Abstract
Nowadays, humanity is facing one of the most dangerous pandemics known as COVID-19. Due to its high inter-person contagiousness, COVID-19 is rapidly spreading across the world. Positive patients are often suffering from different symptoms that can vary from mild to severe including cough, fever, sore throat, and body aches. In more dire cases, infected patients can experience severe symptoms that can cause breathing difficulties which lead to stern organ failure and die. The medical corps all over the world are overloaded because of the exponentially myriad number of contagions. Therefore, screening for the disease becomes overwrought with the limited tools of test. Additionally, test results may take a long time to acquire, leaving behind a higher potential for the prevalence of the virus among other individuals by the patients. To reduce the chances of infection, we suggest a prediction model that distinguishes the infected COVID-19 cases based on clinical symptoms and features. This model can be helpful for citizens to catch their infection without the need for visiting the hospital. Also, it helps the medical staff in triaging patients in case of a deficiency of medical amenities. In this paper, we use the non-dominated sorting genetic algorithm (NSGA-II) to select the interesting features by finding the best trade-offs between two conflicting objectives: minimizing the number of features and maximizing the weights of selected features. Then, a classification phase is conducted using an AdaBoost classifier. The proposed model is evaluated using two different datasets. To maximize results, we performed a natural selection of hyper-parameters of the classifier using the genetic algorithm. The obtained results prove the efficiency of NSGA-II as a feature selection algorithm combined with AdaBoost classifier. It exhibits higher classification results that outperformed the existing methods.
Keywords: COVID-19 prediction, Machine learning, Feature selection, AdaBoost, NSGA-II, Hyper-parameters optimization
Introduction
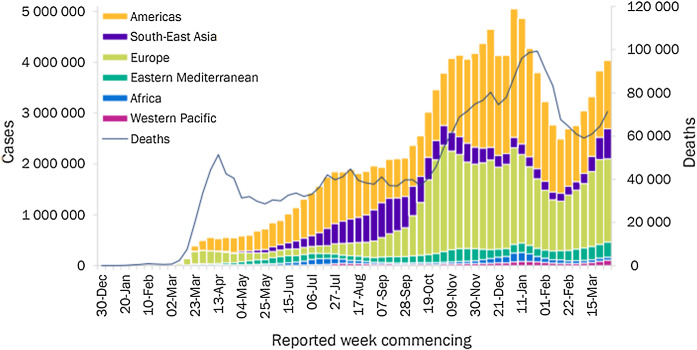
In December 2019, an atypical pneumonia case was discovered in the Hubei province of Wuhan, China. Immediately after this revelation, thousands of other cases have been detected worldwide and soon the situation evolved exponentially to become a global pandemic. On 11th February 2020 [1], the World Health Organization (W.H.O) named this unfamiliar pneumonia as COVID-19. It has created immense chaos erupting worldwide, leading to affect people’s lives and cause a huge number of deaths. As of April 12, 2021, the global epidemiological situation determines that confirmed COVID-19 cases have reached 135,646,617 and 2,930,732 confirmed deaths since the first case was detected. As shown in Fig. 1, the number of COVID-19 cases has increased to reach over 4 million new cases in the past week. A sharp increase of 11% in the number of new deaths compared to the previous week to reach over 71,000 deaths.
Fig. 1.
Number of COVID-19 confirmed cases and total deaths for the period (30 December 2019–12 April 2021) reported weekly by WHO (https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---6-april-2021 (Accessed 12 April 2021))
The WHO reported that, in the case of mild to moderate contamination, the symptoms are dry cough, fatigue, and fever. While, in some dangerous cases, dyspnea, fever, and tiredness can occur [2]. Furthermore, hospitalized patients with COVID-19 suffered from other symptoms like shortness of breath or difficulty breathing, muscle aches, chills, sore throat, runny nose, headache, chest pain, and pink eye. Diabetics, asthmatics, and heart disease patients are more susceptible to the virus [3]. Unlike the other viruses, COVID-19 has a long incubation period varying from 3 to 13 days, while on average, the time between the exposure and the appearance of the symptoms is about 5–6 days. This duration makes COVID-19 more infectious, as positive people continue to communicate with others without knowing about their contamination, which will lead to more infections. Moreover, several infected individuals are asymptomatic and can get the virus without showing any symptom which making the detection, tracking, and containing this disease more challenging. The two aforementioned characteristics of COVID-19 have been contributing to its rapid spread. Phenotypes of this viral infection are distinct in terms of observable characteristics. It can range from no or fairly milder symptoms and quiet recovery without any health issues. Additionally, it drives in certain cases to swift deterioration and failure in the multi-organ system and death.
This epidemic has not only engrossed the scientific community in the search for drug treatment and vaccine but also, in the struggles to examine statistics and meteorological variables to investigate the foremost factors that might contribute to its rapid spread. According to [4], one of the main factors of the sharp rise in transmission of this deadly virus is the weather. In this context, Li et al. [5] aims to investigate the relationship between temperature and sunshine duration with the COVID-19 cases for the country of China. It explored that the temperature has an important yet negative correlation, unlike the sunshine duration which has an inverse association. A recent study showed that episodes of fine particle (PM) pollution play a key role in the rise of the number of contaminated cases [6]. Gupta et al. [7] studied the impact of the most important parameters of weather (maximum, minimum and mean temperature, temperature range, average humidity, humidity range, and wind speed) on the prevalence of the COVID-19 cases in more populated countries, such as India. Moreover, temperature, rainfall, and humidity are explored to be reliable signs to anticipate the number of COVID-19 cases in the coming days [6–8].
The relation between geography and the COVID-19 phenomena has attracted diverse global health specialists because the prevalence of this type of dangerous virus, in particular, is inevitably spatial. They try to identify the infection movement using local or global transmissions based on the contact trajectories within the population network. To this end, experts toward to use Geographical Information System (GIS) and spatial statistics to capture and analyze spatial and geographic data. Different studies have been implemented based on a geographical and geo-spatial analysis in order to understand the locations and the distribution patterns of COVID-19. Some of these studies have focused on specific countries, such as Oman [9], China [10], Indonesia [11], South Korea [12], India [13], United States [14], etc, while others focused at the globe scale [15, 16].
As this virus is a new and fatal strain that has never infected people before, it continues to overload the medical corps because of the exponentially myriad number of infections all over the world and challenge them in many aspects. These challenges including the sharp increases in demands for hospital beds and the overwhelming need for medical resources while many medical staffs have themselves been infected. Currently, reverse transcription-polymerase chain reaction (RT-PCR) has been used as the most validated diagnostic test for COVID-19 infection. Nevertheless, it has long been in shortage in many countries around the world particularly the developing ones. Additionally, test results may take a long time to acquire, leaving behind a higher potential for the prevalence of the virus among other individuals by the patients. Therefore, the development of an automatic diagnosis system became paramount to assist clinicians in triaging infected patients and thus help reduce the infection spread rate. The most common performed methods to detect the virus are based on blood tests [17, 18] that may take a long turnaround time to generate results approximately 3–4 h or imaging modalities including X-ray images and CT scans [19, 20] that could be unavailable in certain hospitals and laboratories. This highlights the critical need to a simple, accurate, and fast Artificial Intelligence (AI) model that can be very useful in these tough times in hopes of early detect the contaminated patients and control the epidemic infestation.
The use of AI in medicine has recently demonstrated prodigious popularity by becoming an adjunct tool for clinicians [21]. Machine learning (ML), which is a well-known research field of AI, enables creating models that reach promised outcomes for the automatic diagnosis of several diseases. ML is often applied for the classification task which tends to categorize data on a basis of a certain number of features. Making use of a small training set can increase the risk of facing the overfitting problem, and therefore, negatively affects the model’s generalizability. Additionally, these samples can be probably non-gaussian noise-contaminated [22, 23]. The number of features also can reflect some issues. It is known that the dataset may contain irrelevant and noisy features that may have an impact on the results [22] and the uncertainties of the model [24]. Thus, the selection of a really important set of features can be an optimal solution to overcome these issues. Moreover, it improves the performance of the classification model and makes it faster in terms of execution time.
In order to develop an optimal classification model, some components must be taken into considerations. Among these components, the configuration of hyper-parameters which is considered a fundamental key to build an effective model [25]. However, the search space of potential combinations of parameter values is probably infinite, and thereby tuning manually becomes impractical, ineffective, time-consuming, and often needs deep knowledge of models. To this end, automatic hyper-parameter optimization is a critical need. Several techniques are existing in this context in which each technique has its strengths and drawbacks [26]. Recently, meta-heuristic algorithms have shown a great outstanding in solving hyper-parameter problems. Genetic algorithm (GA) and particle swarm optimization (PSO) are the most widely used algorithms [26]. To this end, GA is used to select the optimal combination of hyper-parameters. In fact, this technique has been commonly used in different areas [27–29] which provide outstanding results in maximizing the results. It also has revealed to be more efficient compared to other techniques in searching parameters [26]. The main objective of the present paper is to develop a machine learning model for COVID-19 prediction based on clinical symptoms and features. This model can be vastly and rapidly applied as needed during the pandemic.
In summary, our main contributions are as following:
We conducted various experiments to compare the obtained results after running eight machine learning classifiers (Support Vector Machine (SVM), Logistic Regression (LR), Decision Tree (DT), Multi-Layer Perception (MLP), Random Forest (RF), eXtreme Gradient Boosting (XGBoost), Adaptive Boosting (AdaBoost), and Gradient Boosting (GB)), and four feature selection algorithms (SFS, SFFS, SBS, and NSGA-II).
To evaluate the proposed model, we used two different datasets in terms of size, number of symptoms.
We highlight the significant role of feature selection in enhancing the efficiency of the studied classifiers.
The empirical findings confirmed that NSGA-II as feature selection algorithm combined with the AdaBoost classifier can predict COVID-19 patients with an accuracy of 85% for the dataset 1 and 95.56% for the dataset 2.
The rest of this paper is organized as follows. Section 2 reviews state of the art techniques employed for COVID-19 diagnosis. Section 3 presents the machine-learning and feature selection algorithms. Section 4 introduces the proposed model, Sect. 5 reports our evaluation results. Finally, in Sect. 7, we summarize our contribution and we present our future work.
