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim M, Lee J, Jeong CW, Ku JH, Kim HH, Kwak C. Prostate-specific antigen kinetic profiles during androgen deprivation therapy as prognostic factors in castration-resistant prostate cancer. Urol Oncol Semin Orig Investig. 2015;33:203, e1–9. doi: 10.1016/j.urolonc.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 92.Lorente D, Mateo J, Perez-Lopez R, de Bono JS, Attard G. Sequencing of agents in castration-resistant prostate cancer. Lancet Oncol. 2015;16:e279–e292. doi: 10.1016/S1470-2045(15)70033-1. [DOI] [PubMed] [Google Scholar]
- 93.Singla N, Ghandour RA, Raj GV. Investigational luteinizing hormone releasing hormone (LHRH) agonists and other hormonal agents in early stage clinical trials for prostate cancer. Expert Opin Investig Drugs. 2019;28:249–259. doi: 10.1080/13543784.2019.1570130. [DOI] [PubMed] [Google Scholar]
- 94.Ross RW, Oh WK, Xie W, Pomerantz M, Nakabayashi M, Sartor O, Taplin ME, Regan MM, Kantoff PW, Freedman M. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J. Clin. Oncol. 2008;26:842–847. doi: 10.1200/JCO.2007.13.6804. [DOI] [PubMed] [Google Scholar]
- 95.Chang B, Zheng SL, Hawkins GA, Isaacs SD, Wiley KE, Turner A, Carpten JD, Bleecker ER, Walsh PC, Trent JM. Joint effect of HSD3B1 and HSD3B2 genes is associated with hereditary and sporadic prostate cancer susceptibility. Cancer Res. 2002;62:1784–1789. [PubMed] [Google Scholar]
- 96.True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E, Knudsen B, Lange P, Vessella R, Lin D, Hood L, Nelson PS. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci. 2006;103:10991–10996. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin HJ, Jung S, DebRoy AR, Davuluri RV. Identification and validation of regulatory SNPs that modulate transcription factor chromatin binding and gene expression in prostate cancer. Oncotarget. 2016;7:54616. doi: 10.18632/oncotarget.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teixeira AL, Ribeiro R, Morais A, Lobo F, Fraga A, Pina F, Calais-da-Silva FM, Calais-da-Silva FE, Medeiros R. Combined analysis of EGF+ 61G> A and TGFB1+ 869T> C functional polymorphisms in the time to androgen independence and prostate cancer susceptibility. Pharmacogenomics J. 2009;9:341–346. doi: 10.1038/tpj.2009.20. [DOI] [PubMed] [Google Scholar]
- 99.Yang M, Xie W, Mostaghel E, Nakabayashi M, Werner L, Sun T, Pomerantz M, Freedman M, Ross R, Regan M. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J. Clin. Oncol. 2011;29:2565. doi: 10.1200/JCO.2010.31.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teixeira AL, Gomes M, Nogueira A, Azevedo AS, Assis J, Dias F, Santos JI, Lobo F, Morais A, Maurício J. Improvement of a predictive model of castration-resistant prostate cancer: functional genetic variants in TGFβ1 signaling pathway modulation. PLoS One. 2013;8:e72419. doi: 10.1371/journal.pone.0072419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kohli M, Riska SM, Mahoney DW, Chai HS, Hillman DW, Rider DN, Costello BA, Qin R, Lamba J, Sahasrabudhe DM. Germline predictors of androgen deprivation therapy response in advanced prostate cancer. Mayo Clin Proc. 2012;87:240–246. doi: 10.1016/j.mayocp.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, Lan YH, Lu TL, Lee HZ, Juang SH, Chen LM. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin Cancer Res. 2011;17:928–936. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 103.Huang CN, Huang SP, Pao JB, Chang TY, Lan YH, Lu TL, Lee HZ, Juang SH, Wu PP, Pu YS. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2012;23:707–713. doi: 10.1093/annonc/mdr264. [DOI] [PubMed] [Google Scholar]