Related works
Depending on the features and symptoms used to perform COVID-19 prediction, the related studies can be divided into two major categories: symptoms-based for COVID-19 prediction and blood-based test for COVID-19 diagnosis.
Symptoms-based for COVID-19 prediction
Zoabi et al. [30] aims to determine the number of infectious cases based on symptoms and other demographic features. The idea is to develop a machine learning model based on gradient boosting intends to classify the cases. This study focused on eight features: gender, whether age is above 60, known contact with an infected individual, and five initial clinical symptoms; cough, fever, sore throat, shortness of breath, headache. They mentioned a flaw in the used data that has shortcomings and biases. The AUC of the proposed model decreased to 0.862 if this bias is eliminated [31].
Khandy et al. [3] aims to classify clinical reports retrieved from doctor’s notes into four categories of disease: COVID, SARS, ARDS, and both (COVID, ARDS). The data used in this study is accessed from the metadata of these X-ray images of John Hopkins University. It contains 24 attributes namely patient id, offset, sex, age, finding, survival, intubated, wenticu, neededsupplementalO2, extubated, temperature, pO2 saturation, leukocytecount, neutrophil count, lymphocyte count, view, modality, date, location, folder, filename, DOI, URL, License, Clinical notes, and other notes. A feature engineering step is conducted to extract 40 features from the textual attribute “Clinical notes”. The proposed model is based on Logistic Regression (LR) and Multinomial Naïve Bayes (MNB). This model is assessed using a small dataset that holds 212 patients. It can help analyze the clinical reports and make suitable recommendations for battling this epidemic.
Banik et al. [32] aims to estimate the probability of person infection by the COVID-19 virus. To this end, several machine learning algorithms are studied such as LR, MNB, Linear SVM, and DT to build an accurate model to predict the probability that a patient is infected based on clinical symptoms of patients. This model considered many features that have the same importance. However, there are the set of characteristics having more importance to identify COVID-19 cases.
Blood-based test for COVID-19 diagnosis
Batista et al. [33] conducted a prediction model to diagnose COVID-19 using features of blood test based on the SVM algorithm. The used data derived from emergency care admission exams in Brazil. The outcome of this work is to detect the risk of positive COVID-19 diagnosis. A total of 15 predictors are used to train this model such as age, sex, haemoglobin, platelets, red blood cells, Mean Corpuscular Haemoglobin Concentration (MCHC), Mean Corpuscular Haemoglobin (MCH), Red Cell Distribution width (RDW), Mean Corpuscular Volume (MCV), leukocytes, lymphocytes, monocytes, basophils, eosinophils, C-Reactive Protein (CRP). The advantage is that would be very beneficial in assigning testing priorities in case of a shortage of equipment.
Mondal et al. [34] developed a classification model to recognize positive cases from all suspected patients based on the dataset collected from hospital Albert Einstein, Brazil. To this end, various machine learning algorithms are used. MLP, LR, and XGBoost that showed promising results over the remaining algorithms. The best accuracy rate is provided by MLP. This work outperforms the work of [33] in terms of performance due to using more data and features (61 attributes against 15 used by [33]).
Brinati et al. [17] built a diagnosis tool for SARS-CoV-2 detection. It allows distinguishing infected persons based on hematochemical values from routine blood exams. The designed tool is based on a random forest algorithm. Multiple features are used to carry out this work such as C-reactive protein (CRP), Aspartate Aminotransferase (AST), Alanine Amino Transferase (ALT), Gamma Glutamil Transferasi (GGT), Lactate Dehydrogenase (LDH), Leukocyte Count (WBC), Plate- lets, Neutrophils, Lymphocytes, Monocytes, Eosinophils, and Basophils. The proposed model can be a good alternative to rRT-PCR test for recognizing COVID-19 infected patients. Particularly, it can be very useful for developing countries that are facing problems with medical resources and specialized laboratories. Nevertheless, this study suffers from some shortcomings. The training process is based on a reduced number of samples. According to [35], the precision of this analysis test may be highly influenced by problems like inadequate methods for collection, transport, handling, sample contamination, and existence of interfering substances, etc.
Wu el al. [18] developed a diagnostic tool to determine COVID-19 confirmed cases from a variety of suspected patients. This work focused on 11 top-ranking clinical blood indices and built using a random forest algorithm. Although more clinical research is required to validate the tool, it can offer some new insights to ensure the rapid diagnosis of COVID-19 infection in order to deal with serious situations caused by the dangerous characteristics of human-to-human transmission.
Kukar et al. [36] established a diagnostic tool to analyze blood exams and find the suspect cases of COVID-19. It used XGBoost as a machine learning classifier. It employs five blood parameters as features, which are Mean Corpuscular Hemoglobin Concentration (MCHC), eosinophil count, albummin, International Normalized Ratio (INR), and prothrombin activity percentage. This study holds some limitations. First, the analysis was conducted in a single center from which the data collected. While this can restrict generalizability, they expect similar laboratory blood test findings in other centers by using incorporated and advisable reagents, procedures, and technology. Second, a limited number of positive cases was involved in this work.
The main limitation of these studies is that patients should move to the hospital to carry out the test, which may take a long time to acquire, leaving behind a higher potential for the prevalence of the virus among other individuals by the patients.
Background
Artificial intelligence is the ability of machine to mimic cognitive functions associated with human capacity such as, learning, perceiving, solving the problem. In this section, we review some algorithms and techniques that are prerequisites for our work.
Machine learning algorithms
Machine learning is a research area evolving several induction algorithms that training machines to analyze data and acquire information from it. This section includes the background details of supervised machine learning algorithms.
Classical machine learning algorithms
Logistic Regression (LR)
A LR model is a supervised learning algorithm that is used to predict the class membership probability of a given variable based on its relationship with the label [37]. One of the properties of this model is that the final probability prediction may be excessively influenced by a small shift in the input value. In addition, the dimension of the input vector (number of predictors) should be low, as this can affect the cost of the model training and may lead to overfitting. Besides, It can be resulting in the poor generalizability of the model. In fact, LR can be a valuable model to be chosen when there are various data sources that have to be incorporated into a binary classification task, and low complexity is needed.
-
(b)
Support Vector Machine (SVM)
SVM is a supervised machine learning algorithm designed for classification and regression. The aim of SVM is to find the optimal linear or nonlinear boundary that separating data into two or more classes [38]. Before applying SVM, it is necessary to select the function responsible for data separation, called Kernel. The linear function or the Gaussian function is the most frequently used kernel. The remaining parameters are empirically selected by training a variety of models and preserving the model settings providing the lowest error rate. The most commonly used SVM classifier is a linear one. It attempts to predict the class of the test sample between two possible classifications. It works as follows; take a specified number of features with the class label and trying to search for the optimal linear separating hyper-plane in an N-dimensional space, where N defines the number of features. The best hyper-plane is selected using the support vectors and margins. It is based on the distance between the two classes. One of SVM’s main drawbacks is its need for high computational cost when dealing with a large number of data.
-
(c)
Decision Tree (DT)
A DT is a predictive model, capable of giving coherent classification rules. It repeatedly divides the data depending on particular criteria that maximize the separation of the data, producing a tree-structured like [38]. The main idea behind the decision tree algorithm is to select the most important attributes using Attribute Selection Measures (ASM) to split the records. Make that attribute a decision node and breaks the dataset into smaller subsets. The tree starts to build through repeating this process recursively for each leaf until one of the conditions satisfies the termination criteria and all the tuples belong to the same attribute value and no more remaining attributes or instances. To predict a class label, starting from the root and compare the test sample with the values of the root attribute. The comparison consists of following the branch corresponding to the value and jump to the next node. This process repeats until reaches the leaf node with predicted class value.
-
(d)
Multi-Layer Perceptron (MLP)
MLP is a model of Neural Networks (NN), inspired from the structure and function of the brain that learns from data and specializes in pattern recognition. MLP is based on a feed-forward algorithm. The input feature vector is entered into the neurons for training. A back-propagation algorithm is applied to train the neurons, with the flow in the forward direction. Next, the generated output is compared to the desired output using a cost function such as the Mean Squared Error (MSE) function. If the outputs do not match, an error is produced. This error propagates in the backward direction. In this case, the weights are adjusted in order to reduce the error. This processing is repeated until the error becomes zero [39]. There is a layered structure in the NNs with the number of interconnected nodes. Among each one of these nodes, there is an activation function which can be a tangent hyperbolic function, sigmoid function, piece-wise linear function, and threshold function. In the case of binary classification, the neural network is built with a single output node.
Ensemble learning methods
Ensemble learning method refers to combine a set of weak classifiers to produce a single strong classifier in order to improve the overall performance [37].
Bagging techniques
Bagging also known as, Bootstrap aggregating, is a broadly useful ensemble machine learning paradigm. It allows improving the performance of machine learning models and helps also avoid overfitting. The main idea behind bagging is to combine the results-producing by multiple models to get a generalized result. One of the well-versed techniques that following bagging technique is Random Forest (RF).
Random forest is one of the most commonly used supervised machine learning due to its simplicity and diversity as it can be used in both classification and regression tasks. RF aggregates a set of DT that produces a forest [40]. Better prediction results are often obtained often when incorporating more trees in the forest. Each DT is a set of rules that are based on the values retrieved from the input features and optimized to be reliable for classify all the examples of the training set. If the DT built in a deep manner, it can lead to encounter some problems such as over-fitting due to irregularities in the training set. This issue can be solved using Random Forest whereby apply the training phase on multiple train samples. Accordingly, as the number of DT increased as the variance is reduced, therefore minimize the generalization error and turning a strong classifier. Prior to application RF, there are two parameters to be tuning such as the number of trees and the depth level for each tree. Nevertheless, it should be borne in mind that if the discriminatory power over the training dataset increases as DT increases in depth. It often comes at the expense of the loss of generalization performance. The RF is chosen to transform the problem into a collection of hierarchical requests represented as DT. However, RF is not very immune to noisy data.
-
(b)
Boosting techniques
Boosting is an ensemble method designed for enhancing the prediction rate of a machine learning algorithm. The main idea is to train weak learners in a sequential way, where each trying to correct the previous model [41].
One of the most popular ensembles boosting classifier is Adaptive Boosting known as AdaBoost. It aims to combine multiple weak learners into a strong learner to enhance the performance of the prediction model [42]. Basically, the concept of AdaBoost consists of setting the weights of poorly performing classifiers and training the samples in each iteration. The process of generating a weak learner consists of taking equal weights for each sample and trains the weak learner using the weighted data. A coefficient should be chosen based on the performance of this weak learning classifier. In the case of misclassified points, the weights are increased and the weights of correctly classified points are reduced. Then, the weak learning algorithms are run again to obtain a weak classifier for the new weighted data. Repeating this process until all the data points have been correctly classified, or the maximum iteration level has been reached.
Gradient Boosting is another well-known algorithm that belongs to the boosting family. It is an ensemble machine learning method that combines a set of weak learners sequentially with some shrinkage on them [43]. It identifies the weakness of each poor learner using gradients in the loss function. GB conducts variables selection to improve the predictors of the tree as its based-on decision tree-like AdaBoost. Therefore, the model is a combination of two trees. Then, it calculates the difference between these two trees and produces a third tree in order to predict the revised residuals. A number of iterations should be specified to repeat the process. The subsequent trees help to classify the observations which are not well classified by the preceding trees. The predictions of the final model are therefore the weighted sum of the predictions derived by the previous tree model.
A well-known computational speed and model performance is XGBoost, short of eXtreme Gradient Boosting. It is a scalable machine learning algorithm for tree boosting that is commonly used in different fields. It is based on the idea of “Boosting”, which consists of integrating multiple predictors of an ensemble of “weak learners” to build a powerful learner [44]. This process can be done through additive learning strategies during the training phase. The process consists initially of fitted the first learner with the entire input data, and the second model with the residuals to fix the shortcomings of the weak learner. This fitting method is repeated several times until satisfies the stopping criterion. The final prediction of the model is achieved by the sum of the predictions of each learner. In this way, XGBoost helps to reduce overfitting and optimize the computational resources thanks to supporting a variety of regularization techniques.
Features selection algorithms
Selecting the most relevant features that are used for the training phase is deemed to be an essential step for many pattern recognition problems. Accordingly, the key issue resides in how to find the most adequate set of features that match with data classes and can provide an enhancement in the model performance. To deal with this challenging task, many feature selection algorithms have been reported. The main idea resides in removing the irrelevant and repetitive features from the original dataset and keep only the important ones. In this way, feature selection can be a beneficial step in terms of enhancing the performance of the model, reducing the dimension of the dataset in case of handling a large number of features, and avoiding the over-fitting problem. In our work, we aim to apply feature selection algorithms to better discriminate the symptoms of patients infected with COVID-19 virus and thereby improve the efficiency of the model. In this work, we investigate four feature selection algorithms that are considered the most widely used: Sequential Forward Selection (SFS), Sequential Forward Floating Selection (SFFS), Sequential Backward Selection (SBS), and Non-dominated Sorting Genetic Algorithm II (NSGA-II) to choose the optimal subset of features.
Sequential forward selection (SFS)
SFS is a wrapper-based algorithm based on a bottom-up search procedure that allows generating an optimal subset of features [45]. Initially, it is empty and gradually adds features, that are selected upon an evaluation criterion function, one by one until reaching the best subset. Due to its simplicity and speed, SFS is a widely used sequential algorithm. Also, it is very suitable with smaller datasets [46]. Algorithm 1 is the pseudo-code for the SFS algorithm.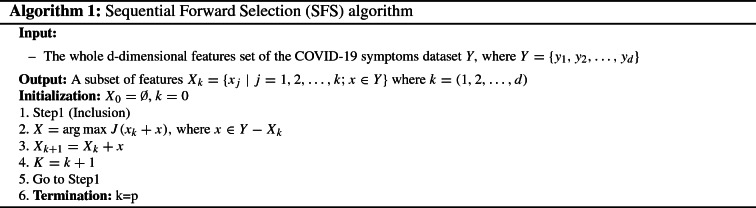
As illustrated in Algorithm 1, the SFS algorithm takes the whole d-dimensional feature set of the COVID-19 symptoms dataset Y as input. It initializes with two empty variables, denotes the set of features to be generated, and a variable k (line 1). At each inclusion step, the algorithm selects the feature x that maximizes the criterion function based on the evaluation of a classification algorithm and adding it to the subset (line 3). This feature is relevant to boost the efficiency of the model. In this way, each feature incorporated into the subset , the variable K will be incremented (line 4). This process is repeated until k equal to p where all desired features are added (lines 5 and 6). The ultimate output of the SFS algorithm is a subset containing the most significant features.
Sequential backward selection (SBS)
From SFS algorithm, we can toggle to SBS algorithm as it is works in the backward direction. It starts by initializes the algorithm with the whole set of features and starts to remove irrelevant features greedily until getting the desired number of features to generate the optimal subset. Algorithm 2 describes how the SBS process works.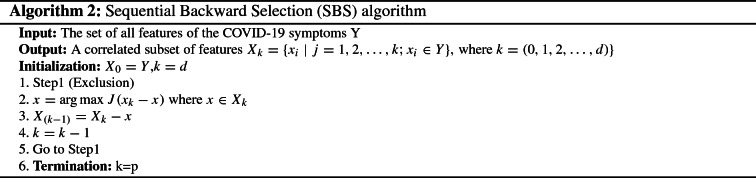 As illustrated in Algorithm 2, the SBS algorithm takes the COVID-19 symptoms dataset Y as input. It initializes the variable with the entire feature set and the size K equal to d. At each step of exclusion, it omits the feature whose dropping yields the maximal performance improvement on the basis of criterion function J (lines 1 and 2). Then, the variable k is decremented (line 4). This process continues sequentially until obtaining the desired number of features (lines 5 and 6). The SBS algorithm generates the interesting features as output.
As illustrated in Algorithm 2, the SBS algorithm takes the COVID-19 symptoms dataset Y as input. It initializes the variable with the entire feature set and the size K equal to d. At each step of exclusion, it omits the feature whose dropping yields the maximal performance improvement on the basis of criterion function J (lines 1 and 2). Then, the variable k is decremented (line 4). This process continues sequentially until obtaining the desired number of features (lines 5 and 6). The SBS algorithm generates the interesting features as output.
Sequential forward floating selection (SFFS)
SFFS algorithm is based on forward and backward steps [47]. It aims to adjust the trade-off between these two steps. SFFS allows a “self-controlled backtracking” that can eventually find better solutions.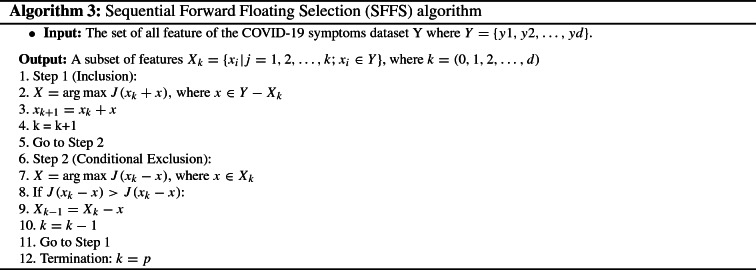 As illustrated in algorithm 3, the SFFS takes the entire feature set as input. The process starts with initializing with an empty set and it is composed of two steps, inclusion and conditional exclusion. At each inclusion step, the feature x that maximizes the criterion is appended to the current feature subset (lines 2, 3 and 4) based on the evaluation of a classification algorithm. After the addition of a feature, a conditional exclusion is examined (lines 5 and 6). At this step, the feature that maximizes the criterion function on the new subset of feature is sought. If the removes of this feature resulting in a gain rise in the performance, it is removed completely from the feature subset (lines 8, 9 and 10). Otherwise, go back to step 1 (line 11). The whole process stops when k equals the number of features (line 12). In the end, the SFFS algorithm yields a subset of the most important features that are beneficial for improving the classification performance.
As illustrated in algorithm 3, the SFFS takes the entire feature set as input. The process starts with initializing with an empty set and it is composed of two steps, inclusion and conditional exclusion. At each inclusion step, the feature x that maximizes the criterion is appended to the current feature subset (lines 2, 3 and 4) based on the evaluation of a classification algorithm. After the addition of a feature, a conditional exclusion is examined (lines 5 and 6). At this step, the feature that maximizes the criterion function on the new subset of feature is sought. If the removes of this feature resulting in a gain rise in the performance, it is removed completely from the feature subset (lines 8, 9 and 10). Otherwise, go back to step 1 (line 11). The whole process stops when k equals the number of features (line 12). In the end, the SFFS algorithm yields a subset of the most important features that are beneficial for improving the classification performance.
Proposed approach
In this section, we introduce our proposed machine learning model which is based on a feature selection phase and a classification phase to predict COVID-19 patient infection. An overview of the proposed approach is illustrated in the Fig. 2. The first phase uses NSGA-II algorithm to select the optimal subset of features that satisfies the conflicting objectives for the training. While, the second phase is to train the model based on the AdaBoost classifier to predict each target class. Finally, a testing phase is conducted to assess the efficiency of the model using the test data.
Fig. 2.