- 104.Huang SP, Bao BY, Hour TC, Huang CY, Yu CC, Liu CC, Lee YC, Huang CN, Pao JB, Huang CH. Genetic variants in CASP3, BMP5, and IRS2 genes may influence survival in prostate cancer patients receiving androgen-deprivation therapy. PLoS One. 2012;7:e41219. doi: 10.1371/journal.pone.0041219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsuchiya N, Narita S, Inoue T, Saito M, Numakura K, Huang M, Hatakeyama S, Satoh S, Saito S, Ohyama C. Insulin-like growth factor-1 genotypes and haplotypes influence the survival of prostate cancer patients with bone metastasis at initial diagnosis. BMC Cancer. 2013;13:1–9. doi: 10.1186/1471-2407-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fraga A, Ribeiro R, Coelho A, Vizcaíno JR, Coutinho H, Lopes JM, Príncipe P, Lobato C, Lopes C, Medeiros R. Genetic polymorphisms in key hypoxia-regulated downstream molecules and phenotypic correlation in prostate cancer. BMC Urol. 2017;17:1–12. doi: 10.1186/s12894-017-0201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Negoita S, Feuer EJ, Mariotto A, Cronin KA, Petkov VI, Hussey SK, Benard V, Henley SJ, Anderson RN, Fedewa S. Annual report to the nation on the status of cancer, part II: recent changes in prostate cancer trends and disease characteristics. Cancer. 2018;124:2801–2814. doi: 10.1002/cncr.31549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lancee M, Tikkinen KA, De Reijke TM, Kataja VV, Aben KK, Vernooij RW. Guideline of guidelines: primary monotherapies for localised or locally advanced prostate cancer. BJU Int. 2018;122:535–548. doi: 10.1111/bju.14237. [DOI] [PubMed] [Google Scholar]
- 109.Arsov C, Winter C, Rabenalt R, Albers P. Current second-line treatment options for patients with castration resistant prostate cancer (CRPC) resistant to docetaxel. Urol Oncol Semin Orig Investig. 2012;30:762–771. doi: 10.1016/j.urolonc.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 110.Lee DJ, Cha EK, Dubin JM, Beltran H, Chromecki TF, Fajkovic H, Scherr DS, Tagawa ST, Shariat SF. Novel therapeutics for the management of castration-resistant prostate cancer (CRPC) BJU Int. 2012;109:968–985. doi: 10.1111/j.1464-410X.2011.10643.x. [DOI] [PubMed] [Google Scholar]
- 111.Rah B, Amin H, Yousuf K, Khan S, Jamwal G, Mukherjee D, Goswami A. A novel MMP-2 inhibitor 3-azidowithaferin A (3-azidoWA) abrogates cancer cell invasion and angiogenesis by modulating extracellular par-4. PLoS One. 2012;7:e44039. doi: 10.1371/journal.pone.0044039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rah B, ur Rasool R, Nayak D, Yousuf SK, Mukherjee D, Kumar LD, Goswami A. PAWR-mediated suppression of BCL2 promotes switching of 3-azido withaferin A (3-AWA)-induced autophagy to apoptosis in prostate cancer cells. Autophagy. 2015;11:314–331. doi: 10.1080/15548627.2015.1017182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.ur Rasool R, Rah B, Amin H, Nayak D, Chakraborty S, Rawoof A, Mintoo MJ, Yousuf K, Mukherjee D, Kumar LD. Corrigendum: dual modulation of Ras-Mnk and PI3K-AKT-mTOR pathways: a novel c-FLIP inhibitory mechanism of 3-AWA mediated translational attenuation through dephosphorylation of eIF4E. Sci Rep. 2016;6:21054. doi: 10.1038/srep21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nayak D, Amin H, Rah B, ur Rasool R, Sharma D, Gupta AP, Kushwaha M, Mukherjee D, Goswami A. A therapeutically relevant, 3, 3’-diindolylmethane derivative NGD16 attenuates angiogenesis by targeting glucose regulated protein, 78 kDa (GRP78) Chem Biol Interact. 2015;232:58–67. doi: 10.1016/j.cbi.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 115.Chakraborty S, ur Rasool R, Kumar S, Nayak D, Rah B, Katoch A, Amin H, Ali A, Goswami A. Cristacarpin promotes ER stress-mediated ROS generation leading to premature senescence by activation of p21 waf-1. Age. 2016;38:1–14. doi: 10.1007/s11357-016-9922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rah B, Lone SH, Rasool RU, Farooq S, Nayak D, Chikan NA, Chakraborty S, Behl A, Mondhe DM, Goswami A. Design and synthesis of antitumor heck-coupled Sclareol analogues: modulation of BH3 family members by SS-12 in autophagy and apoptotic cell death. J Med Chem. 2015;58:3432–3444. doi: 10.1021/jm501942m. [DOI] [PubMed] [Google Scholar]