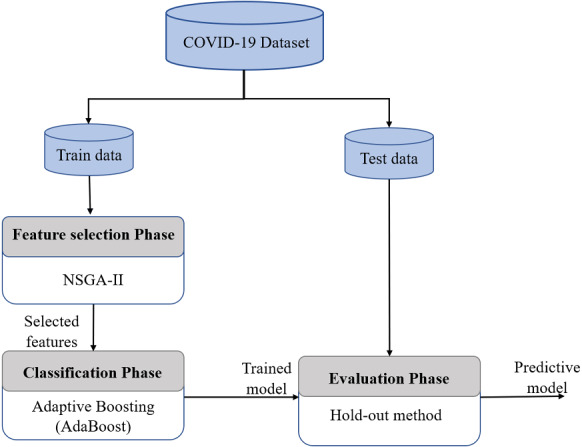
Overview of proposed approach
Feature selection phase
This part aims to identify the optimal set of features needed for COVID-19 prediction. It is expected that the selected features improve the performance of the model and speeding up the training process. To this end, we employed NSGA-II to extract the relevant features. It is a multi-objective search algorithm that has been used to solve several real-world optimization problems [48]. It is considered as one of the most widely used and successful algorithms to find non-dominated solutions. NSGA-II aims to find the trade-off between objectives that are often conflicting and generates a set of optimal solutions called Pareto front or non-dominated solutions. A high-level view of NSGA-II is depicted in Algorithm 4. We define the feature selection phase as a problem that includes two conflicting objectives functions that aim to find the optimal subset of features. The two conflicting objectives are outlined in the following Eq. 1. The solution is represented as a simple coding scheme where a binary chromosome representation is used. Each feature is represented as zero or one based on the objective functions.
| 1 |
The first objective consists of minimizing the number of features in the generated subset . The optimization process aims to reduce the complexity of the input variables by minimizing the number of features for the classifier. The second objective aims to maximize the weight of the selected features. To this end, we use the Information Value (IV) which is one of the most useful techniques that intends to select the essential features. It helps to rank the features based on their importance in the dataset and their relationship with the class label which can be achieved using the weight. Therefore, each feature takes a weight “w” which is ranged between and 1. The goal of IV is to distinguish between the features that have a strong relationship and the ones not. IV measures the difference between the percentage of COVID-19 infected persons (C) and the percentage of NON COVID-19 infected persons (NC) multiplied by the WOE for each respective features. The IV is measured as follows:
| 2 |
where
| 3 |
, the weight of each feature in the obtained subset and “n” denotes the subset size. C, the total number of COVID-19 instances. NC, the total number of NON COVID-19 instances. , the number of COVID-19 within the feature i. the number of NON COVID-19 within the feature i.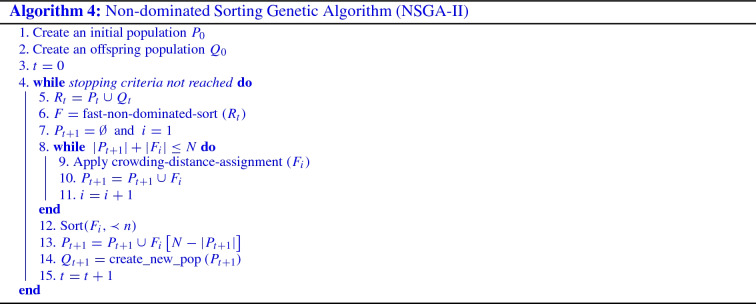
As illustrated in Algorithm 4, it starts by creating an initial population of individuals (line 1). Then, based on crossover and mutation, a child population is generated using the parent population previously created (line 2). The two created populations ( and ) are merged to construct an initial population of size N (line 5). A fast-non-dominated sort is applied which is the main technique used by NSGA-II to classify the solutions into different dominance levels (line 6). In fact, this technique consists of comparing the solutions (individuals) of the population. In this way, all the evaluated solutions based on the objective functions are sorted into different fronts (line 6). Accordingly, solutions that are found in the first Pareto-front F0 were assigned a dominance level of 0 and the fast non-dominated-sort continue to calculate the remaining population. Solutions that are found in this second front were assigned dominance level of 1, and so on. Then, the next population is generated using the dominance level of solutions . NSGA-II relies basically on the crowding distance when making a selection of a subset of solutions that are belong to the same dominance level (line 8). Crowding distance is used to raise variety in the population. To split the front , the solutions should be sorted in descending order (line 12), and the first () elements of are chosen (line 13). Finally, selection, crossover, and mutation are applied to create a new population (line 14). This procedure continue until satisfies the termination criteria.
Classification phase
This phase is designed to classify the suspected infected patients into two classes. Before starting the training process, we split the used datasets into two subsets: training and testing data by applying the hold-out method. We recall that dataset 1 contains 1495 samples, 70% of them are devoted to train (1046 samples) while the remaining 30% (449 samples) are dedicated to test the model. The second dataset includes 99,232 samples that are divided as follows: 69,463 samples for the training and 29,769 samples for the testing. Our goal is to classify the patients into two classes: COVID-19 or NON-COVID-19. To this end, we use the AdaBoost classifier that aims to reduce the misclassification rate of a weak learner and create a strong classifier by combining a set of weak learners.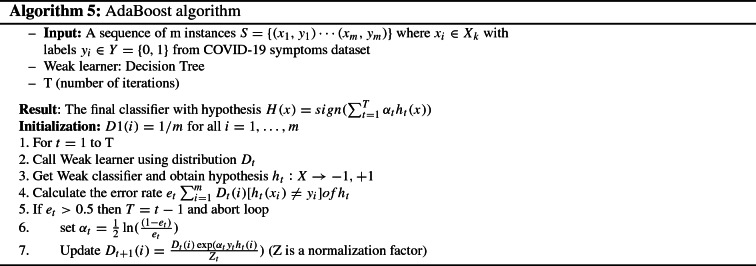
As illustrated in algorithm 5, AdaBoost takes as input the set of training instances m which contains , where each denotes the example and is a binary value label referring to whether is a positive or negative sample. The principle of AdaBoost consists of assigning weights for each sample to increase or decrease this weight later to lessen the classification error. To this end, it initializes the weights of data points according to the function , where m defines the number of samples. It assigns the same weights to all samples. In each iteration t, the weak classifier is trained and used to predict the samples to calculates the error rate (lines 2 and 3) of the misclassified samples. The higher the error, the more the corresponding learner will be weighted when assigning weights to samples (line 4). If the error greater than 0.5, it adjusting the value of and the distribution by putting more weights on the incorrectly classified training samples and fewer weights on the correctly classified (lines 5, 6, and 7) to better classified in the next iteration. This reweighing step allows increasing the importance of examples that were wrongly classified by the previous weak classifier. This process repeats until reaches the desired number of iterations T. Finally, the algorithm yields a strong classifier derived from a weighted combination of weak learners. Typically, the outcome of AdaBoost can negatively affected due to the use of noisy data because each learner is too dependent on the output of its predecessors. In this case, the learner will not capable of correcting the misclassified instances which are noisy data. To this end, we give importance to the verification step which consists of checking the quality of used datasets. In our work, since the dataset 2 is imbalanced, the sampling technique (SMOTE) is used to balance the class distribution before starting the training process.
Evaluation phase
After training the model with the relevant features, it is crucial to check the efficiency of the obtained model. Basically, this phase is considered as a fundamental step that permits the assessment of the built model against data that has never been used in the training phase. It is examined based on the test data (30% of the original dataset) that provided using the hold-out method for both datasets.
Validation
Description of the experimental datasets
In our experimentation, two datasets were considered. These datasets have several symptoms and different sizes. The first dataset (Dataset 1) was obtained from the Wolfram Data Research Repository (2020).1 It contains 1495 patients cases of whom 757 patients are infected with the COVID-19 virus. A total of 12 clinical symptoms covered in the used dataset along with the three other demographic features. The second dataset (Dataset 2)2 was obtained from the study of [30]. It includes a total of 99,232 samples in which 8393 COVID-19 cases. This dataset contains eight features of whom five initial clinical symptoms. Customarily, data preprocessing is a fundamental step before training the model in case the data is inconsistent or incomplete. Indeed, this is not the case with the dataset 1, as we skip this step due to the well-structured binary values. Nevertheless, with the dataset 2, we fall into the problem of imbalanced data where the class NON-COVID-19 outnumber the class COVID-19. To address this problem, we apply one of the most widely used techniques to synthesizing new examples which is the Synthetic Minority Oversampling TEchnique (SMOTE) [49]. This technique of oversampling consists of generating new synthetic samples for the minority class. In this way, the total number of samples in the training set for the two classes is equals [50]. Tables 1 and 2 give a comprehensive description of the features held in the used datasets.
Table 1.
Dataset 1 description
| Indicators | Column | Description | Code description | Type |
|---|---|---|---|---|
| Demographic characteristics | Age | Patient’s age | [1,98] | Numeric |
| Gender | Patient’s gender | 1: Male 0: female | ||
| Clinical symptoms | Fever | Patient fever | 1: Yes 0: No | |
| Cough | Patient dry cough | 1: Yes 0: No | ||
| Fatigue | Patient fatigue | 1: Yes 0: No | ||
| Pains | Patient pains | 1: Yes 0: No | ||
| Nasal congestion | Patient nasal congestion | 1: Yes 0: No | ||
| Shortness of breath | Patient breathing problem | 1: Yes 0: No | ||
| Runny nose | Patient runny nose | 1: Yes 0: No | ||
| Sore Throat | Patient sore throat | 1: Yes 0: No | ||
| Diarrhea | Patient diarrhea | 1: Yes 0: No | ||
| Chills | Patient chills | 1: Yes 0: No | ||
| Headache | Patient headache | 1: Yes 0: No | ||
| Vomiting | Patient vomiting | 1: Yes 0: No | ||
| Other information | Lives in affected area | Patient is from COVID-19 affected area or not | 1: Yes 0: No |
Table 2.
Dataset 2 description
| Indicators | Column | Description | Code description | Type |
|---|---|---|---|---|
| Demographic characteristics | Age | Age 60 years or above | 1: Yes 0: No | Numeric |
| Sex | Patient’s sex | 1: Male 0: female | ||
| Clinical symptoms | Cough | Patient cough | 1: Yes 0: No | |
| Fever | Patient fever | 1: Yes 0: No | ||
| Sore throat | Patient sore throat | 1: Yes 0: No | ||
| Shortness of breath | Patient breathing problem | 1: Yes 0: No | ||
| Headache | Patient headache | 1: Yes 0: No | ||
| Other information | Known with confirmed | Known contact with an individual confirmed to have COVID-19 | 1: Yes 0: No |
Research questions
We defined the following two research questions to evaluate the efficiency of our proposed model:
RQ1. To what extent is the feature selection techniques contributing in improving the performance of the studied classifiers?
RQ2. How does the proposed approach perform compared to similar existing works?
To answer RQ1, we conducted a comparative performance evaluation of various machine learning algorithms (MLP, SVM, LR, DT, GB, RF, XGBoost, and AdaBoost) in order to thoroughly find a useful model to distinguish the COVID-19 cases among suspected individuals. The evaluation step encompasses two phases: with and without applying feature selection. These phases are very important to better investigate the role of feature selection in enhancing the performance of the predictive model. Evaluating the classification model is a fundamental step to test its efficiency. In the literature, several assessment metrics have been proposed which are different according to the nature of the dataset (balanced, imbalanced) [51]. Generally, the most commonly used performance evaluation indicator is the accuracy of the model [52]. However, this metric is not enough to truly judge the model. This highlights the critical need for other assessment parameters to select the well-performing model. To this end, we considered five evaluation metrics as well the accuracy: precision, sensitivity, specificity, f1-score, and AUC.
These metrics can be derived from the following confusion matrix outlined in Table 3.
Table 3.
Confusion matrix
| Predicted class | ||
|---|---|---|
| Actual class | COVID-19 | NON-COVID-19 |
| COVID-19 | True positive (TP) | False positive (FP) |
| NON-COVID-19 | False negative (FN) | True negative (TN) |
- Accuracy: represents the percentage of correctly predicted cases relative to the whole dataset. It is calculated as follows:
4 - Precision: is the exactness which represent the number of positive class predictions that actually belong the positive class. It is a measure of the proportion of patients detected by the classifier to have the COVID-19 that actually had the virus. It is the ratio of the True Positive (TP) instances to the sum of True Positive (TP) and False Positive (FP) instances:
5 - Sensitivity (Recall): represents the percentage of COVID-19 cases that have been predicted correctly (probability of positive test given the suspected patients that have the COVID-19 virus). It is computed as follows:
6 - Specificity: represents the percentage of NON-COVID-19 cases that have been predicted incorrectly (probability of negative test given the suspected patients that have not the COVID-19 virus). It is calculated as follows:
7 - F1-Score: is a harmonic mean of Recall and Precision value. It strikes the perfect balance between Precision and Recall thereby providing a correct evaluation of the model’s performance in classifying COVID-19 patients.
8 - AUC: it allows to compute how much the model is capable of distinguishing between patients infected by COVID-19 or not. It is calculated as follows:
9
To answer RQ2, we sought to compare the obtained results of our proposed model to different existing work based on three performance criteria: Accuracy, Precision and Recall.
Results and discussions
Results for research question 1
Our research study used two COVID-19 symptoms data-sets. Three experiments are conducted in which the SFS, SFFS, SBS and NSGA-II algorithms are used to select the optimal subset of features for COVID-19 prediction. We respectively present the results of the studied machine learning algorithms.
Experimental results with full dataset
In this part, we introduce an empirical evaluation of the studied classifiers that are trained using the full features of the used datasets. As reports in Table 4, for dataset 1, the random forest outperforms all the other used classifiers. It provides the highest AUC value of 81.46%, accuracy of 81.51%, precision of 83.19%, sensitivity of 82.16%, specificity of 80.77%, and F1-score of 82.67%. For dataset 2, Logistic regression achieves the best results with an accuracy of 92.88%, precision of 92.88%, sensitivity of 92.88%, specificity of 94.28% and AUC of 93.58%. Gradient boosting generates closet result to logistic regression with an accuracy of 92.41% and AUC of 93.18%.
Table 4.
Classification results with full datasets
| Dataset | Classifier | Accuracy (%) | Precision (%) | Sensitivity (%) | Specificity (%) | F1-score (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| Dataset 1 | MLP | 79.96 | 82.40 | 79.67 | 80.29 | 81.01 | 79.98 |
| SVM | 75.72 | 78.70 | 75.10 | 76.44 | 76.86 | 75.77 | |
| LR | 77.51 | 80.70 | 76.35 | 78.85 | 78.46 | 77.60 | |
| Decision tree | 79.69 | 79.38 | 84.65 | 74.52 | 81.93 | 79.58 | |
| Gradient boosting | 80.40 | 81.22 | 82.57 | 77.88 | 81.89 | 80.23 | |
| Random forest | 81.51 | 83.19 | 82.16 | 80.77 | 82.67 | 81.46 | |
| XGboost | 80.40 | 81.22 | 82.57 | 77.88 | 81.89 | 80.23 | |
| AdaBoost | 79.96 | 81.86 | 80.50 | 79.33 | 81.17 | 79.91 | |
| Dataset 2 | MLP | 89.36 | 89.36 | 89.36 | 90.01 | 89.36 | 89.68 |
| SVM | 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| LR | 92.88 | 92.88 | 92.88 | 94.28 | 92.88 | 93.58 | |
| Decision tree | 85.96 | 85.96 | 85.96 | 86.27 | 85.96 | 86.12 | |
| Gradient boosting | 92.41 | 92.41 | 92.41 | 93.95 | 92.41 | 93.18 | |
| Random forest | 89.36 | 89.36 | 89.36 | 90.01 | 89.36 | 89.68 | |
| XGboost | 92.36 | 92.36 | 92.36 | 93.94 | 92.36 | 93.15 | |
| AdaBoost | 89.35 | 89.35 | 89.35 | 90.01 | 89.35 | 89.68 |
Bold values highlight the best results for the two studied datasets
Experimental results with filtered dataset
In this part, we present the experimental results of the studied classifiers that are trained based on the attributes extracted using the feature selection algorithms. As reported in Tables 5 and 6, the performance of the majority of classifiers for both datasets is experienced a considerable enhancement due to the feature selection algorithms application.
Table 5.
Experimental results of studied feature selection algorithms for dataset 1
| Feature selection classifier | Accuracy (%) | Precision (%) | Sensitivity (%) | Specificity (%) | F1-score (%) | AUC (%) |
|---|---|---|---|---|---|---|
| 82.41 | 84.32 | 82.57 | 82.21 | 83.44 | 82.39 | |
| 75.72 | 78.7 | 75.1 | 76.44 | 76.86 | 75.77 | |
| 79.29 | 85.58 | 73.86 | 85.58 | 79.29 | 79.72 | |
| 80.4 | 80.97 | 82.99 | 77.4 | 81.97 | 80.2 | |
| 81.07 | 84.21 | 79.67 | 82.69 | 81.88 | 80.97 | |
| 81.96 | 83.19 | 82.16 | 80.77 | 82.67 | 81.46 | |
| 80.85 | 81.38 | 83.4 | 77.88 | 82.38 | 80.64 | |
| 81.07 | 81.97 | 82.99 | 78.85 | 82.47 | 80.92 | |
| 81.96 | 83.61 | 82.57 | 81.25 | 83.09 | 81.91 | |
| 75.72 | 78.7 | 75.1 | 76.44 | 76.86 | 75.77 | |
| 79.73 | 86.41 | 73.86 | 86.54 | 79.64 | 80.2 | |
| 80.4 | 80.97 | 82.99 | 77.4 | 81.97 | 80.2 | |
| 80.62 | 82.63 | 80.91 | 80.29 | 81.76 | 80.6 | |
| 82.41 | 84.32 | 82.57 | 82.21 | 83.44 | 82.39 | |
| 82.85 | 84.75 | 82.99 | 82.69 | 83.86 | 82.84 | |
| 81.07 | 81.45 | 83.82 | 77.88 | 82.62 | 80.85 | |
| 80.18 | 81.15 | 82.16 | 77.88 | 81.65 | 80.02 | |
| 75.72 | 78.7 | 75.1 | 76.44 | 76.86 | 75.77 | |
| 77.73 | 80.26 | 77.59 | 77.88 | 78.9 | 77.74 | |
| 80.62 | 81.3 | 82.99 | 77.88 | 82.14 | 80.44 | |
| 81.07 | 84.21 | 79.67 | 82.69 | 81.88 | 81.18 | |
| 82.63 | 84.98 | 82.16 | 83.17 | 83.54 | 82.67 | |
| 82.85 | 84.75 | 82.99 | 82.69 | 83.86 | 82.84 | |
| 83.3 | 85.17 | 83.4 | 83.17 | 84.28 | 83.29 | |
| 83.52 | 85.09 | 82.91 | 84.19 | 83.98 | 83.55 | |
| 79.51 | 79.83 | 81.20 | 77.67 | 80.51 | 79.44 | |
| 83.07 | 4.96 | 82.05 | 84.19 | 83.48 | 83.12 | |
| 84.51 | 82.8 | 88.46 | 80 | 85.54 | 84.23 | |
| 84.41 | 82.28 | 89.32 | 79.07 | 85.66 | 84.19 | |
| 83.74 | 85.78 | 82.48 | 85.12 | 84.10 | 83.8 | |
| 83.52 | 85.71 | 82.05 | 85.12 | 83.84 | 83.58 | |
| 85 | 90 | 89.32 | 85 | 86.01 | 87.16 |
Bold value indicates the highest result
Table 6.