- 117.Nevedomskaya E, Baumgart SJ, Haendler B. Recent advances in prostate cancer treatment and drug discovery. Int J Mol Sci. 2018;19:1359. doi: 10.3390/ijms19051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Varnai R, Koskinen LM, Mäntylä LE, Szabo I, FitzGerald LM, Sipeky C. Pharmacogenomic biomarkers in docetaxel treatment of prostate cancer: from discovery to implementation. Genes. 2019;10:599. doi: 10.3390/genes10080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nuhn P, De Bono JS, Fizazi K, Freedland SJ, Grilli M, Kantoff PW, Sonpavde G, Sternberg CN, Yegnasubramanian S, Antonarakis ES. Update on systemic prostate cancer therapies: management of metastatic castration-resistant prostate cancer in the era of precision oncology. Eur Urol. 2019;75:88–99. doi: 10.1016/j.eururo.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 120.Pastina I, Giovannetti E, Chioni A, Sissung TM, Crea F, Orlandini C, Price DK, Cianci C, Figg WD, Ricci S. Cytochrome 450 1B1 (CYP1B1) polymorphisms associated with response to docetaxel in Castration-Resistant Prostate Cancer (CRPC) patients. BMC Cancer. 2010;10:1–9. doi: 10.1186/1471-2407-10-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sissung TM, Danesi R, Price DK, Steinberg SM, De Wit R, Zahid M, Gaikwad N, Cavalieri E, Dahut WL, Sackett DL. Association of the CYP1B1* 3 allele with survival in patients with prostate cancer receiving docetaxel. Mol Cancer Ther. 2008;7:19–26. doi: 10.1158/1535-7163.MCT-07-0557. [DOI] [PubMed] [Google Scholar]
- 122.Sissung TM, Baum CE, Deeken J, Price DK, Aragon-Ching J, Steinberg SM, Dahut W, Sparreboom A, Figg WD. ABCB1 genetic variation influences the toxicity and clinical outcome of patients with androgen-independent prostate cancer treated with docetaxel. Clin Cancer Res. 2008;14:4543–4549. doi: 10.1158/1078-0432.CCR-07-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Van den Broeck T, Joniau S, Clinckemalie L, Helsen C, Prekovic S, Spans L, Tosco L, Van Poppel H, Claessens F. The role of single nucleotide polymorphisms in predicting prostate cancer risk and therapeutic decision making. Biomed Res Int. 2014;2014:627510. doi: 10.1155/2014/627510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ingrosso G, Detti B, Scartoni D, Lancia A, Giacomelli I, Baki M, Carta G, Livi L, Santoni R. Current therapeutic options in metastatic castration-resistant prostate cancer. Semin Oncol. 2018;45:303–315. doi: 10.1053/j.seminoncol.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 125.Eeles R, Raghallaigh HN. Men with a susceptibility to prostate cancer and the role of genetic based screening. Transl Androl Urol. 2018;7:61. doi: 10.21037/tau.2017.12.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, Heckmann D, Sidiropoulos N, Waszak SM, Hübschmann D. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell. 2018;34:996–1011. e8. doi: 10.1016/j.ccell.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reese AC, Casey G, Witte JS. Management of Prostate Cancer. Springer; 2012. Hereditary prostate cancer and genetic risk; pp. 79–101. [Google Scholar]
- 128.Nurminen R. Genetic susceptibility factors for prostate cancer at chromosomal region 11q13. 5. 2016 [Google Scholar]
- 129.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN. Deleterious-and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D’Amato M, Adolfsson J, Grönberg H. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–8. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perez CA, Chen H, Shyr Y, Courtney R, Zheng W, Cai Q, Hwang M, Jaboin J, Schleicher S, Moretti L, Wills M, Smith JA, Lu B. The EGFR polymorphism rs884419 is associated with freedom from recurrence in patients with resected prostate cancer. J Urol. 2010;183:2062–9. doi: 10.1016/j.juro.2009.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morote J, Del Amo J, Borque A, Ars E, Hernández C, Herranz F, Arruza A, Llarena R, Planas J, Viso MJ, Palou J, Raventós CX, Tejedor D, Artieda M, Simón L, Martínez A, Rioja LA. Improved prediction of biochemical recurrence after radical prostatectomy by genetic polymorphisms. J Urol. 2010;184:506–11. doi: 10.1016/j.juro.2010.03.144. [DOI] [PubMed] [Google Scholar]