Experimental results of studied feature selection algorithms for dataset 2
| Feature selection classifier | Accuracy (%) | Precision (%) | Sensitivity (%) | Specificity (%) | F1-score (%) | AUC (%) |
|---|---|---|---|---|---|---|
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 92.88 | 92.88 | 92.88 | 94.91 | 92.88 | 93.89 | |
| 93.75 | 93.75 | 93.75 | 94.90 | 93.75 | 94.32 | |
| 86.47 | 86.47 | 86.47 | 87.24 | 86.47 | 86.86 | |
| 92.51 | 92.51 | 92.51 | 93.92 | 92.51 | 93.22 | |
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 92.51 | 92.51 | 92.51 | 93.94 | 92.51 | 93.23 | |
| 92.52 | 92.52 | 92.52 | 93.91 | 92.52 | 93.22 | |
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 94.01 | 94.01 | 94.01 | 99.60 | 94.01 | 96.81 | |
| 95.31 | 95.31 | 95.31 | 98.2 | 95.31 | 96.75 | |
| 92.61 | 92.61 | 92.61 | 93.90 | 92.61 | 93.26 | |
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 91.68 | 91.68 | 91.68 | 92.77 | 91.68 | 92.23 | |
| 95.10 | 95.10 | 95.10 | 98.25 | 95.10 | 96.67 | |
| 95.34 | 95.34 | 95.34 | 98.23 | 95.34 | 96.79 | |
| 91.68 | 91.68 | 91.68 | 92.77 | 91.68 | 92.23 | |
| 92.88 | 92.88 | 92.88 | 94.91 | 92.88 | 93.89 | |
| 95.12 | 95.12 | 95.12 | 98.20 | 95.12 | 96.66 | |
| 86.47 | 86.47 | 86.47 | 87.24 | 86.47 | 86.86 | |
| 92.51 | 92.51 | 92.51 | 93.92 | 92.51 | 93.22 | |
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 95.34 | 95.34 | 95.34 | 98.23 | 95.34 | 96.79 | |
| 91.68 | 91.68 | 91.68 | 92.77 | 91.68 | 92.23 | |
| 91.64 | 91.64 | 91.64 | 95.26 | 91.64 | 93.45 | |
| 95.10 | 95.10 | 95.10 | 98.25 | 95.10 | 96.67 | |
| 94.76 | 94.76 | 94.76 | 98.24 | 94.76 | 96.50 | |
| 92.52 | 92.52 | 92.52 | 93.91 | 92.52 | 93.22 | |
| 95.34 | 95.34 | 95.34 | 98.23 | 95.34 | 96.79 | |
| 92.44 | 92.44 | 92.44 | 93.91 | 92.44 | 93.17 | |
| 92.62 | 92.62 | 92.62 | 93.90 | 92.62 | 93.26 | |
| 95.56 | 95.56 | 95.56 | 98.19 | 95.56 | 96.87 |
Bold value indicates the highest result
-
First experiment
In the first conducted experiment, features are selected using the SFS algorithm as described in Algorithm 1. As depicted in Table 5, the MLP classifier achieved the best classification rate with an average 82.39% of AUC, accuracy of 82.41%, precision of 84.32%, sensitivity of 82.57%, specificity of 83.44%, and F1-score of 83.44%. In fact, the SFS confirmed its efficiency with MLP regarding the result of the full dataset whereas the accuracy rate is increased by 2.45%. Additionally, Random forest, Decision Tree, and XGBoost yielded a slight increase in their accuracy rate that reaches, 81.96%, 80.4%, and 80.85% respectively. For the AdaBoost classifier, the classification accuracy is improved to achieve 81.07% with SFS selection algorithm instead of 79.96% when using all the features. Besides, the Gradient Boosting classifier generates a slight rise. It provides an accuracy rate of 81.07% instead of 80.4% with the full dataset. As illustrated in Table 6, Logistic regression attains the highest performance results with the dataset 2. It provided an AUC value of 94.32%, accuracy of 93.75%, precision of 93.75%, Sensitivity of 93.75%, Specificity of 94.9%, and f1-score of 93.75%. Moreover, Random forest achieved a significant increase compared to its results without feature selection application. It provided 92.62% instead of 89.36% as accuracy. XGBoost, Gradient boosting, and MLP achieved a slight improvement in their classification performance compared to their results without feature selection. These classifiers yielded an accuracy rate of 92.51%, 92.51%, and 92.62%, respectively. To conclude, the best subset of features is extracted using MLP classifier for the first dataset 1. It contains the following features: Age, Gender, Fever, Pains, Nasal congestion, Chills, and Vomiting. While, Logistic regression achieved the highest results for the dataset 2 with the following subset: Cough, Sore Throat, Fever, and Known with confirmed.
-
Second experiment
In the second experiment, the SFFS is used for feature selection as described in Algorithm 3. As shown in Table 5, XGBoost has proved its mettle in terms of performance regarding the remaining classifiers. It generates the best AUC value of 82.84%, accuracy of 82.85%, precision of 84.75%, sensitivity of 82.99%, specificity 82.69%, and f1-score of 83.86%. A slight increase is achieved by decision tree and gradient boosting. The decision tree attains an accuracy of 79.69% with the whole features, whereas, 80.4% with the picked features. The gradient boosting provides 80.62% after feature selection instead of 80.40% as accuracy. Moreover, AdaBoost increased by 1.11% in terms of accuracy rather than the result achieved using the full dataset. The results listed in Table 6 shows that AdaBoost provides the highest classification rate for the dataset 2. It reaches an AUC value of 96.79%, accuracy of 95.34%, precision of 95.34%, Sensitivity of 95.34%, Specificity of 98.23%, and f1- score of 95.34%. A significant increase is attained by the XGBoost classifier. It yields an accuracy of 95.1% with the subset generated using the SFFS feature selection instead of 92.36% when using the whole features. Additionally, decision tree and SVM are improved to reach 92.61% and 94.01% respectively compared to their results without feature selection application, 85.96% and 92.62% respectively. To conclude, the best subset of features for dataset 1 is extracted using XGBoost classifier. It contains the following features: Age, Gender, Fever, Fatigue, Pains, Headache, and Lives in affected area. For the dataset 2, AdaBoost achieved the highest classification results with the following generated subset: Fever, Sore throat, Shortness of breath, and Known with confirmed.
-
Third experiment
In the third experiment, feature selection is carried out using SBS as described in Algorithm 2. The results from Table 5 show that the highest classification rate is scored by the AdaBoost classifier. It yielded the best AUC value of 83.29%, accuracy of 83.3%, precision of 85.17%, sensitivity of 83.4%, specificity of 83.17% and f1-score of 84.28%. XGBoost makes a significant rise, it gives an average accuracy of 82.85% instead of 82.40% when SBS is employed. For random forest, gradient boosting, decision tree, a slightly increase is achieved in terms of accuracy rate: 82.63%, 81.07%, 80.62% respectively. As depicted in Table 6, XGBoost achieved the highest classification results with an AUC value of 96.79%, accuracy of 95.34%, precision of 95.34%, sensitivity of 95.34%, specificity of 98.23%, and f1-score of 95.34% for dataset 2. Additionally, logistic regression attains a considerable increase in its performance compared to the obtained results with full dataset. It generates 95.12% instead of 92.88%. XGBoost and Gradient boosting achieves a slight improvement. It provides 92.62% and 92.51% instead of 92.36% and 92.41% respectively. To conclude, the best subset of features for dataset 1 is extracted using the AdaBoost classifier. It contains the following features: Age, Gender, Fever, Cough, Chills, Headache, and Vomiting. While, XGBoost classifier achieved the best results with the following extracted features for the dataset 2: Fever, Sore Throat, Shortness of breath, headache, and Known with confirmed.
-
Fourth experiment
In this experiment, feature selection is performed using NSGA-II algorithm. Table 5 shows that the highest classification rate is provided by the AdaBoost classifier for dataset 1. It yielded an AUC value of 87.16%, accuracy of 85%, precision of 90%, sensitivity 89.32%, specificity of 85%, and f1-score of 86.01%. Additionally, decision tree and gradient boosting performing well with the subset generated by NSGA-II. It provided as accuracy rate of 84.51% and 84.41% respectively. Besides, the results derived by MLP is improved compared to its results without feature selection. It achieved 83.52% as accuracy rate instead of 79.96%. SVM also generates a noticeable increase. It yielded as accuracy 79.51% instead of 75.72% with all the features. The results described in Table 6 prove the efficiency of AdaBoost with NSGA-II as it achieved the best classification performance for the dataset 2. It generated 96.87% as AUC value, 95.56% of accuracy, 95.56% of precision, sensitivity of 95.56%, specificity of 98.19%, and 95.56% of f1-score. Furthermore, gradient boosting and SVM achieved a high classification accuracy of 95.34% and 95.1% respectively. To conclude, the optimal subset of features for dataset 1 that satisfies the two conflicting objectives included the following features: Fever, Cough, Fatigue, Nasal congestion, Diarrhea, Headache, and Lives in affected area. While the optimal subset of features for dataset 2 contained: Cough, Fever, Sore Throat, Shortness of breath, headache, and Known with confirmed.
Parameter tuning and statistical tests
The process of hyper-parameters tuning for machine learning models has a significant impact on the performance of the model. It is anticipated that the optimal model is obtained after this process. In our proposed model, we choose the natural selection of parameters based on one of the most widely known techniques known as Genetic Algorithms (GA).In this way, we configured the parameters of the NSGA-II for the feature selection algorithm, and also, we adjusted the parameters of the AdaBoost classifier using the technique of hyper-parameters optimization using GA with an objective function: maximizing the accuracy rate. NSGA-II and GA have several parameters which have different effects on their performance. Among these parameters, the population size, the number of generations, and the three basic operations: selection, mutation, and crossover. Additionally, we highlight the major parameters to be tuned in AdaBoost which are max_depth of the base classifier, learning rate (lr), and the number of estimators. The max_depth is the most important parameter in the DT, which controls the maximum depth of the tree.
The learning rate refers to how each tree contributes to the outcomes while the number of estimators indicates the number of learners.
The whole parameters used in this study are described in Table 7. We defined a search space for each parameter and the optimal combination of parameters that gives the best accuracy rate is represented in the Table 7 for the two datasets. For NSGA-II and GA, we used the same values of parameters for both datasets.
Table 7.
Parameters setting
| Algorithms | Parameters |
|---|---|
| NSGA-II | Population size: 100 |
| Selection: binary tournament selection | |
| Crossover: single point crossover, | |
| Mutation: polynomial mutation, | |
| Genetic algorithm | Population size: 100 |
| Mutation: 0.1 | |
| Crossover: 0.9 | |
| AdaBoost (dataset1) | |
| Max_depth=6 | |
| AdaBoost (dataset2) | |
| Max_depth=11 | |
For both datasets, the NSGA-II with AdaBoost classifier is significantly better than the other models. To statistically confirm the hypothesis, a paired t-test is used to evaluate the importance of the obtained results. The null hypothesis deems that there is no statistically significant difference between the two models means “The accuracy of model A = accuracy of model B”. is complimented by the alternate hypothesis which deems that there is a statistically significant difference between the two models means “accuracy of model A accuracy of model B”. From Tables 8 and 9, it is observed that the p-value is less than 0.05 for all the cases. Therefore, the null hypothesis is rejected, and it is wrapped up that there is a statistically significant improvement in predicting the COVID-19 infected patients by using NSGA-II for feature selection with the AdaBoost classifier. Thus, it is drawn to conclude that has outstanding performance than the other studied models.
Table 8.