- 134.Audet-Walsh E, Bellemare J, Lacombe L, Fradet Y, Fradet V, Douville P, Guillemette C, Levesque E. The impact of germline genetic variations in hydroxysteroid (17-beta) dehydrogenases on prostate cancer outcomes after prostatectomy. Eur Urol. 2012;62:88–96. doi: 10.1016/j.eururo.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 135.Jaboin JJ, Hwang M, Lopater Z, Chen H, Ray GL, Perez C, Cai Q, Wills ML, Lu B. The matrix metalloproteinase-7 polymorphism rs10895304 is associated with increased recurrence risk in patients with clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 2011;79:1330–5. doi: 10.1016/j.ijrobp.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bachmann HS, Heukamp LC, Schmitz KJ, Hilburn CF, Kahl P, Buettner R, Nückel H, Eisenhardt A, Rübben H, Schmid KW, Siffert W, Eggert A, Schramm A, Schulte JH. Regulatory BCL2 promoter polymorphism (-938C>A) is associated with adverse outcome in patients with prostate carcinoma. Int J Cancer. 2011;129:2390–9. doi: 10.1002/ijc.25904. [DOI] [PubMed] [Google Scholar]
- 137.Huang SP, Ting WC, Chen LM, Huang LC, Liu CC, Chen CW, Hsieh CJ, Yang WH, Chang TY, Lee HZ, Bao BY. Association analysis of Wnt pathway genes on prostate-specific antigen recurrence after radical prostatectomy. Ann Surg Oncol. 2010;17:312–22. doi: 10.1245/s10434-009-0698-8. [DOI] [PubMed] [Google Scholar]
- 138.Chang CF, Pao JB, Yu CC, Huang CY, Huang SP, Yang YP, Huang CN, Chang TY, You BJ, Lee HZ, Hour TC, Bao BY. Common variants in IGF1 pathway genes and clinical outcomes after radical prostatectomy. Ann Surg Oncol. 2013;20:2446–52. doi: 10.1245/s10434-013-2884-y. [DOI] [PubMed] [Google Scholar]
- 139.Borque Á, del Amo J, Esteban LM, Ars E, Hernández C, Planas J, Arruza A, Llarena R, Palou J, Herranz F, Raventós CX, Tejedor D, Artieda M, Simon L, Martínez A, Carceller E, Suárez M, Allué M, Sanz G, Morote J. Genetic predisposition to early recurrence in clinically localized prostate cancer. BJU Int. 2013;111:549–58. doi: 10.1111/j.1464-410X.2012.11333.x. [DOI] [PubMed] [Google Scholar]
- 140.Langsenlehner T, Renner W, Gerger A, Hofmann G, Thurner EM, Kapp KS, Langsenlehner U. Association between single nucleotide polymorphisms in the gene for XRCC1 and radiation-induced late toxicity in prostate cancer patients. Radiother Oncol. 2011;98:387–93. doi: 10.1016/j.radonc.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 141.De Langhe S, De Ruyck K, Ost P, Fonteyne V, Werbrouck J, De Meerleer G, De Neve W, Thierens H. Acute radiation-induced nocturia in prostate cancer patients is associated with pretreatment symptoms, radical prostatectomy, and genetic markers in the TGFβ1 gene. Int J Radiat Oncol Biol Phys. 2013;85:393–9. doi: 10.1016/j.ijrobp.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 142.Fachal L, Gómez-Caamaño A, Peleteiro P, Carballo A, Calvo-Crespo P, Sánchez-García M, Lobato-Busto R, Carracedo Á, Vega A. Association of a XRCC3 polymorphism and rectum mean dose with the risk of acute radio-induced gastrointestinal toxicity in prostate cancer patients. Radiother Oncol. 2012;105:321–8. doi: 10.1016/j.radonc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 143.Fachal L, Gómez-Caamaño A, Sánchez-García M, Carballo A, Peleteiro P, Lobato-Busto R, Carracedo Á, Vega A. TGFβ1 SNPs and radio-induced toxicity in prostate cancer patients. Radiother Oncol. 2012;103:206–9. doi: 10.1016/j.radonc.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 144.Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, Wilkinson JS, Tyrer J, Misra V, Platte R, Gulliford SL, Sydes MR, Hall E, Bentzen SM, Dearnaley DP, Burnet NG, Pharoah PD, Dunning AM, West CM. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 145.Teixeira AL, Ribeiro R, Morais A, Lobo F, Fraga A, Pina F, Calais-da-Silva FM, Calais-da-Silva FE, Medeiros R. Combined analysis of EGF+ 61G> A and TGFB1+ 869T> C functional polymorphisms in the time to androgen independence and prostate cancer susceptibility. Pharmacogenomics J. 2009;9:341–6. doi: 10.1038/tpj.2009.20. [DOI] [PubMed] [Google Scholar]
- 146.Huang CN, Huang SP, Pao JB, Hour TC, Chang TY, Lan YH, Lu TL, Lee HZ, Juang SH, Wu PP, Huang CY, Hsieh CJ, Bao BY. Genetic polymorphisms in oestrogen receptor-binding sites affect clinical outcomes in patients with prostate cancer receiving androgen-deprivation therapy. J Intern Med. 2012;271:499–509. doi: 10.1111/j.1365-2796.2011.02449.x. [DOI] [PubMed] [Google Scholar]