Significant test results of paired t-test () for Dataset 1
| Model A | Model B | t-test | p-value |
|---|---|---|---|
| 9.1444 | 3.74E06 | ||
| 24.1853 | 8.44E10 | ||
| 10.4965 | 1.19E06 | ||
| 13.2775 | 1.61E07 | ||
| 3.562 | 3.00E03 | ||
| 7.0801 | 2.89E05 | ||
| 7.6714 | 1.54E05 | ||
| 7.9099 | 1.21E05 | ||
| 8.6199 | 6.06E06 | ||
| 15.9806 | 3.25E08 | ||
| 8.6415 | 5.94E06 | ||
| 13.1475 | 1.76E07 | ||
| 10.256 | 1.44E06 | ||
| 8.6415 | 5.94E06 | ||
| 8.7993 | 5.13E06 | ||
| 13.9716 | 1.04E07 | ||
| 7.8576 | 1.27E05 | ||
| 24.1853 | 8.44E10 | ||
| 17.8533 | 1.23E08 | ||
| 10.4523 | 1.23E06 | ||
| 5.6934 | 1.00E04 | ||
| 9.0294 | 4.15E06 | ||
| 7.341 | 2.18E05 | ||
| 6.0921 | 9.04E05 | ||
| 6.863 | 3.68E05 | ||
| 12.397 | 2.31E07 | ||
| 7.765 | 1.40E05 | ||
| 6.7806 | 4.03E05 | ||
| 5.7425 | 1.30E04 | ||
| 8.448 | 7.14E06 | ||
| 7.8391 | 1.30E05 |
Table 9.
Significant test results of paired t-test (=0.05) for Dataset 2
| Model A | Model B | t-test | p-value |
|---|---|---|---|
| 17.5025 | 1.47E08 | ||
| 9.4181 | 2.94E06 | ||
| 8.6483 | 5.91E06 | ||
| 27.6612 | 2.56E10 | ||
| 13.5235 | 1.38E07 | ||
| 14.2987 | 8.54E08 | ||
| 9.7485 | 2.21E06 | ||
| 16.324 | 2.70E08 | ||
| 10.5152 | 1.18E06 | ||
| 7.0754 | 2.91E05 | ||
| 6.3859 | 6.37E05 | ||
| 16.3742 | 2.63E08 | ||
| 13.6838 | 1.25E07 | ||
| 21.7618 | 2.15E09 | ||
| 5.3533 | 2.00E04 | ||
| 4.3751 | 8.00E04 | ||
| 24.8923 | 6.53E10 | ||
| 14.0697 | 9.82E08 | ||
| 4.6437 | 6.00E04 | ||
| 31.4675 | 8.11E11 | ||
| 15.2167 | 4.98E08 | ||
| 13.8253 | 1.14E07 | ||
| 5.6214 | 1.00E04 | ||
| 32.9086 | 5.44E11 | ||
| 19.0164 | 7.08E09 | ||
| 8.8527 | 4.88E08 | ||
| 8.7921 | 5.17E06 | ||
| 17.191 | 1.72E08 | ||
| 5.2141 | 2.00E04 | ||
| 18.3929 | 9.49E09 | ||
| 16.1819 | 2.91E08 |
Results for research question 2
The performance of the proposed model compared to other existing models is presented in this part. From Table 10, it is clearly seen that our proposed model: NSGA-II with AdaBoost classifier achieved promising results compared to existing models proposed by Banik et al. [32] and Zoabi et al. [30] for both dataset 1 and dataset 2. For dataset 1, NSGA-II+AdaBoost provides 85% as accuracy rate, 90% of precision, and 84% of sensitivity, 85% of specificity, 86.01% of f1-score, and 87.16% of AUC, followed by the logistic regression model proposed by [32] which provided 81.2% as accuracy, 79.7% of precision, and 79.7% of sensitivity, and 79.7% of f1-score. For dataset 2, NSGA-II+AdaBoost yielded an accuracy of 95.56%, precision of 95.56%, sensitivity of 95.56%, specificity of 98.1%, and AUC rate of 96.87% followed by Gradient boosting model proposed by [30] that achieved a sensitivity rate of 87.3%, specificity of 71.98% and AUC value of 90%.
Table 10.
Comparison of our model with similar works for COVID-19 prediction
| Dataset | Reference | Model | Accuracy (%) | Precision (%) | Sensitivity (%) | Specificity (%) | F1-score (%) | AUC (%) |
|---|---|---|---|---|---|---|---|---|
| Dataset 1 | Our model | 85 | 90 | 89.32 | 85 | 86.01 | 87.16 | |
| Banik et al. [32] | Logistic Regression | 81.2 | 79.7 | 79.7 | – | 79.7 | – | |
| Naive Bayesian | 75.9 | 73.9 | 73.9 | – | 73.9 | – | ||
| Decision Tree | 71.9 | 70.4 | 67.3 | – | 68.8 | – | ||
| LinearSVM | 80.2 | 77.6 | 80.4 | – | 85 | – | ||
| Random Forest | 80.6 | 77.8 | 84 | – | 80.8 | – | ||
| Dataset 2 | Our model | 95.56 | 95.56 | 95.56 | 98.19 | – | 96.87 | |
| Zoabi et al. [30] | Gradient boosting | – | – | 87.3 | 71.98 | – | 90 |
Bold values highlight the best results for the two studied datasets
The strength of AdaBoost resides in combining weak learners into a powerful learner based on the adjustments of weights. These weights are mainly related to samples that are used by the learner in the training phase. This phase can generate a set of misclassified samples by the learners. In this case, AdaBoost tries to address the wrongly classified samples by empowering them with suitable weights. The large weight is assigned to samples that are misclassified and the small weight is assigned to samples that are already handled well. This ability to identify the misclassify samples and attempt to correct them to be feed to the next learner until an accurate predictor model is built makes AdaBoost considered as one of the most powerful models in binary classification. Additionally to the strength of AdaBoost in our proposed model, the NSGA-II algorithm has also shown a significant performance in solving multi-objective problems. It successfully found the trade-off between two objectives: maximizing the weights of selected features and minimizing the number of features that yielded an optimal subset of features. These features have positively influenced the performance of the predictive model.
Conclusion
At present, the emergence of COVID-19 pandemic is a dangerous menace to global health. Lack of care facilities and rapid spread of the virus has reduced the possibility of corralling this outbreak. Given the severity of these circumstances and the exponential growth of confirmed cases, an automatic detection tool is a pressing need that can bring new helpful avenues for healthcare systems. Application of Machine learning (ML) and Artificial Intelligence (AI) methods gives an assuring solution to assist medical staff in clinical decision making. This perspective highlights the benefits of these tools observed in a diversity of clinical environments and shows the importance of ML and AI algorithms in building these models. The focus of this study is to develop a machine-learning model to diagnose COVID-19 infection based on clinical symptoms and features. Our work is based on three main steps: The first step is feature selection which aims to select the optimal set of features using the NSGA-II algorithm. While the second step is the classification which consists of training the model using the AdaBoost classifier. Finally, an evaluation step is carried out to assess the efficiency of the proposed model based on the test data. We conducted a set of experiments that prove the significant role of NSGA-II in selecting the most important features which improve the classification performance. The proposed model (NSGA-II+AdaBoost) can potentially be useful for early virus prediction. In fact, the use of automatic diagnosis models in clinical decisions could include some issues. These issues reside on that the model could fail to generalize to different patient populations and might provide incorrect decisions which adverse effects on several patient outcomes. Due to the severity of this disease, the diagnosis should be precise and reliable as much as possible. Currently, several positive COVID-19 patients are experiencing two new symptoms including loss of smell and taste, and the possibility of being contaminated without carrying the most common symptoms such as fever and cough, is highly anticipated. Additionally, the daily increase of asymptomatic individuals’ rate makes the diagnosis of COVID-19 infection becoming laborious. This set of circumstances can be possible limitations of our model. In addition, the size of the used datasets was not extensive enough which does not include the two aforementioned symptoms. Hence, these symptoms are hard to skip to obtain a good prediction. Meanwhile, it can lead to model uncertainty. To overcome some of these issues, we suggest incorporating more robust data that is highly recommended to be collected from different countries. Meanwhile, integrate more symptoms and gathering equal quantities of samples for each class as much as a possible. Providing the model with more robust data can be very helpful in reducing uncertainty. Besides, we highlight the need for more performant model results to be more accurate in prediction. This can be achieved by using advanced techniques such as deep learning algorithms. Moreover, we intend to validate the model externally before undergo any clinical usage. This step is invaluable as it is correctly evaluating the model. The results of external validation will determine whether the model performs to a satisfactory degree. We plan also to extend our study by developing a publicly available online tool and to use the Xray images for COVID-19 detection.
Acknowledgements
The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number 7848.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Makram Soui, Email: m.soui@seu.edu.sa.
Nesrine Mansouri, Email: nesrine.mansouri@isimg.tn.
Raed Alhamad, Email: ralhamad@seu.edu.sa.
Marouane Kessentini, Email: marouane@umich.edu.
Khaled Ghedira, Email: khaledghedira3@gmail.com.
References
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;44(59):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanday AMUD, Rabani ST, Khan QR, Rouf N, Din MUM. Machine learning based approaches for detecting COVID-19 using clinical text data. Int. J. Inf. Technol. 2020;12(3):731–739. doi: 10.1007/s41870-020-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selcuk, M., Gormus, S., Guven, M.: Impact of weather parameters and population density on the COVID-19 transmission: evidence from 81 provinces of Turkey. Earth Syst. Environ. 1–14 (2021) [DOI] [PMC free article] [PubMed]
- 5.Li W, Thomas R, El-Askary H, Piechota T, Struppa D, Ghaffar KAA. Investigating the significance of aerosols in determining the coronavirus fatality rate among three European Countries. Earth Syst. Environ. 2020;4(3):513–522. doi: 10.1007/s41748-020-00176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohrer, M., Flahault, A., Stoffel, M.: Peaks of fine particulate matter may modulate the spreading and virulence of COVID-19. Earth Syst. Environ. 1–8 (2020) [DOI] [PMC free article] [PubMed]
- 7.Gupta A, Pradhan B, Maulud KNA. Estimating the impact of daily weather on the temporal pattern of COVID-19 outbreak in India. Earth Syst. Environ. 2020;4(3):523–534. doi: 10.1007/s41748-020-00179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Jing W, Liu J, Ma Q, Yuan J, Wang Y, Du M, Liu M. Effects of temperature and humidity on the daily new cases and new deaths of COVID-19 in 166 countries. Sci. Total Environ. 2020;729:139051. doi: 10.1016/j.scitotenv.2020.139051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Kindi KM, Alkharusi A, Alshukaili D, Al Nasiri N, Al-Awadhi T, Charabi Y, El Kenawy AM. Spatiotemporal assessment of COVID-19 spread over Oman using GIS techniques. Earth Syst. Environ. 2020;4(4):797–811. doi: 10.1007/s41748-020-00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, Su F, Pei T, Zhang A, Du Y, Luo B, Xiao H. COVID-19: challenges to GIS with big data. Geogr. Sustain. 2020;1(1):77–87. [Google Scholar]
- 11.Sarwar S, Waheed R, Sarwar S, Khan A. COVID-19 challenges to Pakistan: Is GIS analysis useful to draw solutions? Sci. Total Environ. 2020;730:139089. doi: 10.1016/j.scitotenv.2020.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezaei, M., Nouri, A.A., Park, G.S., Kim, D.H.: Application of geographic information system in monitoring and detecting the COVID-19 outbreak. Iran. J. Public Health (2020) [DOI] [PMC free article] [PubMed]
- 13.Murugesan B, Karuppannan S, Mengistie AT, Ranganathan M, Gopalakrishnan G. Distribution and trend analysis of COVID-19 in India: geospatial approach. J. Geogr. Stud. 2020;4(1):1–9. doi: 10.21523/gcj5.20040101. [DOI] [Google Scholar]
- 14.Desjardins MR, Hohl A, Delmelle EM. Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: detecting and evaluating emerging clusters. Appl. Geogr. 2020;118:102202. doi: 10.1016/j.apgeog.2020.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha A, Gupta K, Patil M. Monitoring and epidemiological trends of coronavirus disease (COVID-19) around the world. Matrix Sci. Med. 2020;4(4):121. doi: 10.4103/MTSM.MTSM_16_20. [DOI] [Google Scholar]
- 16.Pourghasemi, H. R., Pouyan, S., Farajzadeh, Z., Sadhasivam, N., Heidari, B., Babaei, S., Tiefenbacher, J. P.: Assessment of the outbreak risk, mapping and infestation behavior of COVID-19: application of the autoregressive and moving average (ARMA) and polynomial models. medRxiv (2020) [DOI] [PMC free article] [PubMed]
- 17.Brinati D, Campagner A, Ferrari D, Locatelli M, Banfi G, Cabitza F. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J. Med. Syst. 2020;44(8):1–12. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu, J., Zhang, P., Zhang, L. et al.: Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. medRxiv (Preprint) (2020)
- 19.Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for imagebased diagnosis of COVID-19. PLoS ONE. 2020;15(6):e0235187. doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butt, C., Gill, J., Chun, D., Babu, B.A.: Deep learning system to screen coronavirus disease 2019 pneumonia. Applied (2020). (Intelligence) [DOI] [PMC free article] [PubMed]
- 21.Arasi MA, Babu S. Survey of machine learning techniques in medical imaging. Int. J. Adv. Trends Comput. Sci. Eng. 2019;8(5):210–2116. doi: 10.30534/ijatcse/2019/39852019. [DOI] [Google Scholar]
- 22.Stojanovic V, Prsic D. Robust identification for fault detection in the presence of non-Gaussian noises: application to hydraulic servo drives. Nonlinear Dyn. 2020;100(3):2299–2313. doi: 10.1007/s11071-020-05616-4. [DOI] [Google Scholar]
- 23.Stojanovic V, He S, Zhang B. State and parameter joint estimation of linear stochastic systems in presence of faults and non-Gaussian noises. Int. J. Robust Nonlinear Control. 2020;30(16):6683–6700. doi: 10.1002/rnc.5131. [DOI] [Google Scholar]
- 24.Dong X, He S, Stojanovic V. Robust fault detection filter design for a class of discrete-time conic-type non-linear Markov jump systems with jump fault signals. IET Control Theory Appl. 2020;14(14):1912–1919. doi: 10.1049/iet-cta.2019.1316. [DOI] [Google Scholar]
- 25.Elshawi, R., Maher, M., Sakr, S.: Automated machine learning: state-of-the-art and open challenges. ArXiv (2019)
- 26.Yang L, Shami A. On hyperparameter optimization of machine learning algorithms: theory and practice. Neurocomputing. 2020;415:295–316. doi: 10.1016/j.neucom.2020.07.061. [DOI] [Google Scholar]
- 27.Pršić D, Nedić N, Stojanović V. A nature inspired optimal control of pneumatic-driven parallel robot platform. Proc. Inst. Mech. Eng. Part C J. Mech. Eng. Sci. 2017;231(1):59–71. doi: 10.1177/0954406216662367. [DOI] [Google Scholar]
- 28.Olivares R, Munoz R, Soto R, Crawford B, Cárdenas D, Ponce A, Taramasco C. An optimized brain-based algorithm for classifying Parkinson’s disease. Appl. Sci. Switzerland. 2020;10(5):1827. doi: 10.3390/app10051827. [DOI] [Google Scholar]
- 29.Munoz R, Olivares R, Taramasco C, Villarroel R, Soto R, Barcelos TS, Merino E, Alonso-Sánchez MF. Using black hole algorithm to improve EEG-based emotion recognition. Comput. Intell. Neurosci. 2018 doi: 10.1155/2018/3050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoabi Y, Deri-Rozov S, Shomron N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. npj Digit. Med. 2021;4:1–5. doi: 10.1038/s41746-020-00372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enughwure A, Febaide I. Applications of artificial intelligence in combating Covid-19: a systematic review. Open Access Library J. 2020;7:1–12. [Google Scholar]
- 32.Banik S, Banik S, Ghosh A, Mukherjee A. Probabilistic estimation of COVID-19 using patient’s symptoms. In: Singh TP, Tomar R, Choudhury T, Perumal T, Mahdi HF, editors. Data Driven Approach Towards Disruptive Technologies Studies in Autonomic, Data-Driven and Industrial Computing. Singapore: Springer; 2021. [Google Scholar]
- 33.de Batista, A.F.M., Miraglia, J.L., Donato, T.H.R., Chiavegatto Filho, A.D.P.: COVID-19 diagnosis prediction in emergency care patients: a machine learning approach (2020)
- 34.Mondal MRH, Bharati S, Podder P, Podder P. Data analytics for novel coronavirus disease. Inform. Med. Unlock. 2020;20:100374. doi: 10.1016/j.imu.2020.100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (covid-19) Clin. Chem. Lab. Med. CCLM. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 36.Kukar, M., Gunčar, G., Vovko, T., Podnar, S., Černelč, P., Brvar, M., Zalaznik, M., Notar, M., Moškon, S., Notar, M.: COVID-19 diagnosis by routine blood tests using machine learning. 1–11 (2020) [DOI] [PMC free article] [PubMed]
- 37.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J. Biomed. Inform. 2002;35(5–6):352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal, S.: Data mining: data mining concepts and techniques. In: Proceedings—2013 International Conference on Machine Intelligence Research and Advancement, ICMIRA 2013 (2014)
- 39.Kim, P.: MATLAB Deep Learning: With Machine Learning, 1st edn. Neural Networks and Artificial Intelligence. Apress, Berkely, CA, USA (2017)
- 40.Ao Y, Li H, Zhu L, Ali S, Yang Z. The linear random forest algorithm and its advantages in machine learning assisted logging regression modeling. J. Petrol. Sci. Eng. 2019;174:776–789. doi: 10.1016/j.petrol.2018.11.067. [DOI] [Google Scholar]
- 41.Liu S, Xiao J, Liu J, Wang X, Wu J, Zhu J. Visual diagnosis of tree boosting methods. IEEE Trans. Vis. Comput. Graph. 2018;24(1):163–173. doi: 10.1109/TVCG.2017.2744378. [DOI] [PubMed] [Google Scholar]
- 42.Singer G, Marudi M. Ordinal decision-tree-based ensemble approaches: the case of controlling the daily local growth rate of the COVID-19 epidemic. Entropy. 2020;22(8):871. doi: 10.3390/e22080871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman JH. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002;38(4):367–378. doi: 10.1016/S0167-9473(01)00065-2. [DOI] [Google Scholar]
- 44.Chen, T., Guestrin, C.: XGBoost: A scalable tree boosting system. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 13–17-August-2016, pp. 785–794 (2016)
- 45.Pohjalainen, J., Räsänen, O., Kadioglu, S.: Feature selection methods and their combinations in high-dimensional classification of speaker likability, intelligibility and personality traits. Comput. Speech Lang. 29(1), 145–171 (2015)
- 46.Fan J, Wang X, Wu L, Zhou H, Zhang F, Yu X, Lu X, Xiang Y. Comparison of support vector machine and extreme gradient boosting for predicting daily global solar radiation using temperature and precipitation in humid subtropical climates: a case study in China. Energy Convers. Manage. 2018;164(January):102–111. doi: 10.1016/j.enconman.2018.02.087. [DOI] [Google Scholar]
- 47.Miao J, Niu L. A survey on feature selection. Procedia Comput. Sci. 2016;91(Itqm):919–926. doi: 10.1016/j.procs.2016.07.111. [DOI] [Google Scholar]
- 48.Deb K, Pratap A, Agarwal S, Meyarivan T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002;6(2):182–197. doi: 10.1109/4235.996017. [DOI] [Google Scholar]
- 49.Kovács G. An empirical comparison and evaluation of minority oversampling techniques on a large number of imbalanced datasets. Appl. Soft Comput. J. 2019;83:105662. doi: 10.1016/j.asoc.2019.105662. [DOI] [Google Scholar]
- 50.Blagus R, Lusa L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinform. 2013 doi: 10.1186/1471-2105-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tharwat A. Classification assessment methods. Appl. Comput. Inform. 2018 doi: 10.1016/j.aci.2018.08.003. [DOI] [Google Scholar]
- 52.Ferri C, Hernández-Orallo J, Modroiu R. An experimental comparison of performance measures for classification. Pattern Recogn. Lett. 2009;30(1):27–38. doi: 10.1016/j.patrec.2008.08.010. [DOI] [Google Scholar]